Abstract
Genetic modification of specific genes is emerging as a useful tool to enhance the functions of antitumor T cells in adoptive immunotherapy. Current advances in CRISPR/Cas9 technology enable gene knockout during in vitro preparation of infused T-cell products through transient transfection of a Cas9-guide RNA (gRNA) ribonucleoprotein complex. However, selecting optimal gRNAs remains a major challenge for efficient gene ablation. Although multiple in silico tools to predict the targeting efficiency have been developed, their performance has not been validated in cultured human T cells. Here, we explored a strategy to select optimal gRNAs using our pooled data on CRISPR/Cas9-mediated gene knockout in human T cells. The currently available prediction tools alone were insufficient to accurately predict the indel percentage in T cells. We used data on the epigenetic profiles of cultured T cells obtained from transposase-accessible chromatin with high-throughput sequencing (ATAC-seq). Combining the epigenetic information with sequence-based prediction tools significantly improved the gene-editing efficiency. We further demonstrate that epigenetically closed regions can be targeted by designing two gRNAs in adjacent regions. Finally, we demonstrate that the gene-editing efficiency of unstimulated T cells can be enhanced through pretreatment with IL-7. These findings enable more efficient gene editing in human T cells.
Graphical Abstract
Graphical Abstract.
Introduction
Adoptive T-cell therapy, such as chimeric antigen receptor (CAR)-T-cell therapy, has emerged as a potentially curative therapy for relapsed and refractory cancer (1). However, its long-term efficacy is still insufficient, especially against solid tumors (2). Potentiating the functions of antitumor T cells through gene editing is a promising strategy for next-generation CAR-T-cell therapy (3). We and other groups previously reported that the knockout of specific genes, including PRDM1 (4), PDCD1 (5), TGFBR2 (6) and DNMT3A (7), can substantially improve the therapeutic efficacy of CAR-T cells. To this end, efficient and specific knockout of the target genes is essential.
Clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology has revolutionized the area of gene editing (8). The simplicity of the CRISPR/Cas9 system compared with zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) has made it the most widely used technique for gene knockout (9). One of the key processes in using the CRISPR/Cas9 system is to select the optimal sequences targeted by guide RNAs (gRNAs) (10). Although several design tools have been developed to predict the on-target efficiency of gRNAs (11–14), their prediction accuracy is still insufficient and inconsistent among different experimental settings (15).
The delivery method of Cas9 and gRNAs has an influence on the knockout efficacy (16). While stable viral transduction with Cas9 and gRNAs is a standard method for cell lines, electroporating Cas9-ribonucleoprotein (RNP) complexes has been established as an efficient delivery strategy for cultured T cells (17). Since a lot of prediction tools to design gRNAs have been constructed based on the data obtained in the viral transduction approach, it is uncertain whether the findings can be applicable in the case of RNP electroporation. Moreover, optimal target regions may be different between cancer cell lines and primary T cells (18).
We have been investigating the genetic engineering of CAR-T cells to enhance their functions using transient electroporation of a Cas9-gRNA RNP complex (4,19,20). In this study, we explored a strategy to identify targets with high gene-editing efficiency using accumulated data on the indel percentage of individual gRNAs. While multiple studies already investigated the effect of chromatin accessibility on CRISPR/Cas9-mediated gene-editing efficacy (21–23), we demonstrate that combining cultured T cell-specific epigenetic data with a prediction algorithm significantly improves gRNA design.
Materials and methods
Human T-cell culture
Peripheral blood mononuclear cells (PBMCs) derived from healthy donors (Cellular Technology Limited, Cleveland, OH) were stimulated with mitomycin C-treated K562 cells (JCRB, Osaka, Japan) expressing anti-CD3 mAb (clone OKT3)-derived single-chain variable fragment (scFv) and CD80 on the cell surface at an effector to target ratio of 7:1 (4). Stimulated T cells were cultured with RPMI-1640 medium containing 10% fetal bovine serum, 1% penicillin/streptomycin, and recombinant IL-2 (100 IU/ml, Nipro, Osaka, Japan). Unstimulated T cells were maintained in the presence of recombinant IL-7 (10 ng/ml, PeproTech, Waltham, MA). When indicated, naïve T cells were enriched by magnetically isolating CD45RO− cells using CD45RO MicroBeads (Miltenyi Biotech, Bergisch Gladbach, Germany).
To generate CAR-T cells, T cells were retrovirally transduced with CD19- or GD2-targeting CAR genes two days after stimulation. We used PG13 packaging cells for retrovirus production. The CAR constructs consisted of a single-chain variable fragment (scFv) derived from the clone FMC63 (targeting CD19) (24) or 14g2a with E101K high-affinity mutation (targeting GD2) (25) linked to CD28 and CD3z signaling domains.
CRISPR/Cas9-based gene editing
CRISPR/Cas9-mediated knockout was performed using the Alt-R CRISPR/Cas9 system provided by Integrated DNA Technologies (IDT, Coralville, IA) as previously described (4). The electroporation of a ribonucleoprotein (RNP) complex was performed using a NEPA 21 electroporator (Nepa Gene, Ichikawa, Japan) on days 1–5 after T cell stimulation. Chemically synthesized crRNA and tracrRNA (IDT) were mixed and annealed at 95°C for 5 min followed by gradual cooling to room temperature. An RNP complex was generated by incubating the Alt-R Cas9 Nuclease V3 (25 μg, IDT) with the annealed guide RNAs (300 pmol) at 37°C for 10 to 15 minutes. For simultaneous introduction of dual gRNAs, Cas9 Nuclease V3 (25 μg) and two separately prepared gRNAs (300 pmol each) were mixed in the same tube, and the generated RNP complexes were used for electroporation. The Alt-R Cas9 Electroporation Enhancer was added before electroporation at a final concentration of 2 μM. The following parameters were used for electroporation: poring pulse: voltage, 275 V; pulse length, 1 msec; pulse interval, 50 ms; number of pulses, 2; decay rate, 10%; polarity, + and transfer pulse: voltage, 20 V; pulse length, 50 msec; pulse interval, 50 ms; number of pulses, ±5; decay rate, 40%; polarity, +/–.
Genomic DNA was extracted after 48–72 h using NucleoSpin DNA Rapid Lyse kit (Macherey Nagel, Düren, Germany). Genomic regions containing the target sites were amplified by PCR, and the amplicons were submitted to Sanger sequencing. The editing efficiency of each gRNA was calculated using the Inference of CRISPR Edits (ICE) analysis (26). We used indel percentages to evaluate the efficiency of CRISPR/Cas9-mediated gene editing, which does not necessarily indicate knockout unless it induces a frameshift mutation. When indicated, T cells were pretreated with romidepsin (10 nM, Selleck Chemicals, Houston, TX) for 8 h before electroporation. To analyze transfection efficacy of RNP complexes, TYE 563-labeled duplex RNA (#51-01-20-19, IDT) was used according to the manufacturer's instruction.
Viral transduction
Jurkat cells were retrovirally transduced with Cas9-P2A-truncated NGFR using Retronectin (Takara Bio, Kusatsu, Japan). Plat-A packaging cells (provided by Dr. Kitamura) were used for retrovirus production. We performed transfection using CalPhos Mammalian Transfection Kit (Takara Bio). Virus supernatants were collected 2 days after transfection and directly used for transduction. For lentiviral transduction, Lenti-X 293T cells (Takara Bio) were transfected with pLKO5.sgRNA.EFS.tRFP (a gift from Benjamin Ebert (Addgene plasmid # 57823; http://n2t.net/addgene:57823;RRID: Addgene_57823). Virus supernatants were collected 2 days after transfection and concentrated using Lenti-X concentrator (Takara Bio).
Flow cytometry analysis
Flow cytometry analysis was conducted using FACSCanto II and LSRFortessa (BD Biosciences, Franklin Lakes, NJ), and CytoFLEX S (Beckman Coulter, Brea, CA). The following antibodies were used: PerCP/Cyanine5.5-anti-human CD271 (ME20.4, BioLegend, San Diego, CA), APC-anti-CD8 (RPA-T8, BioLegend), Pacific blue-anti-CCR7 (G043H7, BioLegend), APC-Cy7-anti-CD62L (DREG-56, BioLegend), PE-Cy7-anti-CD27 (M-T271, BioLegend), PE-anti-CD28 (CD28.2, BioLegend), BV510-anti-CD45RA (HI100, BioLegend), and FITC-anti-CD45RA (HI100, BD Biosciences). Data were analyzed with FlowJo v10 software (BD Biosciences).
ATAC-sequencing analysis
ATAC-sequencing was performed as previously described (4). Briefly, CD8+ CAR-T cells cultured for 7 days were incubated for 30 min at 37 ºC in complete culture media supplemented with 100x DNase buffer (250mM MgCl2 and 50mM CaCl2) and 100x Dnase solution (Hanks Balanced Salt Solution with 20 000 IU/ml Turbo Dnase). The cells were then cryopreserved in growth media with 10% DMSO and submitted to sequencing analysis (Genewiz, Plainfield, NJ). Sequencing adapters and low-quality bases were trimmed using Trimmomatic 0.38, and cleaned reads were aligned to the reference genome hg38 using bowtie2. PCR or optical duplicates were removed using Picard 2.18.26. The bigwig files for Integrative Genomics Viewer (IGV) visualization were generated using the bamCoverage function with the following parameters: –normalizeUsing CPM –binSize 1. The data obtained from the donor #1 (D1) were used for analysis. The fastq file of GSE231309 (GSM7256892) was used for the analysis of publicly available ATAC-seq data of CD8+ CAR-T cells. To analyze ATAC-seq data of K562 cells and human mesenchymal stem cells derived from bone marrow (hMSC-BMs), bigwig files were directly obtained from GSE137647 (GSM4083680) and GSE94272 (GSM2472047), respectively. To calculate the ATAC-seq score of gRNAs for hMSC-BMs, read counts in the bigwig file deposited in GSE94272 (GSM2472047; normalized to 10 million reads) were divided by 10 to normalize to CPM.
RNA-sequencing analysis
PBMCs derived from two different donors were used for RNA-sequencing analysis (Rhelixa, Tokyo, Japan). CD8+ primary naïve T cells (CD8+CD45RA+CCR7+), CD8+ unstimulated T cells cultured in the presence of IL-7 (CD8+CD45RA+CCR7+), and CD8+ T cells that were stimulated and cultured in the presence of IL-2 and IL-15 for 3 days (CD8+) were isolated by flow cytometry for collection of total RNA. Sequencing libraries were prepared using SMART-Seq HT PLUS Kit (Takara Bio). Base calling was performed by the Illumina RTA software. Further demultiplexing was performed using Illumina bcl2fastq software. Trimming, mapping, and calculation of read counts were performed using Trim-galore, Hisat2 and HTSeq-count, respectively. Principal component analysis, hierarchical clustering, and pathway analysis were performed using the iDEP tool (27). Genes were ranked by their standard deviation across all samples, and the top 1000 genes with variable expression levels were used for hierarchical clustering analysis. For pathway analysis, differentially expressed genes between naïve T cells and IL-7 treated unstimulated T cells were extracted using DESeq2 (false discovery rate < 0.1 and fold change > 2) and analyzed for enrichment using GO Biological Process gene sets.
Culture of human mesenchymal stem cells from bone marrow
Human mesenchymal stem cells from bone marrow (hMSC-BM) derived from healthy donors (PromoCell, Heidelberg, Germany) were expanded with Mesenchymal Stem Cell Growth Medium 2 (PromoCell) containing 1% penicillin/streptomycin. Electroporation of Cas9/gRNA complexes was performed using the same parameters as those used for T cells.
Statistical analysis
Significance of differences between two groups was assessed with unpaired or paired two-tailed t-test. The differences among three or more groups were compared using one-way ANOVA. When indicated, the data were compared with nonparametric test: Mann-Whitney test for two groups and Kruskal–Wallis test for three or more groups. Correlation between two data was assessed with Spearman's rank correlation coefficient. Differences were considered statistically significant when p value was <0.05. Significant correlation was defined as P value <0.05. The GraphPad Prism 8 software was used for statistical analysis and graphical design.
Results
Currently available gRNA design tools to predict the efficiency of CRISPR/Cas9-mediated gene editing in human T cells
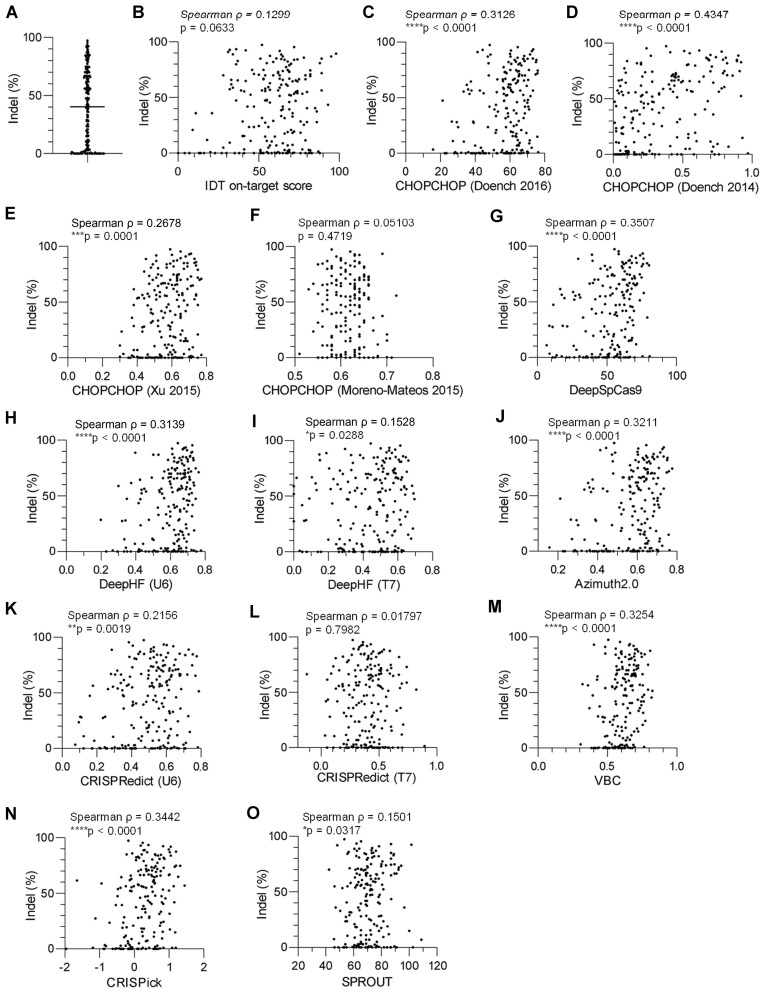
The stable transduction of primary T cells with Cas9 and gRNAs is inefficient due to the large size of the Cas9 cDNA while off-target effects increase during prolonged expression (28). Recently, electroporating Cas9 protein/gRNA complexes into stimulated T cells was established as an efficient delivery method (17). Using this strategy, we have accumulated data on the indel percentages for 205 gRNAs against 110 genes and found that the efficiency was highly variable among different target sites (Figure 1A; Supplementary Table S1).
Figure 1.
Validation of gRNA design algorithms to predict CRISPR/Cas9-mediated gene-editing efficiency in human T cells. (A) T cells were individually electroporated with a Cas9-gRNA complex and analyzed for gene-editing efficiency (n = 205 targets). (B–O) The estimated indel percentages and on-target scores based on the software provided by IDT (B), CHOPCHOP (Doench 2016, C), CHOPCHOP (Doench 2014, D), CHOPCHOP (Xu 2015, E), CHOPCHOP (Moreno-Mateos 2015, F), DeepSpCas9 (G), DeepHF (U6 promoter, H), DeepHF (T7 promoter, I), Azimuth2.0 (J), CRISPRedict (U6 promoter, K), CRISPRedict (T7 promoter, L), VBC (M), CRISPick (N) and SPROUT (O) were plotted for individual gRNA targets. Correlation between the two data was assessed based on Spearman's rank correlation coefficient (n = 205 targets). The indel percentages were obtained at least in duplicate, and the mean values were used for correlation analysis.
We further explored whether the gene-editing efficiency in human T cells can be estimated accurately using currently available online prediction tools, including IDT on-target score, CHOPCHOP web tool (29,30) based on Doench et al. (31), Doench et al. (32), Xu et al. (33) and Moreno-Mateos et al. (34), DeepSpCas9 (35), DeepHF (U6 promoter and T7 promoter) (36), Azimuth 2.0 (31), CRISPRedict (U6 promoter and T7 promoter) (37), Vienna Bioactivity CRISPR score (VBC) (38), CRISPick (39) and SPROUT (40). The majority of the gRNAs were designed regardless of the score calculated by any specific prediction software (Figure 1B–O). Among these tools, the scores calculated by CHOPCHOP (Doench 2014) and DeepSpCas9 achieved the highest correlation with the indel percentage. These scores showed a significant correlation with each other (Supplementary Figure S1). Although multiple prediction scores were significantly correlated with indel percentages, gRNAs with high scores did not always result in efficient indel formation, which highlights the necessity of an additional method to further improve prediction accuracy.
T-cell-specific epigenetic profiles affect gene-editing efficiency
The above results suggest that no single algorithm can precisely predict gene-editing efficiency by transient transfection of Cas9-gRNAs in T cells. The primary method of validation for these algorithms is based on data using cell lines. In addition to the effect of target DNA sequences, chromatin density has been reported to be associated with efficient targeting (41). Since epigenetic architecture is highly variable between cell types, we speculated that T-cell-specific epigenetic status aids in designing optimal gRNA targets.
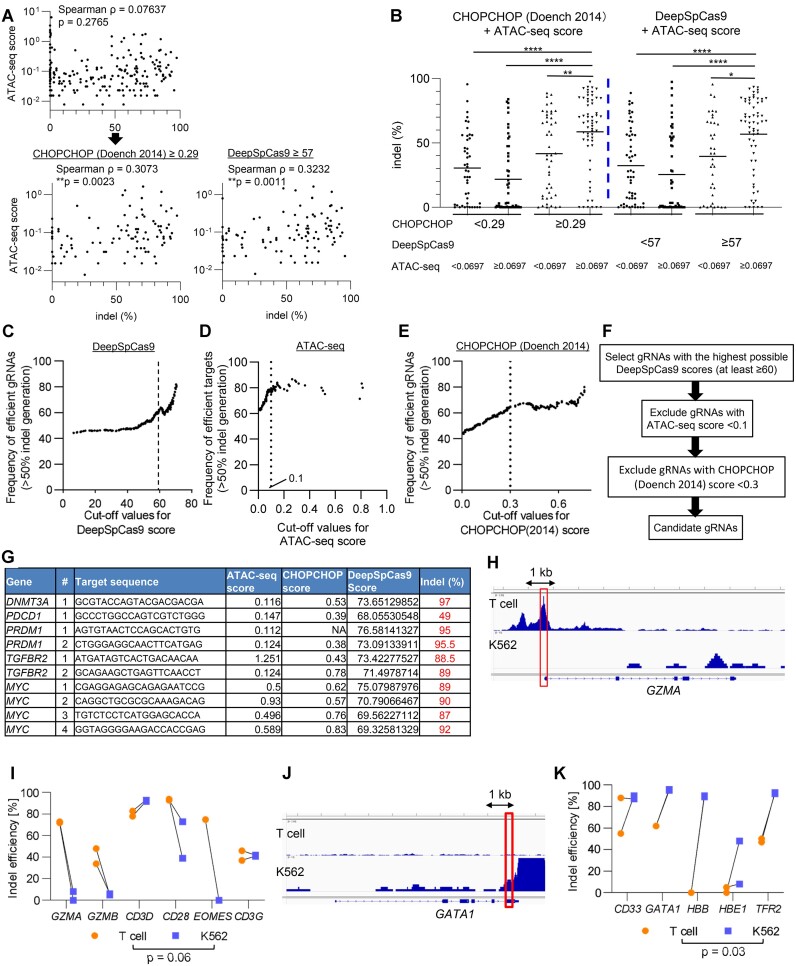
To investigate the correlation between the chromatin accessibility of target sites and gene-editing efficiency in T cells, we used the ATAC-seq data that we previously generated from CAR-T cells (4). The ATAC-seq score of gRNAs was defined as the median value of the read counts of their target regions, and thus is associated with accessibility to the targeted regions. As shown in Figure 2A, the ATAC-seq score alone did not show a significant correlation with the efficiency of indel generation. However, high ATAC-seq scores were significantly associated with efficient indel formation among gRNAs with above-median scores in CHOPCHOP (Doench 2014) or DeepSpCas9 (Figure 2A). The gRNAs with high scores in both ATAC-seq and CHOPCHOP or DeepSpCas9 analysis induced significantly higher indel percentages than the other groups (Figure 2B). The ATAC-seq scores did not show significant correlation with the CHOPCHOP or DeepSpCas9 scores, suggesting that the chromatin accessibility of the target region affects CRISPR/Cas9-mediated gene-editing efficiency independent of the sequence patterns of gRNAs (Supplementary Figure S2A, B).
Figure 2.
Chromatin accessibility of the target region affects the gene-editing efficiency. (A) The indel percentages and chromatin accessibility based on ATAC-seq data were plotted for individual gRNA targets (Spearman's rank correlation analysis) (n = 205). The lower panels represent plots among the gRNAs with above-median values of CHOPCHOP (Doench 2014) score (≥0.29, left panel) or DeepSpCas9 score (≥57, right panel). Correlation was evaluated by the Spearman's rank correlation coefficient. (B) The indel percentages of gRNAs among the four groups classified according to the ATAC-seq values and CHOPCHOP scores or DeepSpCas9 scores. Median values were used as cut-off points for each score (one-way ANOVA with Kruskal–Wallis multiple comparison test, *P < 0.05, **P < 0.01, ****P < 0.0001). (C) The frequency of efficient gRNAs (defined as >50% indel generation) was plotted against the threshold values set for the DeepSpCas9 score (C), ATAC-seq score (D) and CHOPCHOP score (E). For (D), only gRNAs with the DeepSpCas9 score of ≥60 were used for calculation. (F) Selection diagram of gRNAs based on the DeepSpCas9, ATAC-seq and CHOPCHOP scores. (G) Individual indel efficiency of 10 gRNAs designed for five genes (DNMT3A, PDCD1, PRDM1, TGFBR2, and MYC). (H–K) Indel percentages were compared between T cells and K562 cells against the genes with different chromatin accessibility. (H, J) Representative ATAC-seq signal tracks in GZMA and GATA1 gene locus were depicted. Red rectangles denote the regions containing target sites. (I, K) Indel frequency of gRNAs targeting T cell-associated (I) and myeloerythroid-associated genes (K) were compared between T cells and K562 cells (n = 5 or 6 genes, paired two-tailed t-test).
These findings led us to hypothesize that the use of epigenetic information in combination with prediction tools may help to identify optimized gRNA targets. To obtain more practical metrics to optimize the selection process of gRNAs, we investigated whether we could set appropriate cut-off values for each score. When we calculated the frequency of gRNAs that achieved more than 50% indel formation for each cut-off value, the frequency of efficient gRNAs progressively increased along with higher cut-off values in the DeepSpCas9 score, especially at 60 or more as a threshold score (Figure 2C). We then selected gRNAs with the DeepSpCas9 score ≥ 60 and explored the optimal threshold of the ATAC-seq score among them. As shown in Figure 2D, most of the gRNAs with ATAC-seq score of ≥ 0.1 achieved efficient indel formation. While the gRNA efficiency was also improved by setting a threshold for the CHOPCHOP (Doench 2014) score, it became almost constant beyond the cut-off score of ≥0.3 (Figure 2E).
Based on these results, we have set selection flow of gRNAs as follows (Figure 2F). We first choose gRNAs with the highest possible DeepSpCas9 scores (at least more than 60). We then exclude the gRNAs with < 0.1 ATAC-seq score and also those with < 0.3 CHOPCHOP score. We newly designed 10 gRNAs targeting five genes (DNMT3A, PDCD1, PRDM1, TGFBR2 and MYC) that are essentially associated with T cell functions using this strategy (Figure 2G). Although the number of candidate gRNAs that passed the criteria were variable among the target genes, we found that all the selected gRNAs achieved efficient gene editing, which validated the effectiveness of this approach. We also used another publicly available ATAC-seq data of CAR-T cells (GSM7256892) to calculate the ATAC-seq scores. Using the same cutoff of 0.1, the ATAC-seq scores calculated from this dataset also helped to identify efficient gRNA targets when combined with the DeepSpCas9 or CHOPCHOP scores (Supplementary Figure S3A). The scores calculated from both sets of ATAC-seq data showed a significant correlation (Supplementary Figure S3B).
Considering that epigenetic profiles are highly variable among different cell types (e.g. T cells and myeloid cells), gRNA sequences that can efficiently cleave the targets in T cells may not work in different types of cells, and vice versa. To ascertain this possibility, we designed several gRNAs against genes with distinct epigenetic profiles between T cells and the erythroleukemia cell line K562 (Supplementary Table S2). We first selected gRNAs with high ATAC-seq scores in T cells but low ATAC-seq scores in K562 cells (GZMA, GZMB, CD3D, CD3G, CD28 and EOMES) (Figure 2H; Supplementary Figure S4A–E). Consistent with our hypothesis, these gRNAs achieved efficient gene editing in T cells, whereas the majority of them showed poor efficiency in K562 cells (Figure 2I). Conversely, gRNAs targeting myeloid- or erythroid-associated genes, which are epigenetically open in K562 cells (GATA1, CD33, HBB, HBE1 and TFR2), selectively induced successful indel formation in K562 cells (Figure 2J, K; Supplementary Figure S4F–I). These results further corroborate the importance of chromatin accessibility in efficient cleavage by the CRISPR/Cas9 system.
Targeting with two gRNAs enhances the gene-editing efficiency
We then explored how we can improve the gene-editing efficiency of gRNAs with low ATAC-seq scores. Histone deacetylase (HDAC) inhibitors induce genome-wide chromatin remodeling and have been reported to enhance knockout efficiency in HeLa and HT29 cells (42). However, pretreatment with romidepsin, an HDAC inhibitor, did not improve the indel percentage of the tested gRNAs (Supplementary Figure S5A). In addition, romidepsin was highly toxic to human T cells (Supplementary Figure S5B, C) (43), making it impractical to use to enhance gene-editing efficiency.
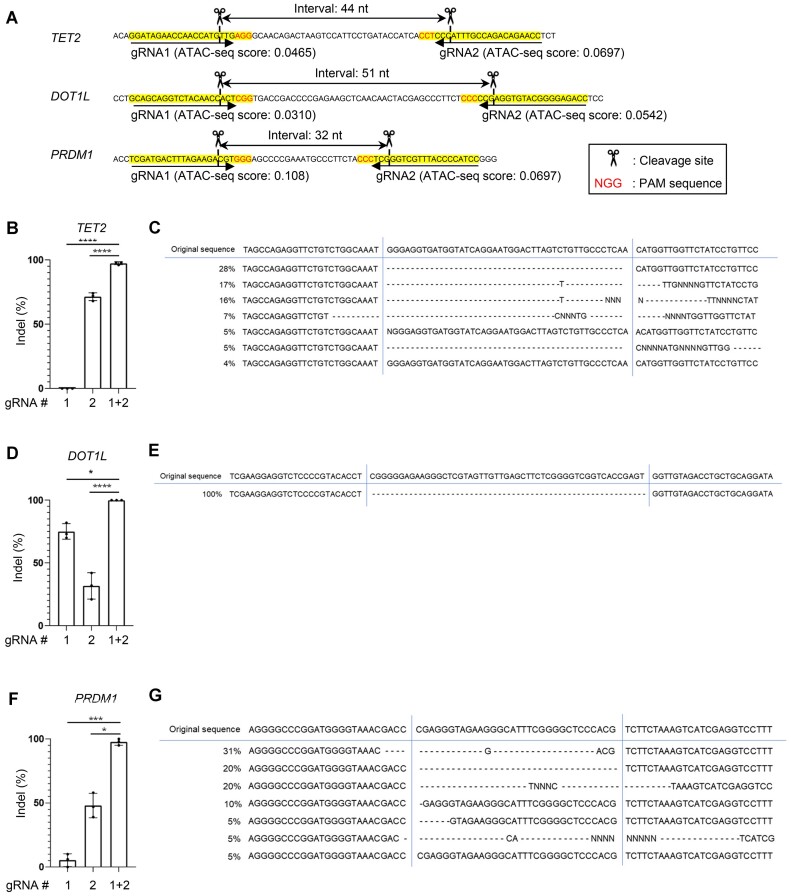
Another approach is targeting two adjacent sites, which can potentially improve gene-editing efficiency (28). We tested whether this strategy can augment the cleavage of epigenetically closed regions (Figure 3A). Although gRNA #1 targeting an epigenetically closed region in the TET2 gene poorly produces cleavage, the concomitant use of gRNA #2 significantly improved the gene-editing efficiency (Figure 3B). The majority of the cells showed deletion between gRNA#1 and gRNA#2 target sites, suggesting their synergistic effects (Figure 3C). Likewise, dual targeting of the adjacent regions in DOT1L or PRDM1 successfully edited the genes by inducing a deletion between the two target sites (Figure 3D–G). These results suggest that the double-targeting strategy can enhance the gene-editing efficiency of epigenetically closed regions for at least some of the targets.
Figure 3.
Dual targeting of adjacent regions can overcome the poor chromatin accessibility. (A) The gRNAs were designed at two proximal regions against TET2, DOT1L, and PRDM1. (B–G) Indel percentage was calculated when two gRNAs were electroporated individually or in combination against TET2 (B), DOT1L (D), and PRDM1 (F) (one-way ANOVA with multiple comparison test; *P < 0.05, ***P < 0.001, ****P < 0.0001, n = 3 experiments). Generated indels through dual gRNA targeting against TET2 (C), DOT1L (E) and PRDM1 (G) were estimated using the ICE algorithm.
We further explored how a pair of gRNAs can be designed to enhance gene-editing efficiency in more detail. Although the PAM sequences following the gRNA targets faced inward in the above designs, we found that any other orientation of the two gRNAs (facing outward or aligned in the same direction) allowed them to delete the intervening region (Figure 3A; Supplementary Figure S6A, B). For the distance between two targets, gRNAs located at least about 500 bp apart can efficiently delete the intervening DNA sequences (Supplementary Figure S6C, D). We further tested if gRNA pairs with longer distance can also delete DNA between the target sites. Surprisingly, we detected a deletion between gRNAs that are apart from each other up to 70 000 nucleotides (Supplementary Figure S7A, B). Although the deletion efficiency cannot be precisely quantified due to the more efficient amplification of shorter fragments by PCR, these results suggest that substantial variability can be accepted for the distance between two gRNA targets.
Efficient gene editing is essentially associated with the modification of T cell functions. We previously reported that PRDM1 knockout in T cells enhances the maintenance of an early memory phenotype (4). As expected, the double-targeting strategy enhanced the gene-editing efficiency in PRDM1 (Figure 3F, G), which resulted in the maintenance of a central memory phenotype (CD45RA−CCR7+) better than single-targeting approaches (Supplementary Figure S8A-C).
Stable expression of Cas9 and gRNAs can induce cleavage at epigenetically closed regions
Electroporation delivers Cas9 protein and gRNAs in a transient manner. We investigated whether constitutively expressed Cas9 and gRNAs can eventually cleave target sites with closed chromatin status. The human T-cell leukemia cell line Jurkat was stably transduced with Cas9 and gRNAs that target epigenetically closed regions within 9 genes and open regions within 2 genes as positive controls (Supplementary Figure S9A–E). While RNP transfection poorly induced cleavage in most of the closed chromatin regions, we observed a gradual increase in the indel percentages in Jurkat cells with stable expression of Cas9 and gRNAs, which reached 60–100% by day 14. These results suggest that gene knockout by constitutively expressed Cas9 and gRNAs is less hampered by closed epigenetic status compared with the transient delivery system.
Short-term culture with IL-7 improves gene-editing efficiency in naïve T cells
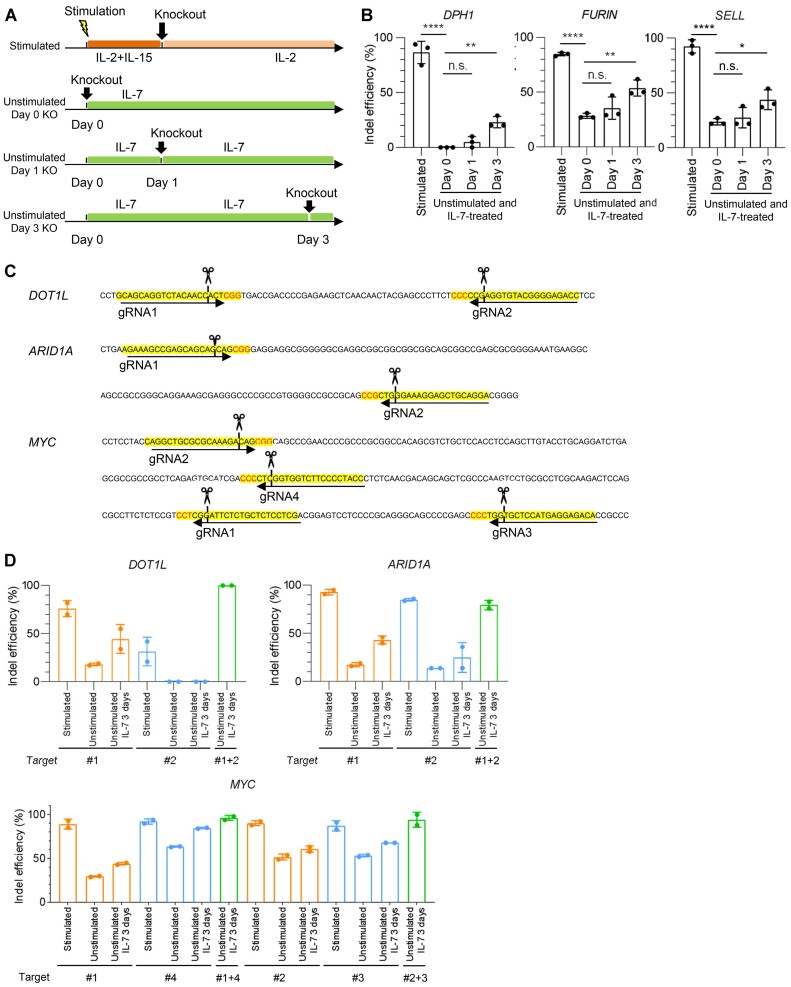
The above data were obtained in activated T cells. Unstimulated naïve T cells are known to be highly resistant to gene knockout (44). We confirmed that the gRNAs that efficiently target DPH1, FURIN and SELL for activated T cells were less efficient when electroporated into T cells soon after thawing (Figure 4A, B). Since T-cell activation inevitably induces naïve T-cell differentiation into memory or effector T cells, the present protocol cannot evaluate the effect of genetic modification in naïve T cells. IL-7 is essential for the maintenance and persistence of naïve and early memory T cells (45). As previously described, IL-7 treatment maintained the naïve T-cell population (CD45RA+ CCR7+ CD62L+) in vitro, which was in contrast to the rapid loss of this subset upon anti-CD3 mAb stimulation (Supplementary Figure S10A–C) (46).
Figure 4.
Gene-editing efficiency is augmented upon IL-7 pretreatment in unstimulated T cells. (A) Schematic diagram of experimental protocols. T cells were electroporated with Cas9/gRNA after stimulation. Alternatively, unstimulated T cells were incubated in the presence of IL-7 for the indicated time periods and electroporated with Cas9/gRNA. (B) The data shown are the estimated indel percentages of gRNAs targeting DPH1, FURIN and SELL for each condition (one-way ANOVA with multiple comparison test; *P < 0.05, **P < 0.01, ****P < 0.0001, n.s., not significant, n = 3 experiments). (C, D) The gRNAs were designed at two proximal regions against DOT1L, ARID1A and MYC. T cells cultured at the indicated conditions were electroporated with single or dual gRNAs. The data shown are the estimated indel percentage for each condition (D, n = 2 experiments).
We examined the influence of IL-7 pre-culture on gene expression profiles of naïve T cells by RNA-sequencing analysis. IL-7-treated T cells showed altered gene expression profiles compared to primary naïve T cells (Supplementary Figure S11A, B). However, gene ontology enrichment analysis of differentially expressed genes between naïve and IL-7-treated T cells demonstrated that IL-7 treatment mainly affected genes associated with cell cycle regulation and DNA repair (Supplementary Figure S11C). IL-7-treated T cells maintained the expression of multiple naïve-associated genes (LEF1, TCF7, KLF2) and did not upregulate effector-associated genes (GZMA, GZMB, and PRF1) (Supplementary Figure S11D). These results suggest that IL-7 treatment at least partly preserved the core gene expression signatures of naïve T cells.
We examined whether preculture with IL-7 affects the gene-editing efficiency of unstimulated T cells (Figure 4A). Intriguingly, the indel percentages were progressively improved after IL-7 treatment in all of the tested genes (Figure 4B). These results suggest that IL-7 pretreatment enables genetic modification of unstimulated T cells. The combinatorial use of IL-7 pretreatment and dual gRNA targeting further improved the gene-editing efficiency in unstimulated T cells (Figure 4C, D). One of the mechanisms for the enhanced gene editing may be related to the efficient entry of RNP complexes into T cells. In fact, IL-7 preculture increased uptake of the fluorescent dye TYE 563-labeled duplex RNA within T cells, suggesting elevated transfection efficiency (Supplementary Figure S12A). In general, cells with a smaller radius need a larger external electric field to achieve permeabilization (47). Naïve T cells become increased in the size of cells after IL-7 preculture, which may partly contribute to the efficient transfection of RNP complexes (Supplementary Figure S12B).
Knockout of PRDM1 in human naïve T cells
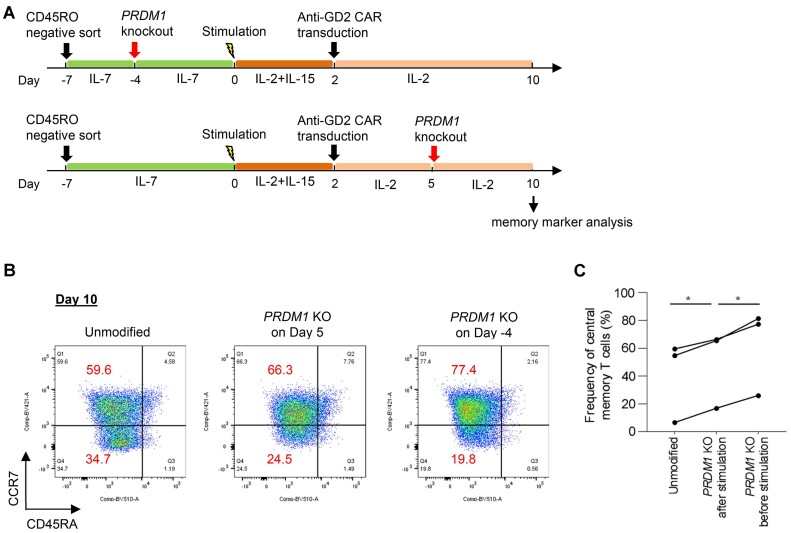
The above strategy enables to ablate a gene before T-cell stimulation and expansion. Although we previously reported that PRDM1 knockout helped to maintain an early memory phenotype in cultured T cells, RNP electroporation was performed around 5 days after stimulation (4), which might have induced precocious T-cell differentiation prior to genetic modification. Knockout at earlier time points may further enhance the effect of PRDM1 disruption. To this end, we enriched naïve T cells by isolating CD45RO-negative cells (Day -7), cultured them without stimulation in the presence of IL-7, and ablated PRDM1 3 days later (Day –4) (Figure 5A; Supplementary Figure S13A, B). T cells were then stimulated and expanded after transduced with anti-GD2 CAR, which was reported to accelerate terminal T-cell differentiation through tonic signaling (48). As comparison, we also knocked out PRDM1 3 days after CAR transduction. Memory marker analysis on Day 10 demonstrated that CAR-T cells ablated with PRDM1 prior to stimulation maintained a central memory phenotype (CD45RA−CCR7+) significantly better than T cells that underwent electroporation after stimulation (Figure 5B, C). These results suggest that genetic modification of T cells prior to activation further reinforces its effect on T cell properties.
Figure 5.
PRDM1 knockout in human naïve T cells enhances the maintenance of central memory CAR-T cells. (A) Naïve T cells were enriched by isolation of CD45RO-negative cells and cultured without stimulation in the presence of IL-7 (Day –7). PRDM1 knockout was performed on Day –4. T cells were then stimulated, transduced with anti-GD2 CAR, and further expanded. For comparison, PRDM1 knockout was performed 5 days after stimulation. (B) Flow cytometry plots analyzing the expression of CCR7 and CD45RA in human CD8+ CAR-T cells on Day 10. (C) The proportion of CD45RA−CCR7+ cells in the CD8+ CAR-T cell population (paired one-way ANOVA with multiple comparison test; *P < 0.05, n = 3 independent experiments).
Epigenetic profiles affect gene-editing efficiency in mesenchymal stem cells
We then investigated if our strategy to use chromatin accessibility data for optimal gene editing can be applicable to other cell types. Mesenchymal stem cells (MSCs) possess pleiotropic immunoregulatory functions and ameliorate severe graft-versus-host disease (49). Since a genetic engineering approach is potentially useful to further enhance their functions (50), we attempted to apply CRISPR/Cas9 gene editing in human bone marrow-derived MSCs (hMSC-BMs). A total of 30 gRNAs targeting three genes (PTEN, KEAP1 and TSC1) were selected, including targets with both high and low ATAC-seq scores calculated based on the data of cultured hMSC-BMs (GSE94272). As shown in Supplementary Figure S14A and B, the ATAC-seq and CHOPCHOP (Doench 2014) scores were significantly correlated with gene editing efficiency in hMSC-BMs (correlation between gene-editing efficiency and scores from other prediction tools are shown in Supplementary Figure S14C–N). We selected two candidate targets for each gene using the workflow constructed in T cells (Figure 2F) and confirmed that all of them achieved sufficient indel percentages (Supplementary Figure S15A). Deletion of PTEN, KEAP1 and TSC1 has been reported to enhance the proliferation of MSC (51–53). We confirmed that the hMSC-BMs ablated with PTEN and KEAP1, but not TSC1, acquired enhanced proliferation compared with the unmodified cells (Supplementary Figure S15B), suggesting that our approach can be exploited in cells other than T cells to modify cellular functions.
Discussion
Several prediction tools to design optimal gRNA targets have been developed. Starting with a concise tool that simply extracts the regions containing PAM sequences, hypothesis-driven tools such as CHOPCHOP have been developed to extract candidate target regions based on DNA sequence features associated with efficient targeting (33,54,55). Thereafter, machine learning-based prediction tools have emerged, including IDT and Azimuth 2.0, which exploit previously accumulated data on the knockout efficiency of individual gRNAs. These methods were further refined along with the innovation of computing technology, which resulted in the development of deep-learning tools, including DeepHF and DeepSpCas9 (13,18).
However, these prediction tools cannot necessarily apply to all experimental settings. First, there are different approaches to CRISPR/Cas9 delivery. Most of the prediction tools were developed based on knockout efficiency data where Cas9 and gRNAs were stably transduced. While transient transfection of Cas9 and/or gRNAs minimizes risk for off-target cleavage (56) and is easily available to primary cells with limited duration of culture, it requires more efficient cleavage compared to a stable expression approach. Some of the prediction tools such as DeepHF and CRISPRedict calculate scores differently according to whether gRNAs are introduced as DNA (expressed under the U6 promoter) or transcribed in vitro by the T7 promoter and transfected into the cell (36,37). These findings suggest that the prediction algorithms established under stable Cas9 and gRNA expression may not always be applicable to the context of transient RNP delivery. The editing efficiency of RNP electroporation can also be affected by multiple factors, such as the parameters of electroporation (voltage and duration), the amount of RNP input, the ratio of Cas9 protein to gRNAs, and the version of Cas9, as previously demonstrated (28).
Second, the knockout efficiency can be significantly affected by cell type. Considering that the majority of the previous data have been obtained using immortalized cell lines, these findings cannot be readily translated to cultured T cells. We examined how precisely these tools can predict the targeting efficiency using our data of human T cells electroporated with a Cas9/gRNA RNP complex. Although some of the tools helped to identify optimal targets, they were still insufficient for accurate prediction. The prediction algorithm provided by IDT was constructed using the data collected through Cas9/gRNA electroporation-mediated knockout, but it did not show a significant correlation in our dataset, suggesting the importance of accumulating T-cell-specific data.
We demonstrated that epigenetic features have a significant impact on gene-editing efficiency. Several groups previously reported that condensed nucleosomes impede Cas9 binding to the target region and inhibit DNA cleavage (21,23). In HEK293T cells, chromatin accessibility in the target locus as measured by DNase I hypersensitivity and histone 3 lysine 4 trimethylation (H3K4me3) deposition was significantly correlated with gene-editing efficiency (41,57).
On the other hand, Barkal et al. reported that Cas9 itself was able to open the closed chromatin region (58). Similar findings were also reported by another group (59). Since these studies introduced Cas9 in different approaches, the findings cannot readily be applicable to T cells electroporated with Cas9/gRNA complexes. In fact, we found that the stable expression of Cas9 and gRNAs effectively targeted epigenetically closed regions (Supplementary Figure S9). These results suggest that chromatin accessibility of the target site is less important when cells constitutively express Cas9 and gRNAs. Leenay et al. explored multiple chromatin features of the target locus including epigenetic profiles that affect the efficiency of indel generation and did not eventually incorporate the chromatin accessibility data in their scoring system (40). While they evaluated indel generation in activated CD4+ T cells, the chromatin accessibility data were obtained from primary T cells or other cell types. Considering that epigenetic profiles are distinct not only among different cell types but also among T cells in different conditions (e.g. memory versus effector cells, primary vs cultured cells) (60,61), the referred epigenetic profiles may have been different from the tested T cells in at least a part of the regions. The unique feature of our study is that we quantified chromatin accessibility at the target locus using ATAC-seq data obtained in T cells in the same conditions (stimulated and cultured CD8+ T cells), which helped to identify optimized targets. A comparison of the gene-editing efficiency between T cells and the K562 cell line showed that the efficiency substantially varied depending on the cell type. These results corroborate the importance of referring to the cell-type specific epigenetic architecture. However, further investigations will be required to explain the data obtained in different platforms in an integrative manner.
Dual gRNA targeting is a promising approach to improve the knockout efficiency. As discussed above, Chen et al. demonstrated that catalytically dead Streptococcus pyogenes Cas9 can alter local chromatin structures and make epigenetically closed sites accessible to the other Cas9 targeting proximal regions (59). Barkal et al. also revealed that Cas9 opens the chromatin region within the surrounding 100 bp (58). These results suggest that using two Cas9/gRNAs targeting close regions may synergistically enhance each other's access to the target site, which was compatible with our data.
Gene editing in naïve T cells rather than stimulated T cells would be important to elucidate the role of genes in effector differentiation and memory formation. However, naïve T cells are highly resistant to gene knockout. We demonstrated that IL-7 preculture enhanced the electroporation efficiency of naïve T cells. Also, the resistance of naïve T cells to CRISPR/Cas9 knockout may be associated with their epigenetic profiles, which differ from those of memory and effector T cells (62). In fact, naïve T cells display a more closed chromatin structure than activated T cells, which can be remodeled via IL-7 treatment (63–65).
In addition to human T cells, human MSCs have been intensely investigated for clinical application against degenerative, inflammatory, and autoimmune diseases. Genetic modification in human MSCs can be a useful strategy to enhance their functions. We confirmed that cell type-specific chromatin accessibility was also related to the gene editing efficiency in hMSC-BMs, and the combination of ATAC-seq scores and CHOPCHOP (Doench 2014) scores further improved the prediction accuracy. Further studies are needed to determine whether our workflow is more generally applicable to other cell types.
In conclusion, we demonstrated the importance of considering epigenetic profiles in selecting optimal CRISPR/Cas9 targets for human T cells. Combining ATAC-seq data and available prediction tools supported the identification of efficient gRNA targets. These findings are highly informative for gene editing in human T cells.
Supplementary Material
Acknowledgements
We thank American Journal Experts (AJE) for English language editing.
Contributor Information
Yusuke Ito, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan.
Satoshi Inoue, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan.
Takahiro Nakashima, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan; Department of Hematology and Oncology, Nagoya City University Graduate School of Medical Sciences, Nagoya, Japan.
Haosong Zhang, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan; Division of Cellular Oncology, Department of Cancer Diagnostics and Therapeutics, Nagoya University Graduate School of Medicine, Nagoya, Japan.
Yang Li, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan; Division of Cellular Oncology, Department of Cancer Diagnostics and Therapeutics, Nagoya University Graduate School of Medicine, Nagoya, Japan.
Hitomi Kasuya, Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan.
Tetsuya Matsukawa, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan; Department of Obstetrics and Gynecology, Nagoya University Graduate School of Medicine, Nagoya, Japan.
Zhiwen Wu, Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan.
Toshiaki Yoshikawa, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan.
Mirei Kataoka, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan.
Tetsuo Ishikawa, Department of Extended Intelligence for Medicine, The Ishii-Ishibashi Laboratory, Keio University School of Medicine, Tokyo, Japan; Advanced Data Science Project, RIKEN Information R&D and Strategy Headquarters, RIKEN, Yokohama, Japan; Collective Intelligence Research Laboratory, Graduate School of Arts and Sciences, The University of Tokyo, Tokyo, Japan.
Yuki Kagoya, Division of Tumor Immunology, Institute for Advanced Medical Research, Keio University School of Medicine, Tokyo, Japan; Division of Immune Response, Aichi Cancer Center Research Institute, Nagoya, Japan; Division of Cellular Oncology, Department of Cancer Diagnostics and Therapeutics, Nagoya University Graduate School of Medicine, Nagoya, Japan.
Data availability
Our ATAC-seq data and RNA-seq data analyzed in this study are publicly available in the Gene Expression Omnibus under accession numbers GSE221788 and GSE237310, respectively. The source code underlying this article is available in Figshare at https://doi.org/10.6084/m9.figshare.24428509.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Japan Agency for Medical Research and Development [JP22bm0704066 to Y.K., JP22ama221303 to Y.K.]; Japan Science and Technology Agency, Fusion Oriented Research for Disruptive Science and Technology program [JPMJFR2060 to Y.K.]; Japan Society for the Promotion of Science, KAKENHI [JP20H03543 to Y.K., JP21K19422 to Y.K., JP22K15575 to Y.I., JP22H02910 to S.I., JP20K21837 to T.I., JP21K02356 to T.I.]; Aichi Cancer Center Joint Research Project on Priority Areas (to Y.K.); Princess Takamatsunomiya Cancer Research Foundation (to Y.K.); Takeda Science Foundation (to Y.K.); The Cell Science Research Foundation (to Y.K.); Uehara Memorial Foundation (to Y.K.); Astellas Foundation for Research on Metabolic Disorders (to Y.K.); SGH Foundation (to Y.K.); Kobayashi Foundation for Cancer Research (to Y.K.); Project Mirai Cancer Research Grants (to Y.K.).
Conflict of interest statement. Y. Kagoya received a commercial research grant from Takara Bio and Kyowa Kirin.
References
- 1. Hou A.J., Chen L.C., Chen Y.Y.. Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug. Discov. 2021; 20:531–550. [DOI] [PubMed] [Google Scholar]
- 2. Shah N.N., Fry T.J.. Mechanisms of resistance to CAR T cell therapy. Nat. Rev. Clin. Oncol. 2019; 16:372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hong M., Clubb J.D., Chen Y.Y.. Engineering CAR-T cells for next-generation cancer therapy. Cancer Cell. 2020; 38:473–488. [DOI] [PubMed] [Google Scholar]
- 4. Yoshikawa T., Wu Z., Inoue S., Kasuya H., Matsushita H., Takahashi Y., Kuroda H., Hosoda W., Suzuki S., Kagoya Y.. Genetic ablation of PRDM1 in antitumor T cells enhances therapeutic efficacy of adoptive immunotherapy. Blood. 2022; 139:2156–2172. [DOI] [PubMed] [Google Scholar]
- 5. Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A.. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017; 7:737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tang N., Cheng C., Zhang X., Qiao M., Li N., Mu W., Wei X.F., Han W., Wang H.. TGF-beta inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI Insight. 2020; 5:e133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prinzing B., Zebley C.C., Petersen C.T., Fan Y., Anido A.A., Yi Z., Nguyen P., Houke H., Bell M., Haydar D.et al.. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci. Transl. Med. 2021; 13:eabh0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janik E., Niemcewicz M., Ceremuga M., Krzowski L., Saluk-Bijak J., Bijak M.. Various aspects of a gene editing system-CRISPR-Cas9. Int. J. Mol. Sci. 2020; 21:9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y., Wang X.. Evaluation of efficiency prediction algorithms and development of ensemble model for CRISPR/Cas9 gRNA selection. Bioinformatics. 2022; 38:5175–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elkayam S., Orenstein Y.. DeepCRISTL: deep transfer learning to predict CRISPR/Cas9 functional and endogenous on-target editing efficiency. Bioinformatics. 2022; 38:i161–i168. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Kim H.K., Min S., Song M., Jung S., Choi J.W., Kim Y., Lee S., Yoon S., Kim H.H.. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol. 2018; 36:239–241. [DOI] [PubMed] [Google Scholar]
- 13. Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D., Zhou C., Zhu C., Chen K., Duan B.et al.. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018; 19:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xue L., Tang B., Chen W., Luo J.. Prediction of CRISPR sgRNA activity using a deep convolutional neural network. J. Chem. Inf. Model. 2019; 59:615–624. [DOI] [PubMed] [Google Scholar]
- 15. Wang J., Zhang X., Cheng L., Luo Y.. An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol. 2020; 17:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atsavapranee E.S., Billingsley M.M., Mitchell M.J.. Delivery technologies for T cell gene editing: applications in cancer immunotherapy. EBioMedicine. 2021; 67:103354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B.et al.. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015; 33:985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Konstantakos V., Nentidis A., Krithara A., Paliouras G.. CRISPR-Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning. Nucleic Acids Res. 2022; 50:3616–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacobi A.M., Rettig G.R., Turk R., Collingwood M.A., Zeiner S.A., Quadros R.M., Harms D.W., Bonthuis P.J., Gregg C., Ohtsuka M.et al.. Simplified CRISPR tools for efficient genome editing and streamlined protocols for their delivery into mammalian cells and mouse zygotes. Methods. 2017; 121-122:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kagoya Y., Guo T., Yeung B., Saso K., Anczurowski M., Wang C.H., Murata K., Sugata K., Saijo H., Matsunaga Y.et al.. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to Be used for adoptive T-cell therapy. Cancer Immunol. Res. 2020; 8:926–936. [DOI] [PubMed] [Google Scholar]
- 21. Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E., Torigoe S.E., Tjian R., Weissman J.S.. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016; 5:e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh R., Kuscu C., Quinlan A., Qi Y., Adli M.. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015; 43:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:9351–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicholson I.C., Lenton K.A., Little D.J., Decorso T., Lee F.T., Scott A.M., Zola H., Hohmann A.W.. Construction and characterisation of a functional CD19 specific single chain fv fragment for immunotherapy of B lineage leukaemia and lymphoma. Mol. Immunol. 1997; 34:1157–1165. [DOI] [PubMed] [Google Scholar]
- 25. Richman S.A., Nunez-Cruz S., Moghimi B., Li L.Z., Gershenson Z.T., Mourelatos Z., Barrett D.M., Grupp S.A., Milone M.C.. High-affinity GD2-specific CAR T cells induce fatal encephalitis in a preclinical neuroblastoma model. Cancer Immunol. Res. 2018; 6:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conant D., Hsiau T., Rossi N., Oki J., Maures T., Waite K., Yang J., Joshi S., Kelso R., Holden K.et al.. Inference of CRISPR edits from Sanger trace data. CRISPR J. 2022; 5:123–130. [DOI] [PubMed] [Google Scholar]
- 27. Ge S.X., Son E.W., Yao R.. iDEP: an integrated web application for differential expression and pathway analysis of RNA-seq data. BMC Bioinf. 2018; 19:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seki A., Rutz S.. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 2018; 215:985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E.. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014; 42:W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E.. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019; 47:W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R.et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016; 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E.. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu H., Xiao T., Chen C.H., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S.et al.. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015; 25:1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J.. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015; 12:982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim H.K., Kim Y., Lee S., Min S., Bae J.Y., Choi J.W., Park J., Jung D., Yoon S., Kim H.H.. SpCas9 activity prediction by DeepSpCas9, a deep learning-based model with high generalization performance. Sci. Adv. 2019; 5:eaax9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang D., Zhang C., Wang B., Li B., Wang Q., Liu D., Wang H., Zhou Y., Shi L., Lan F.et al.. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 2019; 10:4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Konstantakos V., Nentidis A., Krithara A., Paliouras G.. CRISPRedict: a CRISPR-Cas9 web tool for interpretable efficiency predictions. Nucleic Acids Res. 2022; 50:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michlits G., Jude J., Hinterndorfer M., de Almeida M., Vainorius G., Hubmann M., Neumann T., Schleiffer A., Burkard T.R., Fellner M.et al.. Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat. Methods. 2020; 17:708–716. [DOI] [PubMed] [Google Scholar]
- 39. DeWeirdt P.C., McGee A.V., Zheng F., Nwolah I., Hegde M., Doench J.G.. Accounting for small variations in the tracrRNA sequence improves sgRNA activity predictions for CRISPR screening. Nat. Commun. 2022; 13:5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leenay R.T., Aghazadeh A., Hiatt J., Tse D., Roth T.L., Apathy R., Shifrut E., Hultquist J.F., Krogan N., Wu Z.et al.. Large dataset enables prediction of repair after CRISPR-Cas9 editing in primary T cells. Nat. Biotechnol. 2019; 37:1034–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jensen K.T., Floe L., Petersen T.S., Huang J., Xu F., Bolund L., Luo Y., Lin L.. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 2017; 591:1892–1901. [DOI] [PubMed] [Google Scholar]
- 42. Liu B., Chen S., Rose A., Chen D., Cao F., Zwinderman M., Kiemel D., Aissi M., Dekker F.J., Haisma H.J.. Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res. 2020; 48:517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valdez B.C., Brammer J.E., Li Y., Murray D., Liu Y., Hosing C., Nieto Y., Champlin R.E., Andersson B.S.. Romidepsin targets multiple survival signaling pathways in malignant T cells. Blood Cancer J. 2015; 5:e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nussing S., House I.G., Kearney C.J., Chen A.X.Y., Vervoort S.J., Beavis P.A., Oliaro J., Johnstone R.W., Trapani J.A., Parish I.A.. Efficient CRISPR/Cas9 gene editing in uncultured naive mouse T cells for In vivo studies. J. Immunol. 2020; 204:2308–2315. [DOI] [PubMed] [Google Scholar]
- 45. Bradley L.M., Haynes L., Swain S.L.. IL-7: maintaining T-cell memory and achieving homeostasis. Trends Immunol. 2005; 26:172–176. [DOI] [PubMed] [Google Scholar]
- 46. Gattinoni L., Speiser D.E., Lichterfeld M., Bonini C.. T memory stem cells in health and disease. Nat. Med. 2017; 23:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gehl J. Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol. Scand. 2003; 177:437–447. [DOI] [PubMed] [Google Scholar]
- 48. Long A.H., Haso W.M., Shern J.F., Wanhainen K.M., Murgai M., Ingaramo M., Smith J.P., Walker A.J., Kohler M.E., Venkateshwara V.R.et al.. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 2015; 21:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ringden O., Uzunel M., Rasmusson I., Remberger M., Sundberg B., Lonnies H., Marschall H.U., Dlugosz A., Szakos A., Hassan Z.et al.. Mesenchymal stem cells for treatment of therapy-resistant graft-versus-host disease. Transplantation. 2006; 81:1390–1397. [DOI] [PubMed] [Google Scholar]
- 50. Hodgkinson C.P., Gomez J.A., Mirotsou M., Dzau V.J.. Genetic engineering of mesenchymal stem cells and its application in human disease therapy. Hum. Gene Ther. 2010; 21:1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shen Y., Zhang J., Yu T., Qi C.. Generation of PTEN knockout bone marrow mesenchymal stem cell lines by CRISPR/Cas9-mediated genome editing. Cytotechnology. 2018; 70:783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hazrati A., Malekpour K., Soudi S., Hashemi S.M.. CRISPR/Cas9-engineered mesenchymal stromal/stem cells and their extracellular vesicles: a new approach to overcoming cell therapy limitations. Biomed. Pharmacother. 2022; 156:113943. [DOI] [PubMed] [Google Scholar]
- 53. Guijarro M.V., Danielson L.S., Canamero M., Nawab A., Abrahan C., Hernando E., Palmer G.D.. Tsc1 Regulates the proliferation capacity of bone-marrow derived mesenchymal stem cells. Cells. 2020; 9:2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Corsi G.I., Qu K., Alkan F., Pan X., Luo Y., Gorodkin J.. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat. Commun. 2022; 13:3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Labuhn M., Adams F.F., Ng M., Knoess S., Schambach A., Charpentier E.M., Schwarzer A., Mateo J.L., Klusmann J.H., Heckl D. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 2018; 46:1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith R.H., Chen Y.C., Seifuddin F., Hupalo D., Alba C., Reger R., Tian X., Araki D., Dalgard C.L., Childs R.W.et al.. Genome-wide analysis of off-target CRISPR/Cas9 activity in single-cell-derived Human hematopoietic stem and progenitor cell clones. Genes (Basel). 2020; 11:1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chari R., Mali P., Moosburner M., Church G.M.. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015; 12:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barkal A.A., Srinivasan S., Hashimoto T., Gifford D.K., Sherwood R.I.. Cas9 Functionally opens chromatin. PLoS One. 2016; 11:e0152683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen F., Ding X., Feng Y., Seebeck T., Jiang Y., Davis G.D.. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 2017; 8:14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crompton J.G., Narayanan M., Cuddapah S., Roychoudhuri R., Ji Y., Yang W., Patel S.J., Sukumar M., Palmer D.C., Peng W.et al.. Lineage relationship of CD8(+) T cell subsets is revealed by progressive changes in the epigenetic landscape. Cell. Mol. Immunol. 2016; 13:502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nestor C.E., Ottaviano R., Reinhardt D., Cruickshanks H.A., Mjoseng H.K., McPherson R.C., Lentini A., Thomson J.P., Dunican D.S., Pennings S.et al.. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015; 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rezalotfi A., Fritz L., Forster R., Bosnjak B.. Challenges of CRISPR-based gene editing in primary T cells. Int. J. Mol. Sci. 2022; 23:1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Iqbal M.M., Serralha M., Kaur P., Martino D. Mapping the landscape of chromatin dynamics during naive CD4+ T-cell activation. Sci. Rep. 2021; 11:14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Scott-Browne J.P., Lopez-Moyado I.F., Trifari S., Wong V., Chavez L., Rao A., Pereira R.M.. Dynamic changes in chromatin accessibility occur in CD8(+) T cells responding to viral infection. Immunity. 2016; 45:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Frumento G., Verma K., Croft W., White A., Zuo J., Nagy Z., Kissane S., Anderson G., Moss P., Chen F.E.. Homeostatic cytokines drive epigenetic reprogramming of activated T cells into a “naive-memory” phenotype. Iscience. 2020; 23:100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our ATAC-seq data and RNA-seq data analyzed in this study are publicly available in the Gene Expression Omnibus under accession numbers GSE221788 and GSE237310, respectively. The source code underlying this article is available in Figshare at https://doi.org/10.6084/m9.figshare.24428509.