ABSTRACT
We previously conducted a multicenter surveillance study on Candida epidemiology and antifungal resistance in Madrid (CANDIMAD study; 2019–2021), detecting an increase in fluconazole-resistant Candida parapsilosis. We here present data on isolates collected in 2022. Furthermore, we report the epidemiology and antifungal resistance trends during the entire period, including an analysis per ward of admission. Candida spp. incident isolates from blood cultures and intra-abdominal samples from patients cared for at 16 hospitals in Madrid, Spain, were tested with the EUCAST E.Def 7.3.2 method against amphotericin B, azoles, micafungin, anidulafungin, and ibrexafungerp and were molecularly characterized. In 2022, we collected 766 Candida sp. isolates (686 patients; blood cultures, 48.8%). Candida albicans was the most common species found, and Candida auris was undetected. No resistance to amphotericin B was found. Overall, resistance to echinocandins was low (0.7%), whereas fluconazole resistance was 12.0%, being higher in blood cultures (16.0%) mainly due to fluconazole-resistant C. parapsilosis clones harboring the Y132F-R398I ERG11p substitutions. Ibrexafungerp showed in vitro activity against the isolates tested. Whereas C. albicans was the dominant species in most hospital wards, we observed increasing C. parapsilosis proportions in blood. During the entire period, echinocandin resistance rates remained steadily low, while fluconazole resistance increased in blood from 6.8% (2019) to 16% (2022), mainly due to fluconazole-resistant C. parapsilosis (2.6% in 2019 to 36.6% in 2022). Up to 7 out of 16 hospitals were affected by fluconazole-resistant C. parapsilosis. In conclusion, rampant clonal spreading of C. parapsilosis fluconazole-resistant genotypes is taking place in Madrid.
KEYWORDS: Candida, antifungal resistance, C. parapsilosis, Y132F, Madrid
INTRODUCTION
Some institutions have reported changes in Candida species distribution alongside an increase in antifungal resistance rates in recent years, mainly because of the emergence of Candida auris or fluconazole-resistant Candida parapsilosis, which partially intensified during the COVID-19 pandemic (1 – 3). To date, Spain, France, Italy, Greece, Austria, Switzerland, Czech Republic, Germany, Poland, Belgium, the Netherlands, the UK, Ireland, Denmark, Norway, Sweden, Finland, Russia, and Turkey are the European countries that have reported C. auris infections (4, 5). Moreover, fluconazole-resistant C. parapsilosis isolates harboring the Y132F ERG11p substitution have also been recently reported in Turkey, France, Italy, Slovakia, and Spain (1, 3, 6).
Prospective surveillance studies on invasive Candida isolates are key to monitoring resistance rates, studying local epidemiology, and detecting clones or species of particular interest. Unfortunately, such studies are rarely conducted in Spain (7 – 10). Therefore, we conducted a multicenter surveillance study (named CANDIMAD, CANDIdaemia in MADrid) on Candida isolates collected from patients admitted to the main hospitals located in Madrid, Spain, which cover an area of around seven million people. The main observation from isolates collected between 2019 and 2021 was the emergence of fluconazole-resistant C. parapsilosis causing candidemia in patients admitted to five of the participating hospitals. All resistant isolates harbored the Y132F (in four hospitals) or G458S (in a fifth hospital) ERG11p substitutions (11 – 13). The rate of fluconazole resistance in C. parapsilosis at that time showed an upward trend suggesting that the problem in Madrid was increasing in magnitude (13).
Most surveillance studies assessed antifungal resistance during a limited period of time, thus lacking a long-term perspective, which is particularly required after the outbreak of the COVID-19 pandemic (14, 15). Here, we report data from isolates collected in the CANDIMAD study in 2022, the year in which the COVID-19 pandemic started to recede, and assessed the trend of antifungal resistance since the beginning of the project in 2019. Furthermore, we provide additional detailed information concerning Candida spp. epidemiology and rates of resistance per type of ward of admission of patients.
RESULTS
Species epidemiology and antifungal resistance rates found in isolates collected in 2022
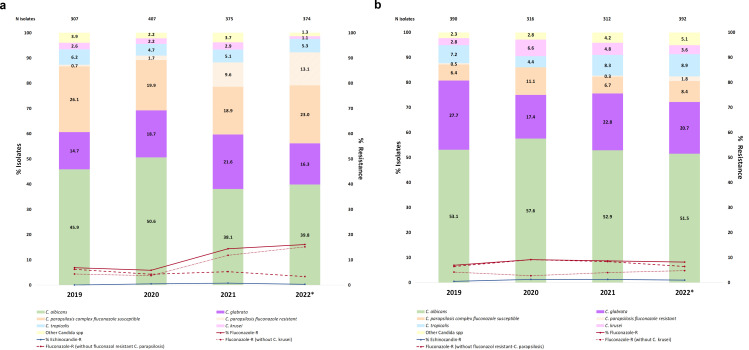
We collected 766 Candida sp. isolates (n = 686 patients) from blood cultures (n = 374, 48.8%) and intra-abdominal samples [n = 392, 51.2%; peritoneal fluid (n = 175, 44.6%), liver samples, (n = 98, 25.0%), peritoneal abscess (n = 88, 22.5%), abdominal drainage (n = 22, 5.6%), spleen (n = 6, 1.5%), abdominal wound exudate (n = 2, 0.5%), and peritoneal biopsy (n = 1, 0.3%)]. Most patients yielded one isolate each (n = 622), but 9.3% of patients (n = 64) yielded ≥2 isolates. Species distribution is shown in Fig. 1. We found that proportions of C. albicans, C. krusei, and other Candida spp. were lower in blood cultures than in intra-abdominal samples (39.8% versus 51.4%, 1.1% versus 3.6%, and 1.3% versus 5.1%, respectively; P < 0.05), whereas C. parapsilosis complex proportions were higher in blood cultures than in intra-abdominal samples (36.1% versus 10.2%; P < 0.05). We did not detect C. auris.
Fig 1.
Species distributions and resistance rates per year in Candida sp. isolates from (a) blood cultures and (b) intra-abdominal samples. *Isolates collected in 2022. Blood cultures: C. parapsilosis complex (C. parapsilosis sensu stricto, n = 134; and C. orthopsilosis, n = 1); Candida spp. (C. lusitaniae, n = 3; C. guilliermondii, n = 1; and C. pararugosa, n = 1); non-Candida (Saccharomyces cerevisiae, n = 2; Rhodotorula mucilaginosa, n = 2; and Cryptococcus neoformans, n = 1), which represented 1.3% of blood isolates. Intra-abdominal samples: C. parapsilosis complex (C. parapsilosis sensu stricto, n = 39; and C. orthopsilosis, n = 1); Candida spp. (C. lusitaniae, n = 11; C. dubliniensis, n = 4; C. guilliermondii, n = 3; C. pararugosa, n = 1; and C. inconspicua, n = 1); non-Candida (Pichia manshurica, n = 1; Wickerhamomyces onychis, n = 1), which represented 0.5% of intra-abdominal isolates. Non-Candida isolates were excluded from the analysis. Adapted from a previously reported publication (12); reprinted with the journal’s permission (license number: 5623520933072); data from 2022 are here newly reported.
No resistance to amphotericin B was found. Overall, echinocandin and fluconazole resistance rates in Candida were 0.7% (n = 5/766) and 12.0% (n = 92/766) of isolates and 0.7% (n = 5/686) and 13.1% (n = 90/686) of patients, respectively. Resistant isolates came from 12/16 hospitals, and no cross-resistance between azoles and echinocandins was found.
A total of 0.3% (n = 1/374; C. glabrata, P633T FKS1 HS1) of blood culture isolates and 1.0% [n = 4/392; C. glabrata (FKS sequence wild type, F659S FKS2 HS1, D666E FKS2 HS1, and D666G FKS2 HS1)] of intra-abdominal isolates were echinocandin resistant (Fig. 1A and B). Per-patient echinocandin resistance rates were 0.3% and 1.5% in blood cultures and intra-abdominal samples (P > 0.05), respectively. Ibrexafungerp showed in vitro activity against the isolates tested (Tables S1 and S2), as we only found three ibrexafungerp non-wild-type isolates from intra-abdominal samples: two C. albicans isolates (echinocandin susceptible and harboring FKS wild-type sequence) and one C. glabrata isolate (echinocandin resistant and harboring an F659S FKS2 HS1 substitution).
A total of 16.0% (n = 60/374) of blood culture isolates were fluconazole resistant (C. parapsilosis, n = 49; C. glabrata, n = 6; C. krusei, n = 4; and C. pararugosa, n = 1). Excluding C. krusei isolates from the analysis, fluconazole resistance rate was 15.1% (Fig. 1A). Likewise, a total of 8.2% (n = 32/392) of isolates from intra-abdominal samples were fluconazole resistant (C. krusei, n = 14; C. parapsilosis, n = 7; C. glabrata, n = 5; C. albicans, n = 1; C. lusitaniae, n = 2; C. guilliermondii, n = 1; C. inconspicua, n = 1; C. tropicalis, n = 1); excluding C. krusei isolates from the analysis, fluconazole resistance rate decreased to 4.8% (Fig. 1B). Per-patient fluconazole resistance rates were 16.5% and 9.5% in blood cultures and intra-abdominal samples (P < 0.05) and 15.6% and 5.5% excluding C. krusei isolates (P < 0.05), respectively. Isolates from blood/intra-abdominal samples that were non-wild type to voriconazole (15.0%/2.8%), posaconazole (2.4%/1.5%), or isavuconazole (5.6%/1.8%) were also fluconazole non-wild type (Tables S1 and S2), except for an isavuconazole-non-wild-type but fluconazole-susceptible C. glabrata isolate from blood.
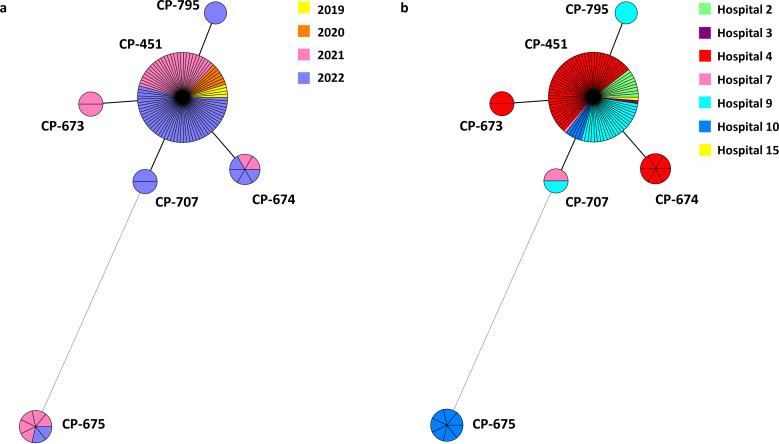
The fluconazole-resistant C. parapsilosis isolates (n = 56) were sourced from 54 patients (two patients simultaneously harbored isolates from blood cultures and intra-abdominal samples) cared for at six hospitals. Isolates were detected in four of the five hospitals previously reported and were newly reported in Hospital 3 and Hospital 7. Most resistant isolates harbored the Y132F-R398I ERG11p substitutions (n = 53) and were grouped into four clonally related genotypes [CP-451 (n = 46 isolates from n = 44 patients cared for at six hospitals), CP-674 (n = 4 isolates from one patient each, all of them cared for at a single hospital), CP-707 (n = 2 isolates from one patient each from two hospitals), and CP-795 (n = 1)]. The remaining resistant isolates harbored the G458S ERG11p substitution (n = 3 isolates from one patient each) and the CP-675 genotype and were exclusively found in one hospital (Fig. 2). We found the V437I ERG11 gene substitution in a C. albicans isolate.
Fig 2.
Minimum spanning tree showing fluconazole-resistant C. parapsilosis genotypes found during the study period (2019–2022) per year of detection (A) or per hospital (B) Circles represent different genotypes, and circle size, the number of isolates belonging to the same genotype. Connecting lines between the circles show profile similarities. The solid bold line indicates differences in only one marker, and the dotted line indicates differences in four or more markers. Genotypes CP-673, CP-674, CP-707, and CP-795 differ from CP-451 at microsatellite markers B, CP6, CP4a, and CP6, respectively.
Trends in species epidemiology over time
Since 2020, in blood cultures, we observed increasing proportions of C. parapsilosis (2020 = 21.6% < 2021 = 28.5% < 2022 = 36.1%; P < 0.05) and decreasing proportions of C. albicans (2020 = 50.6% > 2021 = 38.1%; P < 0.05). We detected an increase in C. glabrata from 2019 to 2021 (2019 = 14.7% > 2021 = 21.6%; P < 0.05) and a decrease in other Candida spp. in 2022 (1.3%) compared with 2019 (3.9%) and 2021 (3.7%) (P < 0.05) (Fig. 1). Few differences were found in species distributions in intra-abdominal samples over time; we found decreasing proportions of C. glabrata (2019 = 27.7% > 2020 = 17.4%; P < 0.05) and increasing proportions of C. krusei (2019 = 2.8% > 2020 = 6.6%; P < 0.05) from 2019 to 2020. We also observed an increase in other Candida spp. between 2019 and 2022 (2019 = 2.3% > 2022 = 5.1%; P < 0.05).
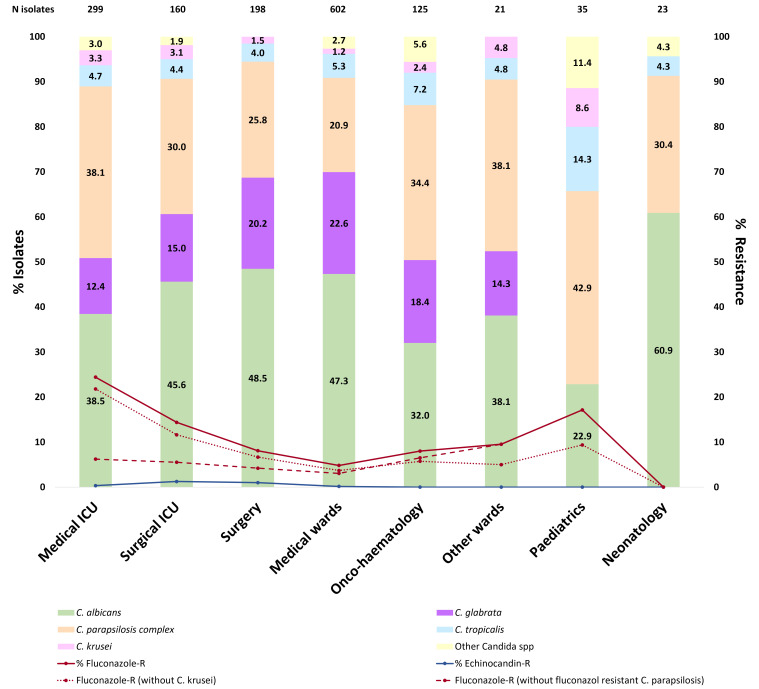
Species distributions are hospital dependent (Fig. S1). We studied the epidemiology of Candida spp. from blood cultures per hospital ward. C. albicans accounted for 22.9% to 60.9% of isolates, being the dominant species in most wards, except for the following wards: pediatrics, oncology-hematology, and other wards. C. parapsilosis complex accounted for 20.9% to 42.9% of cases, being particularly frequent in pediatric wards. C. glabrata was frequent in medical (22.6%) and surgical wards (20.2%) but undetected in pediatrics and neonatology. C. tropicalis proportions were particularly high in pediatrics (14.3%) and accounted for 4.0%–7.2% in other areas. Finally, C. krusei and other Candida spp. were especially frequent in pediatrics (8.6% and 11.4%, respectively) (Fig. 3). Species distribution in intra-abdominal samples was more homogeneous, and isolates mainly came from surgical and medical wards. C. albicans was the dominant species in all hospital wards followed by C. glabrata, except for pediatrics and neonatology (data not shown).
Fig 3.
Species distributions and resistance rates of Candida spp. isolates from blood cultures per ward of admission during the 2019–2022 period.
Trends in antifungal resistance rates over time
Overall, echinocandin resistance rates were low and steady, regardless of the sample type (Fig. 1). Fluconazole resistance rates in blood cultures are shown in Fig. 1A. Resistance to fluconazole increased from 6.8% in 2019 to 16% in 2022 and was mainly impacted by fluconazole-resistant C. parapsilosis, whose rates of fluconazole resistance have been on the rise over the years (2019 = 2.6% < 2020 = 8.0% < 2021 = 34.0% < 2022 = 36.6%; P < 0.05). In fact, the exclusion of fluconazole-resistant C. parapsilosis resulted in significantly lower resistance rates (2019 = 6.2%; 2020 = 4.3%; 2021 = 5.3%; 2022 = 3.4%; P < 0.05). Such an impact was not observed when excluding C. krusei (2019 = 4.3%; 2020 = 3.8%; 2021 = 11.8%; 2022 = 15.1%; P > 0.05) (Fig. 1A). Resistance rates are hospital dependent, as shown by the increasing rate of C. parapsilosis from 2020 to 2021 in Hospitals 2, 4, 9, and 10 (Table 1 and Fig. S1). In addition to the presence of fluconazole-resistant C. parapsilosis isolates in two newly affected hospitals (Hospitals 3 and 7) in 2022, the most relevant observation was the dramatic increase in the number of such isolates in Hospital 4, where resistance rates in blood isolates significantly increased between 2020 and 2022 (Table 1; P < 0.05). In intra-abdominal samples, fluconazole resistance rates did not change over the years and were hugely affected by C. krusei, since its exclusion lowered rates of resistance especially in 2020 and 2021 (P < 0.05) (2019 = 4.2%; 2020 = 2.7%; 2021 = 4.0%; 2022 = 4.8%) (Fig. 1B). In contrast, an increase in fluconazole-resistant C. parapsilosis isolates was detected in intra-abdominal samples in 2022 in Hospital 9 (Fig. S2).
TABLE 1.
Per-year C. parapsilosis fluconazole resistance rates at the seven hospitals affected c
| Fluconazole-resistant C. parapsilosis, N (%) | ||||
|---|---|---|---|---|
| Hospital | 2019 | 2020 | 2021 | 2022 |
| 2 | 0 (0) | 1 (16.7) | 1 (7.7) | 7 (33.3) |
| 3 | 0 (0) | 0 (0) | 0 (0) | 1 (4.3) |
| 4 | 0 (0) | 3 (17.6) | 20 (66.7) | 30 (76.9) |
| 7 | 0 (0) | 0 (0) | 0 (0) | 2 (9.5) |
| 9 | 4 (36.4) | 3 (27.3) | 10 (55.6) | 8 (36.4) |
| 10 | 0 (0) | 0 (0) | 5 (71.4) a | 8 (88.9) b |
| 15 | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) |
All resistant isolates harbored the G458S ERG11p substitution.
A total of 55.6% and 33.3% of resistant isolates harbored the Y132F and G458S ERG11p substitutions, respectively.
Numbers in bold indicate differences reaching statistical significance (2020 versus 2021 and 2020 versus 2022; P < 0.05).
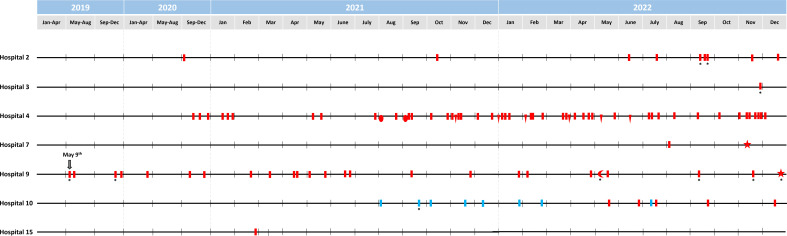
The timeline of the fluconazole-resistant C. parapsilosis isolates detected over the 4-year period is shown in Fig. 4. Resistant C. parapsilosis isolates were found in seven hospitals (Table 1) and were mainly driven by the presence of genotype CP-451, which has been constantly spreading across hospitals over the years; from time to time and to a lesser extent, clonally related fluconazole-resistant genotypes were also detected (Fig. 2 and 4).
Fig 4.
Timeline of the detection of fluconazole-resistant C. parapsilosis isolates at each affected hospital over the 4-year study period. Colored symbols refer to fluconazole-resistant C. parapsilosis isolates from blood cultures, an asterisk represents isolates from intra-abdominal samples. Red indicates isolates within the clonal complex (CP-451, bar; CP-673, oval; CP-674, triangle; CP-707, star; and CP-795, moon) of genotypes harboring the Y132F-R398I ERG11p substitution. Blue indicates isolates within the CP-675 genotype harboring the G458S substitution. Arrow indicates the first time resistant isolates were detected. Adapted from a previously reported figure (11); reprinted with the journal’s permission (order license ID: 1397047–1); data from 2022 are here newly reported.
DISCUSSION
Our study demonstrates rampant clonal spreading of C. parapsilosis fluconazole-resistant genotypes in Madrid, a spread that gained traction at some hospitals in 2022. C. auris remains undetected in blood and intra-abdominal samples in Madrid’s hospitals.
The species distribution in blood isolates per ward of admission was as expected. Whereas C. albicans was usually the most common species, C. parapsilosis was frequent in intensive care unit (ICU) wards and neonatology (16), C. glabrata was associated with abdominal surgery and elderly patients (17), and C. tropicalis was frequent in patients with hematological malignancies (18). Species distribution in pediatrics was dominated by non-albicans Candida spp.; neither C. glabrata nor C. krusei isolates were found in neonatology (19, 20).
The distribution of Candida species in isolates collected during the study period, in both blood cultures and intra-abdominal samples, was comparable to previous studies (9, 21). C. auris has been detected in some hospitals in the Mediterranean area of Spain (22 – 24); however, it remains undetected in invasive isolates in Madrid. In contrast, since 2019, we have detected an increase in fluconazole-resistant C. parapsilosis isolates from blood cultures along with a decrease in C. albicans proportions, as reported elsewhere (25, 26). This fluconazole-resistant C. parapsilosis increase overtook overall fluconazole resistance rates in blood cultures, mainly in patients cared for in ICU wards. In fact, fluconazole-resistant C. parapsilosis in blood cultures increased 14-fold between 2019 and 2022; by excluding such isolates, fluconazole resistance rates would have been low and steady over the study period in Madrid (27). In a recent study conducted in European countries, Arendrup and collaborators detected a high fluconazole resistance rate (17%) in C. parapsilosis blood isolates. Those isolates sourced from Greece, Turkey, and Italy, countries where the fluconazole resistance rates in C. parapsilosis was up to 37%, a figure that is line with the fluconazole resistance rates reported in Madrid between 2021 and 2022 (26).
The increasing detection of fluconazole-resistant C. parapsilosis in Madrid can be attributed to the presence of genotypes harboring the Y132F ERG11p substitution, which could be more associated with patient-to-patient transmission rather than prior azole exposure (1). The fluconazole-resistant C. parapsilosis isolates detected in Madrid mostly belonged to the dominant CP-451 genotype, accounting for 53.5% of all fluconazole-resistant isolates, which has become endemic in many of the hospitals affected. The CP-451 genotype was detected for the first time in 2019; since then, its presence has gained traction and two hospitals became newly affected in 2022. The fact that fluconazole-resistant C. parapsilosis increased in blood cultures and emerged in intra-abdominal samples in one hospital in 2022 is a matter of concern. Furthermore, the presence of three C. parapsilosis fluconazole-resistant isolates from Hospital 3 (two rectal swabs and one skin catheter) and two isolates from Hospital 10 (a wound exudate and a catheter tip) suggests that it might be the tip of the iceberg (data not shown). In contrast, echinocandin resistance was steadily low throughout the entire study period and was mainly due to C. glabrata isolates from the abdominal cavity.
C. parapsilosis shows intrinsic low susceptibility to echinocandins, thus making the emergence of fluconazole resistance a matter of concern. Ibrexafungerp, a new inhibitor of (1, 3)-β-D-glucan synthase, has partial activity against FKS-mutant C. glabrata isolates (28) and here exhibited potent activity against all isolates tested, including fluconazole-resistant C. parapsilosis. Our in vitro observations open the door to future clinical evaluations of the efficacy of ibrexafungerp for the treatment of invasive infections caused by fluconazole-resistant C. parapsilosis.
This study is subject to limitations. We did not collect colonizing samples from patients or environmental samples. Since we were unaware of the nosocomial infection control policies at each hospital, we could not explain the different patterns of fluconazole-resistant C. parapsilosis spreading. Finally, we did not collect the clinical data of infected patients; such analysis will form part of a future study.
In conclusion, our study demonstrates rampant clonal spreading of C. parapsilosis fluconazole-resistant genotypes in Madrid; this spreading gained traction at some hospitals in 2022. Hospital control measures should urgently be taken to bar further spreading and prevent new hospitals from becoming affected. C. auris remains undetected in blood and intra-abdominal samples in Madrid’s hospitals.
MATERIALS AND METHODS
Study period and isolate selection
We studied Candida spp. isolates from blood cultures and intra-abdominal samples sourcing from patients cared for at 16 hospitals in Madrid, Spain (CANDIMAD study) collected from 1 January 2019 to 31 December 2022. As previously reported, one available incident isolate per species, patient, and compartment (blood culture and/or any intra-abdominal samples) was studied (12).
Species identification and antifungal susceptibility testing
Isolates were molecularly identified (12), and we assessed antifungal susceptibilities to amphotericin B, fluconazole, voriconazole, posaconazole, micafungin, and anidulafungin (Sigma-Aldrich, Madrid, Spain), isavuconazole (Basilea Pharmaceutica, Basel, Switzerland), and ibrexafungerp (Scynexis, Inc., Durham, NC, USA) using the EUCAST E.Def 7.3.2 broth dilution method (12, 29). Isolates were categorized as resistant/non-wild type as previously reported (12). Resistant isolates were re-tested.
Molecular characterization of resistant isolates
The FKS1 and FKS2 genes were sequenced in either echinocandin-resistant or ibrexafungerp-non-wild-type isolates (12). We sequenced the ERG11 gene in fluconazole-resistant C. albicans, C. tropicalis, and C. parapsilosis isolates (12). Fluconazole-resistant C. parapsilosis isolates were genotyped by means of species-specific microsatellite markers (CP1, CP4a, CP6, and B) (11, 13) that were highly discriminatory, since the probability of identity for C. parapsilosis was 1.2 × 106 (30). Two or more isolates were considered genotypically identical when they presented the same alleles with all markers. Clonally related genotypes were those that differed in one microsatellite marker (13).
Data analysis
We here report new data regarding the epidemiology and antifungal resistance rates found in isolates collected in 2022. Comparisons of species epidemiology and antifungal resistance rates over time (from 2019 to 2022) were assessed per isolate, sourcing blood cultures or intra-abdominal samples separately, per patient, and also broken down per ward of patient admission. We calculated fluconazole resistance rates per year and compartment considering overall isolates or excluding fluconazole-resistant C. parapsilosis or C. krusei. We also calculated fluconazole-resistant C. parapsilosis proportions exclusively in affected hospitals. Proportions were compared using Epidat v.4.2 (Consellería de Sanidade, Xunta de Galicia, Spain).
ACKNOWLEDGMENTS
We are grateful to the CANDIMAD study group for their participation in the study and to Helena Kruyer for editing assistance.
This study was supported by grants PI18/01155 and PI19/00074 from the Fondo de Investigación Sanitaria (FIS. Instituto de Salud Carlos III; Plan Nacional de I+D+I 2017-2020). The study was co-funded by the European Regional Development Fund (FEDER) "A way of making Europe." This study was partially funded by Scynexis, Inc., Durham, NC, USA. P.E. (CPII20/00015) is a recipient of a Miguel Servet contract supported by FIS. J.D.G. (FI19/00021) holds a predoctoral grant from the FIS.
J.D.G., investigation, methodology, formal analysis, validation, visualization, writing of the original draft, and review and editing. M.M., L.A., E.R., A.P.A., E.G.G.D.P., F.G.R., M.S.C., C.G.E., I.Q.M., N.D.Z., M.M.A., M.T.D.V., A.S.G., and P.M., resources (samples), writing and review and editing. P.E. and J.G., conceptualization, funding acquisition, project administration, resources, supervision, validation, visualization, writing of the original draft, and review and editing.
This study was partially presented at the 33rd European Congress of Clinical Microbiology and Infectious Diseases (ECCMID; oral presentation MEM0103), Copenhagen, Denmark 2023; and at the XXVI Congress of the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC; Oral presentation 438), Santiago de Compostela, Spain 2023.
Contributor Information
Jesus Guinea, Email: jguineaortega@yahoo.es.
Andreas H. Groll, University Children's Hospital Münster, Münster, Germany
on behalf of the CANDIMAD study group:
Judith Díaz-García, Aina Mesquida, Ana Gómez, Marina Machado, Luis Alcalá, Elena Reigadas, Carlos Sánchez-Carrillo, Patricia Muñoz, Pilar Escribano, Jesús Guinea, Ana Pérez-Ayala, Rosaura Pérez-Muñoz, María del Carmen Vera-González, Elia Gómez-García De La Pedrosa, Fernando González-Romo, Paloma Merino-Amador, María Soledad Cuétara, Aída Sánchez-García, Coral García-Esteban, Oscar Cuevas-Lobato, Guadalupe Bernal, Nelly Daniela Zurita, Ainhoa Gutiérrez-Cobos, María Muñoz-Algarra, Isabel Sánchez-Romero, Inmaculada Quiles-Melero, Florinda San Juan-Delgado, María Teresa Durán-Valle, Yolanda Gil-Romero, and Arturo Manuel Fraile Torres
ETHICS APPROVAL
This study was approved by the Ethics Committee of the Gregorio Marañón Hospital (CEim; study no. MICRO.HGUGM.2019-001).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aac.00986-23.
Additional tables and figures complementary to the main document.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Escribano P, Guinea J. 2022. Fluconazole-resistant Candida parapsilosis: a new emerging threat in the fungi arena. Front. Fungal Biol 3:9. doi: 10.3389/ffunb.2022.1010782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ceballos-Garzon A, Peñuela A, Valderrama-Beltrán S, Vargas-Casanova Y, Ariza B, Parra-Giraldo CM. 2023. Emergence and circulation of azole-resistant C. albicans, C. auris and C. parapsilosis bloodstream isolates carrying Y132F, K143R or T220L Erg11P substitutions in Colombia. Front Cell Infect Microbiol 13:1136217. doi: 10.3389/fcimb.2023.1136217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Presente S, Bonnal C, Normand AC, Gaudonnet Y, Fekkar A, Timsit JF, Kernéis S. 2023. Hospital clonal outbreak of fluconazole-resistant Candida parapsilosis harboring the Y132F ERG11p substitution in a french intensive care unit. Antimicrob Agents Chemother 67:e0113022. doi: 10.1128/aac.01130-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geremia N, Brugnaro P, Solinas M, Scarparo C, Panese S. 2023. Candida auris as an emergent public health problem: a current update on European outbreaks and cases. Healthcare (Basel) 11:425. doi: 10.3390/healthcare11030425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kömeç S, Karabıçak N, Ceylan AN, Gülmez A, Özalp O. 2021. Three Candida auris case reports from Istanbul, Turkey. Mikrobiyol Bul 55:452–460. doi: 10.5578/mb.20219814 [DOI] [PubMed] [Google Scholar]
- 6. Štefánek M, Wenner S, Borges V, Pinto M, Gomes JP, Rodrigues J, Faria I, Pessanha MA, Martins F, Sabino R, Veríssimo C, Nogueira ID, Carvalho PA, Bujdáková H, Jordao L. 2022. Antimicrobial resistance and biofilms underlying catheter-related bloodstream coinfection by Enterobacter cloacae complex and Candida parapsilosis. Antibiotics (Basel) 11:1245. doi: 10.3390/antibiotics11091245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuenca-Estrella M, Rodriguez D, Almirante B, Morgan J, Planes AM, Almela M, Mensa J, Sanchez F, Ayats J, Gimenez M, Salvado M, Warnock DW, Pahissa A, Rodriguez-Tudela JL, Barcelona Candidemia Project Study Group . 2005. In vitro susceptibilities of bloodstream isolates of Candida species to six antifungal agents: results from a population-based active surveillance programme. J Antimicrob Chemother 55:194–199. doi: 10.1093/jac/dkh548 [DOI] [PubMed] [Google Scholar]
- 8. Flórez C, Martín-Mazuelos E, Ruiz M, Cisneros JM, Herrero M, García MV, Márquez M, Porras J, Martín P, Gamero C, Castón JJ, Grupo de Estudio de las Candidemias en Andalucía (Andalusian Study Group for Candidemia) . 2009. In vitro susceptibilities of bloodstream isolates of Candida spp.: results from a multicenter active surveillance program in Andalusia. Enferm Infecc Microbiol Clin 27:518–522. doi: 10.1016/j.eimc.2008.09.013 [DOI] [PubMed] [Google Scholar]
- 9. Guinea J, Zaragoza Ó, Escribano P, Martín-Mazuelos E, Pemán J, Sánchez-Reus F, Cuenca-Estrella M, CANDIPOP Project, GEIH-GEMICOMED (SEIMC), and REIPI . 2014. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob Agents Chemother 58:1529–1537. doi: 10.1128/AAC.02155-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nieto MC, Tellería O, Cisterna R. 2015. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagn Microbiol Infect Dis 81:34–40. doi: 10.1016/j.diagmicrobio.2014.05.021 [DOI] [PubMed] [Google Scholar]
- 11. Díaz-García J, Gómez A, Alcalá L, Reigadas E, Sánchez-Carrillo C, Pérez-Ayala A, Gómez-García de la Pedrosa E, González-Romo F, Merino-Amador P, Cuétara MS, García-Esteban C, Quiles-Melero I, Zurita ND, Muñoz-Algarra M, Sánchez-Romero I, Durán-Valle MT, Sánchez-García A, Alcoceba E, Muñoz P, Escribano P, Guinea J, CANDIMAD study group . 2022. Evidence of fluconazole-resistant Candida parapsilosis genotypes spreading across hospitals located in Madrid, Spain and Harboring the Y132F ERG11p substitution. Antimicrob Agents Chemother 66:e0071022. doi: 10.1128/aac.00710-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Díaz-García J, Gómez A, Machado M, Alcalá L, Reigadas E, Sánchez-Carrillo C, Pérez-Ayala A, Gómez-García De La Pedrosa E, González-Romo F, Cuétara MS, García-Esteban C, Quiles-Melero I, Zurita ND, Muñoz-Algarra M, Durán-Valle MT, Sánchez-García A, Muñoz P, Escribano P, Guinea J, CANDIMAD Study Group . 2022. Blood and intra-abdominal Candida spp. from a multicentre study conducted in Madrid using EUCAST: emergence of fluconazole resistance in Candida parapsilosis, low echinocandin resistance and absence of Candida auris. J Antimicrob Chemother 77:3102–3109. doi: 10.1093/jac/dkac288 [DOI] [PubMed] [Google Scholar]
- 13. Díaz-García J, Gómez A, Machado M, Alcalá L, Reigadas E, Sánchez-Carrillo C, Pérez-Ayala A, de la Pedrosa E-G, González-Romo F, Cuétara MS, García-Esteban C, Quiles-Melero I, Zurita ND, Algarra MM, Durán-Valle MT, Sánchez-García A, Muñoz P, Escribano P, Guinea J, on behalf of the CANDIMAD Study Group . 2022. Candida genotyping of blood culture isolates from patients admitted to 16 hospitals in Madrid: genotype spreading during the COVID-19 pandemic driven by fluconazole-resistant C. parapsilosis. J Fungi (Basel) 8:1228. doi: 10.3390/jof8111228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arastehfar A, Yazdanpanah S, Bakhtiari M, Fang W, Pan W, Mahmoudi S, Pakshir K, Daneshnia F, Boekhout T, Ilkit M, Perlin DS, Zomorodian K, Zand F. 2021. Epidemiology of candidemia in Shiraz, southern Iran: a prospective multicenter study (2016-2018). Med Mycol 59:422–430. doi: 10.1093/mmy/myaa059 [DOI] [PubMed] [Google Scholar]
- 15. Won EJ, Choi MJ, Jeong SH, Kim D, Shin KS, Shin JH, Kim YR, Kim HS, Kim YA, Uh Y, Ryoo N, Park JS, Park KU, Byun SA, Lee GY, Kim SH, Shin J. 2022. Nationwide surveillance of antifungal resistance of Candida bloodstream isolates in south Korean hospitals: two year report from Kor-GLASS. J Fungi (Basel) 8:996. doi: 10.3390/jof8100996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soulountsi V, Schizodimos T, Kotoulas SC. 2021. Deciphering the epidemiology of invasive candidiasis in the intensive care unit: is it possible? Infection 49:1107–1131. doi: 10.1007/s15010-021-01640-7 [DOI] [PubMed] [Google Scholar]
- 17. Glöckner A, Cornely OA. 2015. Candida glabrata-unique features and challenges in the clinical management of invasive infections. Mycoses 58:445–450. doi: 10.1111/myc.12348 [DOI] [PubMed] [Google Scholar]
- 18. Fernández-Ruiz M, Puig-Asensio M, Guinea J, Almirante B, Padilla B, Almela M, Díaz-Martín A, Rodríguez-Baño J, Cuenca-Estrella M, Aguado JM. 2015. Candida tropicalis bloodstream infection: Incidence, risk factors and outcome in a population-based surveillance. J Infect 71:385–394. doi: 10.1016/j.jinf.2015.05.009 [DOI] [PubMed] [Google Scholar]
- 19. Steinbach W 2. 2016. Pediatric invasive candidiasis: epidemiology and diagnosis in children. J Fungi (Basel) 2:5. doi: 10.3390/jof2010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weimer KED, Smith PB, Puia-Dumitrescu M, Aleem S. 2022. Invasive fungal infections in neonates: a review. Pediatr Res 91:404–412. doi: 10.1038/s41390-021-01842-7 [DOI] [PubMed] [Google Scholar]
- 21. Bassetti M, Righi E, Ansaldi F, Merelli M, Scarparo C, Antonelli M, Garnacho-Montero J, Diaz-Martin A, Palacios-Garcia I, Luzzati R, Rosin C, Lagunes L, Rello J, Almirante B, Scotton PG, Baldin G, Dimopoulos G, Nucci M, Munoz P, Vena A, Bouza E, de Egea V, Colombo AL, Tascini C, Menichetti F, Tagliaferri E, Brugnaro P, Sanguinetti M, Mesini A, Sganga G, Viscoli C, Tumbarello M. 2015. A multicenter multinational study of abdominal candidiasis: epidemiology, outcomes and predictors of mortality. Intensive Care Med 41:1601–1610. doi: 10.1007/s00134-015-3866-2 [DOI] [PubMed] [Google Scholar]
- 22. Viñuela-Sandoval L, Falces-Romero I, García-Rodríguez J, Eiros-Bouza JM. 2018. Candidemia and colonization by Candida auris, a diagnostic challenge. Enferm Infecc Microbiol Clin (Engl Ed) 36:253–255. doi: 10.1016/j.eimc.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 23. Ruiz-Gaitán A, Moret AM, Tasias-Pitarch M, Aleixandre-López AI, Martínez-Morel H, Calabuig E, Salavert-Lletí M, Ramírez P, López-Hontangas JL, Hagen F, Meis JF, Mollar-Maseres J, Pemán J. 2018. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses 61:498–505. doi: 10.1111/myc.12781 [DOI] [PubMed] [Google Scholar]
- 24. Mulet Bayona JV, Tormo Palop N, Salvador García C, Herrero Rodríguez P, Abril López de Medrano V, Ferrer Gómez C, Gimeno Cardona C. 2020. Characteristics and management of candidaemia episodes in an established Candida auris outbreak. Antibiotics (Basel) 9:558. doi: 10.3390/antibiotics9090558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mamali V, Siopi M, Charpantidis S, Samonis G, Tsakris A, Vrioni G, on behalf of the Candi-Candi Network . 2022. Increasing incidence and shifting epidemiology of candidemia in Greece: results from the first nationwide 10-year survey. J Fungi (Basel) 8:116. doi: 10.3390/jof8020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arendrup MC, Arikan-Akdagli S, Jørgensen KM, Barac A, Steinmann J, Toscano C, Arsenijevic VA, Sartor A, Lass-Flörl C, Hamprecht A, Matos T, Rogers BRS, Quiles I, Buil J, Özenci V, Krause R, Bassetti M, Loughlin L, Denis B, Grancini A, White PL, Lagrou K, Willinger B, Rautemaa-Richardson R, Hamal P, Ener B, Unalan-Altintop T, Evren E, Hilmioglu-Polat S, Oz Y, Ozyurt OK, Aydin F, Růžička F, Meijer EFJ, Gangneux JP, Lockhart DEA, Khanna N, Logan C, Scharmann U, Desoubeaux G, Roilides E, Talento AF, van Dijk K, Koehler P, Salmanton-García J, Cornely OA, Hoenigl M. 2023. European candidaemia is characterised by notable differential epidemiology and susceptibility pattern: results from the ECMM candida III study. J Infect:S0163-4453. doi: 10.1016/j.jinf.2023.08.001 [DOI] [PubMed] [Google Scholar]
- 27. Díaz-García J, Mesquida A, Sánchez-Carrillo C, Reigadas E, Muñoz P, Escribano P, Guinea J. 2021. Monitoring the epidemiology and antifungal resistance of yeasts causing fungemia in a tertiary care hospital in Madrid, Spain: any relevant changes in the last 13 years? Antimicrob Agents Chemother 65:e01827-20. doi: 10.1128/AAC.01827-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mesquida A, Díaz-García J, Sánchez-Carrillo C, Martín-Rabadán P, Alcalá L, Muñoz P, Escribano P, Guinea J. 2022. ΔF659 and F659S substitutions at the HS1 of FKS2 gene, along with E655A and W715L upstream and downstream substitutions, correlate with high Ibrexafungerp MICs against Candida glabrata. Clin Microbiol Infect 28:1154. doi: 10.1016/j.cmi.2022.04.003 [DOI] [PubMed] [Google Scholar]
- 29. Arendrup MC, Meletiadis J, Mouton JW, Lagrou K, Hamal P, Guinea J. 2020. EUCAST definitive document E.DEF 7.3.2. method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7.3.2_Yeast_testing_definitive_revised_2020.pdf. [DOI] [PubMed]
- 30. Guinea J, Arendrup MC, Cantón R, Cantón E, García-Rodríguez J, Gómez A, de la Pedrosa EGG, Hare RK, Orden B, Sanguinetti M, Pemán J, Posteraro B, Ruiz-Gaitán A, Parisi G, Da Matta DA, Colombo AL, Sánchez-Carrillo C, Reigadas E, Muñoz P, Escribano P. 2020. Genotyping reveals high Clonal diversity and widespread Genotypes of Candida causing Candidemia at distant geographical areas. Front Cell Infect Microbiol 10:166. doi: 10.3389/fcimb.2020.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional tables and figures complementary to the main document.