Abstract
Aberrant DNA/RNA hybrids (R-loops) formed during transcription and replication disturbances pose threats to genome stability. DHX9 is an RNA helicase involved in R-loop resolution, but how DHX9 is regulated in response to genotoxic stress remains unclear. Here we report that DHX9 is phosphorylated at S321 and S688, with S321 phosphorylation primarily induced by ATR after DNA damage. Phosphorylation of DHX9 at S321 promotes its interaction with γH2AX, BRCA1 and RPA, and is required for its association with R-loops under genotoxic stress. Inhibition of ATR or expression of the non-phosphorylatable DHX9S321A prevents DHX9 from interacting with RPA and R-loops, leading to the accumulation of stress-induced R-loops. Furthermore, depletion of RPA reduces the association between DHX9 and γH2AX, and in vitro binding analysis confirms a direct interaction between DHX9 and RPA. Notably, cells with the non-phosphorylatable DHX9S321A variant exhibit hypersensitivity to genotoxic stress, while those expressing the phosphomimetic DHX9S321D variant prevent R-loop accumulation and display resistance to DNA damage agents. In summary, we uncover a new mechanism by which ATR directly regulates DHX9 through phosphorylation to eliminate stress-induced R-loops.
Graphical Abstract
Graphical Abstract.
Introduction
The DNA damage response (DDR) is an intricate and crucial system designed to safeguard the integrity of the contents and organization of genetic materials by countering both external and internal threats. Central to this defense mechanism are three structurally related phosphoinositide 3-kinase (PI3K)-related kinases (PIKKs), ATM, ATR and DNA-PKcs, which are activated by DNA damage (1,2). While ATM and DNA-PKcs primarily respond to and repair DNA double-stranded breaks (DSBs), ATR is activated by a broad range of DNA lesions that induce stalled or terminated DNA synthesis, termed replication stress. Like many cellular signaling cascades, DNA damage signaling is conducted by a series of protein phosphorylation events (1). While DNA-PKcs primarily modulates proteins involved in DNA repair through non-homologous end joining (NHEJ) (3), ATM and ATR act as master regulators of cellular responses to DNA double-strand breaks (DSBs) and replication stress, respectively (1,2). ATM and ATR phosphorylate a multitude of target proteins, coordinating various responses, including DNA damage signaling, cell cycle control, apoptosis and DNA repair mechanisms to effectively deal with DNA damage. ATM, in conjunction with its regulators MRE11-Rad50-NBS1 (MRN), is recruited to DNA DSBs, undergoes activation and phosphorylates H2AX at Ser 139 and Chk2 at Thr 68, thereby activating the Chk2-regulated cell cycle checkpoint (1,4). Conversely, ATR, along with ATRIP, is recruited to single-stranded DNA (ssDNA) structures coated with replication protein A (RPA) and becomes fully activated with the assistance of co-regulators such as TopBP1, ETAA1, Rad17 and the Rad9-Rad1-Hus1 (9-1-1) complex. Activated ATR subsequently phosphorylates Chk1 at Ser 317 and Ser 345, resulting in the activation of Chk1-mediated cell cycle checkpoints (1,5). In addition to Chk1 and Chk2, a large number of proteins, including BRCA1, 53BP1, MDC1, RPA32, NBS1 and CtIP, localized to sites of DNA damage, are phosphorylated by either ATM or ATR to facilitate DNA repair (2,6,7). Previous large-scale studies have identified numerous proteins that undergo phosphorylation by ATM/ATR after DNA damage (7–10). However, the full understanding of their specific functions following phosphorylation needs to be further explored.
In addition to external threats that induce damage to DNA, the occurrence of unscheduled R-loops resulting from interference during DNA replication and transcription is a major cause of DNA damage and genome instability (11–15). Typically, the R-loop is a nucleic acid structure of DNA/RNA hybrid with a displaced single-stranded DNA transiently formed during RNA transcription (11–13,16,17). R-loop structures are naturally formed in organisms from bacteria to humans and play significant roles in various cellular processes (14,17). In human cells, R-loops facilitate class switching when formed over switch regions of the immunoglobulin locus (18), and promote efficient transcriptional termination when formed at the termination regions of genes (19). Initially considered harmless within cells, accumulating evidence now suggests that the persistence of R-loops detrimentally affects genome integrity. One substantial indication of R-loops causing DNA damage is the depletion of RNase H1 (RNH1), an enzyme responsible for degrading the RNA strand in DNA/RNA hybrids (20,21). This removal leads to the accumulation of nuclear foci composed of DDR proteins and subsequent rearrangement of the genome. Moreover, when RNH1 is overexpressed in cells harboring mutations that promote R-loops, it effectively represses genomic instability associated with increased R-loop formation (22–24). A recent study further confirms that RPA complex enhances the association between RNH1 and R-loops and stimulates RNH1 activity in vitro (22). In addition to RNH1, several RNA binding proteins and RNA processing factors, such as splicing factors and RNA helicases, possess functions that suppress R-loop formation (11). Proteins involved in DNA repair and the Fanconi Anemia pathway, including BRCA1, BRCA2, FANCD2 and FANCM, are also required for resolving R-loops (25–30). Earlier investigations have highlighted the role of Senataxin (SETX), a nuclear protein that resolves R-loops to facilitate transcription termination and during the conflict of DNA replication and transcription in yeast and mammalian cells (19,31,32). Recent data show that targeting SETX to R-loop requires BRCA1 (27). Additionally, Aquarius (AQR), an RNA helicase, has been implicated in R-loop resolution through the unwinding of DNA/RNA hybrids (33). Notably, emerging evidence demonstrates that a specific group of RNA helicases, such as DDX17, DDX19 and DDX21 with DEAD-box motifs, or DHX9 with a DExH-box motif, actively participate in unwinding DNA/RNA hybrids and effectively suppress the accumulation of R-loops at specific genomic locations (34–37). However, not all R-loop suppressors are well-studied for their regulatory mechanisms.
DHX9, also known as nuclear DNA helicase II (DNH II) and RNA helicase A (RHA), is an evolutionarily conserved DExH-box RNA helicase (38). Like many other R-loop suppressors that accompany the transcription complex during transcription, DHX9 interacts with RNA polymerase II and contributes to the co-transcriptional processing of pre-mRNAs (39,40). Biochemical studies have confirmed that DHX9 possesses helicase activity and binds to single-stranded DNA and RNA, similar to BLM and WRN, to resolve abnormal secondary structures of nucleic acids that arise during DNA replication and transcription (41,42). These structures include DNA/RNA hybrids, D-loops, as well as RNA and DNA guanine quadruplexes (G4-DNA/RNA), all of which are potential sources of genomic instability. Recent investigations have shed light on the crucial role of DHX9 in RNA processing and genome stability. DHX9 also interacts with DDR proteins such as BRCA1, γH2AX and NHEJ factors, including DNA-PKcs and Ku70 (43,44), implying its involvement in DNA repair processes. The study of the DNA/RNA hybrid interactome has demonstrated that DHX9 interacts with PARP1, which is critical in repairing single-stranded breaks and base excision repair. Depletion of DHX9 leads to the accumulation of R-loops in response to camptothecin (CPT), an inhibitor of Topoisomerase I. Both DHX9 and PARP1 are necessary to prevent R-loop-associated DNA damage (35). Furthermore, recent research indicates that loss of DHX9 impairs the ATR-Chk1 signaling, and DHX9 is implicated in the homologous recombination (HR) repair of DNA DSBs by recruiting BRCA1 to DSBs and promoting resection (43). Nevertheless, whether DHX9 is bound to DSBs in cells and how DHX9 is specifically regulated in response to genotoxic stress to resolve R-loops is not understood.
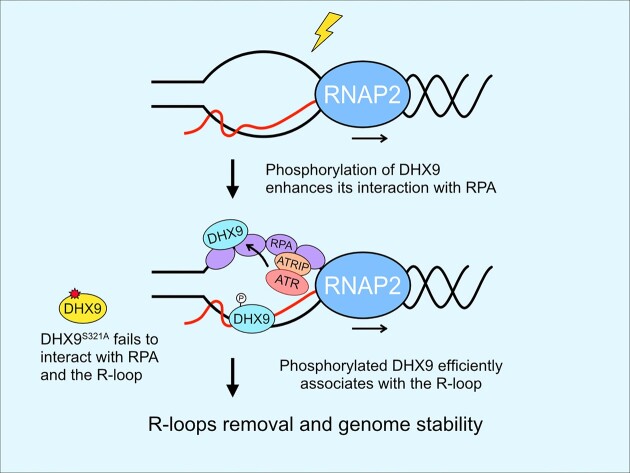
Here, we uncover that ATR phosphorylates DHX9 to promote R-loops resolution during genotoxic stress. DHX9 is phosphorylated at Ser 321 and Ser 688, with Ser 321 phosphorylation being induced by ATR in response to DNA damage. DHX9 phosphorylation at S321 promotes its interaction with γH2AX, BRCA1 and RPA32 and its association with R-loops in response to DNA damage. More importantly, when DHX9 phosphorylation at S321 is compromised, cells display increased R-loops and are hypersensitive to genotoxic drugs. Conversely, a phosphomimetic mutant prevents the accumulation of unscheduled R-loops and restores viability. These results establish a connection between ATR, RPA and DHX9 in responding to DNA damage-induced R-loops, highlighting that ATR directly regulates RNA helicase DHX9 to prevent genome instability.
Materials and methods
Cell culture and inhibitors
HeLa and U2OS cells were obtained from the ATCC and cultured in Dulbecco's modified Eagle's medium (DMEM, ThermoFisher) supplemented with l-glutamine and 10% fetal bovine serum (FBS, ThermoFisher) and maintained at 37°C with 5% CO2. All cells were routinely monitored for mycoplasma contamination using MycoAlert PLUS Mycoplasma detection assay (LT07-710, Lonza). The following inhibitors were used where indicated. KU55933, NU7441, VE822, CPT, olaparib, doxorubicin, cisplatin and gemcitabine (purchased from Selleckchem). AZD6738 (MedChemExpress); and caffeine and hydroxyurea (HU) (Sigma-Aldrich) were kindly provided by Dr Jing-Jer Lin.
Plasmids and inducible cell lines
To clone the full-length cDNA of DHX9 into an ENTRY vector for Gateway cloning, human DHX9 cDNA was amplified using Nested PCR. Nested PCR involved two sequential PCR reactions using different sets of primers. The amplified full-length DHX9 cDNA was cloned into the pENTRY-DHX9 vector using the pENTR/D-TOPO cloning kit (45-0218, ThermoFisher). To generate a siDHX9-1 resistant construct, silent mutations were introduced into the pENTRY-DHX9 vector without altering the protein sequences. The siDHX9-1 resistant vector, designated as pENTRY-DHX9-siR, was subsequently used for site-directed mutagenesis to substitute S321 and S688 on DHX9 with alanines or aspartic acid using PCR and a KLD enzyme mix (M0554, NEB) following the manufacturer's protocol. The primers used are listed below: DHX9 Nested PCR primers, forward (5′-GAGTCACACACGGTCCTAAG-3′) and reverse (5′-AACAAACAAACTACACGGCA-3′). DHX9 entry clone primers, forward (5′-CACCATGGGTGACGTTAAAAATTTTCTG-3′) and reverse (5′-TTAATAGCCGCCACCTCCTCTTC-3′). DHX9 siRNA resistant primers, forward (5′-GTCGTAACTCAGCCCAGAAGAATC-3′) and reverse (5′-AATATTACACTCTGCTGCTCGG-3′). S321A primers, forward (5′-GTTCGAACCAGCTCAGCGACA-3′) and reverse (5′-TGAGCCAATTTGCCAATGTTG-3′). S321D primers, forward (5′-GTTCGAACCAGATCAGCGACAAAAC-3′) and reverse (5′-TGAGCCAATTTGCCAATG-3′). S688A primers, forward (5′-ACCCCTGCATGCTCAGATTCC-3′) and reverse (5′-AGAATCTGATACCGATGGC-3′).
To construct different SFB-fused DHX9 variants, the individual pENTRY-DHX9 clone was subjected to the LR reaction with the destination vector containing the SFB tag using Gateway LR Clonase II enzyme mix (11791020, ThermoFisher). To generate TetOn-regulated SFB-DHX9, desired SFB-DHX9 fragments were obtained by PCR using primers (forward 5′-ATCCGCTAGCGCTACCGGA-3′ and reverse 5′-TAACCGGTTTAATAGCCGCCACCTCCTC-3′). The PCR product of SFB-DHX9 fusions contained a NheI site at the N-terminus and an AgeI site at the C-terminus. The NheI and AgeI-digested PCR fragment was then ligated into the pTetOn3G vector (from Dr. Hsueh-Ping Catherine Chu). To establish inducible cell lines, HeLa cells were transfected with linearized pTetOn3G-SFB-DHX9 vectors or the corresponding empty vector followed by hygromycin (250 μg/ml, ThermoFisher) selection for up to 12 days. Each candidate clone was verified by Western blotting.
Small interfering RNAs (siRNAs) and transfection
For silencing DHX9, RPA32 and ATR, pre-designed Silencer Select siRNA targeting DHX9 (DHX9-1, s4019 and DHX9-2, s4020), RPA32 (s12132) and ATR (s536) and negative control #1 siRNA (4390843) were purchased from ThermoFisher. Typically, 3 nM of siRNA was used for transfection in HeLa and U2OS cells using Lipofectamine RNAiMax (13778150, ThermoFisher) according to the manufacturer's protocol. Unless otherwise specified, cells were subjected to various downstream experiments after 64 h. For transient expression of different SFB-DHX9 fusions in cells, Lipofectamine 2000 (11668019, ThermoFisher) was particularly used for complementary experiments, while polyethyleneimine (PEI) (Sigma-Aldrich) was used for the rest experiments. The sequences of siRNAs are as follows: siDHX9-1 (GAGUGUAACAUCGUAGUAAdTdT), siDHX9-2 (CCCUGUCACUUGUCAGACAdTdT), ATR (UUGUAGAAAUGGAUACUGAdTdT) and RPA32 (GCACCUUCUCAAGCCGAAAdTdT).
Antibodies
The antibodies were used where indicated. pChk1 S317(#12302), pChk1 S345 (#2348), pChk2(#2661), Chk2 (#3440), [pS/pT] QG (#6966) and γH2AX (#9718) were obtained from Cell Signaling Technology; ATR (A300-138A), ATM (A300-299A), Rad17 pS645 (A300-153A), RPA32 (A300-244A), pRPA32 S33 (A300-246A) and pRPA32 S4/8 (A300-245A) antibodies were from Bethyl Lab; BRCA1 (sc-6954), DHX9 (sc-137232), FLAG (sc-166355), Chk1 (sc-8408), PARP1 (sc-8007), PRP19 (sc-514338), PCNA (sc-56) and GAPDH (sc-47724) antibodies from Santa Cruz; DHX9 (ab26271) were purchased from abcam; FLAG (F7425), γH2AX (05–636), α-Tubulin (T6074) and S9.6 (MABE1095) were purchased from Merck; RPA32 (MA1-26418), goat anti-mouse-HRP (G21040), goat anti-rabbit-HRP (G21234), goat anti-mouse Alexa Fluor-594 (A-11005) and goat anti-rabbit Alexa Fluor-488 (A-11034) were from ThermoFisher; pATR (GTX128145), pATM (GTX132146), RPA70 (GTX108749) were obtained from GeneTex; HRP conjugated anti-mouse (115-035-174) and anti-rabbit (211-032-171), light chain specific antibodies were obtained from Jackson ImmunoResearch; affinity-purified DHX9 pS321 and pS688 were customized by GeneTex.
Immunoprecipitation (IP), mass spectrometry and Western blotting
To identify the phosphorylation sites on DHX9, HeLa cells treated with UV or left untreated were directly lysed on culture dishes in 1× sample buffer containing 1× protease and phosphatase inhibitor cocktails (78440, ThermoFisher). The lysates were briefly sonicated, followed by denaturing the lysates at 95°C for 10 min. Debris was removed by centrifugation at 16000 × g for 10 minutes. The resulting supernatants were diluted 10-fold in NETN buffer (20 mM Tris–HCl pH 8.0, 0.5 mM EDTA, 150 mM NaCl, 0.5% Igepal, 10% glycerol, 5 mM NaF) and mixed with either mouse IgG (sc-2025, Santa Cruz) or anti-DHX9 (sc-137232, Santa Cruz) at 4°C for overnight to IP endogenous DHX9. The next day 30 μl of protein G-conjugated magnetic beads (10004D, ThermoFisher) were added to the overnight mixture and incubated for 1 h. Following IP, the beads were washed three times with NETN buffer, and the captured proteins were lysed in 2× SDS sample buffer (100 mM Tris–HCl, 2% SDS, 200 mM DTT, 0.04% Bromophenol blue and 20% glycerol) for subsequent assessment by Western blot and mass spectrometry.
For the mass spectrometry analysis, the affinity-purified DHX9 was separated by SDS-PAGE and subjected to in-gel digestion by trypsin. The resulting peptides were purified and analyzed by LC–MS/MS. Mass spectrometry data were acquired at the Common Mass Spectrometry Facilities for Proteomics and Protein Modification Analysis, Institute of Biological Chemistry, Academia Sinica, Taiwan. To determine the phosphorylation of SFB-DHX9 (WT, S321A and AA) by anti-pS/TQ antibody, HeLa cells were transfected with SFB-DHX9 plasmids by PEI. Two days later, cell lysis and IP were performed the same as described above, except for the primary antibody/beads replaced with the anti-DYKDDDDK (anti-FLAG) magnetic agarose beads (A36798, ThermoFisher). Following IP, the beads were washed three times with NETN buffer, and the captured proteins were lysed in 2× SDS sample buffer for subsequent assessment by Western blot.
To determine the interaction between endogenous DHX9, SFB-DHX9 and RPA proteins in cells after DNA damage, nuclear fractions from cells were collected using cellular fractionations as previously described (45). In brief, the nuclear fractions were resuspended in NETN buffer supplemented with 1× protease and phosphatase inhibitor cocktails (ThermoFisher), sonicated and incubated with or without Universal Nuclease (88700, ThermoFisher) for 30 min at room temperature, followed by centrifugation at 4°C, 16000 × g for 10 min. The resulting supernatants were divided into two parts, with one for the IP of SFB-DHX9 proteins using anti-FLAG magnetic agarose beads and the other for the IP of endogenous RPA32 using anti-RPA32 antibody and protein G-conjugated magnetic beads. After overnight incubation at 4°C, the beads were washed three times with NETN buffer, and the captured proteins were eluted in 2× SDS sample buffer following the final wash.
For western blotting, cell lysates in 2× sample buffer were subjected to electrophoresis by SDS-PAGE and transferred onto PVDF membranes (Immobilon-P, Millipore). Membranes were blocked in TBS-T (1× TBS, 0.1% Tween-20) containing 5% skim milk for 30 min, followed by incubation with primary antibodies for 2 h at room temperature or overnight at 4°C. After the incubation, membranes were washed three times with TBS-T, incubated with secondary antibodies at room temperature for 1 h and washed three more times in TBS-T. The signals were visualized using chemiluminescence reagents from ThermoFisher (#34580) and Bio-Rad (#170562) and detected by the ChemiDoc imaging system (Bio-Rad).
In vitro DHX9 and RPA binding assay
To explore a direct interaction between DHX9 and the RPA complex, an in vitro binding assay was performed. HeLa cells transfected with SFB-DHX9 for 30 h were treated with CPT (1 μM, 1 h), HU (4 mM, 2 h), or left untreated. Cells were lysed in NETN buffer supplemented with 1× protease and phosphatase inhibitors. After brief sonication, lysates were incubated with Universal Nuclease (40 U per sample) at room temperature for 15 min before centrifugation at 16000 × g for 10 min at 4°C. The resulting supernatants were used for IP of SFB-DHX9 proteins with anti-FLAG magnetic agarose beads. After IP, beads were washed sequentially with NETN buffers containing increasing salt concentrations (150, 300, 500 and 750 mM), and then in decreasing order. The beads were then resuspended in 100 μl NETN buffer and subjected to SDS-PAGE/silver staining (24612, ThermoFisher) and Western blotting to confirm the purification of SFB-DHX9 and ensure no residual RPA and other DHX9 interacting proteins before the in vitro RPA binding assay.
To assess the direct interaction between DHX9 and RPA, 10 μl and 30 μl of bead-bound SFB-DHX9 (approximately 80 and 240 ng of SFB-DHX9, respectively) were incubated with 100 ng of the bacterially purified recombinant RPA complex (RPA70-32-14) (46) at 4°C for 1 h. After incubation, the beads were washed in regular NETN buffer. The samples were assessed by Western blot with indicated antibodies.
Immunofluorescence staining of PCNA and DHX9 pS321
HeLa cells on coverslips treated with DMSO or CPT were permeabilized with 0.5% Triton X-100 in PBS for 3 min on ice, washed with PBS, fixed in 100% ice-cold methanol at –20°C for 15 min, followed by PBS wash. Cells were incubated with a blocking solution (3% BSA and 0.05% Tween20 in PBS) at room temperature for 30 min. The fixed cells were then incubated with anti-PCNA and anti-DHX9 pS321 antibodies diluted in blocking solution at room temperature. Following incubation, coverslips were washed 3 × 5 min with PBST (0.05% Tween20 in PBS) and incubated with anti-mouse Alexa Fluor-594 and anti-rabbit Alexa Fluor-488 secondary antibodies at room temperature in the dark for 1 h. Nuclei were counterstained with DAPI (D9542, Sigma-Aldrich) for 10 min, and coverslips were mounted using ProLong Gold Antifade Mountant (P36930, ThermoFisher), imaged with Olympus IX83 inverted microscopy.
Proximity ligation assay (PLA)
In situ PLA was performed using the Duolink PLA kit (Merck). Cells grown on round cover glasses (Φ15 mm) were pre-extracted with PBS containing 0.5% Triton X-100 on ice for 5 min, followed by the fixation in 4% paraformaldehyde/PBS for 15 min at room temperature plus an additional 2 min in PBS-0.5% Triton X-100. The coverslips were incubated in a blocking solution (DUO82007) for 1 h at 37°C. After blocking, primary antibodies were applied to samples for overnight incubation at 4°C. The next day, samples were washed twice and incubated with Duolink PLA probes (anti-rabbit minus (DUO92005) and anti-mouse plus (DUO92001)) for 1 h at 37°C. Subsequent ligation and amplification were carried out using the Detection Reagents Red (DUO92008) following the manufacturer's instructions. Nuclei were stained with DAPI, and images were collected using the EVOS M7000 microscope (ThermoFisher). The antibodies used in PLA are listed as follows: DHX9 (ab26271, abcam), FLAG (F7425, Merck), S9.6 (MABE1095, Merck), RPA32 (MA1-26418, ThermoFisher), γH2AX (05–636, Merck), BRCA1 (sc6954, Santa Cruz).
DNA/RNA hybrids immunoprecipitation (DRIP)-qPCR
To determine whether DHX9 phosphorylation is involved in the suppression of R-loops under genotoxic stress, DRIP was performed with some modifications to the protocol as previously described (47). HeLa cells transfected with control and DHX9 siRNAs for 24 h were introduced with SFB-DHX9 variants (WT, S321A and S321D) using lipofectamine 2000. Two days later, cells were treated with DMSO or CPT (10 μM) for 1 h, harvested by trypsinization, washed with ice-cold 1× PBS and centrifuged at 300 × g for 4 min. The resulting cell pellets were resuspended in lysis buffer (5 mM PIPES pH 7.4, 80 mM KCl and 0.5% Igepal) at 4°C for 30 min and centrifuged at 1000 × g at 4°C for 10 min to obtain the nuclei pellets. The nuclei pellets were then resuspended in the nuclear lysis buffer (25 mM Tris–HCl pH 8.0, 5 mM EDTA and 1% SDS), incubated on ice for 30 min, followed by proteinase K (60 μg) digestion at 55°C for 3 h. Nucleic acids were extracted using phenol/chloroform/isoamyl alcohol (25:24:1), applied to tubes containing phase-lock gel, and centrifuged at 1500 × g for 10 min. The aqueous phase obtained was mixed with 1/10 volume of 3M sodium acetate (pH 5.2) and 2 volumes of ice-cold 100% ethanol for DNA precipitation.
The purified DNA pellets were resuspended in elution buffer and sonicated using an ultrasonicator (Covaris) to generate DNA fragments ranging from 200 to 500 bp. Fragment size was confirmed by agarose gel electrophoresis. To IP DNA/RNA hybrids, 5 μg of sheared nucleic acid was diluted in 1× DRIP binding buffer (10 mM sodium phosphate, pH 7.0, 0.14 M NaCl and 0.05% Triton X-100) and treated with or without RNH (M0297, NEB) at 37°C overnight before IP using 2 μg of S9.6 antibody. The mixture was incubated overnight at 4°C and then bound to protein G dynabeads (ThermoFisher) for 2 h. Dynabeads were subsequently washed and incubated with 140 μg of proteinase K in elution buffer (50 mM Tris, pH 8.0, 10 mM EDTA, 0.5% SDS) at 55°C for 45 min to separate captured DNA/RNA hybrids. The eluates were extracted with phenol/chloroform/isoamyl alcohol and DNA was harvested using an ethanol precipitation protocol. To assess the enrichment of R-loops across the β-actin and γ-actin genes, the purified DNA was utilized for RT-qPCR using specific primer sets as follows: β-actin gene in1 (forward 5′-CGGGGTCTTTGTCTGAGC-3′ and reverse 5′-CAGTTAGCGCCCAAAGGAC-3′), in3 (forward 5′-TAACACTGGCTCGTGTGACAA-3′ and reverse 5′-AAGTGCAAAGAACACGGCTAA-3′), in5 (forward 5′-GGAGCTGTCACATCCAGGGTC-3′ and reverse 5′-TGCTGATCCACATCTGCTGG-3′), and 5′ pause (forward 5′-TTACCCAGAGTGCAGGTGTG-3′ and reverse 5′-CCCCAATAAGCAGGAACAGA-3′). γ-actin gene in1 (forward 5′-CCGCAGTGCAGACTTCCGAG-3′ and reverse 5′-CGGGCGCGTCTGTAACACGG-3′), A (forward 5′-TTCGTGGGCTGGTGAGAAAA-3′ and reverse 5′-CTCCAACACCCAAACCCACT-3′), and B (forward 5′-GGGTCAAGGGATCGTTCTG-3′ and reverse 5′-GCCTGGAGCTCAGTAAGC-3′). RT-qPCR analysis was conducted with a Bio-Rad CFX Connect instrument and SYBR-Green reagents (BP170-8882, Bio-Rad). Each assay was performed in triplicate, and 10% of the sheared nucleic acid was used as input. The DRIP signal at a specific gene region was calculated as a percentage of the input and normalized to the in1 sample of the corresponding undamaged sample.
Slot blot assay
To determine whether loss of DHX9 and expressing phospho-deficient DHX9 in DHX9-depleting cells lead to the accumulation of R-loops in cells under genotoxic stress. The slot blot assay was performed with some modifications to a previously described protocol (33). In brief, cells treated with CPT or left untreated were harvested for the preparation of genomic DNA, which was prepared following the DRIP protocol, with the exception of IP by the S9.6 antibody. Purified nucleic acids (400 ng), treated with or without 5 U RNase H (M0297L, NEB), were applied to a Hybond-N+ hybridization membrane (Cytiva) using a Bio-Dot SF apparatus (1706542, Bio-Rad). The membranes were UV cross-linked at 0.12 J/m2 (UVP CL-1000) and subsequently blocked in TBS-T containing 5% skim milk at room temperature for 1 h. For the detection of dsDNA, the membrane was stained with methylene blue (50484, Sigma-Aldrich) at room temperature for 10 min. To detect DNA/RNA hybrids, the membranes were incubated with the S9.6 antibody overnight at 4°C, followed by three washes with TBS-T. Then, a goat anti-mouse-HRP secondary antibody was applied and incubated for 1 h, followed by three additional washes in TBS-T. Finally, the chemiluminescence signal was visualized using the ChemiDoc imaging system (Bio-Rad).
Annexin V-APC analysis by flow cytometry
To monitor cell apoptosis, HeLa inducible clones were transfected with control or DHX9 siRNA for 24 h before inducing SFB-DHX9 proteins by doxycycline (200 ng/ml). After that, cells were treated with DMSO or 1 μM cisplatin for 24 h. Floating and attached cells were combined for the Annexin V-APC analysis (Biolegend) according to the manufacturer's instructions. Data acquisition was performed using an LSRFortessa Flow cytometer (BD Bioscience). Data analysis and plotting were performed using Kaluza software (Beckman Coulter).
ATP-based cell viability assay
HeLa inducible cell lines transfected with control and DHX9 siRNA for 48 h were seeded in 96-well plates (4000 cells per well) containing vehicle or doxycycline (200 ng/ml) to induce SFB-DHX9 proteins. The next day cells were treated with DMSO, CPT or HU for 24 h, and the cell viability was determined using the Cell Titer-Glo 2.0 luminescent viability assay (Promega) following the manufacturer's protocol. The values were acquired using an ELISA reader (BioTek) and normalized to DMSO samples in each condition. Data are presented in mean ± SD.
Statistical analyses
For ATP-based cell viability, cell number counting, Annexin V staining and DRIP assay, data were analyzed with the two-sided, unpaired Student's t-test. For PLA foci, data were analyzed by the two-sided, unpaired Mann–Whitney U test. Statistical analyses and data presentations were using GraphPad Prism 9. A P value <0.05 was considered statistically significant.
Results
DHX9 is phosphorylated at S321 and S688, with S321 phosphorylation induced by DNA damage
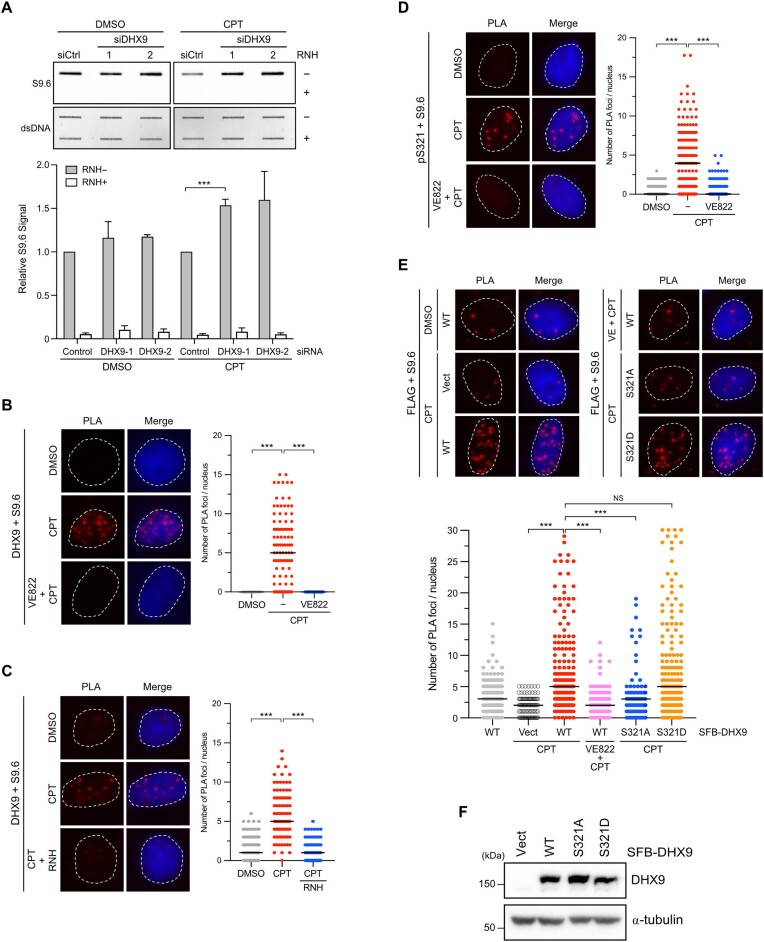
DHX9 is an RNA helicase implicated in the suppression of R-loops due to the interference between replication and transcription caused by CPT (35). However, the precise mechanisms of how DHX9 is targeted to R-loops at DNA damage sites remain unclear. We employed proximity ligation assay (PLA) to visualize the interactions between endogenous DHX9 and DDR proteins in cells. In Figure 1A and Supplementary Figure S1A, we observed fluorescent PLA foci primarily in the nucleus of HeLa cells treated with CPT, representing the close association of DHX9 with γH2AX and BRCA1. To verify the specificity of PLA signals, we depleted endogenous DHX9 using DHX9-1 siRNA that has been used in various studies (35,48). These results confirm the specificity of PLA signals and provide the first evidence of interactions between DHX9 and γH2AX, as well as DHX9 and BRCA1, in cells. Furthermore, our findings validate the interactions of DHX9 with these tested proteins, as previously demonstrated by affinity-purification analysis in other studies (43,44). During the DDR, the RPA complex is a crucial sensor of ssDNA, coordinating the ATR-Chk1 pathway and DNA repair. In a previous study by Zou's group, RPA-ssDNA was used as a bait to identify candidates involved in the DDR (49). DHX9 was identified as one of the interacting candidates with RPA-ssDNA. We next asked if DHX9 localizes with RPA. The PLA analysis revealed that endogenous DHX9 was closely associated with RPA32 at a basal level, and such interaction was profoundly increased following CPT treatment in HeLa cells (Figure 1B). Moreover, Supplementary Figure S1B and S1C demonstrated reproducible associations of DHX9 with γH2AX and RPA32, respectively, in a different cancer cell line, U2OS cells. Concerning that PLA signals after CPT treatment were consistently increased in all combination of antibodies, we further performed a PLA analysis with single DHX9 antibody in HeLa cells treated with DMSO or CPT, and examined levels of BRCA1, DHX9 and RPA proteins after genotoxic stresses using Western blot (Supplementary Figure S1D, E). The results confirmed that single DHX9 antibody did not yield positive PLA signals, and CPT used in PLA analyses as well as other genotoxic stress conditions used in this study did not increase levels of tested proteins. The above findings confirm the specificity of PLA signals and suggest that the interaction between DHX9 and RPA after DNA damage might regulate DHX9’s function at DNA damage sites.
Figure 1.
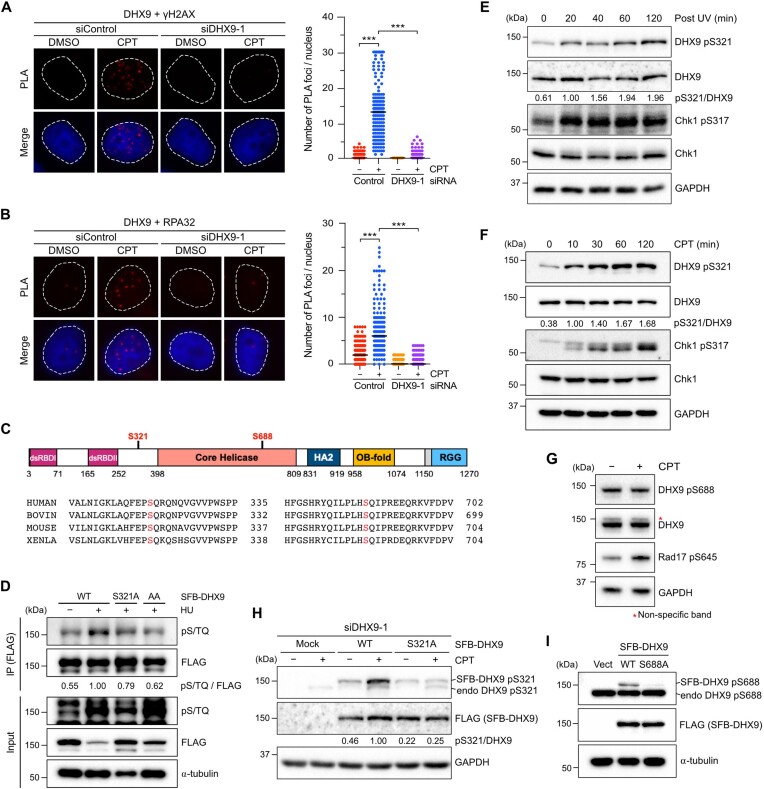
DHX9 is phosphorylated at S321 and S688, with S321 phosphorylation induced by DNA damage. (A, B) The association between DHX9/γH2AX (A) and DHX9/RPA32(B) in HeLa cells. Cells transfected with control or DHX9 siRNA were treated with DMSO or CPT (1 μM, 1 h), analyzed by PLA with indicated antibodies. Representative images were shown, and the numbers of PLA foci were obtained from at least 150 nuclei in each condition. Data are presented in scatter plots with the black line indicating the median, and statistical significance was assessed by the Mann–Whitney test (two-sided). *** P < 0.001. (C) A primary structure of DHX9 and sequence alignment of conserved S321 and S688 across human, bovine, mouse and Xenopus. (D) HeLa cells transiently expressed SFB-DHX9 variants (WT, S321A and AA) were subjected to treatment with or without HU (2 mM) for 1 h. SFB-DHX9 (WT, S321A and AA) were immunoprecipitated by an anti-FLAG antibody under a denaturing condition. Levels of DHX9 phosphorylation at S/TQ sites and protein expression were determined by Western blot with indicated antibodies. (E, F) Time course of DHX9 phosphorylation at S321 was determined by a phospho-specific antibody to this site. HeLa cells irradiated by UV (20 J/m2) (E) or treated with CPT (10 μM) (F) were collected at a series of time points, and phosphorylation of DHX9 pS321 and Chk1 pS317 after DNA damage was accessed by Western blot with indicated antibodies. (G) HeLa cells treated with CPT (1 μM) or left untreated were analyzed by Western blot with indicated antibodies. (H) Validation of the specificity of anti-DHX9 pS321 antibody. HeLa cells devoid of DHX9 by siRNA were transfected with siDHX9-1 insensitive SFB-DHX9WT or SFB-DHX9S321A. Cells were treated with or without CPT (1 μM, 1 h), and the phosphorylation of S321 was determined by Western blot with indicated antibodies. (I) Phosphorylation at S688 was determined by a phospho-specific antibody to this site. HeLa cells transfected with SFB-DHX9WT, SFB-DHX9S688A or the corresponding empty vector were analyzed by Western blot with indicated antibodies.
To further investigate the role of DHX9 in response to DNA damage, we examined whether DHX9 is a target of ATM/ATR signaling. Endogenous DHX9 immunoprecipitated from ultraviolet (UV)-irradiated or untreated HeLa cells were analyzed by mass spectrometry to identify phosphorylation sites (Supplementary Figure S1F). The analysis identified that DHX9 was phosphorylated at Ser 321 upon UV irradiation (Supplementary Figure S1G and Supplementary Table S1). Additionally, we looked at PhosphoSitePlus (https://www.phosphosite.org/), a comprehensive database of protein phosphorylation information. Among the thirteen documented phosphorylation sites of DHX9, we focused on those located within a consensus S/TQ motif, which is preferentially phosphorylated by ATM/ATR. S321 and S688 were the only two S/TQ sites uncovered by at least five high-throughput papers (Supplementary Figure S1H). Furthermore, the alignment of DHX9 across different species revealed that Ser 321 and Ser 688 are highly conserved in many vertebrate species (Figure 1C), suggesting their functional importance. Despite being documented in several large-scale studies (7,8), the functions of phosphorylated S321 and S688 remain uncharacterized. This knowledge gap prompted us to ask whether these sites are indeed phosphorylated by PIKKs in cells after DNA damage. To address this question, immunoprecipitated SFB (S6-FLAG-Streptavidin binding peptide)-tagged DHX9 from hydroxyurea (HU)-treated HeLa cell lysates were analyzed by Western blot using an anti-pS/TQ antibody. In Figure 1D, we observed a background signal of wild-type DHX9 (DHX9WT) detected by the pS/TQ antibody. However, the pS/TQ signal of DHX9WT was significantly increased after HU treatment. Ablation of S321 phosphorylation by substituting an alanine (DHX9S321A) showed a moderate reduction in the pS/TQ signal, while simultaneous substitution of both S321 and S688 with alanines (DHX9AA) further decreased the pS/TQ signal to background levels. These results confirm the phosphorylation of DHX9 at S321 and S688.
To directly characterize DHX9 phosphorylation at S321 and S688, we generated antibodies specifically targeting phosphorylated S321 and S688. Like Chk1 phosphorylation at S317, DHX9 phosphorylation at S321 was quickly detected 20 min after UV treatment, persisting for up to 120 min (Figure 1E). Similarly, upon treatment with 10 μM CPT, S321 phosphorylation was detected as early as 10 min and reached its maximum level at 60 min (Figure 1F). Remarkably, not only UV and CPT but also genotoxic drugs such as gemcitabine, olaparib, doxorubicin and cisplatin, which induce DNA damage by diverse mechanisms, triggered S321 phosphorylation (Supplementary Figure S1I). In contrast to S321 phosphorylation, an intriguing observation emerged when we exposed cells to the same aforementioned genotoxic drugs. A robust S688 phosphorylation was detected in undamaged cells and remained unaffected, implying that S688 phosphorylation was not induced by DNA damage (Figure 1G and Supplementary Figure S1I).
To verify the specificity of phospho-antibodies, we expressed SFB-DHX9WT and SFB-DHX9S321A in cells with endogenous DHX9 depletion using DHX9 siRNA. The SFB-DHX9 constructs contained three silent mutations in DHX9, which did not alter the protein sequences and rendered them insensitive to siDHX9-1 siRNA. In cells treated with CPT, the anti-pS321 antibody specifically recognized DHX9WT but not DHX9S321A confirming its specificity (Figure 1H). Similarly, the anti-pS688 antibody detected DHX9WT but not DHX9S688A when SFB-DHX9 variants were expressed in cells. (Figure 1I). Taken together, our results provide compelling evidence that DHX9 undergoes phosphorylation at both S321 and S688. Importantly, only S321 phosphorylation is triggered by DNA damage, while S688 phosphorylation appears to be constant and unrelated to DNA damage.
ATR phosphorylates DHX9 at S321 in response to genotoxic stress
ATM, ATR and DNA-PKcs are kinases that conduct the DDR by phosphorylating numerous target proteins. We next aimed to identify the specific kinases involved in DHX9 phosphorylation following DNA damage. To do this, we treated cells with inhibitors that selectively target certain kinases, followed by treatment with either CPT or HU (Figure 2A). We used phosphorylation of Chk1 at S317, Chk2 at T68 and RPA32 at S4/8 as indicators of activated ATR, ATM and DNA-PKcs, respectively. Pre-treatment with caffeine, a pan-inhibitor of all three kinases, largely abolished CPT-induced phosphorylation of Chk1 pS317 and RPA32 pS4/8, indicating the effectiveness of caffeine in inhibiting DDR kinases. In parallel, we also observed that the phosphorylation of DHX9 at S321 was significantly diminished (Figure 2B). We then employed inhibitors selectively targeting individual DDR kinases to characterize the kinase responsible for S321 phosphorylation. Our findings revealed that the ATR inhibitor VE822 strongly inhibited S321 phosphorylation, while the ATM inhibitor KU55933 and the DNA-PKcs inhibitor NU7441 had negligible effects on S321 phosphorylation (Figure 2B). In addition to CPT treatment, we applied a 2-h exposure of 2 mM HU to HeLa cells, which primarily activated the ATR-Chk1 signaling pathway but not the ATM and DNA-PKcs. Phosphorylation of S321 was markedly diminished in cells pre-treated with caffeine and VE822, while KU55933 and NU7441 had no impact (Figure 2C). Consistently, in UV-irradiated U2OS cells, both VE822 and another selective ATR inhibitor AZD6738, but not KU55933 and NU7441, substantially inhibited S321 phosphorylation, similar to the effects of caffeine (Supplementary Figure S2A). These findings strongly suggest that ATR is likely the primary kinase responsible for S321 phosphorylation under genotoxic stress.
Figure 2.
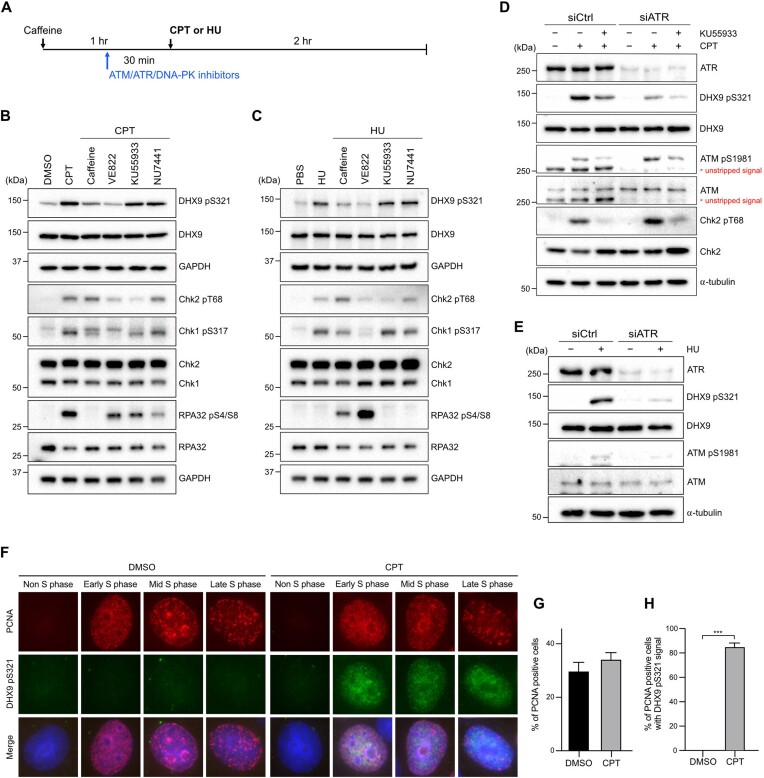
ATR phosphorylates DHX9 at S321 in response to genotoxic stress. (A–C) A schematic diagram illustrated the experimental sequence designed to investigate the kinase responsible for DHX9 phosphorylation. All indicated inhibitors (caffeine 5 mM, VE822 1 μM, KU55933 10 μM, NU7441 10 μM) were administered for 30 min prior to CPT (1 μM) or HU (2 mM) treatments, except for caffeine, which was administered for 1 h (A). The impact of different inhibitors on the CPT-induced (B) and HU-induced (C) phosphorylation of DHX9 S321 in U2OS cells was assessed by Western blot with indicated antibodies. (D, E) Knockdown of ATR impeded DNA damage-induced S321 phosphorylation. HeLa cells transfected with control and ATR siRNA were treated with CPT (1 μM), CPT plus KU55933 (10 μM) or left untreated for 1 h. Phosphorylation of DHX9, ATM and Chk2 was determined by Western blot (D). The status of S321 phosphorylation in HU-treated cells was determined by Western blot with indicated antibodies (E). (F–H) Immunostaining of PCNA and DHX9 pS321 demonstrated the phosphorylation of DHX9 S321 in S phase. HeLa cells treated with DMSO or CPT (1 μM, 1 h) were subjected to pre-extraction in PBS containing 0.5% Triton X-100, followed by MeOH fixation and immunostaining with indicated antibodies. (F) Representative images of immunostaining. (G, H) Quantification results of PCNA positive cells (G) and PCNA positive cells with DHX9 pS321 signal (H). At least 500 nuclei were quantified and data are presented as the mean ± SD of three independent experiments. Statistical significance was assessed by the Student's t-test (two-sided). *** P < 0.001.
To further test if ATR is responsible for S321 phosphorylation, we used siRNA to deplete endogenous ATR before the induction of DNA damage. As shown in Figure 2D, S321 phosphorylation induced by CPT was partially suppressed in siControl cells treated with KU55933. Conversely, S321 phosphorylation was drastically suppressed in cells with ATR depletion and almost completely abolished when KU55933 was additionally administered. Given that CPT treatment activates both ATM and ATR, and that suppression of ATM by KU55933 also affects ATR activation, we next treated cells with HU for 1 h, during which ATM was not activated (Figure 2E). Our results showed a near absence of S321 phosphorylation in ATR-depleted cells (Figure 2E). In contrast to S321 phosphorylation, we found no impact on S688 phosphorylation in cells devoid of ATR, regardless of the treatment with KU55933 (Supplementary Figure S2B, C). To understand if levels of DHX9 S321 phosphorylation vary under different stresses, we compared the S321 phosphorylation triggered by CPT, HU and UV (Supplementary Figure S2D). While UV induced the highest phosphorylation at S321, HU-induced phosphorylation was the lowest. The observed difference in levels of DHX9 pS321 between examined genotoxic agents indicates that timing and dosing of various types of DNA damage influence the level of DHX9 phosphorylation. Furthermore, since CPT-induced damage is S-phase specific and our data indicate that ATR is responsible for DHX9 S321 phosphorylation, we investigated whether the phosphorylation of DHX9 at S321 is cell cycle regulated by co-staining DHX9 pS321 and PCNA, a DNA replicating protein with well-studied cell cycle-dependent features (50). The results confirmed that DHX9 pS321 signal predominantly exists in PCNA positive cells after CPT treatment (Figure 2F–H). In summary, our findings strongly suggest that DHX9 phosphorylation at S321 after DNA damage is primarily mediated by ATR in the S phase.
Phosphorylation of DHX9 promotes its interaction with RPA
Hence DHX9 was associated with DDR proteins and phosphorylated by ATR under genotoxic stress, we next asked the role of DHX9 phosphorylation in its interaction with γH2AX and RPA. PLA analysis revealed a close association between DHX9 pS321 and γH2AX, as well as RPA32 after CPT treatment (Figure 3A and B). To examine whether DNA damage promotes the interaction between DHX9 and RPA, we immunoprecipitated endogenous RPA32 from a nuclear fraction of HeLa cells transfected with SFB-DHX9 or a corresponding empty vector. We found that a small fraction of SFB-DHX9 was coimmunoprecipitated by RPA32, with the coprecipitation notably enhanced after CPT treatment (Figure 3C). Likewise, by IP of endogenous DHX9 from HeLa nuclear extract, our data demonstrated that the interaction between DHX9 and RPA32 was increased in response to UV irradiation (Supplementary Figure S3A), indicating a DNA damage-dependent enhancement of the DHX9–RPA32 interaction.
Figure 3.
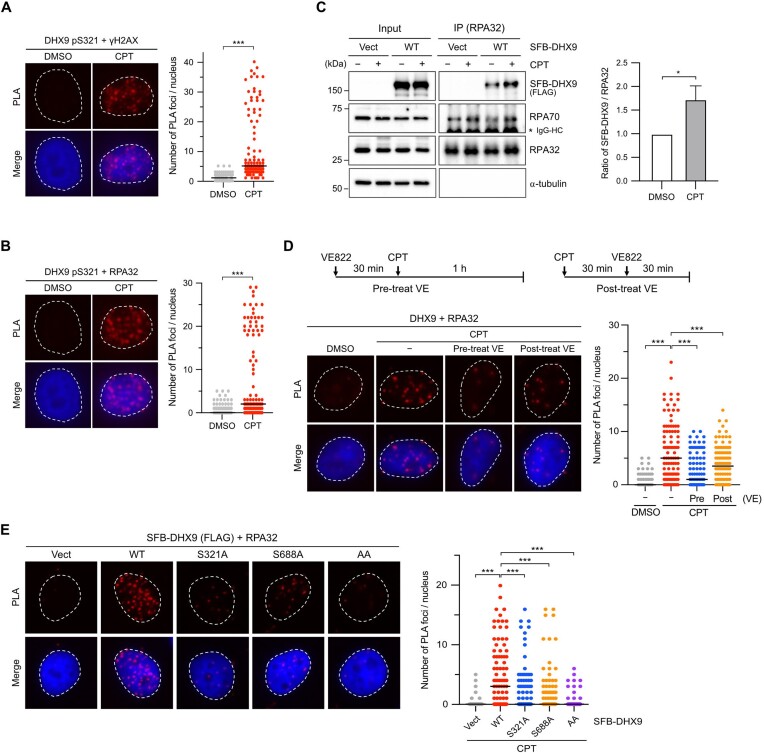
Phosphorylation of DHX9 promotes its interaction with RPA. (A, B) The association between DHX9 pS321/γH2AX (A) and DHX9 pS321/RPA32 (B) in HeLa cells treated with CPT (1 μM, 1 h) or left untreated were analyzed by PLA with indicated antibodies. Representative images were shown, and numbers of PLA foci per cell were counted from at least 150 nuclei in each condition. Data are presented in scatter plots with the black line indicating the median, and statistical significance was assessed by the Mann–Whitney test (two-sided). *** P < 0.001. (C) Interaction between RPA and DHX9 was enhanced after DNA damage. HeLa cells transfected with SFB-DHX9 or the corresponding empty vector were treated with DMSO or CPT (1 μM, 1 h). The nuclei were extracted, followed by the IP of nuclear RPA32 using the anti-RPA32 antibody. Co-purification of SFB-DHX9 and RPA70 by RPA32 was determined by Western blot with the designated antibodies. The levels of SFB-DHX9 co-purified were quantified and data are presented as the mean ± SD of three independent experiments. Statistical significance was assessed by the Student's t-test (two-sided). * P < 0.05. (D) The impact of ATR-mediated DHX9 phosphorylation on the DHX9/RPA32 association was analyzed by PLA. HeLa cells either treated with 1 μM of VE822 30 min before or 30 min after CPT (1 μM) treatment were analyzed by PLA with indicated antibodies. (E) Substitution of phosphorylated serine with alanine for single or double mutations (S321A, S688A, AA) impeded the DHX9/RPA32 interaction after CPT treatment. HeLa cells transfected with the corresponding empty vector (Vect) or SFB-DHX9 variants (WT, S321A, S688A and AA) were treated with CPT (1 μM) for 1 h, and the PLA signal was determined. Representative images were shown, and numbers of PLA foci per cell were counted from at least 150 nuclei in each condition. Data are presented in scatter plots with the black line indicating the median, and statistical significance was assessed by the Mann–Whitney test (two-sided).*** P < 0.001.
To determine whether the association between DHX9 and DDR proteins depends on ATR-mediated phosphorylation in cells, we treated HeLa cells with VE822 either 30 min before or after CPT treatment and analyzed the association of DHX9/RPA32 using PLA. The results showed that pre-incubation of VE822 markedly inhibited the CPT-induced association of DHX9/RPA32, while addition of VE822 30 min after CPT treatment had a mild effect (Figure 3D). A similar finding of DHX9/γH2AX interaction was observed by PLA analysis (Supplementary Figure S3B), suggesting that DHX9 interacts with DDR proteins in an ATR-dependent manner. To further confirm if DHX9 phosphorylation is critical for its interaction with RPA32, we used phospho-deficient DHX9 for PLA analysis. Initially, we examined whether transiently expressed SFB–DHX9 interacts with RPA32 in cells. Like endogenous DHX9, ectopically expressed SFB-DHX9 exhibited an interaction with RPA32 after CPT treatment, which was significantly reduced by pre-incubation with VE822 (Supplementary Figure S3C). Subsequently, we used PLA to analyze the interaction between phospho-deficient DHX9S321A and RPA32 in cells treated with CPT. Figure 3E showed that DHX9S321A failed to interact with RPA32 efficiently compared to DHX9WT. Although phosphorylation at S688 was not induced by DNA damage, we explored if pS688 also contributed to the association with RPA. Surprisingly, DHX9S688A showed a similar extent of impairment in association with RPA32 as DHX9S321A. Further examination of the DHX9AA mutant revealed an additional reduction in the association with RPA32 (Figure 3E). Our observations suggest that DHX9 phosphorylation at S321 and S688 is equally crucial for interacting with RPA after DNA damage. However, since there is limited information available regarding the kinase responsible for S688 phosphorylation, our focus in the following investigations will be on elucidating the role of S321.
Phosphorylation of DHX9 is critical for R-loop association in cells
One of the primary functions attributed to DHX9 is its role in suppressing the accumulation of R-loops during genotoxic stress. Based on the above findings that DHX9 was associated with RPA following DNA damage, we hypothesized that the phosphorylation of DHX9 might play a crucial functional role in the elimination of R-loops during genotoxic stress. We performed a slot blot assay to examine whether loss of DHX9 leads to the accumulation of DNA/RNA hybrids in cells. HeLa cells transfected with control or DHX9 siRNAs were treated with CPT, followed by genomic DNA extraction and transfer to a nylon membrane. DNA/RNA hybrids on the membrane were assessed using the S9.6 antibody, which exhibited a high affinity for DNA/RNA hybrids (35,51). As shown in Figure 4A, cells lacking DHX9 showed a significant increase in DNA/RNA hybrids after CPT treatment, consistent with previous studies (35,43). Analyzed by PLA, we next observed a close association between endogenous DHX9 and DNA/RNA hybrids during CPT treatment. Intriguingly, pre-treating cells with VE822 remarkably diminished the association between DHX9 and DNA/RNA hybrids after CPT treatment (Figure 4B), suggesting a critical involvement of ATR in the interaction between DHX9 and DNA/RNA hybrids.
Figure 4.
Phosphorylation of DHX9 is critical for R-loop association in cells. (A) Depletion of DHX9 led to the accrual of DNA/RNA hybrids in cells. HeLa cells transfected with Control or DHX9 siRNA were treated with DMSO or CPT (1 μM) for 1 h. Genomic DNA extracted from treated cells was analyzed using slot blot assay with the S9.6 antibody. The relative S9.6 signal was quantified, and the data are presented as the mean ± SEM of three independent experiments. *** P < 0.001. (B–D) The inhibition of ATR resulted in the disassociation of endogenous DHX9 from DNA/RNA hybrids. HeLa cells were subjected to different treatments, including CPT (1 μM) alone, 1 μM of VE822 administered 30 min prior to CPT treatment (B, D), or RNH applied after CPT treatment (C). These treated cells were evaluated using PLA with the designated antibodies. (E, F) Replacing phosphorylated serine with alanine (S321A) or asparagine (S321D) had different impacts on the interaction between DHX9 and DNA/RNA hybrids after CPT treatment. HeLa cells depleted with DHX9 were transfected with the corresponding empty vector (Vect) or SFB-DHX9 variants (WT, S321A and S321D). Cells were treated with CPT (1 μM) or left untreated, followed by the PLA analysis (E). Levels of SFB-DHX9 variants (WT, S321A and S321D) in cells were examined by Western blot using indicated antibodies (F). (B–E) Representative images were shown, and the numbers of PLA foci were quantified by analyzing at least 150 nuclei in each condition. Data are presented in scatter plots with the black line indicating the median, and statistical significance was assessed by the Mann–Whitney test (two-sided). *** P < 0.001.
To verify the specificity of DHX9/S9.6 PLA signal and the S9.6 antibody in detecting DNA/RNA hybrids formed after CPT treatment, we repeated PLA analysis with the S9.6 single antibody in HeLa cells or a combination of DHX9/S9.6 antibodies in siDHX9 transfected cells (Supplementary Figure S4). The data validated the specificity of the DHX9/S9.6 PLA signal, as shown in Figure 4B. To further confirm that the PLA signals from DHX9/S9.6 were specific to R-loops, we treated cells with RNH (35). The PLA signal indicating the association of DHX9/S9.6 in CPT-treated cells significantly decreased upon RNH introduction (Figure 4C), thereby confirming the interaction of DHX9 and R-loops. Next, we asked whether the phosphorylation of DHX9 at S321 is necessary for its interaction with R-loops. We repeated the PLA experiment using antibodies specific to phosphorylated S321 and R-loops. As shown in Figure 4D, cells treated with CPT displayed a notable increase in PLA foci, indicating the association between DHX9 pS321 and DNA/RNA hybrids. In contrast, cells pre-treated with VE822 before CPT treatment barely detected PLA signal. Furthermore, we expressed SFB–DHX9 variants in HeLa cells lacking endogenous DHX9 to directly examine whether DHX9 pS321, induced by DNA damage, is crucial for its association with R-loops. Consistent with endogenous DHX9, we observed a robust PLA signal of DHX9WT/S9.6 but not the corresponding empty vector/S9.6 after CPT treatment (Figure 4E). Pre-treating cells with VE822 before CPT treatment noticeably abolished the PLA signal of DHX9WT/S9.6. Unlike DHX9WT, DHX9S321A failed to interact with DNA/RNA hybrids efficiently. Interestingly, the phosphomimetic mutant DHX9S321D displayed a strong interaction with DNA/RNA hybrids comparable to that of DHX9WT. To rule out the possibility that the differences in PLA signals between cells expressing WT and mutant DHX9 were due to uneven protein expression, we determined the protein levels in the duplicated cells used for PLA. The results in Figure 4F confirmed that the DHX9 variants introduced into cells were expressed at similar levels, thereby validating that the observed PLA data were not influenced by variations in DHX9 protein expression. Overall, these findings provide evidence supporting the compromised interaction of DHX9S321A with R-loops.
Phospho-deficient DHX9 fails to suppress R-loops under genotoxic stress
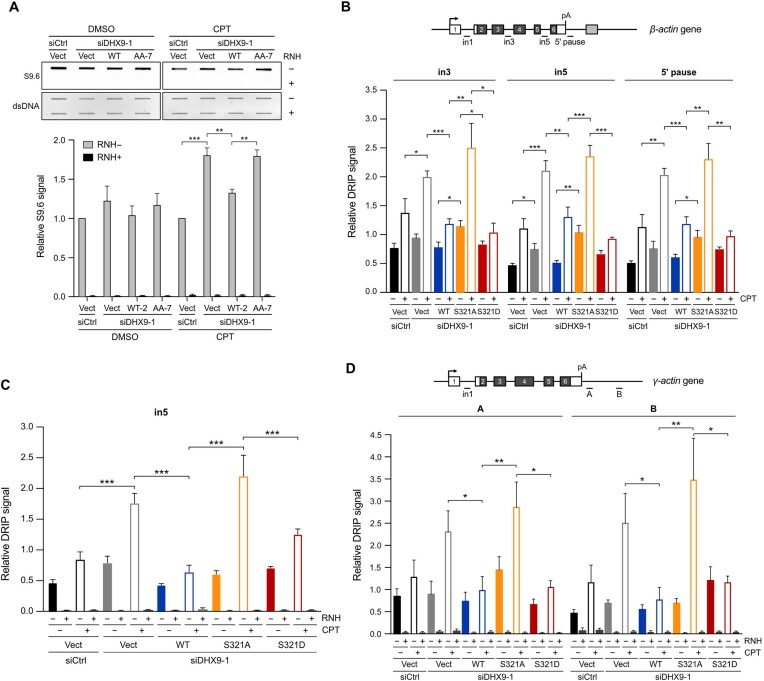
We next asked whether cells expressing phospho-deficient DHX9 that fail to associate with RPA are defective in resolving R-loops. To address this, we established cell lines that stably expressed DHX9WT and phospho-deficient DHX9AA under the control of the doxycycline-regulated Tet-On system and assessed R-loop accumulation using slot blot assay. As shown in Figure 5A, compared to control siRNA transfected cells carrying the empty vector (Vect), silencing endogenous DHX9 in the Vect clone resulted in increased R-loops after CPT treatment. Upon doxycycline induction in DHX9-depleted stable lines, DHX9WT, but not DHX9AA, attenuated CPT-induced R-loops. Since S321 phosphorylation is specifically regulated by DNA damage, we next asked whether the S321A mutation alone impairs the resolution of R-loops. HeLa cells transfected with SFB-DHX9WT, SFB-DHX9S321A or the corresponding empty vector were used to analyze the R-loop profiles of the β-actin gene following the knockdown of DHX9. The DNA/RNA immunoprecipitation (DRIP)-qPCR results revealed that R-loops across tested regions of the β-actin gene (in3, in5 and 5′ pause) in the DHX9-depleted cells carrying the corresponding empty vector were moderately enriched before CPT treatment and profoundly increased after CPT treatment. This observation is consistent with the findings in a previous study (35). Additionally, we observed that DHX9WT effectively suppressed R-loop accumulation after CPT treatment, while DHX9S321A did not display the same suppressive effect. Introducing DHX9S321D reduced R-loop accumulation to levels similar to those seen with DHX9WT (Figure 5B), highlighting the importance of S321 phosphorylation in R-loop suppression. To ascertain that the elevated DRIP signal in DHX9S321A was R-loop dependent, we executed another DRIP analysis focusing on the in5 region of the β-actin gene. Here, purified R-loops underwent treatment with or without RNH prior to RT-qPCR for DRIP signal detection (Figure 5C). The data revealed that RNH treatment completely eliminated the augmented R-loop signal, confirming that the DRIP-qPCR signal specifically originated from R-loops. Furthermore, we investigated R-loop accumulation at the γ−actin gene, another R-loop forming gene being well studied previously. The results mirrored those seen in Figure 5B and C (Figure 5D). In summary, our findings confirm that DHX9 phosphorylation is crucial for the effective suppression of CPT-induced R-loops.
Figure 5.
Phospho-deficient DHX9 fails to suppress R-loops under genotoxic stress. (A) Disruption of DHX9 phosphorylation hindered R-loop suppression. HeLa stable lines were transfected with control siRNA or siDHX9 to silence endogenous DHX9, followed by the induction of SFB-DHX9WT, SFB-DHX9AA or the corresponding empty vector using doxycycline (200 ng/ml). The cell lines were treated with DMSO or CPT (1 μM) for 1 h. Genomic DNA extracted from treated cells was analyzed using slot blot assay with the S9.6 antibody. The relative S9.6 signal was quantified, and the data are presented as the mean ± SEM of three independent experiments. ** P < 0.01; *** P < 0.001. (B) Phosphomimetic DHX9 S321D suppressed the accumulation of R-loops in HeLa cells devoid of DHX9 after CPT treatment. Cells transfected with control or DHX9 siRNA were transiently expressed with SFB-DHX9 (WT, S321A and S321D), followed by CPT treatment (10 μM, 1 h). DNA/RNA hybrids purified using the S9.6 antibody were analyzed by RT-qPCR for the abundance of R-loops over β−actin gene. Values relative to in1 in each condition were plotted as the mean ± SEM of three experiments. Statistical significance was assessed by the Student's t-test (two-sided). * P < 0.05; ** P < 0.01; *** P < 0.001. (C) R-loop levels at the in5 of β−actin gene were suppressed by RNH. Experiments followed the protocol from (B) with the addition of RNH treatment to confirm that the increased DRIP signal was derived from R-loops. Values relative to in1 under each condition were plotted as the mean ± SEM of three experiments. Statistical significance was assessed by the Student's t-test (two-sided). *** P < 0.001. (D) Phosphomimetic S321D reduced R-loop accumulation over the γ-actin gene. Experiments were repeated as in (B), and RNH treatment was incorporated. Values relative to in1 in each condition were plotted as the mean ± SEM of three experiments. Statistical significance was assessed by the Student's t-test (two-sided). * P < 0.05; ** P < 0.01.
DHX9 phosphorylation at S321 enhances its interaction with RPA
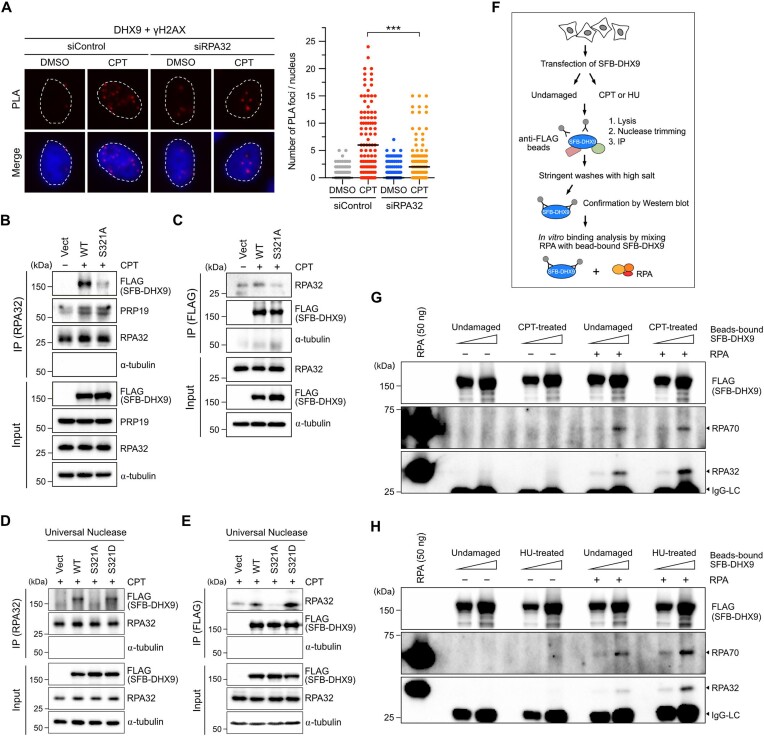
The RPA complex is demonstrated as a sensor for R-loops (22). Given the importance of DHX9 phosphorylation in its association with RPA and R-loops, we speculated that RPA-ssDNA might serve as a platform for recruiting DHX9 to counter DNA damage-induced R-loops. Cells transfected with control or RPA32 siRNA were treated with CPT or left untreated, and PLA was used to assess the association between DHX9 and γH2AX. While the interaction between endogenous DHX9 and γH2AX remained consistent in response to CPT, cells lacking RPA32 exhibited a significant reduction in PLA foci of DHX9/γH2AX (Figure 6A). To further examine if DHX9 phosphorylation contributes to its interaction with RPA, we performed coimmunoprecipitation (Co-IP) to capture endogenous RPA32 from nuclear extracts of cells transfected with DHX9WT or DHX9S321A after CPT treatment. In contrast to PRP19, which was coprecipitated at a similar level between cells expressing DHX9WT and DHX9S321A by RPA32, we found efficient coprecipitation of DHX9WT but not DHX9S321A with RPA32 (Figure 6B). Similarly, when SFB-DHX9 variants were captured using the anti-FLAG antibody, efficient coprecipitation of RPA32 was observed only with DHX9WT but not DHX9S321A (Figure 6C). Previous studies have demonstrated that DHX9 possesses the ability to associate with dsRNA, DNA/RNA hybrids and dsDNA. To determine whether the interaction between DHX9 and RPA is mediated by nucleic acids, we repeated the same Co-IP experiments as described in Figure 6B and C, but included additional treatments with Universal Nuclease, same as Benzonase, to remove DNA and RNA before the IP of RPA32 or SFB-DHX9. In Figure 6D, we observed efficient coprecipitation of DHX9WT, but not DHX9S321A, with RPA32. Interestingly, the IP of RPA32 from HeLa nuclear extracts containing SFB-DHX9S321D treated with Universal Nuclease successfully coprecipitated DHX9S321D. On the other hand, when SFB-DHX9 variants were purified from nuclear extracts treated with Universal Nuclease using anti-FLAG beads, RPA32 was coprecipitated with both DHX9WT and DHX9S321D, but not with DHX9S321A (Figure 6D and E). These results suggest that the interaction between DHX9 and RPA is not mediated by the binding of DNA or RNA.
Figure 6.
DHX9 phosphorylation at S321 enhances its association with RPA. (A) Depletion of RPA impeded the association between DHX9 and γH2AX. HeLa cells transfected with control or RPA32 siRNA were then treated with DMSO or CPT (1 μM) for 1 h. The association between DHX9 and γH2AX was assessed by PLA using the indicated antibodies. Representative images were shown, and numbers of PLA foci were quantified from at least 150 nuclei in each condition. Data are presented in scatter plots with the black line indicating the median, and statistical significance was assessed by Student's t-test (two-sided). *** P < 0.001. (B, C) Ablation of S321 phosphorylation attenuated the interaction between DHX9 and RPA. HeLa cells transfected with SFB-DHX9WT and SFB-DHX9S321A were treated with CPT (1 μM) for 1 h. Endogenous RPA32 and SFB-DHX9 were immunoprecipitated from nuclear extracts with anti-RPA32 and anti-FLAG antibodies, respectively. Proteins coprecipitated by RPA32 (B) and SFB-DHX9 (C) were assessed by Western blot with indicated antibodies. (D, E) The interaction between DHX9 and RPA was not dependent on DNA or RNA. As in (B) and (C), nuclear extracts were pre-incubated with Universal Nuclease before IP of RPA32 (D) and SFB-DHX9 (E). Proteins coprecipitated by RPA32 (D) and SFB-DHX9 (E) were assessed by Western blot with indicated antibodies. (F) A workflow of SFB-DHX9 purification and in vitro SFB-DHX9/RPA binding analysis. (G, H) Purified SFB-DHX9 directly interacted with recombinant RPA. HeLa cells transiently expressing SFB-DHX9 were treated with vehicle, CPT (1 μM, 1 h) or HU (4 mM, 2 h) prior to the purification of SFB-DHX9 using anti-FLAG beads. The purified SFB-DHX9 proteins were incubated with 100 ng of the RPA complex for in vitro binding assay. The interaction between SFB-DHX9 and RPA was determined by Western blot with indicated antibodies.
To elucidate whether DHX9 directly interacts with RPA, we immunoprecipitated SFB-DHX9 from cells treated with vehicle, CPT or HU using anti-FLAG magnetic beads in NETN buffers supplemented with Universal Nuclease. After IP, several rounds of stringent washes were performed to ensure the purity of SFB-DHX9. Western blot analysis confirmed that the purified SFB-DHX9 did not associate with either RPA or PARP1, a previously known DHX9 interactor (35) (Figure 6F and Supplementary Figure S5). Next, the bead-bound SFB-DHX9 was mixed with RPA complex (RPA70-32-14), which was purified from E. coli for in vitro binding assays (46). Notably, the results showed that SFB-DHX9 purified from both undamaged and CPT-treated cells coprecipitated RPA, with the interaction being stronger for the SFB-DHX9 from CPT-treated cells (Figure 6G). A consistent result was observed with SFB-DHX9 purified from HU-treated cells (Figure 6H). While our results suggest that SFB-DHX9 can directly engage with RPA, the efficiency of this binding was not particularly high. It remains unknown whether other mediators or modifications to DHX9 and RPA play roles in the interaction between the two. Collectively, these observations strongly suggest that DHX9 can directly interact with RPA and that this interaction is significantly enhanced upon DHX9 phosphorylation after DNA damage.
Cells expressing phospho-deficient DHX9 are hypersensitive to DNA damage
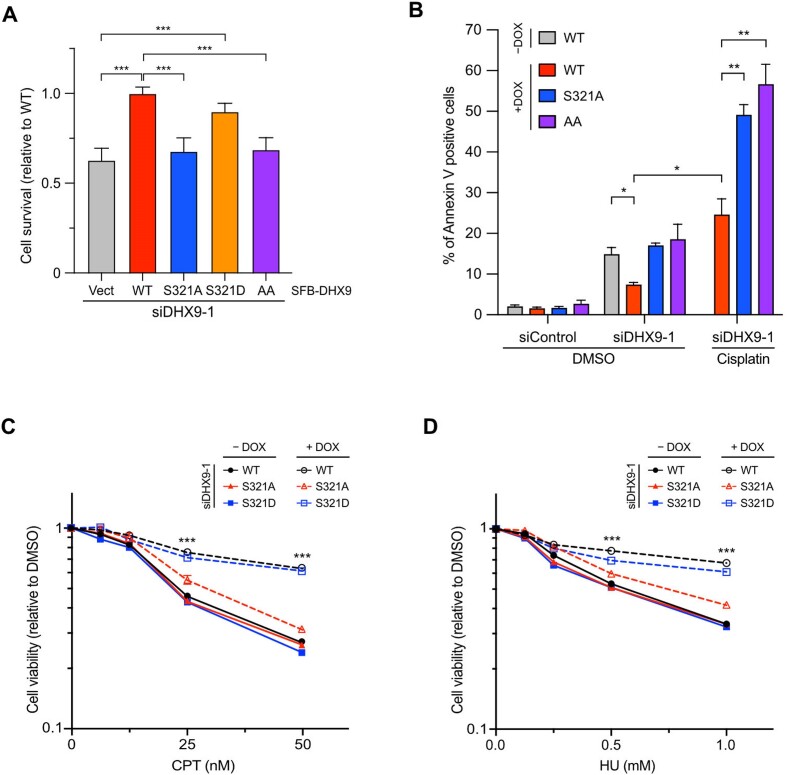
DHX9 plays a pivotal role in suppressing R-loops and maintaining genome stability. Since phospho-deficient DHX9 fails to associate with RPA and R-loops in response to DNA damage, we next investigated whether cells expressing phospho-deficient DHX9 exhibit defects in cell proliferation. In Figure 6A, consistent with previous studies, we found that cells carrying the corresponding empty vector showed viability defects when DHX9 was depleted. In marked contrast to DHX9WT, DHX9S321A failed to rescue the viability defects even when DNA damage was absent. Interestingly, the expression of phosphomimetic DHX9S321D in cells effectively supported cell viability, approaching the level of DHX9WT, indicating its ability to compensate for the absence of endogenous DHX9. Moreover, compared to DHX9S321A, we did not observe a further reduction in cell viability when cells expressed DHX9AA, suggesting that phosphorylation of S321 and S688 might function in the same pathway. Notably, the findings suggest that DHX9 phosphorylation at S321 might be important for supporting cell proliferation even in the absence of genotoxic stress.
To validate whether the decreased cell viability observed above was due to increased cell death, we conducted Annexin V-APC analysis to detect apoptotic cells. We noticed a significant fraction of apoptotic cells in stable cell lines expressing DHX9S321A and DHX9AA upon deletion of endogenous DHX9 (Figure 6B). Additionally, when we treated inducible cell lines with cisplatin, which causes DNA damage and R-loop accumulation, the cell lines carrying DHX9S321A and DHX9AA exhibited hypersensitivity to cisplatin treatment, suggesting that cancer cells expressing phospho-deficient DHX9 might be more susceptible to DNA damage. To extend this finding, we examined the sensitivity of inducible cell lines to different genotoxic drugs. Without doxycycline, all inducible cell lines with DHX9 knockdown displayed similar sensitivity in cell viability after CPT treatment. Whereas the cell line expressing DHX9S321A failed to complement the loss of DHX9 in terms of cell viability, the cell line expressing DHX9S321D effectively restored cell viability to the level of DHX9WT (Figure 6C). In addition, similar results were observed when we treated the inducible cell lines with HU (Figure 6D). Collectively, our findings demonstrate that phosphorylation of DHX9 at S321 promotes the elimination of R-loops after DNA damage, thereby ensuring genome stability and enhancing resistance against genotoxic agents.
Discussion
In this study, we present compelling evidence of the phosphorylation of DHX9 at S321 and S688, with S321 phosphorylation primarily induced by ATR in response to DNA damage. We reveal that DHX9 relies on its association with RPA and phosphorylation by ATR to suppress aberrant R-loops formed during genotoxic stress. The DDR is orchestrated by three key PIKKs, including ATM, ATR and DNA-PKcs, which phosphorylate substrate proteins involved in diverse cellular responses. Based on the association between DHX9 and DDR proteins such as BRCA1 and RPA32 after CPT treatment, we hypothesized that DHX9 could be a potential substrate of the PIKKs. Upon examining data from the PhosphoSitePlus website, we observed the top thirteen phosphorylated sites on DHX9 from system-wide studies, with S321 and S688 located within S/TQ motifs (Supplementary Figure S1H). However, the functional roles of these phosphorylated sites in the DDR remain uncharacterized. Notably, using phospho-specific antibodies, non-phosphorylatable mutants, and siRNA targeting ATR, we provide the first evidence demonstrating that S321 phosphorylation is primarily conducted by ATR in response to a variety of genotoxic agents in HeLa and U2OS cancer cell lines. On the other hand, the regulatory mechanisms governing S688 phosphorylation remain to be elucidated. It is worth noting that phosphorylated S688 is in immediate proximity to histidine, a basic residue that typically acts as a negative determinant for targeting by PIKKs (52). This may explain why the phosphorylation of S688 is not regulated by DNA damage. Additionally, a recent study suggested that oxaliplatin induces phosphorylation of DHX9 at S279 and S321 in colorectal cancer cell lines (53). However, direct evidence of DHX9 phosphorylation at S279 and S321 was lacking in that study. Emerging evidence suggests that oxaliplatin, rather than targeting DNA, plays a more direct role in affecting ribosomal RNA biogenesis (54,55). Nevertheless, whether the phosphorylation at S279 exists under different contexts or in different cell types needs further investigation.
DHX9 plays a crucial role in pre-mRNA processing during transcription and is essential for suppressing RNA processing defects including the occurrence of unscheduled R-loops (35,43,48,56,57). Our results propose a model in which activated ATR, by replication stress or other DNA damage, phosphorylates DHX9 at S321, thereby enhancing DHX9’s interaction with RPA32. This enhanced interaction promotes DHX9 to suppress genotoxic stress-triggered R-loops effectively. RPA senses DNA replication stress and DNA damage and binds to ssDNA serving as a platform for recruiting and activating ATR (5). Recent studies have unveiled an additional role of RPA as a sensor for R-loops (22,58), contributing to the recruitment and activation of RNH1 (22). Although RNH1 has been identified as an R-loop suppressor associated with RPA, previous mass spectrometry analysis showed that the RPA-ssDNA complex pulled down both RNH1 and DHX9, with DHX9 exhibiting a higher number of identified peptides (49). This raises questions about the relative affinity of DHX9 compared to RNH1 for RPA, and whether DHX9 and RNH1 work together to suppress R-loop accumulation requires further investigation. The phosphorylation of DHX9 at S321 occurs rapidly following DNA damage (Figure 1E and F), suggesting that DHX9 may swiftly bind to R-loops as it emerges and/or before the threats progress into deleterious DSBs. On the other hand, the observation of sustained S321 phosphorylation for up to 2 h following UV irradiation suggests that the dephosphorylation mechanism that removes pS321 may not be activated until the R-loops are fully resolved. Moreover, in the absence of DNA damage, the basal level interaction between DHX9 and RPA32 implies a weak or unstable association. However, upon treatment with CPT, this interaction is drastically enhanced, as depicted in Figure 3B and C, suggesting that phosphorylation on DHX9 stabilizes its interaction with RPA. This assumption gains further support from the observation that the ablation of S321 phosphorylation markedly attenuates the association of DHX9 with RPA32 and R-loops (Figures 3E, 4E, 6B–E). In contrast, the S321D mutation mimicking phosphorylation at S321 maintains the interaction of DHX9 with RPA32 and R-loops in cells, thus effectively suppressing aberrant R-loops formation following DNA damage (Figures 4E and 5B–D). Taken together, these results support the notion that DHX9 is not targeted to damage sites by the R-loop, but instead relies on its phosphorylation status for a strong association with RPA and R-loop.
Interestingly, similar defects to DHX9S321A are observed in the DHX9S688A mutation. Furthermore, the double mutations of S321 and S688 to alanines (DHX9AA) lead to almost complete disruption in RPA32 association (Figure 3E), suggesting that phosphorylation on both S321 and S688 is equally critical for the interaction with RPA32. Although DHX9AA exhibits more profound defects in interacting with RPA compared to any single mutant, the cell viability and Annexin V analyses of DHX9AA cells are comparable to those of DHX9S321A. It is possible that the ablation of any phosphorylation site is sufficient to disrupt the majority of the interaction with RPA, leaving R-loops unresolved. However, due to the limited information available regarding the kinase responsible for S688 phosphorylation and a fully resolved structure of hDHX9, it remains challenging to speculate on how the phosphorylation of S321 and S688 on DHX9 spatiotemporally affects its interaction with RPA32. More experiments will be needed to elucidate the kinase responsible for S688 phosphorylation and the role of pS688 in RPA association. A yeast study has demonstrated that DNA damage-induced phosphorylation circuit on Ddc2 (hATRIP) facilitates the recruitment of Mec1-Ddc2 to RPA-ssDNA, enabling the assembly of higher-ordered macromolecules of Mec1-Ddc2-RPA for cell cycle checkpoint activation (59). Additionally, the recruitment of PRP19 to RPA depends on PIKKs-mediated phosphorylation on RPA, which may stably interact through a positively charged pocket in PRP19 (60). In contrast, the recombinant RPA complex used in our in vitro DHX9 and RPA interaction assay was purified from E. coli, lacking critical post-translational modifications (PTMs) induced by DNA damage. Whether PTMs on RPA also play a crucial role in its interaction with phosphorylated DHX9 remains an open question. Further investigations are required to determine if the association between phosphorylated DHX9 and RPA is enhanced or stabilized via charge-charge interactions or in conjunction with different types of PTMs on both DHX9 and RPA. It is also important to consider that additional factors such as RNA processing factors or RPA-interacting partners, may be involved in the interplay between phosphorylated DHX9 and RPA32 following DNA damage. Such complexities cannot be simply recapitulated by in vitro binding analysis.
One fundamental function of DHX9 in preserving genome stability is to eliminate the aberrant R-loops. Upon treatment with CPT, DHX9 and PARP1 work in concert to counteract R-loop-associated DNA damage (35). In BRCA1/2-deficient breast cancer cells, RNF168 ubiquitylated DHX9 to facilitate its recruitment to R-loop-prone genomic loci, preventing the accumulation of R-loops, thereby promoting tumor growth (61). In line with the previous studies, we add an additional regulatory mechanism on DHX9. Cancer cells expressing phospho-deficient DHX9 not only fail to associate with R-loops but also display growth defects, increased apoptosis and hypersensitivity to genotoxic drugs (Figure 7A–D). Apparently, the R-loop suppressing function of DHX9 can be regulated by different PTMs. It is not known whether different PTMs on DHX9 affect each other or work together to promote R-loop resolution. In summary, RNA helicase DHX9 is a versatile protein that plays pivotal roles in RNA metabolism and genome stability. Understanding the molecular mechanisms underlying DHX9 function and its dysregulation in diseases could provide valuable insights into the development of targeted therapeutic interventions and diagnostic approaches.
Figure 7.
Cells expressing phospho-deficient DHX9 are hypersensitive to DNA damage. (A) Cells expressing phospho-deficient DHX9 showed defects in cell survival. HeLa cells devoid of endogenous DHX9 by siRNA were transiently expressed with SFB-tagged DHX9 (WT, S321A, S321D and AA) or the corresponding empty vector. Viable cell numbers were counted and expressed as the ratio of cell survival relative to DHX9WT. Data are presented as the mean ± SD of three independent experiments. *** P < 0.001. (B) Cells expressing phospho-deficient DHX9 led to increased apoptotic cells. HeLa inducible cell lines were transfected with control or DHX9 siRNA, and the expression of SFB-DHX9 (WT, S321A and AA) was induced with doxycycline. Cells treated with DMSO or 1 μM cisplatin for 24 h were collected for Annexin V analysis using Flow cytometry. Data are presented as the mean ± SEM of three independent experiments. * P < 0.05; ** P < 0.01. (C, D) Ablating S321 phosphorylation sensitized cells to DNA damage agents. HeLa inducible cell lines were transfected with DHX9 siRNA to deplete endogenous DHX9. The expression of SFB-DHX9 (WT, S321A and S321D) was induced with doxycycline. The cell lines were incubated with CPT (C) or HU (D) at the indicated concentrations for 24 h, and the cell viability was determined using Cell TiterGlo 2.0. Values of cell viability are relative to vehicle-treated cells and presented as the mean ± SD. All statistical significance was determined by Student's t-test (two-sided). *** P < 0.001.
Supplementary Material
Acknowledgements
The authors want to thank Drs Jing-Jer Lin and Shu-Chun Teng for reagents in the initial stages of establishing the laboratory and helpful discussions on this study. We thank Dr Kuen-Phon Wu for suggestions on our manuscript. We thank Yuen Chen of H.-P.C. Chu's lab for the technical support on DRIP and slot blot assays. We thank Dr Peter Chi for the recombinant human RPA complex. We would like to acknowledge the service provided by the Flow Cytometric Analyzing and Sorting Core of the First Core Laboratory, National Taiwan University College of Medicine, and the Common Mass Spectrometry Facilities for Proteomics and Protein Modification Analysis, Institute of Biological Chemistry, Academia Sinica. We thank all members of the Wu lab for their valuable contributions throughout this project.
Author contributions: M.-Y.L. conducted the majority of experiments throughout the entire study. K.-R.L. performed the experiments in Figures 1E, H, S1F, 2B and 2C. Y.-L.C. and M.-Y.L. co-worked on Figures 1F, I, S1E, S1I, 2D and E, S2B-D, 3C, and 6B, G. L.-Y.T. performed experiments in Figures 3E, S3B and 6C. K.-R.L., B.-Z.Y. and Y.-L.C. worked on Figure S3A. M.-Y.L. and B.-Z.Y. co-worked on experiments in Figures 1A–B, S1A–D, 2F–H, and 5A–D. C.-S.P.W. conducted Figure S2A. H.-P.C.C. provided reagents, technical support, and interpreted data. M.-Y.L. and C.-S.P.W. analyzed the data and wrote the manuscript. C.-S.P.W. conceived the study and supervised the project.
Contributor Information
Mei-Yin Liu, Department and Graduate Institute of Pharmacology, College of Medicine, National Taiwan University, Taipei 100233, Taiwan.
Keng-Ru Lin, Department and Graduate Institute of Pharmacology, College of Medicine, National Taiwan University, Taipei 100233, Taiwan.
Yuh-Ling Chien, Department and Graduate Institute of Pharmacology, College of Medicine, National Taiwan University, Taipei 100233, Taiwan.
Bing-Ze Yang, Department and Graduate Institute of Pharmacology, College of Medicine, National Taiwan University, Taipei 100233, Taiwan.
Li-Yu Tsui, Department and Graduate Institute of Pharmacology, College of Medicine, National Taiwan University, Taipei 100233, Taiwan.
Hsueh-Ping Catherine Chu, Institute of Molecular and Cellular Biology, National Taiwan University, Taipei 106319, Taiwan.
Ching-Shyi Peter Wu, Department and Graduate Institute of Pharmacology, College of Medicine, National Taiwan University, Taipei 100233, Taiwan.
Data availability
The data underlying this article are available in the article and in its online supplementary material. The phosphorylation sites underlying this article are available in ProteomeXchange at http://www.proteomexchange.org/ under accession code PXD043743. Further data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
NSTC, National Science and Technology Council [110-2628-B-002-037, 111-2628-B-002-018 to C.-S.P.W.]; NTU, National Taiwan University [110L7755, 111L7737, 112L7720 to C.-S.P.W.]; M.-Y.L. was supported by NTU [110L4000, 111L4000]. Funding for open access charge: National Taiwan University College of Medicine and NSTC.
Conflict of interest statement. None declared.
References
- 1. Blackford A.N., Jackson S.P.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davis A.J., Chen B.P., Chen D.J.. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst.). 2014; 17:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee J.H., Paull T.T.. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005; 308:551–554. [DOI] [PubMed] [Google Scholar]
- 5. Marechal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Traven A., Heierhorst J.. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005; 27:397–407. [DOI] [PubMed] [Google Scholar]
- 7. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y.et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 8. Stokes M.P., Rush J., Macneill J., Ren J.M., Sprott K., Nardone J., Yang V., Beausoleil S.A., Gygi S.P., Livingstone M.et al.. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:19855–19860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Munk S., Sigurethsson J.O., Xiao Z., Batth T.S., Franciosa G., von Stechow L., Lopez-Contreras A.J., Vertegaal A.C.O., Olsen J.V.. Proteomics reveals global regulation of protein SUMOylation by ATM and ATR kinases during replication stress. Cell Rep. 2017; 21:546–558. [DOI] [PubMed] [Google Scholar]
- 10. Bennetzen M.V., Larsen D.H., Bunkenborg J., Bartek J., Lukas J., Andersen J.S.. Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell. Proteomics. 2010; 9:1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brickner J.R., Garzon J.L., Cimprich K.A.. Walking a tightrope: the complex balancing act of R-loops in genome stability. Mol. Cell. 2022; 82:2267–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia-Muse T., Aguilera A.. R loops: from physiological to pathological roles. Cell. 2019; 179:604–618. [DOI] [PubMed] [Google Scholar]
- 13. Skourti-Stathaki K., Proudfoot N.J.. A double-edged sword: r loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014; 28:1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 15. Groh M., Gromak N.. Out of balance: r-loops in human disease. PLoS Genet. 2014; 10:e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sollier J., Cimprich K.A.. Breaking bad: r-loops and genome integrity. Trends Cell Biol. 2015; 25:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 18. Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R.. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003; 4:442–451. [DOI] [PubMed] [Google Scholar]
- 19. Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan Y.A., Aristizabal M.J., Lu P.Y., Luo Z., Hamza A., Kobor M.S., Stirling P.C., Hieter P.. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014; 10:e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M.. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014; 5:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nguyen H.D., Yadav T., Giri S., Saez B., Graubert T.A., Zou L.. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell. 2017; 65:832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., Hieter P.. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012; 26:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E.et al.. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009; 35:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwab R.A., Nieminuszczy J., Shah F., Langton J., Lopez Martinez D., Liang C.C., Cohn M.A., Gibbons R.J., Deans A.J., Niedzwiedz W.. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell. 2015; 60:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia-Rubio M.L., Perez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., Aguilera A.. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015; 11:e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L.et al.. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 2015; 57:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shivji M.K.K., Renaudin X., Williams C.H., Venkitaraman A.R.. BRCA2 Regulates transcription elongation by RNA polymerase II to prevent R-loop accumulation. Cell Rep. 2018; 22:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bhatia V., Barroso S.I., Garcia-Rubio M.L., Tumini E., Herrera-Moyano E., Aguilera A.. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014; 511:362–365. [DOI] [PubMed] [Google Scholar]
- 30. Chiang H.C., Zhang X., Li J., Zhao X., Chen J., Wang H.T., Jatoi I., Brenner A., Hu Y., Li R.. BRCA1-associated R-loop affects transcription and differentiation in breast luminal epithelial cells. Nucleic Acids Res. 2019; 47:5086–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mischo H.E., Gomez-Gonzalez B., Grzechnik P., Rondon A.G., Wei W., Steinmetz L., Aguilera A., Proudfoot N.J.. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 2011; 41:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alzu A., Bermejo R., Begnis M., Lucca C., Piccini D., Carotenuto W., Saponaro M., Brambati A., Cocito A., Foiani M.et al.. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell. 2012; 151:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sollier J., Stork C.T., Garcia-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A.. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014; 56:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boleslavska B., Oravetzova A., Shukla K., Nascakova Z., Ibini O.N., Hasanova Z., Andrs M., Kanagaraj R., Dobrovolna J., Janscak P.. DDX17 helicase promotes resolution of R-loop-mediated transcription-replication conflicts in human cells. Nucleic Acids Res. 2022; 50:12274–12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cristini A., Groh M., Kristiansen M.S., Gromak N.. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018; 23:1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song C., Hotz-Wagenblatt A., Voit R., Grummt I.. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017; 31:1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hodroj D., Recolin B., Serhal K., Martinez S., Tsanov N., Abou Merhi R., Maiorano D.. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017; 36:1182–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee T., Pelletier J.. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016; 7:42716–42739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anderson S.F., Schlegel B.P., Nakajima T., Wolpin E.S., Parvin J.D.. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998; 19:254–256. [DOI] [PubMed] [Google Scholar]
- 40. Nakajima T., Uchida C., Anderson S.F., Lee C.G., Hurwitz J., Parvin J.D., Montminy M.. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997; 90:1107–1112. [DOI] [PubMed] [Google Scholar]
- 41. Chakraborty P., Grosse F.. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.). 2011; 10:654–665. [DOI] [PubMed] [Google Scholar]
- 42. Chakraborty P., Grosse F.. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010; 38:4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chakraborty P., Hiom K.. DHX9-dependent recruitment of BRCA1 to RNA promotes DNA end resection in homologous recombination. Nat. Commun. 2021; 12:4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mischo H.E., Hemmerich P., Grosse F., Zhang S.. Actinomycin D induces histone gamma-H2AX foci and complex formation of gamma-H2AX with Ku70 and nuclear DNA helicase II. J. Biol. Chem. 2005; 280:9586–9594. [DOI] [PubMed] [Google Scholar]
- 45. Floro J., Dai A., Metzger A., Mora-Martin A., Ganem N.J., Cifuentes D., Wu C.S., Dalal J., Lyons S.M., Labadorf A.et al.. SDE2 is an essential gene required for ribosome biogenesis and the regulation of alternative splicing. Nucleic Acids Res. 2021; 49:9424–9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lei K.H., Yang H.L., Chang H.Y., Yeh H.Y., Nguyen D.D., Lee T.Y., Lyu X., Chastain M., Chai W., Li H.W.et al.. Crosstalk between CST and RPA regulates RAD51 activity during replication stress. Nat. Commun. 2021; 12:6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanz L.A., Chedin F.. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019; 14:1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aktas T., Avsar Ilik I., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A.. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017; 544:115–119. [DOI] [PubMed] [Google Scholar]
- 49. Marechal A., Li J.M., Ji X.Y., Wu C.S., Yazinski S.A., Nguyen H.D., Liu S., Jimenez A.E., Jin J., Zou L.. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell. 2014; 53:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Celis J.E., Celis A.. Cell cycle-dependent variations in the distribution of the nuclear protein cyclin proliferating cell nuclear antigen in cultured cells: subdivision of S phase. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:3262–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J.. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986; 89:123–130. [DOI] [PubMed] [Google Scholar]
- 52. Kim S.T., Lim D.S., Canman C.E., Kastan M.B.. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 1999; 274:37538–37543. [DOI] [PubMed] [Google Scholar]
- 53. Lin Y.C., Yu Y.S., Lin H.H., Hsiao K.Y.. Oxaliplatin-induced DHX9 phosphorylation promotes oncogenic circular RNA CCDC66 expression and development of chemoresistance. Cancers (Basel). 2020; 12:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sutton E.C., DeRose V.J.. Early nucleolar responses differentiate mechanisms of cell death induced by oxaliplatin and cisplatin. J. Biol. Chem. 2021; 296:100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bruno P.M., Liu Y., Park G.Y., Murai J., Koch C.E., Eisen T.J., Pritchard J.R., Pommier Y., Lippard S.J., Hemann M.T.. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med. 2017; 23:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yuan W., Al-Hadid Q., Wang Z., Shen L., Cho H., Wu X., Yang Y.. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res. 2021; 49:8573–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jain A., Bacolla A., Del Mundo I.M., Zhao J., Wang G., Vasquez K.M.. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013; 41:10345–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Feng S., Manley J.L.. Replication protein A associates with nucleolar R loops and regulates rRNA transcription and nucleolar morphology. Genes Dev. 2021; 35:1579–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yates L.A., Tannous E.A., Morgan R.M., Burgers P.M., Zhang X.. A DNA damage-induced phosphorylation circuit enhances Mec1(ATR) Ddc2(ATRIP) recruitment to Replication Protein A. Proc. Natl. Acad. Sci. U.S.A. 2023; 120:e2300150120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dubois J.C., Yates M., Gaudreau-Lapierre A., Clement G., Cappadocia L., Gaudreau L., Zou L., Marechal A.. A phosphorylation-and-ubiquitylation circuitry driving ATR activation and homologous recombination. Nucleic Acids Res. 2017; 45:8859–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patel P.S., Abraham K.J., Guturi K.K.N., Halaby M.J., Khan Z., Palomero L., Ho B., Duan S., St-Germain J., Algouneh A.et al.. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J. Clin. Invest. 2021; 131:e140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. The phosphorylation sites underlying this article are available in ProteomeXchange at http://www.proteomexchange.org/ under accession code PXD043743. Further data underlying this article will be shared on reasonable request to the corresponding author.