Abstract
Ethanol metabolism plays an essential role in how the body perceives and experiences alcohol consumption, and evidence suggests that modulation of ethanol metabolism can alter the risk for alcohol use disorder (AUD). In this review, we explore how ethanol metabolism, mainly via alcohol dehydrogenase and aldehyde dehydrogenase 2 (ALDH2), contributes to drinking behaviors by integrating preclinical and clinical findings. We discuss how alcohol dehydrogenase and ALDH2 polymorphisms change the risk for AUD, and whether we can harness that knowledge to design interventions for AUD that alter ethanol metabolism. We detail the use of disulfiram, RNAi strategies, and kudzu/isoflavones to inhibit ALDH2 and increase acetaldehyde, ideally leading to decreases in drinking behavior. In addition, we cover recent preclinical evidence suggesting that strategies other than increasing acetaldehyde-mediated aversion can decrease ethanol consumption, providing other potential metabolism-centric therapeutic targets. However, modulating ethanol metabolism has inherent risks, and we point out some of the key areas in which more data are needed to mitigate these potential adverse effects. Finally, we present our opinions on the future of treating AUD by the modulation of ethanol metabolism.
Keywords: alcohol metabolism, alcohol use disorder, ethanol
Introduction
Unhealthy alcohol drinking includes any alcohol use that puts your health or safety at risk (Witkiewitz et al. 2019). One such risk is alcohol use disorder (AUD), a medical condition characterized by the inability to control alcohol use despite physical and social consequences. Deaths due to AUD have risen in recent years, increasing the need to prevent or treat unhealthy drinking patterns (Da et al. 2020). Research over the past century has demonstrated that ethanol metabolites substantially contribute to the psychological and physical effects of alcohol, and therefore ethanol metabolism is important to understand in the context of AUD. The major metabolite of ethanol, acetaldehyde (AcH), is a toxic compound, which induces aversive reactions such as flushing, vasodilation, palpitations, and bronchoconstriction at high systemic concentrations (Quertemont and Didone 2006). However, AcH also plays key roles in the motivational, locomotor, and anxiolytic effects of ethanol (Correa et al. 2012, Peana and Acquas 2013). AcH is further metabolized to acetate, which mediates similar motor inhibitory effects as ethanol and functions as an alternative cellular energy source to glucose as a substrate of the tricarboxylic acid (TCA) cycle (Peana et al. 2016). Acetate can also be a precursor for molecules such as gamma-aminobutyric acid (GABA), which can mediate neuronal effects of ethanol, and acetyl-CoA, which can be used for lipogenesis or histone acetylation (Bradshaw 2021). Clearly, AcH and acetate play a major role in the effect that ethanol has on the brain and therefore, altering the concentrations of ethanol and its metabolites may represent a therapeutic opportunity for AUD treatment.
Ethanol is metabolized to AcH mainly by alcohol dehydrogenase (ADH), while the microsomal ethanol oxidizing system (MEOS) and catalase also play organ-specific roles in ethanol clearance (Gil-Mohapel et al. 2019). The predominant route for ingested ethanol metabolism is through gut and liver ADH1, and the resulting AcH is further metabolized to acetate mainly by aldehyde dehydrogenase 2 (ALDH2) (Cederbaum 2012). Much of the acetate produced by the oxidation of AcH leaves the liver and circulates to peripheral tissues where it is then activated to Acetyl CoA (Cederbaum 2012). In addition, Cytochrome P450 2E1 (CYP2E1) is an enzyme in the MEOS which detoxifies ethanol and is commonly induced after chronic ethanol consumption. CYP2E1 is mainly expressed in the liver, but is also expressed in extra-hepatic tissues including certain brain regions, but catalase is the main ethanol metabolizing enzyme in the brain (Gil-Mohapel et al. 2019). Several polymorphisms in ADH and ALDH genes are associated with the risk of AUD, indicating that ethanol metabolism plays a key role in the development of these ailments. In this review, we will highlight the roles of ethanol-metabolizing enzymes in AUD and discuss the therapeutic potential of ADH1 and ALDH2 modulators.
Alcohol dehydrogenase
ADH is the most important enzyme in ethanol clearance, and ADH mainly in the gut and liver clears over 90% of systemic alcohol (Fig. 1) (Haseba et al. 2020). There are seven human ADH genes (ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, and ADH7), but only Class I, Class IV, and to a lesser extent, Class II and Class III enzymes have shown specificity for ethanol in vitro or in vivo (Table 1) (Deltour et al. 1999, Haseba et al. 2019, Tsermpini et al. 2022). Two polymorphisms at Class I ADH gene ADH1B produce the ADH1B*2 and ADH1B*3 enzymes which have higher activity for alcohol oxidation, increased AcH production, and confer a protective effect on AUD (Li et al. 2008, Evsyukov and Ivanov 2013). Conversely, people with the ADH1B*1 or ADH1C*2 alleles, which exhibit significantly slower ethanol metabolism than the corresponding alleles, exhibit a higher risk of developing AUD (Edenberg 2007). In all, these data suggest that ADH polymorphisms that increase the rate of ethanol metabolism may be protective against AUD, likely due to the uncomfortable effects of increased AcH.
Figure 1.
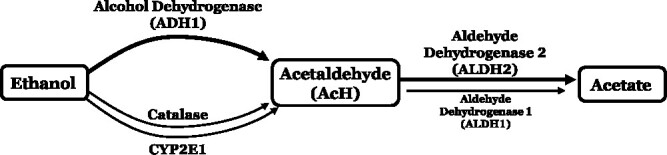
The enzymes alcohol dehydrogenase (ADH1), CYP2E1, and catalase all contribute to oxidative metabolism of ethanol, with ADH1 playing a predominant role; AcH is mainly metabolized by ALDH2 in the mitochondria, with a small contribution from aldehyde dehydrogenase 1 (ALDH1), to form acetate
Table 1.
Human ADH and aldehyde dehydrogenase polymorphisms.
| Description | Official Gene Name | Allele Differences |
Activity (vs. *1) |
Alternate Names |
|---|---|---|---|---|
| Alcohol dehydrogenase 1A (Class I), α polypeptide | ADH1A | ADH1 | ||
| Alcohol dehydrogenase 1B (Class I), β polypeptide | ADH1B*1 | Arg47, Arg369 | ADH2*1 | |
| ADH1B*2 | His47, Arg369 | ↑ | ADH2*2 Rs1229984 |
|
| ADH1B*3 | Arg47, Cys369 | ↑ | ADH2*3 Rs2066702 |
|
| Alcohol dehydrogenase 1C (Class I), γ polypeptide | ADH1C*1 | Arg272, Ile350 | ADH3*1 | |
| ADH1C*2 | Gln272, Val350 | ↓ | ADH3*2 Rs1693482 |
|
| Alcohol dehydrogenase 4 (Class II), π polypeptide | ADH4 | ADH2 HEL-S-4 |
||
| Alcohol dehydrogenase 5 (Class III), χ polypeptide | ADH5 | ADH3 FDH ADHX, AMEDS BMFS7 FALDH, GSNOR, GSH-FDH, HEL-S-60p |
||
| Alcohol dehydrogenase 6 (Class V) | ADH6 | ADH5 | ||
| Alcohol dehydrogenase 7 (Class IV), μ or σ polypeptide | ADH7 | ADH4 | ||
| Aldehyde dehydrogenase 1 family member A1 | ALDH1A1 | ALDH1 RALDH1 |
||
| Aldehyde dehydrogenase 2 | ALDH2*1 | ALDH2 | ||
| ALDH2*2 | ALDH2*Glu504Lys | ↓ | ALDH2*2 rs671 |
Although the enhanced ethanol metabolism of human ADH variants is known to decrease the risk of AUD, inhibition of ADH1 in preclinical models has also decreased ethanol consumption. One recent study has found that treatment of ethanol-dependent rats with ADH1 inhibitor fomepizole consistently reduced ethanol self-administration likely due to changes in ethanol metabolism (Peana et al. 2017). Studies in our laboratory have validated this finding, showing that Adh1-KO mice consume less ethanol than their WT counterparts in drinking-in-the-dark and 2-bottle choice experiments (unpublished observations). Since inhibition of ADH1 leads to higher ethanol and lower AcH and acetate levels, it is difficult to determine the mechanism driving the decreased drinking behavior, but ethanol satiation due to higher ethanol concentrations is likely involved to some extent. However, more research is needed to determine how alterations in the peripheral metabolism of ethanol affect neural reward and aversive feedback mechanisms after ethanol consumption. Both the mouse and human data reinforce that ADH1-mediated changes in ethanol, AcH, and acetate levels are important in regulating drinking behavior in mice and humans.
Aldehyde dehydrogenase
There are two main ALDH enzymes that metabolize ethanol-derived AcH: mitochondrial ALDH2 and cytosolic ALDH1 (encoded by the ALDH1A1 gene) (Table 1) (Edenberg 2007). ALDH2 plays a major role in metabolizing AcH, while ALDH1 plays a less important role, and therefore, we will focus on ALDH2 going forward (Fig. 1). A coding variant known as the ALDH2*2 allele that affects an estimated 8% of the world population leads to the substitution of lysine for glutamate, which disrupts the ALDH2 tetramer and results in ~80% or 100% reduction in AcH clearance for heterozygous or homozygous carriers of the allele, respectively (Edenberg 2007, Chen et al. 2014). Accordingly, the buildup of AcH in people with this variant after ethanol consumption leads to aversive effects such as nausea, headache, and palpitations, which are thought to contribute to the 4-fold decrease in AUD risk for carriers of the ALDH2 polymorphism (Yang et al. 2010, Wang et al. 2021). However, there is a stark difference between the homozygote and heterozygote mutants of the ALDH2 gene. When measuring AcH metabolism in the liver of both mutants, the results indicated that only the homozygote cannot tolerate any amount of alcohol intake, indicating that AUD can develop in people heterozygous for the ALDH2*2 allele even when moderate amounts of alcohol are consumed (Enomoto et al. 1991). Due to the unpleasant symptoms after ethanol consumption, ALDH2-deficient individuals are at lower risk of developing alcohol-related diseases and therefore, therapeutics that inhibit ALDH2 have been of interest in the treatment of AUD (Wang et al. 2020).
ALDH2 exhibits wide expression throughout the body, but previous studies have theorized that ~90% of ethanol-derived AcH is cleared by the liver (Gil-Mohapel et al. 2019). One study used global- and tissue-specific Aldh2-deficient mice to clarify how ALDH2 in different organs metabolize systemic AcH (Guillot et al. 2019). Unexpectedly, this study found that the liver (hepatocytes) only contributes about 30%–40% of blood AcH clearance, suggesting that other ALDH2 expressing organs must play a role in clearing systemic AcH (Guillot et al. 2019). In addition, liver-specific Aldh2 deletion (Aldh2Hep−/−) only decreased excessive ethanol intake without affecting light to moderate ethanol consumption, while global Aldh2−/− mice exhibit drastically reduced ethanol intake (Guillot et al. 2019). It remains unclear which other organs are playing a role in AcH metabolism observed in Aldh2Hep−/− mice, but ALDH2 activity in several organs could be involved (Guillot et al. 2019). In fact, adipose tissue ALDH2 may also be involved in AcH clearance as Aldh2Adipo−/− mice had 1.7-fold higher levels of AcH than their WT mice after acute ethanol gavage. Several other organs also exhibit ALDH2 protein expression/enzymatic activity, including intestine, lungs, brain, and spleen, and future studies should investigate these organs in AcH clearance (Guillot et al. 2019). These results suggest that the liver is not the sole organ responsible for AcH metabolism and ALDH2 activity in organs other than the liver influences drinking behavior.
Catalase and cytochrome P450 2E1
Two possible hypotheses for AcH accumulation in the brain have been proposed. First, peripherally generated AcH passes through the blood–brain barrier (BBB) to influence brain AcH concentrations. Second, local ethanol metabolism by catalase produces AcH in the brain (Jamal et al. 2007). Catalase is the main metabolizer of ethanol in the brain, and catalase-derived AcH is thought to be involved in ethanol-mediated behavioral changes, but it remains a matter of controversy (Fig. 1) (Jamal et al. 2007). Hydrogen peroxide (H2O2) is the major substrate of catalase, and catalase-mediated ethanol metabolism in the brain is dependent upon H2O2 concentration. Several studies have shown that H2O2 is likely involved in behavioral effects like voluntary alcohol intake. Early studies found that increasing H2O2 in the brain via hyperoxia increased ethanol-induced changes in movement and that different levels of H2O2 in the central nervous system resulted in differing amounts of ethanol consumed, implying that the brain catalase-H2O2 system could mediate alcohol self-administration (Pastor et al. 2002, Ledesma et al. 2014). There are also other enzymes present in the brain that may contribute to brain ethanol oxidation such as CYP2E1 and other forms of ADH, but more evidence is required to determine whether these enzymes play an appreciable role in brain ethanol clearance.
Modulators of alcohol metabolism as therapeutics for AUD
Development of AUD therapeutics has focused on several aspects of AUD, which include decreasing alcohol craving and reward, increasing the aversive effects of ethanol, and altering the interoceptive effects of ethanol. Most research on modulating ethanol metabolism for AUD has focused on increasing the aversive effects of ethanol consumption by increasing AcH levels since the protective effects of this strategy are known due to genetic variants (Yang et al. 2010). Both ALDH2*2 and ADH1B*2 are protective against AUD due to increased AcH, and both alleles together offer a synergistic protective effect (Higuchi et al. 1995, Luczak et al. 2009).
Disulfiram
One therapeutic targeting this pathway is disulfiram, a FDA-approved medication for AUD treatment (Table 2). Disulfiram is a non-selective and irreversible ALDH2 and ALDH1 inhibitor, causing AcH to accumulate after alcohol intake (Haass-Koffler et al. 2017). The buildup of toxic AcH results in headaches, nausea, vomiting, and other unpleasant symptoms which is thought to decrease the desire to consume alcohol (Steckler et al. 2013). This was the original premise for the use of disulfiram, but it was found later that disulfiram also increases dopamine concentrations in the brain, which can help prevent cravings in some individuals (Steckler et al. 2013). Disulfiram is administered orally where it is then absorbed in the adipose tissue, gastrointestinal tract, and finally crosses the BBB where it affects dopamine concentrations (Lanz et al. 2023). Although daily doses range from 250 to 500 mg, studies have shown that there is substantial variability in disulfiram responses and that lower doses do not lead to disulfiram–ethanol reactions in close to half of the tested subjects (Lanz et al. 2023). This variability between patients poses serious risks when prescribing disulfiram as some may have toxic reactions even at the lowest dose, and not enough pharmacokinetic studies have been performed in humans to understand disulfiram’s efficacy and safety amongst patient variability. Although disulfiram has shown promise in the treatment of AUD, conflicting efficacy results in clinical practice combined with its adverse effects indicate that it may be best as an adjunct to other therapeutic measures (Hughes and Cook 1997, Haass-Koffler et al. 2017).
Table 2.
Therapeutic potential of selected alcohol metabolism modulators in AUD.
| Therapeutic | Mechanism | FDA-Approved? | PROS | CONS |
|---|---|---|---|---|
| Disulfiram | ALDH inhibitor | Yes | Improves alcohol avoidance | Off-target effects High levels of AcH |
| ANS-6637 | ALDH2 inhibitor | No | Decreases alcohol liking Specific and reversible inhibition |
ANS-6637 withdrawn due to liver toxicity |
| DCR-AUD (Dicerna) | ALDH2 mRNA inhibitor | No | Uses RNAi to specifically target ALDH2 | Still in clinical trial phase Only targets the liver |
| Isoflavones/Kudzu (puerarin, daidzin, daidzein) | ALDH inhibitor | Yes (puerarin–heart disease) |
Potential to utilize dietary intervention | Higher blood alcohol level |
| Alda-1 | ALDH2 activator | No | Enhances clearance of AcH and other toxic aldehydes | Could reverse protective effect of ADH1/ALDH2 polymorphisms on AUD |
| Fomepizole | ADH inhibitor | Yes (methanol/ ethylene glycol poisoning) |
Approved for use in ethylene glycol and methanol toxicity | Increase in ethanol concentration |
| Metadoxine | Increases alcohol clearance | No (approved in certain European countries) |
Decreases intoxication Potentially beneficial for treating ALD |
Mechanism not entirely clear |
Selective and reversible aldehyde dehydrogenase 2 inhibitors
ANS-6637 (originally CVT-10216), a selective and reversible ALDH2 inhibitor, showed clear efficacy in preclinical models of alcohol consumption and has since been developed as an orally active treatment for AUD by Amygdala Neurosciences, Inc. (Arolfo et al. 2009). A Phase 1b study of ethanol interactions with a single dose of ANS-6637 in healthy men showed promising results with the most common adverse events being flushing and increased heart rate (NCT03203499) (O’Malley et al. 2020). Participants receiving ANS-6637 also reported decreased liking of alcohol compared to those who received placebo (O’Malley et al. 2020). However, in a Phase 2 study lasting 5-weeks, there were significant safety concerns related to liver toxicity that led to early termination of the study (NCT03970109). The company has since identified a new lead compound (ANS-858) that is currently in preclinical development (Amygdala Neurosciences 2023). If newer ALDH2-specific inhibitors under investigation can decrease alcohol consumption and craving with less variability and side effects compared to disulfiram, it may open new avenues for AUD treatment.
Aldehyde dehydrogenase 2 RNAi
Another approach to target ALDH2 is genetic targeting, using RNA technology to decrease ALDH2 expression. The efficacy of such technologies has been validated in preclinical models, showing that antisense RNA can decrease ALDH2 expression and ethanol consumption in rats (Ocaranza et al. 2008). Gene targeting technologies have improved significantly over the last few years, and Dicerna (a Novo Nordisk Company) has employed its GalXC™ small interfering RNA technology to develop a candidate for AUD treatment, DCR-AUD (Table 2). DCR-AUD targets ALDH2 messenger RNA (mRNA) specifically in the liver to decrease liver ALDH2 protein expression and increase systemic AcH levels after ethanol consumption (Zhang et al. 2022). The Phase 1 clinical trial for DCR-AUD in healthy volunteers to determine whether ALDH2 knockdown was efficient and safe was recently completed, but no efficacy results are publicly available (NCT05021640) (Zhang et al. 2022). A Phase 1b clinical trial assessing repeat dosing of DCR-AUD is currently ongoing (NCT05845398). It will be interesting to see whether this strategy with specific inhibition of liver ALDH2 in humans replicates the preclinical studies with Aldh2Hep−/− mice, with only slight increases in circulating AcH and reductions only in excessive drinking (Guillot et al. 2019).
Kudzu and isoflavones
Traditional Chinese medicine has used the kudzu plant to treat AUD and combat intoxication for centuries (Table 2) (Liang and Olsen 2014, Swift and Aston 2015). Research into the protective mechanisms of this plant suggests that its flavonoids (i.e. puerarin, daidzin, daidzein) may inhibit the ALDH2 pathway to decrease alcohol consumption (Swift and Aston 2015). One study in alcohol-preferring and non-preferring rats showed that kudzu decreased ethanol intake/withdrawal and a clinical study demonstrated that 1 week of kudzu treatment in heavy drinking individuals decreased their ethanol consumption, validating the use of this traditional medicine for AUD (Liang and Olsen 2014, Swift and Aston 2015). Recent studies have focused on evaluating the individual effects of the flavones puerarin, daidzin, and daidzein on alcohol consumption. Daidzin cut alcohol consumption by half in hamsters, while puerarin was able to decrease ethanol consumption in female alcohol-preferring rats by >50% (Penetar et al. 2012). Although the kudzu-based isoflavones are not FDA-approved, isoflavone puerarin is already FDA-approved to treat coronary heart disease, so puerarin will likely be the isoflavone investigated for use in AUD clinical studies (Penetar et al. 2012). It is important to note that dietary isoflavones and administered isoflavones may play diverging roles in alcohol consumption, based on data showing that low isoflavone-containing diets consume less alcohol (Eduardo and Abrahao 2022). In all, further studies are needed to clarify the roles and toxicity of isoflavones in alcohol consumption.
Alda-1
In contrast to the aforementioned ALDH inhibitor drugs, Alda-1 is the first identified selective activator of ALDH2 (Table 2), acting as a molecular chaperone of the enzyme (Virgolini and Pautassi 2022). Although an activator of ALDH2 would be expected to increase drinking behavior since ALDH2 deficiency decreases ethanol consumption, a recent study showed that Alda-1 can decrease ethanol intake in ethanol-preferring rats without changing sucrose preference (Rivera-Meza et al. 2019). This suggests that an increased capacity to eliminate AcH may not lead to increases in ethanol consumption for people with normal ALDH2 function; however, ALDH2 activation in people with ALDH2 deficiency would be expected to clear excess AcH and remove the aversive phenotype that protects against AUD after ethanol consumption. It is also possible that the Alda-1-mediated decrease in drinking behavior is due to the enhanced capacity of ALDH2 to metabolize other toxic aldehydes generated during alcohol consumption such as 4-hydroxynonenal and may contribute to neuroinflammation-induced drinking (Rivera-Meza et al. 2019). Since Alda-1 is a small molecule, it is also possible that the effects on drinking behavior are off-target effects. Regardless, the potential risks to increase drinking and reverse the protective effects of the ADH1B*2 and ALDH2*2 alleles for AUD make development of this compound for AUD treatment extremely difficult. However, Alda-1-mediated decreases in drinking behavior should be further studied in preclinical models to determine whether decreases in AcH in animals with normal ALDH2 are due to decreases in reward, craving, or other mechanisms as this is important knowledge when developing ethanol metabolism modulators for AUD treatment.
Alcohol dehydrogenase modulation
Preclinical data using ADH-inhibitor fomepizole (Table 2) showed decreases in drinking behavior for ethanol-preferring rats, providing an interesting possibility of treating AUD by modulating ADH-mediated ethanol metabolism (Peana et al. 2017). Fomepizole is approved for use in ethylene glycol and methanol toxicity by competitively inhibiting ADH (Sande et al. 2012). Further preclinical studies should be completed to determine how fomepizole-mediated decrease in ethanol consumption is related to, but it is also possible to try this therapy in humans since fomepizole is safe for short-term use and has been used to study human ethanol metabolism and reverse the disulfiram ethanol reaction (Sande et al. 2012). Immunization against ADH in rat models of ethanol consumption has also shown that decreases in ADH-mediated metabolism can decrease drinking behavior, through modulating ethanol metabolism as well as dopamine levels (Pshezhetsky et al. 1993, Mitkin et al. 2020). These findings indicate that ADH-mediated metabolism is important in the acquisition and maintenance of ethanol preference, but much more investigation is required to determine whether ADH inhibition is a potential therapeutic target for AUD. Since inhibition of ADH will increase ethanol levels, this could cause further damage if continued heavy drinking occurs, and ADH1-deficient animals are known to be more susceptible to ethanol-mediated toxicity (Haseba et al. 2019). Therefore, caution must be taken when trying to translate these findings to human subjects.
Metadoxine
Metadoxine is approved to treat acute alcohol intoxication in certain European countries, but not the United States, and works by increasing alcohol clearance as demonstrated in a randomized, double-blind, placebo-controlled study (Shpilenya et al. 2002). Metadoxine increases urine and plasma clearance of alcohol and its metabolites, potentially through increased ALDH activity, preventing decreased ADH activity, and protection against liver oxidative stress, but the exact mechanism is unclear (Addolorato et al. 2003). Metadoxine has also been investigated for the treatment of AUD. One study in alcohol-dependent patients found that metadoxine improved abstinence compared to patients receiving no pharmacotherapy, but these findings must be further confirmed in randomized studies (Guerrini et al. 2006). In a retrospective analysis, patients with AUD given metadoxine for alcohol intoxication exhibited decreased drinks per week compared to baseline and many (2/3) were abstinent (Leggio et al. 2011). Given that metadoxine may also be beneficial for the treatment of alcohol-associated liver disease (ALD) (Higuera-de la Tijera et al. 2015), further studies should be completed to determine the efficacy in large-scale trials.
Discussion
Although investigating ethanol pharmacokinetics represents an exciting approach to investigate novel medications for AUD, there are several challenges that must be considered (Haass-Koffler et al. 2017). The primary focus of the several preceding therapeutics is to inhibit ALDH2 activity in the body to increase AcH and create unpleasant symptoms when consuming alcohol. However, this mechanism to prevent drinking is a double-edged sword—the aversive effects of AcH may prevent drinking, but people who are exposed to large amounts of AcH are at greater risk for developing cancer (Fig. 2). AcH promotes cancer in several ways—it can interfere with the copying of DNA or inhibit the process of damaged DNA repair (Zhai et al. 2023). This theory has been tested in numerous ALDH2-deficient heavy drinkers for the effects of long-term AcH exposure. Individuals with ALDH2 deficiency have extremely high concentrations of AcH in their mouth and aerodigestive tract, leading to much higher risks of head and neck, esophageal, and colon cancer (Seitz and Becker 2007). To emphasize the severity of increased levels of AcH, the International Agency for Research on Cancer asserts that AcH should be classified as a carcinogen and suggests that the risk for developing certain cancers is lower in drinkers who are exposed to less AcH during alcohol metabolism (Seitz and Becker 2007). In all, these data suggest that treating AUD by inhibiting ALDH2 could lead to higher risks of cancer if patients continue to drink while taking such inhibitors, and this should be considered when developing pharmacological approaches.
Figure 2.
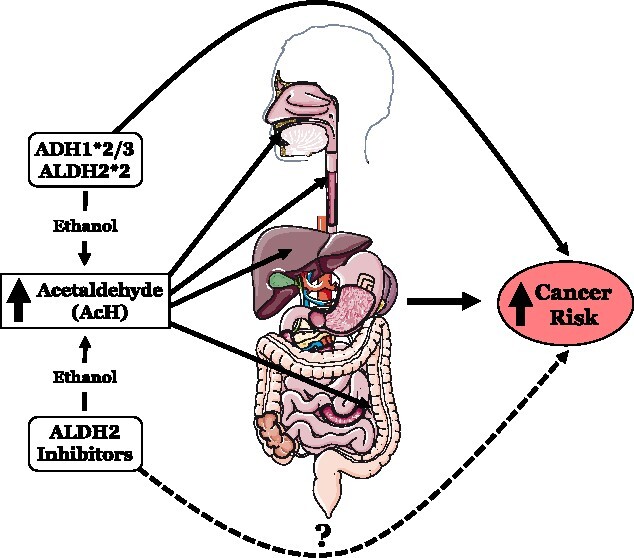
Depiction of the potential harmful effects that increased AcH after ethanol consumption may have on the body; Alcohol/aldehyde dehydrogenase polymorphisms are known to increase the risk of cancer of the mouth, esophagus, liver, and colon, but whether ALDH2 inhibitors also increase cancer risk by increasing AcH is unknown
Preclinical evidence also points toward targeting catalase to decrease ethanol consumption by inhibiting AcH production in the brain. However, inhibition of catalase would also cause an excess of H2O2, which would likely lead to detrimental levels of oxidative stress and overt toxicity over time (Zhu et al. 2005). These potential downsides, coupled with the fact that catalase has wide systemic expression, make the development of catalase inhibitors for AUD extremely unlikely. However, H2O2 scavengers may be therapeutically beneficial in AUD to prevent brain AcH accumulation and H2O2-mediated cell damage and should be further studied.
Outside of increasing AcH as a mechanism to decrease drinking behavior, modulation of ethanol metabolism in other ways may also be beneficial for AUD. The fact that metadoxine, Alda-1, and fomepizole are all able to decrease ethanol consumption highlights a knowledge gap in the contribution of AcH and acetate to drinking behavior that requires further study. Since these are small molecules, they could potentially have off-target effects that may lead to further AUD target discovery. However, using genetic models to confirm these findings would indicate that decreases in AcH below normal levels may be beneficial for AUD treatment. In addition, since the inhibition of ADH or ALDH2 leads to a decrease in acetate accumulation, it suggests that acetate may play an important role in ethanol consumption. There is still much work to be done to fully understand the contributions of organ-specific ethanol metabolism to drinking behaviors, and pursuing these questions may provide new therapeutic approaches for AUD.
There are currently only three drugs approved to treat AUD in the United States—disulfiram, naltrexone, and acamprosate. Efforts to further understand and characterize how modulation of ethanol metabolism can be harnessed for AUD treatment are crucial to provide a wider range of options for patients with AUD. We believe there are several knowledge gaps that must be addressed going forward. First, few studies examine how alcohol metabolism interacts with other systems that regulate ethanol intake, and this should be a priority going forward. For instance, the hormone fibroblast growth factor 21 (FGF21) is induced by ethanol consumption and acts on specific populations of neurons to repress further ethanol drinking (Schumann et al. 2016, Talukdar et al. 2016). However, it is not known how modulating alcohol metabolism would affect the induction of FGF21 and the subsequent repression of ethanol intake. People harboring ALDH2*2 have been shown to have altered cortisol and corticosterone levels after ethanol administration (Gao et al. 2019), demonstrating that altered ethanol metabolism could change the production and action of hormones that regulate drinking behavior. In addition, further research is needed to elucidate the timespan in which ADH and ALDH can be inhibited during ethanol consumption without causing harm. In our view, targeting ADH and ALDHs should be designed as a short-term transitional therapy that is combined with psychosocial interventions and medical management, the current standard of AUD care (Addolorato et al. 2016). Co-treatment of ethanol metabolism modulators with other approved pharmacological therapies for AUD should be investigated to determine whether combination treatment will provide better outcomes. Overall, we believe that a better understanding of how ethanol metabolism drives drinking behavior will provide more therapeutic interventions for AUD in the future.
Acknowledgements
This review covers a very broad topic, and we apologize to colleagues whose research may not have been discussed or cited due to space and citation limitations.
Contributor Information
Taylor Lehner, Laboratory of Liver Diseases, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, 5625 Fishers Lane, Bethesda, MD 20892, United States.
Bin Gao, Laboratory of Liver Diseases, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, 5625 Fishers Lane, Bethesda, MD 20892, United States.
Bryan Mackowiak, Laboratory of Liver Diseases, National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, 5625 Fishers Lane, Bethesda, MD 20892, United States.
Authors’ contributions
T.L. searched the data for the article and wrote the manuscript; B.G. supervised the review and edited the manuscript; B.M. supervised the whole review and edited the manuscript.
CRediT authors’ contributions
Taylor Lehner (Conceptualization [lead], Writing—original draft [lead], Writing—review & editing [equal]), Bin Gao (Funding acquisition [lead], Project administration [equal], Resources [lead], Supervision [supporting], Writing—original draft [supporting], Writing—review & editing [supporting]), Bryan Mackowiak (Conceptualization [equal], Project administration [equal], Supervision [lead], Writing—original draft [equal], Writing—review & editing [equal])
Conflict of interest: None declared.
Funding
This work described from Dr Bin Gao’s lab in this review article was supported by the intramural program of National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health (B.G.).
Data availability
All data from the review is present in the current manuscript.
References
- Addolorato G, Ancona C, Capristo E. et al. Metadoxine in the treatment of acute and chronic alcoholism: a review. Int J Immunopathol Pharmacol 2003;16:207–14. [DOI] [PubMed] [Google Scholar]
- Addolorato G, Mirijello A, Barrio P. et al. Treatment of alcohol use disorders in patients with alcoholic liver disease. J Hepatol 2016;65:618–30. [DOI] [PubMed] [Google Scholar]
- Amygdala Neurosciences Incorporated . Amygdala Neurosciences Awarded $2.0 Million NIH Grant to Conduct IND Enabling Studies [Press Release], 2023. https://amygns.com/news/amygdala-neurosciences-awarded-2-0-million-nih-grant-to-conduct-ind-enabling-studies/.
- Arolfo MP, Overstreet DH, Yao L. et al. Suppression of heavy drinking and alcohol seeking by a selective ALDH-2 inhibitor. Alcohol Clin Exp Res 2009;33:1935–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw PC. Acetyl-CoA metabolism and histone acetylation in the regulation of aging and lifespan. Antioxidants (Basel) 2021;10:532. 10.3390/antiox10040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum AI. Alcohol metabolism. Clin Liver Dis 2012;16:667–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Ferreira JC, Gross ER. et al. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev 2014;94:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa M, Salamone JD, Segovia KN. et al. Piecing together the puzzle of acetaldehyde as a neuroactive agent. Neurosci Biobehav Rev 2012;36:404–30. [DOI] [PubMed] [Google Scholar]
- Da BL, Im GY, Schiano TD. Coronavirus disease 2019 hangover: a rising tide of alcohol use disorder and alcohol-associated liver disease. Hepatology 2020;72:1102–8. [DOI] [PubMed] [Google Scholar]
- Deltour L, Foglio MH, Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1,Adh3, and Adh4 null mutant mice: overlapping roles of Adh1 AND Adh4 in ethanol clearance and metabolism of retinol to retinoic acid*. J Biol Chem 1999;274:16796–801. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism. Alcohol Res Health 2007;30:9–13. [PMC free article] [PubMed] [Google Scholar]
- Eduardo PMC, Abrahao KP. Food composition can influence how much alcohol your animal model drinks: a mini-review about the role of isoflavones. Alcohol Clin Exp Res 2022;46:6–12. [DOI] [PubMed] [Google Scholar]
- Enomoto N, Takase S, Takada N. et al. Alcoholic liver disease in heterozygotes of mutant and normal aldehyde dehydrogenase-2 genes. Hepatology 1991;13:1071–5. [PubMed] [Google Scholar]
- Evsyukov A, Ivanov D. Selection variability for Arg48His in alcohol dehydrogenase ADH1B among Asian populations. Hum Biol 2013;85:569–77. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhou Z, Ren T. et al. Alcohol inhibits T-cell glucose metabolism and hepatitis in ALDH2-deficient mice and humans: roles of acetaldehyde and glucocorticoids. Gut 2019;68:1311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Mohapel J, Bianco CD, Cesconetto PA. et al. Chapter 51 – ethanol exposure during development, and brain oxidative stress. In: Preedy VR (ed). Neuroscience of Alcohol. Academic Press, 2019, 493–503. 10.1016/B978-0-12-813125-1.00051-9. [DOI] [Google Scholar]
- Guerrini I, Gentili C, Nelli G. et al. A follow up study on the efficacy of metadoxine in the treatment of alcohol dependence. Subst Abuse Treat Prev Policy 2006;1:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot A, Ren T, Jourdan T. et al. Targeting liver aldehyde dehydrogenase-2 prevents heavy but not moderate alcohol drinking. Proc Natl Acad Sci U S A 2019;116:25974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass-Koffler CL, Akhlaghi F, Swift RM. et al. Altering ethanol pharmacokinetics to treat alcohol use disorder: can you teach an old dog new tricks? J Psychopharmacol 2017;31:812–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseba T, Okuda T, Maruyama M. et al. Roles of two major alcohol dehydrogenases, ADH1 (class I) and ADH3 (class III), in the adaptive enhancement of alcohol metabolism induced by chronic alcohol consumption in mice. Alcohol Alcohol 2019;55:11–9. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Murayama M. et al. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry 1995;152:1219–21. [DOI] [PubMed] [Google Scholar]
- Higuera-de la Tijera F, Servín-Caamaño AI, Serralde-Zúñiga AE. et al. Metadoxine improves the three- and six-month survival rates in patients with severe alcoholic hepatitis. World J Gastroenterol 2015;21:4975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JC, Cook CCH. The efficacy of disulfiram: a review of outcome studies. Addiction 1997;92:381–95. [PubMed] [Google Scholar]
- Jamal M, Ameno K, Uekita I. et al. Catalase mediates acetaldehyde formation in the striatum of free-moving rats. Neurotoxicology 2007;28:1245–8. [DOI] [PubMed] [Google Scholar]
- Lanz J, Biniaz-Harris N, Kuvaldina M. et al. Disulfiram: mechanisms, applications, and challenges. Antibiotics (Basel) 2023;12:524. 10.3390/antibiotics12030524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma JC, Baliño P, Aragon CMG. Reduction in central H2O2 levels prevents voluntary ethanol intake in mice: a role for the brain catalase-H2O2 system in alcohol binge drinking. Alcohol Clin Exp Res 2014;38:60–7. [DOI] [PubMed] [Google Scholar]
- Leggio L, Kenna GA, Ferrulli A. et al. Preliminary findings on the use of metadoxine for the treatment of alcohol dependence and alcoholic liver disease. Hum Psychopharmacol 2011;26:554–9. [DOI] [PubMed] [Google Scholar]
- Li H, Gu S, Cai X. et al. Ethnic related selection for an ADH class I variant within East Asia. PLoS One 2008;3:e1881. 10.1371/journal.pone.0001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Olsen RW. Alcohol use disorders and current pharmacological therapies: the role of GABAA receptors. Acta Pharmacol Sin 2014;35:981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TL. Meta-Analyses of ALDH2 and ADH1B with Alcohol Dependence in Asians. Washington, DC: American Psychological Association, 2009. 10.1037/11855-026. [DOI] [PubMed] [Google Scholar]
- Mitkin NA, Anokhin PK, Belopolskaya MV. et al. Active immunization against serum alcohol dehydrogenase normalizes brain dopamine metabolism disturbed during chronic alcohol consumption. Alcohol 2020;83:17–28. [DOI] [PubMed] [Google Scholar]
- O’Malley SS, Shram MJ, Levy-Cooperman N. et al. Interaction of ethanol and oral ANS-6637, a selective ALDH2 inhibitor in males: a randomized, double-blind, placebo-controlled, single-ascending dose cohort study. Alcohol Clin Exp Res 2020;44:1885–95. [DOI] [PubMed] [Google Scholar]
- Ocaranza P, Quintanilla ME, Tampier L. et al. Gene therapy reduces ethanol intake in an animal model of alcohol dependence. Alcohol Clin Exp Res 2008;32:52–7. [DOI] [PubMed] [Google Scholar]
- Pastor R, Sanchis-Segura C, Aragon CM. Ethanol-stimulated behaviour in mice is modulated by brain catalase activity and H2O2 rate of production. Psychopharmacology (Berl) 2002;165:51–9. [DOI] [PubMed] [Google Scholar]
- Peana AT, Acquas E. Behavioral and biochemical evidence of the role of acetaldehyde in the motivational effects of ethanol. Front Behav Neurosci 2013;7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peana AT, Rosas M, Porru S. et al. From ethanol to salsolinol: role of ethanol metabolites in the effects of ethanol. J Exp Neurosci 2016;10:137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peana AT, Pintus FA, Bennardini F. et al. Is catalase involved in the effects of systemic and pVTA administration of 4-methylpyrazole on ethanol self-administration? Alcohol 2017;63:61–73. [DOI] [PubMed] [Google Scholar]
- Penetar DM, Toto LH, Farmer SL. et al. The isoflavone puerarin reduces alcohol intake in heavy drinkers: a pilot study. Drug Alcohol Depend 2012;126:251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pshezhetsky AV, Danilova RA, Fedorova IM. et al. Influence of the immunization against heterologous alcohol dehydrogenase on liver alcohol dehydrogenase isozymes and alcohol abuse of rats. Eur J Biochem 1993;212:757–61. [DOI] [PubMed] [Google Scholar]
- Quertemont E, Didone V. Role of acetaldehyde in mediating the pharmacological and behavioral effects of alcohol. Alcohol Res Health 2006;29:258–65. [PMC free article] [PubMed] [Google Scholar]
- Rivera-Meza M, Vásquez D, Quintanilla ME. et al. Activation of mitochondrial aldehyde dehydrogenase (ALDH2) by ALDA-1 reduces both the acquisition and maintenance of ethanol intake in rats: a dual mechanism? Neuropharmacology 2019;146:175–83. [DOI] [PubMed] [Google Scholar]
- Sande M, Thompson D, Monte AA. Fomepizole for severe disulfiram-ethanol reactions. Am J Emerg Med 2012;30:262.e3–5. [DOI] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P. et al. KLBis associated with alcohol drinking, and its gene product β-klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A 2016;113:14372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health 2007;30:44–7. [PMC free article] [PubMed] [Google Scholar]
- Shpilenya LS, Muzychenko AP, Gasbarrini G. et al. Metadoxine in acute alcohol intoxication: a double-blind, randomized, placebo-controlled study. Alcohol Clin Exp Res 2002;26:340–6. [PubMed] [Google Scholar]
- Steckler G, Witkiewitz K, Marlatt GA. Chapter 13 – relapse and lapse. In Miller PMs (ed.), Principles of Addiction. San Diego: Academic Press, 2013, 125–32. 10.1016/B978-0-12-398336-7.00013-9. [DOI] [Google Scholar]
- Swift RM, Aston ER. Pharmacotherapy for alcohol use disorder: current and emerging therapies. Harv Rev Psychiatry 2015;23:122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S, Owen BM, Song P. et al. FGF21 regulates sweet and alcohol preference. Cell Metab 2016;23:344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsermpini EE, Plemenitaš Ilješ A, Dolžan V. Alcohol-induced oxidative stress and the role of antioxidants in alcohol use disorder: a systematic review. Antioxidants 2022;11:1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgolini MB, Pautassi RM. Chapter two – converging mechanisms in ethanol neurotoxicity. In: Slikker W, Aschner M, Costa LGS (eds). Advances in Neurotoxicology, Vol. 8. Academic Press, 2022, 49–92. 10.1016/bs.ant.2022.06.002. [DOI] [Google Scholar]
- Wang W, Wang C, Xu H. et al. Aldehyde dehydrogenase, liver disease and cancer. Int J Biol Sci 2020;16:921–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Chang B, Li X. et al. Role of ALDH2 in hepatic disorders: gene polymorphism and disease pathogenesis. J Clin Transl Hepatol 2021;9:90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewitz K, Litten RZ, Leggio L. Advances in the science and treatment of alcohol use disorder. Sci Adv 2019;5:eaax4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Yokoyama A, Yokoyama T. et al. Relationship between genetic polymorphisms of ALDH2 and ADH1B and esophageal cancer risk: a meta-analysis. World J Gastroenterol 2010;16:4210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Yamauchi T, Shangraw S. et al. Ethanol metabolism and melanoma. Cancers (Basel) 2023;15:1258. 10.3390/cancers15041258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XYF, Gertsik L, Hanrahan J. First-in-human clinical trial of DCR-AUD: evaluation of siRNA knockdown of hepatic acetaldehyde dehydrogenase 2 using a novel bioanalytic assay. Alcohol Clin Exp Res 2022;46:39A. [Google Scholar]
- Zhu D, Tan KS, Zhang X. et al. Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J Cell Sci 2005;118:3695–703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from the review is present in the current manuscript.


