Abstract
Oral administration facilitates the direct delivery of drugs to lesions within the small intestine and colon, making it an ideal approach for treating patients with inflammatory bowel disease. However, multiple physical barriers impede the delivery of oral RNA drugs through the gastrointestinal tract. Herein, we developed a novel oral siRNA delivery system that protects nucleic acids in extreme environments by employing exosomes derived from milk to encapsulate tumor necrosis factor-alpha (TNF-α) siRNA completely. The remarkable structural stability of milk-derived exosomes (M-Exos), as opposed to those from HEK293T cells, makes them exceptional siRNA carriers. Results demonstrate that milk exosomes loaded with TNF-α siRNA (M-Exo/siR) can effectively inhibit the expression of TNF-α-related inflammatory cytokines. Moreover, given that milk exosomes are composed of unique lipids with high bioavailability, orally administered M-Exo/siR effectively reach colonic tissues, leading to decreased TNF-α expression and successful alleviation of colitis symptoms in a dextran sulfate sodium-induced inflammatory bowel disease murine model. Hence, milk-derived exosomes carrying TNF-α siRNA can be effectively employed to treat inflammatory bowel disease. Indeed, using exosomes naturally derived from milk may shift the current paradigm of oral gene delivery, including siRNA.
Keywords: Milk-derived exosome, Oral gene delivery, siRNA, Inflammatory bowel disease, TNF-α
Graphical abstract
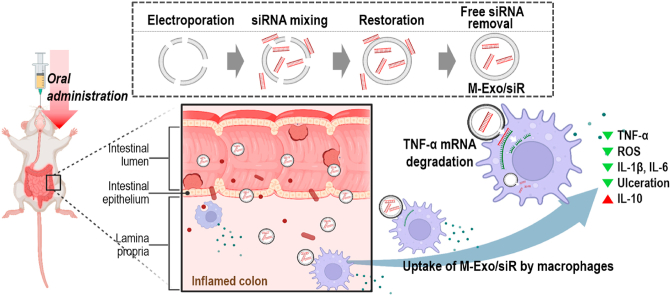
Schematic diagram of the colitis treatment process through the oral administration of M-Exo/siR. TNF-α siRNA was loaded into M-Exo via electroporation. Orally administered M-Exo/siR successfully reached the inner wall of the colon in IBD mice and reduced TNF-α expression. This led to the regulation of the expression of inflammatory cytokines and the reduction of ulcers.
Highlights
-
•
Milk-derived exosomes (M-Exos)' superior structural stability makes it an efficient siRNA carrier.
-
•
M-Exo loaded with TNF-α siRNA (M-Exo/siR) effectively suppresses TNF-α expression and related pro-inflammatory cytokines.
-
•
The unique lipid composition of M-Exos underlies the exceptional stability in the GI tract.
-
•
Orally administered M-Exo/siR alleviates IBD-related symptoms by reducing TNF-α expression after reaching the colon.
1. Introduction
Inflammatory bowel disease (IBD) is an inflammatory disease of the intestine [1], typically divided into Crohn's disease (CD), accompanied by inflammatory skip lesions throughout the digestive tract from the mouth to the anus, and ulcerative colitis (UC), characterized by continuous inflammation throughout the colon [2]. The etiology of IBD is complex, making treatment difficult and the rate of recurrence high [3]. Given that the main pathological manifestations of IBD are limited to the intestinal tissues, and non-targeted therapies promote systemic absorption, leading to side effects and reduced efficacy, oral administration is considered the optimal route for ensuring direct delivery of drugs to the pathogenic site within the intestines. However, the oral administration of biologics, particularly RNA drugs, is hindered by their susceptibility to the gastrointestinal environment [4]; moreover, their size impedes their transport across the intestinal epithelium [5,6]. Therefore, formulation-based delivery technologies are required to improve the performance of orally administered biological drugs.
One such technology includes using exosomes, which have a critical physiological role in intercellular communication and the transport of macromolecules between cells and tissues. Exosomes can also serve as vehicles for administering various drug payloads, particularly nucleic acids, as their composition provides superior tolerability [7,8]. In particular, exosomes derived from milk (M-Exos) are a unique class of evolutionarily conserved extracellular vesicles that maintain the integrity of packaged nucleic acids and other biomacromolecules during transit through the stomach and gastrointestinal (GI) tract [9]. Additionally, M-Exos possesses the feature of being actively transported and absorbed into small intestinal and colonic cells [10]. While most mammalian cell-derived exosomes are not suitable or available as vehicles for the oral administration of drugs owing to their labile nature under harsh physiological conditions [11], milk exosomes are robust and have demonstrated stability under acidic and other severe conditions. Therefore, M-Exos may offer a unique opportunity to overcome the challenge surrounding the oral delivery of drugs, including oligonucleotides.
TNF-α is a key cytokine in IBD [12,13], the signaling of which orchestrates the production of various inflammatory cytokines in the acute phase, affecting cell proliferation, apoptosis, and immunomodulation, resulting in an imbalanced colonic microenvironment [14]. However, clinically available anti-TNF-α antibody drugs, such as Adalimumab, Infliximab, and Golimumab, can cause patient discomfort as well as off-target systemic side effects, and their long-term use can lead to serious complications [15]. Moreover, monoclonal antibodies, generally intended to inhibit or block protein function, must recognize the complicated spatial conformation of certain proteins, preventing them from identifying a target molecule with high activity, affinity, or specificity. However, siRNA modalities have inherent advantages over antibody drugs, as any gene of interest can be targeted, provided the correct nucleotide sequence is selected along the target mRNA. To address these issues, we have developed a new platform for siRNA delivery that can locally target the inflamed colonic site via oral administration to modulate the anti-inflammatory immune response.
In the current study, we loaded M-Exos with TNF-α siRNA via electroporation (M-Exo/siR) and assessed its efficacy in treating colitis in a murine model. From a lipidomics point of view, M-Exos exhibit excellent stability due to the lipid composition, which is fundamentally different from cell-derived exosomes. Moreover, following oral administration, the M-Exo carrier stably passes through the GI tract to the colitis lesions (Fig. 1A), where they induce the degradation of TNF-α mRNA. Accordingly, M-Exos hold great promise as oral gene delivery vehicles for clinical applications. In particular, TNF-α siRNA-loaded M-Exos might represent an effective biologic treatment modality for IBD.
Fig. 1.
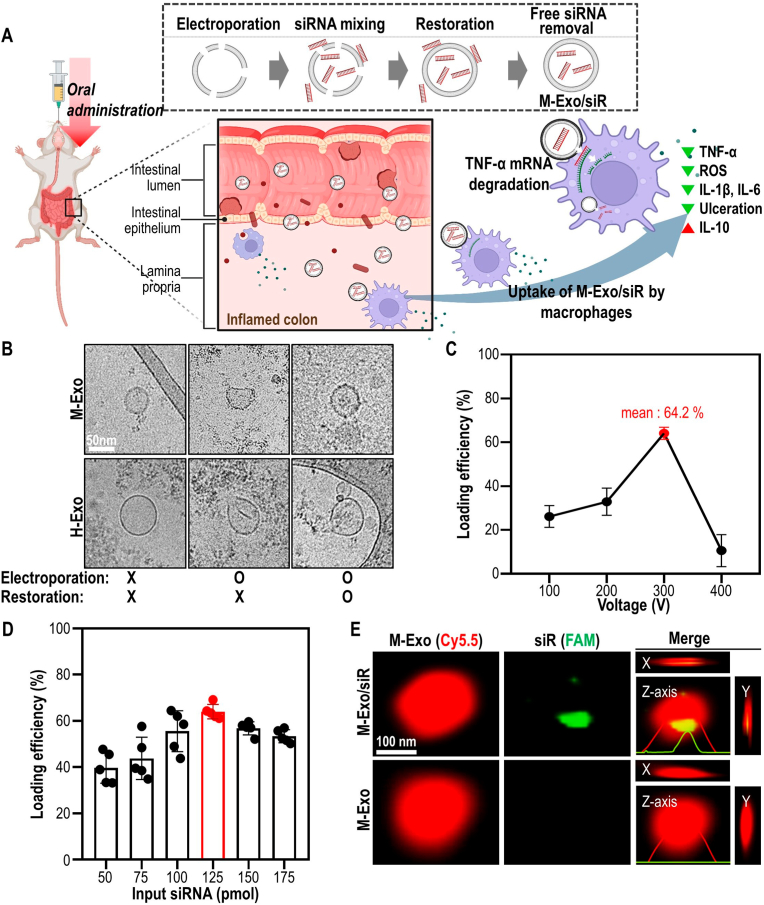
Preparation of M-Exo/siR and optimization of electroporation conditions for loading TNF-α siRNA into M-Exos. (A) Schematic of the colitis treatment process through oral administration of M-Exo/siR. (B) Comparative changes of morphology of exosomes according to electroporation conditions. (C) Loading efficiency of TNF-α siRNA into M-Exos according to voltage. The mixing ratio of M-Exos and siRNA is the same in all conditions. Data are mean ± SD (n = 9). (D) Loading efficiency according to the M-Exo and TNF-α ratio. The voltage, pulse number, and pulse length used for electroporation are consistent in all conditions. Data are mean ± SD (n = 5). (E) Representative SRM images were measured in the x-, y-, and z-axis directions of M-Exo/siR. Red (Cyanine 5.5-labeled M-Exos); Green (5′-Fluorescein phosphoramidite-labeled TNF-α siRNA).
2. Results
2.1. M-Exos maintain their structure upon electroporation for complete siRNA encapsulation
For RNA drugs that are easily degraded, reaching the target site is a major bottleneck in determining therapeutic output [16]. One way to overcome this is to encapsulate siRNA therapeutics in a carrier to protect them from immune cells and RNA-degrading enzymes [17]. Loading siRNA into exosomes requires the membrane to be compromised to allow macromolecules to enter; however, the membranes must be subsequently restored to ensure complete drug encapsulation. To achieve this, we encapsulated TNF-α siRNA in M-Exo by modifying conventional electroporation methods [18,19] with a “restoration step” to stabilize the M-Exo membrane at 4 °C for 3 h following electroporation and incubation with the TNF-α siRNA mixture (Fig. 1A).
Electroporation during siRNA encapsulation should not alter their physicochemical properties, thus ensuring that the exosomes stably deliver intact drugs to the desired location. To ensure this, changes in the morphology of the exosomes before and after electroporation were closely examined. For comparison, we prepared another exosome derived from HEK293T cells culture medium (H-Exo). Although milk yielded approximately 5-fold more exosomes than HEK293T cells (Figs. S1A–B), both M − and H-Exos expressed exosome protein markers (TSG101 and CD9), regardless of electroporation. As in previous studies [20], Alix was not detected in M-Exos. In addition, GM130 (Golgi maker) and Calnexin (endoplasmic reticulum marker) — negative markers of exosomes — were detected only in HEK293T cell lysates (Fig. S1C). However, cryo-transmission electron microscopy (cryo-TEM) confirmed that cracks and structural deformations occurred on the surfaces of M-Exos and H-Exos immediately after electroporation (Fig. 1B, S1D). Meanwhile, only M-Exos were restored to their spherical shape following incubation of electroporated M-Exos at 4 °C for 3 h. In contrast, H-Exos continued to exhibit a rough shape even after the restoration step.
Next, to optimize the electroporation conditions for M-Exo/siR, the siRNA loading efficiency was determined by altering the voltage (Fig. 1C). When M-Exos were electroporated at 300 V with five pulses, the loading efficiency was the highest at 64.2 % (Figs. S1E–F). In addition, TNF-α siRNA was loaded with the highest efficiency in M-Exos at a ratio of 125 pmol per 1010 M-Exo particles (Fig. 1D). Therefore, all electroporation experiments comprised 300 V and five pulses with an optimized siRNA and M-Exo ratio.
Complete encapsulation of siRNA by exosomes is required to protect orally administered drugs from the harsh environment in the GI tract, including low pH and endonuclease degradation. Thus, 3D plots of exosome images obtained by super-resolution microscopy (SRM) were used to determine the distribution of TNF-α siRNA in M-Exos (Fig. 1E). Images captured in x, y, and z directions revealed that the 6-FAM (5′-fluorescein phosphoramidite)-labeled siRNA signal was located near the center of M-Exos. Additionally, siRNA encapsulated within M-Exo/siR was considerably more stable in the serum than free siRNA (Fig. S1G). Taken together, these results indicate that M-Exo possesses appropriate stability and can protect encapsulated drugs from external stimuli.
2.2. M-Exo/siR ameliorates inflammation by downregulating TNF-α without inducing cytotoxicity
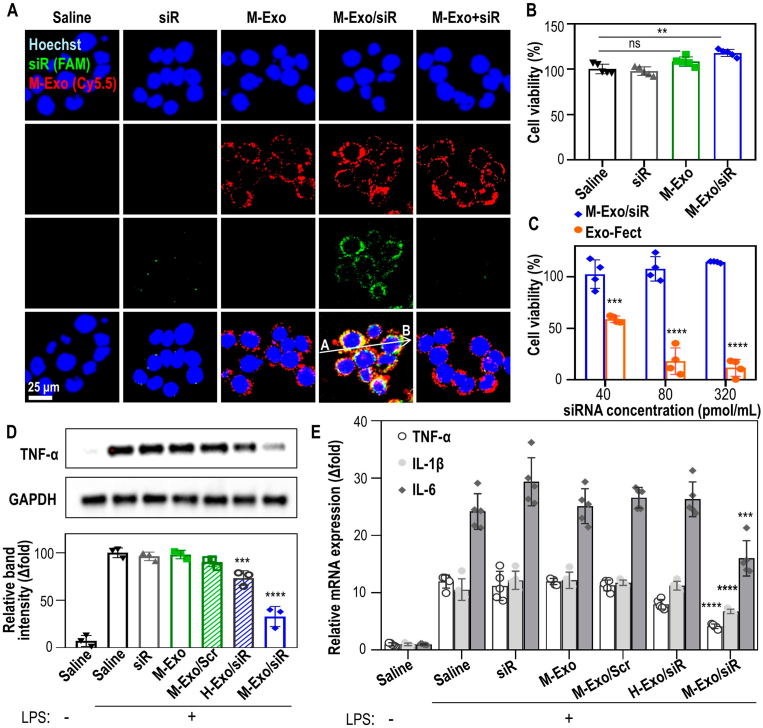
Prior to examining the anti-inflammatory effects of M-Exo/siR in vitro, we confirmed the cellular uptake of siRNAs by NCM460 cells (Fig. 2A). In brief, after treating NCM460 cells with fluorescently labeled M-Exo/siR for 24 h, the correlation between M-Exos and siRNA fluorescence was measured to determine whether the siRNA was sufficiently released into the cytoplasm (Fig. S2A). The calculated correlation coefficient was 0.1923, indicating that most siRNA was released from the M-Exos. In contrast, a simple mixture of M-Exos and siRNA (M-Exo + siR), in which siRNA was not encapsulated by the exosome, failed to deliver siRNA into the cytoplasm (Fig. 2A). The effective cellular uptake of M-Exo/siR by RAW264.7 cells was also observed (Fig. S2B).
Fig. 2.
Cytotoxicity and anti-inflammatory efficacy of M-Exo/siR in vitro. (A) Intracellular TNF-α siRNA delivery via M-Exo/siR in NCM460 cells. Blue (Hoechst 33342 fluorescence staining of nuclei); Green (5′-Fluorescein phosphoramidite-labeled TNF-α siRNA); Red (Cyanine 5.5-labeled M-Exos). (B) Cytotoxicity of M-Exo/siR in NCM460. Data are mean ± SD (n = 5); ns = not significant, **p < 0.01. (C) Comparison of NCM460 cell viability according to the concentration of M-Exo/siR and Exo-Fect™. Data are mean ± SD (n = 4); ***p < 0.001, ****p < 0.0001. (D) TNF-α protein down-regulation effect of M-Exo/siR in RAW 264.7 cells. Each group was treated with 100 ng/mL LPS for 8 h, followed by a 24 h incubation with their respective substances. Data are mean ± SD (n = 3); ***p < 0.001, ****p < 0.0001. (E) Relative mRNA expression of pro-inflammatory cytokines. Each group was treated with 100 ng/mL LPS for 8 h, followed by a 24 h incubation with their respective substances. Data are mean ± SD (n = 5); ***p < 0.001, ****p < 0.0001.
Treatment of NCM460 cells with siR, M-Exo, or M-Exo/siR did not induce cellular toxicity, but a slight increase in cell proliferation was observed in the M-Exo/siR-treated group (Fig. 2B); nor was cytotoxicity observed in cells transfected with up to 320 pmol/mL siRNA. In contrast, Exo-Fect™—a commercially available transfection kit for inserting small RNA into isolated exosomes—elicited significant cytotoxicity in a concentration-dependent manner (Fig. 2C).
Next, we investigated whether intracellular delivery of siRNA by M-Exo/siR reduced the abundance of TNF-α at the mRNA and protein levels in RAW264.7 cells. Lipopolysaccharide (LPS)-mediated TNF-α expression was significantly reduced following M-Exo/siR treatment by 67.2 %, whereas no inhibition was observed following treatment with M-Exo loaded with scrambled siRNA (M-Exo/Scr). Meanwhile, treatment with H-Exo/siR induced a slight reduction in TNF-α protein levels (Fig. 2D). This reduced level of inhibition was likely due to alterations in the membrane structure of H-Exos during electroporation, resulting in lower siRNA encapsulation efficiency. (Fig. 1B, S1F).
We also assessed the expression of colitis-associated pro-inflammatory cytokines at the mRNA level in LPS-stimulated Raw 264.7 cells (Fig. 2E). The expressions of Interleukin (Il)1b, Il6, and Tnfa mRNAs were significantly suppressed in the M-Exo/siR-treated groups compared to the LPS-stimulated saline groups. In particular, Tnfa mRNA expression decreased in an M-Exo/siR concentration-dependent manner, with the most significant inhibitory effect (70 %) observed at a concentration of 320 pmol/mL siRNA (Fig. S2C). These findings suggest that in vitro cellular uptake of M-Exo/siR elicits an anti-inflammatory effect without inducing cellular cytotoxicity.
2.3. Successful siRNA delivery reduces colitis pathology by downregulating TNF-α expression
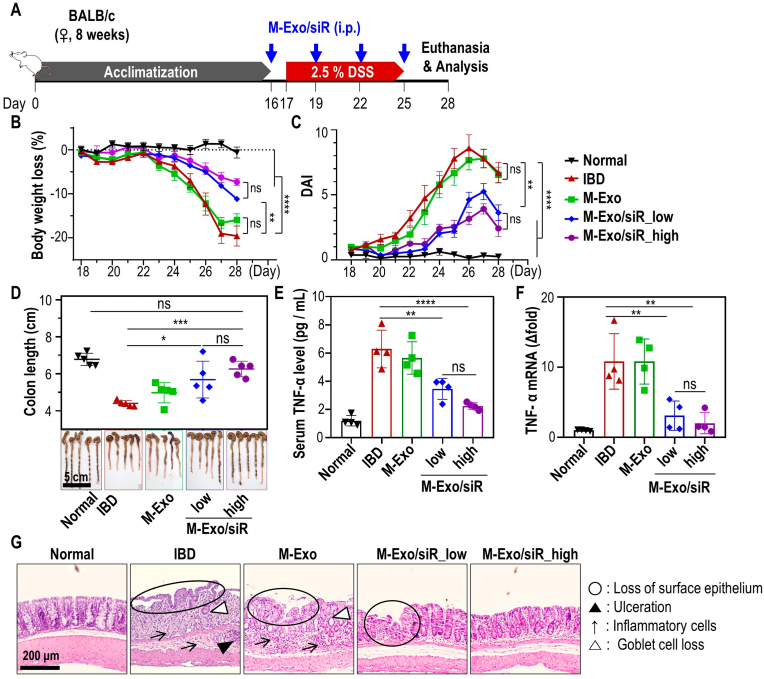
To demonstrate the in vivo therapeutic effect of M-Exo/siR on colitis, DSS-treated mice exhibiting clinical hallmarks of colitis, including weight loss, colonic shortening, and inflammation in the colon were used. In brief, 8-week-old female BALB/c mice were acclimatized for 16 days, and colitis was induced from days 17–25 via 2.5 % DSS oral administration. From day 16, mice were intraperitoneally (i.p.) injected with M-Exo/siR carrying siRNA at low (16 nmol/kg) and high (32 nmol/kg) concentrations at 3-day intervals (Fig. 3A). Mice with DSS-induced colitis treated with M-Exo/siR at low and high concentrations recovered their body weight and disease activity index (DAI) scores. In contrast, mice treated with M-Exo did not exhibit improved IBD symptoms (Fig. 3B and C). In addition, colon length in the low- and high-concentration M-Exo/siR-treated group was significantly longer than that of the IBD or M-Exo-treated groups (Fig. 3D). The expression of serum TNF-α protein and Tnfa mRNA in colonic tissue was also significantly decreased in M-Exo/siR groups compared to the control groups (Fig. 3E–F). Hematoxylin and eosin (H&E) staining of colonic tissues collected from mice euthanized on day 28 revealed infiltration of inflammatory cells, loss of surface epithelium, ulceration, and goblet cell loss within colitis ulcers in IBD- and M-Exo-treated mice. In contrast, a mild loss of the surface epithelium was observed in the M-Exo/siR-treated groups (Fig. 3G). The M-Exo/siR_high group exhibited slightly more favorable therapeutic effects than the M-Exo/siR_low group, however, there was no statistically significant difference between the two groups. (Fig. 3). Collectively, these results indicate that TNF-α siRNA encapsulated in M-Exos has the potential to functionally modulate inflammation by silencing Tnfa in a DSS-induced colitis model.
Fig. 3.
In vivo therapeutic effect of intraperitoneally (i.p.) injected M-Exo/siR against colitis. (A) Treatment schedule of dextran sulfate sodium (DSS) colitis induction and i.p. injection of M-Exo/siR. After the acclimatization period, 8-week-old female BALB/c mice were i.p. injected with M-Exo/siR four times at three-day intervals beginning one day before 2.5 % DSS administration. (B) Daily body weight loss of mice. Data are mean ± SD (n = 8); ns = not significant, **p < 0.01, ****p < 0.0001. (C) Daily disease activity index (DAI) score. Data are mean ± SD (n = 8); ns = not significant, **p < 0.01, ****p < 0.0001. (D) Representative extracted colon image and colon length on day 28. Data are mean ± SD (n = 8); ns = not significant, *p < 0.05, ***p < 0.001. (E) Serum TNF-α cytokine level on day 28. Data are mean ± SD (n = 4); ns = not significant, **p < 0.01, ****p < 0.0001. (F) Tnfa mRNA level in colitis colon tissue on day 28. Data are mean ± SD (n = 4); ns = not significant, **p < 0.01. (G) Representative histopathological images of colon tissues stained with hematoxylin and eosin (H&E) on day 28. Open black circles (loss of surface epithelium); black arrows (infiltration of inflammatory cells); black triangles (ulceration); open black triangles (loss of goblet cells).
2.4. Orally administered TNF-α siRNA in M-Exos efficiently reaches the intestines of mice
Next, we assessed whether the exosomes were highly stable against the acidic pH of the GI tract. The dynamic light scattering (DLS) results revealed that the size of M-Exos remained relatively constant, even with pH changes. However, the size of H-Exos decreased under acidic conditions; their size decreased further when the pH increased from 2 to 6 (Fig. S3A).
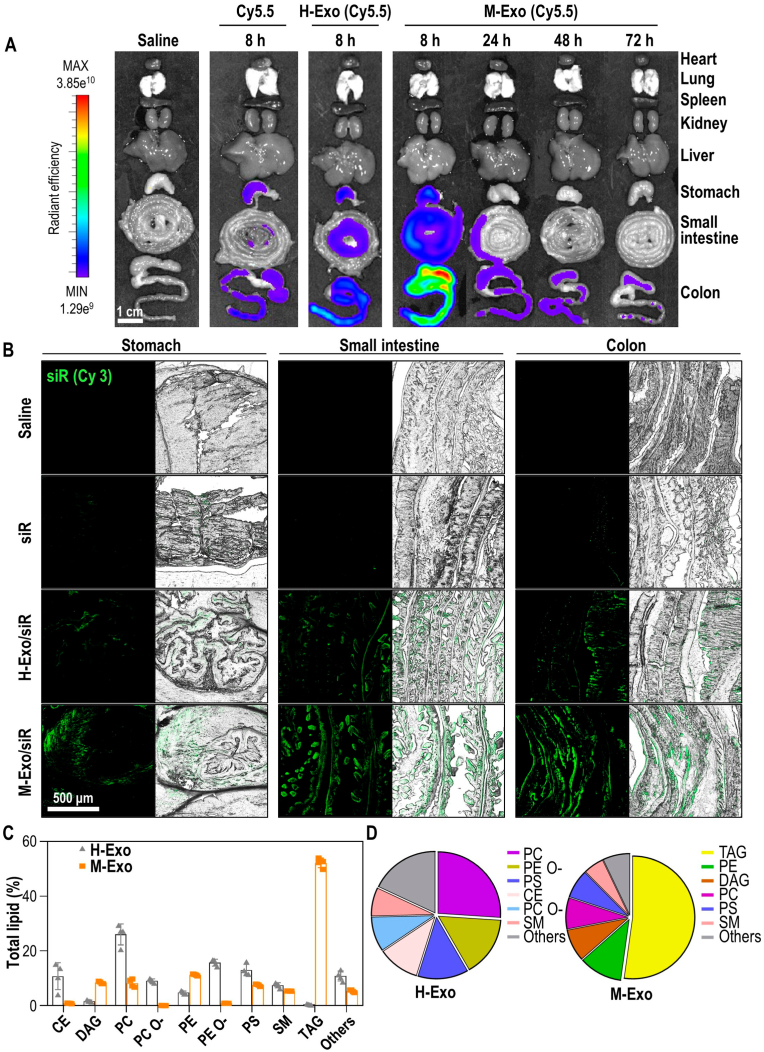
We also evaluated the bioavailability and tissue biodistribution of M-Exos labeled with Cy5.5 in mice after oral gavage. The in vivo biodistribution results for the major organs (heart, lungs, spleen, kidneys, and liver) and the GI tract clearly showed fluorescence signals of orally administered M-Exos only within the GI tract (Fig. 4A). M-Exos were absorbed through the GI tract within 8 h of oral administration, after which the signal gradually decreased until 72 h. Fluorescence signals were also confirmed in the GI tract of mice fed H-Exos; however, the intensity was weak.
Fig. 4.
Stability and lipid composition analysis of M-Exos and H-Exos. (A) Bio-distribution of Cy5.5-labeled M-Exos. After oral administration of saline, Cyanine 5.5, Cyanine 5.5-labeled H-Exos, and Cyanine 5.5-labeled M-Exos to 8-week-old female BALB/c, radiant efficiency was measured at 8 h, 24 h, 48 h, or 72 h. (B) Tissue images of Cy3-labeled siRNA absorbed into the stomach, small intestine, and colon. Green (Cyanine 3-labeled TNF-α siRNA). (C) Lipidomics analysis of M-Exos compared to H-Exos. Data are mean ± SD (n = 4). (D) Lipid composition ratio constituting H-Exos and M-Exos.
To confirm that siRNA was delivered to the intestine by M-Exos, exosomes encapsulating Cy3-labeled siRNA were orally administered to mice (Fig. 4B). Relatively no signals were observed in the tissues of mice administered siR or H-Exo/siR, whereas strong siRNA signals were observed in the stomach, small intestine, and large intestine of mice administered M-Exo/siR.
Differences in siRNA delivery were further evaluated based on the physicochemical properties of M-Exos and H-Exos from a lipidomic perspective (Fig. 4C and D). Phosphatidylcholine (PC), phosphatidylethanolamine (-ether) (PE O-), phosphatidylserine (PS), cholesterol esters (CE), and phosphatidylcholine (-ether) (PC O-) comprised the primary lipid components of H-Exos, whereas those of M-Exos were triglyceride (TAG), phosphatidylethanolamine (PE), diacylglycerol (DAG), and PC. In particular, TAG—three fatty acids esterified to a glycerol backbone—accounted for more than half of the lipids in M-Exos. A similar trend was observed when comparing M-Exos to mesenchymal stem cell-derived exosomes (MSC-Exos), which had CE, PC, PE O– PS, SM, and PC O- as their primary components (Fig. S3B). Moreover, the PC/PE ratio of the lipids within M-Exos was significantly lower than that of H-Exos or MSC-Exos (Fig. S3C). Collectively, our findings indicate that the unique lipid composition of M-Exos underlies the exceptional stability in the GI tract.
2.5. Oral gavage of M-Exo/siR reverses the inflammatory cytokine imbalance caused by colitis
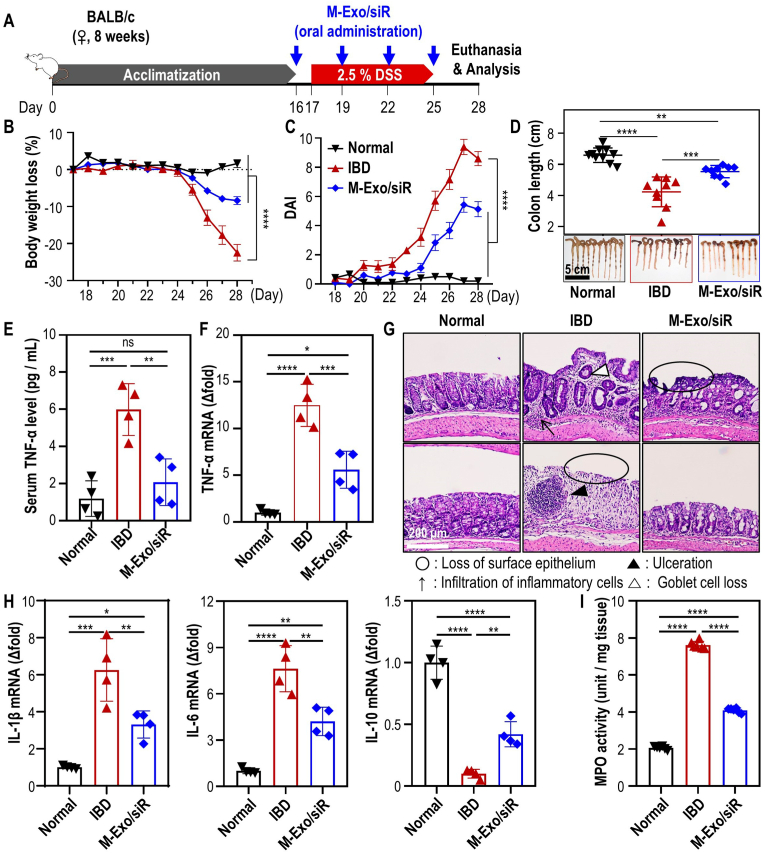
To assess the impact of orally administered M-Exo/siR on the DSS-inflamed colonic epithelium with disrupted intestinal barrier function, mice were orally administered an M-Exo and siR dose that was 1/5th lower than the high dose-M-Exo/siR-treated group in the i.p. experiment (final siRNA concentration: 6.42 nmol/kg, Fig. 5A). Orally administered M-Exo/siR efficiently ameliorated colitis-associated inflammation, as evidenced by body weight recovery, improved DAI scores, and restoration of the colon length (Fig. 5B–D). Moreover, serum TNF-α and Tnfa expression in colonic tissues were markedly reduced in the M-Exo/siR-treated mice compared to the DSS-colitis mice. Additionally, colonic tissue sections stained H&E revealed significantly reduced ulceration, goblet cell loss, and superficial epithelial loss in the M-Exo/siR-treated group compared with the IBD group (Fig. 5G), indicating that M-Exo/siR treatment protected the colonic epithelium against pathological damage.
Fig. 5.
In vivo colitis treatment efficacy of M-Exo/siR through oral gavage. (A) Treatment schedule of DSS colitis induction and oral administration of M-Exo/siR. After the acclimatization period, 8-week-old female BALB/c mice were orally administered M-Exo/siR four times at three-day intervals beginning one day before 2.5 % DSS administration. (B) Daily body weight loss of mice. Data are mean ± SD (n = 10); ****p < 0.0001. (C) Daily disease activity index (DAI) score. Data are mean ± SD (n = 10); ****p < 0.0001. (D) Representative extracted colon image and colon length on day 28. Data are mean ± SD (n = 8); ***p < 0.001 and ****p < 0.0001. (E) Serum TNF-α cytokine level on day 28. Data are mean ± SD (n = 4); ns = not significant, **p < 0.01, ***p < 0.001 (F) Tnfa mRNA level in colitis colon tissue on day 28. Data are mean ± SD (n = 4); *p < 0.05, ***p < 0.001, ****p < 0.0001. (G) Representative histopathological images of colon tissues stained with hematoxylin and eosin (H&E) on day 28. Open black circles (loss of surface epithelium); black arrows (infiltration of inflammatory cells); black triangles (ulceration); open black triangles (loss of goblet cells). (H) Analysis of inflammatory cytokine mRNA expression in colonic tissues. Data are mean ± SD (n = 4); *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (I) Colonic myeloperoxidase (MPO) activity. Data are mean ± SD (n = 6); ****p < 0.0001.
Analysis of the expression of genes associated with the TNF-α cascade showed that M-Exo/siR treatment decreased the levels of pro-inflammatory cytokines (Il1b and Il6) while increasing that of an anti-inflammatory cytokine, Il10 (Fig. 5H). Similarly, the abundance of inflammation-related proteins (TNF-α, IL-1β, and IL-6) was significantly decreased in colonic tissues following the oral administration of M-Exo/siR, while IL-10 was increased (Fig. S4). Measurement of myeloperoxidase (MPO) activity, as a representative of reactive oxygen species (ROS) levels, revealed that M-Exo/siR treatment mitigated IBD-associated MPO activity in colonic tissues (Fig. 5I).
We also evaluated the efficacy of M-Exo/siR after delayed treatment. To this end, colitis was induced in BALB/c mice via administration of 3 % DSS from day 17 to day 23, followed by oral administration of M-Exo/siR every 3 days (Fig. S5A). The group orally administered M-Exo/siR exhibited improved body weight, DAI score, and colon length. In addition, the high colon weight/colon length ratio induced by DSS was reduced by M-Exo/siR treatment (Figs. S5B–E). M-Exo/siR also increased IL-10 protein abundance while decreasing TNF-⍺, IL-1β, and IL-6 in colonic tissues (Fig. S5F). Finally, organ H&E staining confirmed that treatment with M-Exo/siR did not cause systemic inflammation or toxicity (Fig. S5G). Overall, our findings indicate that oral delivery of TNF-α siRNA by M-Exos effectively and safely treats colitis in a murine model.
3. Discussion
In this study, we investigated whether oral gavage of TNF-⍺ siRNA via M-Exos could alleviate colitis symptoms in the inflamed colon. Oral delivery systems that are advantageous for treating IBD must protect siRNAs to withstand the harsh conditions in the GI environment [5,21,22]. Indeed, a non-invasive oral administration system is attractive in terms of patient convenience and reduced side effects [23]. However, only four oral biologics for IBD therapy have entered clinical trials; none have been approved [24]. Two major challenges hinder the development of these oral biologic delivery systems. First, the complexity of the delivery system impedes the transition from clinical to procedural development. Second, most biologics currently under development are antibody-driven and require high drug doses to overcome the effects of drugs compromised by encapsulation procedures. Therefore, the oral delivery of siRNAs that exhibit therapeutic efficacy at low doses could be a breakthrough in IBD treatment.
These aspects have led many researchers to consider exosomes as carriers of RNA. In particular, M-Exos can be optimized as naturally derived carriers for the oral delivery of RNA. Milk contains approximately 6 trillion exosomes per fluid ounce [25]. When milk is consumed, the M-Exos exhibit high stability against various digestive enzymes and pH conditions in the body [26], enabling their delivery of various biomacromolecules to the liver, brain, placenta, and gut [27].
Indeed, the lipidomics results of exosomes suggest that the structural stability of M-Exos facilitates the efficient delivery of siRNA to intestinal lesions, even under extremely acidic conditions. The stability and size of the pores in the exosome membrane bilayer generated by an electric field strongly depend on the resting transmembrane voltage [[28], [29], [30], [31]]. The transmembrane voltage differs depending on the membrane lipid composition, while the electropermeabilization of membrane lipids is altered by electroporation [32]. As evidenced by the PC/PE ratio—a critical factor in membrane dynamics—the high PE ratio in M-Exos created a thickened membrane, increasing their stability compared to cell-derived exosomes [33,34]. Moreover, the relatively high abundance of TAG in M-Exos may act as a lubricant for the brushing motion of the two membrane leaflets, leading to rapid material exchange between the membranes. In addition to rapid shape dynamics, TAG-mediated reduction in bending stiffness and increased steric entropic repulsion may have enabled the successful encapsulation of siRNAs through rapid membrane structure restoration [35]. However, a more detailed analysis of membrane dynamics is required to investigate the unique features of M-Exos.
Furthermore, M-Exos that possess innate microRNAs, mRNAs, or proteins could elicit anti-inflammatory effects that aid in alleviating IBD symptoms. Indeed, exosomes derived from colostrum contain numerous factors that promote cell regeneration and anti-inflammation [20,36]. Thus, it is crucial to consider the possible effects of these endogenous components when utilizing M-Exos as a means of orally delivering nucleic acids.
4. Conclusion
In this study, we demonstrated that inhibition of TNF-α expression improves IBD symptoms. Based on these results, we propose a new and robust platform based on M-Exos that enables the oral administration of gene therapy for colitis. Due to the unique membrane lipid composition of M-Exo, orally administered M-Exo/siR stably pass through the GI tract to the colitis lesions with excellent stability, and induce degradation of Tnfa mRNA within the inflamed lesions of mice. Ultimately, this downregulation in TNF-α expression reduces pro-inflammatory cytokine and ROS levels in the inflamed colon, alleviating intestinal colitis. We believe that our research will have a significant impact on the development of new oral gene therapeutics. Moreover, this M-Exo-based oral drug delivery system will prove useful in the clinical treatment of various inflammatory diseases, including IBD.
5. Materials and methods
5.1. Isolation of exosomes from bovine milk and HEK293T cells
M-Exos were extracted from commercial milk (pasteurized low-fat milk from a farm in Maeil, Jongno-gu, Republic of Korea). Milk was continuously centrifuged at 5000 × g for 30 min and 12 000 × g for 1 h at 4 °C (Avanti J-E, Beckman Coulter, Brea, USA). The supernatant was filtered with a 40-μm strainer (93040, SPL, Pocheon-si, Republic of Korea) and centrifuged using an ultracentrifuge (Optima XE-100, Beckman Coulter, Brea, USA) at 35 000 × g for 1 h and 70 000 × g for 3 h at 4 °C. Among the three separated layers, the middle layer was sequentially filtered with 0.8, 0.45, and 0.2-μm syringe filters (16592-K, 16555-K, 16534-K, Sartorius AG, Göttingen, Germany). To collect the M-Exo pellets, the filtered solution was centrifuged at 100 000 × g for 1 h. The exosome pellet was re-suspended with Dulbecco's phosphate-buffered saline (DPBS; CA008-050, GenDEPOT, Katy, USA) and mixed overnight at 4 °C.
HEK293T cells were cultured in serum-free media for 2 days. Thereafter, the cell culture supernatant was collected and serially centrifuged at 300 × g for 10 min, 2000 × g for 10 min, and 10 000 × g for 30 min at 4 °C to remove cells and cell debris. The supernatant was then harvested and centrifuged at 150 000 × g for 90 min at 4 °C to collect the HEK298T cell exosome (H-Exo) pellets. The H-Exo pellet was washed and dissolved with DPBS (CA008-050, GenDEPOT, Katy, USA).
5.2. Electroporation of M-Exo and siRNA loading efficiency
For electroporation, RNA-free 10 × PBS (AM9625, Invitrogen, Waltham, USA) was diluted 1/50 in diethylpyrocarbonate (DEPC)-treated water (WR2004-050-00, Biosesang, Seongnam-si, Republic of Korea) and used as the electroporation buffer. M-Exos and H-Exos were added to the electroporation buffer to prepare a solution with 1.6 × 1011 particles/mL. All electroporation experiments were performed using Gene Pulser Xcell Electroporation Systems (1652660, BIO-RAD, Hercules, USA) and Gene Pulser/MicroPulser Electroporation Cuvettes with a 0.4 cm gap (1652088, BIO-RAD, Hercules, USA) according to the electroporation conditions. After electroporation, the required siRNA concentrations were added to the M-Exos and H-Exos to prepare M-Exo/siR and H-Exo/siR, respectively. Each Exo/siR was then incubated at 4 °C for 3 h with mild shaking to facilitate restoration. To remove unloaded free siRNA, centrifugation was performed at 12 000 × g for 10 min using a 30 kDa-Amicon Ultra-0.5 centrifugal filter unit (UFC503096, Merck Millipore, Burlington, USA). The unloaded free siRNA was separated from the centrifugal filter and quantified with the RiboGreen™ RNA Quantitation Kit (R11490, Invitrogen, Waltham, USA) according to the manufacturer's protocol; the loaded siRNA was inversely estimated. We used AccuTarget α Negative Control siRNA (SN-1003, Bioneer, Daejeon, Republic of Korea) as a scrambled siRNA. The sequence for TNF-α siRNA (Bioneer, Daejeon, Republic of Korea) is shown in Table 1 and was prepared based on previous siRNA studies [37].
Table 1.
siRNA sequences.
| Tnfa | Sense | 5′-GGGUGUUCAUCCAUUCUCU-3′ |
| Antisense | 5′-AGAGAAUGGAUGAACACCC-3′ |
5.3. M-Exo characterization
5.3.1. Dynamic light scattering (DLS) analysis
M − and H-Exos were diluted to 4 × 109 particles/mL in DPBS (CA008-050, GenDEPOT, Katy, USA), and the samples were placed in disposable cuvettes (art. 01960-00, Kartell LABWARE, Noviglio, Italy). The size distribution of each sample was measured by DLS (Zetasizer Nano ZS; Malvern Instruments, Malvern, UK) at a fixed angle of 173°. Each sample was subjected to three independent measurements and averaged using the Zetasizer software v7.13 (Malvern Instruments, Malvern, UK).
5.3.2. Nanoparticle tracking analysis (NTA)
To numerically analyze M-Exo and H-Exo nanoparticles, each sample was mixed in distilled water. Diluted samples were video-recorded using a NanoSight (LM10, Malvern Instruments, Malvern, UK). The recorded video was analyzed using NanoSight NTA v2.3 software (Malvern Instruments, Malvern, UK)
5.3.3. Cryo-transmission electron microscopy (Cryo-TEM)
The morphologies of M − and H-Exos were analyzed via TEM. Each sample was fixed overnight in 0.5 % glutaraldehyde. The fixed samples were centrifuged at 150 000 × g for 30 min. The M − and H-Exo pellets were then resuspended in absolute ethanol, placed on a lacey grid, and maintained at −180 °C using liquid nitrogen. Each sample was vitrified using the Vitrobot System (MARKII FP 5350/60; FEI, Hillsboro, OR, USA). Vitrified M − and H-Exos were visualized using a cryo-TEM (Tecnai G2 F20, FEI Company, Oregon, USA).
5.3.4. M-exo labeling and super-resolution microscopy (SRM)
TNF-α siRNA was labeled with 5-fluorescein phosphoramidite (6-FAM) (Bioneer, Daejeon, Republic of Korea) and loaded into M-Exos via electroporation. To label the M-Exos of M-Exo/siR, M-Exo/siR was mixed with Cyanine5.5-N-hydroxysuccinimide (Cy5.5-NHS) ester (#67020, Lumiprobe, Maryland, USA) overnight at 4 °C according to the manufacturer's protocol. Free dye and unloaded siRNA were removed by centrifugation at 12 000 × g for 10 min in a 30 kDa-Amicon Ultra-0.5 Centrifugal Filter Unit (UFC503096, Merck Millipore, Burlington, USA). Stained M-Exo/siR was mixed with Fluoromount-G™ mounting medium and added dropwise to a glass slide (S9911, MATSUNAMI glass, Bellingham, USA). The samples were then covered with a cover slip (0107032, ZEISS, Oberkochen, Germany) and stored in darkness for more than 2 days for fixation. The prepared samples were analyzed using a super-resolution microscope (Elyra7, ZEISS, Oberkochen, Germany), and the image resolution was improved using Lattice SIM2 software.
5.3.5. Western blot analysis
M-Exos and H-Exos were lysed with radioimmunoprecipitation (RIPA) buffer (89900, Thermo Fisher Scientific, Waltham, USA) containing 1 % protease inhibitor at 4 °C for 30 min. The supernatant was separated from the lysate by centrifugation at 14 000 × g for 15 min and quantified using a bicinchoninic acid protein assay kit (23227, Thermo Fisher Scientific, Waltham, USA). Proteins in the separated supernatant were denatured using a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (SF2002-110-00; Biosesang, Seongnam-si, Republic of Korea). Samples were electrophoresed on 10 % SDS-polyacrylamide gel. After the separated proteins were transferred onto a nitrocellulose membrane, the membrane was blocked with 5 % skim milk for 30 min at 25 °C. Blocked membranes were incubated with primary antibodies overnight at 4 °C. The membrane was rinsed several times for 15 min each with TBS-T buffer (Tris-buffered saline with 0.1 % Tween 20). The washed membrane was incubated in a 5 % skim milk solution with an HRP-tagged second antibody for 1 h at 25 °C. The labeled proteins were visualized using a luminescent image analyzer (iBright CL750, Invitrogen, Waltham, USA). The following primary antibodies used were Alix (1:500, NB100-65678, Novus Biologicals, Colorado, USA), Tsg-101 (1:1000, ab83, Abcam, Cambridge, UK), CD9 (1:1000, NB500-494, Novus Biologicals, Colorado, USA), GM130 (1:500, PA1077, Thermo Fisher Scientific, Waltham, USA), Calnexin (1:1000, ab227310, Abcam, Cambridge, UK), TNF-α (1:1000, ab1793, Abcam, Cambridge, UK), and GAPDH (1:1000, ab8245, Abcam, Cambridge, UK). Meanwhile, goat anti-mouse IgG (HRP) (1:1000, 1706516, BIO-RAD, Hercules, USA), and goat anti-rabbit IgG (HRP) (1:1000, 1706515, BIO-RAD, Hercules, USA) secondary antibodies were used.
5.3.6. Lipidomics analysis
After independently extracting more than 6 × 109 particles of M-Exo and H-Exo four times each, the collected samples were stored at −80 °C, and lipidomics analysis (Lipotype GmbH, Dresden, Germany) was performed. Samples were lipid extracted using chloroform/methanol and analyzed by direct infusion on a QExactive mass spectrometer (Thermo Scientific) mounted with a TriVersa NanoMate ion source (Advion Biosciences). Samples were analyzed with a resolution of Rm/z = 200 = 17 500 for MSMS experiments and Rm/z = 200 = 280 000 for MS. Additional enzymatic fluorometric assay was conducted to quantify PC and PE lipids, as previously described with minor modifications [38]. The lipid class abbreviations were as follows: CE, cholesterol esters; Cer, ceramide; CL, cardiolipin; DAG, diacylglycerol; HexCer, hexosylceramide; LPA, lyso-phosphatidate; LPC O-, lyso-phosphatidylcholine (-ether); LPE O-, lyso-phosphatidylethanolamine (-ether); LPI, lyso-phosphatidylinositol; LPS, lyso-phosphatidylserine; PA, phosphatidate; PC O-, phosphatidylcholine (-ether); PE O-, phosphatidylethanolamine (-ether); PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol.
5.3.7. Serum stability of siRNA
Blood was extracted from the heart of mice and separated by centrifugation at 2000 × g for 10 min at 4 °C to obtain serum. The separated serum was mixed with M-Exo/siR solution (or siRNA solution) at a 9:1 ratio. These samples were incubated at 37 °C for 0, 2, 4, 8, 12, and 24 h. Afterward, 1 % Triton X-100 (TR-1020-500-00, Biosesang, Seongnam-si, Republic of Korea) was added, and siRNA was concentrated by centrifugation at 12 000 × g for 10 min using a 3 kDa-Amicon Ultra-0.5 centrifugal filter unit (UFC500396, Merck Millipore, Burlington, USA). The concentrated siRNA was loaded onto a 1 % agarose gel, and electrophoresis was performed to measure the intensity of RNA bands. The gel was analyzed using an agarose gel analyzer (iBright CL750, Invitrogen, Waltham, USA).
5.4. In vitro investigation
5.4.1. Cell culture
HEK293T cells (CRL-3216, American Type Culture Collection, Manassas, USA), RAW264.7 cells (40071, Korean Cell Line Bank, Republic of Korea), and NCM460 cells (NCM460, INCELL, San Antonio, USA) were cultured in Dulbecco's modified eagle medium (DMEM) with high glucose (CM002-050, GenDEPOT, Katy, USA) supplemented with 10 % fetal bovine serum (FBS) (16000044, Gibco, Grand Island, USA) and 1 % antibiotic-antimycotic (CA002-010, GenDEPOT, Katy, USA). All cells were cultured at 37 °C in a 5 % CO2 atmosphere.
5.4.2. Cell viability assay
NCM460 cells were seeded in 96-well plates (1.0 × 104 cells/well) for 24 h. After replacing the old media with fresh media, siR (TNF-α siRNA; 160.5 pmol/mL), M-Exo (M-Exo; 2 × 1010 particles/mL), M-Exo/siR (TNF-α siRNA; 160.5 pmol/mL, M-Exo; 2 × 1010 particles/mL), or Exo-Fect™ (TNF-α siRNA; 160.5 pmol/mL, M-Exo; 2 × 1010 particles/mL) were added to each well, and the plates were incubated for 24 h. Cells were then washed with fresh culture media, replaced with media containing 10 % CCK-8 solution (CK04, Dojindo, Kumamoto, Japan), and incubated for 45 min at 37 °C; the absorbance was detected at 450 nm using a microplate reader (SpectraMAX 340, Molecular Devices, San Jose, USA).
5.4.3. Cellular uptake analysis
NCM460 cells and RAW264.7 cells were seeded in confocal dishes (3 × 105 cells/dish and 5 × 105 cells/dish, respectively) for 24 h. After replacing the old media with fresh media, siR (TNF-α siRNA; 160.5 pmol/mL), M-Exo (M-Exo; 2 × 1010 particles/mL), M-Exo/siR (w/electroporation) (TNF-α siRNA; 160.5 pmol/mL, M-Exo; 2 × 1010 particles/mL), or M-Exo + siR (w/o electroporation) (TNF-α siRNA; 160.5 pmol/mL, M-Exo; 2 × 1010 particles/mL) were added to each dish and incubated for 24 h; Cy5.5-labeled M-Exos and 6-FAM-labeled siRNA were used. The cells were fixed with formaldehyde and treated with Hoechst 33342 solution (62249, Thermo Fisher Scientific) for 15 min to stain nucleic acids. Correlation images were obtained using a confocal microscope (Leica TCS SP6; Leica, Wetzlar, Germany). The fluorescence correlation intensities and correlation coefficients were calculated using the Leica Application Suite X software.
5.5. Quantitative real-time polymerase chain reaction (qRT-PCR)
Raw 264.7 cells were seeded in 35-mm culture dishes (3 × 105 cells/dish); 24 h after seeding, the cells were treated with 100 ng/mL LPS for 8 h. The cells were washed twice with DPBS, and the culture medium was replaced with serum-free media. Each dish was treated with siR (TNF-α siRNA; 160.5 pmol/mL), M-Exo (M-Exo; 2 × 1010 particles/mL), M-Exo/Scr (scramble siRNA; 160.5 pmol/mL, M-Exo; 2 × 1010 particles/mL), H-Exo/siR (TNF-α siRNA; 160.5 pmol/mL, H-Exo; 2 × 1010 particles/mL), or M-Exo/siR (TNF-α siRNA; 160.5 pmol/mL, M-Exo; 2 × 1010 particles/mL) for 24 h. Total RNA was extracted from tissues and cells using the QIAzol lysis reagent (79306, QIAGEN, Hilden, Germany). Complementary DNA (cDNA) was synthesized from the extracted RNA using AccuPower® RT PreMix (K-2041-B, Bioneer, Daejeon, Republic of Korea) according to the manufacturer's protocol. Quantitative real-time PCR was performed using TOPreal™ qPCR 2X PreMIX (SYBR Green with high ROX) (RT501 M; Enzynomics, Daejeon, Republic of Korea), and the amplified target genes were monitored with a real-time PCR device (QuantStudio 1 Real-Time PCR, Applied Biosystems, Waltham, USA). Gapdh was used as an internal control gene. All sequences used in this experiment are listed in Table 2.
Table 2.
qRT-PCR primer sequences.
| Gene | Sequence (5′–3′) |
|---|---|
|
Tnfa |
F: AGTGGAGGAGCAGCTGGAGT |
| R: TCCCAGCATCTTGTGTTTCTG | |
|
Il1b |
F: ATGGCAACTGTTCCTGAACTCAACT |
| R: CAGGACAGGTATAGATTCTTTCCTTT | |
|
Il6 |
F: GAGGATACCACTCCCAACAGACC |
| R: TTCACAGAGGATACCACTCC | |
|
Il10 |
F: CTATGCAGTTGATGAAGATGTCAAA |
| R: CCTGGTAGAAGTGATGCCCCAGGCA | |
| Gapdh | F: CATGCCGCCTGGAAACCTGCCA |
| R: TGGGCTGGGTGGTCCAGGGGTTTC |
(F: Forward; R: Reverse).
5.6. In vivo murine investigation
5.6.1. In vivo M-Exo/siR efficacy evaluation in a colitis mouse model
All mouse experiments were approved by the Korea Institute of Science and Technology (KIST; Approved number: 2022-086-2) and performed in accordance with the relevant guidelines of the Institutional Animal Care and Use Committee (IACUC). BALB/c mice (female, 8-week-old) were purchased from Orient Bio (Seongnam-si, Republic of Korea) to generate the ulcerative colitis model.
To assess the efficacy of i.p. injection, mice were randomly divided into five groups: (1) Normal (Saline); (2) IBD (Saline); (3) M-Exo (M-Exo; 4 × 1012 particles/kg); (4) M-Exo/siR low dose (TNF-α siRNA; 16.1 nmol/kg, M-Exo; 2 × 1012 particles/kg); (5) M-Exo/siR high dose (TNF-α siRNA; 32.1 nmol/kg, M-Exo; 4 × 1012 particles/kg). To assess the efficacy of oral administration, mice were randomly divided into three groups: (1) Normal (Saline), (2) IBD (Saline), and (3) M-Exo/siR (M-Exo: 8 × 1011 particles/kg, TNF-α siRNA: 6.42 nmol/kg). Each group was acclimated for 16 days. From day 16 to day 25, each group was i.p. injected or orally administered each substance according to its dose at 3-day intervals. From days 17–25, all groups except the normal group were administered a 2.5 % DSS solution via autoclaved tap water. The weight and feces volume of each mouse were monitored daily. On day 27, the mice were euthanized via cervical dislocation, and colon length was assessed.
In addition, to assess the delayed therapy of M-Exo/siR in IBD mice, the mice were randomly divided into three groups: (1) Normal (Saline), (2) IBD (Saline), and (3) M-Exo/siR (M-Exo: 8 × 1011 particles/kg, TNF-α siRNA: 6.42 nmol/kg). After the acclimatization period, a 3.0 % DSS solution was administered via autoclave tap water to all groups except for the normal group from day 17 to day 23. From day 23, mice were orally administered the drugs three times every 3 days. The body weight and the volume of feces of each mouse were measured daily. On day 30, the mice were euthanized via cervical dislocation, and colon length and colon weight/length were assessed. The colon was opened longitudinally and washed with PBS. Excess fluid was removed from the colon and weighed per unit length.
5.6.2. DAI scoring
The DAI was calculated by three independent observers daily from days 18–28 based on the following characteristics: weight loss, stool bleeding, and stool consistency (Table 3). The total score ranged from 0 to 12. Higher scores indicated higher disease severity.
Table 3.
Disease Activity index.
| Score | Weight loss (%) | Stool bleeding | Stool consistency |
|---|---|---|---|
| 0 | None | Negative hemoccult | Normal |
| 1 | 1–5% | Negative hemoccult | Soft but still formed |
| 2 | 6–10 % | Positive hemoccult | Soft |
| 3 | 11–18 % | Blood trace in stool visible | Very soft; wet |
| 4 | >18 % | Gross rectal bleeding | Watery diarrhea |
5.6.3. Ex vivo bio-distribution analysis
Eight-week-old female BALB/c mice were orally administered Cy5.5-labeled M-Exos (4 × 1012 particles/kg) using an oral zone. Mice were euthanized at 8, 24, 48, and 72 h after oral administration, and the major visceral organs (lungs, heart, kidneys, spleen, and liver) and gastrointestinal tract were harvested. The fluorescence intensities of the collected organs were analyzed using an in vivo imaging system (IVIS) (Lumina Series III; PerkinElmer, Waltham, USA).
5.6.4. Analysis of siRNA uptake in the GI tract
Saline, TNF-α siRNA (Cy3 labeled-TNF-α siRNA; 32.1 nmol/kg), H-Exo/siR (Cy3 labeled-TNF-α siRNA; 32.1 nmol/kg, H-Exo; 4 × 1012 particles/kg), or M-Exo/siR (Cy3 labeled-TNF-α siRNA; 32.1 nmol/kg, M-Exo; 4 × 1012 particles/kg) were orally administered to mice; the stomach, small intestine, and colon were harvested 8 h later. Each organ was dehydrated and embedded in paraffin. Subsequently, all organs were sectioned at a thickness of 5 μm and observed using a confocal microscope (Leica TCS SP6, Leica, Wetzlar, Germany).
5.6.5. Enzyme-linked immunosorbent assay
To measure the abundance of TNF-α in the blood, blood was collected from the heart of a mouse. The collected blood was centrifuged at 2000×g for 20 min at 4 °C to separate the plasma. Plasma TNF-α was quantified using the Mouse TNF-α high sensitivity ELISA kit (BMS607HS, Invitrogen, Waltham, USA) according to the manufacturer's protocol. Colon tissues were homogenized with RIPA buffer (89900, Thermo Fisher Scientific, Waltham, USA) containing 1 % protease inhibitor at 4 °C for 5 min to measure the abundance of inflammation-related proteins. The lysate was centrifuged at 14 000×g for 15 min at 4 °C to separate the supernatant. ELISA analysis for each protein was conducted following the manufacturer's protocol, and the information regarding the ELISA kit is as follows. ELISA analysis for each protein was conducted following the manufacturer's protocol, and the information regarding the ELISA kit is as follows; TNF-α (430907, BioLegned, San Diego, USA), IL-1β (MLB00C, R&D Systems, Minneapolis, USA), IL-6 (M6000B-1, R&D Systems, Minneapolis, USA), and IL-10 (M1000B-1, R&D Systems, Minneapolis, USA).
5.6.6. MPO activity
MPO activity was measured using an MPO activity assay kit (ab105136; Abcam, Cambridge, UK) according to the manufacturer's protocol. Briefly, the colons (10 mg) of mice euthanized on day 28 were homogenized with MPO assay buffer, and the sample was centrifuged at 13 000 × g for 10 min. After collecting the supernatant, the MPO activity assay was performed. MPO activity of the samples was calculated by measuring the absorbance at 412 nm using a microplate reader (SpectraMAX 340, Molecular Devices, San Jose, USA).
5.7. Statistical analysis
A two-tailed Student's t-test was used to confirm the statistical significance of the differences between the two groups. One-way analysis of variance (ANOVA) with the Tukey–Kramer post hoc test was used for multi-group comparisons. All results were presented as mean ± standard deviation (SD). All data were analyzed using the Prism software (version 9.0). Statistical significance was set as follows: not significant (ns) > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
CRediT authorship contribution statement
Geonhee Han: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation. Hyosuk Kim: Writing – original draft, Validation, Methodology, Data curation. Hochung Jang: Methodology, Data curation. Eun Sun Kim: Writing – review & editing, Methodology, Conceptualization. Sun Hwa Kim: Writing – review & editing, Supervision, Resources, Project administration, Funding acquisition, Conceptualization. Yoosoo Yang: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
This study was supported by the Bio & Medical Technology Development Program (NRF-2022M3E5F2018170) and the Intramural Research Program of the Korea Institute of Science and Technology (KIST).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2023.12.010.
Contributor Information
Sun Hwa Kim, Email: sunkim@kist.re.kr.
Yoosoo Yang, Email: ysyang@kist.re.kr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Faye A.S., Hung K.W., Cheng K., Blackett J.W., McKenney A.S., Pont A.R., Li J., Lawlor G., Lebwohl B., Freedberg D.E. Minor hematochezia decreases use of venous thromboembolism prophylaxis in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2020;26(9):1394–1400. doi: 10.1093/ibd/izz269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogler G., Singh A., Kavanaugh A., Rubin D.T. Extraintestinal manifestations of inflammatory bowel disease: current concepts, treatment, and implications for disease management. Gastroenterology. 2021;161(4):1118–1132. doi: 10.1053/j.gastro.2021.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan Q. A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol. Res. 2019;2019 doi: 10.1155/2019/7247238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Driscoll C.M., Bernkop-Schnurch A., Friedl J.D., Preat V., Jannin V. Oral delivery of non-viral nucleic acid-based therapeutics - do we have the guts for this? Eur. J. Pharmaceut. Sci. 2019;133:190–204. doi: 10.1016/j.ejps.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Duran-Lobato M., Niu Z., Alonso M.J. Oral delivery of biologics for precision medicine. Adv. Mater. 2020;32(13) doi: 10.1002/adma.201901935. [DOI] [PubMed] [Google Scholar]
- 6.Madani F., Hsein H., Busignies V., Tchoreloff P. An overview on dosage forms and formulation strategies for vaccines and antibodies oral delivery. Pharmaceut. Dev. Technol. 2020;25(2):133–148. doi: 10.1080/10837450.2019.1689402. [DOI] [PubMed] [Google Scholar]
- 7.Duan L., Xu L., Xu X., Qin Z., Zhou X., Xiao Y., Liang Y., Xia J. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale. 2021;13(3):1387–1397. doi: 10.1039/d0nr07622h. [DOI] [PubMed] [Google Scholar]
- 8.Fu P., Zhang J., Li H., Mak M., Xu W., Tao Z. Extracellular vesicles as delivery systems at nano-/micro-scale. Adv. Drug Deliv. Rev. 2021;179 doi: 10.1016/j.addr.2021.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong J., Xia B., Shan S., Zheng A., Zhang S., Chen J., Liang X.J. High-quality milk exosomes as oral drug delivery system. Biomaterials. 2021;277 doi: 10.1016/j.biomaterials.2021.121126. [DOI] [PubMed] [Google Scholar]
- 10.Wolf T., Baier S.R., Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma caco-2 cells and rat small intestinal IEC-6 cells. J. Nutr. 2015;145(10):2201–2206. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Chen D., Ho E.A. Challenges in the development and establishment of exosome-based drug delivery systems. J. Contr. Release. 2021;329:894–906. doi: 10.1016/j.jconrel.2020.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Neurath M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014;14(5):329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen O.H., Ainsworth M.A. Tumor necrosis factor inhibitors for inflammatory bowel disease. N. Engl. J. Med. 2013;369(8):754–762. doi: 10.1056/NEJMct1209614. [DOI] [PubMed] [Google Scholar]
- 14.Beutler B., Greenwald D., Hulmes J.D., Chang M., Pan Y.C., Mathison J., Ulevitch R., Cerami A. Identity of tumour necrosis factor and the macrophage-secreted factor cachectin. Nature. 1985;316(6028):552–554. doi: 10.1038/316552a0. [DOI] [PubMed] [Google Scholar]
- 15.A.S.o.N.C. From the American Association of Neurological Surgeons. C.I.R.A.C.o.N.S.E.S.o.M.I.N.T.E.S.o.N.E.S.O.S.f.C.A. Interventional Radiology Society of Europe. S.o.I.R.S.o.N.S. Interventions. World Stroke O., Sacks D., Baxter B., Campbell B.C.V., Carpenter J.S., Cognard C., Dippel D., Eesa M., Fischer U., Hausegger K., Hirsch J.A., Shazam Hussain M., Jansen O., Jayaraman M.V., Khalessi A.A., Kluck B.W., Lavine S., Meyers P.M., Ramee S., Rufenacht D.A., Schirmer C.M., Vorwerk D. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int. J. Stroke. 2018;13(6):612–632. doi: 10.1177/1747493018778713. [DOI] [PubMed] [Google Scholar]
- 16.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20(8):629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Y., Siegwart D.J., Anderson D.G. Strategies, design, and chemistry in siRNA delivery systems. Adv. Drug Deliv. Rev. 2019;144:133–147. doi: 10.1016/j.addr.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W., Qu M., Li L., Liu T., Lin M., Yu X. SiRNA in MSC-derived exosomes silences CTGF gene for locomotor recovery in spinal cord injury rats. Stem Cell Res. Ther. 2021;12(1):334. doi: 10.1186/s13287-021-02401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren M.R., Zhang C., Vedadghavami A., Bokvist K., Dhal P.K., Bajpayee A.G. Milk exosomes with enhanced mucus penetrability for oral delivery of siRNA. Biomater. Sci. 2021;9(12):4260–4277. doi: 10.1039/d0bm01497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han G., Cho H., Kim H., Jang Y., Jang H., Kim D.E., Kim E.S., Kim E.H., Hwang K.Y., Kim K., Yang Y., Kim S.H. Bovine colostrum derived-exosomes prevent dextran sulfate sodium-induced intestinal colitis via suppression of inflammation and oxidative stress. Biomater. Sci. 2022;10(8):2076–2087. doi: 10.1039/d1bm01797g. [DOI] [PubMed] [Google Scholar]
- 21.Cao Y., Rewatkar P., Wang R., Hasnain S.Z., Popat A., Kumeria T. Nanocarriers for oral delivery of biologics: small carriers for big payloads. Trends Pharmacol. Sci. 2021;42(11):957–972. doi: 10.1016/j.tips.2021.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Vela Ramirez J.E., Sharpe L.A., Peppas N.A. Current state and challenges in developing oral vaccines. Adv. Drug Deliv. Rev. 2017;114:116–131. doi: 10.1016/j.addr.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C., Merlin D. Nanoparticle-mediated drug delivery systems for the treatment of IBD: current perspectives. Int. J. Nanomed. 2019;14:8875–8889. doi: 10.2147/IJN.S210315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W., Michalowski C.B., Beloqui A. Oral delivery of biologics in inflammatory bowel disease treatment. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.675194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wijenayake S., Eisha S., Tawhidi Z., Pitino M.A., Steele M.A., Fleming A.S., McGowan P.O. Comparison of methods for pre-processing, exosome isolation, and RNA extraction in unpasteurized bovine and human milk. PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0257633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vashisht M., Rani P., Onteru S.K., Singh D. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl. Biochem. Biotechnol. 2017;183(3):993–1007. doi: 10.1007/s12010-017-2478-4. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X., You L., Zhang Z., Cui X., Zhong H., Sun X., Ji C., Chi X. Biological properties of milk-derived extracellular vesicles and their physiological functions in infant. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.693534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho M.C., Casciola M., Levine Z.A., Vernier P.T. Molecular dynamics simulations of ion conductance in field-stabilized nanoscale lipid electropores. J. Phys. Chem. B. 2013;117(39):11633–11640. doi: 10.1021/jp401722g. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez M.L., Risk M., Reigada R., Vernier P.T. Size-controlled nanopores in lipid membranes with stabilizing electric fields. Biochem. Biophys. Res. Commun. 2012;423(2):325–330. doi: 10.1016/j.bbrc.2012.05.122. [DOI] [PubMed] [Google Scholar]
- 30.Sengel J.T., Wallace M.I. Imaging the dynamics of individual electropores. Proc. Natl. Acad. Sci. U.S.A. 2016;113(19):5281–5286. doi: 10.1073/pnas.1517437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casciola M., Kasimova M.A., Rems L., Zullino S., Apollonio F., Tarek M. Properties of lipid electropores I: Molecular dynamics simulations of stabilized pores by constant charge imbalance. Bioelectrochemistry. 2016;109:108–116. doi: 10.1016/j.bioelechem.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Kotnik T., Rems L., Tarek M., Miklavcic D. Membrane electroporation and electropermeabilization: mechanisms and models. Annu. Rev. Biophys. 2019;48:63–91. doi: 10.1146/annurev-biophys-052118-115451. [DOI] [PubMed] [Google Scholar]
- 33.Caillon L., Nieto V., Gehan P., Omrane M., Rodriguez N., Monticelli L., Thiam A.R. Triacylglycerols sequester monotopic membrane proteins to lipid droplets. Nat. Commun. 2020;11(1):3944. doi: 10.1038/s41467-020-17585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawaliby R., Trubbia C., Delporte C., Noyon C., Ruysschaert J.M., Van Antwerpen P., Govaerts C. Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 2016;291(7):3658–3667. doi: 10.1074/jbc.M115.706523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pakkanen K.I., Duelund L., Qvortrup K., Pedersen J.S., Ipsen J.H. Mechanics and dynamics of triglyceride-phospholipid model membranes: implications for cellular properties and function. Biochim. Biophys. Acta. 2011;1808(8):1947–1956. doi: 10.1016/j.bbamem.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Han G., Kim H., Kim D.E., Ahn Y., Kim J., Jang Y.J., Kim K., Yang Y., Kim S.H. The potential of bovine colostrum-derived exosomes to repair aged and damaged skin cells. Pharmaceutics. 2022;14(2) doi: 10.3390/pharmaceutics14020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Tang Y., Cai D. IKKbeta/NF-kappaB disrupts adult hypothalamic neural stem cells to mediate a neurodegenerative mechanism of dietary obesity and pre-diabetes. Nat. Cell Biol. 2012;14(10):999–1012. doi: 10.1038/ncb2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita S.Y., Tsuji T., Terada T. Protocols for enzymatic fluorometric assays to quantify phospholipid classes. Int. J. Mol. Sci. 2020;21(3) doi: 10.3390/ijms21031032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.