Abstract
Compared with proteins, DNA and RNA are more difficult languages to interpret because four-letter coded DNA/RNA sequences have less information content than 20-letter coded protein sequences. While BERT (Bidirectional Encoder Representations from Transformers)-like language models have been developed for RNA, they are ineffective at capturing the evolutionary information from homologous sequences because unlike proteins, RNA sequences are less conserved. Here, we have developed an unsupervised multiple sequence alignment-based RNA language model (RNA-MSM) by utilizing homologous sequences from an automatic pipeline, RNAcmap, as it can provide significantly more homologous sequences than manually annotated Rfam. We demonstrate that the resulting unsupervised, two-dimensional attention maps and one-dimensional embeddings from RNA-MSM contain structural information. In fact, they can be directly mapped with high accuracy to 2D base pairing probabilities and 1D solvent accessibilities, respectively. Further fine-tuning led to significantly improved performance on these two downstream tasks compared with existing state-of-the-art techniques including SPOT-RNA2 and RNAsnap2. By comparison, RNA-FM, a BERT-based RNA language model, performs worse than one-hot encoding with its embedding in base pair and solvent-accessible surface area prediction. We anticipate that the pre-trained RNA-MSM model can be fine-tuned on many other tasks related to RNA structure and function.
Graphical Abstract
Graphical Abstract.
Introduction
Three essential biomacromolecules in living organisms are DNA, RNA and proteins, all of which are linear polymers, whose components are denoted by a fixed number of letters of the alphabet (typically 4 for DNA and RNA and 20 for proteins). The sequences of different letter combinations encode their biological functions, very much like meaningful sentences in human language. As a result, language models such as BERT (Bidirectional Encoder Representations from Transformers) (1) and GPT (Generative Pre-trained Transformer) (2,3), originally developed for natural language processing, found their ways into dissecting the sequence–structure–function relationship of biological sequences (4,5).
Most previous efforts have been focused on proteins. Examples are UniRep (6), UDSMProt (7), ESM-1b (8), TAPE (9), ProteinBERT (10) and ProtTrans (11). UniRep (6) applied a recurrent neural network to learn a unified representation of proteins and examined its ability to predict protein stability and mutation effects. UDSMProt (7) developed a universal deep sequence model based on Long Short-Term Memory (LSTM) cells and demonstrated it by using several classification tasks, including enzyme class prediction, Gene Ontology prediction and remote homology detection. ESM-1b (8) is a deep transformer model trained on 250 million protein sequences and tested on secondary structure, tertiary contact and mutation–effect prediction. TAPE (9) systematically evaluates different self-supervised learning methods on protein sequences by utilizing five downstream tasks. ProteinBert (10) combined a bidirectional language modeling task with a Gene Ontology annotation prediction task to pre-train a protein sequence language model. Its performance was evaluated on nine downstream tasks associated with protein structure, post-translational modifications and biophysical attributes. ProtTrans (11) compared six natural language models (i.e. Transformer-XL, XLNet, BERT, Albert, Electra and T5) pre-trained on UniRef (12) and BFD (13) protein sequence databases, and suggested that the embedding of these protein language models have led to them learning some ‘grammar’ from protein sequences.
More recent attention has been placed on DNAs and RNAs. These include Bert-like models for DNA, including DNABert (14) with the downstream prediction task of promoters, splice sites and transcription factor-binding sites, iEnhancer-BERT (15) with a focus on enhancer identification and BERT6mA (16) for predicting N6-methyladenine sites. For RNAs, preMLI (17) trained an rna2vec model (18) to obtain the RNA word vector representation for predicting microRNA–long non-coding RNA (lncRNA) interactions. RNA-FM (19) constructed a BERT-based RNA foundation model on large unannotated RNA sequences with applications in prediction of structural and functional properties.
However, unlike human languages, whose evolution is mostly lost in history, the evolutionary history of biological sequences is well preserved through the appearance of proteins (or DNAs/RNAs) with the same function but different sequences in different species. Such an evolutionary history of sequences, revealed from multiple sequence alignment (MSA), allows an in-depth analysis of sequence conservation and mutational coupling due to structural and functional requirements (20–22). Indeed, employing homologous sequences was one of the key factors for the recent success of AlphaFold2 (13) for highly accurate prediction of protein structures. Similarly, incorporating evolutionary information in a language model (MSA transformer) allows improved performance in protein contact map and secondary structure prediction over a single-sequence-based language model with fewer parameters (23).
To build a similar MSA transformer for RNA, one must first obtain RNA homologs. Searching for RNA homologs, however, is more challenging than searching for protein homologs due to the isosteric nature of RNA base pairs and because sequence identity is much easier to lose for a sequence denoted by fewer letters of the alphabet (4 versus 20). A sequence-based search such as BLAST-N (24) against a nucleotide database often leads to only a few homologs, if any. A more sensitive approach is to search for homologous sequences compatible with a defined secondary structure because secondary structure is more conserved than sequence (25). Currently the best secondary-structure-based approach (26,27) is Infernal (28), which is based on a covariance model. Infernal was employed to build RNA families (Rfam) by manual curation with known or predicted experimental secondary structures (29). However, Rfam updates slowly and only ∼4000 families have been curated so far. Moreover, using experimental secondary structures for alignment in some RNAs prohibits the application of Rfam MSAs to ab initio structure prediction. This problem was solved with the development of RNAcmap (30), which integrates BLAST-N, Infernal and a secondary structure predictor such as RNAfold (31) for a fully automatic homology search. RNAcmap was further improved with additional iteration (32) and a large expansion of the sequence database (33).
One task for examining the usefulness of a language model is base pair prediction. Most previous techniques were built on benchmark databases of secondary structure from comparative analysis, which may not be accurate or complete (34). Examples of these benchmarks are RNA STRAND (35), ArchiveII (36) and bpRNA (37). SPOT-RNA was the first end-to-end prediction of RNA secondary structure and tertiary base pairs by initial learning on bpRNA and transfer learning on 3D structure-derived base pairs (34). Using base pairs derived from RNA 3D structures is necessary because experimentally determined RNA structures are the only gold standard dataset for base pair annotations at a single nucleotide resolution. Moreover, many base pairs such as non-canonial base pairs and pseudoknots are stabilized by tertiary interactions. Although a few deep learning methods have been published since SPOT-RNA [e.g. DMfold (38), MXfold2 (39) and Ufold (40)], SPOT-RNA and its latest improvement with evolutionary information [SPOT-RNA2 (41)] remain the only two methods trained and tested purely with RNA structure-derived base pairs. Another downstreaming task for language model testing is solvent accessibility prediction. Three methods, RNAsnap (42), RNAsol (43) and RNAsnap2 (44), were developed. The first method is based on support vector machines, whereas the latter two are deep learning techniques, all with evolutionary profiles as input.
This work reports an RNA MSA-transformer language model (RNA-MSM) based on homologous sequences generated from RNAcmap3, which has the advantage of using predicted secondary structure for homology search and possesses significantly more homologous sequences than Rfam for training a more expressive language model. RNA-MSM applied one learnable position-embedding layer to encode the row and column information, separately. Our results show that RNA structure information emerges directly in the output attention maps and embeddings. Moreover, further fine-tuning led to significantly improved performance on base pair prediction and solvent accessibility prediction over existing state-of-the-art techniques. in comparison, RNA-FM, a BERT-based RNA language model, performs worse than one-hot encoding with its embedding in base pair and solvent-accessible surface area (ASA) prediction.
Materials and methods
MSA generation and the training set for unsupervised learning
We downloaded 4069 RNA families (version 14.7) from https://rfam.xfam.org on 09/04/2022. The fully automatic RNAcmap3 for homolog search and sequence alignment (33) was employed for these 4069 RNA families by using their covariance models (CMs) for each family. Although the language model is unsupervised learning, we excluded the Rfam families which contain RNA sequences with experimentally determined structures in order to minimize potential overfitting for structural inference. This leads to a total of 3932 Rfam families. The median value for the number of MSA sequences for these families by RNAcmap3 is 2184. All sequences were pre-processed by replacing Ts with Us in RNA sequences and substituting ‘R’, ‘Y’, ‘K’, ‘M’, ‘S’, ‘W’, ‘B’, ‘D’, ‘H’, ‘V’ and ‘N’ with ‘X’, similar to previous work (19). The final vocabulary of our model contains six letters: ‘A’, ‘G’, ‘C’, ‘U’, ‘X’ and ‘–’. This dataset is called TR0 for unsupervised learning.
Training and test sets for downstream models
To perform downstream tasks, we prepared two datasets based on experimentally determined RNA structures, which have the gold standard information for base pairs and solvent accessibility. All RNA-containing structures were initially downloaded from the PDB (45) website on 9 August 2021. They were randomly split into training (TR1, 80%), validation (VL1, 10%) and independent test (TS1, 10%), respectively. Sequences with similarity of >80% in VL1 and TS1 were first removed using CD-HIT-EST (46) and then any sequences in TR1 with similar structures to the sequences in VL1 and TS1 [TM-score > 0.45, according to RNA-align (47)] were also removed. Similarly, any sequences in VL1 having similar structures to sequences in TS1 were also removed. Finally, we obtained 405 RNAs for TR1, 40 RNAs for VL1 and 39 RNAs for TS1. We updated the test set by downloading RNA structures deposited between 9 August 2021 and 14 July 2022 from the PDB website. We removed sequences by using CD-HIT-EST (80%) and RNA-align (TM-score of 0.45) against TR1, VL1 and TS1. This led to 31 RNA tertiary structures for TS2. The base pairs of all datasets (TR1, VL1, TS1 and TS2) were derived from their respective tertiary structures by using DSSR software (48). We combined TS1 and TS2 to make the final independent test set with 70 RNAs (TS). It should be noted that the training set of RNA-MSM has been filtered by excluding the Rfam families which contains RNA sequences with experimentally determined structures. Therefore, the RNAs used in downstream models would have no overlap with those used for training RNA-MSM. Moreover, we further prepared a HardTS set by testing our method on unseen structures based on a TM-score threshold of 0.3, against our training and validation sets. It includes nine newly solved RNA structures (deposited between 14 July 2022 and 1 June 2023 from the PDB website) and 21 RNA structures contained in the TS set.
Network architecture
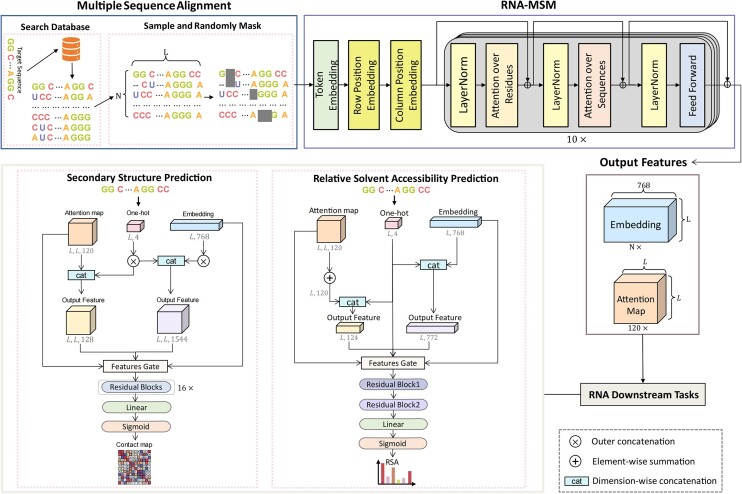
As shown in Figure 1, RNA-MSM mainly consists of two modules: embedding and MSA transformer, as in the protein MSA transformer work (23). The embedding module consists of one initial embedding layer and two learnable position-embedding layers (Figure 1). The position-embedding layers encode rows (number of entries in the MSA) and columns (sequence length) of the MSA separately. A 1D sequence position embedding is applied to the row of the MSA, allowing the model to recognize the sequential order of nucleotides. In addition, each column of the MSA is embedded with a separate positional embedding, allowing the model to perceive the entire MSA as a series of ordered sequences.
Figure 1.
The network architecture of RNA-MSM with downstream tasks for predicting secondary structure and RNA solvent accessibility for one RNA sequence of length L along with N– 1 homologous sequences. The RNA-MSM model is stacked with 10 transformer blocks, each containing 12 attention heads. The output embedding identifies all sequences in a 768-dimensional space. A total of 120 attentional maps were constructed by stitching the attention scores among the residues learned by the 12 attention heads. The feature gate used in the downstream tasks means a feature combination selection gate.
We employed a similar configuration of the MSA transformer module (Figure 1) to the protein MSA transformer work (23). Briefly, this module is made of a stack of MSA transformer blocks. Each MSA transformer block has a residue and sequence attention layer with 12 attention heads with embedding size of 768, followed by a feedforward layer. LayerNorm is applied either before or after the attention layer. The residue attention layer captures interactions between nucleotides and integrates all the sequence attention maps within the MSA, resulting in a single attention map being shared by all sequences to reduce memory usage. Moreover, sharing one attention map among all input sequences might lead to learning the inherent structural information (23). Through the feedforward layer, the input is passed over fully connected layers and activated using GELU activation functions. Specially, we modified a few specific parameters as follows. The number of blocks was changed from 12 to 10 due to the graphics processing unit (GPU) memory limitation. We also changed the training precision from half-precision to 32-bit precision, which demonstrated an improved performance.
We define an input MSA as a matrix, N× L where N represents the number of entries in the MSA and L represents the sequence length. After the embedding module, it was embedded into a tensor N× L × 768 and fed into the MSA transformer module. The final output contains two features: an embedding of N×L × 768 that represents the last MSA transformer block's output and an attention map L×L × 120 derived from all residue attention layers where 120 was from multiplication of the number of attention heads by the number of MSA transformer blocks (10).
RNA-MSM model training and inference
RNA-MSM was trained with a probability of 0.1 dropout after each layer. A total of 300 epochs were trained using eight 32G GTX V100 GPUs with Adam optimizer set to 0.0003, warmup step set to 16 000, weight decay set to 0.0003, batch size set to 1 and learning rate set to 0.0003. The training stage stopped when the F1 value on validation set VL1 does not increase in 10 consecutive epochs. For each input MSA, 1024 RNAs are randomly selected in addition to the query sequence if the number of sequences of input MSA is larger than 1024. It should be noted that the representative sequence is picked every time.
The maximum number of tokens is set to 16 384 because of the memory limitation of 32G of each V100 GPU. We employed the BERT masked language modeling objective function for training. Briefly, 20% tokens of the input MSA were substituted by a special token [MASK] at random. Of this 20%, 10% will be replaced by other words at random, 10% will be changed back to the original words and 80% will remain as [MASK]. The model was trained to predict the original tokens of the masked ones based on those tokens that are not masked in the sequence. During the inference, we employ the hhfilter (49) to sample the RNA sequences in the maximum diversity manner, as the number of RNA sequences in some MSAs is huge. We experimented with the number of sampled sequences from all MSAs and found that sampling 512 sequences is a good balance of performance and computational cost.
RNA secondary structure prediction
To establish a downstream task for RNA-MSM, we employed a simple ResNet (50) similar to that utilized in Singh et al. (34). As shown in Supplementary Figure S1, the architecture of the model consists of 16 residual blocks followed by a fully connected (FC) block. Each residual block consists of two convolutional layers with a kernel size of 3 × 3 and 5 × 5, and with a filter size of 48. We tested one-hot encoding, the embeddings as well as the attention map obtained from RNA-MSM as the input for the ResNet16 model. These input embeddings were converted into a 3D tensor by the outer concatenation function as described in RaptorX-Contact (51). The model was implemented in the Pytorch framework and uses the Nvidia GPU to speed up training. The model trained by using the Adam optimization function with a learning rate of 0.001, and the cross entropy was used as loss function. We trained the model on TR1 and the trained model was chosen based on the performance on VL1.
RNA solvent accessibility prediction
The ASA reflects the extent that a nucleotide in an RNA chain is exposed to solvents or other functional biomolecules. Unlike secondary structure dominated by local interactions, solvent accessibility is a measure of 3D structure in one dimension. The ASA labels of TR1, VL1 and TS are calculated from their individual 3D chain structures (instead of protein–RNA complex structures) by the POPS package (52) with a probe radius of 1.4 Å. All ASAs were normalized to relative accessible surface areas (RSAs). To be specific, we divided the ASA values by the maximum ASA of each corresponding nucleotide (i.e. A, G = 400 Å2, U, C = 350 Å2) as mentioned in Yang et al. (42).
We employed a simple network based on ResNet architecture to construct the RNA solvent accessibility prediction model. As shown in Supplementary Figure S2, this network mainly contains two blocks. The first block contains two convolutional layers (53) to capture high-level feature maps and one standard ‘Squeeze-and-Excitation’ (SE) module (54), which defines model interdependencies between channels. The second block contains one self-attention layer (55) and a simple multilayer perceptron (MLP) layer. The self-attention layer is a core module in transformer architectures (55), which jointly attend to different representation subspaces, and the MLP layer fuses all the features and generates the outputs.
The outputs yielded by the model are values ranging from 0 to 1 as predicted relative solvent accessibility (RSA). The predicted ASA values were obtained by converting the RSA values to actual ASAs with the above-mentioned normalization factors. The mean absolute error between the predicted RSA and the actual RSA was used as a loss function. The model was trained on TR1 and validated on VL1. During the training, the learning rate scheduler of ‘Cosine Annealing Warm Restarts’ (56) was used to adjust the learning rate. The training iteration stops when the loss on the VL1 no longer decreases.
We determined the hyperparameters (number of repetitions for each type of residual block = 1, width of the network = 64, learning rate = 0.005, batch size = 16, head of self-attention layers = 8) of the network by optimizing the performance on the VL1 dataset using one-hot encoding. After designing the network, we performed a number of experiments to examine whether the language embedding can improve the representations of sequence in ASA prediction. Different inputs for the ASA prediction model, including the same sequence profile as RNAsnap2, one-hot encoding, embeddings and attentions from the language models, were tested.
Results
RNA-MSM model
The MSA transformer (23) originally developed for MSAs of homologous protein sequences was modified for MSAs of RNA sequences. The network architecture of RNA-MSM, along with its downstream tasks, is illustrated in Figure 1. Specifically, the MSA generated from the RNA query sequence by RNAcmap3 was used as an input for the network. We did not employ Rfam MSAs because the median number of sequences in Rfam families is only 45. Instead, we employed representative sequences from 3932 Rfam families, excluding those with experimentally determined structures (see the Materials and methods for more details). The two-dimensional input passes through the embedding layer and the series of axial transformer modules, where the attention operates over both rows and columns of the input. The final outputs are a 1D MSA sequence representation and a 2D residue attention map that were employed as the input for downstream tasks. The model was trained by the masked language modeling objective function (see the Materials and methods for more details).
Direct base pairing information in attention map
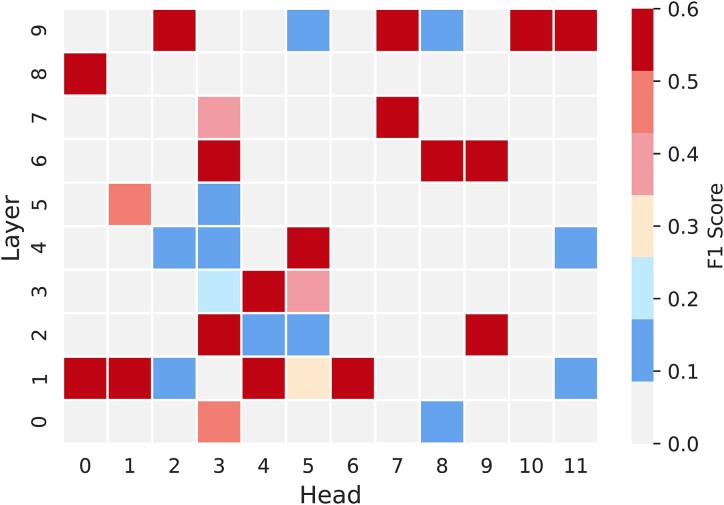
With the self-attention mechanism of the transformer model, pairwise interactions can be established between any positions within the sequence. In principle, multiple attention heads employed in the attention layers of the transformer module should capture a variety of features from the input sequence by focusing on different portions of the input sequence simultaneously as demonstrated for proteins (57). Here, we examined if the attention maps derived from the RNA-MSA transformer module can serve as a base pairing probability map in the RNA sequence, using an independent TS test set.
Figure 2 shows that some attention maps can be directly employed for predicting secondary structure with reasonable accuracy (the average harmonic mean of precision and recall, F1 score, all larger than 0.5). Furthermore, top-K attention maps were selected by F1 scores and combined to predict the RNA secondary structure even more accurately. For example, Top-2 attention maps lead to an impressive performance with F1 score at 0.563 and the Matthews correlation coefficient (MCC) at 0.562 (Supplementary Table S1). These unsupervised results provided the evidence that some structural information was captured by some layers in the attention maps.
Figure 2.
The normalized F1 scores of 120 attention maps from different layers and attention heads of RNA-MSM as input of RNA secondary structure prediction on the independent TS test set.
RNA secondary structure prediction
Model training
To further improve the secondary structure prediction beyond unsupervised learning, secondary structure prediction models were trained, validated and tested on the secondary structure dataset derived from known 3D RNA structures. Briefly, the validation and test sets were structurally different (TM-score < 0.45) from each other and from the training set. In addition, the sequences in the test and validation sets were refined with sequence identity of < 80% within each set. The training (TR1), validation (VL1) and test (TS) sets have 405, 40 and 70 RNAs, respectively. The secondary structure prediction model was designed based on the ResNet model as shown in Supplementary Figure S1 (details can be found in the Materials and methods).
Feature comparison
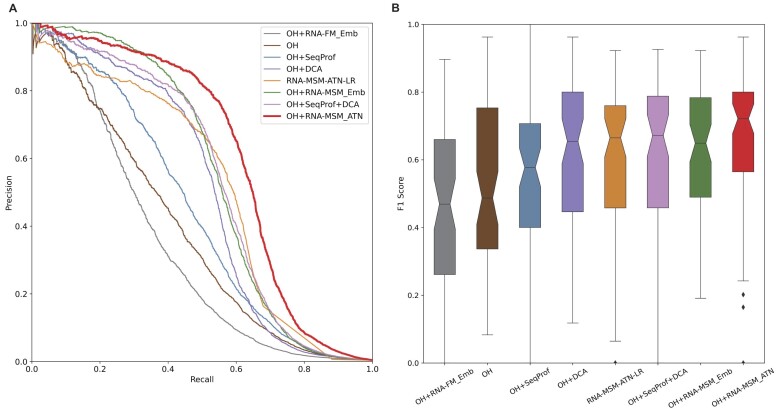
To illustrate the usefulness of the features from the language models, we compared it with various previously employed features for secondary structure prediction. These features include one-hot encoding (OH), contact maps from direct coupling analysis (DCA) by gremlin (58), sequence profiles from MSA (SeqProf) (28), embedding from the RNA foundation model (RNA-FM_Emb) (19), the logistic regression of the attention map (RNA-MSM-ATN-LR), embedding (RNA-MSM_Emb) and attention map (RNA-MSM_ATN) from our RNA-MSM model. Some feature combinations were also examined. Figure 3A compares precision–recall curves of different input feature by using the same network on the independent test set TS. The precision–recall curves were drawn by moving the threshold value for defining base pairs according to the probability output from the RNA-MSM model and plotted by putting all RNAs together as a single set for examining the performance of various methods. OH can be considered as the baseline model for all features compared. It is a bit surprising that the RNA-FM_Emb alone or RNA-FM_Emb combined with OH was even worse than OH alone when employed as the features for secondary structure prediction. The AUC_PR values for the TS set are 0.340 for RNA-FM_Emb, 0.334 for OH + RNA-FM_Emb and 0.377 for OH, respectively (Supplementary Table S2). In comparison, both sequence profile (SeqProf) and DCA provide a boost in model performance when combined with OH, with AUC_PR values at 0.377 for OH, 0.438 (OH + SeqProf), 0.501 for OH + DCA and 0.535 for OH + SeqProf + DCA, respectively (Figure 3A; Supplementary Table S2). The same is true for the features from the current model (RNA-MSM_Emb and RNA-MSM_ATN). Combining them with OH leads to AUC_PR values of 0.547 and 0.610, respectively; i.e. combining OH with the attention map from RNA-MSM yields the best model for secondary structure prediction. Interestingly, the simple logistic regression training on attention maps can achieve AUC_PR at 0.516, only worse than the models trained with attention maps (RNA-MSM_ATN) or embedding (RNA-MSA_Emb). Model performance can also be evaluated according to F1 score and MCC (1 for perfect prediction and 0 for random prediction). The overall trends are the same (Supplementary Table S2). Figure 3B further shows the distributions of F1 score for each predicted RNA secondary structure by different features. The thresholds for these F1 scores were generated for the best F1 scores on the validation set. The model based on OH + RNA-MSM_ATN not only has the best average F1 score, but also has the narrowest distribution, with few poor predictions. This indicates that the model based on OH + RNA-MSM_ATN has the best performance for the majority of RNAs.
Figure 3.
Performance comparison on the test set (TS) given various features or feature combinations according to the precision–recall curve (A) and the distribution of F1 scores (B) for each RNA given by various features all trained with the same model network (Supplementary Figure S1). The features shown here are one-hot encoding (OH), sequence profiles generated by multiple sequence alignment (SeqProf), direct coupling analysis of covariation by gremlin (DCA), the embedding of RNA foundation model (RNA-FM_Emb), the logistic regression of the attention map (RNA-MSM-ATN-LR), and the embedding and attention map from this work (RNA-MSM_Emb and RNA-MSM_ATN). The best model is the combination OH + RNA-MSM_ATN based on either area under the precision–recall curve (A) or F1 score (B).
Comparison with traditional methods
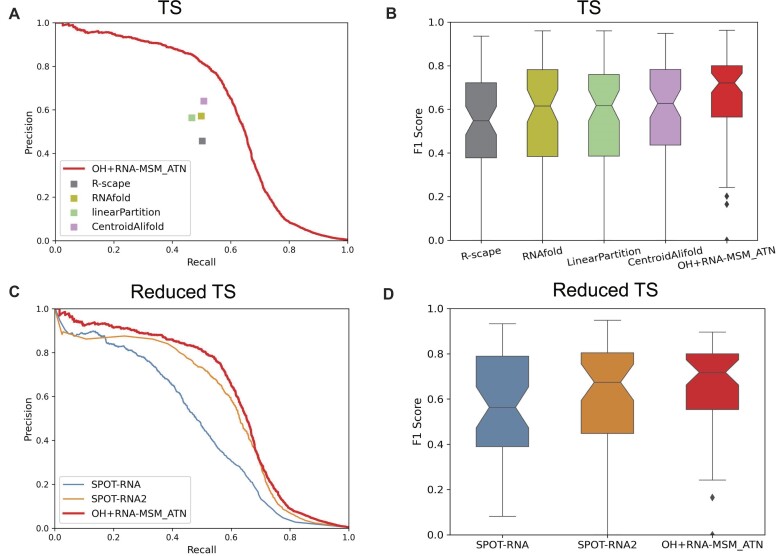
We compared our best secondary structure predictor (OH + RNA-MSM_ATN) with traditional folding-based single [RNAfold (59) and linearPartitition (60)] and multi-sequence-based [CentroidAlifold (61)] techniques in Figure 4A and B and Supplementary Table S2. Based on the same MSAs generated by RNAcmap3, the alignment-based CentroidAlifold with the CONTRAfold inference engine (IE) and a gamma value of 16 were employed for its best performance as before (41). According to the precision–recall curves, OH + RNA-MSM_ATN has the best performance in AUC_PR (Figure 4A) and in the distribution of F1 scores (Figure 4B) for the TS set, as well as the highest F1 score and MCC value (Supplementary Table S2).
Figure 4.
(A) Comparison of precision–recall curves given by OH + RNA-MSM_ATN, R-scape, RNAfold, LinearPartition and CentroidAlifold on the TS test set. (B) Comparison of F1 score distributions given by OH + RNA-MSM_ATN, R-scape, RNAfold, linearPartition and CentroidAlifold. (C) Comparison of precision–recall curves given by OH + RNA-MSM_ATN, SPOT-RNA and SPOT-RNA2 using a reduced TS set after excluding similar structures to the training set in SPOT-RNA/SPOT-RNA2. (D) Comparison of F1 score distributions given by OH + RNA-MSM_ATN, SPOT-RNA and SPOT-RNA2.
Comparison with SPOT-RNA and SPOT-RNA2
Figure 4C compares the best secondary structure predictor (OH + RNA-MSM_ATN) with two other RNA structure-trained secondary structure predictors (SPOT-RNA and SPOT-RNA2) using a reduced test set [48 RNAs, after removing those RNA structures similar (TM-score > 0.45) to the training set and validate set employed in SPOT-RNA and SPOT-RNA2]. According to the precision–recall curves, SPOT-RNA2 has the second best performance, with AUC_PR at 0.56, among the existing methods compared (Supplementary Table S2), compared with 0.6 by OH + RNA-MSM_ATN for the reduced set. The conclusion is also true for the distribution of F1 scores (Figure 4D). Supplementary Table S2 provides a detailed comparison for AUC_PR, F1 scores and MCC. Here, we did not attempt to compare with other deep-learning techniques because they were not trained by 3D structure-derived base pairs.
The factors controlled the performance
The wide distribution of F1 score shown in Figures 3 and 4 indicates that the model OH + RNA-MSM_ATN performs well on some RNAs but not others. To provide a better understanding, we examined the dependence of F1 scores on the sequence length (Supplementary Figure S3A) and on the performance of RNAfold (Supplementary Figure S3B), because RNAfold was employed as the initial secondary structure for homologous sequence search in RNAcmap3. We did not find any significant correlation between the F1 scores and sequence lengths. However, there is a strong correlation between F1 scores given by OH + RNA-MSM_ATN and that given by RNAfold, with Pearson's correlation coefficient (PCC) of 0.774. Despite the influence from RNAfold prediction, most F1 scores given by OH + RNA-MSM_ATN are an improvement over those from RNAfold (Supplementary Figure S3B), except for those that already achieved a high performance (F1 score > 0.7 by RNAfold).
Base pairs due to tertiary interactions
Supplementary Table S3 further examines the performance of different methods on canonical and non-canonical base pairs according to F1 score, precision and recall. It shows that the method based on the MSM language model remains the best for both canonical and non-canonical base pairs. Although predicting non-canonical base pairs is challenging, 82% improvement in F1 score over one-hot encoding is observed. When compared with SPOT-RNA and SPOT-RNA2 for the reduced independent test set, the model OH + RNA-MSM_ATN improves over SPOT-RNA by 28% and over SPOT-RNA2 by 7% in F1 score for canonical base pairs and over SPOT-RNA by 136% and SPOT-RNA2 by 4% in F1 score for non-canonical base pairs. Supplementary Table S4 further compares the performance on pseudoknots, lone base pairs and triplets. Interestingly, OH + RNA-MSM-ATN is the best for lone base pairs but OH + SeqProf + DCA (F1 score = 0.278) or SPOT-RNA (F1 score = 0.274 for the reduced TS set) is the best for pseudoknots, whereas F1 score = 0.243 for OH + RNA-MSM_ATN (F1 score = 0.176 for the reduced TS set).
Ensemble model
An ensemble of four models (OH + SeqProf + DCA, OH + RNA-MSM_Emb, RNA-MSM_ATN and OH + RNA-MSM_ATN) can improve the performance over OH + RNA-MSM_ATN by another 2% in terms of F1 score. The improvement for canonical, non-canonical and pseudoknot base pairs is shown in Supplementary Table S5. The largest improvement is on pseudoknot base pairs, which now has the best performance even on the reduced TS set, when compared with SPOT-RNA and SPOT-RNA2 (Supplementary Table S4).
Test on the HardTS set
To further ensure the generalizable performance, we tested the downstream base pair prediction model using the HardTS set after excluding those RNAs having a TM-score > 0.3 with those RNAs in the training set and validate set. RNA-MSM continues to have the best performance when compared with SPOT-RNA, SPOT-RNA2, R-scape, RNAfold, LinearPartition and CentroidAlifold, as shown in Supplementary Figure S4 and Supplementary Table S6. The results confirm the generalizability of the model in predicting unseen base pairing structures.
Direct solvent accessibility information in embedding
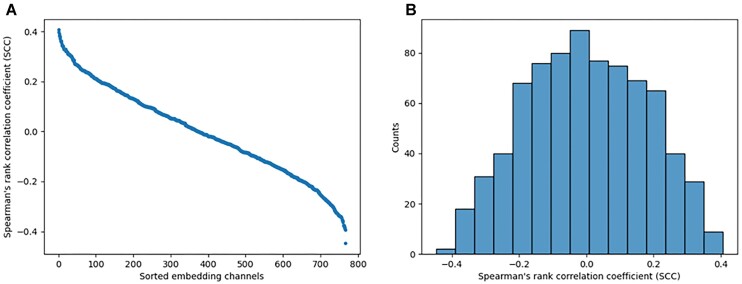
Given the fact that unsupervised attention maps contain base pairing information (Figure 2), it is of interest to examine if the RNA-MSM model can capture solvent accessibility directly from the original MSA without supervised training. To answer this question, we employed the original embeddings of RNA-MSM for this analysis. The embedding represents the MSA of a target sequence as L × 768 matrices where L is denoted as the length of RNA chain sequence. Each nucleotide is represented by a vector of 768 channels. Here we examined the correlation between weights in channels with relative solvent accessibility (RSA) based on Spearman's rank correlation coefficient (SCC).
Figure 5A shows the sorted mean SCC values for the TS set for different embedding channels. The result shows that the highest positive SCC score is 0.407 and the lowest SCC score is –0.448. The distribution of correlation coefficients shown in Figure 5B further reveals that there are quite a few channels with weak correlations to RSA, indicating that some embedding channels did capture the structural information of RNAs without training.
Figure 5.
(A) The mean Spearman correlation coefficients (SCCs, y-axis) on the test set of 768 embedding channels (x-axis) from the last layers of RNA-MSM, sorted according to the SCC values. (B) The distribution of SCC values.
RNA solvent accessibility prediction
Model training
The training, validation and test sets for secondary structure predictions were employed for solvent accessibility prediction. The metrics for the performance of different predictors are the same as those used in RNAsnap2 (44), including the PCC of individual chains between predicted and actual RSA values and mean absolute error (MAE) of individual chains between predicted and actual ASA values. Nucleotides that are missing their neighbors in sequence were excluded during evaluation.
Comparison with previous methods
Supplementary Table S7 summarizes the performance comparison among previous work and different feature combinations. OH alone yields a reasonable performance with PCC = 0.387 and MAE of 31.85 on TS. Adding the sequence profile generated from MSA further improves the performance. Surprisingly, the embedding from a previous language model RNA-FM performed even worse than OH. This is also true for the attention map from RNA-MSM. RNA-MSM_ATN alone as a feature does not do as well as OH, but its combination with OH is useful for going beyond OH. The best performing method is based on the RNA-MSM embedding (RNA-MSM_Emb). Its combination with OH yields the lowest MAE and the highest PCC. RNA-MSM_Emb has a slightly lower PCC value and worse MAE than OH + RNA-MSM_Emb. Thus, we will choose OH + RNA-MSM_Emb as our final model for ASA prediction. The performance of the OH + RNA-MSM embedding alone is a 7% improvement in PCC and 3% in MAE over the previously developed method RNAsnap2_pro, which is based on sequence profile information. The improvement is statistically significant according to the distribution of PCC and MAE values for individual RNAs (Supplementary Figure S5A, B) and P-values (Supplementary Figure S5C–F). In order to compare our model with RNAsnap2 and M2pred fairly, we refined the test set TS by removing similar structures to the sequences in the training and validating set from RNAsnap2 and M2pred, with TM-score > 0.45 or TM-score > 0.3. Therefore, new test sets TS* and TS** were obtained. On both new test sets, the model OH + RNA-MSM_Emb still had the best performance (Supplementary Table S8). This result confirmed that our model for ASA prediction can be applied to unseen structures.
Ensemble model
We further examined the usefulness of an ensemble model by simply averaging the outputs of individual models (62). We test two strategies for the ensemble. The Ensemble_T model is made of the top three models produced by the training stage according to the PCC on VL1. The Ensemble_F model is made of OH, RNA-MSM_Emb and OH + RNA-MSM_Emb. As shown in Supplementary Table S8, both ensemble models show improved performance on the testing set, with Ensemble_F having a slight edge. We found that Ensemble_F yielded a PCC of 0.447 and an MAE of 31.10 for the TS set, exhibiting 3% improvement in PCC and a 3% improvement in MAE over the best single model OH + RNA_MSM_Emb, separately. Ensemble_T achieved a PCC of 0.449 and an MAE of 31.23 on the TS set, exhibiting 4% and 2% improvement over the model OH + RNA_MSM_Emb, separately. In addition, both models exhibited varying degrees of enhancement in their performance on the TS* and TS** set (Supplementary Table S8). They also have a narrower distribution of PCC and MAE on the TS set than the single model (Supplementary Figure S6).
The factors controlling the performance
The MAE of different RNA chains varies over a wide range when predicting their ASA; this phenomenon may be controlled by some factors. The length of RNA chains may affect the performance of the predictor. In addition, RNAfold was employed to search and generate homologous sequences by RNAcmap3. Thus, we examined the relationship between ASA prediction performance and F1 score of RNAfold. We found that the PCC has a negative correlation with RNA length (Supplementary Figure S7A) but a positive correlation with RNAfold F1 score (Supplementary Figure S7B).
Generalization beyond families trained
One interesting question is whether or not the prediction performance is robust for those families that are not in training or validation sets. Here, we examined the Rfam families in the TS set that were not in both the training set and validation set. There were a total of eight families. Supplementary Table S9 shows the results of RNAs in these eight families (RF01998, RF00080, RF00075, RF00028, RF01786, RF01084, RF00061 and RF01051). For secondary structure prediction, the majority (6/8) have a high prediction accuracy (F1 score > 0.64). Those with a poor secondary structure prediction have a poor RNA-fold prediction (F1 score by RNAfold < 0.26). For ASA, all have MAE values that are lower than the average performance (33.45). This result indicates that the method developed here can be generalized to families outside the training and validation sets.
Visualization
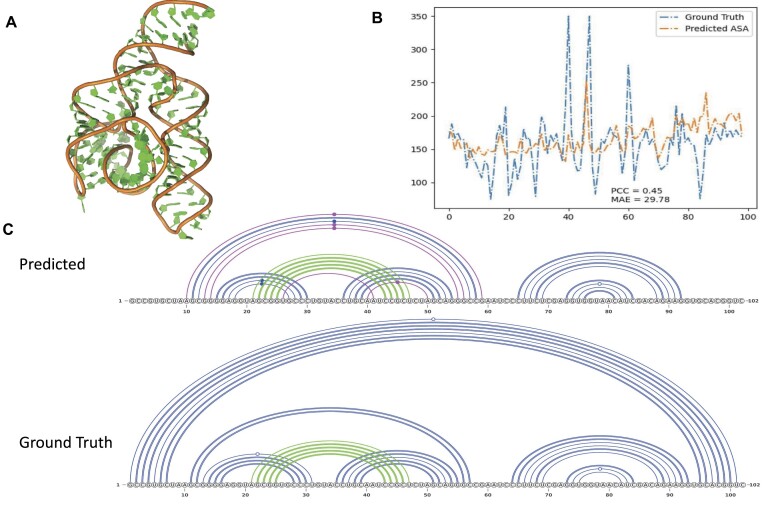
Figure 6 shows one example with the average performance of the secondary structure prediction by model OH + RNA-MSM_ATN (Figure 6C) and ASA prediction by model OH + RNA-MSM_Emb (Figure 6B) for the bacterial SRP Alu domain (PDB 4WFM, Chain A). As shown in Figure 6, this RNA has a well-folded structure with a unique topology. The predicted ASA has a PCC value of 0.454 with MAE = 29.78, whereas the predicted secondary structure captured most inner base pairs, including the pseudoknots. The main missing helix strand is the long-range contacts for |i–j|>87. The F1 score is 0.74.
Figure 6.
An example (chain A in PDB ID 4WFM) for ASA and secondary structure prediction with average performance. (A) The 3D structure of 4WFM_A. (B) The result of ASA prediction. The blue line denotes the actual ASA values calculated from the native structure. The yellow line denotes the ASA values predicted by the model OH + RNA-MSM_Emb. (C). The base pairs of 4WFM_A represented by arc diagrams with canonical base pairs in blue color, pseduoknot base pairs in green color and wrongly predicted base pairs in magenta color (false positives). The predicted secondary structure by model OH + RNA-MSM_ATN with F1 score 0.714, as compared with the native base pairing structure for 4WFM_A RNA (Ground Truth).
Discussion
We presented an RNA MSA-transformer language model (RNA-MSM), based on homologous RNA sequences. RNA-MSM takes the multiple aligned sequences as an input, and outputs corresponding embeddings and attention maps. We have demonstrated that these outputs contain the structural information of the query sequence of the input MSA and evaluated our model on two structure-related downstream tasks, namely RNA secondary structure prediction and RNA solvent accessibility prediction. Interestingly, we found that different downstream tasks prefer different representation features. Specifically, the attention maps outperformed embeddings for RNA secondary structure prediction, while the opposite is true for the RNA ASA prediction task. It is likely that the attention maps of the 2D form contain the information on the connections between nucleotides as demonstrated in Figure 2, whereas the embedding compressed the structural information into one dimension, which is more suitable for 1D structural properties such as ASA.
We employed RNAcmap3 to search and generate homologous sequences. Our original plan was to use Rfam families (29) but the median number of sequences in an Rfam family (44) is too low. Moreover, some RNA families relied on experimentally determined secondary structure for seed alignment, which would interfere with our goal to generate MSAs in the absence of any known experimental information. Thus, we employed RNAcmap3, which produced many more homologous sequences by including not only the nucleotide sequences in NCBI, but also those from RNAcentral (63), the genomic sequences from Genome Warehouse (GWH) (64) and the genomic sequences from MGnify (65). Indeed, the median value for the number of MSA sequences for Rfam families expands to > 2000. Significantly more homologous sequences allow a better training of the present RNA-MSM model. However, one limitation of RNAcmap3 is the time-consuming nature of obtaining the MSAs. It takes on average 9 h to obtain an MSA for one RNA of length 60. Moreover, the method needs a secondary structure predictor to provide an initial secondary structure for building the covariance model. RNAfold was used here. This leads to the overall performance being somewhat dependent on the performance of RNAfold. Further studies are required to avoid the influence of the use of an initial secondary structure predictor.
Previous methods RNAsnap2_seq and RNAsnap2_pro made use of RNA secondary structure as a feature to input. We also examined the possibility of using predicted secondary structure as an additional feature for improving RNA-MSM-based models. We did find that some improvement was made when combining the secondary structure with one-hot encoding, but no significant improvement was observed when combining with the feature of OH + RNA-MSM_Emb. This result may be because the RNA-MSM model implicitly contains information on the secondary structure. Therefore, we did not include predicted secondary structure as a feature for our final model.
After this work was completed, we noticed a new method called M2pred published on RNA solvent accessibility prediction, which utilized multi-scale context features via a multi-shot neural network (66). We downloaded this evolution profile-based method from https://github.com/XueQiangFan/M2pred/ and compared it with RNA-MSM. The result shows that M2pred performs worse than our method (OH + RNA-MSM_Emb) not only on the newly released structures in the PDB (TS2) but also on the TS*, a subset of TS which removed structures similar to the sequences (TM-score > 0.45, according to RNA-align) in the training and validating set from their benchmark dataset. For this reduced set (TS*), the average correlation coefficient is 0.389 by M2pred and 0.436 by OH + RNA-MSM_Emb. The MAE is 32.56 by M2pred and 31.64 by OH + RNA-MSM_Emb. OH + RNA-MSM_Emb also shows a better distribution on TS2 and TS* in Supplementary Figure S5. This is true even after removing similar structures according to TM-score > 0.3 (the TS** set, Supplementary Table S8). Thus, OH + RNA-MSM_Emb retains the best performance, when compared with the latest method.
One common problem of RNA language models including RNA-MSM is a limited sequence length due to the limited memory capacity of GPUs. The common reason for the high memory consumption of these RNA language models is because the transformer requires a space of the magnitude of the square of the input length. Even though we employed relatively new GPUs V100, the sequence length of the input can only be set to 1024. This leads to the loss of relationships between long-distance nucleotides. The next version of the model should allow a better handing of sequences of arbitrary length using a method such as asymmetric cross-attention (67).
One pressing problem is whether deep learning models can be generalized to the families that were not in the training or validation sets (68). We have made our best effort to separate training from validation and test by excluding structurally similar RNAs based on RNA-align with a strict threshold of TM-score > 0.3. We further showed that the accuracy of models based on RNA-MSM features for several Rfam families unseen by the secondary structure predictor and the ASA predictor during training are as high as other RNAs. This suggests that the model proposed here is able to go beyond the families contained in the training.
Given recent success in protein structure prediction by AlphaFold2 (13), it is of great interest to extend this success to RNA structure prediction. Several efforts have been applied in bioRxiv (69). However, some of these efforts have failed in CASP 15 RNA structure prediction, where the top four predictors remained traditional energy-based techniques without using deep learning techniques. In particular, the top method AIchemy_RNA2 (70) is based on a statistical energy function tailored to RNA with orientation-dependent base pairing and stacking interactions plus a rotameric backbone (71). The poor performance of AI-based techniques in tertiary structure prediction is largely due to poor capability for going beyond known structures. Given the robustness of our model for predicting unseen structures on base pairs and solvent accessibility, we are currently applying the RNA-MSM model to RNA tertiary structure.
Supplementary Material
Acknowledgements
We acknowledge the support of the Shenzhen Bay supercomputing facility and Pengcheng Cloudbrain.
Author contributions: Ya. Z., J.C. and J.Z. conceived and supervised the study. Yi. Z., M.L. J.J. and Z.G. implemented the algorithms and performed the data analysis. F.X., T.L., K.C., J.S. and X.H. contributed to the protocol optimization and experimental design. G.S. and Y.T. provided scientific guidance and contributed to study supervision. Ya. Z, Yi. Z, M.L., J.J. and Z.G. wrote the manuscript, and all authors read and contributed to the editing of the manuscript and approved the final version.
Contributor Information
Yikun Zhang, School of Electronic and Computer Engineering, Peking University, Shenzhen 518055, China; AI for Science (AI4S)-Preferred Program, Peking University Shenzhen Graduate School, Shenzen 518055, China.
Mei Lang, Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China.
Jiuhong Jiang, Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China.
Zhiqiang Gao, Shanghai Artificial Intelligence Laboratory, Shanghai 200232, China; Peng Cheng Laboratory, Shenzhen 518066, China.
Fan Xu, Peng Cheng Laboratory, Shenzhen 518066, China.
Thomas Litfin, Institute for Glycomics, Griffith University, Parklands Dr, Southport, QLD 4215, Australia.
Ke Chen, Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China.
Jaswinder Singh, Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China.
Xiansong Huang, Peng Cheng Laboratory, Shenzhen 518066, China.
Guoli Song, Peng Cheng Laboratory, Shenzhen 518066, China.
Yonghong Tian, Peng Cheng Laboratory, Shenzhen 518066, China.
Jian Zhan, Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China.
Jie Chen, School of Electronic and Computer Engineering, Peking University, Shenzhen 518055, China; Peng Cheng Laboratory, Shenzhen 518066, China.
Yaoqi Zhou, Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China; Institute for Glycomics, Griffith University, Parklands Dr, Southport, QLD 4215, Australia.
Data availability
All source codes, models and datasets of RNA-MSM along with the RNA secondary structure predictor and ASA predictor are publicly available in Zenodo at https://doi.org/10.5281/zenodo.8280831.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
The Natural Science Foundation of China [22350710182]; the Shenzhen Science and Technology Program [KQTD20170330155106581 to Y.Z.]; the National Key R&D Program of China [2022ZD0160100 to Z.G and 2022ZD0118201 to J.C.]; Griffith University Postdoctoral Fellowship to TL; and the Natural Science Foundation of China [61972217, 32071459, 62176249, 62006133 and 62271465 to J.C.].
Conflict of interest statement. The authors declare no competing financial interest.
References
- 1. Devlin J., Chang M.-W., Lee K., Toutanova K.. BERT: pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of the 2019 Conference of the North. 2019; Stroudsburg, PA: Association for Computational Linguistics; 4171–4186. [Google Scholar]
- 2. Radford A., Narasimhan K., Salimans T., Sutskever I.. Improving language understanding by generative pre-training. 2018; https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf.
- 3. Brown T., Mann B., Ryder N., Subbiah M., Kaplan J.D., Dhariwal P., Neelakantan A., Shyam P., Sastry G., Askell A.et al.. Larochelle H., Ranzato M., Hadsell R., Balcan M.F., Lin H.. Language models are few-shot learners. Advances in Neural Information Processing Systems. 2020; NY: 33. Curran Associates, Inc; 1877–1901. [Google Scholar]
- 4. Ofer D., Brandes N., Linial M.. The language of proteins: NLP, machine learning & protein sequences. Comput. Struct. Biotechnol. J. 2021; 19:1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee D., Xiong D., Wierbowski S., Li L., Liang S., Yu H.. Deep learning methods for 3D structural proteome and interactome modeling. Curr. Opin. Struct. Biol. 2022; 73:102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alley E.C., Khimulya G., Biswas S., AlQuraishi M., Church G.M.. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods. 2019; 16:1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Strodthoff N., Wagner P., Wenzel M., Samek W.. UDSMProt: universal deep sequence models for protein classification. Bioinformatics. 2020; 36:2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., Guo D., Ott M., Zitnick C.L., Ma J.et al.. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA. 2021; 118:e2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rao R., Bhattacharya N., Thomas N., Duan Y., Chen X., Canny J., Abbeel P., Song Y.S.. Evaluating protein transfer learning with TAPE. Adv. Neural Inf. Process. Syst. 2019; 32:9689–9701. [PMC free article] [PubMed] [Google Scholar]
- 10. Brandes N., Ofer D., Peleg Y., Rappoport N., Linial M.. ProteinBERT: a universal deep-learning model of protein sequence and function. Bioinformatics. 2022; 38:2102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elnaggar A., Heinzinger M., Dallago C., Rehawi G., Wang Y., Jones L., Gibbs T., Feher T., Angerer C., Steinegger M.et al.. ProtTrans: towards understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2021; 44:7112–7127. [DOI] [PubMed] [Google Scholar]
- 12. Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H.. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007; 23:1282–1288. [DOI] [PubMed] [Google Scholar]
- 13. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ji Y., Zhou Z., Liu H., Davuluri R.V.. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome. Bioinformatics. 2021; 37:2112–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luo H., Chen C., Shan W., Ding P., Luo L.. Huang D.-S., Jo K.-H., Jing J., Premaratne P., Bevilacqua V., Hussain A.. iEnhancer-BERT: a novel transfer learning architecture based on DNA-language model for identifying enhancers and their strength. ICIC 2022. Intelligent Computing Theories and Application. 2022; Cham: Springer International Publishing; 153–165. [Google Scholar]
- 16. Tsukiyama S., Hasan M.M., Deng H.-W., Kurata H.. BERT6mA: prediction of DNA N6-methyladenine site using deep learning-based approaches. Brief. Bioinform. 2022; 23:bbac053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu X., Jiang L., Jin S., Zeng X., Liu X.. preMLI: a pre-trained method to uncover microRNA–lncRNA potential interactions. Brief. Bioinform. 2022; 23:bbab470. [DOI] [PubMed] [Google Scholar]
- 18. Yi H.C., You Z.H., Cheng L., Zhou X., Jiang T.H., Li X., Wang Y.-B.. Learning distributed representations of RNA and protein sequences and its application for predicting lncRNA-protein interactions. Comput. Struct. Biotechnol. J. 2020; 18:20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J., Hu Z., Sun S., Tan Q., Wang Y., Yu Q., Zong L., Hong L., Xiao J., King I.et al.. Interpretable RNA foundation model from unannotated data for highly accurate RNA structure and function predictions. 2022; arXiv doi:08 August 2022, preprint: not peer reviewedhttps://arxiv.org/abs/2204.00300.
- 20. Whisstock J.C., Lesk A.M.. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003; 36:307–340. [DOI] [PubMed] [Google Scholar]
- 21. Lobb B., Doxey A.C.. Novel function discovery through sequence and structural data mining. Curr. Opin. Struct. Biol. 2016; 38:53–61. [DOI] [PubMed] [Google Scholar]
- 22. Wright E.S. RNAconTest: comparing tools for noncoding RNA multiple sequence alignment based on structural consistency. RNA. 2020; 26:531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rao R.M., Liu J., Verkuil R., Meier J., Canny J., Abbeel P., Sercu T., Rives A.. Meila M., Zhang T.. MSA Transformer. Proceedings of the 38th International Conference on Machine Learning, Proceedings of Machine Learning Research. 2021; 139:ICML; 8844–8856. [Google Scholar]
- 24. Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menzel P., Gorodkin J., Stadler P.F.. The tedious task of finding homologous noncoding RNA genes. RNA. 2009; 15:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freyhult E.K., Bollback J.P., Gardner P.P.. Exploring genomic dark matter: a critical assessment of the performance of homology search methods on noncoding RNA. Genome Res. 2007; 17:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vasavada M., Byron K., Song Y., Wang J.T.L.. Elloumi M., Iliopoulos C.S., Wang J.T.L., Zomaya A.Y.. Genome-wide search for pseudoknotted noncoding RNA: a comparative study. 2015; 155–164.Pattern Recognition in Computational Molecular Biology. [Google Scholar]
- 28. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., Griffiths-Jones S., Toffano-Nioche C., Gautheret D., Weinberg Z.et al.. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021; 49:D192–D200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang T., Singh J., Litfin T., Zhan J., Paliwal K., Zhou Y.. RNAcmap: a fully automatic pipeline for predicting contact maps of RNAs by evolutionary coupling analysis. Bioinformatics. 2021; 37:3494–3500. [DOI] [PubMed] [Google Scholar]
- 31. Lorenz R., Hofacker I.L., Stadler P.F.. RNA folding with hard and soft constraints. Algorithms Mol. Biol. 2016; 11:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh J., Paliwal K., Singh J., Litfin T., Zhou Y.. Improved RNA homology detection and alignment by automatic iterative search in an expanded database. 2022; bioRxiv doi:07 October 2022, preprint: not peer reviewed 10.1101/2022.10.03.510702. [DOI]
- 33. Chen K., Litfin T., Singh J., Zhan J., Zhou Y.. The master database of all possible RNA Sequences and its integration with RNAcmap for RNA Homology Search. 2023; bioRxiv doi:03 February 2023, preprint: not peer reviewed 10.1101/2023.02.01.526559. [DOI] [PMC free article] [PubMed]
- 34. Singh J., Hanson J., Paliwal K., Zhou Y.. RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning. Nat. Commun. 2019; 10:5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andronescu M., Bereg V., Hoos H.H., Condon A.. RNA STRAND: the RNA secondary structure and statistical analysis database. BMC Bioinformatics. 2008; 9:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sloma M.F., Mathews D.H.. Exact calculation of loop formation probability identifies folding motifs in RNA secondary structures. RNA. 2016; 22:1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Danaee P., Rouches M., Wiley M., Deng D., Huang L., Hendrix D.. bpRNA: large-scale automated annotation and analysis of RNA secondary structure. Nucleic Acids Res. 2018; 46:5381–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L., Liu Y., Zhong X., Liu H., Lu C., Li C., Zhang H.. DMfold: a novel method to predict RNA secondary structure with pseudoknots based on deep learning and improved base pair maximization principle. Front. Genet. 2019; 10:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato K., Akiyama M., Sakakibara Y.. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 2021; 12:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu L., Cao Y., Wu J., Peng Q., Nie Q., Xie X.. UFold: fast and accurate RNA secondary structure prediction with deep learning. Nucleic Acids Res. 2022; 50:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singh J., Paliwal K., Zhang T., Singh J., Litfin T., Zhou Y.. Improved RNA secondary structure and tertiary base-pairing prediction using evolutionary profile, mutational coupling and two-dimensional transfer learning. Bioinformatics. 2021; 37:2589–2600. [DOI] [PubMed] [Google Scholar]
- 42. Yang Y., Li X., Zhao H., Zhan J., Wang J., Zhou Y.. Genome-scale characterization of RNA tertiary structures and their functional impact by RNA solvent accessibility prediction. RNA. 2017; 23:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun S., Wu Q., Peng Z., Yang J.. Enhanced prediction of RNA solvent accessibility with long short-term memory neural networks and improved sequence profiles. Bioinformatics. 2019; 35:1686–1691. [DOI] [PubMed] [Google Scholar]
- 44. Hanumanthappa A.K., Singh J., Paliwal K., Singh J., Zhou Y.. Single-sequence and profile-based prediction of RNA solvent accessibility using dilated convolutional neural network. Bioinformatics. 2020; 36:5169–5176. [DOI] [PubMed] [Google Scholar]
- 45. Rose P.W., Prlić A., Altunkaya A., Bi C., Bradley A.R., Christie C.H., Costanzo L.D., Duarte J.M., Dutta S., Feng Z.et al.. The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017; 45:D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu L., Niu B., Zhu Z., Wu S., Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gong S., Zhang C., Zhang Y.. RNA-align: quick and accurate alignment of RNA 3D structures based on size-independent TM-scoreRNA. Bioinformatics. 2019; 35:4459–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lu X.-J., Bussemaker H.J., Olson W.K.. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015; 43:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steinegger M., Meier M., Mirdita M., Vöhringer H., Haunsberger S.J., Söding J.. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics. 2019; 20:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He K., Zhang X., Ren S., Sun J.. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition. 2016; Las Vegas, NV, USA: 770–778. [Google Scholar]
- 51. Wang S., Sun S., Li Z., Zhang R., Xu J.. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 2017; 13:e1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cavallo L., Kleinjung J., Fraternali F.. POPS: a fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res. 2003; 31:3364–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krizhevsky A., Sutskever I., Hinton G.E.. Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q.. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems 25. 2012; NY: Curran Associates, Inc; 1097–1105. [Google Scholar]
- 54. Hu J., Shen L., Albanie S., Sun G., Wu E.. Squeeze-and-Excitation networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020; 42:2011–2023. [DOI] [PubMed] [Google Scholar]
- 55. Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., Garnett R.. Attention is All you Need. Advances in Neural Information Processing Systems. 2017; 30:NY: Curran Associates, Inc. [Google Scholar]
- 56. Loshchilov I., Hutter F.. SGDR: stochastic gradient descent with warm restarts. International Conference on Learning Representations. 2017; [Google Scholar]
- 57. Rao R., Meier J., Sercu T., Ovchinnikov S., Rives A.. Transformer protein language models are unsupervised structure learners. International Conference on Learning Representations. 2021; [Google Scholar]
- 58. De Leonardis E., Lutz B., Ratz S., Cocco S., Monasson R., Schug A., Weigt M.. Direct-coupling analysis of nucleotide coevolution facilitates RNA secondary and tertiary structure prediction. Nucleic Acids Res. 2015; 43:10444–10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lorenz R. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang H., Zhang L., Mathews D.H., Huang L.. LinearPartition: linear-time approximation of RNA folding partition function and base-pairing probabilities. Bioinformatics. 2020; 36:i258–i267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hamada M., Sato K., Asai K.. Improving the accuracy of predicting secondary structure for aligned RNA sequences. Nucleic Acids Res. 2011; 39:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ganaie M.A., Hu M., Malik A.K., Tanveer M., Suganthan P.N.. Ensemble deep learning: a review. Eng. Appl. Artif. Intell. 2022; 115:105151. [Google Scholar]
- 63. The RNAcentral Consortium RNAcentral: a hub of information for non-coding RNA sequences. Nucleic Acids Res. 2019; 47:D221–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., Xu X., Hao L., Li Z., Gong Z.et al.. Genome Warehouse: a public repository housing genome-scale data. Genomics. Proteomics Bioinformatics. 2021; 19:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J.et al.. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 2020; 48:D570–D578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fan X.-Q., Hu J., Tang Y.-X., Jia N.-X., Yu D.-J., Zhang G.-J.. Predicting RNA solvent accessibility from multi-scale context feature via multi-shot neural network. Anal. Biochem. 2022; 654:114802. [DOI] [PubMed] [Google Scholar]
- 67. Jaegle A., Gimeno F., Brock A., Vinyals O., Zisserman A., Carreira J.. Meila M., Zhang T.. Perceiver: general perception with iterative attention. Proceedings of the 38th International Conference on Machine Learning, Proceedings of Machine Learning Research. 2021; 139:PMLR; 4651–4664. [Google Scholar]
- 68. Szikszai M., Wise M., Datta A., Ward M., Mathews D.H.. Deep learning models for RNA secondary structure prediction (probably) do not generalize across families. Bioinformatics. 2022; 38:3892–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Das R., Kretsch R.C., Simpkin A., Mulvaney T., Pham P., Rangan R., Bu F., Keegan R., Topf M., Rigden D.et al.. Assessment of three-dimensional RNA structure prediction in CASP15. Proteins. 2023; 10.1002/prot.26602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen K., Zhou Y., Wang S., Xiong P.. RNA tertiary structure modeling with BRiQ potential in CASP15. Proteins. 2023; 10.1002/prot.26574. [DOI] [PubMed] [Google Scholar]
- 71. Xiong P., Wu R., Zhan J., Zhou Y.. Pairing a high-resolution statistical potential with a nucleobase-centric sampling algorithm for improving RNA model refinement. Nat. Commun. 2021; 12:2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source codes, models and datasets of RNA-MSM along with the RNA secondary structure predictor and ASA predictor are publicly available in Zenodo at https://doi.org/10.5281/zenodo.8280831.