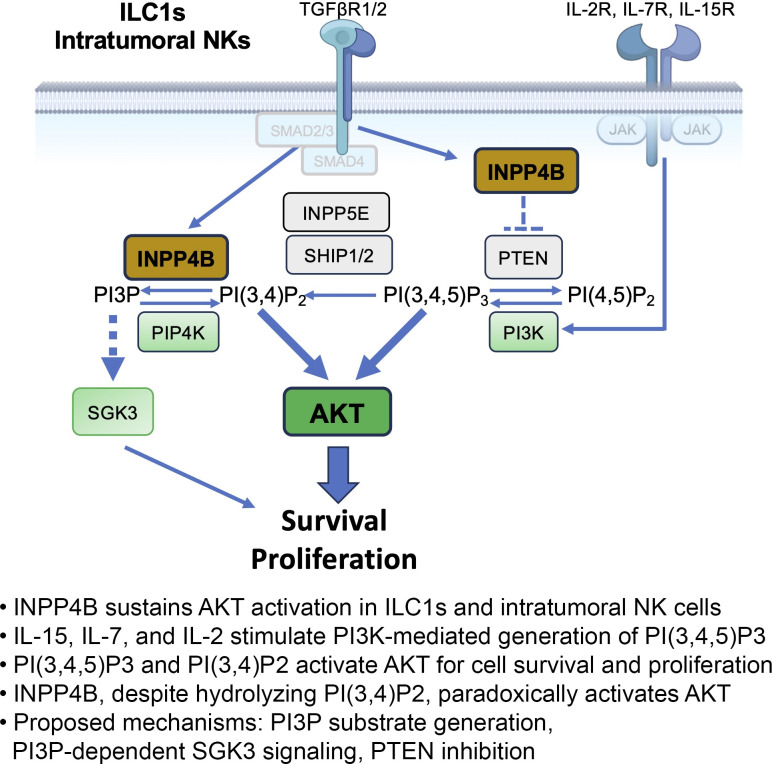
In this study, it was found that the inositol phosphatase INPP4B is required to sustain ILC1s in peripheral tissues and NK cells in the tumor microenvironment through activation of AKT.
Abstract
Innate lymphoid cells (ILCs) are a heterogeneous population of lymphocytes that coordinate early immune responses and maintain tissue homeostasis. Type 1 innate immune responses are mediated by natural killer (NK) cells and group 1 ILCs (ILC1s). Despite their shared features, NK cells and ILC1s display profound differences among various tissue microenvironments. Here, we identify the inositol polyphosphatase INPP4B as a hallmark feature of tissue-resident ILC1s and intratumoral NK cells using an scRNA-seq atlas of tissue-associated and circulating NK/ILC1s. Conditional deletion of Inpp4b in ILC1s and NK cells reveals that it is necessary for the homeostasis of tissue-resident ILC1s but not circulating NK cells at steady-state. Inpp4b-deficient cells display increased rates of apoptosis and reduced activation of the prosurvival molecule AKT. Furthermore, expression of Inpp4b by NK/ILC1s is necessary for their presence in the intratumoral environment, and lack of Inpp4b impairs antitumor immunity. These findings highlight INPP4B as a novel regulator of tissue residency and antitumor function in ILC1s and NK cells.
Graphical Abstract
Introduction
Innate lymphoid cells (ILCs) are a heterogeneous population of lymphocytes that share a common ID2+ progenitor, a reliance on common γ chain cytokines for their maturation and survival, and a lack of somatically rearranged T and B cell antigen receptors. Natural killer (NK) cells were first characterized based on their remarkable cytotoxic potential against tumor cells and were later shown to play important roles in antiviral immunity. Three additional groups of ILCs have been identified based on the expression of signature cytokines and master transcriptional regulators (Peng and Colonna, 2023; Vivier et al., 2018). Group 1 ILCs (ILC1s) produce IFNγ and TNFα and express the transcription factor T-BET. Group 2 ILCs (ILC2s) produce IL-5 and IL-13 and express high levels of GATA-3. Group 3 ILCs (ILC3s) produce IL-22 and IL-17 and depend on the transcription factor RORγt.
The phenotypes of NK cells and ILC1s share many important similarities. Both populations are identified by surface expression of NK1.1 and NKp46 in C57BL/6 mice and lack traditional lineage markers (CD3, CD5, CD19). However, ILC1s and NK cells differ in several notable aspects. In contrast to NK cells, which circulate freely throughout the body, ILC1s are largely tissue resident (Gasteiger et al., 2015). Moreover, ILC1s develop independently of the transcription factors EOMES and NFIL3, which are both necessary for NK differentiation (Cortez et al., 2014; Sojka et al., 2014). Conversely, the transcription factor HOBIT is critical for the development of hepatic ILC1s while being dispensable for NK cell differentiation (Friedrich et al., 2021; Yomogida et al., 2021).
The tissue residency program of ILC1s is associated with a discrete set of phenotypic and transcriptional markers. In contrast to circulating NK cells, which express the integrin CD49b and the homing molecule CD62L, ILC1s express CD49a and CD69, which are both associated with retention in the tissue. Previous work has demonstrated an important role for TGFβ imprinting in establishing the phenotype and transcriptional programs of ILC1s in the salivary gland and tumor microenvironment (TME; Cortez et al., 2016; Gao et al., 2017). TGFβ signaling through SMAD2/3 induces the transcription of HOBIT, markers of tissue residency such as CD49a, and other ILC1 markers, such as CD73 and TRAIL. Concurrently, SMAD4-deficient NK cells, which display unrestrained TGFβ signaling, acquire an ILC1-like phenotype and transcriptome (Cortez et al., 2017). While the role of TGFβ family signaling is well recognized in promoting the differentiation of ILC1s and tissue-resident memory CD8+ T (CD8+ TRM) cells (Cortez et al., 2016; Mackay et al., 2015), the downstream signaling pathways that are induced by TGFβ are less well defined.
To address this gap in knowledge, we conducted a comprehensive analysis of existing bulk and single-cell transcriptomic datasets, comparing ILC1s and NK cells (McFarland et al., 2021). Through this analysis, we identified the inositol phosphatase INPP4B as a consistent and robust feature of tissue-resident ILC1s as well as intratumoral NK cells. INPP4B, along with INPP4A, belongs to a family of mammalian inositol polyphosphate 4-phosphatase enzymes (Ferron and Vacher, 2006). The function of INPP4B has largely been characterized in cancer biology where it has been shown to metabolize phosphoinositide (PI) species, catalyzing the metabolism of PtdIns(3,4)P2 to PtdIns(3)P to regulate AKT signaling (Agoulnik et al., 2011; Fedele et al., 2010; Hamila et al., 2021; Hodgson et al., 2011). Alternatively, INPP4B may negatively regulate Protein Kinase C signaling through the dephosphorylation of PtdIns(4,5)P2 (Hodgson et al., 2014). While these functions suggest an inhibitory role for INPP4B, it has paradoxically been shown to positively regulate AKT phosphorylation and act as an oncogenic driver (Hamila et al., 2021). Aside from its role in AKT signal transduction, INPP4B has also been demonstrated to have an AKT-independent role through its product PtdIns(3)P, which activates serum/glucocorticoid-regulated kinase family member 3 (SGK3) to promote cell proliferation and survival (Gasser et al., 2014; Pokorny et al., 2021). PtdIns(3)P also has an important role in shaping early endosomes and cargo trafficking, and INPP4B has been shown to be involved in the recycling of membrane receptors such as TGFβRII and EGFR (Aki et al., 2020; Rodgers et al., 2021). Inpp4b−/− mice are viable but exhibit decreased bone mass and osteoporosis attributed to dysregulated osteoclast differentiation (Ferron et al., 2011).
We utilized a mouse model of conditional deletion of Inpp4b in NKp46-expressing cells to study its function in ILC1s and NK cells. We observed a selective reduction of Inpp4b-deficient ILC1s in the peripheral tissues, consistent with the high expression of Inpp4b in these cells. Biochemical analysis showed that lack of Inpp4b impaired the induction of the prosurvival molecule AKT, which was paralleled by a proapoptotic transcriptional signature. Lastly, we observed a reduction of Inpp4b-deficient NK cells in tumor infiltrates, leading to impaired antitumor immunity against B16 melanoma metastases. These findings elucidate a novel role for INPP4B in supporting the maintenance of tissue-resident ILC1s and intratumoral NK cells.
Results and discussion
Inpp4b is a transcriptional hallmark of tissue-resident lymphocytes
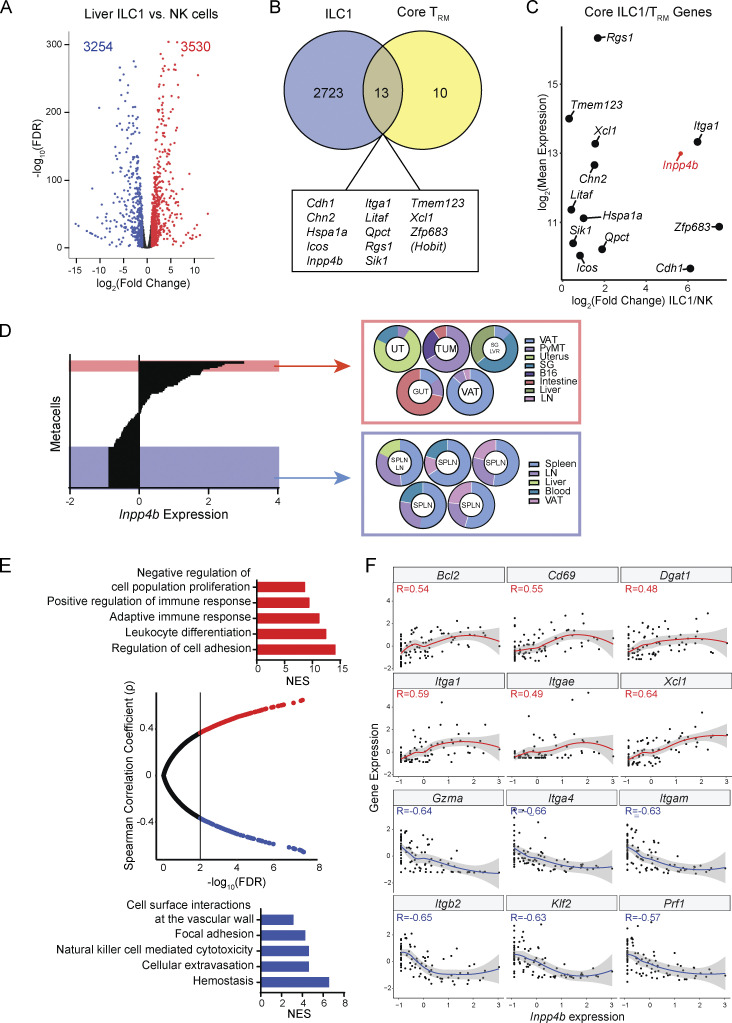
Despite overlapping surface markers, the phenotype and function of ILC1s and NK cells exhibit major differences. To gain an understanding of the molecular features of ILC1s that separate this lineage from NK cells, we examined previously generated bulk RNA sequencing (RNA-seq) transcriptomes of both cell types from the murine liver, where ILC1s are most abundant (Weizman et al., 2017). Differential expression analysis between these two cell types revealed 6,784 differentially expressed genes (DEGs), suggesting major transcriptional differences between these two cell types (Fig. 1 A). One major hallmark of ILC1s is that they are largely tissue resident whereas NK cells are freely circulating. This tissue-resident program of ILC1s shares many similarities with that of CD8+ TRM cells including the transcriptional regulator HOBIT, which is necessary for tissue residency of CD8+ TRM cells and liver ILC1s (Mackay et al., 2016). We reasoned that with regard to the tissue-resident phenotype of ILC1s, the most robust hallmarks of this program would also be shared with CD8+ TRM transcriptional programs. Thus, we overlapped our ILC1-enriched gene set with genes enriched in TRM from multiple tissues (Mackay et al., 2013; Fig. 1 B). From this approach, we derived a core set of genes that were universally enriched among ILC1s and CD8+ TRM cells, irrespective of tissue origin. In validation of our approach, we noticed several genes in our core set that had known significance in ILC1 development and function (e.g., Hobit, Itga1, Icos, Xcl1). In addition to these known hallmarks, we observed several genes with no known function in immunity such as Qpct, Tmem123, and Inpp4b. Out of these molecules, the inositol phosphatase Inpp4b stood out as both highly expressed and highly enriched in ILC1s (Fig. 1 C). As our initial approach relied on bulk transcriptomics from a limited set of tissues, we turned to a recently generated single-cell atlas of ILC1s and NK cells from 12 different tissues, including two different TMEs (McFarland et al., 2021). Cells from this study were clustered into metacells based on transcriptional similarity, allowing the deconvolution of consistent transcriptional programs from diverse tissues (Fig. 1 D). We first ranked metacells by Inpp4b expression and observed a sharp separation along this parameter. We then sampled the topmost Inpp4b-expressing metacells and compared the tissue distribution of these clusters against those with the least Inpp4b expression (Fig. 1 D). We observed that Inpp4b-enriched metacells were predominantly found in almost all peripheral tissues as well as TMEs from B16 and PyMT breast TMEs. In contrast, Inpp4b-low-expressing metacells were largely depleted from tissues and enriched in spleen and blood. Having established an association between Inpp4b expression and distribution in non-lymphoid tissues, we then asked what other transcriptional features were coexpressed or negatively associated with Inpp4b expression. We performed gene correlation analysis and segregated genes based on how their expression correlated with that of Inpp4b among the 88 identified metacells (Fig. 1 E). We then performed gene ontology (GO) enrichment analysis on the positively correlated and negatively correlated gene sets. Consistent with our observations on tissue distribution, we found GO terms related to cell adhesion and leukocyte maturation enriched among genes positively correlated with Inpp4b expression. In contrast, we found terms related to vascular adhesion, circulation, and cytotoxicity enriched with the negative correlation module. We next examined the most strongly correlated or anticorrelated genes from each module that belonged to these enriched GO terms (Fig. 1 F). Within the positive correlation module, we found many known markers of tissue residence such as Itga1 (CD49a), Itgae (CD103), and Cd69. We also observed enrichment of Bcl2, suggesting that Inpp4b-high-expressors may be more resilient against apoptosis. Within the negative correlation module, we observed markers of circulating lymphocytes including Itgam (CD11b), Klf2, and Itgb2 (CD18). Additionally, we noticed that Gzma and Prf1 were negatively associated with Inpp4b expression, suggesting a regulatory role for this phosphatase in cytotoxic function. Collectively, our bioinformatic analysis of both population-based and single-cell transcriptomes identified Inpp4b as a universal marker of tissue-resident ILC1s and CD8+ TRM and suggested that this gene may play an important role in the homeostasis of these populations.
Figure 1.
Inpp4b is a molecular hallmark of ILC1s and CD8+ TRM transcriptional programs. (A) Volcano plot of DEGs between liver ILC1s and NK cells. Raw data were extracted from GSE103901. (B) Overlap between ILC1 transcriptional signature with core CD8+ TRM profiles. (C) Mean expression and fold change magnitude of shared ILC1/CD8+ TRM genes between liver ILC1s and NK cells. (D) Metacells ranked by Inpp4b expression. The top 5 Inpp4b-expressing metacells are highlighted in red. The lowest Inpp4b-expressing metacells are highlighted in blue (28 in total). Pie charts showing tissue distribution of highlighted metacells (five randomly selected of bottommost 28). Processed data were extracted from GSE158547. Metacells are designated based on the predominant distribution. (E) Spearman correlation of Inpp4b expression with other genes and GO enrichment of positively and negatively correlated gene modules. (F) Individual gene expression profiles as a function Inpp4b expression. SG = salivary gland, UT = uterus, TUM = tumor, SPLN = spleen, LN = lymph node, LVR = liver, VAT = visceral adipose tissue, NES = normalized enrichment score.
INPP4B is required for the cell-intrinsic maintenance of tissue ILC1s
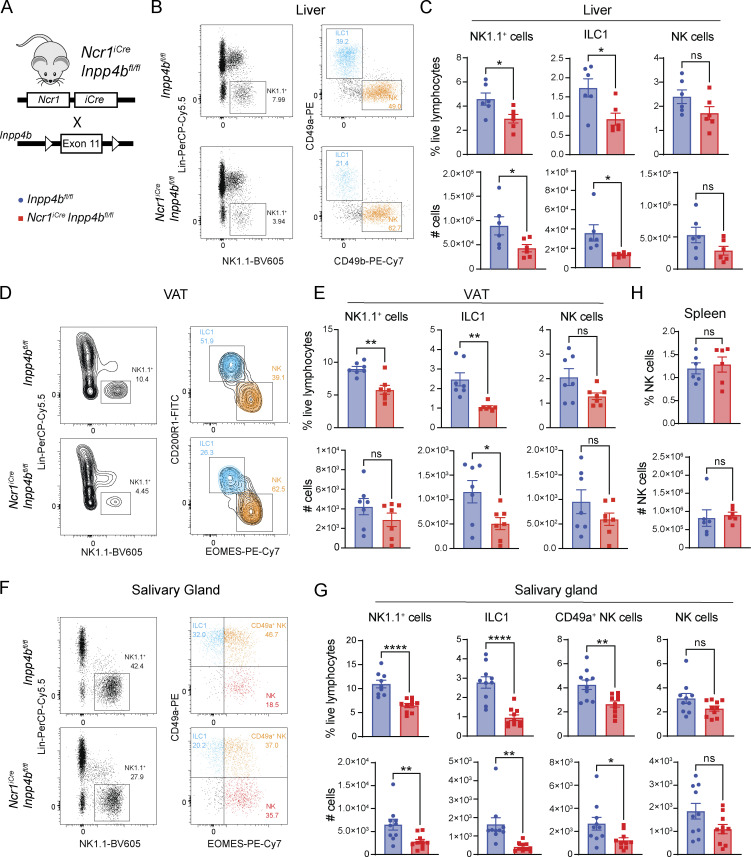
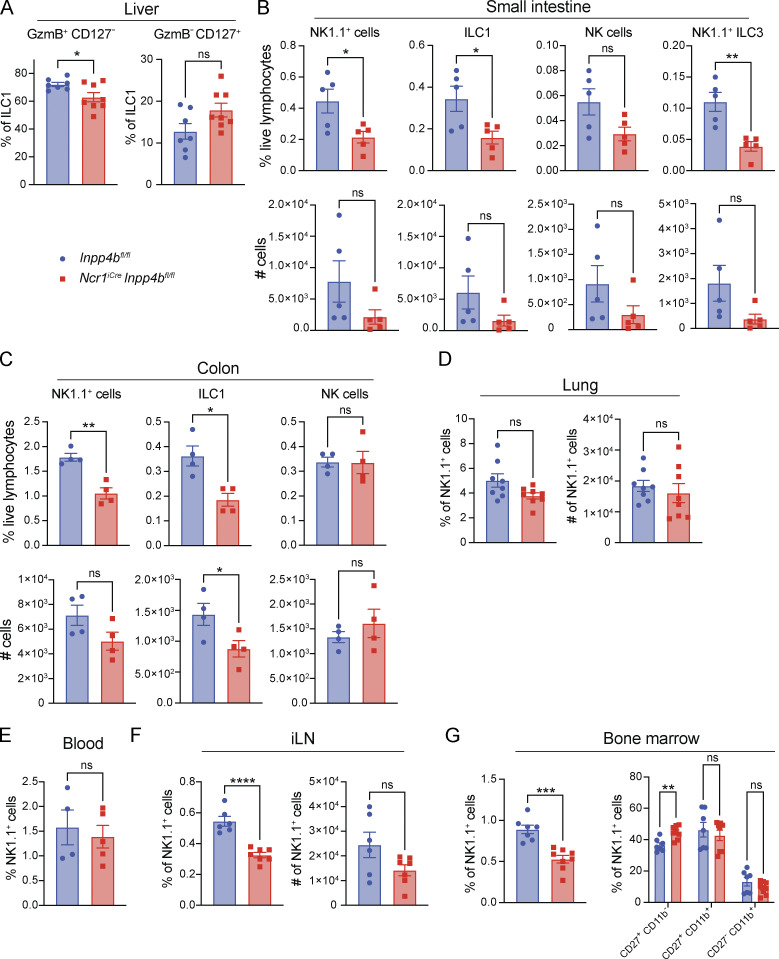
To further assess the function of INPP4B, we crossed Ncr1iCre and Inpp4bfl/fl mice to generate a mouse model of cell-intrinsic deletion of Inpp4b in ILC1s and NK cells (NcriCreInpp4bfl/fl; Fig. 2 A). Given the association between Inpp4b expression and ILC1s, we first examined the liver since it is the largest reservoir of ILC1s in the mouse. We observed a reduction in total liver Lin− NK1.1+ cells. Further inspection of ILC1s and NK populations using the classical markers CD49a and CD49b revealed that this reduction was primarily due to a decrease in ILC1s in terms of relative frequency and total number (Fig. 2, B and C). Liver ILC1s comprise a CD127+ subset that is prone to IFN-γ production and a CD127− subset that is prone to cytotoxicity, characterized by Granzyme B expression (Friedrich et al., 2021; Yomogida et al., 2021). We noted that Inpp4b deficiency led to a mild reduction of the cytotoxic subset (Fig. S1 A). We then examined the visceral adipose tissue (VAT), which was described to house CD200R1+ EOMES− ILC1s and CD200R1− EOMES+ NK cells (O’Sullivan et al., 2016). As in the liver, VAT NK1.1+ cells were reduced, which was largely due to a reduction in CD200R1+ ILC1s (Fig. 2, D and E). The salivary gland harbors ILC1s, NK cells, as well as a distinctive population of CD49a+ EOMES+ NK1.1+ NK cells that share some phenotypic features with ILC1s, such as CD49a (further in text: CD49a+ NK cells). We observed a reduction in total NK1.1+ cells, which was due to a major decrease of CD49a+ EOMES− ILC1s, and a moderate reduction of CD49a+ NK cells, while NK cells proper were not significantly affected (Fig. 2, F and G). ILC1 populations in the small intestine and colon were also reduced (Fig. S1, B and C). Such reduction extended to small intestinal ILC3 that express NK1.1, which are thought to be undergoing a process of conversion into ILC1s (Viant et al., 2016). Inpp4b deficiency did not affect NK1.1+ cells in the lung (Fig. S1 D). Frequency and total numbers of splenic and blood NK cells were also unaffected (Fig. 2 H and Fig. S1 E). Altogether, these data suggest that INPP4B is more crucial for the maintenance of tissue-resident ILC1s than for circulating NK cells. We did observe a reduced frequency of NK cells in the inguinal lymph node and bone marrow (Fig. S1, F and G). Moreover, the bone marrow’s immature CD27+ NK cells were slightly increased, possibly indicating a maturation defect in the bone marrow. However, this defect seems to be compensated for when NK cells enter the periphery (Fig. S1 G). Heightened sensitivity of lymph nodes and bone marrow NK cells to Inpp4b deficiency compared with blood and spleen NK cells suggests that a subset of cells in these tissues may share similar features to peripheral tissue-resident lymphocytes.
Figure 2.
Ncr1iCreInpp4bfl/fl mice have reduced ILC1s in peripheral tissues. (A) Diagram showing genetic construct for conditional deletion of Inpp4b. (B and C) (B) Representative flow cytometry plots and (C) quantification of liver NK1.1+ cells, ILC1s, and NK cells in Inpp4bfl/fl and NcriCreInpp4bfl/fl mice (n = 6). Data are pooled from two independent experiments. (D and E) (D) Representative flow cytometry plots and (E) quantification of VAT NK1.1+ cells, ILC1s, and NK cells (n = 7). The experiment is representative of four independent experiments. (F and G) (F) Representative flow cytometry plots and (G) quantification of salivary gland NK1.1+, ILC1s, CD49a+ NK cells, and NK cells proper (n = 10). Data are pooled from three independent experiments. (H) Quantification of splenic NK cells (n = 6). Data are representative of two independent experiments. Results for C, E, G, and H are shown as mean ± SEM. P values were calculated using unpaired two-tailed Student’s t test; *P < 0.05, **P < 0.01, ****P < 0.0001, ns = not significant.
Figure S1.
Quantification of Inpp4bfl/fl and NcriCreInpp4bfl/fl NK1.1+ cells, ILC1s, and NK cells in various tissues. (A) Liver CD127+ and CD127− ILC1s (n = 7–8). Data are pooled from two independent experiments. (B–F) Small intestine (n = 5), colon (n = 4), lung (n = 8), blood (n = 4–5), and inguinal lymph nodes (iLN) (n = 6) NK1.1+ cells, ILC1s, and NK cells. For lung, iLN, and bone marrow, data are pooled from two independent experiments. For small intestine, colon, and blood, data are representative of two independent experiments. Results are shown as mean ± SEM. P values were calculated using unpaired Student’s t test. (G) Bone marrow NK1.1+ cells (mean ± SEM; unpaired Student’s t test) and their CD27+CD11 b−, CD27+CD11b+, and CD27−CD11b+ subsets (n = 7–8; mean ± SEM; multiple unpaired two-tailed Student’s t test with multiple test correction using the Holm–Sidak method). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant.
INPP4B promotes survival of ILC1s through phosphorylation of AKT
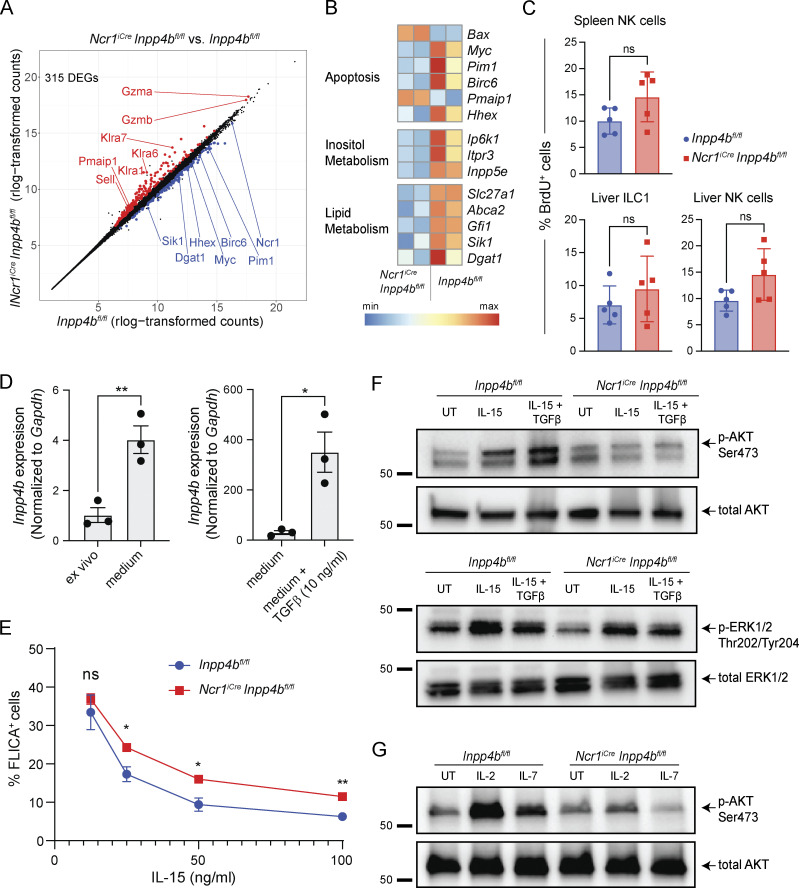
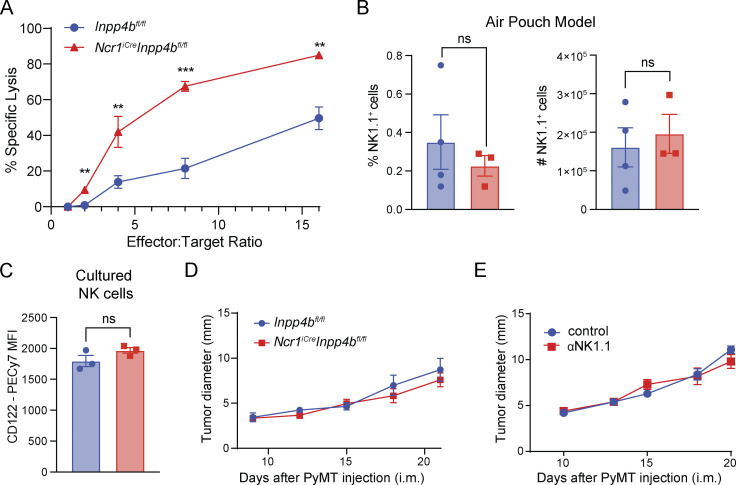
We then sought to define the mechanism by which INPP4B impacts ILC1 numbers. We used bulk RNA-seq to characterize the transcriptomes of sorted liver ILC1s from Inpp4bfl/fl and NcriCreInpp4bfl/fl mice. Using a false discovery rate (FDR) cutoff of 0.05, we identified 315 DEGs. Inspection of DEGs revealed upregulation of genes related to cytotoxicity (Gzma, Gzmb) in Inpp4b-deficient cells (Fig. 3 A). Apoptosis-related genes were also differentially regulated, including induction of pro-apoptotic molecules (Bax, Pmaip1) and downregulation of proliferative and anti-apoptotic genes (Myc, Pim1, Birc6, Hhex). We also observed downregulation of genes related to inositol metabolism (Ip6k1, Itpr3, Inpp5e) and lipid metabolism (Slc27a1, Abca1, Gfi1, Sik1, Dgat1; Fig. 3 B). Given the downregulation of Myc, we investigated whether the reduction of ILC1s may be due to reduced proliferation. However, BrdU incorporation in vivo by these cells was unchanged between Inpp4bfl/fl and NcriCreInpp4bfl/fl mice (Fig. 3 C). We next examined whether cells lacking Inpp4b showed an increased tendency for apoptosis. Due to the scarcity of ILC1s and the difficulty of identifying dying ILC1s in tissues, we instead explored whether INPP4B influenced the cellular response to survival signals in vitro. To guarantee an adequate number of cells for this evaluation, we utilized NK cells expanded in vitro with IL-2. We found that these culture conditions significantly induced Inpp4b expression compared with freshly isolated NK cells (Fig. 3 D). Evidently, the minimal levels of TGFβ present in the serum of culture medium were adequate to induce Inpp4b transcription. Further addition of exogenous TGFβ to the culture medium further increased the expression of Inpp4b, aligning with findings from our prior studies implicating Inpp4b as a TGFβ-inducible gene (Cortez et al., 2016, 2017). NK cells were examined for caspase activity in response to decreasing concentrations of IL-15. In comparison to cultured control NK cells, those deficient in Inpp4b consistently displayed higher levels of caspase activation across a range of IL-15 concentrations (Fig. 3 E). Thus, Inpp4b deficiency reduces cell survival in response to IL-15. Conversely, Inpp4b deficiency increased the ability of cultured NK cells to lyse a tumor target cell (YAC-1; Fig. S2 A), consistent with a regulatory role for INPP4B in the expression of cytotoxic mediators observed by transcriptome analyses. We also examined whether Inpp4b deficiency may impact cell migration in the tissues. However, the capacity of NK cells to migrate in an air pouch model of inflammation was unaffected (Fig. S2 B).
Figure 3.
Inpp4b-deficient cells exhibit reduced survival and reduced p-AKT in response to IL-15. (A) DEGs between hepatic ILC1s from Inpp4bfl/fl and NcriCreInpp4bfl/fl mice. (B) Heatmaps of DEGs arranged by pathways. (C) Frequency of BrdU+ cells in spleen and liver (n = 5). Data are representative of two independent experiments. (D) RT-PCR analysis of Inpp4b expression in NK cells cultured in vitro with or without TGFβ stimulation. The experiments are representative of two independent experiments. (E) Frequency of FLICA+ cells over indicated concentrations of IL-15. Two technical replicates from two biological replicates were used for each concentration (n = 4). Data are representative of two independent experiments. (F and G) Western blot of p-AKT, total AKT, p-ERK1/2, and total ERK1/2 in control and Inpp4b-deficient cells under specified conditions. Molecular weight markers are indicated in kD. Data are representative of two to four independent experiments. (C–E) Results are shown as mean ± SEM. P values were calculated using unpaired two-tailed Student’s t test (C and D) or multiple unpaired two-tailed Student’s t test with multiple test correction using the Holm–Sidak method (E). *P < 0.05, **P < 0.01, ns = not significant. Source data are available for this figure: SourceData F3.
Figure S2.
INPP4B promotes tissue residency of NK1.1+ cells and restrains their cytotoxic potential. (A) YAC-1 target cell lysis assay. Technical replicates (n = 2–3) were used for each E:T ratio. (B) Percentage and total numbers of NK1.1+ cells in the air pouch. (C) CD122 expression on Inpp4b-deficient and control NK cells cultured as in Fig. 3 G. (D and E) Growth curve of intramammary (i.m.) implanted PyMT in Inpp4bfl/fl and NcriCreInpp4bfl/fl mice (n = 5) (D), and in untreated or α-NK1.1-treated Inpp4bfl/fl mice (n = 3–4) (E). (A–E) Data are representative of two independent experiments. Results are shown as mean ± SEM. P values are calculated using unpaired Student’s t test (B and C) or using multiple unpaired two-tailed Student’s t test with multiple test correction using the Holm–Sidak method (A, D, and E). MFI = mean fluorescent intensity. **P < 0.01, ***P < 0.001, ns = not significant.
In NK cells, AKT plays a pleiotropic role but is generally considered to promote NK cell survival, proliferation, and activation through multiple downstream pathways (Ali et al., 2015). AKT activation is most robustly observed downstream of either IL-15 signaling or engagement of activating receptors. Thus, we assessed phosphorylation of AKT (p-AKT) in cultured NK cells from control and NcriCreInpp4bfl/fl mice stimulated in vitro with IL-15. As Inpp4b expression appears to be associated with TGFβ imprinting, we also examined p-AKT under IL-15 and TGFβ cotreatment. We found that Inpp4b-deficient cells displayed defective p-AKT under both IL-15–stimulated and IL-15 plus TGFβ conditions (Fig. 3 F). Additionally, we uncovered a similar deficit in p-ERK1/2, albeit less pronounced. Inpp4b-deficient and control cells showed similar expression of the receptor for IL-15 (CD122; Fig. S2 C), corroborating that INPP4B sustains CD122 downstream signaling rather than its expression. We finally assessed whether INPP4B enhanced p-AKT signaling by IL-15 only or also by other common γ chain cytokines, supporting development, proliferation, and survival of ILC1s and NK cells, such as IL-2 and IL-7 (Fig. 3 G). Indeed, Inpp4b-deficiency impaired AKT phosphorylation in response to both IL-2 and IL-7 (Fig. 3 G). Collectively, these results demonstrate a requirement of INPP4B in IL-15–, IL-2–, and IL-7–mediated phosphorylation of AKT.
INPP4B is required for tumor-associated NK cells and NK-dependent antitumor immunity
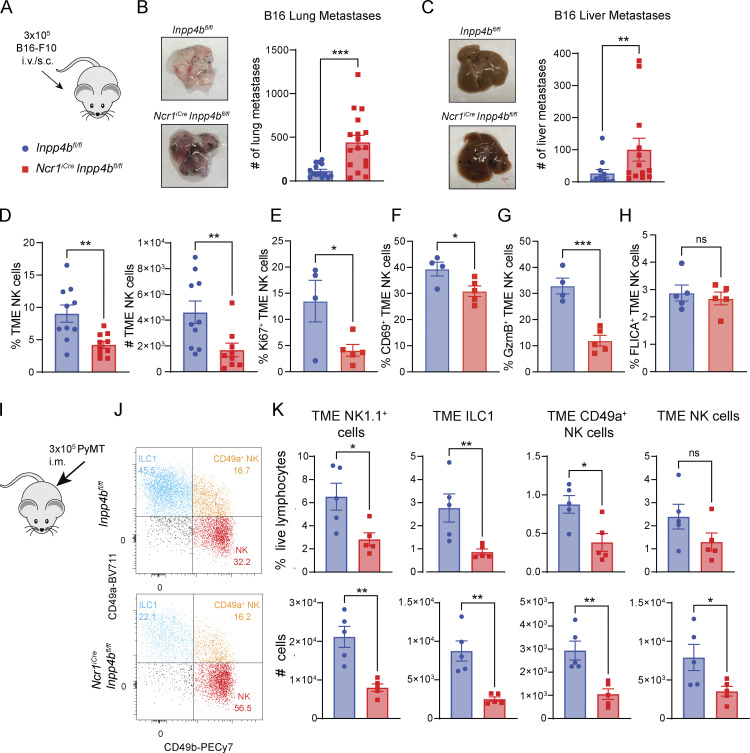
Examination of Inpp4b expression in our single-cell RNA-seq (scRNA-seq) atlas revealed enrichment of Inpp4b not only in peripheral tissue ILC1s but also in NK cells within the TME. Thus, we investigated the effect of Inpp4b deficiency on the function of tumor NK cells in a model of B16-F10 melanoma metastasis, where NK cells play an essential role in tumor control (Gorelik et al., 1982; Fig. 4 A). 2 weeks after injection, Inpp4bfl/fl and NcriCreInpp4bfl/fl mice were sacrificed and organs were collected for analysis of metastases. NcriCreInpp4bfl/fl animals displayed a profound failure to control B16 metastasis in the lung compared with controls (Fig. 4 B). B16-F10 melanoma has also been reported to colonize the liver, which we observed in our experiments (Gorelik et al., 1982). B16-F10 liver metastases were also significantly increased in NcriCreInpp4bfl/fl mice, reflecting a significant failure of NK cells and ILC1s in limiting tumor burden (Fig. 4 C). To determine if this failure in antitumor immunity was due to a reduction in intratumoral NK cells, we turned to a subcutaneous model of the same tumor line to cleanly profile the TME (Fig. 4, D–H). B16-F10 tumors were implanted in the hind flank and allowed to grow to a moderate size to prevent incorporation of surrounding lymph nodes. When we examined the TME of B16-F10 tumors, we did observe a drastic reduction of tumor NK cells from NcriCreInpp4bfl/fl mice, further reflecting a failure of Inpp4b-deficient NK cell maintenance in the TME (Fig. 4 D). This was accompanied by reduced proliferation capacity (Fig. 4 E), activation (Fig. 4 F), and cytotoxicity of tumor NK cells (Fig. 4 G); however, we did not observe a difference in apoptotic NK cells at least in the early phase of tumor growth (Fig. 4 H).
Figure 4.
INPP4B is required for maintenance of intratumoral NK cells and ILC1s. (A) Diagram showing B16-F10 experimental setup. (B and C) Tumor cells were injected either i.v. (B and C) or subcutaneously (s.c.; D–H). (B) Quantification of B16-F10 lung metastases (n = 15–17). Data are pooled from three independent experiments. (C) Quantification of B16-F10 liver metastases (n = 13–14). Data are pooled from two independent experiments. (D–H) intratumoral NK1.1+ cells in subcutaneous B16-F10 melanoma (n = 10) were analyzed for frequency and total numbers (D), Ki67 expression (E), CD69 expression (F), GzmB production capacity (G), and apoptosis (H). (I) Diagram showing PyMT experimental setup; i.m. = intramammary. (J) Representative flow cytometry plot of intratumoral ILC1s, CD49a+ NK cells, and NK cells in PyMT from control and NcriCreInpp4bfl/fl mice. (K) Quantification of tumor-infiltrating NK1.1+ cells, ILC1s, CD49a+ NK cells, and NK cells (n = 5). Data from D–H and J and K are representative of two independent experiments. (B–H and K) Results are shown as mean ± SEM. P values for D–H and K were calculated using unpaired Student’s t test. P values for B and C were calculated using Mann–Whitney test; *P < 0.05, **P < 0.01, ***P < 0.001, ns = not significant.
As Inpp4b was shown to be strongly enriched in the breast cancer TME, we also examined an orthotopic model of triple-negative breast cancer (PyMT) to further explore the role of Inpp4b on the biology of tumor-associated NK cells (Fig. 4 I; Guy et al., 1992). Following intramammary injection, PyMT tumors were allowed to grow to a moderate size but preceding involvement of peripheral lymph nodes. NK cells did not have a major role in the control of the growth of PyMT tumor (Fig. S2, D and E). However, Inpp4b deficiency impacted the phenotype of NK1.1+ cells infiltrating PyMT. We identified three distinct populations of NK1.1+ cells that mirrored those observed in the salivary gland: ILC1s, NK cells, and CD49a+ CD49b+ NK cells (Fig. 4, J and K). PyMT tumors from NcriCreInpp4bfl/fl animals displayed a significant reduction in total tumor NK1.1+ cells. Recapitulating our prior observations in the salivary gland, this reduction was primarily due to a progressive loss of intratumoral CD49a+ CD49b− ILC1s, followed by CD49a+ CD49b+ NK and then CD49a− CD49b+ NK cells. Altogether, our data reflect a profound deficit of Inpp4b-deficient NK cells and ILC1s in the TME. When these cells play an important role in tumor elimination, such as in the B16 metastatic model, Inpp4b deficiency results in significant impairment in antitumor immunity.
Concluding remarks
In this study, we integrate multiple bulk and single-cell transcriptomic analyses to identify Inpp4b as a novel regulator of tissue-resident and intratumoral ILC1s and NK cells. We positively associate Inpp4b expression with known cell adhesion molecules and antiapoptotic and proliferative pathways. Conversely, Inpp4b is negatively associated with genes associated with circulating lymphocytes. Using a mouse model of conditional Inpp4b deletion, we show a general reduction of tissue-resident ILC1s in the liver, salivary gland, VAT, small intestine, and colon; NK cells in lymph node and bone marrow are mildly affected, but completely unaffected in the blood and spleen. We demonstrate that INPP4B promotes cell survival through phosphorylation of AKT while restraining NK cell–mediated cytotoxicity. In addition to promoting tissue-resident cell homeostasis, INPP4B is also necessary for the accumulation of intratumoral NK cells in models of B16-F10 melanoma and triple-negative breast cancer. This holds true regardless of whether NK cells actively contribute to the elimination of the tumor. NcriCreInpp4bfl/fl mice display a profound impairment in NK-mediated antitumor control of B16-F10 metastases. While reduction of intratumoral NK cells did not alter triple-negative breast cancer growth, other types of breast cancer may be more sensitive to NK cell–mediated control.
Proper regulation of PI3K signaling is critical for the development and function of immune cells. 5′ phosphatases, such as SHIP1, have been well-characterized in regulating immune activation (Gumbleton and Kerr, 2013). Our findings reveal a novel role for the 4′ phosphatase INPP4B in regulating tissue-resident and antitumor immunity. Specifically, we show that INPP4B facilitates phosphorylation and activation of AKT, a downstream target of PI3K that promotes NK cell survival, proliferation, and activation through multiple downstream pathways (Ali et al., 2015). While INPP4B has been shown to play context-dependent roles in cellular proliferation and survival, our findings are consistent with work demonstrating a positive role for INPP4B in growth of acute myeloid leukemia, colon cancer, and breast cancer cells either through positive regulation of AKT or SGK3 (Chi et al., 2015; Gasser et al., 2014; Guo et al., 2016; Rijal et al., 2015). Our findings also align with a prior report in cancer cells whereby Inpp4b deficiency blocks AKT phosphorylation through enhanced phosphatase and tensin homolog deleted on chromosome 10 activity (Guo et al., 2016). Our studies do not exclude a parallel role for SGK3 in mediating INPP4B-dependent survival. SGK3 is expressed at moderate levels in ILC1s and may represent an additional AKT-independent pathway for INPP4B to act through.
In the context of the current knowledge of immunity and ILCs, our work clarifies the role of INPP4B in the innate immune system. Our findings suggest an impactful role for the pathway of 4′ inositol metabolism in shaping the nature of the immune response, particularly that of ILC1s and NK cells embedded in tissues. Furthermore, expression of INPP4B is induced by tissue factors like TGFβ, illustrating a generalizable mechanism whereby the tissue microenvironment rewires universal signaling pathways, such as PI3K signaling, to diversify lymphocyte phenotypes and sustain cell survival. As INPP4B is a feature of CD8+ TRM cells in addition to ILC1s, it would be of great interest to investigate this signaling mediator in adaptive immune cells. Further characterization of INPP4B-mediated pathways may allow the development of therapies specifically targeting tissue-resident lymphocytes in the context of tissue inflammation and solid tumor immunotherapy.
Materials and methods
Mice
Mice were bred and maintained in specific pathogen–free facilities at Washington University in Saint Louis. Ncr1iCre mice were a generous gift from E. Vivier (Center d’Immunologie de Marseille-Luminy, Marseille, France), and Inpp4bfl/fl mice were provided by J. Vacher (Institut de Recherches Cliniques de Montreal, Montreal, Canada). Mice used for experiments were between 8 and 16 wk of age and were on a C57BL/6 background. Control mice were sex-matched littermates. C57BL/6J mice were purchased from the Jackson Laboratory. All studies were conducted in accordance with the Washington University Animal Studies Committee.
Tissue dissociation and cell isolation
Tissues were isolated and processed into single-cell suspensions as previously described (McFarland et al., 2021). Mice were transcardially perfused with cold 1× PBS before tissue extraction. Single-cell suspensions of spleen and lymph nodes were achieved by dissociating tissues through a 70-μM filter. Blood was collected from anesthetized mice via cardiac puncture. Single-cell suspensions of liver, salivary gland, VAT, lung, and tumors were obtained by finely mincing tissues and digesting them for 30 min at 37°C with shaking in collagenase D and DNase (both MilliporeSigma). Small intestine and colon were cleaned of Payer’s patches and/or luminal content and opened lengthwise. Intestines were incubated in Hank’s balanced salt solution containing 10% FCS and 5 mM EDTA for 2 × 20 min on the rotator, each time followed by vigorous vortexing. Intestinal tissue was further digested in the 10% RPMI medium containing collagenase IV (C-5138; MilliporeSigma) for 40 min with vigorous shaking at 37°C. Lymphocytes were purified using Percoll (Cytiva) gradients (40/80 for liver, 40/70 for salivary gland, VAT, intestines, and tumors). Before proceeding with antibody staining, cells were preincubated with mouse Fc receptor–blocking antibody (2.4G2). For experiments requiring cultured NK cells, NK cells were purified from splenocytes using DX5+ MACS (magnetic-activated cell sorting) positive selection columns (Miltenyi).
Flow cytometry
Lineage cocktail, unless otherwise specified, consisted of CD3, CD5, and CD19. For surface stain–only panels, 7-AAD or DAPI were added to assess viability. Prior to fixation for intracellular stains, cells were stained with LIVE/DEAD fixable aqua. Cells were fixed using Invitrogen eBioscience FOXP3 kit (for transcription factors) or BD Cytofix/Cytoperm kit (for cytosolic proteins). Cell counting was performed using counting beads (eBioscience). Cells were acquired on FACS Symphony system (BD Biosciences) and analyzed using FlowJo software. The following antibodies were used for FACS: NK1.1 (PK136; #108710), CD3e (145-2C11; #100328), CD19 (6D5, #115534), CD45 (30-F11; #103116), CD5 (53-7.2; #100624), NKp46 (29A1.4; #137616), CD49a (HMα1; #142604), CD49b (HMα2; #103504/103522), CD200R1 (OX-110; #123909), CD69 (H1.2F3; #104545), Ki67 (11F6; #151206), GzmB (GB11; #515403), Streptavidin (#405206; all Biolegend), EOMES (Dan11mag, #25-4875-82; Thermo Fisher Scientific), CD49a (Hα31/8; #564863), and CD122 (TM-B1; #559884; both BD Biosciences).
Cell lines
B16-F10 was derived from a C57BL/6 male mouse melanoma tumor established with the parental line B16-F0 (Nakamura et al., 2002). PY8119 breast cancer cells used for PyMT studies were obtained from ATCC (CRL-3278). B16-F10, PY8119, and YAC-1 cells were maintained in complete RPMI supplemented with 10% bovine calf serum.
BrdU proliferation assay
Mice were fed with BrdU (BD Biosciences) in drinking water for 7 d. Mice were sacrificed and cells were assessed for BrdU incorporation by BD Pharmingen BrdU APC Flow Kit per manufacturer protocol.
FLICA (fluorochrome-labeled inhibitors of caspases assay)
NK cells were purified and cultured with indicated concentrations of IL-15 overnight. Caspase activity and apoptosis were assessed by flow cytometry using the FAM-FLICA Poly Caspase Assay (Immunochemistry Technologies) per manufacturer protocol.
Yac-1 cytotoxicity assay
Splenic NK cells were purified and cultured in a complete 10% RPMI medium supplemented with IL-2 for 5 d. Lymphokine-activated killer cells were cocultured with CFSE-labeled YAC-1 target cells at indicated E:T (effector:target) ratios for 6 h in V-bottom plates. Frequency of PI−CFSE+ cells was measured at 0 and 6 h of coculture. Specific lysis was calculated as (Frequencyt0 − Frequency6 h)/Frequencyt0. YAC-1 cells alone were measured to estimate the basal rate of cell death.
Quantitative RT-PCR
Splenic NK cells were single-cell sorted as live, lymphocyte-sized cells that were CD3ε− CD19− NK1.1+. Cells were either immediately lysed for RNA extraction or seeded at a density of 300,000 cells per well and cultured for 6 d in a complete 10% RPMI medium supplemented with IL-2. On day 6, 100,000 cells were plated in either 10% RPMI or 10% RPMI and 10 ng/ml of TGFβ (PeproTech) for 16 h. Total RNA was extracted from samples using an Rneasy Micro Plus Kit (Qiagen). Complementary DNA (cDNA) was synthesized using iScript cDNA synthesis kit (Bio-Rad). RT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) on the CFX Connect Real-Time PCR Detection System (Bio-Rad). The expression of Inpp4b was normalized to the expression of the housekeeping gene Gapdh. The following primers were used: Gapdh: For-5′-GAGCCAAAAGGGTCATC-3′; Rev-5′-CCATCCACAGTCTTCTGGG-3′; Inpp4b: For-5′-ACAGCACCAGAAAGTCTGAGC-3′, and Rev-5′-CTTTCTGACATCTGCTCACGGA-3′.
Air pouch model of inflammation
The assessment of migration capacity of NK cells via the air pouch method was performed as previously described (Duarte et al., 2012). In short, 3 ml of sterilized air was injected subcutaneously into the back skin of anesthetized mice. On day 3 after injection, an additional 3 ml of sterilized air was injected into the air pouch. On day 6, 1 ml of 200 µg/ml poly(I:C) was injected into the pouch to induce inflammation. After 6 h, mice were sacrificed and air pouch was washed with 3 × 1 ml of PBS to collect inflammatory exudate.
Tumor studies
Adherent B16F10 melanoma cells were removed from tissue culture plates using a Trypsin plus 1 mM EDTA solution and washed twice in PBS before twice passing through a 70-μM cell filter and resuspended in PBS. To establish subcutaneous tumors in mice, 3 × 105 cells in 100 μl of sterile PBS were implanted into the rear hind flanks of mice. Tumors were monitored by caliper measurements and mice were sacrificed when tumor size reached 5–7 mm. For the B16F10 lung metastases model, mice were injected with 3 × 105 B16F10 melanoma cells in 100 μl PBS i.v. On day 14 mice were euthanized, perfused, and lungs and livers were collected. Metastatic foci from both lungs and whole liver were counted manually. For breast cancer tumors, subcutaneous injections were performed using 5 × 105 PY8119 breast cancer cells (CRL-3278; ATCC) suspended in 50 μl of sterile PBS and injected into the mammary fat pad. Tumors were monitored by caliper measurements and mice were sacrificed between day 16 and 18 after tumor inoculation (<10 mm). NK cell depletion was conducted using injections of anti-mouse NK1.1 (Clone PK136; BioXCell) i.p.
Bulk RNA-seq
Liver ILC1s were sort purified using a FACSAria II (BD Biosciences) as live, lymphocyte-sized cells that were negative for CD3ε, CD5, CD19, and CD49b and positive for CD45, NK1.1, NKp46, and CD49a. Total RNA was extracted from samples using an RNeasy Micro Plus Kit (Qiagen). Total RNA integrity was determined using Agilent Bioanalyzer or 4200 TapeStation. Library preparation was performed with 10 ng total RNA with a Bioanalyzer RNA integrity number score >8. Double standard cDNA was prepared using the SMARTer Ultra Low RNA kit for Illumina Sequencing (Takara-Clontech) per the manufacturer’s protocol. cDNA was fragmented using a Covaris E220 sonicator using peak incident power 18, duty factor 20%, and cycles per burst 50 for 120 s. cDNA was blunt ended, had an A base added to the 3′ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were then amplified for 12–15 cycles using primers, incorporating unique dual index tags. Fragments were sequenced on an Illumina HiSeq. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq software and a custom Python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 primary assembly with Spliced Transcripts Alignment to a Reference version 2.5.1a. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.6-p5. Aligned gene counts were processed using the DESeq2 package with R (version 3.5; Love et al., 2014). Genes with fewer than 10 counts among all the samples were excluded. DEGs were defined as protein-coding genes with an average expression >100 counts and an FDR <0.05.
Analysis of bulk RNA-seq and scRNA-seq datasets
Bulk transcriptomes of liver ILC1s and NK cells were retrieved from GSE103901. Read counts were quantified and normalized using R and the DESeq2 package. Differentially expressed genes were identified by log2FoldChange >1 and FDR <0.05. Single-cell transcriptomes of ILC1s and NK cells were retrieved from GSE158547. Cells were normalized and clustered as previously described. For gene correlation analysis, Spearman correlation tests were applied for all genes and Inpp4b expression among the 88 identified metacells, and multiple test adjustment (Benjamini–Hochberg) was performed. Positively and negatively correlated gene modules were submitted to Metascape for GO analysis using default settings (Zhou et al., 2019).
Western blot
Purified NK cells were expanded in 96-well plates in complete RPMI supplemented with IL-2 for 6 d. Cells were then resuspended in complete RPMI with or without 10 ng/ml TGFβ (PeproTech) and plated at a density of 300,000 cells per well for 16 h. Cells were then stimulated with 100 ng/ml IL-15, IL-2, or IL-7 (all PeproTech) for 30 min, washed in cold PBS two times, and lysed in radioimmunoprecipitation assay buffer with 2 mM PMSF for 20 min on ice. Cell lysate was centrifuged at 12,000 rpm for 15 min and supernatant was collected. After protein estimation, samples were mixed in SDS loading buffer and an equal amount of protein was loaded on 4–20% Mini-PROTEAN TGX Precast Protein Gels. Gels were run in Tris-Glycine-SDS buffer and resolved proteins were transferred to polyvinylidene difluoride membrane. After transfer, the membrane was washed in TBS with 0.01% tween 20 (TBS-T) and blocked in 5% BSA or non-fat dried milk for 1 h followed by incubation in primary antibodies for overnight at 4°C. The membrane was washed three to five times in TBS-T followed by incubation in HRP-labeled secondary antibody for 1–2 h. The membranes were developed using Super Signal West Pico PLUS chemiluminescent substrate (#34577; Thermo Fisher Scientific) inside Bio-Rad ChemiDoc MP imaging system. Antibodies used were as follows: p-AKT (Ser473) (#4060S; CST), total AKT (#4691S; CST), p-P44/42 MAPK (ERK1/2) (#9101S; CST), total ERK1/2 (#4995S; CST), anti-rabbit IgG (#7074; CST).
Online supplemental material
We are including two supplemental figures that support and extend the findings and figures contained in the manuscript. Fig. S1 shows the quantification of Inpp4bfl/fl and NcriCreInpp4bfl/fl ILC1s and NK cells in various tissues. Fig. S2 shows the impact of Inpp4b deficiency on NK cell cytotoxicity, migration, and CD122 expression, as well as on the growth of intramammary implanted PyMT tumors.
Supplementary Material
is the source file for Fig. 3.
Acknowledgments
The authors thank the Genome Technology Access Center at McDonnell Genome Institute and the members of the Pathology and Immunology FACS core facility at Washington University School of Medicine.
This work was supported by National Institutes of Health 5F30DK127540, 5T32DK077653 (V. Peng), RO1 DK132327, RO1 AI134035, and RO1 DK124699 (M. Colonna).
Author contributions: Experiments were designed and executed by V. Peng, R. Sudan, T. Trsan, V.S. Cortez, B. Bhattarai, and M. Molgora. The manuscript was written and edited by V. Peng, T. Trsan, and M. Colonna. J. Vacher contributed essential reagents. M. Colonna contributed to funding acquisition and project administration. All authors reviewed and approved the final version of this manuscript.
Data availability
Sequencing data of RNA-seq generated in this study are deposited in the Gene Expression Omnibus (GSE250334). For the identification of INPP4B as an ILC1 hallmark, we used published bulk RNA-seq and single-cell data (GSE103901, GSE158547).
References
- Agoulnik, I.U., Hodgson M.C., Bowden W.A., and Ittmann M.M.. 2011. INPP4B: The new kid on the PI3K block. Oncotarget. 2:321–328. 10.18632/oncotarget.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aki, S., Yoshioka, K., Takuwa, N., Takuwa, Y., 2020. TGFβ receptor endocytosis and Smad signaling require synaptojanin1−, PI3K-C2α−, and INPP4B−mediated phosphoinositide conversions. Mol. Biol. Cell. 31:360–372. 10.1091/mbc.E19-11-0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, A.K., Nandagopal N., and Lee S.-H.. 2015. IL-15-PI3K-AKT-mTOR: A critical pathway in the life journey of natural killer cells. Front. Immunol. 6:355. 10.3389/fimmu.2015.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, M.N., Guo S.T., Wilmott J.S., Guo X.Y., Yan X.G., Wang C.Y., Liu X.Y., Jin L., Tseng H.-Y., Liu T., et al. 2015. INPP4B is upregulated and functions as an oncogenic driver through SGK3 in a subset of melanomas. Oncotarget. 6:39891–39907. 10.18632/oncotarget.5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, V.S., Cervantes-Barragan L., Robinette M.L., Bando J.K., Wang Y., Geiger T.L., Gilfillan S., Fuchs A., Vivier E., Sun J.C., et al. 2016. Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity. 44:1127–1139. 10.1016/j.immuni.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, V.S., Fuchs A., Cella M., Gilfillan S., and Colonna M.. 2014. Cutting edge: Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol. 192:4487–4491. 10.4049/jimmunol.1303469 [DOI] [PubMed] [Google Scholar]
- Cortez, V.S., Ulland T.K., Cervantes-Barragan L., Bando J.K., Robinette M.L., Wang Q., White A.J., Gilfillan S., Cella M., and Colonna M.. 2017. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18:995–1003. 10.1038/ni.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, D.B., Vasko, M.R., Fehrenbacher, J.C., 2012. Models of inflammation: Carrageenan air pouch. Curr Protoc. 1:e183. 10.1002/0471141755.ph0506s56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele, C.G., Ooms L.M., Ho M., Vieusseux J., O’Toole S.A., Millar E.K., Lopez-Knowles E., Sriratana A., Gurung R., Baglietto L., et al. 2010. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc. Natl. Acad. Sci. USA. 107:22231–22236. 10.1073/pnas.1015245107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron, M., Boudiffa M., Arsenault M., Rached M., Pata M., Giroux S., Elfassihi L., Kisseleva M., Majerus P.W., Rousseau F., and Vacher J.. 2011. Inositol polyphosphate 4-phosphatase B as a regulator of bone mass in mice and humans. Cell Metab. 14:466–477. 10.1016/j.cmet.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron, M., and Vacher J.. 2006. Characterization of the murine Inpp4b gene and identification of a novel isoform. Gene. 376:152–161. 10.1016/j.gene.2006.02.022 [DOI] [PubMed] [Google Scholar]
- Friedrich, C., Taggenbrock R.L.R.E., Doucet-Ladevèze R., Golda G., Moenius R., Arampatzi P., Kragten N.A.M., Kreymborg K., Gomez de Agüero M., Kastenmüller W., et al. 2021. Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat. Immunol. 22:1256–1267. 10.1038/s41590-021-01013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Souza-Fonseca-Guimaraes F., Bald T., Ng S.S., Young A., Ngiow S.F., Rautela J., Straube J., Waddell N., Blake S.J., et al. 2017. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18:1004–1015. 10.1038/ni.3800 [DOI] [PubMed] [Google Scholar]
- Gasser, J.A., Inuzuka H., Lau A.W., Wei W., Beroukhim R., and Toker A.. 2014. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol. Cell. 56:595–607. 10.1016/j.molcel.2014.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger, G., Fan X., Dikiy S., Lee S.Y., and Rudensky A.Y.. 2015. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 350:981–985. 10.1126/science.aac9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik, E., Wiltrout R.H., Okumura K., Habu S., and Herberman R.B.. 1982. Role of NK cells in the control of metastatic spread and growth of tumor cells in mice. Int. J. Cancer. 30:107–112. 10.1002/ijc.2910300118 [DOI] [PubMed] [Google Scholar]
- Gumbleton, M., and Kerr W.G.. 2013. Role of inositol phospholipid signaling in natural killer cell biology. Front. Immunol. 4:47. 10.3389/fimmu.2013.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S.T., Chi M.N., Yang R.H., Guo X.Y., Zan L.K., Wang C.Y., Xi Y.F., Jin L., Croft A., Tseng H.-Y., et al. 2016. INPP4B is an oncogenic regulator in human colon cancer. Oncogene. 35:3049–3061. 10.1038/onc.2015.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, C.T., Cardiff R.D., and Muller W.J.. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961. 10.1128/mcb.12.3.954-961.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamila, S.A., Ooms L.M., Rodgers S.J., and Mitchell C.A.. 2021. The INPP4B paradox: Like PTEN, but different. Adv. Biol. Regul. 82:100817. 10.1016/j.jbior.2021.100817 [DOI] [PubMed] [Google Scholar]
- Hodgson, M.C., Deryugina E.I., Suarez E., Lopez S.M., Lin D., Xue H., Gorlov I.P., Wang Y., and Agoulnik I.U.. 2014. INPP4B suppresses prostate cancer cell invasion. Cell Commun. Signal. 12:61. 10.1186/s12964-014-0061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson, M.C., Shao L.J., Frolov A., Li R., Peterson L.E., Ayala G., Ittmann M.M., Weigel N.L., and Agoulnik I.U.. 2011. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 71:572–582. 10.1158/0008-5472.CAN-10-2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, L.K., Minnich M., Kragten N.A.M., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.-L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Wynne-Jones E., Freestone D., Pellicci D.G., Mielke L.A., Newman D.M., Braun A., Masson F., Kallies A., Belz G.T., and Carbone F.R.. 2015. T-Box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity. 43:1101–1111. 10.1016/j.immuni.2015.11.008 [DOI] [PubMed] [Google Scholar]
- McFarland, A.P., Yalin A., Wang S.-Y., Cortez V.S., Landsberger T., Sudan R., Peng V., Miller H.L., Ricci B., David E., et al. 2021. Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity. 54:1320–1337.e4. 10.1016/j.immuni.2021.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., Yoshikawa N., Yamaguchi Y., Kagota S., Shinozuka K., and Kunitomo M.. 2002. Characterization of mouse melanoma cell lines by their mortal malignancy using an experimental metastatic model. Life Sci. 70:791–798. 10.1016/S0024-3205(01)01454-0 [DOI] [PubMed] [Google Scholar]
- O’Sullivan, T.E., Rapp M., Fan X., Weizman O.-E., Bhardwaj P., Adams N.M., Walzer T., Dannenberg A.J., and Sun J.C.. 2016. Adipose-resident group 1 innate lymphoid cells promote obesity-associated insulin resistance. Immunity. 45:428–441. 10.1016/j.immuni.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, V., and Colonna M.. 2023. The biology of natural killer cells and innate lymphoid cells. In Paul’s Fundamental Immunology. Flajnik M.F., Singh N.J., and Holland S.M., editors. Lippincott Williams and Wilkins, Philadelphia, PA. 537–572. [Google Scholar]
- Pokorny, D., Truebestein L., Fleming K.D., Burke J.E., and Leonard T.A.. 2021. In vitro reconstitution of Sgk3 activation by phosphatidylinositol 3-phosphate. J. Biol. Chem. 297:100919. 10.1016/j.jbc.2021.100919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal, S., Fleming S., Cummings N., Rynkiewicz N.K., Ooms L.M., Nguyen N.-Y.N., Teh T.-C., Avery S., McManus J.F., Papenfuss A.T., et al. 2015. Inositol polyphosphate 4-phosphatase II (INPP4B) is associated with chemoresistance and poor outcome in AML. Blood. 125:2815–2824. 10.1182/blood-2014-09-603555 [DOI] [PubMed] [Google Scholar]
- Rodgers, S.J., Ooms L.M., Oorschot V.M.J., Schittenhelm R.B., Nguyen E.V., Hamila S.A., Rynkiewicz N., Gurung R., Eramo M.J., Sriratana A., et al. 2021. INPP4B promotes PI3Kα-dependent late endosome formation and Wnt/β-catenin signaling in breast cancer. Nat. Commun. 12:3140. 10.1038/s41467-021-23241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka, D.K., Plougastel-Douglas B., Yang L., Pak-Wittel M.A., Artyomov M.N., Ivanova Y., Zhong C., Chase J.M., Rothman P.B., Yu J., et al. 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 3:e01659. 10.7554/eLife.01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant, C., Rankin L.C., Girard-Madoux M.J.H., Seillet C., Shi W., Smyth M.J., Bartholin L., Walzer T., Huntington N.D., Vivier E., and Belz G.T.. 2016. Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci. Signal. 9:ra46. 10.1126/scisignal.aaf2176 [DOI] [PubMed] [Google Scholar]
- Vivier, E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate lymphoid cells: 10 years on. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Weizman, O.-E., Adams N.M., Schuster I.S., Krishna C., Pritykin Y., Lau C., Degli-Esposti M.A., Leslie C.S., Sun J.C., and O’Sullivan T.E.. 2017. ILC1 confer early host protection at initial sites of viral infection. Cell. 171:795–808.e12. 10.1016/j.cell.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomogida, K., Bigley T.M., Trsan T., Gilfillan S., Cella M., Yokoyama W.M., Egawa T., and Colonna M.. 2021. Hobit confers tissue-dependent programs to type 1 innate lymphoid cells. Proc. Natl. Acad. Sci. USA. 118:e2117965118. 10.1073/pnas.2117965118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., and Chanda S.K.. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10:1523. 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 3.
Data Availability Statement
Sequencing data of RNA-seq generated in this study are deposited in the Gene Expression Omnibus (GSE250334). For the identification of INPP4B as an ILC1 hallmark, we used published bulk RNA-seq and single-cell data (GSE103901, GSE158547).