Abstract
Generative artificial intelligence can be applied to medical imaging on tasks such as privacy-preserving image generation and superresolution and denoising of existing images. Few prior approaches have used cardiac magnetic resonance imaging (cMRI) as a modality given the complexity of videos (the addition of the temporal dimension) as well as the limited scale of publicly available datasets. We introduce GANcMRI, a generative adversarial network that can synthesize cMRI videos with physiological guidance based on latent space prompting. GANcMRI uses a StyleGAN framework to learn the latent space from individual video frames and leverages the timedependent trajectory between end-systolic and end-diastolic frames in the latent space to predict progression and generate motion over time. We proposed various methods for modeling latent time-dependent trajectories and found that our Frame-to-frame approach generates the best motion and video quality. GANcMRI generated high-quality cMRI image frames that are indistinguishable by cardiologists, however, artifacts in video generation allow cardiologists to still recognize the difference between real and generated videos. The generated cMRI videos can be prompted to apply physiologybased adjustments which produces clinically relevant phenotypes recognizable by cardiologists. GANcMRI has many potential applications such as data augmentation, education, anomaly detection, and preoperative planning.
Keywords: Generative AI, cardiac magnetic resonance imaging, StyleGAN, video generation, physiologic guidance
1. Introduction
Cardiac magnetic resonance imaging (cMRI) is a high-quality imaging modality for visualizing cardiac form and function, offering an unparalleled ability to capture specific details related to cardiovascular disease while sparing patients from the risks of radiation exposure. However, the application of cMRI in routine clinical practice is constrained by its high cost and lengthy acquisition time, making it impractical for sick patients that cannot hold still (‘breath holds’) or serve as the primary modality for serial measurement of disease progression. Imaging research is also hindered by the limited number of publicly available large-volume datasets.
Numerous studies have explored the tradeoff between acquisition time and resolution quality in MRI research, and methods like k-space undersampling approaches including partial Fourier (Noll et al., 1991; Peters et al., 2000; Ahn et al., 1986), sliding window (van Vaals et al., 1993; Foo et al., 1995; Korosec et al., 1996), parallel imaging (Pruessmann et al., 1999; Kozerke et al., 2004), and compressed sensing (Lustig et al., 2007; Usman et al., 2011; Jung et al., 2009) are in use to expedite scans while preserving image quality. These techniques seek to leverage less data but still yield high-quality images using advanced reconstruction algorithms.
These traditional techniques do not utilize previously collected data samples and generative machine learning to facilitate high-resolution reconstruction. However, because of the recent significant advancements in generative deep learning and synthetic data generation, this data-driven approach could pave the way for enhanced accuracy, resolution, and efficiency. Such approaches have been applied in brain MRI and knee MRI (Pinaya et al., 2022; Astuto et al., 2021), and could be similarly leveraged in cardiac MRI analysis and interpretation.
In this paper, we present GANcMRI, a cMRI specific StyleGAN model capable of generating cMRI videos, improving their temporal resolution, and visualizing disease progression with related physiologic prompting. We formulate video generation, similarly to Tian et al. (2020), as the problem of finding a suitable trajectory through the latent space across time of a pretrained image generator. Using our ED-to-ES and frame-to-frame methods, we find a trajectory in the latent space that corresponds to the progression over time. Starting from a random point in the latent space, we use this trajectory to generate a sequence of latent space points that correspond to a sequence of frames comprising a video. In addition to generating synthetic videos, we use Frame-to-frame method to increase the temporal resolution of real videos, by first projecting each of their frames to the latent space and interpolating the intermediate frames. Finally, we also propose a method for synthetic physiological prompting of cMRIs using latent space calculus to manipulate images. To demonstrate the performance of the approach, we verify that the synthetic videos have high quality based on their FID and FVD scores as well as provide blinded images and videos to domain experts (cardiologists) to evaluate whether they can identify generated vs. real images and if there are notable artifacts or changes from GANcMRI.
2. Related Works
Generative AI in MR
Recent advances in deep learning, specifically Generative adversarial networks (GANs) (Goodfellow et al., 2014), and Diffusion models (Dhariwal and Nichol, 2021; Rombach et al., 2022), allowed for high quality 2D medical image generation and the results have proven useful in various medical applications. Generative AI has been used to increase the size of image datasets (Diller et al., 2020; Pinaya et al., 2022), translate from one imaging modality to the other (Osman and Tamam, 2022; Li et al., 2022), privacy preservation, and superresolution (Wahid et al., 2022; Chen et al., 2018a,b)
Video data generation
While the achievements in image synthesis have been noteworthy, video synthesis is more complex because of the need to accurately model dynamics and the progression of time. To avoid expensive conv3d layers, the current state-of-art methods for video synthesis are predominantly leveraging architectures initially designed for image synthesis, such as StyleGAN (Karras et al., 2019) or Latent Diffusion (Ho et al., 2020). These methods are subsequently fine-tuned for video applications (Blattmann et al., 2023; Fox et al.), or undergo slight architectural modification to include the temporal dimension (Brooks et al., 2022; Skorokhodov et al., 2022). To the best of our knowledge, as of the current date, no papers have explored the application of generative AI in producing synthetic medical images with the temporal dimension.
StyleGAN2
StyleGAN introduced by Karras et al. (2019) is an architecture that uses concepts from classical GANs (Goodfellow et al., 2014), variational autoencoders (Kingma and Welling, 2022) and neural style transfer (Gatys et al., 2015) to produce high-quality images while having a well-structured latent space. StyleGAN2 is an improved version that allows for projecting real images to the latent space and semantic editing (Karras et al., 2020). StyleGAN achieved state of the art performance on different medical datasets (Woodland et al., 2022), and has also been used for the generation of volumetric images (Hong et al., 2021).
3. Methods
3.1. Data
We constructed our dataset using imaging data from the UK Biobank (UKBB) cardiac MRI cohort (Littlejohns et al., 2020). Our dataset consists of 45, 531 unique 4-chamber long axis (LAX) cine cMRIs from different UKBB participants. The cMRI videos, originally varying in height (144 − 210, avg ~ 207) and width (114 − 210, avg ~ 169), and with 50 frames, were uniformly resized to [256 × 256 × 50] for robust data processing. We select the first frame of the image to be the end-diastole (ED) frame (as described in UKB protocol by Littlejohns et al. (2020)), and the frame in which the LV area is the smallest (measured by ukbb-cmr (Bai et al., 2018)) as end-systole (ES) frame. 80−10−10 train-val-test split was used giving us 36, 425 videos in training dataset, 4, 553 videos in validation dataset and 4, 553 videos in test dataset. Each frame of the cMRI video was saved as a separate image giving us in total 2, 276, 550 images (1, 821, 250 in train, 227, 650 in val and, 227, 650 in test). While the image generator was trained on the full dataset, the number of video files used to develop video generation methods is given in Table 2.
3.2. Image generation
We train the StyleGAN2 architecture from scratch using TensorFlow’s official implementation (Karras et al.), on grayscale cMRI frames of resolution 256×256. To determine the best training checkpoint and to evaluate the performance of the image generation we use Frechet Inception Distance (FID) introduced by Heusel et al. (2018). Because FID uses the InceptionV3 network pretrained on RGB images, we convert our grayscale images to RGB before computing FID. However, since there is a potential issue with using a RGB network on grayscale images, we introduce a task-specific metric cFID. The only difference between cFID and FID is the backbone. Namely, cFID employs the encoder output (conv4 activations) from ukbb_cmr’s (Bai et al., 2018) pretrained fully convolutional network, trained for four-chamber segmentation in long-axis view. The conv4 layer activations are reshaped from (batch_size,height,width,num_channels) = (batch_size,16, 16, 256) to (batch_size,256, 256), then the mean is taken across channels resulting in the shape (batch_size, 256), i.e. for each frame we get 256 features.
Additionally, synthetic image quality and similarity to real distribution is assessed by a clinical cardiologist and cardiac imager with > 5 years of experience in cMRI. He was presented with a 100 pairs of real and fake frames, and he was asked to select a real one between the two (evaluation UI is shown in Figure 6).
3.3. Video generation
We treat videos as time-continuous signals x(t) in the image space , such that . Let be a latent space learned by StyleGAN2 for image generation, and let F be a function that maps w ∈ W to X. Assuming that F is a time-signal preserving function then ∀ t F(w(t)) = x(t) i.e. every real video can be represented as a sequence of latent points. This relationship allows us to define videos in the latent space learned by StyleGAN as shown in Figure 1. Therefore, we focus on modeling w(t) signal in the latent space W, which is easier than modeling x(t), because any point in W corresponds to a valid cMRI frame. Note the that first frame corresponds to t = 1.
Figure 1:
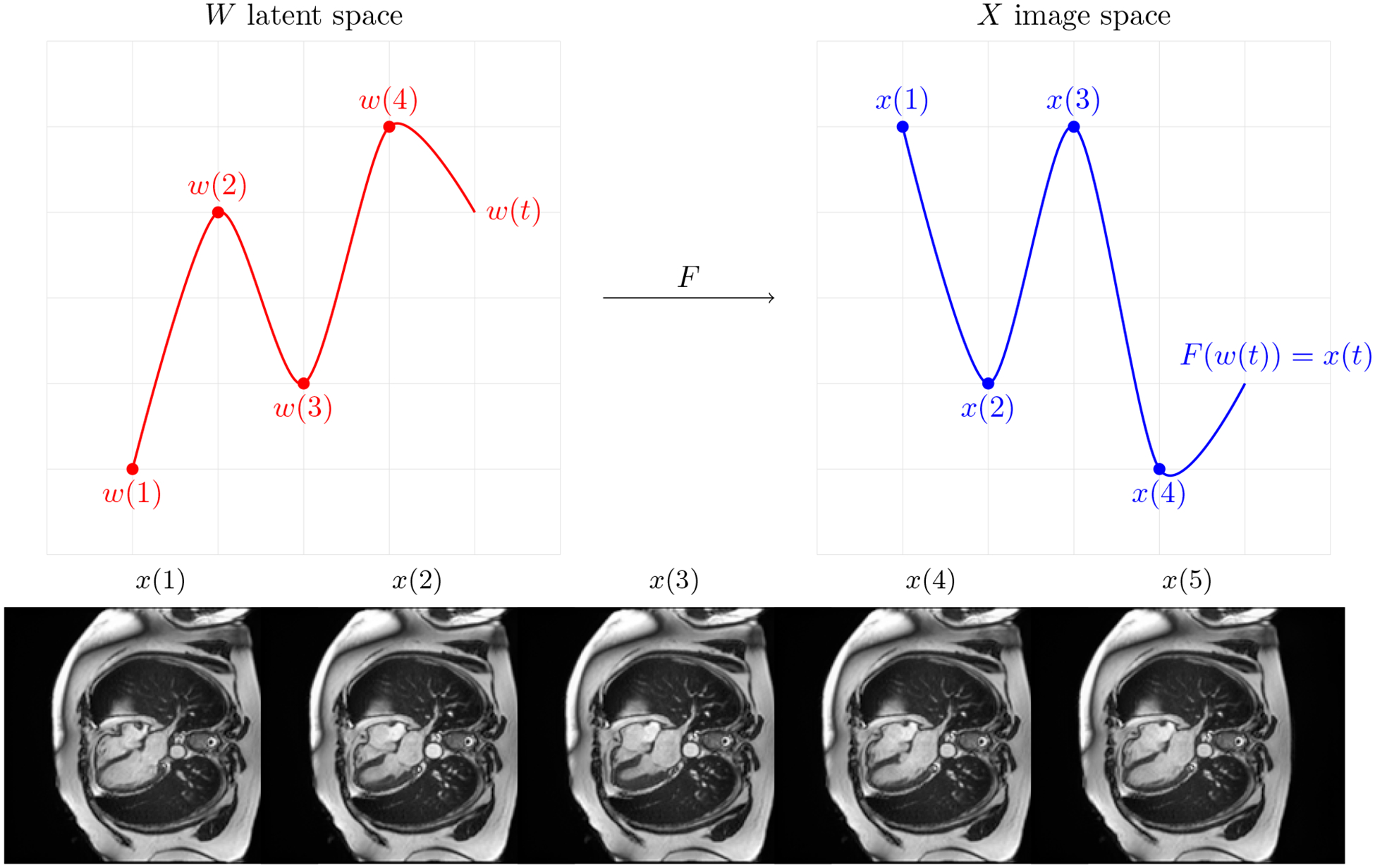
A continuous signal in latent space corresponds to the continuous signal in the image space that represents a video.
3.3.1. ED-to-ES model
Cardiac function is a cyclical filling and pumping process, typically defined by maximum relaxation at end-diastole (ED) and maximum contraction at endsystole (ES). We introduce ED-to-ES method, modeling the time-signal in the latent space as:
Here, kED→ES is the trajectory from ED to ES, and its negative represents the ES to ED trajectory, enabling full cardiac cycle simulation.
To find k explicitly we first need a way to move from the image space to the latent space, in other words, we need to project real images onto the latent space. We use the projector from the original implementation of StyleGAN2 (P : X → W s.t. P(x) = w). Then kED→ES is calculated as follows. For each video, we project ED frame and ES frame to the latent space, and then take a mean (across videos) of the difference between ES latent point and ED latent point, i.e:
Where N is the number of videos, ES_frms is a list of ES frames, ED_frms is a list of ED frames, and ED_frms[j] and ES_frms[j] are from the same video.
3.3.2. Frame-to-frame model
Second, we propose Frame-to-frame model.
In this case, we will have a trajectory k for each movement in time from w(i) to w(i + 1), in other words, we will model movement in time for every frame (50 frames), instead of just ED to ES (Figure 2). Similarly to the first method, we calculate ki→i+1 by taking a mean across videos of the difference between i + 1-th frame corresponding latent point and i-th frame corresponding latent point.
Where videos is a list of videos, and videos[j][i] is the i-th frame from the j-th video. However, when using Frame-to-frame method we have to use multiple trajectories (ki→i+1) and the transition between them might not be smooth, which would result in video jitter. To fix the sharp transition, we fit a PCA model PCA(·), computing 32 principal components of the dataset K = {k1→2 ...ki→i+1 ...k49→50}, and use this pretrained model to update our frame-to-frame trajectories:
Figure 2:
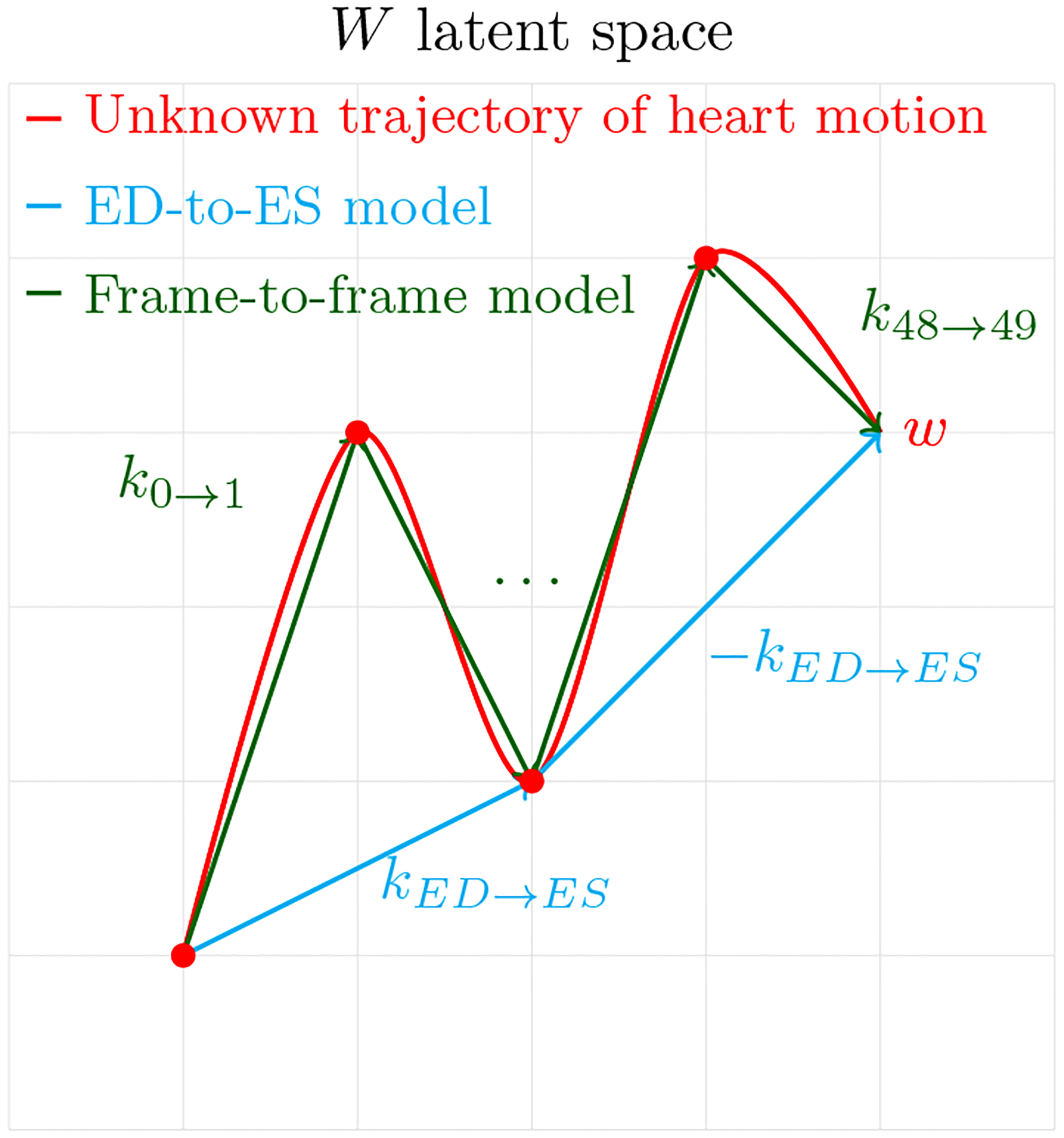
ED-to-ES method finds a latent direction that corresponds to the movement of the heart from end-diastole to end-systole in the image space. Frame-to-frame method finds 49 latent directions corresponding to the movement of the heart from frame to frame in the image space
3.3.3. Applying ED-to-ES and Frame-to-frame for video generation
To generate a synthetic video resembling cMRIs in the UK Biobank, we first need to get the starting latent code w(1) that corresponds to the ED frame in the image space. We do so by picking a random point in the latent space w and then moving along the direction from ED to ES.
Next, we use either ED-to-ES or Frame-to-frame to predict future timesteps resulting in a 50 frame video. When using ED-to-ES method we use kED→ES trajectory to generate the first 25 frames, and −kED→ES to generate the last 25 frames, while for the Frame-to-frame method, we use a different trajectory for every frame transition.
3.3.4. Temporal super-resolution
Using the latent space projection of real cMRI frames we can increase the temporal resolution of cMRI. Given that {w1,...wM} wi = P(i − th frame) are latent codes for all frames of a single cMRI video, (M is the number of frames) we compute the transition trajectories for this single video
Then, we fit a PCA model (and call it tPCA) on the dataset of all transitions calculating the first (M − 6) principal components of the dataset T = {t1→2 ...t49→50}, and update the transition trajectories:
Finally, starting from w1 we can compute latent codes for any number of intermediate frames. For
For example, to compute a frame that comes in between of 4 and 5 we take h = 4.5.
We conduct two experiments with temporal super-resolution. First, we reduce a 50-frame video to 25 frames by removing every second frame, then we use GANcMRI to interpolate intermediate frames and restore it to 50-frames. We also apply the same process to the original 50-frame video to obtain a 100-frame video, that we use for visual quality assessment.
3.3.5. Evaluation
To evaluate the video generation performance of the ED-to-ES and the Frame-to-frame methods, we use Frechet Video Distance (FVD) introduced by Unterthiner et al. (2019) from the implementation by Skorokhodov et al. (2022). When calculating FVD we sample 16 equally spaced frames from both the synthetic and real videos.
We assess temporal super-resolution performance using the mean structural similarity index (meanSSIM). meanSSIM is an average of SSIM computed between the real (removed) and the interpolated frames.
Additionally, a cardiologist conducts visual quality testing on 100 video sets using a custom UI (Figure 7). Each set consists of four cardiac MRI videos: a 50-frame real video, a 100-frame AI-enhanced video (temporal super-resolution), a 50-frame fully AI-generated video using ED-to-ES method, and a 50-frame fully AI-generated video using Frame-to-frame method.
3.4. Physiologic guidance
We aim to find the latent space trajectory kpheno_low→pheno_high (for arbitrary phenotype), such that if we move the latent point along this trajectory the phenotype value in the corresponding image will increase. To do so, we first find the mean of the phenotype values and then classify all the cMRIs with the phenotype value lower than the mean as low, and all the cMRIs with the phenotype value higher than the mean high. Next, we embed ED frames of low cMRIs and high cMRIs and get lists low latents and high latents. Finally, we calculate k as follows:
Given that w is a latent code of any real or the generated image, we can increase the value of the phenotype in w by adjusting it as follows: wadjusted = w + c kpheno_low→pheno_high. To verify the accuracy, we use automated measurement methods on F(wadjusted) and observe that there is a linear relationship between c and a measurement obtained using these methods. We focus on ED frames and the physiologic adjustments of left ventricular sphericity index (calculated in the same manner as Vukadinovic et al. (2023)) and left ventricular area, but the described method can be applied for any phenotype.
4. Results
4.1. Synthetic single frame cMRI images are indistinguishable from the real ones
Image generation performance was evaluated with FID, cFID, and the visual quality check was performed by a cardiologist. FID was computed between 50, 000 random samples from real image distribution and 50, 000 random samples from synthetic image distribution [FID = 92.58 cFID = 20.03].
In the evaluation of 100 pairs of fake and real images, the clinical cardiologist incorrectly identified the fake image as real in 60% of the cases and correctly identified the real image in the remaining 40% of cases. Figure 3 shows a sample of 4 real and 4 fake images.
Figure 3:
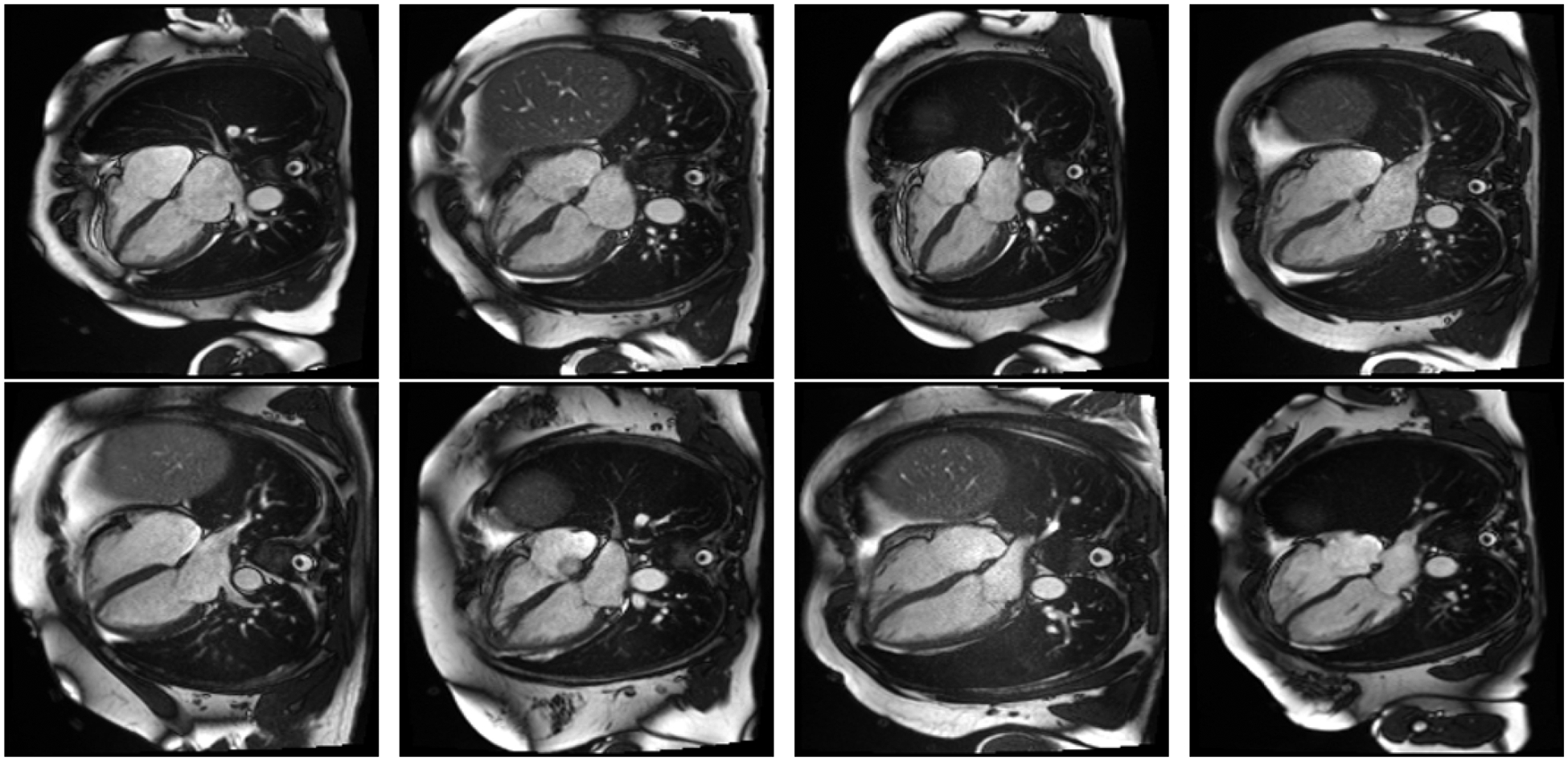
Top row: AI generated images. Bottom row: Real images.
4.2. Frame-to-frame video generation outperforms other approaches
All three methods for video generation (ED-to-ES, Frame-to-frame and, super-resolution) yielded high-quality videos with smooth transitions. We made fully synthetic ED-to-ES generated videos and fully synthetic Frame-to-frame generated videos publicly available in the supplementary material.
Frechet Video Distance (FVD) was computed between each two pairs of distributions (Table 1). FV D gives two indicators that Frame-to-frame model is superior to ED-to-ES model. First,
means that frame-to-frame distribution is closer to the real distribution. And
shows us that the distribution of videos produced by Frame-to-frame method is more stable than ED-to-ES method.
Table 1:
FVD for each two pairs of distributions
| 35.28 | 95.16 | |
The super-resolution quality was assessed separately, as described in methods 3.3.5, with meanSSIM. MeanSSIM computed on 100 video samples is 0.7.
All methods were evaluated in a visual quality test. Frame-to-frame videos were a clear winner, being assigned the highest ranking among synthetic methods in 50% of cases (Figure 4). Interestingly, the superresolution videos with 100 frames performed worse than their 50-frame competitors. While synthetic videos may appear convincingly real upon initial observation, a cardiologist was able to successfully distinguish all real videos from their synthetic counterparts with absolute accuracy.
Figure 4:
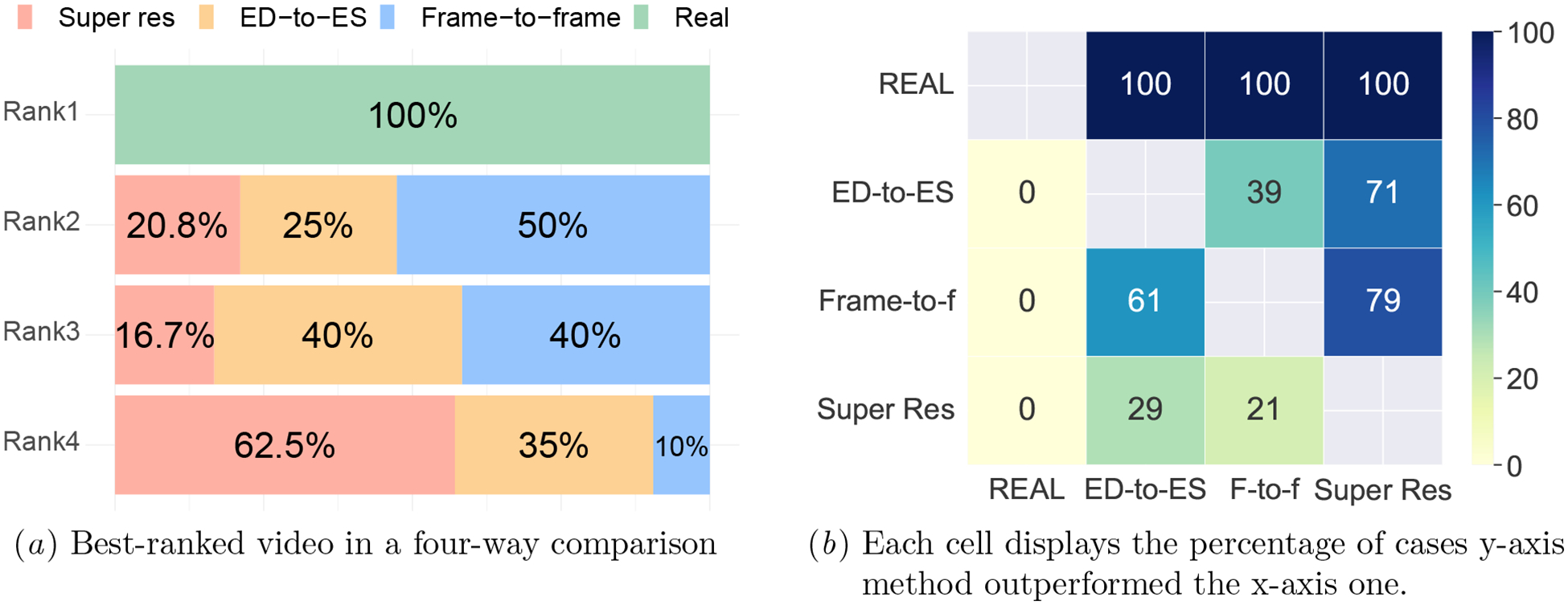
A cardiologist ranked the quality of real and synthetic cMRIs in a side-by-side comparison.
Finally, to showcase our method’s efficiency and scalability, we benchmarked it using an NVIDIA RTX 2080 Ti GPU and i9–9820X CPU. Our frame-to-frame approach took 0.73s ± 0.02s to produce a 50-frame 256×256 grayscale cMRI video in 100 trials and ED-to-ES took 0.71s ± 0.01s.
4.3. GANcMRI accurately reflects physiologic adjustment
As explained in methods 3.4, we perform morphological adjustment by moving along the trajectory kpheno low→pheno high, and we do so by adding a scaled value of the trajectory to the latent code corresponding to cMRI image. In our experiments, we picked a scalar c to be in the interval from [−2, 3], because scalars outside of this range result in abnormally small/big phenotype values. Upon visual inspection, it is easy to see (Figure 5) that moving along the trajectory ksphericity_low→sphericity_high in the latent space GANcMRI generates ED frames ranging from low sphericity index to high sphericity index.
Figure 5:
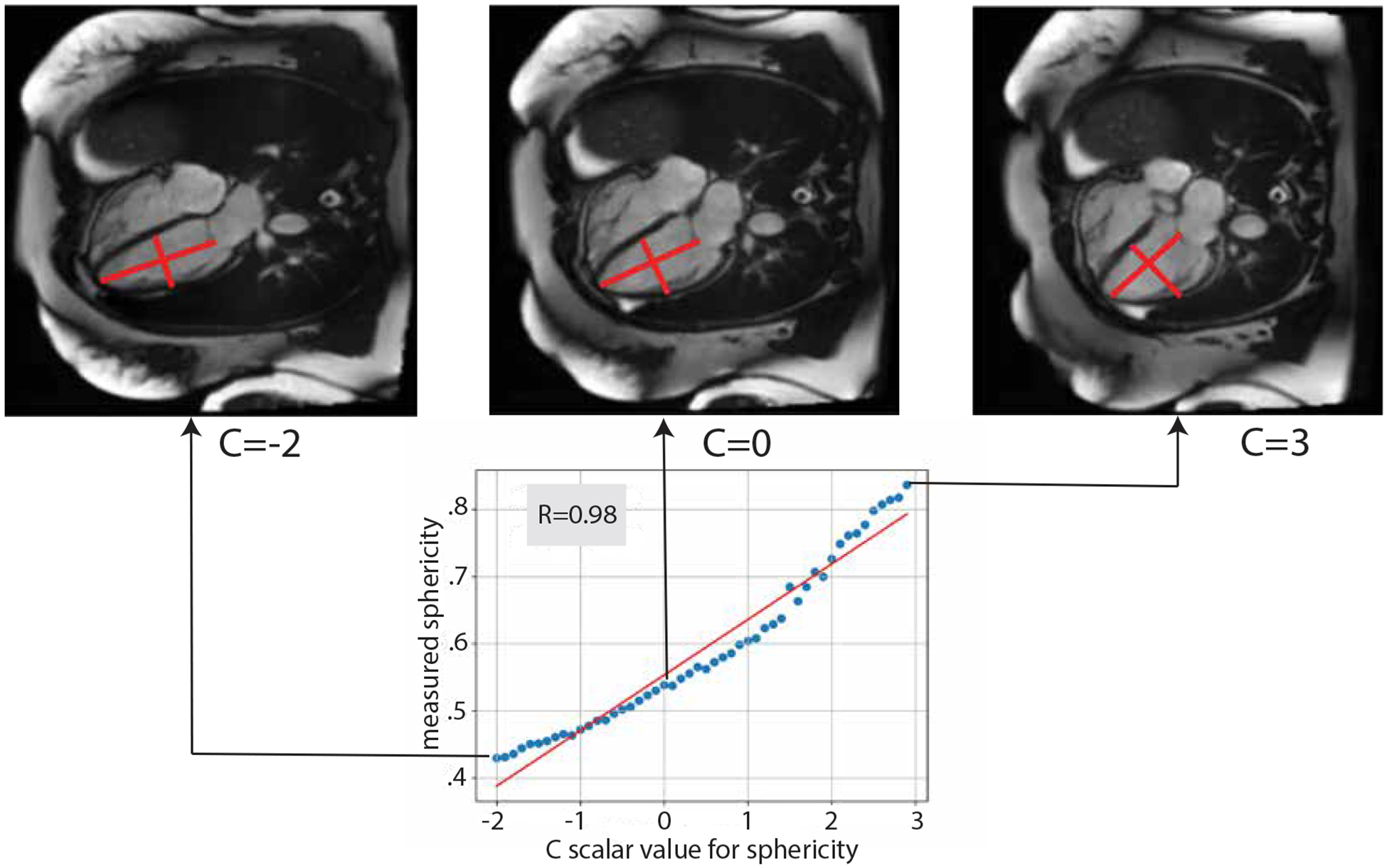
By increasing the scalar C we move along the latent space direction that corresponds to increasing the sphericity index in image space
To confirm the relationship numerically, we perform a pearson R test between the values of c and the actual phenotype value of the synthetic images corresponding to the modified latent vector. Indeed, the correlation is strong, resulting in pearson R = 0.98 and p value = 7.8·10−35 for left ventricular sphericity index, and pearson R = 0.89 and p value = 6.9·10−18 for left ventricular area (Figures 8, 9).
These results enable conditional synthetic cMRI video creation by pre-selecting heart properties. The code and the weights to run conditional cMRI video generation with GANcMRI are available at https://github.com/vukadinovic936/GANcMRI
5. Discussion
We utilized a generative machine learning approach, the substantial UK Biobank cMRI data, and latent space calculus to generate cMRI images and videos. Nearing the level of being indistinguishable by clinicians, optimally generated cMRI videos can open new avenues in cardiac imaging research and clinical practice. We were able to generate individual synthetic cMRI frames of a quality indistinguishable from real ones. By interpreting videos as continuous signals within the image latent space, we model time trajectories on a frame-to-frame basis to produce cMRI videos of higher temporal resolution and superior quality compared to other video generation methods, although they remain distinguishable from the real ones. Finally, we demonstrate physiologic guidance can be applied to synthetic cMRIs to generate clinically relevant changes in the synthetic videos.
GANcMRI is a versatile tool with multiple downstream applications: it enables conditional video generation by tweaking physiological attributes of initial frames and employing Frame-to-frame model for time progression; when combined with language models, like GPT, it facilitates physiologic guidance with natural language prompts; it can produce synthetic datasets, thus mitigating privacy issues; and, it possesses the capacity for spatial super-resolution, transforming low-res MRI scans into higher resolution 256 × 256 resolution images.
A few limitations still persist. While our synthetic image generation methods are sufficient to be indistinguishable from natural images, limitations in generated videos still allow them to be identified. Generated videos present smoother transitions than the actual cardiac motion and lack the Brownian motion of the blood pool, making them distinguishable by cardiologists. Given our limited information approach, Brownian motion was unable to be simulated, however future iterations might be able to more closely replicate actual cardiac motion with cardiacphase specific motion. In our current model, the time progress trajectory is approximated with linear trajectories which may not capture complex temporal dynamics sufficiently. In our experiments, we did not identify any non-physiologic artifacts or unnatural images, however a more thorough evaluation was bottlenecked by cardiologist time. A more detailed evaluation and ablation studies are warranted in the future if ever put into clinical practice.
Further research should be undertaken to optimize the quality of cMRI imaging. Data-driven generative approaches might decrease the amount of necessary data, allowing for faster and cheaper scans. Super-resolution should be approached with caution given the potential for hallucination, however, our physiological prompting within the embedding space allows the generation of reasonably looking attributes consistent with cardiovascular disease. Further work remains to leverage polynomial regression to better approximate trajectories and the architecture to improve the model’s understanding of temporal dimension.
Supplementary Material
Appendix A. Implementation and Evaluation details
Table 2:
Number of files used for each method
| Method | Number of files |
|---|---|
| ED-to-ES | 547 |
| Frame-to-Frame | 151 |
| Super-res | 1 |
| Spher phys | 139 (68 low, 71 high) |
| LV phys | 149 (83 low, 66 high) |
Figure 6:
User interface for evaluating the quality of synthetic cMRI frames. 1 real and 1 fake frame are shown and the evaluator is asked to choose the frame that he/she thinks is real.
Figure 7:

User interface for evaluating the quality of synthetic cMRI videos. The evaluator is shown a real video, ED-to-ES generated video, Frame-to-frame generated video and super-resolution video. They click the radio button next to the video they think is real, and assign rank from 1 to 4 in order of quality to all videos.
Figure 8:
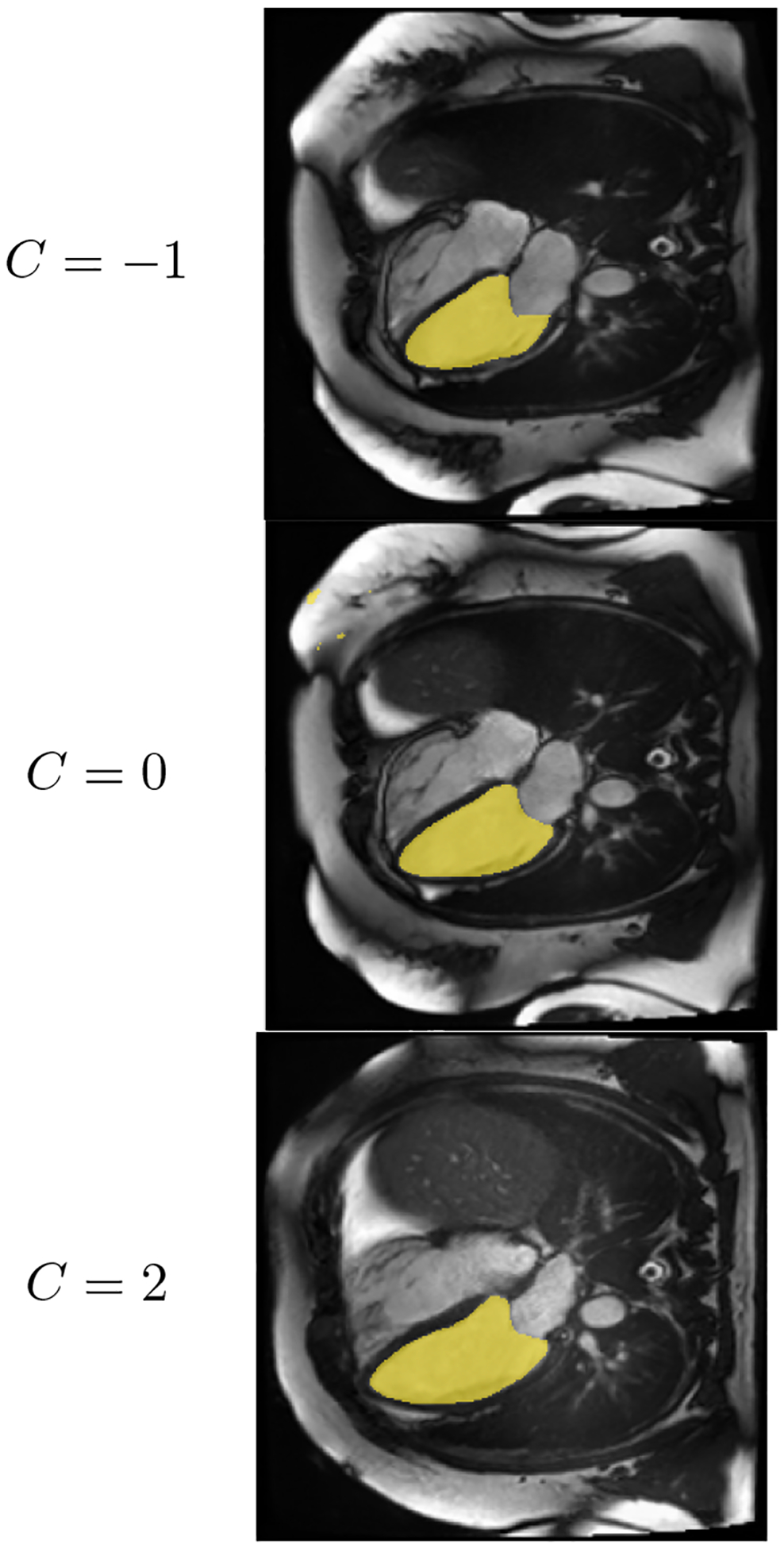
As the scalar C increases, LV area increases
Figure 9:
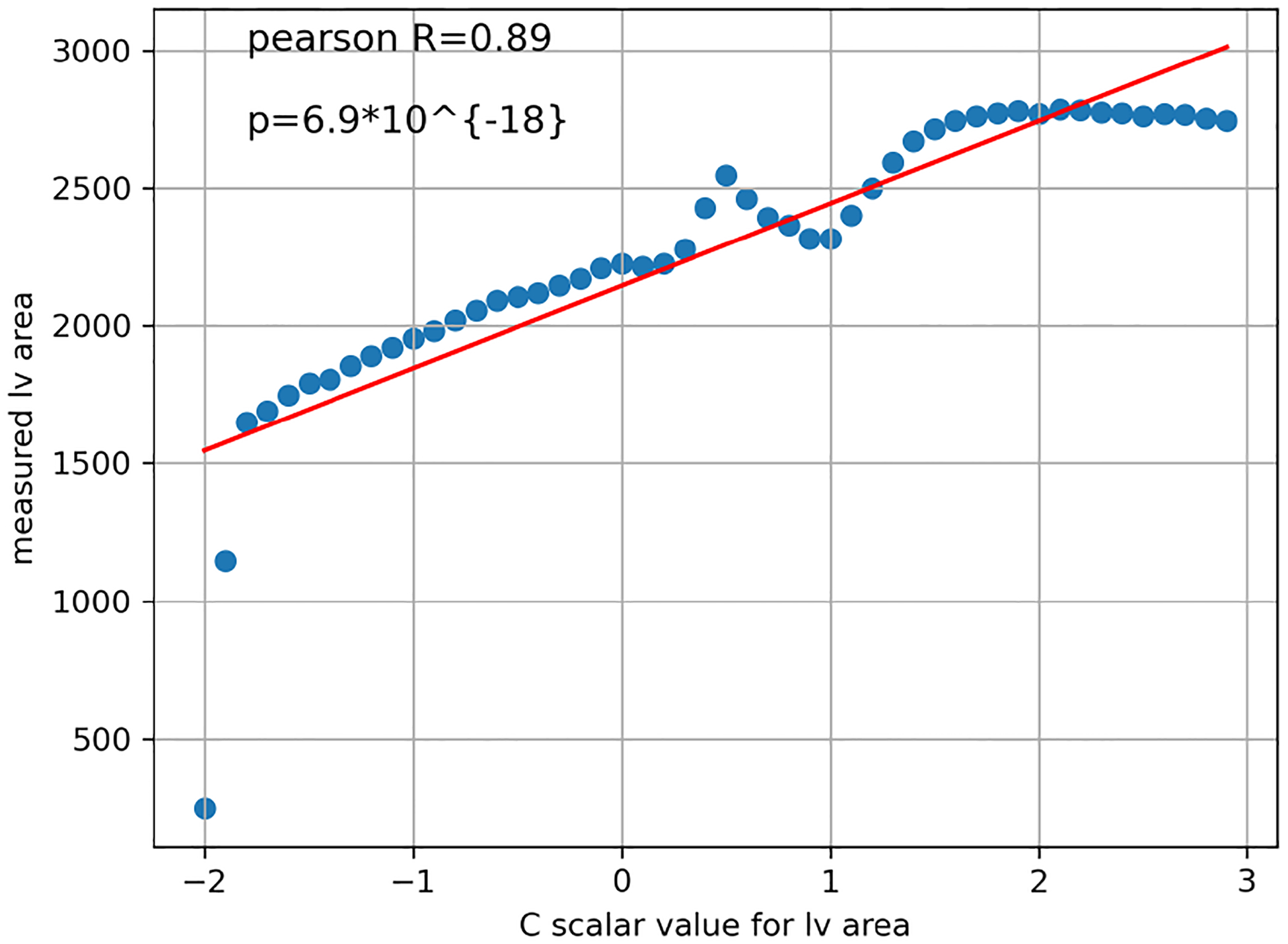
Moving along the direction of ksmall_lv_area→big_lv_area is correlated with the increase in lv area.
Contributor Information
Milos Vukadinovic, University of California, Los Angeles, USA.
Alan C Kwan, Cedars-Sinai Medical Center, USA.
Debiao Li, Cedars-Sinai Medical Center, USA.
David Ouyang, Cedars-Sinai Medical Center, USA.
References
- Ahn CB, Kim JH, and Cho ZH. High-speed spiral-scan echo planar NMR imaging-I. IEEE transactions on medical imaging, 5(1):2–7, 1986. ISSN 0278–0062. doi: 10.1109/TMI.1986.4307732. [DOI] [PubMed] [Google Scholar]
- Astuto Bruno, Flament Io, Namiri Nikan K., Shah Rutwik, Bharadwaj Upasana, Link Thomas M., Bucknor Matthew D., Pedoia Valentina, and Majumdar Sharmila. Automatic Deep Learning–assisted Detection and Grading of Abnormalities in Knee MRI Studies. Radiology: Artificial Intelligence, 3(3):e200165, May 2021. doi: 10.1148/ryai.2021200165. URL https://pubs.rsna.org/doi/10.1148/ryai.2021200165. Publisher: Radiological Society of North America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Wenjia, Sinclair Matthew, Tarroni Giacomo, Oktay Ozan, Rajchl Martin, Vaillant Ghislain, Lee Aaron M., Aung Nay, Lukaschuk Elena, Sanghvi Mihir M., Zemrak Filip, Fung Kenneth, Jose Miguel Paiva Valentina Carapella, Young Jin Kim Hideaki Suzuki, Kainz Bernhard, Matthews Paul M., Petersen Steffen E., Piechnik Stefan K., Neubauer Stefan, Glocker Ben, and Rueckert Daniel. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. Journal of Cardiovascular Magnetic Resonance, 20(1):65, September 2018. ISSN 1532–429X. doi: 10.1186/s12968-018-0471-x. URL 10.1186/s12968-018-0471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattmann Andreas, Rombach Robin, Ling Huan, Dockhorn Tim, Seung Wook Kim Sanja Fidler, and Kreis Karsten. Align your Latents: High-Resolution Video Synthesis with Latent Diffusion Models, April 2023. URL http://arxiv.org/abs/2304.08818. arXiv:2304.08818 [cs]. [Google Scholar]
- Brooks Tim, Hellsten Janne, Aittala Miika, Wang Ting-Chun, Aila Timo, Lehtinen Jaakko, Liu Ming-Yu, Efros Alexei A., and Karras Tero. Generating Long Videos of Dynamic Scenes, June 2022. URL http://arxiv.org/abs/2206.03429. arXiv:2206.03429 [cs]. [Google Scholar]
- Chen Yuhua, Shi Feng, Christodoulou Anthony G., Xie Yibin, Zhou Zhengwei, and Li Debiao. Efficient and Accurate MRI Super-Resolution Using a Generative Adversarial Network and 3D Multilevel Densely Connected Network. In Frangi Alejandro F., Schnabel Julia A., Davatzikos Christos, Alberola-López Carlos, and Fichtinger Gabor, editors, Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, Lecture Notes in Computer Science, pages; 91–99, Cham, 2018a. Springer International Publishing. ISBN 978-3030-00928-1. doi: 10.1007/978-3-030-00928-1_11. [DOI] [Google Scholar]
- Chen Yuhua, Xie Yibin, Zhou Zhengwei, Shi Feng, Christodoulou Anthony G., and Li Debiao. Brain MRI super resolution using 3D deep densely connected neural networks. In 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), pages 739–742. IEEE, 2018b. [Google Scholar]
- Dhariwal Prafulla and Nichol Alexander. Diffusion Models Beat GANs on Image Synthesis. In Advances in Neural Information Processing Systems, volume 34, pages 8780–8794. Curran Associates, Inc., 2021. URL https://papers.nips.cc/paper/2021/hash/49ad23d1ec9fa4bd8d77d02681df5cfa-Abstract.html. [Google Scholar]
- Diller Gerhard-Paul, Vahle Julius, Radke Robert, Vidal Maria Luisa Benesch, Fischer Alicia Jeanette, Bauer Ulrike M. M., Sarikouch Samir, Berger Felix, Beerbaum Philipp, Baumgartner Helmut, Orwat Stefan, and German Competence Network for Congenital Heart Defects Investigators. Utility of deep learning networks for the generation of artificial cardiac magnetic resonance images in congenital heart disease. BMC medical imaging, 20(1):113, October 2020. ISSN 1471–2342. doi: 10.1186/s12880-020-00511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo TK, Bernstein MA, Aisen AM, Hernandez RJ, Collick BD, and Bernstein T. Improved ejection fraction and flow velocity estimates with use of view sharing and uniform repetition time excitation with fast cardiac techniques. Radiology, 195(2):471–478, May 1995. ISSN 0033–8419. doi: 10.1148/radiology.195.2.7724769. [DOI] [PubMed] [Google Scholar]
- Fox Gereon, Tewari Ayush, Elgharib Mohamed, and Theobalt Christian. StyleVideoGAN: A Temporal Generative Model using a Pretrained StyleGAN. URL https://vcai.mpi-inf.mpg.de/projects/stylevideogan/.
- Gatys Leon A., Ecker Alexander S., and Bethge Matthias. A Neural Algorithm of Artistic Style, September 2015. URL http://arxiv.org/abs/1508.06576. arXiv:1508.06576 [cs, q-bio]. [Google Scholar]
- Goodfellow Ian J., Jean Pouget-Abadie Mehdi Mirza, Xu Bing, David Warde-Farley Sherjil Ozair, Courville Aaron, and Bengio Yoshua. Generative Adversarial Networks, June 2014. URL http://arxiv.org/abs/1406.2661. arXiv:1406.2661 [cs, stat]. [Google Scholar]
- Heusel Martin, Ramsauer Hubert, Unterthiner Thomas, Nessler Bernhard, and Hochreiter Sepp. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium, January 2018. URL http://arxiv.org/abs/1706.08500. arXiv:1706.08500 [cs, stat]. [Google Scholar]
- Ho Jonathan, Jain Ajay, and Abbeel Pieter. Denoising Diffusion Probabilistic Models, December 2020. URL http://arxiv.org/abs/2006.11239. arXiv:2006.11239 [cs, stat]. [Google Scholar]
- Hong Sungmin, Marinescu Razvan, Dalca Adrian V., Bonkhoff Anna K., Bretzner Martin, Rost Natalia S., and Golland Polina. 3D-StyleGAN: A StyleBased Generative Adversarial Network for Generative Modeling of Three-Dimensional Medical Images. In Engelhardt Sandy, Oksuz Ilkay, Zhu Dajiang, Yuan Yixuan, Mukhopadhyay Anirban, Heller Nicholas, Huang Sharon Xiaolei, Nguyen Hien, Sznitman Raphael, and Xue Yuan, editors, Deep Generative Models, and Data Augmentation, Labelling, and Imperfections, Lecture Notes in Computer Science, pages 24–34, Cham, 2021. Springer International Publishing. ISBN 978-3030-88210-5. doi: 10.1007/978-3-030-88210-5_3. [DOI] [Google Scholar]
- Jung Hong, Sung Kyunghyun, Nayak Krishna S., Kim Eung Yeop, and Ye Jong Chul. k-t FOCUSS: A general compressed sensing framework for high resolution dynamic MRI. Magnetic Resonance in Medicine, 61(1):103–116, 2009. ISSN 1522–2594. doi: 10.1002/mrm.21757. URL https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.21757. [DOI] [PubMed] [Google Scholar]
- Karras Tero, Laine Samuli, Aittala Miika, Hellsten Janne, Lehtinen Jaakko, and Aila Timo. Analyzing and Improving the Image Quality of StyleGAN, March 2020. URL http://arxiv.org/abs/1912.04958. arXiv:1912.04958 [cs, eess, stat].
- Kingma Diederik P. and Welling Max. Auto-Encoding Variational Bayes, December 2022. URL http://arxiv.org/abs/1312.6114. arXiv:1312.6114 [cs, stat].
- Korosec FR, Frayne R, Grist TM, and Mistretta CA. Time-resolved contrast-enhanced 3D MR angiography. Magnetic Resonance in Medicine, 36 (3):345–351, September 1996. ISSN 0740–3194. doi: 10.1002/mrm.1910360304. [DOI] [PubMed] [Google Scholar]
- Kozerke Sebastian, Tsao Jeffrey, Razavi Reza, and Boesiger Peter. Accelerating cardiac cine 3D imaging using k-t BLAST. Magnetic Resonance in Medicine, 52(1):19–26, July 2004. ISSN 0740–3194. doi: 10.1002/mrm.20145. [DOI] [PubMed] [Google Scholar]
- Li Wen, Xiao Haonan, Li Tian, Ren Ge, Lam Saikit, Teng Xinzhi, Liu Chenyang, Zhang Jiang, Francis Kar-Ho Lee Kwok-Hung Au, Victor HoFun Lee Amy Tien Yee Chang, and Cai Jing. Virtual Contrast-Enhanced Magnetic Resonance Images Synthesis for Patients With Nasopharyngeal Carcinoma Using Multimodality-Guided Synergistic Neural Network. International Journal of Radiation Oncology, Biology, Physics, 112(4): 1033–1044, March 2022. ISSN 1879–355X. doi: 10.1016/j.ijrobp.2021.11.007. [DOI] [PubMed] [Google Scholar]
- Littlejohns Thomas J., Holliday Jo, Gibson Lorna M., Garratt Steve, Oesingmann Niels, Fidel Alfaro-Almagro Jimmy D. Bell, Boultwood Chris, Collins Rory, Conroy Megan C., Crabtree Nicola, Doherty Nicola, Frangi Alejandro F., Harvey Nicholas C., Leeson Paul, Miller Karla L., Neubauer Stefan, Petersen Steffen E., Sellors Jonathan, Sheard Simon, Smith Stephen M., Sudlow Cathie L. M., Matthews Paul M., and Allen Naomi E.. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nature Communications, 11(1): 2624, May 2020. ISSN 2041–1723. doi: 10.1038/s41467-020-15948-9. URL https://www.nature.com/articles/s41467-020-15948-9. Number: 1 Publisher: Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig Michael, Donoho David, and Pauly John M.. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine, 58(6):1182–1195, December 2007. ISSN 0740–3194. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- Noll DC, Nishimura DG, and Macovski A. Homodyne detection in magnetic resonance imaging. IEEE transactions on medical imaging, 10(2):154–163, 1991. ISSN 0278–0062. doi: 10.1109/42.79473. [DOI] [PubMed] [Google Scholar]
- Osman Alexander F. I. and Tamam Nissren M.. Deep learning-based convolutional neural network for intramodality brain MRI synthesis. Journal of Applied Clinical Medical Physics, 23(4):e13530, April 2022. ISSN 1526–9914. doi: 10.1002/acm2.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DC, Korosec FR, Grist TM, Block WF, Holden JE, Vigen KK, and Mistretta CA. Undersampled projection reconstruction applied to MR angiography. Magnetic Resonance in Medicine, 43(1):91–101, January 2000. ISSN 0740–3194. doi: . [DOI] [PubMed] [Google Scholar]
- Pinaya Walter H. L., Tudosiu Petru-Daniel, Dafflon Jessica, da Costa Pedro F., Fernandez Virginia, Nachev Parashkev, Ourselin Sebastien, and Cardoso M. Jorge. Brain Imaging Generation with Latent Diffusion Models, September 2022. URL http://arxiv.org/abs/2209.07162. arXiv:2209.07162 [cs, eess, q-bio].
- Pruessmann KP, Weiger M, Scheidegger MB, and Boesiger P. SENSE: sensitivity encoding for fast MRI. Magnetic Resonance in Medicine, 42(5): 952–962, November 1999. ISSN 0740–3194. [PubMed] [Google Scholar]
- Rombach Robin, Blattmann Andreas, Lorenz Dominik, Esser Patrick, and Ommer Björn. HighResolution Image Synthesis with Latent Diffusion Models, April 2022. URL http://arxiv.org/abs/2112.10752. arXiv:2112.10752 [cs].
- Skorokhodov Ivan, Tulyakov Sergey, and Elhoseiny Mohamed. StyleGAN-V: A Continuous Video Generator with the Price, Image Quality and Perks of StyleGAN2, May 2022. URL http://arxiv.org/abs/2112.14683. arXiv:2112.14683 [cs].
- Tian Yu, Ren Jian, Chai Menglei, Olszewski Kyle, Peng Xi, Metaxas Dimitris N., and Tulyakov Sergey. A Good Image Generator Is What You Need for High-Resolution Video Synthesis. October 2020. URL https://openreview.net/forum?id=6puCSjH3hwA.
- Unterthiner Thomas, Sjoerd van Steenkiste Karol Kurach, Marinier Raphael, Michalski Marcin, and Gelly Sylvain. Towards Accurate Generative Models of Video: A New Metric & Challenges, March 2019. URL http://arxiv.org/abs/1812.01717. arXiv:1812.01717 [cs, stat].
- Usman M, Prieto C, Schaeffter T, and Batchelor PG. k-t Group sparse: a method for accelerating dynamic MRI. Magnetic Resonance in Medicine, 66(4):1163–1176, October 2011. ISSN 1522–2594. doi: 10.1002/mrm.22883. [DOI] [PubMed] [Google Scholar]
- van Vaals JJ, Brummer ME, Dixon WT, Tuithof HH, Engels H, Nelson RC, Gerety BM, Chezmar JL, and den Boer JA. ”Keyhole” method for accelerating imaging of contrast agent uptake. Journal of magnetic resonance imaging: JMRI, 3(4):671–675, 1993. ISSN 1053–1807. doi: 10.1002/jmri.1880030419. [DOI] [PubMed] [Google Scholar]
- Vukadinovic Milos, Kwan Alan C., Yuan Victoria, Salerno Michael, Lee Daniel C., Albert Christine M., Cheng Susan, Li Debiao, Ouyang David, and Clarke Shoa L.. Deep learning-enabled analysis of medical images identifies cardiac sphericity as an early marker of cardiomyopathy and related outcomes. Med, 4(4):252–262.e3, April 2023. ISSN 26666340. doi: 10.1016/j.medj.2023.02.009. URL https://linkinghub.elsevier.com/retrieve/pii/S2666634023000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid Kareem A., Xu Jiaofeng, Dina El-Habashy Yomna Khamis, Abobakr Moamen, Brigid McDonald Nicolette O Connell, Thill Daniel, Ahmed Sara, Christina Setareh Sharafi Kathryn Preston, Travis C Salzillo Abdallah Mohamed, He Renjie, Cho Nathan, Christodouleas John, Fuller Clifton D., and Naser Mohamed A.. DeepLearning-Based Generation of Synthetic HighResolution MRI from Low-Resolution MRI for Use in Head and Neck Cancer Adaptive Radiotherapy. preprint, Radiology and Imaging, June 2022. URL http://medrxiv.org/lookup/doi/10.1101/2022.06.19.22276611. [Google Scholar]
- Woodland McKell, Wood John, Anderson Brian M., Kundu Suprateek, Lin Ethan, Koay Eugene, Odisio Bruno, Chung Caroline, Hyunseon Christine Kang Aradhana M. Venkatesan, Yedururi Sireesha, De Brian, Lin Yuan-Mao, Patel Ankit B., and Brock Kristy K.. Evaluating the Performance of StyleGAN2-ADA on Medical Images. In Zhao Can, Svoboda David, Wolterink Jelmer M., and Escobar Maria, editors, Simulation and Synthesis in Medical Imaging, Lecture Notes in Computer Science, pages 142–153, Cham, 2022. Springer International Publishing. ISBN 978-3-031-16980-9. doi: 10.1007/978-3-031-16980-9_14. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


