The sperm annulus, a septin-based ring structure, migrates distally along the flagellum to form a midpiece/principal piece boundary. Hoque et al. show that the membrane-binding Cby3/ciBAR1 complex is indispensable for the precise stoppage of the annulus at the midpiece/principle piece junction of the sperm tail.
Abstract
Proper compartmentalization of the sperm flagellum is essential for fertility. The annulus is a septin-based ring that demarcates the midpiece (MP) and the principal piece (PP). It is assembled at the flagellar base, migrates caudally, and halts upon arriving at the PP. However, the mechanisms governing annulus positioning remain unknown. We report that a Chibby3 (Cby3)/Cby1-interacting BAR domain-containing 1 (ciBAR1) complex is required for this process. Ablation of either gene in mice results in male fertility defects, caused by kinked sperm flagella with the annulus mispositioned in the PP. Cby3 and ciBAR1 interact and colocalize to the annulus near the curved membrane invagination at the flagellar pocket. In the absence of Cby3, periannular membranes appear to be deformed, allowing the annulus to migrate over the fibrous sheath into the PP. Collectively, our results suggest that the Cby3/ciBAR1 complex regulates local membrane properties to position the annulus at the MP/PP junction.
Introduction
The establishment of proper cellular compartments is vital for fundamental biological processes in eukaryotic cells, permitting the intricate spatial organization of functional components and well-coordinated control of cellular activities. The sperm flagellum, a modified motile cilium with a 9 + 2 microtubule structure, is essential for sperm motility (Griswold, 2016; Teves and Roldan, 2022). It is structurally divided into three compartments: the mitochondrial sheath-containing midpiece (MP), the fibrous sheath-containing principal piece (PP), and the end piece (EP), with no accessory structures. At the junction of MP and PP, the annulus, a septin-based ring structure, plays a key role in the compartmentalization of the sperm tail (Lehti and Sironen, 2017; Pereira and Sousa, 2023). There, the annulus has been shown to act as a structural support as well as a membrane diffusion barrier (Kwitny et al., 2010). Mutations in septins and missing annuli have been reported in infertile men (Dirami et al., 2015; Kuo et al., 2012; Lhuillier et al., 2009; Sugino et al., 2008; Wang et al., 2022) and mice (Ihara et al., 2005; Kissel et al., 2005; Shen et al., 2017), highlighting the importance of the annulus in male fertility.
Spermiogenesis is a complex biological process during which post-meiotic round spermatids differentiate into elongated spermatozoa (Griswold, 2016; Lehti and Sironen, 2017; Teves and Roldan, 2022). Mouse spermiogenesis takes ∼14 d and is divided into 16 different steps (Oakberg, 1956). The annulus starts to form at around step 9 (S9) at the flagellar base in close contact with the flagellar membrane (Avidor-Reiss et al., 2017; Fawcett et al., 1970; Guan et al., 2009; Lehti and Sironen, 2017). At S15, the annulus is fully mature and physically migrates caudally along the flagellum to its destination at the proximal end of the PP (∼20 μm in mice). The annulus is an electron-dense, membrane-bound organelle, comprised of septins (SEPTs) 2, 4, 6, 7, and 12 (Kissel et al., 2005; Kuo et al., 2012, 2015; Shen et al., 2017; Steels et al., 2007; Toure et al., 2011). Septins are highly conserved GTP-binding proteins that form hetero-oligomers, assembling into fibrous ring structures (Kuo et al., 2015; Weirich et al., 2008). Previous studies demonstrated that mice harboring mutations in either SEPT4 or SEPT12 show severe male fertility defects due to abnormal sperm morphology with loss of the annulus and poor motility (Ihara et al., 2005; Kissel et al., 2005; Shen et al., 2017). However, little is known about the molecular components and the underlying mechanisms for assembly, migration, and precise positioning of the annulus.
Cilia and flagella are ubiquitous, microtubule-based protrusions that play crucial roles in various important cellular processes. Genetic defects in their function are associated with pleiotropic disorders termed ciliopathies including male infertility (Derderian et al., 2023; Mill et al., 2023; Wallmeier et al., 2020). Cilia are nucleated from mother centriole–derived basal bodies at the cell surface. Anchoring of basal bodies to the plasma membrane is mediated by fibrous connections termed distal appendages (Kumar and Reiter, 2021; Zhao et al., 2023). These are ninefold symmetric, pinwheel-like structures, radiating from the distal end of basal bodies, that contain several core components including CEP164 and CEP83 (also known as CCDC41; Bowler et al., 2019; Yang et al., 2018; Zhao et al., 2023). However, the molecular basis underlying basal body attachment to the membrane remains to be elucidated.
We previously reported that the 14.5-kD coiled-coil protein Chibby1 (Cby1) localizes to the ciliary base and plays a critical role in the formation and function of cilia (Burke et al., 2014; Steere et al., 2012; Voronina et al., 2009). Germline Cby1−/− mice display several hallmarks of ciliary defects, including chronic upper airway infection (Love et al., 2010), polycystic kidneys (Lee et al., 2014), and exocrine pancreatic insufficiency (Cyge et al., 2021), as well as hydrocephalus and polydactyly at low frequency. Cby1 forms a complex with the Cby1-interacting BAR domain–containing 1 and 2 (ciBAR1 and 2; previously known as FAM92A and B; Li et al., 2016). Bin/Amphiphysin/Rvs (BAR) domain–containing proteins are known to dimerize and form crescent-shaped structures to sculpt and maintain membrane curvatures, and participate in key membrane-associated events such as organelle biogenesis, vesicle trafficking, cell division, and cell polarity (Simunovic et al., 2019; Suetsugu et al., 2014). The Cby1/ciBAR complex is recruited to basal bodies through interactions of Cby1 with the distal appendage protein CEP164 (Burke et al., 2014; Siller et al., 2017). In mature cilia, Cby1 is detected at the distal end of the distal appendages as a ring in close association with ciliary membranes (Burke et al., 2014), suggesting that the Cby1/ciBAR complex binds to lipid membranes and may play a role in local membrane dynamics. Cby1 and ciBAR1 are mutated in human ciliopathies, Joubert syndrome and polydactyly, respectively (Epting et al., 2020; Schrauwen et al., 2019). However, the exact role of the Cby1/ciBAR1 complex in ciliogenesis remains to be elucidated.
In addition to Cby1, there are two other Cby family members, Cby2 (also known as SPERT and Nurit; Feige et al., 2002) and Cby3. Cby2 and Cby3 share 26% and 35% identity to Cby1, respectively. However, their biological roles in mammalian development are unknown. In this study, we show that Cby3 is exclusively expressed in the testis and localizes to the annulus in differentiating spermatids. Like Cby1, Cby3 forms a complex with ciBAR1, and ciBAR1 also localizes to the annulus. We show that mice lacking Cby3 or ciBAR1 exhibit severe male fertility defects with sharply bent sperm tails, reminiscent of sperm devoid of annuli. Remarkably, the annulus is present in Cby3−/− and ciBAR1−/− sperm but misplaced caudally into the PP, indicating that the Cby3/ciBAR1 complex is indispensable for the precise stoppage of the annulus at the MP/PP junction along the sperm flagellum. Furthermore, using ultrastructure expansion microscopy (U-ExM), we demonstrate that Cby3 and ciBAR1 colocalize in close proximity to the curved proximal membranes of the flagellar pocket, a membrane invagination at the base of flagella. Interestingly, in Cby3−/− sperm, periannular membranes appear to be deformed probably due to decreased membrane stiffness, allowing the annulus to migrate past the MP/PP junction into the PP. Collectively, our findings imply that the membrane-binding Cby3/ciBAR1 complex is necessary for the precise positioning of the annulus at the MP/PP junction likely via regulation of local membrane rigidity, thereby contributing to proper compartmentalization of the sperm flagellum.
Results
Cby3 is exclusively expressed in the testis and localizes to the annulus during spermiogenesis
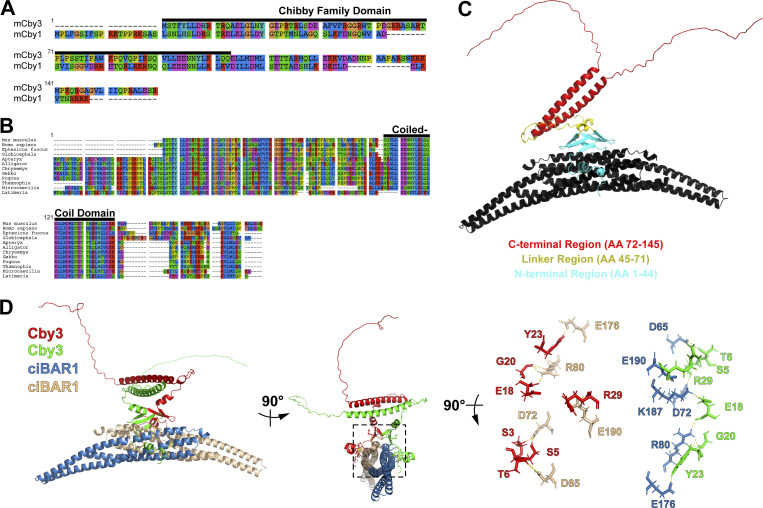
Mouse Cby3 is a 16-kD protein composed of 140 amino acids with a C-terminal coiled-coil domain (Fig. S1 A). Mouse Cby3 shows 35% identity and 57% similarity to mouse Cby1. Cby3 is conserved in mammals and some species of birds, reptiles, and fish with the most distant ancestor being the coelacanth, Latimeria chalumnae (Fig. S1 B).
Figure S1.
Multiple sequence alignment of Cby3 proteins and AlphaFold2-Multimer prediction of the Cby3/ciBAR1 complex. (A) Alignment of mouse Cby3 and Cby1 proteins. (B) Evolutionary conservation of Cby3 protein. Multiple sequence alignments were performed using TreeView. Accession numbers for Cby3 proteins: Mus musculus, NP_083967.1; Homo sapiens, NP_001157916.1; Eptesicus fuscus, XP_008156917.2; Globicephala melas, XP_030700670.1; Apteryx mantelli, XP_013808933.1; Alligator mississippiensis, XP_006263599.1; Chrysemys picta, XP_005295910.1; Gekko japonicus, XP_015281930.1; Pogona vitticeps, XP_020638417.1; Thamnophis sirtalis, XP_013908651.1; Microcaecilia unicolor, XP_030066943.1; and Latimeria chalumnae, XP_005995419. Eight classes of amino acids are shown in different colors. (C) Ribbon diagram of the AlphaFold2-Multimer-predicted structure of a complex of human Cby3 and the BAR domain of human ciBAR1 (aa 10–220). The N-terminal regions of the Cby3 dimer make contact with the BAR domains of ciBAR1 dimer. ciBAR1 is depicted in black, and different domains of Cby3 are shown in different colors as indicated. (D) The interaction interface between the N-terminal regions of Cby3 dimer and the BAR domains of ciBAR1 dimer with each molecule color-coded. The structure is rotated as indicated, and a detailed view of the boxed region in the middle panel is shown in the right panel. Selected side chains are shown as sticks. The dotted yellow lines represent hydrogen bonds.
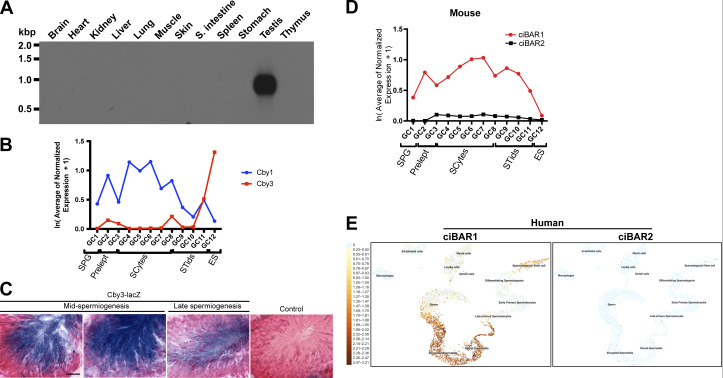
To examine the expression of Cby3 mRNA, we performed a Northern blot analysis of mouse tissues and found that Cby3 is exclusively expressed in the testis (Fig. S2 A). Our analysis of previous single-cell RNA sequencing data for murine spermatogenesis indicated that Cby3 is highly expressed in elongating spermatids, while Cby1 is expressed in various spermatogenic cell types (Fig. S2 B; Green et al., 2018). To understand the physiological role of Cby3, we generated Cby3-KO first mice from targeted embryonic stem cells created by the MRC-Harwell Institute (Fig. S3 B). Since this mouse line carries a lacZ reporter in the Cby3 gene locus, we performed X-gal staining of testis sections from heterozygous Cby3 KO-first mice and found that Cby3 expression peaks in elongating spermatids during mid- to late spermiogenesis (Fig. S2 C).
Figure S2.
Cby3 is highly expressed in elongating spermatids, while ciBAR1, but not ciBAR2, is expressed in the testis. (A) Northern blot analysis of mouse Cby3. 2 µg of polyA+ RNA was loaded in each lane. (B) RNA levels of Cby1 and Cby3 during murine spermatogenesis. The single-cell RNA sequencing datasets were retrieved from Green et al. (2018). GC, germ cell; SPG, spermatogonia; Prelept, preleptotene; SCytes, spermatocytes; STids, round spermatids; ES, elongated spermatids. (C) X-gal staining of adult testis sections from a negative control WT mouse or a mouse heterozygous for the Cby3 KO-first allele that contains a lacZ reporter. Tissues were counterstained with nuclear fast red. Scale bar, 10 µm. (D) RNA levels of ciBAR1 and ciBAR2 during murine spermatogenesis. The single-cell RNA sequencing datasets were retrieved from Green et al. (2018). GC, germ cell; SPG, spermatogonia; Prelept, preleptotene; SCytes, spermatocytes; STids, round spermatids; ES, elongated spermatids. (E) RNA levels of ciBAR1 and ciBAR2 in the cell clusters of the adult human testis. The single-cell RNA sequencing datasets were obtained from UCSC Cell Browser (https://cells.ucsc.edu/) (Guo et al., 2018). Source data are available for this figure: SourceData FS2.
Figure S3.
Generation and initial characterization of Cby3−/− mice. (A) Schematic of Cby3 protein structure. (B) Generation of Cby3−/− mice. (C) PCR genotyping of Cby3 mice. The locations of genotyping primers for WT (P1 and P2, 313 bp) and KO (P3 and P4, 205 bp) alleles are indicated by arrows in B. (D) Testis lysates prepared from adult mice were processed for Western blotting using Cby3 antibody. GAPDH was used as a loading control. (E) Representative adult testis images. (F) Representative images of PAS staining of adult testis sections. (G) Paraffin sections of male efferent ducts were immunostained for A-tub. Nuclei were detected with DAPI. (H) Representative Western blot images of sperm lysates using phospho-PKA substrate antibody (left) and phospho-tyrosine antibody (right) under non-capacitating (NC) and capacitating (CAP) conditions (n = 11–13 mice per genotype). α-Tubulin was used as a loading control. Regions of interest used for quantification are indicated by a vertical bar on the right of Western blots. Scale bars for E, 5 mm; F, 100 µm; and G, 50 µm. Source data are available for this figure: SourceData FS3.
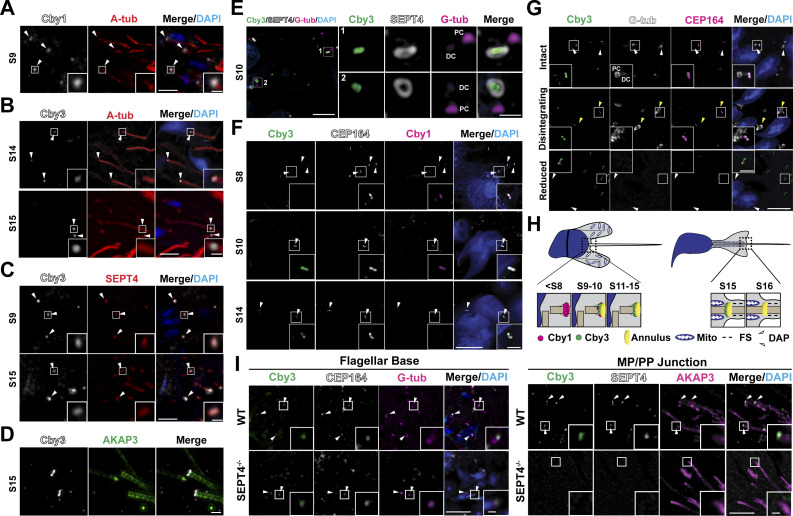
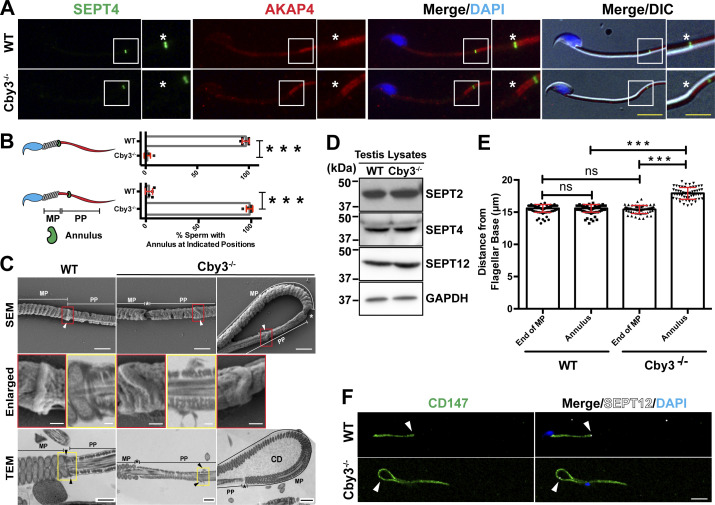
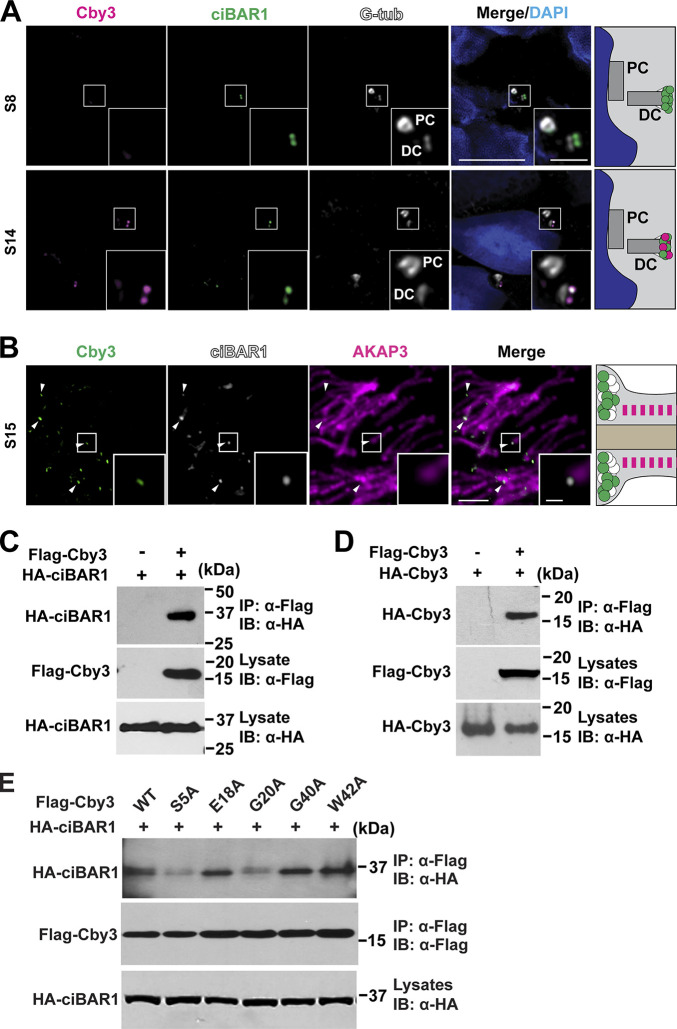
To study the subcellular localization of Cby3, we performed immunofluorescence (IF) staining of adult testis sections (Fig. 1). Consistent with its localization in ciliogenesis (Burke et al., 2014; Steere et al., 2012), Cby1 was detectable at the base of flagella from spermiogenesis S2–3 to around S10 (Fig. 1 A). In marked contrast, Cby3 was apparent at the flagellar base around S9 to S14 and in the middle portion of the flagellum at S15 but was undetectable at S16 (Fig. 1 B). This localization pattern is reminiscent of the annulus that demarcates the midpiece (MP) and principal piece (PP) of the sperm flagellum (Kuo et al., 2015; Kwitny et al., 2010; Shen et al., 2017; Toure et al., 2011). The annulus initially assembles at the flagellar base around S9, continues to mature until S14, and then physically migrates caudally along the flagellum to the MP/PP junction at S15. IF staining of adult testis sections for Cby3 and the annulus marker SEPT4 revealed that Cby3 colocalizes with SEPT4 at the flagellar base and the MP/PP junction (Fig. 1 C). Structured illumination microscopy (SIM) imaging of testis sections immunostained for Cby3 and the fibrous sheath marker AKAP3 further confirmed the annulus localization of Cby3 (Fig. 1 D). Next, we performed triple IF staining of adult testis sections for Cby3, SEPT4, and the centriolar marker γ-tubulin (G-tub) and imaged by SIM (Fig. 1 E). SEPT4-positive annuli were clearly visible at the base of flagella, and Cby3 showed smaller ring-like structures within the annulus. The Cby3/SEPT4 ring was consistently observed slightly distally to the G-tub-positive domain on the distal centriole.
Figure 1.
Localization of Cby1 and Cby3 during spermiogenesis. (A) Adult testis paraffin sections were immunostained for Cby1 (arrowheads) and A-tub. (B) Adult testis paraffin sections were immunostained for Cby3 (arrowheads) and A-tub. (C) Adult testis paraffin sections were immunostained for Cby3 (arrowheads) and SEPT4. (D) Adult testis frozen sections were immunostained for Cby3 and AKAP3. (E) Adult testis frozen sections were immunostained for Cby3, SEPT4, and G-tub. Note that G-tub prominently localizes to the proximal centriolar adjunct, which nucleates an aster of microtubules (Manandhar et al., 1998). (F) P30 testis frozen sections were immunostained for Cby1, Cby3, and CEP164. White arrowheads denote distal appendages. (G) Adult testis frozen sections were immunostained for Cby3, G-tub, and CEP164. The status of G-tub-positive centriolar structures is noted on the left. Yellow arrowheads indicate disintegrating G-tub-positive structures in the process of centrosome reduction. White arrowheads point to the base of flagella. (H) Model for Cby3 localization. Spermatogenic staging was assessed based on PNA lectin labeling of acrosomes and nuclear morphology. Mito, mitochondria; FS, fibrous sheath; DAP, distal appendage. (I) WT and SEPT4−/− adult testis paraffin sections were immunostained for Cby3, CEP164, and G-tub or Cby3, SEPT4, and AKAP3. Nuclei were detected with DAPI. Scale bars for A–C and G, 5 and 1 μm (insets); D, 1 μm; E and F, 10 and 1 μm (enlarged images or insets); I, 10 and 2 µm (insets).
To elucidate the temporally regulated localization patterns of Cby1 and Cby3 during spermiogenesis, we performed IF staining of testis sections at postnatal day (P) 30 when the annulus is still at the base of the flagellum during the synchronized first wave of spermatogenesis (Fig. 1 F; Bellvé et al., 1977; Ernst et al., 2019). Triple IF staining for Cby1, Cby3, and the distal appendage marker CEP164 demonstrated that Cby1 is present at the distal appendages of basal bodies in early round spermatids until around S10 (top panels). We found that there is a narrow time window when both Cby1 and Cby3 coexist (middle panels). In later steps, only Cby3 was observed at the base of flagella (bottom panels). During late spermiogenesis, centrioles, especially the distal centrioles, undergo extensive remodeling and degeneration through a process known as centrosome reduction (Avidor-Reiss et al., 2015; Manandhar and Schatten, 2000; Manandhar et al., 1998). G-tub-positive centriolar structures progressively disintegrate and are undetectable in late-stage testicular sperm (Manandhar et al., 1998). We noticed that CEP164 is clearly detectable when G-tub-positive structures are degenerated but disappears at S15 when Cby3 relocates to the MP/PP junction (Fig. 1 G). This suggests that the initiation of annulus migration may be coupled with centrosome reduction and loss of distal appendages. In sum, these results indicate that Cby1 is present at the flagellar base in round spermatids until around S10, and then Cby3 appears around the time when the annulus starts to assemble at S9 (Fig. 1 H). At S15, Cby3 co-migrates with the annulus to the MP/PP junction. Once annulus migration is complete, Cby3 is no longer detectable at S16.
To explore the role of the annulus in the initial recruitment of Cby3 and its subsequent migration to the MP/PP junction, we performed IF staining of adult SEPT4−/− testis sections lacking the annulus (Kissel et al., 2005) along with wild-type (WT) controls (Fig. 1 I). Triple IF staining of Cby3, CEP164, and G-tub revealed no significant changes in Cby3 localization in SEPT4−/− spermatids at the flagellar base (Fig. 1 I, left). In contrast, triple IF staining of Cby3, SEPT4, and AKAP3 showed that Cby3 is undetectable at the MP/PP junction in SEPT4−/− spermatids (Fig. 1 I, right). Taken together, these results indicate that Cby3 is recruited to the distal region of basal bodies but unable to relocate to the MP/PP junction in the absence of the annulus.
Loss of Cby3 in mice results in male infertility with kinked sperm tail morphology
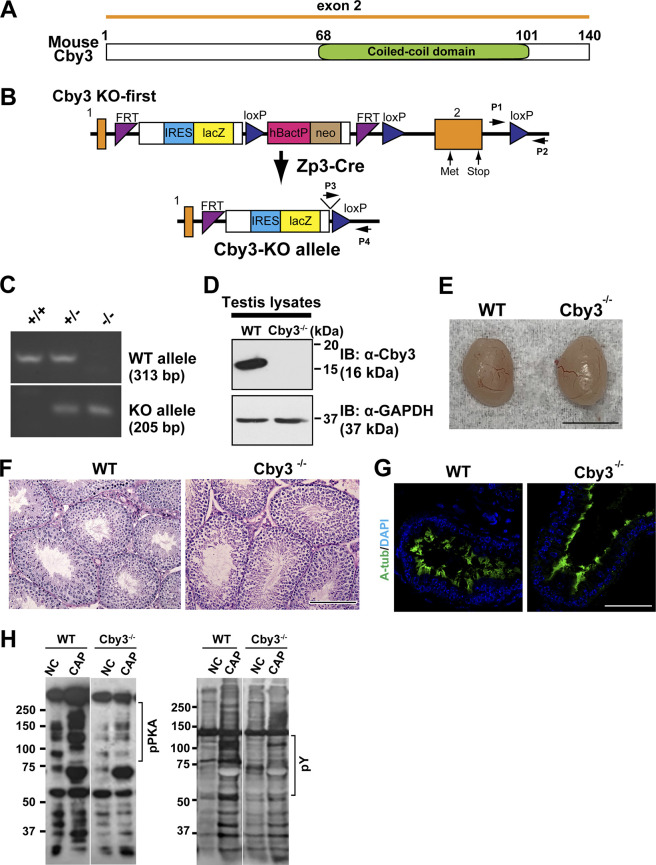
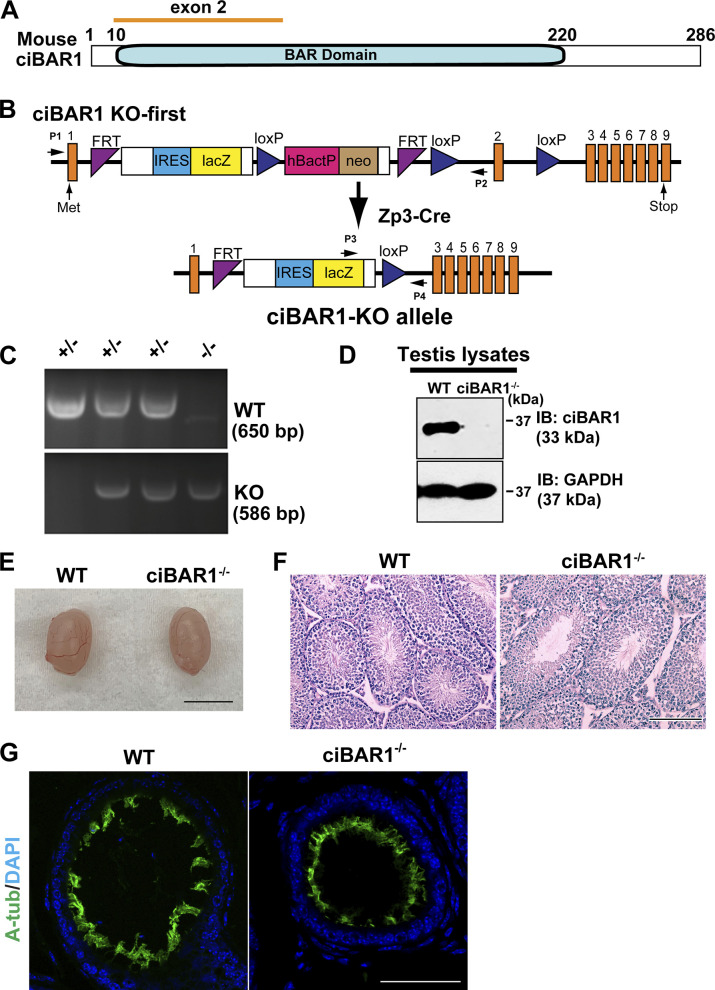
To explore the role of Cby3 in male fertility, we generated germline Cby3−/− mice by mating Cby3-KO first mice with Zp3-Cre transgenic mice, which express Cre recombinase specifically in oocytes (Fig. S3 B; Lewandoski et al., 1997). This results in the excision of exon 2, which contains the entire Cby3-coding sequence (Fig. S3, A and B). PCR analyses verified the correct genotypes (Fig. S3 C) and Western blotting of testis lysates confirmed the complete loss of Cby3 protein in Cby3−/− samples (Fig. S3 D).
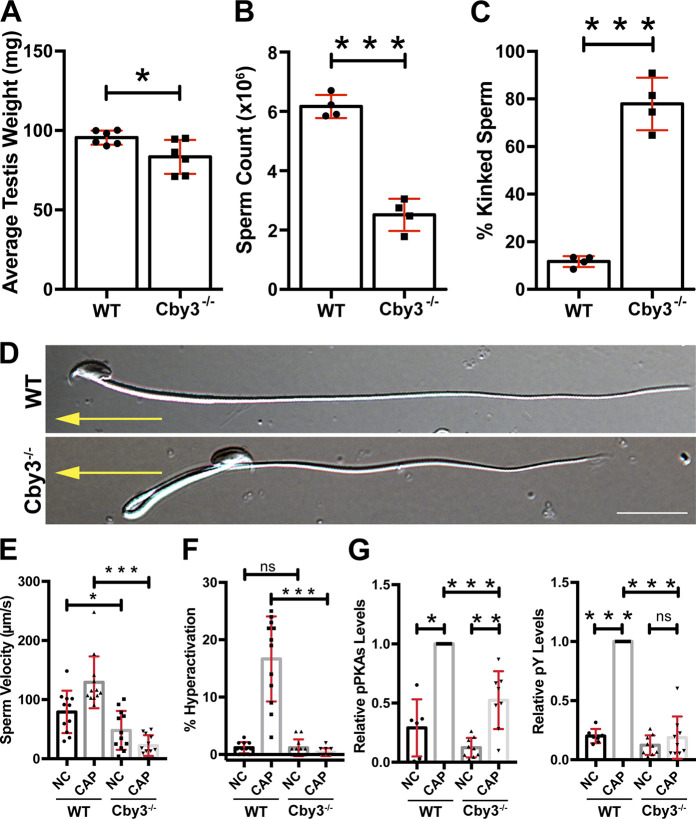
Cby3−/− mice were born at the expected Mendelian ratios (Table S1) and lived to adulthood without any overt phenotypic abnormalities. To assess the biological importance of Cby3 in spermatogenesis, we examined male fertility by mating mice with WT females and found that Cby3−/− males produced no offspring (Table S2). Cby3−/− mice exhibited slightly reduced testis weights (Fig. 2 A and Fig. S3 E). However, no overt histological abnormalities were noticeable on PAS-stained testis sections from Cby3−/− mice (Fig. S3 F). Defective motile cilia in the efferent ducts have also been shown to contribute to male infertility (Hoque et al., 2021, 2022), but no major changes were noticeable in the IF staining for motile cilia in the efferent ducts of Cby3−/− mice (Fig. S3 G).
Figure 2.
Cby3−/− males are infertile with sharply kinked sperm tails. (A) Quantification of testis weights (n = 6 mice per genotype). The average weight of both testes is plotted. (B) Sperm counts (n = 4 mice per genotype). (C) Quantification of sperm with kinked tails (n = 5 mice per genotype). (D) Representative DIC images of cauda epididymal sperm. Yellow arrows point to the direction of sperm movement. Scale bar, 10 μm. (E and F) Sperm velocity and hyperactivation were measured under non-capacitating (NC) and capacitating (CAP) conditions using CASA (n = 11–13 mice per genotype). (G) Quantification of Western blot results for PKA substrate phosphorylation (pPKAs) and tyrosine phosphorylation (pY) under NC and CAP conditions (n = 11–13 mice per genotype). See also Fig. S3 H. Means ± SD are shown. Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns).
To gain insight into the cause of male infertility, we quantified the number of sperm from the cauda epididymis and found that Cby3−/− mice had significantly lower sperm counts in comparison to WT mice (Fig. 2 B). Intriguingly, the vast majority of Cby3−/− sperm possessed sharply kinked tails (Fig. 2, C and D). To evaluate sperm motility, we performed computer-assisted sperm analysis (CASA) under non-capacitating (NC) and capacitating conditions (CAP). Cby3−/− sperm showed decreased sperm velocity under both NC and CAP conditions (Fig. 2 E) and failed to become hyperactivated (Fig. 2 F). Upon capacitation, HCO3- influx upregulates protein kinase A (PKA) phosphorylation and protein tyrosine phosphorylation in sperm (Mohanty et al., 2023; Takei et al., 2021). Consistent with impaired hyperactivation, phosphorylation of both PKA substrates and tyrosine-containing proteins was significantly diminished in Cby3−/− sperm (Fig. 2 G and Fig. S3 H).
Mispositioning of the annulus in the PP of Cby3−/− sperm tails
The kinked tail morphology of Cby3−/− sperm is strikingly similar to that of sperm lacking the annulus (Ihara et al., 2005; Kissel et al., 2005; Shen et al., 2017). The annulus is an essential cellular structure for male fertility and plays a role as a structural support as well as a membrane diffusion barrier (Kwitny et al., 2010). Mice with loss-of-function mutations in the Sept4 or Sept12 gene show male fertility defects due to a complete lack of the annulus and bent sperm tail morphology (Ihara et al., 2005; Kissel et al., 2005; Shen et al., 2017). More importantly, septin mutations have been reported in infertile men (Kuo et al., 2012; Wang et al., 2022), and sperm with missing or defective annuli have been observed in patients with asthenozoospermia (Dirami et al., 2015; Lhuillier et al., 2009; Sugino et al., 2008).
To investigate the status of the annulus, we performed IF staining of cauda epididymal sperm for the annulus marker SEPT4 and the fibrous sheath marker AKAP4. Intriguingly, in Cby3−/− sperm, the annulus was detected but failed to stop at the MP/PP junction and was misplaced in the PP (Fig. 3 A). Quantification of annulus locations based on IF staining for SEPT4 and AKAP4 revealed that nearly all Cby3−/− sperm had an annulus in the PP (Fig. 3 B). To further validate the mispositioning of the annulus in Cby3−/− sperm, we performed scanning and transmission electron microscopy (SEM and TEM) analyses of sperm from the cauda epididymis (Fig. 3 C). The annulus was positioned at the MP/PP junction in WT sperm but mispositioned into the PP in Cby3−/− sperm. Notably, we observed that the annulus of Cby3−/− sperm was frequently deformed or occasionally missing. This implies that the annulus is unstable and degraded or slid off the flagellum in mature Cby3−/− sperm. In testis lysates, however, the annulus septins 2, 4, and 12 were present at comparable levels in WT and Cby3−/− testes as shown by Western blotting (Fig. 3 D). Quantitatively, in Cby3−/− sperm, the annulus is positioned ∼2.5 µm away from the MP/PP junction, whereas no major change was found in MP length (Fig. 3 E). CD147 (Basigin) is a transmembrane glycoprotein that is partitioned to the MP membrane of cauda epididymal sperm, and in SEPT4−/− sperm lacking the annulus, CD147 distributes over the entire flagellar membrane (Kwitny et al., 2010). Similarly, we found that Cby3−/− sperm exhibit a defective diffusion barrier (Fig. 3 F). Collectively, our data suggest that Cby3 plays a role in the precise positioning of the annulus at the MP/PP junction during spermiogenesis as well as in the establishment and/or maintenance of the membrane diffusion barrier either directly or indirectly via regulation of the annulus.
Figure 3.
The annulus is displaced into the principal piece in Cby3−/− sperm. (A) Cauda epididymal sperm were immunostained for SEPT4 and AKAP4. Asterisks indicate the MP/PP junction. (B) Quantification of sperm with the indicated annulus positions (n = 8 fields of view with ≥ 25 annuli per field). (C) SEM and TEM studies of cauda epididymal sperm. Enlarged images of the boxed regions are shown in the middle panels. Arrowheads denote the annulus. Asterisks indicate the MP/PP junction. MP, midpiece; PP, principal piece; CD, cytoplasmic droplet. (D) Testis lysates prepared from adult mice were processed for Western blotting using antibodies against SEPT2, 4, and 12. GAPDH was used as a loading control. (E) Quantification of the distance from the base of the flagellum to the distal end of the MP or the annulus (n = 8 fields of view with ≥60 sperm per field). Mitochondria and the annulus were detected by Biotracker and SEPT4 antibody, respectively. (F) Cauda epididymal sperm were immunostained for CD147 and SEPT12 (arrowheads). Nuclei were detected with DAPI. Scale bars for A, 10 and 5 µm (enlarged images); C, 1 µm and 200 nm (enlarged images); F, 10 μm. Means ± SD are shown. Student’s t test: ***P < 0.001, not significant (ns). Source data are available for this figure: SourceData F3.
ciBAR1 interacts with Cby3 and localizes to the annulus during spermiogenesis
We previously reported that ciBAR1 and 2 interact with Cby1, localize to the ciliary base, and play important roles in ciliogenesis (Li et al., 2016). This raised the possibility that Cby3 might also bind to ciBAR proteins. Our analysis of recent single-cell RNA-sequencing datasets indicated that ciBAR1 is highly expressed, whereas ciBAR2 shows no or little expression during murine and human spermatogenesis (Fig. S2, D and E; Ernst et al., 2019; Guo et al., 2018). To examine the localization of ciBAR1 and 2 proteins in the testis, we performed IF staining of adult testis sections and found that ciBAR1, but not ciBAR2, was present at the flagellar base throughout the round spermatid steps even when Cby3 was undetectable (Fig. 4 A, S8), possibly in a complex with Cby1. In elongating spermatids, ciBAR1 colocalized with Cby3 at the flagellar base (Fig. 4 A, S14). Subsequently, ciBAR1 was observed at the annulus at the MP/PP junction at S15 but not at S16 (Fig. 4 B). These data suggest that ciBAR1 may work in a complex with Cby3 at the annulus during spermiogenesis. To directly test the physical interaction between Cby3 and ciBAR1, we performed coimmunoprecipitation (Co-IP) assays and observed that like Cby1, Cby3 bound to ciBAR1 (Fig. 4 C). Cby1 forms a homodimer through its C-terminal coiled-coil domain (Mofunanya et al., 2009). Co-IP assays demonstrated that Cby3 also interacts with itself (Fig. 4 D).
Figure 4.
ciBAR1 binds to Cby3 and localizes to the annulus. (A) Adult frozen testis sections were immunostained for Cby3, ciBAR1, and G-tub. PC, proximal centriole; DC, distal centriole. (B) Adult testis paraffin sections were immunostained for Cby3, ciBAR1, and AKAP3. (C) Coimmunoprecipitation assays from HEK293T cells expressing Flag-Cby3 and HA-ciBAR1 as indicated. (D) Coimmunoprecipitation assays from HEK293T cells expressing Flag-Cby3 and HA-Cby3 as indicated. (E) Co-immunoprecipitation assays from HEK293T cells expressing Flag-Cby3 mutants and HA-ciBAR1 as indicated. Nuclei were detected with DAPI. Scale bars for A and B, 5 and 1 µm (insets). Source data are available for this figure: SourceData F4.
To better understand the molecular interaction between Cby3 and ciBAR1, we employed AlphaFold2-Multimer to predict the structure of a Cby3 dimer/ciBAR1 dimer complex using the protein sequences of the full-length human Cby3 and the human ciBAR1 BAR domain (aa 10–220; Jumper et al., 2021; Richard et al., 2022 Preprint). In the top predicted model (Fig. S1 C), the N-terminal regions of a Cby3 dimer make extensive interactions with the BAR domains of a ciBAR1 dimer, similar to the predicted structure of a Cby1/ciBAR1 complex (van Breugel et al., 2022). We identified residues S5, T6, E18, G20, Y23, and R29 in both Cby3 molecules that are potentially involved in direct ciBAR1 binding via electrostatic interactions (Fig. S1 D). To validate our structural prediction, we generated alanine substitution mutants for S5, E18, and G20, and G40 and W42 as negative controls. Co-IP experiments indicated the importance of S5 and G20 in ciBAR1 binding (Fig. 4 E). Taken together, these findings suggest that ciBAR1 interacts with Cby3 and together may act in proper compartmentalization of the sperm flagellum.
ciBAR1−/− sperm display kinked tail morphology with mispositioning of the annulus in the PP
Mouse ciBAR1 is a 286 aa protein that contains a membrane-binding BAR domain (Fig. S4 A). To investigate the in vivo function of ciBAR1 during spermatogenesis, we obtained ciBAR1 KO-first mice from the MRC-Harwell Institute (Fig. S4 B). To generate germline ciBAR1−/− mice, ciBAR1 KO-first mice were crossed with Zp3-Cre mice to remove exon 2, leading to a truncation at amino acid position at 9. Genotypes were examined by PCR (Fig. S4 C), and Western blotting demonstrated that ciBAR1 protein is undetectable in the testis lysate from ciBAR1−/− mice (Fig. S4 D).
Figure S4.
Generation and initial characterization of ciBAR1−/− mice. (A) Schematic of ciBAR1 protein structure. (B) Generation of ciBAR1−/− mice. (C) PCR genotyping of ciBAR1 mice. The locations of genotyping primers for WT (P1 and P2, 650 bp) and KO (P3 and P4, 586 bp) alleles are indicated by arrows in B. (D) Testis lysates prepared from adult mice were processed for Western blotting using antibodies against ciBAR1 and loading control GAPDH. (E) Representative adult testis images. (F) Representative images of PAS staining of adult testis sections. (G) Paraffin sections of male efferent ducts were immunostained for A-tub. Nuclei were detected with DAPI. Scale bars for E, 5 mm; F, 100 µm; and G, 50 µm. Source data are available for this figure: SourceData FS4.
We found that ciBAR1−/− mice are born at a lower-than-expected Mendelian ratio (18%; Table S3). However, once born, ciBAR1−/− mice survived to adulthood and appeared grossly normal. After breeding with WT females for 2 mo, ciBAR1−/− males showed subfertility with a 70% reduction in litter size (Table S4). No obvious histological changes were observed on the PAS-stained testis sections from ciBAR1−/− mice (Fig. S4 F). Furthermore, we detected the abundant presence of motile cilia in the efferent ducts of ciBAR1−/− mice (Fig. S4 G).
Similar to Cby3−/− mice (Fig. 2), ciBAR1−/− mice displayed slightly reduced testis weights (Fig. 5 A and Fig. S4 E) and significantly lower sperm counts (Fig. 5 B). About 41% of sperm from the cauda epididymis of ciBAR1−/− mice had a kinked tail morphology (Fig. 5, C and D), phenocopying Cby3−/− sperm. Moreover, we found that the annulus is misplaced into the PP in ciBAR1−/− sperm (Fig. 5 E). Quantification of annulus locations showed that 65% of ciBAR1−/− sperm had the annulus in the PP (Fig. 5 F). In addition, ciBAR1−/− sperm also displayed altered membrane diffusion barrier function (Fig. 5 G). Thus, the majority of ciBAR1−/− sperm, like Cby3−/− sperm, fail to position the annulus precisely at the MP/PP junction. These results raised the interesting possibility that ciBAR1 collaborates with Cby3 during annulus migration and positioning at the MP/PP junction.
Figure 5.
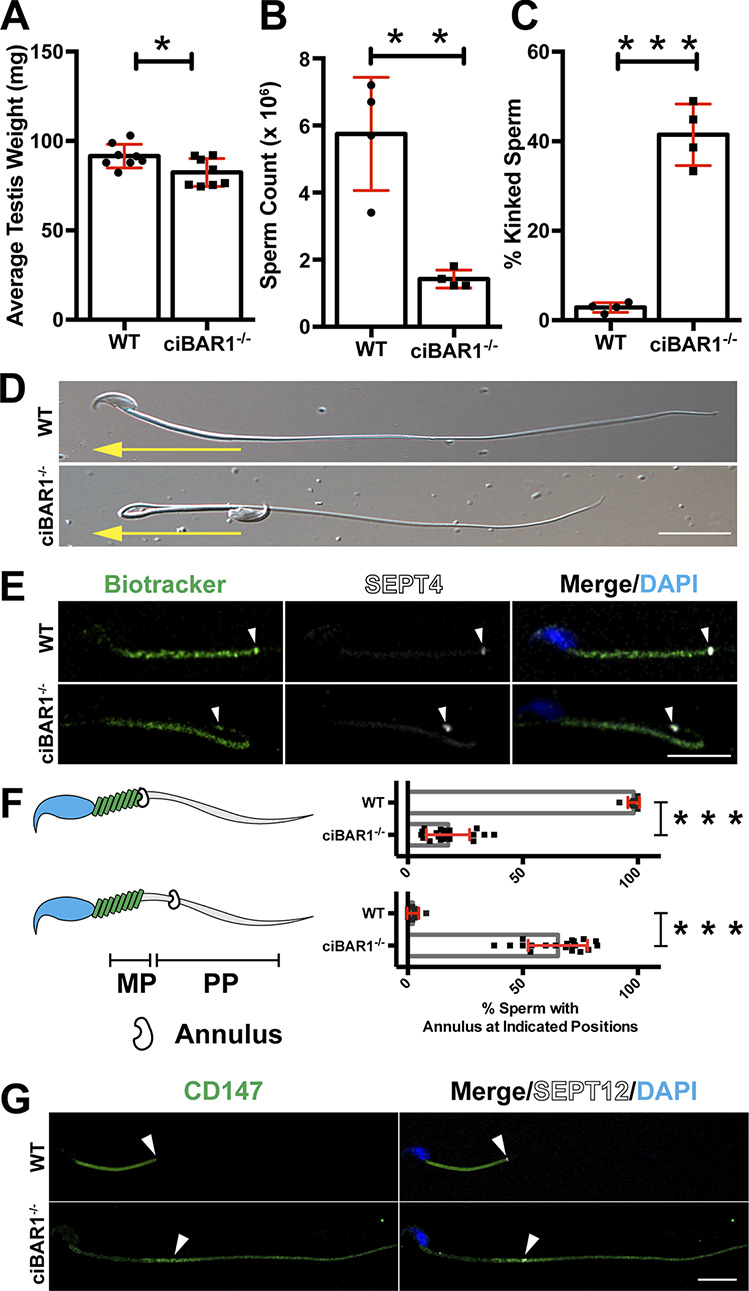
ciBAR1−/− sperm phenocopy Cby3−/− sperm. (A) Quantification of testis weights (n = 8 mice per genotype). (B) Sperm counts (n = 4 mice per genotype). (C) Quantification of sperm with kinked tails (n = 4 mice per genotype). (D) Representative DIC images of cauda epididymal sperm. Yellow arrows denote the direction of sperm movement. (E) Cauda epididymal sperm were immunostained for SEPT4 (arrowheads). Mitochondria and nuclei were visualized with Biotracker and DAPI, respectively. (F) Quantification of sperm with the indicated annulus positions (n = 8 fields of view with ≥25 annuli per field). (G) Cauda epididymal sperm were immunostained for CD147 and SEPT12 (arrowheads). Nuclei were visualized by DAPI. Means ± SD are shown. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars for D, E, and G, 10 µm.
Mutual dependence of Cby3 and ciBAR1 for their annulus localization
To investigate whether Cby3 and ciBAR1 are recruited to the annulus in a mutually dependent manner, we performed IF staining of adult testis sections from the knockout mice. Cby3 was visible at the distal region of the distal centriole in WT spermatids, whereas Cby3 was absent in ciBAR1−/− spermatids (Fig. 6 A). In the reciprocal scenario, ciBAR1 was recruited to the base of flagella prior to annulus migration in Cby3−/− spermatids (Fig. 6 B, top). However, ciBAR1 was undetectable at the MP/PP junction in S15 Cby3−/− spermatids (Fig. 6 B, bottom). Taken together, our results demonstrated that the annulus recruitment of Cby3 absolutely requires the existence of ciBAR1. In marked contrast, ciBAR1 is able to localize to the annulus at the flagellar base in the absence of Cby3, likely through interaction with Cby1, but fails to comigrate with the annulus to the MP/PP junction.
Figure 6.
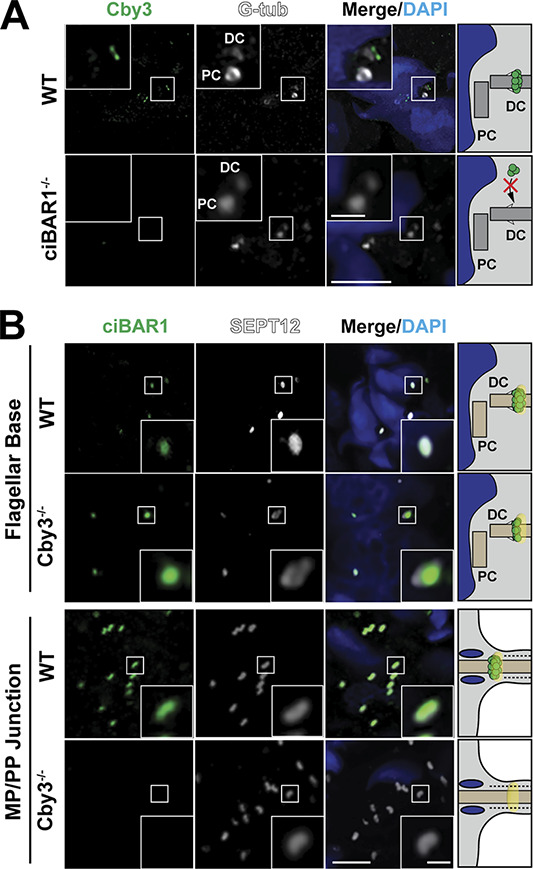
Recruitment of Cby3 and ciBAR1 to the annulus. (A) WT and ciBAR1−/− adult testis frozen sections were immunostained for Cby3 and G-tub. (B) WT and Cby3−/− adult testis frozen sections were immunostained for ciBAR1 and SEPT12. Nuclei were detected with DAPI. PC, proximal centriole; DC, distal centriole. Scale bars for A and B, 5 and 1 µm (insets).
Cby3 and ciBAR1 form a ninefold symmetric structure at the flagellar base
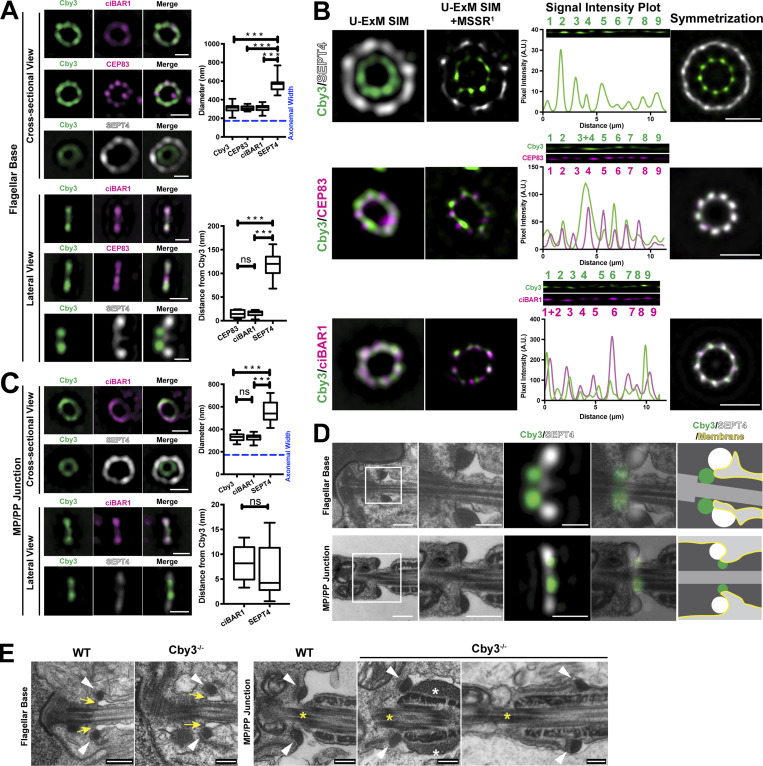
To gain insight into the molecular function of the Cby3/ciBAR1 complex in annulus positioning, we investigated their nanoscale localization at the annulus using ultrastructure expansion microscopy (U-ExM; Gambarotto et al., 2019). Testis sections were isotropically expanded 3.67-fold in each dimension (Fig. S5 A), and images were acquired by SIM. At the base of the flagellum, Cby3 and ciBAR1 showed extensive colocalization both on cross-sectional and lateral views (Fig. 7 A). Several density masses for Cby3 and ciBAR1 were noticeable in cross-sectional ring structures. To elucidate the cellular structure to which Cby3 localizes, we coimmunostained Cby3 and the distal appendage protein CEP83. This CEP83 antibody has been shown to detect the outer region of the distal appendages of basal bodies (Kanie et al., 2023 Preprint). Consistent with being a distal appendage component, CEP83 showed a ninefold symmetrical distribution and Cby3 exhibited substantial colocalization with CEP83 (Fig. 7 A; Tables S5 and S6). On the other hand, SEPT4-ring structures were significantly larger and positioned anterior to the Cby3 rings (Fig. 7 A; Tables S5 and S6).
Figure S5.
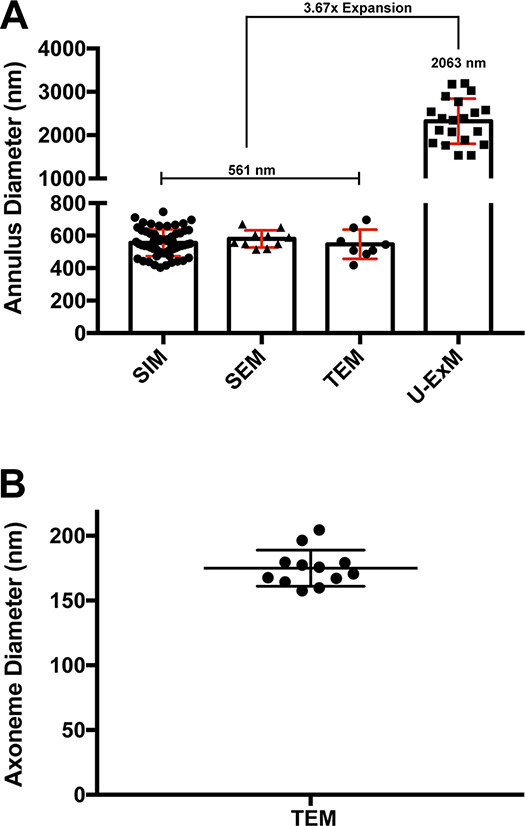
Calculation of expansion factor. (A) Quantification of annulus diameters using various imaging modalities. The annulus was detected with SEPT4 antibody for SIM and ExM (n = 32–40 for SIM and U-ExM, n = 8 for TEM, n = 10 for SEM). The fold expansion was calculated by dividing the average annulus diameter value of U-ExM (2063 nm) by that of SIM, SEM, and TEM (561 nm). (B) Axoneme diameters were determined based on TEM of WT testes (n = 12). The axoneme width is used in Fig. 7, A and C.
Figure 7.
Ultrastructure expansion microscopy reveals a ring-like organization of Cby3- and ciBAR1-asociated structures at the annulus. (A and C) Adult testis sections were expanded via U-ExM and immunostained for Cby3, ciBAR1, and SEPT4, and fluorescence signals at the flagellar base or MP/PP junction were imaged using SIM. The diameter and relative position of ring structures were quantified (n ≥ 32 measurements per each). See also Tables S5 and S6. (B) Adult testis sections were expanded via U-ExM and immunostained for Cby3, ciBAR1, and SEPT4. Fluorescence signals at the flagellar base were imaged using SIM and processed by MSSR. Radial intensity profile plots and ninefold symmetrized images were generated. (D) ExM-SIM images for lateral views of Cby3 and SEPT4 in A and C were overlayed with TEM images. (E) WT and Cby3−/− adult testes were subjected to TEM. The annuli at the flagellar base and the MP/PP junction are indicated by the arrowheads. Yellow arrows point to the membrane invagination of the flagellar pocket. Yellow asterisks indicate the MP/PP junction. White asterisks denote the accumulation of submembranous electron-dense materials in the PP. Blue dashed lines represent an experimentally measured axonemal diameter (See Fig. S5 B). Box and whisker plots represent median and minimum/maximum values. Student’s t test: ***P < 0.001, not significant (ns). Scale bars for A–C, 272 nm; D, 1 µm and 272 nm (enlarged TEM and IF images); E, 250 nm.
To further enhance the spatial resolution of fluorescence images, we took advantage of Mean-Shift Super Resolution (MSSR), which computationally extends the axial resolution up to 1.7-fold (Torres-García et al., 2022), resulting in a near 17-nm resolution. Our MSSR analysis revealed that Cby3 and ciBAR1 exhibit a ninefold symmetric pattern at the flagellar base, overlapping with CEP83 (Fig. 7 B).
We also examined the localization of Cby3, ciBAR1, and SEPT4 at the MP/PP junction by U-ExM (Fig. 7 C). Cby3 and ciBAR1 extensively colocalized at the MP/PP junction but showed no ninefold symmetric distribution patterns. Intriguingly, like at the flagellar base, Cby3 rings were smaller than SEPT4 rings, but both were found at a similar location on the lateral view. Quantification showed no significant changes in the diameter of Cby3, ciBAR1, and SEPT4 rings between the flagellar base and the MP/PP junction (Fig. 7, A and C; Tables S5 and S6). These findings indicate that prior to annulus migration, Cby3 and ciBAR1 localize to the distal portion of the distal appendages, distinct from the annulus, but this ninefold symmetric organization is no longer retained at the MP/PP junction once annulus migration occurs.
Cby3 and ciBAR1 localize near the curved membrane regions of the flagellar pocket
Our U-ExM results above showed that the Cby3 protein positions proximally to the annulus at the flagellar base but in parallel with the annulus at the MP/PP junction (Fig. 7, A and B). To correlate the U-ExM data with ultrastructural features of spermatids, we overlaid the U-ExM SIM images with TEM micrographs (Fig. 7 D). These alignments revealed that Cby3 localizes to the areas of distal appendages in close proximity with the curved membrane regions of the flagellar pocket, a deep membrane invagination at the flagellar base. At the MP/PP junction, it was more evident that Cby3 is concentrated near the area where curved membranes are present.
BAR domain–containing proteins directly bind to lipid membranes to generate and maintain local membrane curvatures (Simunovic et al., 2019; Suetsugu et al., 2014). We hypothesized that the Cby3/ciBAR1 complex binds to the curved membrane of the flagellar pocket to regulate the accurate positioning of the annulus at the MP/PP junction. If so, loss of Cby3 may affect the morphology of flagellar pocket membranes. To this end, we performed a TEM analysis of the testes to examine periannular membranes. The electron-dense annulus was present at the flagellar base in Cby3−/− spermatids (Fig. 7 E, left, arrowheads). At the proximal tip of the membrane invagination, curved membrane structures were observed (Fig. 7 E, left, yellow arrows), and no major difference in the overall morphology of the periannular membrane was noted in Cby3−/− spermatids. At the MP/PP junction of S15 WT spermatids, the annulus migrated and came to rest precisely at the proximal end of the fibrous sheath (Fig. 7 E, right, arrowheads). However, in Cby3−/− spermatids, the periannular membrane appeared to be at least partially extended, allowing the annulus to continue to migrate over the fibrous sheath. These findings suggest that the Cby3/ciBAR1 complex contributes to increased membrane rigidity rather than membrane curvature, and the inner rigid membrane enclosing the flagellar axoneme appears to be essential for the annulus to halt upon reaching the fibrous sheath in the PP.
Discussion
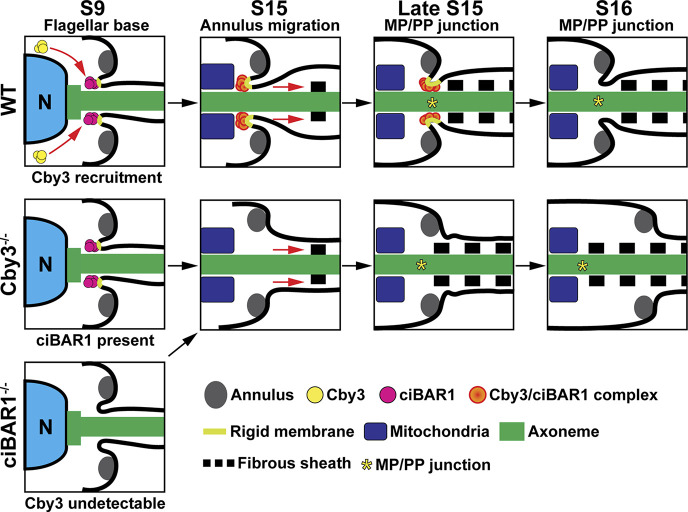
To date, little is known about the molecular components and the underlying mechanisms for the assembly, migration, and positioning of the annulus. We demonstrated that the Cby3/ciBAR1 complex plays a critical role in male fertility. Based on our findings, we propose a model for the migration and positioning of the annulus at the MP/PP junction (Fig. 8). Cby1 and ciBAR1 are detectable at the flagellar base in early spermiogenesis. Around S9, when annulus assembly initiates, Cby3 is recruited to the annulus, while Cby1 expression declines. The recruitment of Cby3 may be achieved through distal appendages, independent of the annulus. As the annulus matures through the continued recruitment of septins, Cby3 and ciBAR1 remain at the flagellar base in a ninefold symmetric pattern in close proximity to flagellar membranes. At S15, the Cby3/ciBAR1 complex comigrates caudally with the annulus along the flagellum and comes to rest upon arriving at its destination. In the absence of the other, ciBAR1, but not Cby3, is recruited to the flagellar base. In both Cby3−/− and ciBAR1−/− sperm, the annulus fails to stop at the MP/PP junction, contributing to their kinked tail morphology. Since ciBAR1 contains a lipid-binding BAR domain, we postulate that the Cby3/ciBAR1 complex directly binds to the curved membranes of the flagellar pocket. Surprisingly, our results suggest that the Cby3/ciBAR1 complex contributes to local membrane rigidity rather than membrane curvature. This function is indispensable for the placement of the annulus precisely at the MP/PP junction. A limitation of this study is that it was difficult to find the annulus migrating through the MP on TEM sections and examine periannular membrane morphology since annulus migration seems to occur in a short timeframe. It is also plausible that Cby3 and ciBAR1 interact with other factors at the PP that are required for annulus positioning.
Figure 8.
Model for annulus positioning at the MP/PP junction by the Cby3/ciBAR1 complex. Prior to S9, ciBAR1 is detectable at the distal appendages of basal bodies. Cby3 is then recruited to the distal appendages around S9, while annulus assembly initiates independently. The Cby3/ciBAR1 complex remains at the flagellar base, and the annulus continues to mature. At S15, the annulus migrates caudally to the MP/PP junction. Throughout this process, the Cby3/ciBAR1 complex localizes to the proximal curved membranes of the flagellar pocket and likely confers local membrane rigidity. When the annulus reaches its destination, the increased membrane rigidity prevents membrane deformation and aids the annulus to halt at the proximal point of the fully developed fibrous sheath. In Cby3−/− spermatids, ciBAR1 is recruited to the flagellar base but fails to stably associate with the periannular membrane during annulus migration, while ciBAR1−/− spermatids show no detectable Cby3 recruitment at the flagellar base. In both mutants, the curved periannular membrane exhibits decreased rigidity, allowing the annulus to migrate over the fibrous sheath into the PP. N, nucleus.
It is puzzling that Cby3−/− mice are infertile, whereas ciBAR1−/− mice retain subfertility with a lower percentage of sperm kinked tail morphology. Although the reason for this is not entirely clear, one possibility is that Cby3 may have additional function(s) in sperm tail morphogenesis and male fertility, independent of ciBAR1. Additionally, there are over 70 different BAR domain-containing proteins in humans and mice (Carman and Dominguez, 2018). Thus, it is also plausible that other BAR-domain protein(s) may compensate for the loss of ciBAR1 in fertility.
Intriguingly, Cby1 is expressed in early-round spermatids and then Cby3 takes over when the annulus starts to form. In agreement with the localization of Cby1 at the distal end of distal appendages (Burke et al., 2014), the recruitment of Cby3 to the flagellar base seems to occur through the distal appendages as it shows a ring-like distribution with ninefold symmetry (Fig. 7). It is possible that Cby3 physically interacts with CEP164 and/or other distal appendage proteins. We reported that Cby1 promotes ciBAR-mediated membrane tubule formation in cultured mammalian cells probably via enhanced or stabilized association with membranes (Li et al., 2016). Similarly, we envision that Cby3 facilitates ciBAR1 binding to flagellar membranes. ciBAR1 belongs to the N-BAR family of the BAR domain superfamily, which also includes F-BAR and I-BAR families (Carman and Dominguez, 2018). N-BAR domain proteins form crescent-shaped dimers, bind to highly curved membrane structures, and play key roles in diverse biological processes such as endocytosis and cytokinesis (Simunovic et al., 2019; Suetsugu et al., 2014). BAR domain–containing proteins typically bind to negatively charged phospholipids such as phosphoinositides (Simunovic et al., 2019; Suetsugu et al., 2014). However, the lipid-binding specificity of the Cby3/ciBAR1 complex is currently unknown. The lipid composition of sperm flagellar membranes is not well understood, but PI(4,5)P2 has been shown to be enriched in ciliary membranes at the base of primary cilia (Conduit et al., 2021; Garcia-Gonzalo et al., 2015). Therefore, it is tempting to speculate that the Cby1/ciBAR1 complex recognizes PI(4,5)P2-enriched membrane regions with appropriate curvature at the base of sperm flagella.
Our U-ExM SIM studies revealed that Cby3 and ciBAR1 localize as ring-like structures that are smaller than the annulus (Fig. 7). Cby3 rings are located anterior to the annulus at the flagellar base but parallel to the annulus at the MP/PP junction. These results are consistent with our model in which the Cby3/ciBAR1 complex localizes to the curved membrane region of the flagellar pocket. By doing so, the Cby3/ciBAR1 complex may provide stability and rigidity to the periannular membrane to prevent membrane deformation. This function is crucial for halting annulus migration at the proximal end of the fibrous sheath-containing PP at S15, by which time PP formation has been completed (Irons and Clermont, 1982). In agreement with the importance of the fibrous sheath in regulating annulus positioning, sperm from mice lacking AKAP4, a major fibrous sheath component, show annulus mispositioning in the PP (Xu et al., 2020). Our data also indicate that Cby3 and ciBAR1 are critical for membrane diffusion barrier function. At present, how the membrane diffusion barrier is established and maintained in the sperm tail is poorly understood. Cby3 and ciBAR1 might be directly involved in the formation of a lateral diffusion barrier at the MP/PP junction. Alternatively, the defects in the diffusion barrier might be caused secondarily by the deformation of the annulus.
The annulus migrates caudally ∼20 µm to the MP/PP junction at some point during S15. What is the cue for the initiation of annulus migration? During late spermiogenesis, centrioles, especially the distal centriole, undergo extensive remodeling and degeneration through a process known as centrosome reduction (Avidor-Reiss et al., 2015; Manandhar and Schatten, 2000; Manandhar et al., 1998). G-tub-positive centriolar structures progressively disintegrate and are undetectable in late-stage testicular sperm (Manandhar et al., 1998). Although the timeframe of centrosome reduction is not defined, our results show a correlation between the disappearance of G-tub-positive centriolar structures and the distal appendage protein CEP164, and the migration of Cby3-positive annulus (Fig. 1 G). Hence, it is possible that the initiation of annulus migration is coupled with centrosome reduction and loss of distal appendages. Clearly, more research is required to understand the initiation timing, molecular motor, and velocity of annulus migration and the potential role of the Cby3/ciBAR1 complex in these processes.
In summary, our work uncovers a novel molecular mechanism governing the precise positioning of the annulus, thereby shedding light on the proper compartmentalization of the sperm flagellum. Although direct membrane binding remains to be elucidated, we speculate that the Cby3/ciBAR1 complex associates with the curved membranes of the flagellar pocket at the annulus and positions the annulus at the MP/PP junction through maintaining membrane rigidity. Since Cby3 expression is restricted to the testis, inhibition of Cby3 functions offers a promising drug target for male contraceptives.
Materials and methods
Mouse models and mouse handling
All mouse experiments were performed in accordance with National Institute of Health (NIH) guidelines and approved by the Institutional Animal Care and Use Committee (IACUC) of Stony Brook University (#245427). Mice were housed in individually ventilated cages in a maximum isolation facility with free access to water and food under a 12-h light and dark cycle. A maximum of four mice were kept in the same cage.
Mouse embryonic stem (ES) cells with validated targeting of the Cby3 locus were purchased from the MRC-Harwell Institute. ES cells were injected into blastocysts by the animal facility at the Cold Spring Harbor Laboratory. Chimeric males were crossed with female C57BL/6J mice (RRID:IMSR_JAX:000664) to generate Cby3 KO-first mice. ciBAR1 KO-first mice were obtained from the MRC Harwell Institute. Cby3 and ciBAR1 KO-first mice were crossed with C57BL/6-Tg(Zp3-cre)93Knw/J mice (RRID:IMSR_JAX:003651) to remove the lacZ and neomycin-resistance cassettes and create germline knockout mice. Cby3 and ciBAR1 heterozygous mice were then backcrossed over eight generations onto FVB/NJ (RRID:IMSR_JAX:001800) and C57BL/6J (RRID:IMSR_JAX:000664) backgrounds, respectively, for phenotypic consistency and reproducibility. Genotyping was performed by PCR with genomic DNA from toe clips using the following primers: Cby3 WT allele, 5′-GAGCATCCGGGTTTTCCTTG-3′ (P1) and 5′-AGGCTAGCTTCAAACTCCAG-3′ (P2; 313 bp); Cby3−/− allele, 5′-CATTCTAGTTGTGGTTTGTC-3′ (P3) and 5′-AGTGCTTGGATTAAAGGCGT-3′ (P4; 205 bp); ciBAR1 WT allele, 5′-CTGAAGGAGGATGCTTGGTGTTCCC-3′ (P1) and 5′-GAAGGGCAACATTCCCAACTCTTCC-3′ (P2; 650 bp); and ciBAR1−/− allele, 5′-GCTACCATTACCAGTTGGTCTGGTGTC-3′ (P3) and 5′-TGGTCACTCAAAGCAAACACACAGC-3′ (P4; 586 bp).
Quantification of sperm
Cauda epididymal sperm were prepared and counted as previously described (Hoque et al., 2021). Briefly, one cauda epididymis was dissected from 2- to 4-mo-old mice, cut four to five times in a 35-mm dish with 1 ml PBS, pH 7.4, and incubated at 37°C for 30 min. The sperm were collected and counted on a hemocytometer with appropriate dilutions.
Northern blotting
A mouse polyA+ RNA Northern blot was purchased from Origene Technologies. The blot was hybridized with mouse full-length Cby3 cDNA probes labeled with 32P by random priming and then exposed to x-ray films.
X-gal staining of testis
Testes were harvested from control or Cby3 KO-first heterozygous mice, embedded in Tissue-Tek optimal cutting temperature (OCT) compound (Sakura Finetek), and snap-frozen in 2-methylburane prechilled in liquid nitrogen. 5-µm-thick sections were cut using a Leica CM1950 cryostat, fixed with 2% paraformaldehyde (PFA), and 0.25% glutaraldehyde in PBS for 10 min at 4°C, and washed twice with wash buffer (0.01% sodium deoxycholate, 2 mM MgCl2, and 0.02% NP-40 in PBS) for 10 min each. Subsequently, sections were incubated with wash buffer containing 1 mg/ml X-gal, 5 mM K3Fe(CN)6, and 5 mM K4Fe(CN)6 for 24 h at room temperature in the dark. Specimens were then washed twice with wash buffer for 10 min each, counterstained with nuclear fast red, and mounted with Permount (Thermo Fisher Scientific).
Histological staining
Testes were dissected from mice, fixed in 4% PFA or Bouin’s solution (Ricca Chemical) overnight at 4°C, and embedded in paraffin blocks. Then, 5-µm sections were cut using a Leica RM2125 microtome and stained with either hematoxylin and eosin (H&E; Poly Scientific R&D Corp.) or periodic Acid-Schiff Kit (PAS; Sigma-Aldrich) and mounted with Permount.
Immunofluorescence staining
Paraffin tissue sections were deparaffinized and processed for antigen retrieval by autoclaving for 17.5 min in a citrate-based antigen unmasking solution (Vector Laboratories). Tissues were then extracted with 0.5% Triton X-100 in PBS for 5 min or 0.5% SDS in PBS for 10 min. Cauda sperm were fixed in 4% PFA overnight at 4°C, carefully spread on glass slides, and extracted with 0.5% Triton X-100 in PBS for 5 min. Samples were blocked with 5% goat serum in antibody dilution buffer (2% BSA in PBS) for 1 h at room temperature. Primary antibodies, listed in Table S7, were diluted in antibody dilution buffer and applied to samples, followed by incubation overnight at 4°C. Samples were then washed three times with PBS for 5 min each and secondary antibodies in antibody dilution buffer were applied for 1 h at room temperature. After extensive washing with PBS, DAPI was applied for 5 min at room temperature, and the specimens were mounted with Fluoromount-G (Southern Biotech). Cby1 was detected using the Alexa Fluor 488 Tyramide SuperBoost Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions.
For frozen sections, testes were embedded in OCT, snap-frozen in 2-methylburane prechilled in liquid nitrogen, and cut into 10-µm-thick sections using a Leica CM1950 cryostat. The sections were rehydrated with PBS and then fixed with 100% acetone for 10 min at −20°C. Subsequent immunostaining procedures were performed as described above.
Image acquisition and analysis
Histology images of testes and DIC images of sperm were acquired on a Leica DMI6000B microscope with either a 20×/0.50 NA, 40×/0.75 NA, or 63×/1.25 NA oil objective equipped with a Leica DFC7000T camera. Fluorescent images were acquired on a Leica SP8X with an HCPL APO 100×/1.4 NA oil objective or a Zeiss LSM 980 with Airyscan 2 in AiryScan mode with Plan-Apochromat 63×/1.40 NA Oil DIC f/ELYRA or 100×/1.4 NA Oil DIC objectives. For structured illumination microscopy (SIM) imaging, specimens were analyzed by using a Nikon SIM (N-SIM) instrument with a 100×/1.49 NA objective. The Zeiss LSM 980 was equipped with T-PMT, MA-PMT, GaAsP-PMT, and Airyscan 2 multiplex with 4Y and 8Y. The N-SIM instrument was equipped with an Andor iXon3 897 electron-multiplying charge-coupled device (EMCCD) camera (Andor Technology).
All images were processed with the Leica Application Suite X, Zeiss Zen Blue, or the Nikon NIS-Elements image analysis software and then further processed with ImageJ, Adobe Photoshop, and Adobe Illustrator.
Generation and purification of Cby3 antibody
An antigen-purified guinea pig anti-Cby3 antibody was generated at Thermo Fisher Scientific against His-tagged full-length mouse Cby3. The antigen was expressed and purified from E. coli under denaturing conditions (8 M urea) using Ni-NTA His•Bind Resin (Millipore Sigma). Subsequently, the antigen was dialyzed against 10 mM sodium acetate buffer pH 5.0 containing 2 M urea.
Transmission electron microscopy (TEM) of testis and sperm samples
TEM was performed as previously described (Hoque et al., 2021; Siller et al., 2017). Briefly, adult testes or cauda epididymal sperm were isolated and immersion-fixed in 2% PFA and 2% glutaraldehyde in PBS overnight at 4°C. Seminiferous tubules were isolated and cut into about 1-mm3 pieces. Samples were washed with PBS, placed in 2% osmium tetroxide in PBS, dehydrated in a graded series of ethanol, and embedded in Embed 812 resin (Electron Microscopy Sciences). Ultrathin sections were cut at 80 nm using a Leica EM UC7 ultramicrotome and placed on Formvar-coated slot copper grids. Sections were then counterstained with uranyl acetate and lead citrate and viewed with a FEI Tecnai12 BioTwinG2 electron microscope. Digital images were acquired with an XR-60 CCD digital camera system (Advanced Microscopy Techniques).
Scanning electron microscopy (SEM) of cauda epididymal sperm
SEM was conducted as described (Korneev et al., 2021). Briefly, sperm were isolated from the cauda epididymis as above, diluted 1:10 in 100 mM sodium cacodylate, spun down, and fixed in 3.5% glutaraldehyde in 100 mM sodium cacodylate for 15 min at room temperature. Sperm were then deposited on a glass coverslip, washed three times in 100 mM sodium cacodylate, and dehydrated in a series of graded ethanol solutions (50%, 70%, 90%, and 100% twice) for 20 min each. The coverslips were subjected to chemical drying using a graded series of hexamethyldisilazane (HDMS; Electron Microscopy Sciences) and ethanol (1:2, 1:1, 2:1, and 100% HDMS twice) for 10 min each and allowed to dry overnight at room temperature. The coverslips were then mounted on a metal SEM stub, coated with a 5-nm-thick carbon layer, and viewed with a Zeiss Crossbeam 340 focused ion beam-scanning electron microscope with an InLens Duo detection system.
AlphaFold2-multimer prediction of the Cby3/ciBAR1 complex
The predicted structures of the Cby3/ciBAR1 were obtained by submitting the full-length human Cby3 sequence (UniProt ID: A6NI87) and the BAR-domain (aa 10–220) of human ciBAR1 (UniProt ID: A1XBS5) to the AlphaFold2-Multimer server (https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb) in a 2:2 ratio. Cross-chain hydrogen bonds critical for Cby3 and ciBAR1 interactions were identified using the Find polar contacts function in PyMol.
Ultrastructure expansion microscopy (U-ExM) of testis sections
U-ExM was performed as previously described with modifications (Gambarotto et al., 2019). In brief, mice were anesthetized with either isoflurane or a cocktail of ketamine and xylazine and perfused with 4% PFA and 20% acrylamide in PBS. Testes were isolated and postfixed in 4% PFA and 20% acrylamide in PBS overnight at 4°C. Testes were then cryoprotected in 30% sucrose in PBS overnight at 4°C, embedded in OCT compound, and snap-frozen in 2-methylburane prechilled in liquid nitrogen. Next, 10-µm-thick sections were cut on a Leica CM1950 cryostat and collected on glass coverslips. The specimens were further fixed in 4% PFA and 20% acrylamide in PBS for 3–4 h at 37°C and incubated in monomer solution (19% sodium acrylate, 10% acrylamide, and 0.1% bis-acrylamide in PBS) overnight at 4°C. Cover glasses containing tissue sections were inverted onto cold gelling solution (monomer solution with 0.5% TEMED and 0.5% APS, with APS added last) for 5 min on ice and then 90 min at 37°C.
After gelation, a 4-mm biopsy punch (Integra Miltex) was used to excise regions of interest, and punches were incubated in denaturation buffer (200 mM SDS, 200 mM NaCl, and 50 mM Tris in water, pH 6.8) in 1.5-ml tubes for 10 min at 65°C and then for 90 min at 95°C. Samples were then pre-expanded three times in water for 30 min at room temperature. Next, samples were washed with PBS three times for 10 min each and blocked with 5% goat serum in antibody dilution buffer (2% BSA in PBS) in a 24-well dish for 1 h at room temperature with gentle rocking. Samples were washed with PBS three times for 10 min each and incubated with primary antibodies in antibody dilution buffer overnight at 4°C. Samples were washed with PBS containing 0.05% Tween-20 three times for 30 min each at room temperature and incubated with secondary antibodies in antibody dilution buffer containing 0.05% DAPI overnight.
For full expansion, samples were incubated in water four times for 15 min each at room temperature and then overnight with gentle rocking. To properly orient the expanded samples, they were transferred to Delta T Culture Dishes with glass bottom (Thermo Fisher Scientific) and mounted under a Leica DMI6000B epifluorescence microscope. For SIM imaging, the samples were transferred to a poly-L-lysine-coated Delta T Culture Dishes, and a small drop of water (∼50 μl) was added to the samples to prevent shrinkage.
To determine the expansion factor of samples, several testicular sperm annuli were imaged by TEM, and the peak-to-peak (P2P) average width of the annulus was quantified and compared with the P2P average width of SEPT4-positive structures in expanded samples.
ExM data processing
For Fig. 7, A and C, N-SIM reconstructed images were imported into Fiji. To determine the diameter of individual ring structures, the line profile tool was used with a line width of 10 pixels. A line was drawn across a region of interest (ROI), and a fluorescence intensity plot profile was generated. The P2P width was then recorded and corrected by the expansion factor. At least 10 images were measured, with each containing at least three ring structures for each protein, and the average diameter was calculated. To analyze the distance between Cby3 rings and those of ciBAR1, CEP83, and SEPT4, only clear lateral views of the ring structures were selected. Reconstructed N-SIM images were imported into Fiji, and a two-color composite image was generated. Profile plots with a line width of 10 pixels were then drawn across ROIs. The positions of the lines were then added to the ROI manager for each channel, and multiplot profiles were generated. The P2P distance between Cby3 peaks and those of ciBAR1, CEP83, and SEPT4 was recorded and corrected by the expansion factor. At least 12 different images were quantified with each containing at least three structures of interest.
For Fig. 7 B, images with apparent ninefold symmetric fluorescent signals of Cby3, CEP83, and ciBAR1 were processed with the mean-shift super-resolution (MSSR) plugin on Fiji with the following parameters: AMP = 1, order = 1, and PSF computationally derived (Torres-García et al., 2022). After applying MSSR, a circle was drawn around the ROIs and the polar-transformed plugin was used to create a linearized readout of the image. A fluorescence intensity plot profile was then generated. For two-color images, each channel was linearized individually, a composite image was created, and a multiplot profile was generated. For symmetrization, MSSR-processed images were duplicated eight times and each duplicate was rotated in 40° intervals. The nine total images were then concatenated and a mean intensity z-stack was generated.
Alignment of TEM/U-ExM images
Representative TEM and U-ExM images with average annulus width were selected and scaled using scale bars. These images were then aligned in Adobe Illustrator using the centroid of the annulus signal as a reference point. Overlay images were then generated by setting the fluorescent image opacity to 40% and placing it on top of the TEM image. For easier visualization, cellular features were traced and rendered using Adobe Illustrator.
Sperm collection and motility assays
Both cauda epididymides were dissected from 2- to 4-mo-old mice, placed in 1 ml of modified Toyoda-Yokoyama-Hoshi (m-TYH) media, incubated for 10 min at 37°C for swim-out, and the cauda epididymides were discarded. The m-TYH medium is a non-capacitating (NC) medium and contains 119.37 mM NaCl, 4.78 mM KCl, 1.19 mM KH2 PO4, 1.19 mM MgSO4, 5.56 mM glucose, 1.71 mM CaCl2, 20 mM HEPES, 0.51 mM Na-pyruvate, 10 µg/ml gentamicin, and 0.0006% phenol red, pH 7.3 (Tourzani et al., 2018). For the capacitating (CAP) medium, the NC medium was supplemented with 15 mM NaHCO3 and 5 mg/ml BSA, pH 7.2–7.4. Both media were equilibrated in a humidified atmosphere of 5% CO2 at 37°C.
For sperm motility and hyperactivation measurements, after 1-h incubation in NC or CAP media in 5% CO2 at 37°C, sperm suspensions were loaded onto 80-µm-depth chamber slides (Leja slides; Spectrum Technologies), placed on a microscope stage at 37°C, and imaged with a 4× dark-field objective (Olympus) on an Axio Lab.A1 microscope (Zeiss). 90-frame videos were recorded at 60 Hz and analyzed using computer-assisted sperm analysis (CASA; CEROS II; Hamilton Throne Research) as described previously (Sánchez-Cárdenas et al., 2018). The settings for cell recognition included a minimum cell size of four pixels, static head size of 0.13–2.43, static head intensity of 0.10–1.52, and static head elongation of 5–100. Sperm with curvilinear velocity (VCL) > 271.00 µm/s, linearity <50.00%, and amplitude of lateral head >3.50 µm were considered hyperactive. At least 5 microscopy fields corresponding to a minimum of 200 spermatozoa were analyzed for each treatment in each experiment. Data were presented as the percentage of hyperactive sperm out of the motile population.
Western blotting of sperm lysates for phospho-PKA substrates and tyrosine phosphorylation
After 1-h incubation in NC or CAP media at 37°C, sperm (1 × 106) were collected by centrifugation at 12,100 × g for 5 min, washed twice with PBS, resuspended in Laemmli SDS samples buffer without β-mercaptoethanol, boiled for 5 min, and centrifuged at 12,100 × g for 5 min. Supernatants were supplemented with 5% β-mercaptoethanol and boiled for an additional 5 min. Protein extracts were analyzed by 8% SDS-PAGE and electrotransferred to PVDF membranes (Bio-Rad). Membranes were blocked with 10% fat-free milk in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST) for 1 h at room temperature and probed with phospho-PKA substrate antibody (1:10,000, 9624, clone 100G7, RRID:AB_331817; Cell Signaling Technology) in TBST containing 1% BSA overnight at 4°C. Membranes were washed with TBST six times for 10 min each and incubated with HRP-conjugated anti-rabbit secondary antibody (1:10,000; Jackson ImmunoResearch Laboratories) in TBST for 1 h at room temperature. Following six washes in TBST for 10 min each, an enhanced chemiluminescence detection kit (ECL Prime; GE Healthcare) was used for the detection of protein bands on x-ray films. After immunoblotting of phospho-PKA substrates, membranes were stripped in TBST supplemented with 0.7% β-mercaptoethanol for 20 min at 60°C with agitation and then washed with TBST six times for 10 min each. Membranes were blocked with TBST containing 20% fish gelatin (G7765; Sigma-Aldrich) for 1 h at room temperature, probed with anti-pY antibody (1:10,000, 05-321, clone 4G10, RRID:AB_309678; Millipore) overnight at 4°C, and incubated with HRP-conjugated anti-mouse secondary antibody (1:10,000; Jackson ImmunoResearch Laboratories) in TBST for 1 h at room temperature, followed by detection as above. Membranes were stripped again and probed with anti-α-tubulin antibody (2144S, RRID:AB_221058; Cell Signaling) as a loading control. Image analysis was performed using ImageJ. In all cases, results were divided by the α-tubulin intensity to account for small differences in loading. Ratios were then normalized to the WT CAP value for each respective replicate. Therefore, the value of each ratio of the WT CAP is 1 and the other ratios were normalized to that WT CAP value. The images shown are representative of experiments repeated three times (n = 5) using three different animals.
Mammalian expression constructs
An expression construct for Flag-Cby1 and HA-ciBAR1 was generated on CMV promoter-driven pCD-betaG-FLAG and pCS2+ backbones, respectively, as described previously (Li et al., 2016; Takemaru et al., 2003). A full-length human Cby3 plasmid was purchased from the DNASU Plasmid Repository (HsCD00865569). For the generation of Flag- and HA-tagged Cby3 plasmids, a Cby3 cDNA insert was amplified by PCR and subcloned into pCD-betaG-Flag and pCS2+HA, respectively. Primers for generating both Flag- and HA-Cby3 plasmids were 5′-TCAGGAATTCAATGCCCTCCCTGTCCACCTT-3′ and 5′-TCAGCTCGAGTCACTGCGAGTCCAGAGCGC-3′. To generate the expression plasmids for Flag-tagged Cby3 point mutants, cDNAs encoding individual Cby3 mutants were created by PCR-based mutagenesis and subcloned into pCD-betaG-Flag. A primer for creating Cby3 S5A mutation was 5′-TCAGGAATTCCATGCCCTCCCTGGCCACCTTCTACCTGCTGG-3′; primers for Cby3 E18A mutation were 5′-ACAACACGCGCCAGGCCGCGCTGGGCCTCGCCTACGG-3′ and 5′-CCGTAGGCGAGGCCCAGCGCGGCCTGGCGCGTGTTGT-3′; primers for Cby3 G20A mutation were 5′-CGCGCCAGGCCGAGCTGGCCCTCGCCTACGGCGCGCC-3′ and 5′-GGCGCGCCGTAGGCGAGGGCCAGCTCGGCCTGGCGCG-5’; primers for Cby3 G40A mutation were 5′- CCTTCGTGTTCCGCGGCGCGCGGTGGACCACTGAGAG-3′ and 5′-CTCTCAGTGGTCCACCGCGCGCCGCGGAACACGAAGG-3′; primers for Cby3 W42A mutation were 5′-TGTTCCGCGGCGGGCGGGCGACCACTGAGAGCCAGCT-3′ and 5′- AGCTGGCTCTCAGTGGTCGCCCGCCCGCCGCGGAACA-3′. The coding regions of all constructs were validated by DNA sequencing.
Cell culture, transfection, and coimmunoprecipitation assays
HEK293T cells were obtained from ATCC (CRL-3216) and maintained in DMEM with 10% FBS and 100 U/ml penicillin-streptomycin. Transfections were performed using Lipofectamine 3000 (Thermo Fisher Scientific). Co-IP and immunoblotting were performed as previously described (Li et al., 2010, 2016). Briefly, transfected HEK293T cells were collected and lysed in ice-cold lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, and 10% glycerol) with protease inhibitor cocktail (Roche) and incubated for 20 min on ice with intermittent agitation, followed by sonication. Cell lysates were cleared by centrifugation at 12,000 rpm for 10 min at 4°C. The supernatants were then incubated with 2 µg of primary antibody for 3 h overnight at 4°C, followed by the addition of 20 μl of a mixture of protein A and G beads (Sigma-Aldrich) and rotation for 3 h at 4°C. The beads were collected and washed three times with 1 ml of ice-cold lysis buffer for 10 min each before SDS-PAGE. The primary antibodies used were mouse Flag M2 (1:1,000, F3165, RRID:AB_259529; Sigma-Aldrich) and rat HA (1:500, 11867423001, clone 3F10, RRID:AB_390918; Roche) antibodies. HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Protein bands were visualized using chemiluminescence (SuperSignal West Pico PLUS Chemiluminescent Substrate; Pierce) with x-ray films, and the developed films were then scanned.
Western blot analysis of testis lysates
Testis samples were harvested from adult WT, Cby3−/−, and ciBAR1−/− mice. Testis lysates were prepared by homogenization at 4°C in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) and supplemented with protease inhibitor cocktail (Roche). Protein concentration was determined using the Bio-Rad DC protein assay and 20 µg of protein per lane was loaded onto a SDS PAGE and subjected to Western blotting. The primary antibodies used were rabbit Cby3 (1:500; a gift from Proteintech), rabbit ciBAR1 (1:500, 26013-1-AP, RRID:AB_2879735; Proteintech), and mouse GAPDH (1:10,000, H86504M; Biodesign International) antibodies. HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Protein bands were visualized using chemiluminescence (SuperSignal West Pico PLUS Chemiluminescent Substrate; Pierce) with x-ray films, and the developed films were then scanned.
Quantification and statistical analysis
Two-tailed Student’s t tests were used to determine statistical significance, and P < 0.05 was considered to be significant. Asterisks were used to indicate P values as follows: *P < 0.05; **P < 0.01; ***P < 0.001; and ns, not significant.
Online supplemental material
This manuscript is accompanied by five supplemental figures. Fig. S1 shows multiple sequence alignments of Cby3 proteins. It also contains AlphaFold2-Miltimer prediction of the Cby3/ciBAR1 complex related to Fig. 4 E. Fig. S2 shows expression of Cby1, Cby3, ciBAR1, and ciBAR2 in the testis. Fig. S3 shows the generation and initial characterization of Cby3−/− mice. It also includes Western blots of sperm lysates using phospho-PKA substrate antibody and phospho-tyrosine antibody, related to Fig. 2 D. Fig. S4 shows the generation and initial characterization of ciBAR1−/− mice. Fig. S5 shows the calculation of the expansion factor, related to Fig. 7. Table S1 shows Mendelian ratios of Cby3−/− mice. Table S2 shows the fertility of Cby3−/− male mice. Table S3 shows Mendelian ratios of ciBAR1−/− mice. Table S4 shows the fertility of ciBAR1−/− male mice. Table S5 contains measurements of diameters and distances from Cby3 at the flagellar base by U-ExM. Table S6 contains measurements of diameters and distances from Cby3 at the MP/PP junction by U-ExM. Table S7 contains a list of primary antibodies used for immunofluorescence staining.
Supplementary Material
shows Cby3−/− mice are born at predicted Mendelian ratios.
shows fertility test of Cby3−/− male mice.
shows ciBAR1−/− mice are born at a lower-than-expected Mendelian ratio.
shows the fertility test of ciBAR1−/− male mice.
shows diameters and distances from Cby3 at the flagellar base as measured by U-ExM.
shows diameters and distances from Cby3 at the MP/PP junction as measured by U-ExM.
shows antibodies used for immunofluorescence staining.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
Acknowledgments
We would like to thank Drs. Jean-Ju Chung, Yaron Fuchs, and Haoyu Qi for reagents and mouse testis paraffin blocks, and Dr. Michael Frohman for critical reading of the manuscript. We acknowledge the Research Histology Core Laboratory in the Department of Pathology at Stony Brook University School of Medicine for assistance with histological preparations and the Central Microscopy Imaging Center Core at Stony Brook University for assistance with confocal microscopy, SIM, TEM, and SEM.
This work was supported by grants from National Institute of Diabetes and Digestive and Kidney Diseases (R01DK123641 to K.-I. Takemaru), National Institute of Child Health and Human Development (R01HD109232 to K.-I. Takemaru, R01HD38082 to P.E. Visconti, F31HD111271 to M. Hoque), and National Heart, Lung, and Blood Institute (F31HL168828 to E.N. Kim).
Author contributions: Conceptualization, M. Hoque, P.E. Visconti, and K.-I. Takemaru; Formal analysis, M. Hoque; Funding acquisition; M. Hoque, E.N. Kim, P.E. Visconti, and K.-I. Takemaru; Investigation, M. Hoque, W.D. Weber, and F.-Q. Li; Methodology, M. Hoque, W.D. Weber, F.-Q. Li, and K.-I. Takemaru; Resources, P.-L. Kuo and P.E. Visconti; Supervision, P.E. Visconti and K.-I. Takemaru; Validation, M. Hoque, F.-Q. Li, E.N. Kim, and J.J. Chen; Writing-original draft, M. Hoque, F.-Q. Li, W.D. Weber, and K.-I. Takemaru; and Writing-review and editing, E.N. Kim, J.J. Chen, P.-L. Kuo, and P.E. Visconti. All authors read and approved the final manuscript.
Data availability
The data are shown in the published article and additional data files are available upon reasonable request.
References
- Avidor-Reiss, T., Ha A., and Basiri M.L.. 2017. Transition zone migration: A mechanism for cytoplasmic ciliogenesis and postaxonemal centriole elongation. Cold Spring Harb. Perspect. Biol. 9:a028142. 10.1101/cshperspect.a028142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss, T., Khire A., Fishman E.L., and Jo K.H.. 2015. Atypical centrioles during sexual reproduction. Front. Cell Dev. Biol. 3:21. 10.3389/fcell.2015.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellvé, A.R., Cavicchia J.C., Millette C.F., O’Brien D.A., Bhatnagar Y.M., and Dym M.. 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 74:68–85. 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler, M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., and Loncarek J.. 2019. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 10:993. 10.1038/s41467-018-08216-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, M.C., Li F.Q., Cyge B., Arashiro T., Brechbuhl H.M., Chen X., Siller S.S., Weiss M.A., O’Connell C.B., Love D., et al. 2014. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J. Cell Biol. 207:123–137. 10.1083/jcb.201406140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman, P.J., and Dominguez R.. 2018. BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys. Rev. 10:1587–1604. 10.1007/s12551-018-0467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, S.E., Davies E.M., Fulcher A.J., Oorschot V., and Mitchell C.A.. 2021. Superresolution microscopy reveals distinct phosphoinositide subdomains within the cilia transition zone. Front. Cell Dev. Biol. 9:634649. 10.3389/fcell.2021.634649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyge, B., Voronina V., Hoque M., Kim E.N., Hall J., Bailey-Lundberg J.M., Pazour G.J., Crawford H.C., Moon R.T., Li F.Q., and Takemaru K.I.. 2021. Loss of the ciliary protein Chibby1 in mice leads to exocrine pancreatic degeneration and pancreatitis. Sci. Rep. 11:17220. 10.1038/s41598-021-96597-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derderian, C., Canales G.I., and Reiter J.F.. 2023. Seriously cilia: A tiny organelle illuminates evolution, disease, and intercellular communication. Dev. Cell. 58:1333–1349. 10.1016/j.devcel.2023.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirami, T., Rode B., Wolf J.P., Gacon G., Dulioust E., and Touré A.. 2015. Assessment of the frequency of sperm annulus defects in a large cohort of patients presenting asthenozoospermia. Basic Clin. Androl. 25:10. 10.1186/s12610-015-0026-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epting, D., Senaratne L.D.S., Ott E., Holmgren A., Sumathipala D., Larsen S.M., Wallmeier J., Bracht D., Frikstad K.M., Crowley S., et al. 2020. Loss of CBY1 results in a ciliopathy characterized by features of Joubert syndrome. Hum. Mutat. 41:2179–2194. 10.1002/humu.24127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, C., Eling N., Martinez-Jimenez C.P., Marioni J.C., and Odom D.T.. 2019. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun. 10:1251. 10.1038/s41467-019-09182-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett, D.W., Eddy E.M., and Phillips D.M.. 1970. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol. Reprod. 2:129–153. 10.1095/biolreprod2.1.129 [DOI] [PubMed] [Google Scholar]
- Feige, E., Chen A., and Motro B.. 2002. Nurit, a novel leucine-zipper protein, expressed uniquely in the spermatid flower-like structure. Mech. Dev. 117:369–377. 10.1016/S0925-4773(02)00217-4 [DOI] [PubMed] [Google Scholar]
- Gambarotto, D., Zwettler F.U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J.G., Reuss M., Unser M., et al. 2019. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods. 16:71–74. 10.1038/s41592-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo, F.R., Phua S.C., Roberson E.C., Garcia G. III, Abedin M., Schurmans S., Inoue T., and Reiter J.F.. 2015. Phosphoinositides regulate ciliary protein trafficking to modulate hedgehog signaling. Dev. Cell. 34:400–409. 10.1016/j.devcel.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, C.D., Ma Q., Manske G.L., Shami A.N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S.J., Moore B.B., et al. 2018. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell. 46:651–667.e10. 10.1016/j.devcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold, M.D. 2016. Spermatogenesis: The commitment to meiosis. Physiol. Rev. 96:1–17. 10.1152/physrev.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, J., Kinoshita M., and Yuan L.. 2009. Spatiotemporal association of DNAJB13 with the annulus during mouse sperm flagellum development. BMC Dev. Biol. 9:23. 10.1186/1471-213X-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J., Grow E.J., Mlcochova H., Maher G.J., Lindskog C., Nie X., Guo Y., Takei Y., Yun J., Cai L., et al. 2018. The adult human testis transcriptional cell atlas. Cell Res. 28:1141–1157. 10.1038/s41422-018-0099-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque, M., Chen D., Hess R.A., Li F.Q., and Takemaru K.I.. 2021. CEP164 is essential for efferent duct multiciliogenesis and male fertility. Reproduction. 162:129–139. 10.1530/REP-21-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque, M., Kim E.N., Chen D., Li F.Q., and Takemaru K.I.. 2022. Essential roles of efferent duct multicilia in male fertility. Cells. 11:341. 10.3390/cells11030341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara, M., Kinoshita A., Yamada S., Tanaka H., Tanigaki A., Kitano A., Goto M., Okubo K., Nishiyama H., Ogawa O., et al. 2005. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell. 8:343–352. 10.1016/j.devcel.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Irons, M.J., and Clermont Y.. 1982. Kinetics of fibrous sheath formation in the rat spermatid. Am. J. Anat. 165:121–130. 10.1002/aja.1001650204 [DOI] [PubMed] [Google Scholar]
- Jumper, J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie, T., Love J.F., Fisher S.D., Gustavsson A.K., and Jackson P.K.. 2023. A hierarchical pathway for assembly of the distal appendages that organize primary cilia. bioRxiv. 10.1101/2023.01.06.522944 (Preprint posted January 10, 2023). [DOI] [Google Scholar]
- Kissel, H., Georgescu M.-M., Larisch S., Manova K., Hunnicutt G.R., and Steller H.. 2005. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev. Cell. 8:353–364. 10.1016/j.devcel.2005.01.021 [DOI] [PubMed] [Google Scholar]
- Korneev, D., Merriner D.J., Gervinskas G., de Marco A., and O’Bryan M.K.. 2021. New insights into sperm ultrastructure through enhanced scanning electron microscopy. Front. Cell Dev. Biol. 9:672592. 10.3389/fcell.2021.672592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., and Reiter J.. 2021. How the centriole builds its cilium: Of mothers, daughters, and the acquisition of appendages. Curr. Opin. Struct. Biol. 66:41–48. 10.1016/j.sbi.2020.09.006 [DOI] [PubMed] [Google Scholar]
- Kuo, Y.-C., Shen Y.-R., Chen H.-I., Lin Y.-H., Wang Y.-Y., Chen Y.-R., Wang C.-Y., and Kuo P.-L.. 2015. SEPT12 orchestrates the formation of mammalian sperm annulus by organizing core octameric complexes with other SEPT proteins. J. Cell Sci. 128:923–934. [DOI] [PubMed] [Google Scholar]
- Kuo, Y.C., Lin Y.H., Chen H.I., Wang Y.Y., Chiou Y.W., Lin H.H., Pan H.A., Wu C.M., Su S.M., Hsu C.C., and Kuo P.L.. 2012. SEPT12 mutations cause male infertility with defective sperm annulus. Hum. Mutat. 33:710–719. 10.1002/humu.22028 [DOI] [PubMed] [Google Scholar]
- Kwitny, S., Klaus A.V., and Hunnicutt G.R.. 2010. The annulus of the mouse sperm tail is required to establish a membrane diffusion barrier that is engaged during the late steps of spermiogenesis. Biol. Reprod. 82:669–678. 10.1095/biolreprod.109.079566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.L., Santé J., Comerci C.J., Cyge B., Menezes L.F., Li F.-Q., Germino G.G., Moerner W.E., Takemaru K., and Stearns T.. 2014. Cby1 promotes Ahi1 recruitment to a ring-shaped domain at the centriole-cilium interface and facilitates proper cilium formation and function. Mol. Biol. Cell. 25:2919–2933. 10.1091/mbc.e14-02-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti, M.S., and Sironen A.. 2017. Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 97:522–536. 10.1093/biolre/iox096 [DOI] [PubMed] [Google Scholar]
- Lewandoski, M., Wassarman K.M., and Martin G.R.. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7:148–151. 10.1016/S0960-9822(06)00059-5 [DOI] [PubMed] [Google Scholar]
- Lhuillier, P., Rode B., Escalier D., Lorès P., Dirami T., Bienvenu T., Gacon G., Dulioust E., and Touré A.. 2009. Absence of annulus in human asthenozoospermia: Case report. Hum. Reprod. 24:1296–1303. 10.1093/humrep/dep020 [DOI] [PubMed] [Google Scholar]
- Li, F.Q., Chen X., Fisher C., Siller S.S., Zelikman K., Kuriyama R., and Takemaru K.I.. 2016. BAR domain-containing FAM92 proteins interact with Chibby1 to facilitate ciliogenesis. Mol. Cell. Biol. 36:2668–2680. 10.1128/MCB.00160-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F.Q., Mofunanya A., Fischer V., Hall J., and Takemaru K.. 2010. Nuclear-cytoplasmic shuttling of Chibby controls beta-catenin signaling. Mol. Biol. Cell. 21:311–322. 10.1091/mbc.e09-05-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, D., Li F.Q., Burke M.C., Cyge B., Ohmitsu M., Cabello J., Larson J.E., Brody S.L., Cohen J.C., and Takemaru K.. 2010. Altered lung morphogenesis, epithelial cell differentiation and mechanics in mice deficient in the Wnt/β-catenin antagonist Chibby. PLoS One. 5:e13600. 10.1371/journal.pone.0013600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar, G., and Schatten G.. 2000. Centrosome reduction during rhesus spermiogenesis: Gamma-tubulin, centrin, and centriole degeneration. Mol. Reprod. Dev. 56:502–511. [DOI] [PubMed] [Google Scholar]
- Manandhar, G., Sutovsky P., Joshi H.C., Stearns T., and Schatten G.. 1998. Centrosome reduction during mouse spermiogenesis. Dev. Biol. 203:424–434. 10.1006/dbio.1998.8947 [DOI] [PubMed] [Google Scholar]
- Mill, P., Christensen S.T., and Pedersen L.B.. 2023. Primary cilia as dynamic and diverse signalling hubs in development and disease. Nat. Rev. Genet. 24:421–441. 10.1038/s41576-023-00587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofunanya, A., Li F.Q., Hsieh J.C., and Takemaru K.. 2009. Chibby forms a homodimer through a heptad repeat of leucine residues in its C-terminal coiled-coil motif. BMC Mol. Biol. 10:41. 10.1186/1471-2199-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, G., Tourzani D.A., Gervasi M.G., Houle E., Oluwayiose O., Suvorov A., Pilsner J.R., and Visconti P.E.. 2023. Effects of preconception exposure to phthalates on mouse sperm capacitation parameters. Andrology. 11:1484–1494. 10.1111/andr.13423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg, E.F. 1956. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 99:507–516. 10.1002/aja.1000990307 [DOI] [PubMed] [Google Scholar]
- Pereira, R., and Sousa M.. 2023. Morphological and molecular bases of male infertility: A closer look at sperm flagellum. Genes. 14:383. 10.3390/genes14020383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, E., Michael O.N., Alexander P., Natasha A., Andrew S., Tim G., Augustin Ž., Russ B., Sam B., Jason Y., et al. 2022. Protein complex prediction with AlphaFold-multimer. bioRxiv. 10.1101/2021.10.04.463034 (Preprint posted March 10, 2022). [DOI] [Google Scholar]
- Sánchez-Cárdenas, C., Montoya F., Navarrete F.A., Hernández-Cruz A., Corkidi G., Visconti P.E., and Darszon A.. 2018. Intracellular Ca2+ threshold reversibly switches flagellar beat off and on. Biol. Reprod. 99:1010–1021. 10.1093/biolre/ioy132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen, I., Giese A.P., Aziz A., Lafont D.T., Chakchouk I., Santos-Cortez R.L.P., Lee K., Acharya A., Khan F.S., Ullah A., et al. 2019. FAM92A underlies nonsyndromic postaxial polydactyly in humans and an abnormal limb and digit skeletal phenotype in mice. J. Bone Miner. Res. 34:375–386. 10.1002/jbmr.3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y.R., Wang H.Y., Kuo Y.C., Shih S.C., Hsu C.H., Chen Y.R., Wu S.R., Wang C.Y., and Kuo P.L.. 2017. SEPT12 phosphorylation results in loss of the septin ring/sperm annulus, defective sperm motility and poor male fertility. PLoS Genet. 13:e1006631. 10.1371/journal.pgen.1006631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller, S.S., Sharma H., Li S., Yang J., Zhang Y., Holtzman M.J., Winuthayanon W., Colognato H., Holdener B.C., Li F.Q., and Takemaru K.I.. 2017. Conditional knockout mice for the distal appendage protein CEP164 reveal its essential roles in airway multiciliated cell differentiation. PLoS Genet. 13:e1007128. 10.1371/journal.pgen.1007128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic, M., Evergren E., Callan-Jones A., and Bassereau P.. 2019. Curving cells inside and out: Roles of BAR domain proteins in membrane shaping and its cellular implications. Annu. Rev. Cell Dev. Biol. 35:111–129. 10.1146/annurev-cellbio-100617-060558 [DOI] [PubMed] [Google Scholar]
- Steels, J.D., Estey M.P., Froese C.D., Reynaud D., Pace-Asciak C., and Trimble W.S.. 2007. Sept12 is a component of the mammalian sperm tail annulus. Cell Motil. Cytoskeleton. 64:794–807. 10.1002/cm.20224 [DOI] [PubMed] [Google Scholar]
- Steere, N., Chae V., Burke M., Li F.Q., Takemaru K., and Kuriyama R.. 2012. A Wnt/beta-catenin pathway antagonist Chibby binds Cenexin at the distal end of mother centrioles and functions in primary cilia formation. PLoS One. 7:e41077. 10.1371/journal.pone.0041077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu, S., Kurisu S., and Takenawa T.. 2014. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol. Rev. 94:1219–1248. 10.1152/physrev.00040.2013 [DOI] [PubMed] [Google Scholar]
- Sugino, Y., Ichioka K., Soda T., Ihara M., Kinoshita M., Ogawa O., and Nishiyama H.. 2008. Septins as diagnostic markers for a subset of human asthenozoospermia. J. Urol. 180:2706–2709. 10.1016/j.juro.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Takei, G.L., Tourzani D.A., Paudel B., and Visconti P.E.. 2021. Activation of cAMP-dependent phosphorylation pathways is independent of ROS production during mouse sperm capacitation. Mol. Reprod. Dev. 88:544–557. 10.1002/mrd.23524 [DOI] [PubMed] [Google Scholar]
- Takemaru, K., Yamaguchi S., Lee Y.S., Zhang Y., Carthew R.W., and Moon R.T.. 2003. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 422:905–909. 10.1038/nature01570 [DOI] [PubMed] [Google Scholar]
- Teves, M.E., and Roldan E.R.S.. 2022. Sperm bauplan and function and underlying processes of sperm formation and selection. Physiol. Rev. 102:7–60. 10.1152/physrev.00009.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-García, E., Pinto-Cámara R., Linares A., Martínez D., Abonza V., Brito-Alarcón E., Calcines-Cruz C., Valdés-Galindo G., Torres D., Jabloñski M., et al. 2022. Extending resolution within a single imaging frame. Nat. Commun. 13:7452. 10.1038/s41467-022-34693-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure, A., Rode B., Hunnicutt G.R., Escalier D., and Gacon G.. 2011. Septins at the annulus of mammalian sperm. Biol. Chem. 392:799–803. 10.1515/BC.2011.074 [DOI] [PubMed] [Google Scholar]
- Tourzani, D.A., Paudel B., Miranda P.V., Visconti P.E., and Gervasi M.G.. 2018. Changes in protein O-GlcNAcylation during mouse epididymal sperm maturation. Front. Cell Dev. Biol. 6:60. 10.3389/fcell.2018.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel, M., Rosa E Silva I., and Andreeva A.. 2022. Structural validation and assessment of AlphaFold2 predictions for centrosomal and centriolar proteins and their complexes. Commun. Biol. 5:312. 10.1038/s42003-022-03269-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina, V.A., Takemaru K., Treuting P., Love D., Grubb B.R., Hajjar A.M., Adams A., Li F.Q., and Moon R.T.. 2009. Inactivation of Chibby affects function of motile airway cilia. J. Cell Biol. 185:225–233. 10.1083/jcb.200809144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeier, J., Nielsen K.G., Kuehni C.E., Lucas J.S., Leigh M.W., Zariwala M.A., and Omran H.. 2020. Motile ciliopathies. Nat. Rev. Dis. Primers. 6:77. 10.1038/s41572-020-0209-6 [DOI] [PubMed] [Google Scholar]
- Wang, G., Zhu X., Gao Y., Lv M., Li K., Tang D., Wu H., Xu C., Geng H., Shen Q., et al. 2022. Biallelic loss-of-function mutations in SEPTIN4 (C17ORF47), encoding a conserved annulus protein, cause thin midpiece spermatozoa and male infertility in humans. Hum. Mutat. 43:2079–2090. 10.1002/humu.24475 [DOI] [PubMed] [Google Scholar]
- Weirich, C.S., Erzberger J.P., and Barral Y.. 2008. The septin family of GTPases: Architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9:478–489. 10.1038/nrm2407 [DOI] [PubMed] [Google Scholar]
- Xu, K., Yang L., Zhang L., and Qi H.. 2020. Lack of AKAP3 disrupts integrity of the subcellular structure and proteome of mouse sperm and causes male sterility. Development. 147:dev181057. 10.1242/dev.181057 [DOI] [PubMed] [Google Scholar]
- Yang, T.T., Chong W.M., Wang W.J., Mazo G., Tanos B., Chen Z., Tran T.M.N., Chen Y.D., Weng R.R., Huang C.E., et al. 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023. 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Khan Z., and Westlake C.J.. 2023. Ciliogenesis membrane dynamics and organization. Semin. Cell Dev. Biol. 133:20–31. 10.1016/j.semcdb.2022.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows Cby3−/− mice are born at predicted Mendelian ratios.
shows fertility test of Cby3−/− male mice.
shows ciBAR1−/− mice are born at a lower-than-expected Mendelian ratio.
shows the fertility test of ciBAR1−/− male mice.
shows diameters and distances from Cby3 at the flagellar base as measured by U-ExM.
shows diameters and distances from Cby3 at the MP/PP junction as measured by U-ExM.
shows antibodies used for immunofluorescence staining.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
Data Availability Statement
The data are shown in the published article and additional data files are available upon reasonable request.