eToc Blurb
C. elegans exhibit stereotyped global brain dynamics, but it was not known what evokes them or whether they are internally driven. Gauthey et al., show that high intensity blue light commonly used in calcium imaging evokes similar global brain dynamics. The dynamics are reduced or absent when imaged at longer wavelengths, and are partly mediated by the light-sensitive gustatory receptor GUR-3.
Stereotyped oscillations in population neural activity recordings from immobilized C. elegans have garnered significant interest for their striking low dimensionality and their evocative state-space trajectories or manifolds. Previously these oscillations have been interpreted as intrinsically driven global motor commands corresponding to spontaneous fictive locomotion. Here we tested whether these oscillations are intrinsic. We show that similar oscillations are evoked by high-intensity blue light commonly used for calcium imaging. Oscillations are reduced or absent and have lower frequency when a longer imaging wavelength is used. Under the original blue light illumination, oscillations are reduced or have lower frequency in animals that lack GUR-3, an endogenous light- and hydrogen-peroxide sensitive gustatory receptor. Consistent with GUR-3’s involvement, adding hydrogen peroxide decreases the proportion of recordings that exhibit no oscillations under longer wavelength illumination, and the power and frequency of the oscillations under hydrogen peroxide treatment were higher in WT animals than in gur-3 mutants. We therefore propose that blue light evokes global oscillations in part through the creation of reactive oxygen species that activate the hydrogen-peroxide sensing receptor GUR-3.
Immobilized C. elegans exhibit stereotyped global brain dynamics in populations of neurons that oscillate out of phase with a period of 30 to 150 s (frequency of 0.007 Hz to 0.033 Hz). A low-dimensional approximation of the time derivative of these dynamics plotted in neural-state space, e.g. via Principal Component Analysis, forms evocative stereotyped trajectories or manifolds1, Figure 1a. Their low dimensionality is striking because it suggests that a large fraction of the worm’s brain uses only three degrees of freedom. These dynamics have been the subject of intense study, for example to reveal connectivity2 or latent factors that may sculpt these dynamics3-6, to explore how the dynamics degrade with age7, or to compare these dynamics to those observed during movement8. Nonetheless, it has remained unclear exactly what these dynamics represent and what evokes them. One interpretation is that these dynamics are internally driven and represent motor commands for a fictive locomotory sequence of forward, backwards and turn movements that the animal would perform if it were not immobilized1. Here we seek to investigate whether these dynamics are internally driven and, if not, what drives them during immobilization.
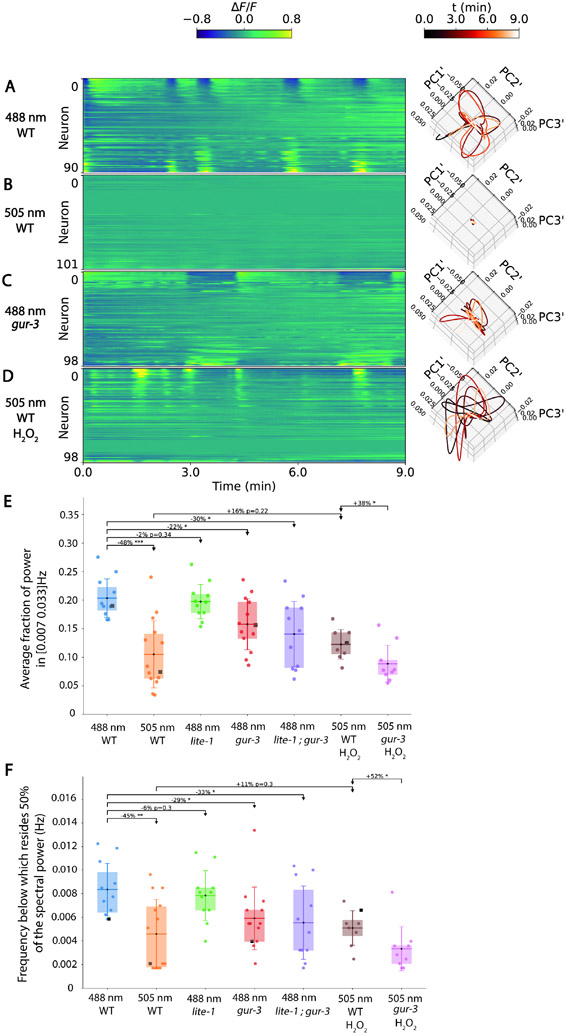
Figure 1. Oscillatory neural dynamics depend on imaging wavelength and GUR-3.
A-D) GCaMP6s population neural activity recordings are shown along with corresponding state-space plots of the derivative of neural dynamics projected onto their first three Principal Components (PCs) A). Wild Type (WT) background animals imaged at 488 nm B) WT at 505 nm C) gur-3 mutant at 488 nm and D) WT at 505 nm in the presence of 10mM H2O2. Each recording corresponds to the median recording of that type in our dataset ranked by the average fraction of power in the 0.007 to 0.033 Hz frequency range. The complete set of all recordings are shown in Figure S1. Neurons are displayed sorted by weight of the first PC. Neuron identity is not preserved across recordings. E) Average fraction of power in the frequency range 0.007 to 0.033 Hz. Each point is the average fraction across all neurons in a single recording. Black square denotes the median recordings shown in panels A-D. N=11, 14, 11, 13, 11, 8 and 9 recordings for each condition, respectively. Boxes extend from first to third quartiles, whiskers show standard deviation from minimum to maximum. Horizontal line indicates mean. F) Frequency below which resides 50% of the average spectral power. Comparisons list percent change in mean and a multiple hypothesis adjusted p-value calculated via the one-sided Welch’s t-test and adjusted in the manner of Benjamini-Hochberg. * p<0.05, ** p<0.01, *** p<0.001.
The GCaMP family of genetically encoded calcium indicators is often imaged using 488 nm light. When imaged at this wavelength, C. elegans population neural activity often exhibits oscillations in the 0.007 Hz to 0.033 Hz range (Figure 1A). We had previously9 found it convenient to image population activity with 505 nm light—a wavelength that is still close to GCaMP6’s excitation peak of 498 nm, and for which GCaMP6s has similar absorption. We noticed that in contrast to recordings made at 488nm, many recordings at 505 nm qualitatively lacked oscillations in the 0.007-0.033 Hz band (Figure 1B, Supplementary Figure S1, Supplementary Figure S2C).
To investigate, we recorded population calcium activity from 11 individuals at 488 nm and 14 individuals at 505 nm, each expressing GCaMP6s in the nucleus of all neurons. We used light intensities of 4 mW/mm2. This was a lower intensity than what we had used previously at 488 nm (see methods), but higher than previously for 505 nm 9. Each recording included on average 95 neurons typically recorded for at least 9 mins.
When imaging at 505 nm, oscillations were visibly absent in many of our recordings, while in other recordings oscillations were present but were weaker or appeared to be at lower frequencies (Supplementary Figure S1, Supplementary Figure S2). We quantified oscillations in each recording three ways: we measured the relative power in the frequency band of interest (0.007-0.033 Hz, Figure 1E); we calculated the proportion of neurons that had at least 20% of its power in that frequency band (Supplementary Figure S2B); and we calculated the frequency below which the majority of oscillations resided7 (Figure 1F). All three approaches yield the same trend: recordings made at 505 nm had dramatically lower power in the frequency band of interest, proportionally fewer neurons contributed, and the frequency below which 50% of the spectral power resided significantly decreased. We also inspected the number of `quiet’ recording that had no oscillations, defined as a recording in which less than 6% of the neurons exhibited at least 20% of their power in the frequency band of interest. A much larger proportion of 505 nm recordings were `quiet’ and lacked oscillations (6 of 14 recordings) compared to 488 nm (0 of 11), Figure S2C.
C. elegans have at least two molecular pathways that interact with high-intensity blue light: the lite-1 pathway and the gur-3 pathway10. In our calcium imaging experiments with 488 nm light, gur-3 and gur-3,lite-1 double mutants, but not lite-1 mutants, showed significantly reduced power in the frequency band compared to WT, had a significantly lower frequency below which 50% of the spectra power resides (Figure 1E,F and Supplementary Figure S1), and had a smaller proportion of neurons with power in the frequency band of interest (Supplementary Figure S2B). This suggests that 488 nm mediated activation of GUR-3 contributes to the oscillations we observe.
GUR-3 had previously been shown to activate neurons when illuminated by blue light of wavelengths and intensities similar to those used for calcium imaging, including 485 nm. By contrast, longer wavelength light, such as 500 nm, required much higher intensity to activate neurons via GUR-3 10. Our measurements are consistent with these prior results and suggest that 488 nm is more efficient than 505 nm light at inducing oscillations via GUR-3.
GUR-3 is a gustatory receptor expressed most notably in sensory neuron I2 but also in I4 and AVD10. GUR-3 mediates response to reactive oxygen species such as hydrogen peroxide10 and is not thought to respond to sunlight at typical intensities found at the Earth’s surface10. High enough intensity blue light, however, creates reactive oxygen species that activate GUR-3. If the global oscillations we observe are partly driven by GUR-3 activation, we would also expect reactive oxygen species to contribute to these oscillations. Consistent with this hypothesis 10 mM hydrogen peroxide delivered to the worm during 505 nm imaging caused a decrease in the proportion of `quiet’ recordings (Figure S2C). Further consistent with GUR-3’s involvement, the power and frequency of the oscillations under hydrogen peroxide treatment were significantly higher in WT animals than in gur-3 defective mutants (Figure 1E,F), and the number of neurons involved in the oscillation was also higher (Figure S2B).
We conclude that blue-light evokes 0.007-to-0.033-Hz stereotyped oscillations like those that are often observed at 488 nm but are less often observed at 505 nm. We propose that the oscillations we observe are driven in part by light-generated reactive oxygen species mediated by GUR-3 expressed in neurons. Without GUR-3, oscillations are diminished, slower, and fewer neurons are involved. The oscillations are therefore not spontaneous or intrinsically driven but reflect, in part, the animal’s response to detected reactive oxygen species, or to other light-evoked phenomenon. We have shown that light is sufficient to evoke the oscillations we observe, but we note that this does not imply that all oscillations in all possible conditions are always light evoked. Our measurements provide insight into what may cause the oscillations but they neither support nor refute the claim that similar oscillations represent motor commands. Future work is needed to understand what properties of the network dictate the timescale of oscillations.
Supplementary Material
C. elegans exhibit stereotyped global brain dynamics, but it was not known what evokes them or whether they are internally driven. Gauthey et al., show that these oscillations are not spontaneous but rather are light evoked by the high intensity blue light commonly used in calcium imaging and mediated, in part, by the gustatory receptor GUR-3.
Acknowledgement:
Worm strain MT21783 and MT21793 were kind gift from Horvitz lab, MIT. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Research reported in this work was supported by the National Institutes of Health, National Institute of Neurological Disorders and Stroke under New Innovator award number DP2-NS116768 to AML; the Simons Foundation under award SCGB #543003 to A.M.L.; by the Swartz Foundation through the Swartz Fellowship for Theoretical Neuroscience to F.R.; and by the National Science Foundation, through the Center for the Physics of Biological Function (PHY-1734030). The content is solely the responsibility of the authors and does not represent the official views of any funding agency.
Inclusion and diversity statement:
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests: The authors declare no competing interests.
References:
- 1.Kato S, Kaplan HS, Schrödel T, Skora S, Lindsay TH, Yemini E, Lockery S, and Zimmer M (2015). Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell 163, 656–669. 10.1016/j.cell.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A, Chandra S, and Ott E (2023). Network inference from short, noisy, low time-resolution, partial measurements: Application to C. elegans neuronal calcium dynamics. Proceedings of the National Academy of Sciences 120, e2216030120. 10.1073/pnas.2216030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linderman S, Nichols A, Blei D, Zimmer M, and Paninski L (2019). Hierarchical recurrent state space models reveal discrete and continuous dynamics of neural activity in C. elegans. bioRxiv, 621540. 10.1101/621540. [DOI] [Google Scholar]
- 4.Costa AC, Ahamed T, and Stephens GJ (2019). Adaptive, locally linear models of complex dynamics. PNAS 116, 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fieseler C, Zimmer M, and Kutz JN (2020). Unsupervised learning of control signals and their encodings in Caenorhabditis elegans whole-brain recordings. Journal of The Royal Society Interface 17, 20200459. 10.1098/rsif.2020.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mudrik N, Chen Y, Yezerets E, Rozell CJ, and Charles AS (2022). Decomposed Linear Dynamical Systems (dLDS) for learning the latent components of neural dynamics. 10.48550/arXiv.2206.02972. [DOI] [Google Scholar]
- 7.Wirak GS, Florman J, Alkema MJ, Connor CW, and Gabel CV (2022). Age-associated changes to neuronal dynamics involve a disruption of excitatory/inhibitory balance in C. elegans. eLife 11, e72135. 10.7554/eLife.72135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallinen KM, Dempsey R, Scholz M, Yu X, Linder A, Randi F, Sharma AK, Shaevitz JW, and Leifer AM (2021). Decoding locomotion from population neural activity in moving C. elegans. eLife 10, e66135. 10.7554/eLife.66135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randi F, Sharma AK, Dvali S, and Leifer AM (2022). Neural signal propogation atlas of C. elegans. 10.48550/arXiv.2208.04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatla N, and Horvitz HR (2015). Light and hydrogen peroxide inhibit C. elegans Feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron 85, 804–818. 10.1016/j.neuron.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.