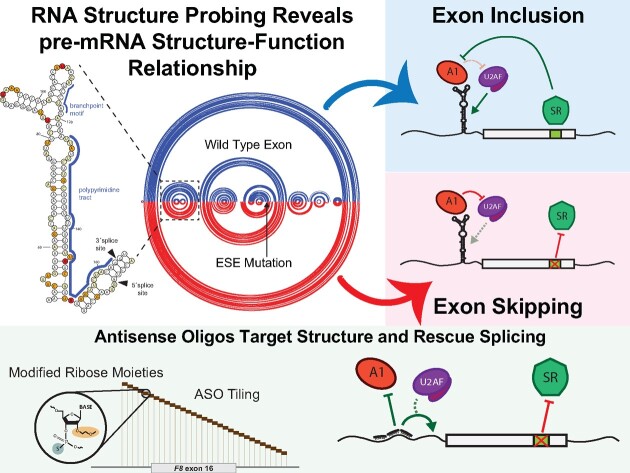
Abstract
Pathogenic variants in the human Factor VIII (F8) gene cause Hemophilia A (HA). Here, we investigated the impact of 97 HA-causing single-nucleotide variants on the splicing of 11 exons from F8. For the majority of F8 exons, splicing was insensitive to the presence of HA-causing variants. However, splicing of several exons, including exon-16, was impacted by variants predicted to alter exonic splicing regulatory sequences. Using exon-16 as a model, we investigated the structure–function relationship of HA-causing variants on splicing. Intriguingly, RNA chemical probing analyses revealed a three-way junction structure at the 3′-end of intron-15 (TWJ-3–15) capable of sequestering the polypyrimidine tract. We discovered antisense oligonucleotides (ASOs) targeting TWJ-3–15 partially rescue splicing-deficient exon-16 variants by increasing accessibility of the polypyrimidine tract. The apical stem loop region of TWJ-3–15 also contains two hnRNPA1-dependent intronic splicing silencers (ISSs). ASOs blocking these ISSs also partially rescued splicing. When used in combination, ASOs targeting both the ISSs and the region sequestering the polypyrimidine tract, fully rescue pre-mRNA splicing of multiple HA-linked variants of exon-16. Together, our data reveal a putative RNA structure that sensitizes F8 exon-16 to aberrant splicing.
Graphical Abstract
Graphical Abstract.
Introduction
Noncoding sequences (introns) interrupt protein-coding information (exons) in most human genes. Conserved sequences known as splice sites (ss) demarcate exon-intron boundaries (1). Messenger RNA (mRNA) biogenesis requires intron removal from precursor mRNA (pre-mRNA) and exon ligation; this step of gene expression is known as pre-mRNA splicing (2,3). The spliceosome, a multi-megadalton ribonucleoprotein (RNP) complex, assembles de novo on every intron to catalyze splicing reactions. This process involves the stepwise assembly of five uracil-rich small nuclear ribonucleoprotein particles (U snRNPs) and hundreds of accessory RNA-binding proteins on a pre-mRNA (4,5). To facilitate spliceosome assembly, exon definition is a critical initial step where exon-intron boundaries are defined (6). In this early spliceosome complex, U1 snRNP recognizes the 5′ss while the U2 snRNP auxiliary factor (U2AF) binds the 3′ss and polypyrimidine (poly-Y) tract (7–10). This initial step is highly regulated in cells by the presence or absence of sequences that can function as exonic splicing enhancers or silencers (11–16). Additionally, structured features within pre-mRNA can regulate splicing (17). For example, structured RNA elements influence splice site accessibility (18,19) by modulating protein–RNA interactions (20–22).
Aberrant splicing contributes to the etiology of many inherited diseases (23). Pathogenic variants impact pre-mRNA splicing through a variety of mechanisms. Most notably, variants remodel the cis-regulatory landscape of pre-mRNAs by ablation or creation of splice sites, and auxiliary splicing regulatory sequences such as exonic or intronic splicing enhancers (ESE and ISE, respectively) and splicing silencers (ESS and ISS, respectively). Splicing-sensitive variants cripple the integrity of the gene, resulting in the production of a faulty message that is either unstable or encodes an internally deleted protein (24–26). Antisense oligonucleotides (ASOs) are a promising therapeutic modality for rescuing pathogenic aberrant splicing patterns as their direct base pairing abilities make them highly customizable and specific to targets. Although challenges such as toxicity, delivery and stability represent barriers to the clinical translation of ASOs (27), solutions to these challenges exist, as exemplified by the recent FDA approval of multiple ASO drugs (28–31).
Our previous work implicated thousands of disease-causing variants in aberrant splicing of both constitutive and alternative exons (23,24). This work exploited the sequence bias of ESEs and ESSs to discover pathogenic genetic variants that cause the loss or gain of these putative functional elements relative to common, high allele frequency polymorphisms. Among these, the F8 gene had the highest frequency of variants predicted to affect splicing regulatory sequences (23). The F8 gene encodes for a procoagulant called Factor VIII (FVIII), which is required for initiating the coagulation cascade to form blood clots in response to wounds (32). Through this cascade, activated FVIII must bind to another procoagulant called Factor IX in its activated form. This initial interaction with FVIII sets off a series of biochemical reactions with additional procoagulants that forms a fibrin scaffold, serving as the basis for blood clotting. Thus, genetic variants can inactivate F8 and cause Hemophilia A (HA), a deadly, X-linked recessive bleeding disorder.
The F8 gene encodes 2351 amino acids split across 26 exons. Although the majority of these exons are constitutively spliced into a canonical 9032 nucleotide transcript, GENCODE (Version 43) contains several isoforms arising from alternative promoter usage and alternative 3′ss selection in exon-16 and intron-22. Despite modest support for alternative splicing, several constitutively spliced F8 exons are aberrantly spliced in HA patients, (33–36). These data suggest the hypothesis that a broad array of pathogenic missense variants in the F8 gene may manifest as splicing defects due to loss of exon identity or activation of cryptic splice sites. To test this hypothesis, we investigated 97 HA-causing variants on F8 pre-mRNA splicing using a heterologous reporter assay. We discovered that among the eleven exons studied, splicing of exon-16 was the most sensitive to HA-causing variants. RNA structure probing revealed a predicted intronic three-way junction that sequesters the exon-16 3′ss. ASOs targeting this structured element rescue diverse splicing-sensitive HA-causing variants of F8 exon-16, in both a heterologous and endogenous splicing context. Together, these data suggest an unexpected role for RNA structure in modulating exon identity and provide a rationale for the development of novel RNA therapeutics to potentially treat a subset of HA patients.
Materials and methods
F8 splicing reporters
The sequences of wild-type (WT) F8 exons and 100–250 nucleotides flanking each exon were amplified from human genomic DNA (Promega) using WT PCR primers shown in Supplementary Table S1. Following gel purification, PCR products were ligated into pACT7_SC14 (HBB minigene reporter as previously described (37)) using homology-based cloning technology (In-Fusion HD Cloning kit, Takara Bio). Following sequence verification, each plasmid was then used as a template for site-directed mutagenesis via overlap-extension PCR using mutagenesis primers shown in Supplementary Table S1. Splicing reporter plasmids were then sequence-validated using Sanger sequencing to confirm successful cloning and identity of splicing reporters. F8 minigene splicing reporters were derived using a similar approach, where the test exon in addition to flanking introns and exons, are cloned in between a strong promoter and polyadenylation site. The naming designation for each F8 variant investigated in this study is based on the Human Genome Variation Society (HGVS) nomenclature. Therefore, each pathogenic variant of an F8 exon presented in this study is based on the nucleotide being mutated (e.g. A > C), and its position within the coding DNA sequence context tested relative to the first ATG start codon (38). Each pathogenic variant’s designation based on the HGVS nomenclature is shown in Supplementary Table S1.
Cell-based in vitro splicing assays
HEK293T cells (ATCC) were cultured in 6-well tissue culture plates (CytoOne, USA Scientific) using Dulbecco’s Modified Eagle Medium (Gibco, supplemented with 10% FBS) at 37°C, 5% CO2. The cells were transiently transfected at ∼60–80% confluency with 2.5 μg of each F8 splicing reporter using Lipofectamine 2000 (Invitrogen). Total RNA was harvested from cells 24 h post-transfection using the Direct-zol RNA Miniprep kits (Zymo Research). Each splicing assay was performed with three independent/biological replicates. To ensure rigor and reproducibility for splicing-sensitive variants identified, splicing assays were performed with a total of nine independent/biological replicates, with the exception of exon-16c.5562G>T which contains 18 independent/biological replicates to assess variability.
ASO walk and combinatorial ASO experiments
2′-methoxyethyl (2′MOE) phosphorothioate substituted ASOs complementary to F8 exon-16 and flanking introns were designed from the reverse complement of the F8 sense sequence, creating non-overlapping 18-mers as shown in Supplementary Table S2. F8 exon-16 ASOs were designed to contiguously tile across the exon and its flanking introns. ASOs were synthesized by Integrated DNA Technologies (IDT). HEK293T cells (ATCC) were cultured in 96-well tissue culture plates (Perkin Elmer) as described above. Cells were transiently transfected with 250 ηg of WT or pathogenic variant splicing reporter and 10 ℘mol of each ASO (final concentration: ∼0.43 ηg/μl) using Lipofectamine 2000 (Invitrogen). 24 h post-transfection, cells were harvested and prepared for total RNA purification using the Quick-DNA/RNA Viral MagBead kit from Zymo Research and an Agilent Bravo NGS A liquid handler (39). Each experiment type (e.g. ASO walk or combinatorial ASO assays) was performed with three independent/biological replicates.
hnRNPA1 overexpression and western blot analysis
HEK293T cells (ATCC) were cultured in 6-well tissue culture plates as described above. Cells were co-transfected with 1.25 μg of the WT splicing reporter, 1.25 μg of either an empty expression vector or a T7-tagged hnRNPA1 expression vector, and 10 ℘mol of ASO(s) as described above. Total RNA and protein were isolated 24 h post-transfection using a RSB lysis buffer (10 mM Tris pH 7.0, 100 mM NaCl, 5 mM MgCl2, 0.5% NP40, 0.5% Triton X-100, and EDTA-free Protease Inhibitor Cocktail [Roche]). Following a 10 min incubation on ice, the cell lysate was then centrifuged at 10 000 × g for 10 min at 4°C. The supernatant was then collected and aliquoted for two separate applications. The first aliquot, comprising ∼90% of the cell lysate, was prepared for total RNA purification using the Direct-zol RNA Miniprep kits from Zymo Research. The remaining ∼10% of the cell lysate was then homogenized into a denaturing buffer solution containing 4X NuPAGE™ LDS Sample Buffer in preparation for polyacrylamide gel electrophoresis (Invitrogen™ NuPAGE™, 4–12%, Bis-Tris, 1.0–1.5 mm, Mini Protein Gels) and subsequent western blots. For western blots, protein samples were transferred to a Immobilon NC membrane (Millipore) using a Genie Blotter (Idea Scientific). Membranes were probed with anti-HSP90 (Santa Cruz Biotech) and anti-T7 (Novagen) monoclonal antibodies and visualized by HRP conjugated secondary antibodies and chemiluminescence (Pierce). These experiments were performed with three independent/biological replicates.
Two-step RT-qPCR and analysis of splicing reporter assays
A minimum of 500 ηg of purified total RNA was used as input for all first-strand cDNA synthesis using random primers and Multiscribe Reverse Transcriptase (Applied Biosystems). The resulting cDNA was then used as a template for endpoint PCR amplification using specific primers that detect our mRNA splicing reporter isoforms, Globin F: 5′-CGCAACCTCAAACAGACACC-3′; Globin R: 5′-TGAGCTGCACTGTGACAAGC-3′. The forward primer of the pair contains a 5′-FAM modification. The resulting amplicons were then analyzed using agarose gel electrophoresis to empirically evaluate mRNA isoforms detected. Intron-spanning primers against SRSF3 mRNA were used as an endogenous internal control (SRSF3 F: 5′-GTAAGAGTGGAACTGTCGAATGG-3′; SRSF3 R: 5′-CGATCTCTCTCTTCTCCTATCTCTAG-3′). The abundance of each 5′-FAM labeled mRNA isoform is quantified using capillary electrophoresis and fragment analysis (UC Berkeley, DNA Sequencing Center). For fragment analysis, each sample is suspended in a formamide solution that contains a proper size standard for sizing detected fragments (GeneScan 1200 Liz, Applied Biosystems). Analysis was performed in PeakScanner (Thermofisher). Quantification of splicing efficiency is achieved by comparing relative fluorescence units (RFU) between 5′-FAM labeled reporter isoforms that include or exclude an exon of interest. The RFU detected for each reporter isoform is then plugged into the following formula to calculate the PSI index, which reflects the splicing efficiency of an exon in either the WT or pathogenic variant context:
 |
The mean PSI for a given reporter context is then calculated using all its respective replicates for a corresponding experiment. Statistical significance in the differences between the mean PSI of the control group(s) versus the experimental group(s) is determined using analysis of variance (ANOVA), and Dunett’s post-hoc test. All statistical tests for PSI analysis were done in GraphPad Prism 9. Values are determined to be statistically significant if the calculated P-value is below an alpha value of ≤0.05.
In vitro transcribed RNA for F8 exon-16 WT and pathogenic variants
Templates for WT or pathogenic variants of F8 exon-16 pre-mRNA sequences corresponding to the reporter plasmid inserts were synthesized by a primer assembly reaction designed using Primerize (40). RNA was purified by denaturing PAGE and eluted from gel slices overnight in 10 mM Tris pH 7.5, 480 mM sodium acetate, 1 mM EDTA, 0.1% SDS. Following ethanol precipitation, transcripts were resuspended in ddH2O and quantified by UV spectrophotometry.
In vitro SHAPE-MaP-seq of F8 targets
F8 exon-16 in vitro transcribed pre-mRNA sequences were first denatured by incubating at 95°C for 3 min in 65 mM Na-HEPES (pH 8.0). The denatured RNA was then allowed to slowly cool to room temperature (RT) for 15 min, after which MgCl2 was supplemented to 1 mM for a total volume of 15 μl and incubated at RT for an additional 5 min. To chemically modify RNA, 2-aminopyridine-3-carboxylic acid imidazolide (2A3) was added to a final concentration of 100 mM and incubated for 2 min at 37°C (41,42). The reaction was then quenched using dithiothreitol (DTT) to a final concentration of 500 mM at RT for 10 min. Reactions which substituted anhydrous dimethyl sulfoxide (DMSO) for 2A3 were used as negative controls. All modified RNAs, including negative controls, were then purified using RNA Clean & Concentrator-5 (Zymo Research). Modified RNAs were fragmented to a median size of 200 nucleotides by incubation at 94°C for 1 min using NEBNext® Magnesium RNA Fragmentation Kit and then purified using NEB’s recommended ethanol precipitation protocol. Purified RNA was then prepared for reverse transcription, incubating the RNA with 1 μl of 10 mM dNTPs and 2 μl of 20 μM random hexamers at 70°C for 5 min, followed by immediate transfer to ice. Reverse transcription reactions were then supplemented with 4 μl of 5X RT buffer (250 mM Tris-HCl pH 8.3, 375 mM KCl), 2 μl of 0.1 M DTT, 1 μl of 120 mM MnCl2, 10 U of SUPERase RNase Inhibitor, and 200 U of Superscript II Reverse Transcriptase (SSII, ThermoFisher Scientific, cat. 18064014) to a final volume of 20 μl. These reactions were then incubated at 25°C for 10 min to allow for partial primer extension, followed by incubation at 42°C for 3 h to enable efficient extension. SSII was then heat-inactivated by incubation at 75°C for 20 min. Reverse transcription reactions were then supplemented with EDTA to a final concentration of 6 mM to chelate Mn2+ ions and incubated at RT for 5 min. MgCl2 was then added to a final concentration of 6 mM for each reaction (41). Reverse transcription reactions were then used as input material for NEBNext® Ultra™ II DNA library Prep Kit for Illumina® (New England Biolabs, cat. E7645L), using NEBNext Multiplex Oligos for Illumina® (Unique Dual index UMI Adaptors DNA Set 1, cat. E7395). Subsequent reactions were performed following manufacturer instructions. All sequencing was performed on an Illumina iSeq100 instrument using a paired-end 2 × 150 sequencing reagent cartridge and flow cell. All SHAPE-MaP-seq experiments were performed in duplicate.
SHAPE-MaP data analysis and RNA structure prediction
All the relevant data analysis steps were conducted using RNA Framework v2.6.9 (43). Reads produced from Illumina libraries were pre-processed and mapped using the rf-map module (parameters: -b2 -mp ‘–no-mixed –no-discordant’ -bs), ensuring only paired-end mates with expected mate orientation were considered with Bowtie2. The mutational signal was obtained using the rf-count module (parameters: -m -pp -nd -ni), enabling mutation counts of reads produced from properly paired mates. Mutational signal was normalized relative to an unmodified control using parameters (-sm 3 -nm 1 -mu 0.05) and further normalized using the 2–8% normalization approach provided by RNA Framework (44). Normalized reactivities were then supplied to RNAstructure to generate data-driven predicted structure models (45).
In silico analysis of splice site strength and protein-RNA interactions
MaxEntScan was used to analyze splice 5′ and 3′ site sequences of exons used in this study (46). To determine 3′ss strength, 23-mer sequences encompassing the last 20 intronic positions and first three exonic positions were selected for analysis. For 5′ss strength, 9-mers consisting of the last three positions of each exon and the first six positions of the downstream intron were selected for analysis. The tool RBPmap was used to identify putative protein-RNA interactions sites within F8 pre-mRNAs (47). In this analysis, we used sequences corresponding to F8 exon-16 and flanking introns from the splicing reporter. We included a high stringency constraint to match known RBP motifs within our input sequence, as well as a conservation filter to selectively identify motifs that best match sequences from the human and mouse genomes.
Results
Discovery of splicing-sensitive HA-causing variants in F8
We previously described inherited disease-causing variants with the potential to alter the landscape of ESEs or ESSs (24). Among all candidate genes, F8 had the highest number of variants per exon and total number of putative splicing-sensitive point variants (23). To determine whether these HA-causing variants can induce aberrant splicing, we analyzed 97 distinct variants across eleven F8 exons by generating heterologous splicing reporters where the wild-type (WT) or pathogenic variant for each F8 exon, plus 100–250 bp of flanking intron sequence, were cloned into the human beta globin (HBB) minigene splicing reporter (Figure 1A and Supplementary Table S1). Following transient transfections of each reporter into HEK293T cells, HBB reporter mRNA isoform levels are quantified by a two-step, semi-quantitative end-labeled RT-qPCR assay and fragment analysis via capillary electrophoresis.
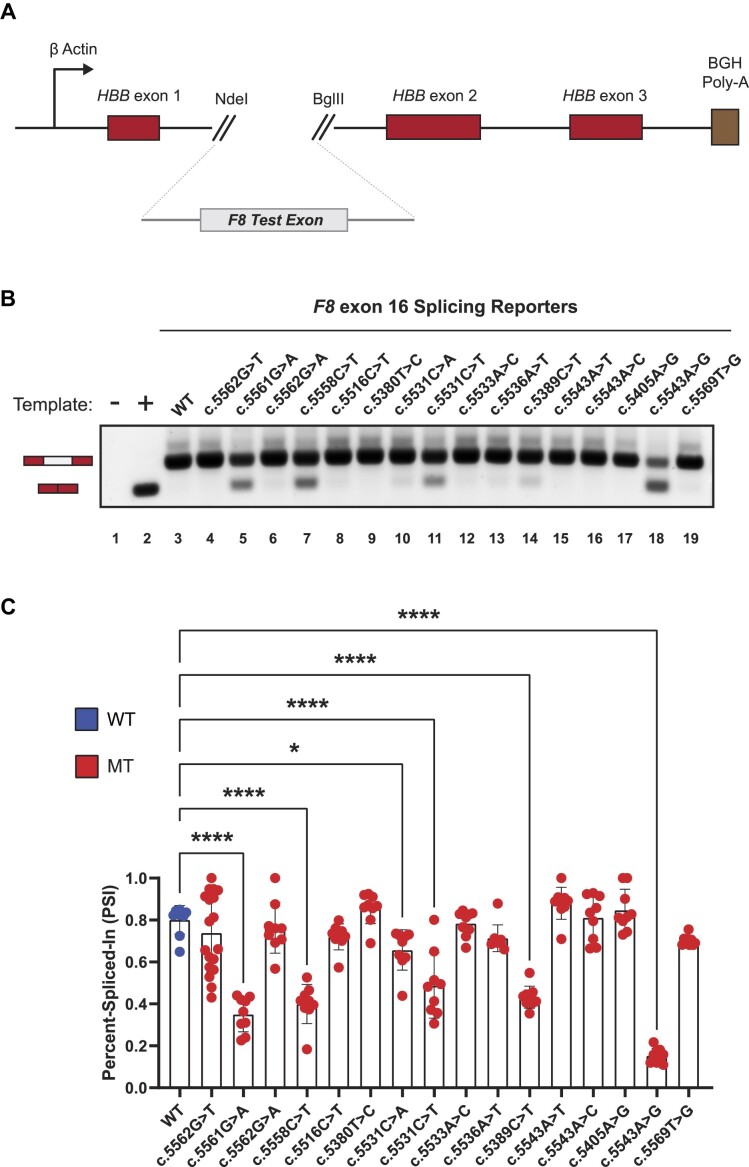
Figure 1.
In vitro cell-based splicing reporter assays reveal F8 exon-16 as a highly fragile exon susceptible to pathogenic variant-induced aberrant splicing. (A) Schematic of the heterologous splicing reporter used to assess the impacts of variants on splicing. Each test exon and flanking intronic sequence of F8 gene was cloned between exon-1 and exon-2 of the HBB minigene. (B) A representative agarose RNA gel showing the effects on splicing by various HA-causing variants in a panel of F8 exon-16 splicing reporters. Controls include a no template reaction (lane 1) and a positive control for exon skipping (lane 2). (C) Quantification of various HA-causing variants on the splicing extent of F8 exon-16. Percent-spliced-in (PSI) refers to the ratio of test exon skipped to test exon included in mRNA. Statistical significance between comparisons is denoted by asterisks that represent P-values with the following range of significance: *P≤ 0.05, and ****P≤ 0.0001. Statistical significance was determined using analysis of variance (ANOVA), and Dunett’s post-hoc test. Each exon-16 splicing reporter context tested contains nine independent/biological replicates; only exon-16c.5562G>T contains 18 independent/biological replicates to assess variability.
Out of the eleven F8 exons tested, HA-causing variants across seven F8 exons did not cause exon skipping (Supplementary Figure S1). In contrast, we found that HA-causing variants in four exons (exon-7, exon-11, exon-16 and exon-18) caused exon skipping in the heterologous reporter context, indicating that the splicing fidelity of these exons may be particularly sensitive to variants (Figure 1B and Supplementary Figure S2). For example, Figure 1B shows splicing assays for 16 HA-causing variants of exon-16. In comparison to the WT exon-16 splicing reporter which is efficiently spliced (lane 3), we found that six pathogenic variants, namely exon-16c.5561G>A, exon-16c.5558C>T, exon-16c.5531C>A, exon-16c.5531C>T, exon-16c.5389C>T and exon-16c.5543A>G significantly reduced exon-16 inclusion (Figure 1B,C). These results show that the in vitro splicing of F8 exon-7, -11, -16 and -18 are sensitive to HA-causing variants.
A predicted three-way junction RNA structure occludes the 3′ss of F8 exon-16
Pathogenic variants have previously been reported to disrupt native RNA structures and as a consequence alter their biological function (48). To determine how splicing-sensitive HA-causing variants influence the structure of fragile exons like F8 exon-16, we performed selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling coupled to high-throughput sequencing (SHAPE-MaP-seq) on in vitro transcribed (IVT) RNA. Both the WT and pathogenic variant containing IVT exon-16 RNA encompasses the same sequence context as the splicing reporters. As such, for all SHAPE probing data presented, all nucleotide position numbering shown is based relative to the IVT RNA template (Supplementary Figure S3A). Accessible nucleotides strongly react with 2-aminopyridine-3-carboxylic acid imidazolide (2A3) in SHAPE-MaP-seq assays (41). Nucleotides acylated by 2A3 can vary in their degree of modification, or SHAPE reactivity, reflecting how accessible the nucleotides may be in the context of the global folding architecture of the RNA (41,44,49).
Using 2A3 in our SHAPE-MaP-seq assays, we first generated SHAPE reactivity profiles for WT exon-16 and the highly splicing-sensitive exon-16c.5543A>G variant (Figure 2A,B). We used arc diagrams to compare SHAPE-driven folding predictions of WT and exon-16c.5543A>G transcripts (Figure 2C). The exon-16c.5543A>G variant induces subtle RNA structural changes, creating several long-range base pairing interactions within the exon and between the flanking introns. Like exon-16c.5543A>G, additional SHAPE probing demonstrates that other splicing-sensitive HA-causing variants in exon-16 also exhibited modest structural changes compared to the WT transcript (Supplementary Figures 3B–G and S4). Taken together, the SHAPE-MaP-seq experiments suggest that splicing-sensitive HA-causing variants modestly impact the secondary structure of exon-16 compared to the WT context.
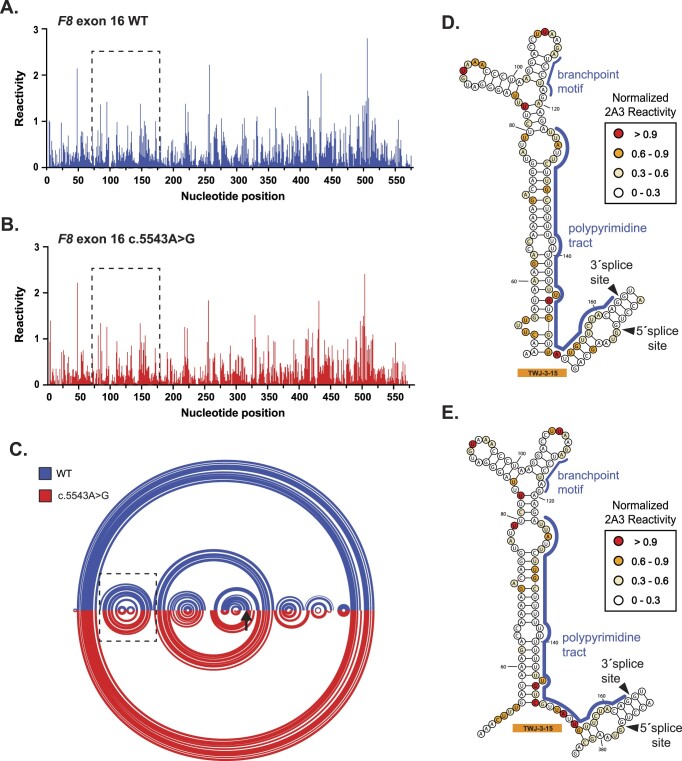
Figure 2.
SHAPE probing identifies a native RNA structure (TWJ-3–15) that is uniquely positioned at the 3′ss of F8 exon-16. (A,B) A normalized SHAPE reactivity plot for WT exon-16 (blue) and the exon-16c.5543A>G pathogenic variant (red), respectively. (C) Intramolecular base pairing interactions, constrained by normalized SHAPE reactivity are represented by arcs joining different regions of the transcript. Arc diagrams for WT exon-16 and exon-16c.5543A>G transcripts are depicted in blue and red, respectively. The broken box indicates the position of TWJ-3–15. The black arrow signifies the position of the c.5543A > G variant (D,E) SHAPE-driven secondary structure prediction of TWJ-3–15 depicted in its 2D state for WT exon-16 and exon-16c.5543A>G transcripts, respectively. Core splicing signals are annotated within the structure. All SHAPE probing data (N = 2) presented were generated in vitro using the SHAPE reagent 2A3, and all subsequent data analysis was performed in RNA Framework. All nucleotide positions numbering shown are based on the IVT RNA template used for SHAPE probing, from the 5′ to 3′ orientation.
When analyzing both the WT and all pathogenic variant SHAPE-driven structure predictions together, we discovered a three-way junction RNA structure involving the 3′ss of F8 exon-16 that is supported by the experimental reactivity patterns but not from in silico RNA folding alone (Figure 2D,E; Supplementary Figures S5 and 6). Our SHAPE-driven structural predictions suggest that an upstream region of F8 intron-15 base pairs with the branchpoint and poly-Y tract, potentially occluding their accessibility to splicing factor 1 (SF1) and U2AF. This 3′ss three-way junction structure, which we will refer to as TWJ-3–15 (Three-Way Junction at the 3′-end of intron-15), may be a potential target for splice-modulating compounds to enhance splice site strength and therefore increase exon-16 inclusion during splicing.
TWJ-3-15 attenuates F8 exon-16 3′ss strength
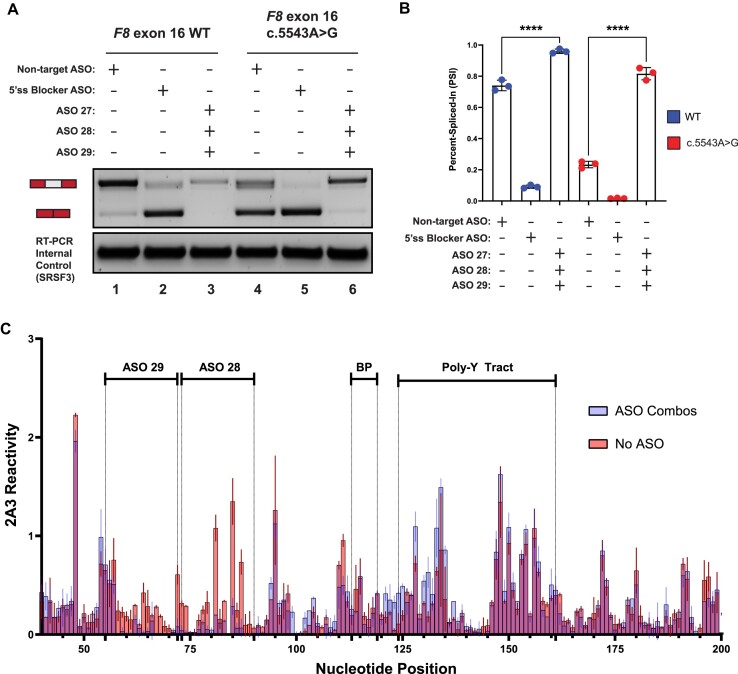
To determine if splicing-sensitive HA-causing variants of exon-16 can be rescued by ASOs, we designed non-overlapping, phosphorothioate-substituted, 2′-methoxyethyl modified 18-mer ASOs that span exon-16 and its flanking introns in a splicing reporter context (Figure 3A; Supplementary Table S2 and Supplementary Figure S7). We include an ASO that has no complementarity to F8 sequences, serving as our non-targeting (NT) control (Figure 3B, lane 1), and an ASO ‘blocker’ that specifically targets the 5′ss of exon-16 to directly inhibit its splicing (Figure 3B, lane 2). Co-transfection of ASO32, ASO30, ASO29, ASO27, ASO6, ASO4 or ASO3 with the exon-16c.5543A>G reporter resulted in a significant increase in exon-16 inclusion relative to the control (Figure 3C). Of note, these ASOs target the upstream and downstream intronic regions of exon-16c.5543A>G. ASO32, ASO30, ASO29 and ASO27 target regions upstream of the 3′ss, including sequences comprising TWJ-3–15. These ASOs significantly increase inclusion of exon-16c.5543A>G (Figure 3C, P-value < 0.0001). ASO6, ASO4 and ASO3 target a region downstream of the 5′ss and also significantly increase exon-16c.5543A>G inclusion (Figure 3C, P-value < 0.0008). We also observed that ASOs targeting exon-16, including the site of the variant at c.5543A > G, inhibited splicing. As a control we repeated the ASO walk on the WT F8 exon-16 reporter. As observed from the exon-16c.5543A>G walk, ASOs directly targeting the exon, except for ASO21, strongly inhibited the inclusion of WT exon-16 during splicing (Figure 3D). By contrast, individual ASOs targeting the flanking introns had little impact on WT exon-16 splicing relative to the NT control. Interestingly, ASO6 and ASO4 which target the intronic region downstream of the 5′ss enhanced exon-16 splicing relative to the NT control (Figure 3D, P-value < 0.0011). Taken together, these ASO walks indicate that targeting regions adjacent to F8 exon-16 with ASOs, but not the exon itself, may rescue inclusion of splicing-sensitive HA-causing exon-16 variants by perturbing the influence of inhibitory elements found in the flanking introns.
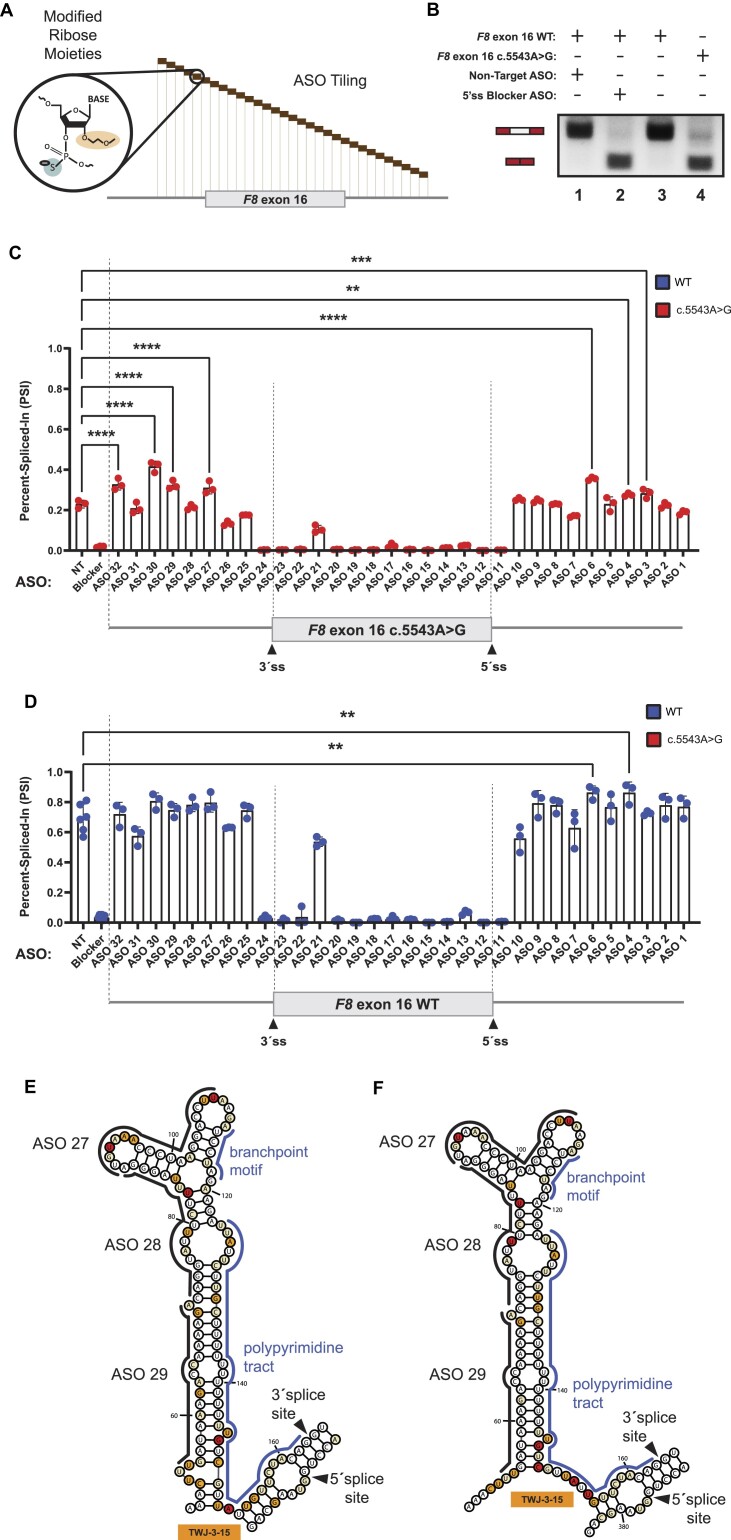
Figure 3.
ASO walk reveals splice-modulating ASOs for the highly splicing-sensitive exon-16c.5543A>G variant. (A) A mock schematic of an ASO walk. Each ASO used in our walks are 18 nucleotides in length and are designed using ribose sugars that are modified with a 2′-methoxyethyl group (2′-MOE, highlighted in light orange), and the phosphate backbone is modified to a phosphorothioate backbone (highlighted in light blue). Each 18-mer ASO is contiguous by design, tiling across exon-16 and its flanking introns with no overlaps between each ASO. (B) Proof-of-concept demonstrating how our ASOs are expected to work in the ASO walk experiments. As shown in the annotative matrix above a representative agarose gel, the first two controls consist of our 5′ss blocker ASO (positive control) and our non-targeting ASO (negative control) being co-transfected with our WT exon-16 splicing reporter to demonstrate that our designed ASOs can modulate splicing. The last two controls consist of our WT exon-16 and exon-16c.5543A>G splicing reporters without ASOs co-transfected to illustrate the typical splicing ratios we may expect to see from their splicing. Expected mRNA isoforms including or excluding the test exon are also annotated to the left of the agarose gel. (C) and (D) show our ASO walk data for the exon-16c.5543A>G variant and WT exon-16, respectively. Data corresponding to the exon-16c.5543A>G variant is annotated by a red color whereas the WT is annotated by a blue color. ASO walk results for both (C) and (D) are quantified using the PSI ratio. Statistical significance between comparisons are denoted by asterisks that represent P-values with the following range of significance: ns, P> 0.05, **P≤ 0.01, ***P≤ 0.001, and ****P≤ 0.0001. Statistical significance was determined using analysis of variance (ANOVA), and Dunett’s post-hoc test. Each exon-16 splicing reporter context and condition tested contains three independent/biological replicates. A schematic model of exon-16 and its flanking introns are shown at the bottom of each plot to illustrate relative positions of ASOs. (E) and (F) respectively depict SHAPE-driven secondary structure predictions of TWJ-3–15 for WT exon-16 and exon-16c.5543A>G, where each (E) and (F) show where promising ASOs are hybridizing to within the structure. The sequence is numbered according to the nucleotide positions of the heterologous splicing reporter, from the 5′ to 3′ orientation.
Comparison of our SHAPE and ASO walk data revealed that ASOs which individually and most significantly improved splicing of exon-16c.5543A>G achieved this effect by directly targeting TWJ-3–15 (Figure 3E,F). These ASOs include ASO29 and ASO27, which had a statistically significant improvement on exon-16c.5543A>G splicing, and ASO28 which also targets the structure but was unable to significantly rescue splicing of exon-16c.5543A>G. Thus, our data supports the hypothesis that co-transfecting more than one ASO targeting TWJ-3–15 may further destabilize this structure to increase 3′ss accessibility and thereby enhance proper splicing of exon-16.
Splice-modulating ASOs alter the predicted structure and function of TWJ-3-15
To test the hypothesis that a combination of ASOs can additively enhance splicing of F8 exon-16c.5543A>G compared to a single ASO when targeting TWJ-3–15, we performed additional cell-based splicing assays where exon-16c.5543A>G was co-transfected with ASO29, ASO28 and ASO27. Due to the observation that WT exon-16 is inefficiently spliced (Figure 1C), we also co-transfected the WT reporter with the trio ASO combination in parallel. Remarkably, for both the WT and exon-16c.5543A>G reporters, we discover that the trio ASO combination had a significant effect on exon-16 inclusion (Figure 4A,B). Relative to WT exon-16 co-transfected with the NT control (Figure 4A,B), the trio ASO combination can enhance the splicing efficiency of WT exon-16 (Figure 4A,B), significantly increasing exon-16 inclusion (P-value < 0.0001). Similarly, we observed that the trio combination significantly rescued exon-16c.5543A>G splicing (Figure 4A,B, P-value < 0.0001) and completely restores splicing of exon-16c.5543A>G to WT levels (Figure 4A,B, compare lanes 1 and 6). Additionally, in comparing this result to a preliminary screen consisting of a duo ASO combination that includes ASO29 and ASO28 which worked the best (Supplementary Figure S8), the trio combination comprising ASO29, ASO28 and ASO27 had a stronger additive effect on rescuing exon-16 splicing, also demonstrating that ASO27 is necessary to improve splicing.
Figure 4.
A combination of ASOs targeting TWJ-3–15 can rescue splicing of the highly splicing-sensitive exon-16c.5543A>G variant by increasing 3′ss accessibility. (A) A representative agarose gel depicting the results from our in vitro cell-based splicing assays testing duo and trio ASO combinations’ ability to modulate reporter splicing (upper panel). The lower panel depicts an internal control corresponding to the SRSF3 mRNA (lower panel). Each splicing assay condition is annotated as shown in the matrix above the gel. Expected mRNA isoforms including or excluding the test exon are also annotated to the left of the agarose gel. (B) A plot quantifying the results from (A) using the PSI ratio. The WT context is annotated by a blue color whereas the exon-16c.5543A>G pathogenic variant is annotated by a red color. The same annotative matrix seen in (A) is used under the plot to label each ASO condition tested for each context. Statistical significance between comparisons are denoted by asterisks that represent P-values with the following range of significance: ns, P> 0.05, and ****P≤ 0.0001. Statistical significance was determined using analysis of variance (ANOVA) and Dunett’s post-hoc test. Each exon-16 splicing reporter context and condition tested contains three independent/biological replicates. (C) An overlay plot comparing normalized 2A3 reactivities between two distinct SHAPE probing conditions used to probe the exon-16c.5543A>G variant. One SHAPE condition probes exon-16c.5543A>G with ASOs present (annotated light blue), and the other condition probes exon-16c.5543A>G without ASOs present (annotated light red). Admixing of colors (indicated by purple hue) where this is indistinguishable overlap represents similar SHAPE reactivity values between the two probing conditions at that nucleotide position. The nucleotide positions where the ASOs bind, in addition to important splicing signals, are annotated in the plot. All SHAPE probing data presented were generated in vitro using the SHAPE reagent 2A3, and all subsequent data analysis was performed in RNA Framework. The sequence is numbered according to the nucleotide positions of the heterologous splicing reporter, from the 5′ to 3′ orientation.
To test the hypothesis that ASOs targeting TWJ-3–15 increases the accessibility of the poly-Y tract by antagonizing the TWJ-3–15 structure, we performed additional chemical probing experiments. Briefly, we adapted the same SHAPE-MaP-seq approach as previously described with the exception that prior to SHAPE probing, the in vitro RNA template was unfolded and then re-folded in the presence of ASOs. In order to draw comparisons between the probing condition with ASOs present and the control condition without ASOs, we calculated the average SHAPE reactivity for each nucleotide from the respective datasets and plotted them together (Figure 4C). Differences between each dataset’s SHAPE reactivities are represented by their distinct color annotation, whereas predominant admixing of colors represent minimal differences. Accordingly, the results from this experiment show that there is an increased shift in SHAPE reactivities for nucleotides that surround or comprise the poly-Y tract in the condition with ASO29 and ASO28 present, compared to the condition with no ASOs present (Figure 4C). The reduced SHAPE reactivities in the regions with ASO29 and ASO28 complementarity provide a direct measure of ASO binding to the intended target sites (Figure 4C). Additionally, a single ASO such as ASO27 is also capable of increasing the SHAPE activities for nucleotides comprising the poly-Y tract (Supplementary Figure S9). These results indicate that appropriately designed ASOs enhance exon-16 splicing in part by destabilizing TWJ-3–15 to increase the accessibility of the 3′ss, likely increasing the accessibility of the poly-Y tract to U2AF.
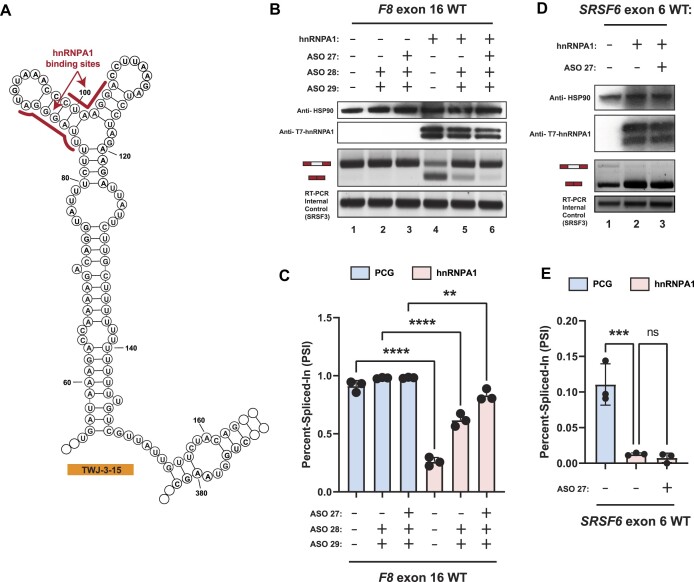
TWJ-3-15 harbors hnRNPA1-dependent splicing silencers
Results of our RNA structure probing experiments in the presence and absence of specific ASO combinations suggest that TWJ-3–15 reduces the strength of F8 exon-16 definition by occluding the poly-Y tract. To determine if TWJ-3–15 may also contain any functional binding sites for RNA-binding proteins (RBPs), we used RBPmap to identify RBP consensus motifs within the structure (47). We found two binding sites for the splicing repressor hnRNPA1 within TWJ-3–15 (Figure 5A, indicated in red). Based on nucleotide position numbering relative to the IVT RNA template used for SHAPE probing (Supplementary Figures S3A, 6 and 7), and subsequent analysis of TWJ-3–15, the first potential binding site is found at nucleotide positions 84–90 (UUAGGGA) and the second motif is found at nucleotide positions 99–105 (CUAAGGA). We term these predicted hnRNPA1 binding sites as ISS-15–1 and ISS-15–2, respectively. Based on published research, these predicted binding sites harbor motifs either identical or highly similar to the hnRNPA1 consensus motif, ‘UAGG’ (50,51). These predicted hnRNPA1 binding sites are positioned within the apical stem loop region of TWJ-3–15. Intriguingly, ASO28 and ASO27 directly bind ISS-15–1 and ISS-15–2, respectively. These ASOs, when used individually or in combination, antagonized the aberrant splicing of the exon-16c.5543A>G variant (Figure 3C and Supplementary Figure S8A). We hypothesize that this structured element weakens the F8 exon-16 3′ss by sequestering the poly-Y tract and recruiting the splicing repressor protein hnRNPA1. To determine if TWJ-3–15 is sufficient to induce hnRNPA1-dependent exon skipping we replaced the robust 3′ss of F8 exon-15 with the 3′ss of exon-16, which includes TWJ-3–15 (see Supplementary Figure S1). By contrast to the WT exon-15 reporter, we found that overexpression of hnRNPA1 induced significant levels of exon skipping in the TWJ-3–15 chimeric reporter (Supplementary Figure S10). These data suggest that TWJ-3–15 is sufficient to induce hnRNPA1-dependent exon skipping in a heterologous context. If hnRNPA1 recognizes ISS-15–1 and ISS-15–2, we predict that ASO28 and ASO27 will block hnRNPA1-dependent exon skipping. To test this hypothesis, we co-transfected the WT F8 exon-16 splicing reporter into HEK293T cells with or without hnRNPA1 overexpression. We performed these experiments with several ASO conditions: (1) transfecting ASO28 and ASO29 to mask one putative hnRNPA1 binding site, ISS-15–1, while exposing the poly-Y track, (2) or transfecting ASO27, ASO28 and ASO29 to mask both putative hnRNPA1 binding sites, ISS-15–1 and ISS-15–2, while simultaneously exposing the poly-Y track in TWJ-3–15. As expected, overexpression of hnRNPA1 strongly inhibits splicing of WT exon-16 (compare lane 4 to lane 1, Figure 5B,C). By contrast, we observed that the combination of ASO28 and ASO29 partially attenuated the effect of hnRNPA1 overexpression, and the combination of all three ASO resulted in the strongest attenuation hnRNPA1-dependent exon-16 skipping (compare lane 4 to lanes 5 and 6). Taken together, these data suggest that ISS-15–1 and ISS-15–2 are recognized by hnRNPA1 and that masking both sites is required to rescue splicing efficiently.
Figure 5.
hnRNPA1 cooperates with TWJ-3–15 to amplify inhibitory effects at the 3′ss of F8 exon-16. (A) Secondary structure model of TWJ-3–15 showing predicted hnRNPA1 binding motifs underscored in red. The sequence is numbered according to the nucleotide positions of the heterologous splicing reporter, from the 5′ to 3′ orientation. (B) Representative Western blot and agarose gel depicting results from our hnRNPA1-ASO competition assay. Upper two panels depict western blots for HSP90 and T7-epitope tagged hnRNPA1, respectively. Lower two panels depict the HBB splicing assay and the SRSF3 internal control for the RT-PCR reaction, respectively. Each condition tested in the assay is annotated as shown in the matrix above the gel. (C) A plot quantifying the results from (B) using the PSI ratio. Co-transfection of the WT exon-16 splicing reporter with either the empty expression vector (PCG) or the hnRNPA1 expression vector is indicated by a light blue or light red color, respectively. The same annotative matrix seen in (B) is used under the plot to label each ASO condition tested for each context. (D) Representative Western blot and agarose gel electrophoresis depicting results from our SRSF6 splicing assay. Each condition tested in the assay is annotated as shown in the matrix above the gel. (E) A plot quantifying the results from (D) using the PSI ratio. Co-transfection of the SRSF6 exon 6 splicing reporter with either the empty expression vector (PCG) or the hnRNPA1 expression vector is indicated by a light blue or light red color, respectively. The same annotative matrix seen in (D) is used under the plot to label each ASO condition tested for each context. Epitopes targeted by specific antibodies in the western blots are indicated to the left of their respective blots. Expected mRNA isoforms including or excluding the test exon are also annotated to the left of the agarose gel. Statistical significance between comparisons are denoted by asterisks that represent P-values with the following range of significance: ns, P> 0.05, **P≤ 0.01, ***P≤ 0.001, and ****P≤ 0.0001. Statistical significance was determined using analysis of variance (ANOVA), and Dunett's post-hoc test. Each exon-16 splicing reporter context and condition tested contains three independent/biological replicates.
ASO27 contains a potential high affinity hnRNPA1 binding site (5′-AGGTCCTTAGGGTTTACA-3′), inviting the hypothesis that ASO27 may attenuate exon-16 splicing in our hnRNPA1-ASO competition assay by directly binding to and sponging hnRNPA1. To test this hypothesis, we repeated the hnRNPA1-ASO competition assay using an orthogonal hnRNPA1-responsive splicing reporter that does not bind ASO27. This splicing reporter contains SRSF6 exon-6, a reporter previously validated to exhibit splicing suppression upon hnRNPA1 overexpression (52). We observe that, relative to the empty expression vector, overexpression of hnRNPA1 caused skipping of SRSF6 exon-6, as expected (Figure 5D, compare lane 1 to lanes 2 and 3; Figure 5E). The effect of hnRNPA1 overexpression is maintained in the presence of ASO27 (Figure 5D, lane 2), suggesting that ASO27 does not attenuate hnRNPA1-directed inhibition of splicing by binding to the splicing factor itself. Taken together, these experiments support our hypothesis that the trio ASO combination rescues exon-16 splicing in part by blocking bona fide hnRNPA1-dependent silencers found within TWJ-3–15 and increasing accessibility of the 3′ss poly-Y tract.
Targeting TWJ-3-15 rescues multiple splicing-sensitive HA-causing variants of F8 exon-16
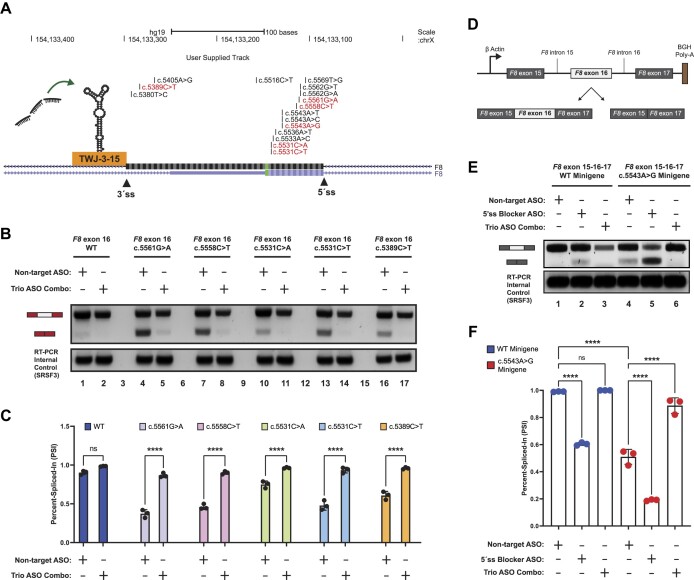
Our experiments indicate that the splicing fidelity of F8 exon-16 appears to be regulated in part by an RNA structure that weakens the 3′ss and recruits hnRNPA1 to further suppress exon definition (Figure 6A). Because this feature is shared across all exon-16 HA-causing variants (Supplementary Figures S11–16) and is targeted by ASOs capable of rescuing exon-16c.5543A>G splicing, we reasoned that targeting TWJ-3–15 might rescue splicing of other splicing-sensitive exon-16 variants. To test this hypothesis, we co-transfected WT exon 16, exon-16c.5561G>A, exon-16c.5558C>T, exon-16c.5531C>A, exon-16c.5531C>T and exon-16c.5389C>T splicing reporters with the NT ASO control or the trio ASO combination. Similar to exon-16c.5543A>G, we observed that co-transfecting ASO29, ASO28 and ASO27 together strongly promotes inclusion of other splicing-sensitive HA-causing pathogenic variants of exon-16 (Figure 6B,C). Taken together, these data demonstrate that ASO29, ASO28 and ASO27 interfere with the function of TWJ-3–15 and can generally rescue a broad array of splicing-sensitive pathogenic alleles implicated in HA.
Figure 6.
A combination of ASOs targeting TWJ-3–15 in a heterologous and endogenous context can rescue splicing for a broad array of HA-associated variants of exon-16 by increasing 3′ss accessibility and blocking hnRNPA1 binding. (A) A UCSC Genome Browser screenshot depicting the F8 exon-16 locus and the positions of HA-causing variants tested in this study. Pathogenic variants demonstrated to be splicing-sensitive from our assays are shown in red, whereas non-sensitive variants are shown in black. The 3′ and 5′ splice sites are annotated in addition to TWJ-3–15. Successful ASOs targeting TWJ-3–15. (B) A representative agarose gel depicting the results from our cell-based splicing assays testing the trio ASO combinations’ ability to rescue splicing of other HA-linked splicing-sensitive exon-16 variants (upper panel). The SRSF3 internal control for the RT-PCR reaction is also shown (lower panel). Each splicing assay condition included in this specific assay is annotated as shown in the matrix above the gel. Expected mRNA isoforms including or excluding the test exon are also annotated to the left of the agarose gel. (C) A plot quantifying the results from (B) using the PSI ratio. Each sequence context tested (WT or pathogenic variant) is annotated by a distinct color. The same annotative matrix seen in (B) is used under the plot to label each ASO condition tested for each context. (D) A schematic depicting the F8 exon-16 minigene splicing reporter to validate aberrant splicing defects in an endogenous context. F8 exon-16, along with its neighboring introns and exons as annotated, are cloned in between a strong promoter and polyadenylation signal. Expected isoforms generated from the splicing reporter are also shown and annotated. (E) A representative agarose gel depicting the results from our exon-16 minigene splicing assays validating aberrant splicing and trio ASO rescue effects in an endogenous context. Each assay condition included in this experiment is annotated as shown in the matrix above the gel. Expected mRNA isoforms including or excluding exon-16 are also annotated to the left of the agarose gel. (F) A plot quantifying the results from (E) using the PSI ratio. Each sequence context tested (WT minigene or exon-16c.5543A>G minigene) is annotated by a distinct color. The same annotative matrix seen in (E) is used under the plot to label each ASO condition tested for each context. Statistical significance between comparisons are denoted by asterisks that represent P-values with the following range of significance: ns, P> 0.05, and ****P≤ 0.0001. Statistical significance was determined using analysis of variance (ANOVA), and Dunett’s post-hoc test. Each exon-16 splicing reporter context and condition tested contains three independent/biological replicates.
Splicing reporters, like those employed in this study, enable the rapid screening and identification of splicing-sensitive variants across a wide array of exons. However, the heterologous sequence contexts do not always accurately reflect the splicing patterns of the endogenous gene. We therefore established an orthogonal splicing reporter to determine whether exon-16c.5543A>G induces exon skipping in a more native context. We substituted the HBB locus in our reporter plasmid with the region of the F8 gene spanning from exon-15 to exon-17, including the full length introns flanking exon-16 (Figure 6D). To make appropriate comparisons, we generated minigenes for both WT exon-16 and the exon-16c.5543A>G pathogenic variant that induces the most severe exon skipping. Once the reporters were generated and sequence-validated, we then repeated splicing assays to determine if the exon-16c.5543A>G variant induces exon skipping, and if the trio ASO combination also rescued this splicing defect. This minigene assay demonstrates that c.5543A > G induces exon-16 skipping in a native context (Figure 6E, compare lanes 1 and 4). As expected, the exon-16 5′ss blocking ASO reduces inclusion of the WT and pathogenic variant minigene reporters (Figure 6E, compare lanes 1 to 2 and 4 to 5), whereas the trio ASO combination significantly increased exon-16c.5543A>G inclusion (Figure 6E, compare lanes 4 and 6, P-value < 0.0001). Together, these data demonstrate that our heterologous reporter assay fully recapitulates the results of an orthogonal model system presenting exon-16 and the c.5543A > G pathogenic variant in an native context.
Discussion
RNA structure–function relationships in exon definition
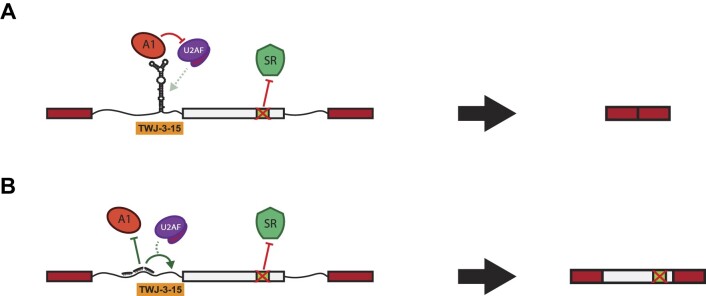
Our results suggest that a putative RNA structure, TWJ-3–15, sensitizes exon-16 to aberrant splicing by attenuating the strength of the 3′ss. This interpretation is supported by several lines of evidence. First, SHAPE data demonstrates that upstream intronic sequences base pair with the poly-Y tract. This base pairing interaction could reduce accessibility of the poly-Y tract to U2AF. A similar mechanism was demonstrated for the 5′ss of Tau exon 10 (53). In this case, the RNA helicase p68 controls accessibility of the 5′ss to U1 snRNP and ASOs stabilizing this structure inhibit splicing (54). Similarly, at least two putative hnRNPA1 binding sites are present in the apical stem of TWJ-3–15. These sequences appear to be functional binding sites as overexpression of hnRNPA1 inhibits splicing of both wild-type and splicing defective exon-16 variants. TWJ-3–15 also imparts hnRNPA1-dependent exon skipping in a heterologous constitutive exon, demonstrating the presence of splicing silencer activity (Supplementary Figure S10). Finally, we also discovered splice-modulating ASOs that rescue exon-16 inclusion by disrupting the function of this RNA structure. ASO27, ASO28 and ASO29 antagonize TWJ-3–15 by occluding hnRNPA1-responsive splicing silencers and by exposing the poly-Y tract, underscoring the functional significance of this structure and its sequence. Despite strong splice site signals (Supplementary Table S3), our data suggest that TWJ-3–15 sensitizes exon-16 to HA-causing variants that cripple ESEs or create ESSs (Figure 7A). Modulating this RNA structure–function relationship with ASOs rescues splicing of a diverse array of splicing-sensitive HA-linked variants of exon-16 (Figure 7B). We note that in the absence of in cellulo chemical probing data, TWJ-3–15 remains a structural model that requires additional experimental validation. However, the impact of our work does not rest solely on this RNA structure. Collectively, our data strongly suggest that this sequence functions as an intronic splicing silencer and that ASOs occluding this region rescues an array of splicing-sensitive variants in exon-16.
Figure 7.
The loss of a critical ESE in F8 exon-16 is hypothesized to amplify the inhibitory nature of TWJ-3–15 to alter exon definition and splicing fidelity. The following models depict experimentally-driven mechanisms on how F8 exon-16 (in light gray) may be aberrantly spliced and rescued as determined in this study. (A) A schematic depicting the loss of a critical ESE in exon-16 due to the A > G pathogenic variant in the exon-16c.5543A>G variant. Losing the ESE diminishes the ability to recruit a positive splicing factor that likely regulates TWJ-3–15 and hnRNPA1 antagonism at the 3′ss of F8 exon-16, leading to decreased 3′ss strength. (B) A schematic depicting the trio ASO combinations’ ability to rescue splicing of exon-16c.5543A>G by destabilizing TWJ-3–15, and preventing the recruitment of hnRNPA1 to the 3′ss. Collectively, our data-supported model indicates that the trio ASOs block the recruitment of a negative splicing factor and increases the accessibility of the 3′ss to the splicing machinery. TWJ-3–15 is annotated by a simplified depiction of the ‘Y-shaped’ RNA secondary structure at the 3′ss of exon-16. RBPs binding to TWJ-3–15 and this region such as hnRNPA1 and U2AF are respectively annotated. The predicted ESE is annotated in light green within exon-16, and its binding partner, presumably an RBP like SR proteins that are known to enhance splicing, is depicted as well.
By contrast to TWJ-3–15, we found that ASOs hybridizing to the exon-16 itself, spare one, inhibit pre-mRNA splicing (Figure 3D). While ASO21 hybridizes to a sequence predicted to function as a splicing silencer and therefore might not be expected to interfere with splicing, it is less clear why other ASOs targeting the exon are deleterious to splicing. One possibility is that the secondary structure of exon-16 is important for exon definition. Indeed, SHAPE-MaP-seq suggests extensive intramolecular pairing juxtaposes the 5′ss and 3′ss (Supplementary Figures S11–16). Perhaps ASOs targeting exon-16 interfere with base pairing interactions within the exon and disrupt splicing. Alternatively, F8 exon-16 may be strongly dependent on exonic splicing enhancers. In this model, ASOs targeting the exon could mask binding sites for splicing factors.
The interplay of genetic variation and exon definition is complex
Interpreting the impact of genetic variation on pre-mRNA splicing is fundamental to understanding genotype-phenotype relationships (25,55,56). Recognition of exon-intron boundaries requires the combinatorial interaction of splicing regulatory proteins with ESEs and ESSs, and is required for both constitutive and alternative splicing (57). Recent high-throughput saturating mutagenesis screens of model human exons demonstrate that constitutive exons are more resilient to substitutions than alternative exons (58). In their study, Baeza-Centurion et al. use human genetic variation to argue that alternative exons are more prone to aberrant splicing than constitutive exons. Surprisingly, our work demonstrates that splicing of F8 exon-16 is vulnerable to pathogenic genetic variants, despite limited evidence for regulated alternative splicing (Figure 1 and Supplementary Figure S2). Interestingly, pathogenic genetic variants in exon-16 and the 5′ss of intron 15 cause aberrant exon-16 splicing in HA patients (59,60).
An important consideration for interpreting this result is that common polymorphisms (minor allele frequency ≥ 5%) are fixed in the population and unlikely to negatively affect fitness. By contrast, rare pathogenic variants can induce aberrant splicing of constitutive exons (61–71). We previously demonstrated that pathogenic variants are more likely to cause the loss or gain of ESEs or ESSs, respectively (24). Despite this observation, we were surprised that for a majority of exons tested in our study, pathogenic variants tested here did not significantly alter splicing efficiency relative to the WT reporters (Supplementary Figure S1). Clearly, additional layers of regulatory information such as splicing factor occupancy maps, chromatin structure, and RNA polymerase kinetics need to be considered in future predictive models (25,55,56,72).
Aberrant splicing of F8 in Hemophilia A
An HA diagnosis causes a severe life expectancy disadvantage. The global prevalence of HA is ∼17.5 cases per 100 000 males, while the prevalence at birth is 26.4 per 100 000 males (73). These prevalence estimates suggest as many as 1 million patients globally are afflicted with HA. Based on the Coagulation Factor Variant Database, exon-16 contains 4.5% of the 3052 unique pathogenic variants in the F8 gene. These data suggest that >40 000 HA patients could harbor exon-16 pathogenic variants. Our work suggests that ∼37.5% of variants tested in exon-16 induce aberrant splicing, suggesting that globally, >15 480 patients could benefit from ASOs targeting TWJ-3–15. In the future, it will be important to create patient-derived models to confirm the role for TWJ-3–15 in exon-16 aberrant splicing in vivo. Better models will facilitate development of singular ASOs targeting TWJ-3–15 capable of fully rescuing exon-16 splicing. F8 exon-16 encodes a portion of the A3 domain which is required for efficient blood clotting and is frequently mutated in HA (74–76). Because exon-16 is divisible by 3, the aberrant skipped isoform is predicted to express an internally deleted protein, rather than undergoing nonsense mediated decay.
Limitations of the study and future directions
There are several limitations of our study. Because we did not have access to patient-derived cell lines and cell lines expressing the canonical full length F8 mRNA isoform are not readily available, we employed the well-established HBB reporter system transfected into HEK293T cells to assay the impact of F8 pathogenic variants on exon identity (77). This minimal system exploits the context dependence of exon definition as most auxiliary cis-regulatory signals are near splice sites (78). However, because this reporter system lacks most of the endogenous F8 sequence, it remains possible that results obtained with the heterologous reporter may differ from results obtained from the endogenous locus. Additionally, other processes such as chromatin modification and transcriptional elongation can regulate splicing (79). While the artificial system used here may differ from the endogenous locus, exon-16 and intron-15 variants that cause aberrant exon-16 splicing have been reported in HA patients (59,60). Despite this uncertainty, our study represents an important step towards correcting splicing-sensitive variants in F8 exon-16. Additional work is required to validate or invalidate aspects of this potential structure–function relationship in exon-16 identity. It will also be important to learn how ESEs antagonize the function of this putative regulatory element.
In the absence of patient-derived cell models, other groups developed CRISPR edited induced pluripotent stem cell models (77,80). In the future, translating our findings into a pre-clinical setting will require the testing of variants and ASOs in the context of the endogenous F8 locus in a disease-in-a-dish preclinical model. Although we have not tested our hypotheses in a patient-derived cell line or in engineered cell models, evidence supporting the fragile nature of exon-16 can be found in HA patient registries. The European Coagulation Factor Variant Database contains at least four unrelated patients with three different synonymous pathogenic variants at position c.5586 causing severe to modest HA phenotypes (75,81,82). Silent pathogenic variants most frequently induce aberrant splicing but can also alter RNA structure and mRNA stability, leading to changes in gene expression. Additionally, variants in both intron-15 and exon-16 cause aberrant splicing of exon-16 in HA patient samples (59,60). These published clinical findings, combined with our in vitro analysis of HA-pathogenic variants in exon-16, support the hypothesis that aberrant splicing of F8 exon-16 occurs in HA patients and represents a potential therapeutic target. Finally, it will be important to determine how the F8 protein tolerates missense variants in exon-16. Recent studies demonstrated that non-conservative substitutions in exon-19, including those associated with aberrant splicing, have modest effects on protein activity when expressed from a cDNA (83). Thus, the potential translation of ASOs targeting TWJ-3–15 requires a similar study of F8 protein function and secretion using a pre-clinical model system focusing on the endogenous F8 locus.
Supplementary Material
Acknowledgements
We thank the Toxic RNA Lab (TRL), a curricular undergraduate research experience led by J.R.S., for their help in generating the F8 splicing reporters. We thank Alexander J. Ritter for writing the Python script that expedited our capacity to quickly design and test antisense oligonucleotides presented in this study. We thank Danny Incarnato for the gift of 2A3 and for their help in getting the RNA Framework analysis pipeline up and running. We thank the Sanford lab, and Gina Mawla for their feedback on manuscript drafts.
Author contributions: V.T., G.C., M.D.S. and J.R.S. conceptualized and led the experiments presented in this study. V.T., H.T., P.H.D., C.O., P.C., I.Q., A.H. and S.L generated the heterologous F8 splicing reporters. V.T. and M.G. performed all in vitro cell-based splicing assays. V.T. developed and established the automation platform used in this study. V.T. and M.G. designed and assayed all antisense oligonucleotides targeting F8 exon-16 using the automation platform. G.C., A.G.J. and V.T. generated the in vitro RNA templates for SHAPE-MaP-seq. G.C. and N.M.F. designed, performed, prepared and analyzed all in vitro SHAPE-MaP-seq experiments and sequencing libraries for subsequent RNA structure predictions. V.T. and M.G. performed the hnRNPA1-ASO competition assay to experimentally validate hnRNPA1 binding sites identified in this study. V.T. and M.G. designed, generated, and assayed the F8 exon-16 minigene reporters used in this study. V.T. and G.C. performed all of the data analysis and visualization presented in this study. V.T. wrote the manuscript, with input and review of successive manuscript drafts by G.C., M.D.S., and J.R.S.
Contributor Information
Victor Tse, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA; Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Guillermo Chacaltana, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, CA, 95064, USA; Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Martin Gutierrez, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA; Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Nicholas M Forino, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA; Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Arcelia G Jimenez, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Hanzhang Tao, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Phong H Do, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Catherine Oh, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Priyanka Chary, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Isabel Quesada, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Antonia Hamrick, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Sophie Lee, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Michael D Stone, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, CA, 95064, USA; Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Jeremy R Sanford, Department of Molecular, Cell and Developmental Biology, University of California Santa Cruz, Santa Cruz, CA, 95064, USA; Center for Molecular Biology of RNA, University of California Santa Cruz, Santa Cruz, CA, 95064, USA.
Data availability
The sequencing data underlying this article are available in the Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ and can be accessed under accession code GSE230495.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R35GM130361 to J.R.S.; R01GM095850 to M.D.S.]; Santa Cruz Cancer Benefit Group to M.D.S.; UCSC Office of Research Seed Funding for the Center for Open Access Splicing Therapeutics. Funding for open access charge: National Institutes of Health [R35GM130361].
Conflict of interest statement. The authors affirm that no conflict of interest exists for this work.
References
- 1. Breathnach R., Benoist C., O’Hare K., Gannon F., Chambon P.. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc. Natl. Acad. Sci. USA. 1978; 75:4853–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chow L.T., Gelinas R.E., Broker T.R., Roberts R.J.. An amazing sequence arrangement at the 5’ ends of adenovirus 2 messenger RNA. Cell. 1977; 12:1–8. [DOI] [PubMed] [Google Scholar]
- 3. Berget S.M., Moore C., Sharp P.A.. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA. 1977; 74:3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konarska M.M., Sharp P.A.. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987; 49:763–774. [DOI] [PubMed] [Google Scholar]
- 5. Kastner B., Will C.L., Stark H., Lührmann R.. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb. Perspect. Biol. 2019; 11: 10.1101/cshperspect.a032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berget S.M. Exon Recognition in Vertebrate Splicing (∗). J. Biol. Chem. 1995; 270:2411–2414. [DOI] [PubMed] [Google Scholar]
- 7. Kohtz J.D., Jamison S.F., Will C.L., Zuo P., Lührmann R., Garcia-Blanco M.A., Manley J.L.. Protein-protein interactions and 5’-splice-site recognition in mammalian mRNA precursors. Nature. 1994; 368:119–124. [DOI] [PubMed] [Google Scholar]
- 8. Graveley B.R., Hertel K.J., Maniatis T.. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA. 2001; 7:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruskin B., Zamore P.D., Green M.R.. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988; 52:207–219. [DOI] [PubMed] [Google Scholar]
- 10. Wu S., Romfo C.M., Nilsen T.W., Green M.R.. Functional recognition of the 3’ splice site AG by the splicing factor U2AF35. Nature. 1999; 402:832–835. [DOI] [PubMed] [Google Scholar]
- 11. Reed R., Maniatis T.. A Role for Exon Sequences and Splice-Site Proximity in Splice Site Selection. Cell. 1987; 46:681–690. [DOI] [PubMed] [Google Scholar]
- 12. Michaud S., Reed R.. A functional association between the 5’ and 3' splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993; 7:1008–1020. [DOI] [PubMed] [Google Scholar]
- 13. Watakabe A., Tanaka K., Shimura Y.. The role of exon sequences in splice site selection. Genes Dev. 1993; 7:407–418. [DOI] [PubMed] [Google Scholar]
- 14. Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B.. Systematic identification and analysis of exonic splicing silencers. Cell. 2004; 119:831–845. [DOI] [PubMed] [Google Scholar]
- 15. Fairbrother W.G., Yeh R.-F., Sharp P.A., Burge C.B.. Predictive identification of exonic splicing enhancers in human genes. Science. 2002; 297:1007–1013. [DOI] [PubMed] [Google Scholar]
- 16. Ke S., Shang S., Kalachikov S.M., Morozova I., Yu L., Russo J.J., Ju J., Chasin L.A.. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011; 21:1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warf M.B., Berglund J.A.. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010; 35:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Graveley B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005; 123:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shepard P.J., Hertel K.J.. Conserved RNA secondary structures promote alternative splicing. RNA. 2008; 14:1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muro A.F., Caputi M., Pariyarath R., Pagani F., Buratti E., Baralle F.E.. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 1999; 19:2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buratti E., Muro A.F., Giombi M., Gherbassi D., Iaconcig A., Baralle F.E.. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004; 24:1387–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones A.N., Graß C., Meininger I., Geerlof A., Klostermann M., Zarnack K., Krappmann D., Sattler M.. Modulation of pre-mRNA structure by hnRNP proteins regulates alternative splicing of MALT1. Sci. Adv. 2022; 8:eabp9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sterne-Weiler T., Sanford J.R.. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol. 2014; 15:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sterne-Weiler T., Howard J., Mort M., Cooper D.N., Sanford J.R.. Loss of exon identity is a common mechanism of human inherited disease. Genome Res. 2011; 21:1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mort M., Sterne-Weiler T., Li B., Ball E.V., Cooper D.N., Radivojac P., Sanford J.R., Mooney S.D.. MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing. Genome Biol. 2014; 15:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J.. Deciphering the splicing code. Nature. 2010; 465:53–59. [DOI] [PubMed] [Google Scholar]
- 27. Gagliardi M., Ashizawa A.T.. The challenges and strategies of antisense oligonucleotide drug delivery. Biomedicines. 2021; 9:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hill S.F., Meisler M.H.. Antisense oligonucleotide therapy for neurodevelopmental disorders. Dev. Neurosci. 2021; 43:247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rigo F., Chun S.J., Norris D.A., Hung G., Lee S., Matson J., Fey R.A., Gaus H., Hua Y., Grundy J.S.et al.. Pharmacology of a central nervous system delivered 2’-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J. Pharmacol. Exp. Ther. 2014; 350:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R.. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010; 24:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J., Hu C., Moufawad El Achkar C., Black L.E., Douville J., Larson A., Pendergast M.K., Goldkind S.F., Lee E.A., Kuniholm A.et al.. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 2019; 381:1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ronayne E.K., Peters S.C., Gish J.S., Wilson C., Spencer H.T., Doering C.B., Lollar P., Spiegel P.C. Jr, Childers K.C.. Structure of Blood Coagulation Factor VIII in Complex With an Anti-C2 Domain Non-Classical, Pathogenic Antibody Inhibitor. Front. Immunol. 2021; 12:697602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang Q., Xiang M., Lu Y., Ruan Y., Ding Q., Wang X., Xi X., Wang H.. Characterisation and quantification of F8 transcripts of ten putative splice site mutations. Thromb. Haemost. 2015; 113:585–592. [DOI] [PubMed] [Google Scholar]
- 34. Jourdy Y., Fretigny M., Nougier C., Négrier C., Bozon D., Vinciguerra C.. Splicing analysis of 26 F8 nucleotide variations using a minigene assay. Haemophilia. 2019; 25:306–315. [DOI] [PubMed] [Google Scholar]
- 35. Zimmermann M.A., Gehrig A., Oldenburg J., Müller C.R., Rost S.. Analysis of F8 mRNA in haemophilia A patients with silent mutations or presumptive splice site mutations. Haemophilia. 2013; 19:310–317. [DOI] [PubMed] [Google Scholar]
- 36. Famà R., Borroni E., Zanolini D., Merlin S., Bruscaggin V., Walker G.E., Olgasi C., Babu D., Agnelli Giacchello J., Valeri F.et al.. Identification and functional characterization of a novel splicing variant in the F8 coagulation gene causing severe hemophilia A. J. Thromb. Haemost. 2020; 18:1050–1064. [DOI] [PubMed] [Google Scholar]
- 37. Rothrock C., Cannon B., Hahm B., Lynch K.W.. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003; 12:1317–1324. [DOI] [PubMed] [Google Scholar]
- 38. den Dunnen J.T., Antonarakis S.E.. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000; 15:7–12. [DOI] [PubMed] [Google Scholar]
- 39. Chang T., Draper J.M., Van den Bout A., Kephart E., Maul-Newby H., Vasquez Y., Woodbury J., Randi S., Pedersen M., Nave M.et al.. A method for campus-wide SARS-CoV-2 surveillance at a large public university. PLoS One. 2021; 16:e0261230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tian S., Yesselman J.D., Cordero P., Das R.. Primerize: automated primer assembly for transcribing non-coding RNA domains. Nucleic Acids Res. 2015; 43:W522–W526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marinus T., Fessler A.B., Ogle C.A., Incarnato D.. A novel SHAPE reagent enables the analysis of RNA structure in living cells with unprecedented accuracy. Nucleic Acids Res. 2021; 49:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morandi E., van Hemert M.J., Incarnato D.. SHAPE-guided RNA structure homology search and motif discovery. Nat. Commun. 2022; 13:1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Incarnato D., Morandi E., Simon L.M., Oliviero S.. RNA Framework: an all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications. Nucleic Acids Res. 2018; 46:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siegfried N.A., Busan S., Rice G.M., Nelson J.A.E., Weeks K.M.. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods. 2014; 11:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 47. Paz I., Kosti I., Ares M. Jr, Cline M., Mandel-Gutfreund Y.. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014; 42:W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Waldern J.M., Kumar J., Laederach A.. Disease-associated human genetic variation through the lens of precursor and mature RNA structure. Hum. Genet. 2022; 141:1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cordero P., Das R.. Rich RNA Structure Landscapes Revealed by Mutate-and-Map Analysis. PLoS Comput. Biol. 2015; 11:e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. An P., Grabowski P.J.. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 2007; 5:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han K., Yeo G., An P., Burge C.B., Grabowski P.J.. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005; 3:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Howard J.M., Lin H., Wallace A.J., Kim G., Draper J.M., Haeussler M., Katzman S., Toloue M., Liu Y., Sanford J.R.. HNRNPA1 promotes recognition of splice site decoys by U2AF2 in vivo. Genome Res. 2018; 28:689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., Wu J.Y.. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5’ splice site. Mol. Cell. Biol. 2011; 31:1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peacey E., Rodriguez L., Liu Y., Wolfe M.S.. Targeting a pre-mRNA structure with bipartite antisense molecules modulates tau alternative splicing. Nucleic Acids Res. 2012; 40:9836–9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Leung M.K.K., Xiong H.Y., Lee L.J., Frey B.J.. Deep learning of the tissue-regulated splicing code. Bioinformatics. 2014; 30:i121–i129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cygan K.J., Sanford C.H., Fairbrother W.G.. Spliceman2: a computational web server that predicts defects in pre-mRNA splicing. Bioinformatics. 2017; 33:2943–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hertel K.J. Combinatorial control of exon recognition. J. Biol. Chem. 2008; 283:1211–1215. [DOI] [PubMed] [Google Scholar]
- 58. Baeza-Centurion P., Miñana B., Valcárcel J., Lehner B.. Mutations primarily alter the inclusion of alternatively spliced exons. eLife. 2020; 9:e59959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tavassoli K., Eigel A., Pollmann H., Horst J.. Mutational analysis of ectopic factor VIII transcripts from hemophilia A patients: identification of cryptic splice site, exon skipping and novel point mutations. Hum. Genet. 1997; 100:508–511. [DOI] [PubMed] [Google Scholar]
- 60. Yenchitsomanus P., Thanootarakul P., Akkarapatumwong V., Oranwiroon S., Pung-Amritt P., Veerakul G., Mahasandana C.. Mutation causing exon 15 skipping and partial exon 16 deletion in factor VIII transcript, and a method for direct mutation detection. Haemophilia. 2001; 7:335–338. [DOI] [PubMed] [Google Scholar]
- 61. Holm L.L., Doktor T.K., Hansen M.B., Petersen U.S.S., Andresen B.S.. Vulnerable exons, like ACADM exon 5, are highly dependent on maintaining a correct balance between splicing enhancers and silencers. Hum. Mutat. 2022; 43:253–265. [DOI] [PubMed] [Google Scholar]
- 62. Cartegni L., Hastings M.L., Calarco J.A., de Stanchina E., Krainer A.R.. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006; 78:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ars E., Serra E., García J., Kruyer H., Gaona A., Lázaro C., Estivill X.. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000; 9:237–247. [DOI] [PubMed] [Google Scholar]
- 64. Kelly D.P., Whelan A.J., Ogden M.L., Alpers R., Zhang Z.F., Bellus G., Gregersen N., Dorland L., Strauss A.W.. Molecular characterization of inherited medium-chain acyl-CoA dehydrogenase deficiency. Proc. Natl. Acad. Sci. USA. 1990; 87:9236–9240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gregersen N., Andresen B.S., Bross P., Winter V., Rüdiger N., Engst S., Christensen E., Kelly D., Strauss A.W., Kølvraa S.. Molecular characterization of medium-chain acyl-CoA dehydrogenase (MCAD) deficiency: identification of a lys329 to glu mutation in the MCAD gene, and expression of inactive mutant enzyme protein in E. coli. Hum. Genet. 1991; 86:545–551. [DOI] [PubMed] [Google Scholar]
- 66. Tubeuf H., Caputo S.M., Sullivan T., Rondeaux J., Krieger S., Caux-Moncoutier V., Hauchard J., Castelain G., Fiévet A., Meulemans L.et al.. Calibration of pathogenicity due to variant-induced leaky splicing defects by using BRCA2 Exon 3 as a Model System. Cancer Res. 2020; 80:3593–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rave-Harel N., Kerem E., Nissim-Rafinia M., Madjar I., Goshen R., Augarten A., Rahat A., Hurwitz A., Darvasi A., Kerem B.. The molecular basis of partial penetrance of splicing mutations in cystic fibrosis. Am. J. Hum. Genet. 1997; 60:87–94. [PMC free article] [PubMed] [Google Scholar]
- 68. Aznarez I., Chan E.M., Zielenski J., Blencowe B.J., Tsui L.-C.. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum. Mol. Genet. 2003; 12:2031–2040. [DOI] [PubMed] [Google Scholar]
- 69. Dietz H.C., Valle D., Francomano C.A., Kendzior R.J. Jr, Pyeritz R.E., Cutting G.R.. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993; 259:680–683. [DOI] [PubMed] [Google Scholar]
- 70. D’Souza I., Poorkaj P., Hong M., Nochlin D., Lee V.M., Bird T.D., Schellenberg G.D.. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc. Natl. Acad. Sci. USA. 1999; 96:5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu H.X., Cartegni L., Zhang M.Q., Krainer A.R.. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 2001; 27:55–58. [DOI] [PubMed] [Google Scholar]
- 72. Holm L.L., Doktor T.K., Flugt K.K., Petersen U.S.S., Pedersen R., Andresen B.S.. All exons are not created equal - Exon vulnerability determines the effect of exonic mutations on splicing. 2023; bioRxiv doi:14 June 2023, preprint: not peer reviewed 10.1101/2023.06.14.544306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iorio A., Stonebraker J.S., Chambost H., Makris M., Coffin D., Herr C., Germini F., Data and Demographics Committee of the World Federation of Hemophilia. Establishing the prevalence and prevalence at birth of hemophilia in males: a meta-analytic approach using national registries. Ann. Intern. Med. 2019; 171:540–546. [DOI] [PubMed] [Google Scholar]
- 74. Giansily-Blaizot M., Rallapalli P.M., Perkins S.J., Kemball-Cook G., Hampshire D.J., Gomez K., Ludlam C.A., McVey J.H.. The EAHAD blood coagulation factor VII variant database. Hum. Mutat. 2020; 41:1209–1219. [DOI] [PubMed] [Google Scholar]
- 75. McVey J.H., Rallapalli P.M., Kemball-Cook G., Hampshire D.J., Giansily-Blaizot M., Gomez K., Perkins S.J., Ludlam C.A.. The European Association for Haemophilia and Allied Disorders (EAHAD) Coagulation Factor Variant Databases: important resources for haemostasis clinicians and researchers. Haemophilia. 2020; 26:306–313. [DOI] [PubMed] [Google Scholar]
- 76. Gouw S.C., Van Der Bom J.G., Van Den Berg H.M., Zewald R.A., Ploos Van Amstel J.K., Mauser-Bunschoten E.P.. Influence of the type of F8 gene mutation on inhibitor development in a single centre cohort of severe haemophilia A patients. Haemophilia. 2011; 17:275–281. [DOI] [PubMed] [Google Scholar]
- 77. Zhou H., Arechavala-Gomeza V., Garanto A.. Experimental model systems used in the preclinical development of nucleic acid therapeutics. Nucleic Acid Ther. 2023; 33:238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fu X.-D., Ares M. Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gehring N.H., Roignant J.-Y.. Anything but ordinary – emerging splicing mechanisms in eukaryotic gene regulation. Trends Genet. 2021; 37:355–372. [DOI] [PubMed] [Google Scholar]
- 80. Tomkiewicz T.Z., Nieuwenhuis S.E., Cremers F.P.M., Garanto A., Collin R.W.J.. Correction of the splicing defect caused by a recurrent variant in ABCA4 (c.769-784C>T) that underlies stargardt disease. Cells. 2022; 11:3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Higuchi M., Kazazian H.H. Jr, Kasch L., Warren T.C., McGinniss M.J., Phillips J.A. 3rd, Kasper C., Janco R., Antonarakis S.E.. Molecular characterization of severe hemophilia A suggests that about half the mutations are not within the coding regions and splice junctions of the factor VIII gene. Proc. Natl. Acad. Sci. USA. 1991; 88:7405–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Johnsen J.M., Fletcher S.N., Huston H., Roberge S., Martin B.K., Kircher M., Josephson N.C., Shendure J., Ruuska S., Koerper M.A.et al.. Novel approach to genetic analysis and results in 3000 hemophilia patients enrolled in the My Life, Our Future initiative. Blood Adv. 2017; 1:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lombardi S., Leo G., Merlin S., Follenzi A., McVey J.H., Maestri I., Bernardi F., Pinotti M., Balestra D.. Dissection of pleiotropic effects of variants in and adjacent to F8 exon 19 and rescue of mRNA splicing and protein function. Am. J. Hum. Genet. 2021; 108:1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data underlying this article are available in the Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/ and can be accessed under accession code GSE230495.