Abstract
Background
Observational studies have previously demonstrated a significant relationship among both metabolic syndrome (Mets) and colorectal cancer (CRC). Whether there is a causal link remains controversial.
Objective
To clarify whether Mets and their components have a causal effect on colorectal cancer, we have carried out a bidirectional Mendelian randomization analysis (MR).
Methods
This study started from genome-wide association data for Mets and its 5 components (hypertension, waist circumference, fasting blood glucose, serum triglycerides, and serum high-density lipoprotein cholesterol) and colorectal cancer. Mendelian randomization (MR) techniques were used in the study to examine their associations.
Results
After Benjamini-Hochberg multiple corrections, genetically predicted significant causal link exists between WC (waist circumference) and CRC. The OR was 1.35 (95 % CI: 1.08–1.69; p = 0.0096). Other Mets components (HBP, FBG, TG, HDL), on the other hand, found no evidence of a genetic link between CRC and Mets. In addition, MR results showed that CRC was not causally related to either Mets or the components. We get the same result in the validated dataset.
Conclusion
According to the bidirectional MR investigation shows a significant causal relationship among obesity and CRC in the Mets component but no causal relationship in the opposite direction.
Keywords: Mendelian randomization (MR), Metabolic syndrome (mets), Colorectal cancer (CRC), Instrumental variant (IV), Single-nucleotide polymorphism (SNP), Waist circumference (WC)
1. Introduction
Colorectal cancer (CRC) already has nearly 900,000 mortalities per year, and it has become the 4th most deadly cancer worldwide [1]. To enhance colorectal cancer prevention and early diagnosis, advanced approaches are considered necessary. While many risk factors were reported for the development of CRC in recent years, there is still a need for further research to better understand the mechanisms underlying CRC development and extend prevention and early diagnosis [[2], [3], [4]]. Metabolic syndrome (Mets) has emerged as a significant risk factor for CRC recently, however, the relationship between Mets and CRC has not been understood.
Evidence from epidemiology indicates that metabolic syndrome tends to increase the risk of colorectal cancer [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]]. This finding has been based on investigations of metabolic syndrome assessment determinants (waist circumference), metabolic syndrome clinical consequences (type 2 diabetes and hypertension), metabolic syndrome serum components (hypertriglyceridemia and low HDL cholesterol), and hyperinsulinemia markers [18]. Moreover, a number of meta-analyses and articles suggest a bidirectional relationship between Mets and CRC [5,6,19]. On the other hand, World Cancer Research Fund and the American Institute for Cancer Research have not identified Mets as a factor that is causally related to colorectal cancer, however, there may be an association between Mets and BMI [20]. The existing observational studies have a number of flaws. Firstly, the evidence for most of other risk factors, however, is not sufficient to demonstrate a causal relationship. Secondly, retrospective surveys are susceptible to selection and information bias. Additionally, the associations observed between colorectal cancer (CRC), metabolic syndrome (MetS), and its components in observational research may be influenced by confounding factors, limited sample size, restricted follow-up duration, and the possibility of reverse causation. These factors could potentially lead to misleading conclusions [21].
Mendelian randomization analysis has been a potent and sophisticated technique that can be used in conjunction with conventional observational studies to investigate the relationship between exposure and disease [22]. It can establish correlations between exposure factors and outcomes, determining a causal relationship between exposure factors and disease. It constitutes a more reliable method of inferring causality. It has the potential to overcome the limitations of observational research, which are assessed by genetic variation as an instrumental variable [23]. Furthermore, MR is derived from the fact that allele frequencies have been assigned from parents to offspring and that genotypes fixed during fertilized egg formation were also unaffected by disease, seeking to avoid mixing bias and reverse causation problems [24]. Genome-wide association studies (GWAS) have identified thousands of variants related to complex exposures, which opens up the possibility of a wide range of applications for MR [25].
We have conducted the first Mendelian randomization study to examine the bidirectional relationship between the metabolic syndrome and each of its components and CRC, which provides a new way of summarizing previously separate associations.
2. Material and methods
2.1. Study designs
Fig. 1 depicts a concise overview of this MR design. As shown in Fig. 1B, the components of Mets were defined by five elements according to the three criteria for diagnosis of metabolic syndrome mentioned earlier [[26], [27], [28]]. We investigated the causal relationship between colorectal cancer and metabolic syndrome and their components. We have used statistics from the most representative of GWASs for Mets, hypertension, waist circumference (WC), fasting blood glucose (FBG), serum triglycerides (TG), and serum high-density lipoprotein cholesterol (HDL-C). And an assessment was also made in the opposite direction. In order to substantiate our findings, we implemented Mendelian randomization (MR) techniques, incorporating genetic data from diverse sources pertaining to the five constituents of metabolic syndrome (MetS) and colorectal cancer (CRC) genome-wide association studies (GWAS). This methodology enabled us to evaluate the connections between MetS components and CRC by utilizing autonomous datasets, thereby validating our outcomes. Through the utilization of MR methods across numerous datasets, our objective was to fortify the resilience and dependability of our deductions concerning the association between MetS components and CRC. Furthermore, for Mendelian randomization (MR) to yield reliable results, the single-nucleotide polymorphisms (SNPs) included in the analysis must satisfy several assumptions. As presented in Fig. 1A, these assumptions are as follows: (1) Robust association: There needs to be a strong and consistent association between the instrumental variants (IVs) and the exposures under investigation. This ensures that the genetic variants serve as valid proxies for the exposure variables. (2) Absence of confounding: There should be no confounding variables that distort the relationship between the genetic variants and the outcome variables. Confounding factors can lead to biased estimates of causal effects. (3) Exclusivity of pathways: The instrumental variants must solely influence the outcomes of interest through their association with the exposures being studied. This assumption implies that there are no alternative pathways or mechanisms through which the genetic variants affect the outcomes [29]. In Supplementary Table 2, the data sources used in this investigation are described in detail. To reduce ethnic mismatches, we restricted our analysis to participants of mostly European origin.
Fig. 1.
Overview of the study design in this bidirectional MR study.
2.2. Selection of genetic components for MR observations
SNPs have been analyzed for each exposure factor based on the three main MR assumptions. We selected SNPs as IVs with independent genetics (p < 5 × 10−8) and without any linkage disequilibrium (LD) r2 < 0.01 and a distance of 1000 kb from the relevant dataset to make sure that our instrumental variables are powerful enough instruments to explain phenotypic variances.
2.3. Data sources and SNP selection of mets
Mets summary-level data has been obtained from the most comprehensive GWAS in the UK Biobank [30]. Contains 291,107 individuals with no missing data for genotype, outcome, and covariates. We also performed an MR analysis of subgroups for each component of the Mets. For waist circumference, we extracted aggregated data from Genetic Investigation of Anthropometric Traits (GIANT) including 224,459 individuals of European ancestry. As to hypertension, the aggregated statistics are available from Medical Research Council Integrative Epidemiology Unit (MRC-IEU) UK Biobank GWAS Pipeline [31]. We collected the most comprehensive summarized-metadata from GWAS for FBG in the Meta-analysis of Glucose and Insulin Related Traits Consortium (MAGIC), which include 281,416 diabetes patients [32]. The statistical results for TG and HDL-C have been obtained from the Global Lipids Genetics Consortium's most representative GWAS of 188,577 subjects (GLGC) [33]. The validation datasets for the components of Mets were obtained from reputable sources such as the MRC-IEU or the UK Biobank. We selected their respective satisfactory variables to construct instrumental variables (Supplementary Table 2).
2.4. Data sources and SNP selection of CRC
CRC summary-level data was currently available in the FinnGen cohort. The FinnGen study combines genomic data with digital health data from Finnish adults over the age of 18 [34]. Prospective epidemiological cohorts, disease-based cohorts, and hospital biobank samples are all included in this resource. Details are available on the organization's webpage (https://www.finngen.fi/fi). We used phenotypic and genetic data from a biological sample pool of 429,209 participants with CRC. The validation data for the CRC comes from the UK Biobank. The specific variable summary selection information can be found in Supplementary Table 2.
2.5. Statistical analyses
We used the random effect model inverse variance weighting (IVW) method as the main methods to determine the possible causal relationship between CRC and Mets and its components, because it provided reliable causal estimates in the lack of directional pleiotropy. In addition, we performed alternative analyses using a weighted median, simple model, weighted model, and MR-Egger methods. Then, we tested for directional level pleiotropy using MR-Egger intercepts [35]. We performed sensitivity analysis and heterogeneity assessment to determine whether heterogeneity and pleiotropy within the genetic instrument skewed the MR results. The degree of heterogeneity among SNPs was calculated using the Cochran Q-statistic. A “leave-one-out” sensitivity analysis and pleiotropy RESidualSum and Outlier (MR-PRESSO) analysis have been employed to determine presence of pleiotropy and to generate outlier-adjusted estimates by removing any outlier SNP of pleiotropy [36]. Odds ratios (ORs) and corresponding 95 % confidence intervals (CIs) for outcomes have been measured to one-SD increase in levels of lipid-related traits. The statistical analysis has been carried out to use R (v.4.2.2) software. The “devtools”, “TwoSampleMR”, and “MRPRESSO” packages of the R statistical program were used for MR. To account for univariable MR analysis, we regarded associations with p values of False Discovery Rate (FDR) to be strong evidence of causal associations. As for sensitivity analyses, the effects were deemed statistically significant at p < 0.05, and all statistical tests were two-sided.
3. Results
3.1. The causal effect of mets and its components on CRC
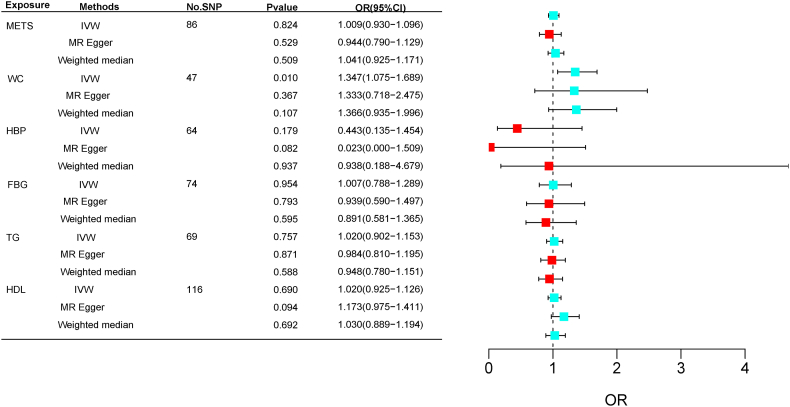
As shown in Table 1, we finally included 87, 46, 170, 75, 72, and 116 SNPs in the MR analyses as genetic instruments for Mets, WC, HBP, FBG, TG, and HDL-C, respectively. In Table 1 and Fig. 2, the results of MR analysis have been displayed. After accounting for multiple testing, for genetically predicted waist circumference, they were significant associated with increased risk of CRC. The OR with 95 % CI of per log-odds increment in WC liability has been 1.35 (95 % CI: 1.08–1.69; p = 0.0096 < P(FDR-correction) = 0.0133) in the IVW model (Fig. 3B). We did not find any heterogeneity or horizontal pleiotropy (Cochran's Q, p > 0.05; Egger intercept = 0.0003, p = 0.705) (Fig. 3C). Moreover, the funnel plot (Supplementary Fig. 2) revealed an invisible asymmetry. In addition, the leave-one-out analysis showed that the identified associations were not substantially altered after removing any single variant (Fig. 3A), indicating that the results are stable.
Table 1.
Genetic predicted MetS and its components on risk of CRC in the MR analysis.
| Exposure | Outcome | No.SNP | Methods | OR | 95%CI | Pvalue | Heterogeneity |
Pleiotropy |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MR-Egger |
IVW |
MR-Egger |
MR-PRESSO |
|||||||||||
| Q | P | Q | P | Intercept | P | No. Of outlier | P | |||||||
| MetS | CRC | 86 | IVW | 1.0093 | 0.9297–1.0958 | 0.8245 | 95.8547 | 0.1773 | 96.6363 | 0.1827 | 0.0050 | 0.4103 | NA | 0.2100 |
| MR Egger | 0.9441 | 0.7898–1.1285 | 0.5291 | |||||||||||
| Weighted median | 1.0406 | 0.9246–1.1711 | 0.5092 | |||||||||||
| Simple mode | 1.1781 | 0.9036–1.5360 | 0.2291 | |||||||||||
| Weighted mode | 1.1555 | 0.9566–1.3959 | 0.1374 | |||||||||||
| WC | CRC | 47 | IVW | 1.3475 | 1.0751–1.6889 | 0.0096 | 41.0972 | 0.6380 | 41.0986 | 0.6773 | 0.0003 | 0.9710 | NA | 0.7050 |
| MR Egger | 1.3331 | 0.7182–2.4745 | 0.3672 | |||||||||||
| Weighted median | 1.3661 | 0.9350–1.9959 | 0.1068 | |||||||||||
| Simple mode | 1.4059 | 0.7259–2.7229 | 0.3177 | |||||||||||
| Weighted mode | 1.3897 | 0.8931–2.1623 | 0.1514 | |||||||||||
| HBP | CRC | 64 | IVW | 0.4428 | 0.1348–1.4542 | 0.1793 | 78.0347 | 0.0822 | 80.6638 | 0.0662 | 0.0166 | 0.1534 | NA | 0.0670 |
| MR Egger | 0.0227 | 0.0003–1.5093 | 0.0820 | |||||||||||
| Weighted median | 0.9375 | 0.1878–4.6793 | 0.9373 | |||||||||||
| Simple mode | 2.6592 | 0.0585–120.7946 | 0.6172 | |||||||||||
| Weighted mode | 2.1504 | 0.1343–34.4244 | 0.5903 | |||||||||||
| FBG | CRC | 74 | IVW | 1.0073 | 0.7875–1.2885 | 0.9536 | 71.8116 | 0.4841 | 71.9314 | 0.5134 | 0.0019 | 0.7303 | NA | 0.5200 |
| MR Egger | 0.9394 | 0.5896–1.4968 | 0.7932 | |||||||||||
| Weighted median | 0.8907 | 0.5811–1.3652 | 0.5952 | |||||||||||
| Simple mode | 1.1206 | 0.5034–2.4949 | 0.7811 | |||||||||||
| Weighted mode | 0.9652 | 0.5991–1.5548 | 0.8845 | |||||||||||
| Triglycerides | CRC | 69 | IVW | 1.0196 | 0.9018–1.1528 | 0.7569 | 71.8533 | 0.3203 | 72.0843 | 0.3444 | 0.0022 | 0.6440 | NA | 0.3570 |
| MR Egger | 0.9840 | 0.8103–1.1950 | 0.8713 | |||||||||||
| Weighted median | 0.9476 | 0.7799–1.1513 | 0.5878 | |||||||||||
| Simple mode | 1.1513 | 0.7607–1.5725 | 0.6302 | |||||||||||
| Weighted mode | 0.9641 | 0.7878–1.1799 | 0.7239 | |||||||||||
| HDL | CRC | 116 | IVW | 1.0202 | 0.9246–1.1258 | 0.6902 | 123.8539 | 0.2487 | 127.1389 | 0.2068 | −0.0077 | 0.0848 | NA | 0.2000 |
| MR Egger | 1.1726 | 0.0748–1.4105 | 0.0940 | 95.8547 | 0.1773 | 96.6363 | 0.1827 | 0.0050 | 0.4103 | NA | 0.2100 | |||
| Weighted median | 1.0303 | 0.8889–1.1943 | 0.6916 | |||||||||||
| Simple mode | 0.8572 | 0.6337–1.1594 | 0.3194 | |||||||||||
| Weighted mode | 1.0068 | 0.8722–1.1622 | 0.9262 | |||||||||||
Fig. 2.
Forest plot of MR analysis results.
Fig. 3.
Visualization of WC results in MR analysis.
In contrast, no significant correlation was found between the other components of the metabolic syndrome and CRC. In terms of heterogeneity, Cochran's Q test revealed no significant heterogeneity (p-values >0.05 for both IVW and MR-Egger), and funnel plots showed no substantiation of heterogeneity as well (Supplementary Fig. 2). In terms of pleiotropy, neither the MR-Egger method nor MR-PRESSO revealed the presence of horizontal pleiotropy. Furthermore, leave-one-out analyses revealed that the outcomes have been consistent. The validation Mendelian randomization (MR) analysis yielded similar results to the aforementioned findings. Specifically, we observed a significant correlation between waist circumference (WC) and colorectal cancer (CRC) in the validation analysis (odds ratio [OR]: 1.002; 95 % confidence interval [CI]: 1.0002–1.0041; p = 0.029< P(FDR-correction) = 0.04). However, no significant associations were found between CRC and the other four components of metabolic syndrome (Supplementary Table 4 and Fig. 4). Supplementary Fig. 1 shows a visualization of the findings of the other components of the MR analysis.
Fig. 4.
Validation of the MR analysis forest plot results for the cohort.
3.2. Causal effect of CRC on mets and their components
In the reverse MR analyses after a rigorous cascade of SNP selection, we finally utilized 8 variants for Mets, 2 variants for WC, 5 variants for HBP, 5 variants for FBG, 2 variants for TG, and 2 variants for HDL as genetic instruments etc. As the number of SNP included was all small, only IVW was used in the MR analysis as a causal analysis. As illustrated in Supplementary Table 3, MR results indicated that CRC has not been causally related to either Mets or the components, with ORs close to 1. Egger's tests indicated that there was no potential of horizontal pleiotropy. In addition, Cochran's Q result demonstrates that there wasn't any substantial heterogeneity.
4. Discussion
This is the first paper to explore causal associations between the Mets and its five components with outcome CRC. And a secondary validation was performed to ensure the reliability of the results. Our bidirectional two-sample Mendelian randomization (MR) analysis unveiled a significant association between genetically predicted waist circumference (WC), a metric for abdominal obesity, and an elevated susceptibility to colorectal cancer (CRC). However, no such causal association was found in the other four components (FBG, HBP, HDL, TG). Our findings did not provide any substantiation for a causal effect in the reverse direction. It shows that the relationship between the two is unidirectional and there is no offset caused by the opposite direction. These results lend support to the notion that individuals with greater adiposity, particularly around the waist, exhibit an increased likelihood of developing CRC. The conclusion drawn from a significant amount of epidemiological evidence is that Mets are linked to an increased risk of CRC [8,13,15,37]. Several meta-analyses of cohort studies have also confirmed Mets as an independent risk factor for CRC, and high WC may explain much of this association, which is consistent with our findings [5,7]. This may be related to the fact that overexpression of the Mets core gene IL6 can promote the malignancy of CRC, which is highly dependent on the mTOR-S6K signaling pathway [12]. Regarding the reverse causal link, observational studies on the occurrence of CRC and Mets identified a higher risk of Mets component in colorectal cancer patients, but it has also been demonstrated that only extremely high quantities of metabolic factors increase the risk [38]. The lack of evidence for a decisive causal relationship between CRC and Mets in MR analysis suggests that the observed association may be the result of confounding factors or that the association may not be strong enough to demonstrate causality. In addition to the need for extremely high quantities of metabolic factors to show the risk of cancer [38], some studies demonstrate that the presence of Mets increases the risk of CRC in women but not in men [14]. Similar to those diagnosed after age 50, Mets and obesity are positively associated with CRC in those who develop the disease before that age [10]. These data suggest that the strength of the connection difference in the association between Mets and CRC depends on the specific definition of Mets and the age and gender of CRC patients.
In terms of Mets components, A link between obesity and colorectal cancer (CRC) was formed among obesity-related diseases [39]. In a retrospective cohort study in Wenzhou, China, differences between groups were statistically significant for TG, HDL, HBP, diabetes mellitus (DM), and body mass index (BMI) by using univariate analysis. However, the multifactorial analysis showed that only BMI, DM, TG, and Mets were significantly and independently associated with OS [12]. In the DM group, high TG and cholesterol levels increased the risk of CRC by 4.118-fold. Insulin resistance and the insulin-like growth factor 1 systems are crucial in the link between Mets and CRC [40]. One prospective cohort study from Taiwan confirmed that high TG and cholesterol level increased the risk of CRC by 4.118-fold in the DM group [11]. Additionally, a meta-analysis concluded that the Mets as a whole did not carry a greater risk than the sum of their parts [7], which is in line with the findings of our MR study. Therefore, it is still unknown how each component will affect the other, and this needs to be researched further. In overview, epidemiological research on the relationship between Mets and CRC has produced mixed results. Only a suggest causal correlation between WC and CRC was present in our MR analysis; no positive correlation between Mets and CRC was found. The outcomes were stronger and there was no chance of heterogeneity or potential horizontal multidirectional. Further investigation is necessary to identify the exact causes of remaining factors and CRC. Our results suggest that controlling abdominal obesity may be more crucial than controlling blood pressure, lipids, and blood glucose in preventing the development of CRC.
Even though the precise mechanism by which Mets causes CRC is unknown, various theories have been put forth. The currently available evidence points to obesity and hyperinsulinemia/insulin resistance as key factors in the association between Mets and cancer [40,41]. Particularly, several adipocytokines, such as TNF-α, IL-6, lipocalin, etc., secreted by adipose tissue (especially VAT, Visceral adipose tissue) in obese individuals can result in insulin resistance syndrome [42]. Well, first of all, the adipose tissue hormone leptin may have an impact on cell proliferation because it activates the MAPK signaling pathway. On the other hand, it might encourage cancer metastasis, angiogenesis, and a rise in the expression of matrix metalloproteinase-2 [43]. Second, angiogenesis is a crucial stage in the development and spread of tumors. The most significant pro-angiogenic factor secreted by adipocytes (VAT only) is VEGF, and insulin, IGF-1, leptin, TNF-α, and hypoxia all promote its secretion [44]. Last but not least, it has been discovered that some pro-inflammatory cytokines, including cytokines, reactive oxygen products, and inflammatory pathways (NF–B), aid in the development of cancer by accelerating the cell cycle, promoting oncogene expression, and decreasing tumor suppression [45].
Our MR analysis identified a significant causal link between WC and CRC, but we did not find causal association between the remaining four exposures and CRC using genetic tools while adhering strictly to the three MR hypotheses. It is worth noting that other MR studies in this field have reported different findings from ours. Since blood lipids, diabetes and obesity are closely related, obesity's inflammatory and endocrine effects have been put forth as the main mechanism to account for this association [46]. The majority of MR analyses have focused on studying individual factors, with the majority examining the relationship between obesity [[47], [48], [49], [50], [51], [52]], diabetes [[53], [54], [55]], blood lipids [[56], [57], [58], [59]] and CRC, did not include hypertension as a factor. High total cholesterol levels were associated with a higher risk of CRC in some studies, though the findings are inconsistent [58]. Fasting insulin has been discovered to be related with the risk of CRC by Han Zhang and Murphy, N. et al. [54,60], and Jung, S. Y. et al. Confirmed a potential causal relationship between insulin resistance and CRC [53], but this association was not found in type 2 diabetes patients [55]. Neither of them explored with respect to fasting glucose. The findings on obesity and CRC are inconsistent [51,52], and most studies use BMI as a proxy for obesity, instead of indicators such as waist circumference to better replace visceral adipose tissue as the main effect molecule of obesity.
This study has several strengths. Compared to observational studies, MR analysis reduces the risk of reverse association bias and minimizes the impact of confounding factors that influence exposure. We were able to evaluate the causal relationship between two distinct sources of Mets and CRC, and the consistency of our results further strengthens our conclusions. Moreover, we have largest and most authoritative data on Mets and its components as well as CRC. Because visceral adipose tissue is increasingly recognized as an endocrine organ for synthesizing obesity-mediated hormones and cytokines, we used WC as a measure of obesity rather than BMI because it is a better proxy for estimating visceral fat, giving the results more validity [61,62]. Furthermore, we conducted sensitivity and heterogeneity analyses, which demonstrated that the MR results we obtained were robust. Furthermore, we used the MR-Egger intercept and MR-PRESSO methods to assess and correct for horizontal pleiotropy in the MR analysis, respectively, ensuring the reliability and robustness of our results. The study we conducted has some drawbacks, though. First, it's unclear how genetic tools work or how they affect risk factors. Second, the pooled data for CRC that we used could not be stratified based on pertinent covariates (e.g., age, sex, smoking, alcohol consumption, and underlying diseases such as inflammatory bowel disease). Therefore, it is not possible to know whether Mets in certain subgroups may promote the risk of CRC.
In summary, waist circumference can causally lead to CRC, which may largely explain the strong clinical association between Mets and CRC. Consequently, it is imperative to prioritize certain aspects in early cancer screening, such as directing CRC screening efforts towards obese individuals and promoting weight loss among this population to mitigate CRC risk.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Only publicly available data were used in this study; the data sources and processing of these data are described in the Materials and Methods. The UK Biobank is an open access resource. Bona fide researchers can apply to use the UK Biobank dataset by registering and applying at this website: http://ukbiobank.ac.uk/register-apply/.
Funding
The National Natural Science Foundation of China. (the funder: Yuping Wang, No. 82260122), the Foundation of The First Hospital of Lanzhou University, China. (the funder: Ya Zheng, No. ldyyyn2021-59).
CRediT authorship contribution statement
Yuhua Chen: Methodology, Writing – original draft. Wanru Kong: Conceptualization. Min Liu: Investigation. Qiang Li: Validation. Yuping Wang: Funding acquisition. Ya Zheng: Funding acquisition, Writing – review & editing. Yongning Zhou: Visualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to express our sincere appreciation to the UK Biobank Consortium, the Genetic Investigation of ANthropometric Traits (GIANT) consortium, A GWAS of 29 studies the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC), Global Lipids Genetics Consortium (GLGC), FINNGEN consortium for making their summary-level statistics public.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23872.
Contributor Information
Yuhua Chen, Email: 978631913@qq.com.
Wanru Kong, Email: kwr1993@163.com.
Min Liu, Email: liuminlzu@sina.com.
Qiang Li, Email: liqldyy@126.com.
Yuping Wang, Email: wangyuping@lzu.edu.cn.
Ya Zheng, Email: zhengya10@126.com.
Yongning Zhou, Email: zhouyn@lzu.edu.cn.
Abbreviations
- BMI
body-mass index
- CI
confidence interval
- CRC
colorectal cancer
- EAF
effect allele frequency
- FBG
fasting blood glucose
- GIANT
Genetic Investigation of Anthropometric Traits
- GLGC
Global Lipids Genetics Consortium
- GWAS
genome-wide association study
- HDL-C
high-density lipoprotein cholesterol
- IGF1
insulin-like growth factor 1
- IGFBP3
insulin-like growth factor binding protein 3
- IV
instrumental variant
- IVW
inverse variance-weighted
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MAGIC
Meta-analysis of Glucose and Insulin Related Traits Consortium
- Mets
metabolic syndrome
- MR
Mendelian randomization
- MRC-IEU
Medical Research Council Integrative Epidemiology Unit
- OR
odds ratio
- PRESSO
pleiotropy residual sum and outlier
- SD
standard deviation
- SE
standard error
- SNP
single-nucleotide polymorphism
- TG
triglycerides
- WC
waist circumference
- WHO
World Health Organization
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Patel S.G., Karlitz J.J., Yen T., Lieu C.H., Boland C.R. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. The Lancet Gastroenterol. & Hepatol. 2022;7(3):262–274. doi: 10.1016/S2468-1253(21)00426-X. [DOI] [PubMed] [Google Scholar]
- 3.Hull M.A., Rees C.J., Sharp L., Koo S. A risk-stratified approach to colorectal cancer prevention and diagnosis. Nat. Rev. Gastroenterol. Hepatol. 2020;17(12):773–780. doi: 10.1038/s41575-020-00368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colorectal cancer screening in average-risk adults. Ann. Intern. Med. 2019;171(9) doi: 10.7326/P19-0015. [DOI] [PubMed] [Google Scholar]
- 5.Shen X., Wang Y., Zhao R., et al. Metabolic syndrome and the risk of colorectal cancer: a systematic review and meta-analysis. 2021;36(10):2215–2225. doi: 10.1007/s00384-021-03974-y. [DOI] [PubMed] [Google Scholar]
- 6.Han F., Wu G., Zhang S., Zhang J., Zhao Y., Xu J. The association of metabolic syndrome and its components with the incidence and survival of colorectal cancer: a systematic review and meta-analysis. Int. J. Biol. Sci. 2021;17(2):487–497. doi: 10.7150/ijbs.52452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esposito K., Chiodini P., Capuano A., et al. Colorectal cancer association with metabolic syndrome and its components. a Systematic Review with Meta-analysis. 2013;44(3):634–647. doi: 10.1007/s12020-013-9939-5. [DOI] [PubMed] [Google Scholar]
- 8.Tran T.T., Gunathilake M., Lee J., Kim J. Association between metabolic syndrome and its components and incident colorectal cancer in a prospective cohort study. Cancer. 2022;128(6):1230–1241. doi: 10.1002/cncr.34027. [DOI] [PubMed] [Google Scholar]
- 9.Liu T., Fan Y., Zhang Q., et al. The combination of metabolic syndrome and inflammation increased the risk of colorectal cancer. Inflamm. Res. : Off. J. European Histamine Res. Soc. [et al] 2022;71(7–8):899–909. doi: 10.1007/s00011-022-01597-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin E.H., Han K., Lee D.H., et al. Association between metabolic syndrome and the risk of colorectal cancer diagnosed before age 50 Years according to tumor location. Gastroenterology. 2022;163(3):637–648. doi: 10.1053/j.gastro.2022.05.032. e632. [DOI] [PubMed] [Google Scholar]
- 11.Hsu S.H., Syu D.K., Chen Y.C., Liu C.K., Sun C.A., Chen M. The association between hypertriglyceridemia and colorectal cancer: a long-term community cohort study in taiwan. Int. J. Environ. Res. Publ. Health. 2022;19(13) doi: 10.3390/ijerph19137804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Zhao J., Wu X., Zhang Y., Jin Y., Cai W. Clinical and genomic characteristics of metabolic syndrome in colorectal cancer. Aging. 2021;13(4):5442–5460. doi: 10.18632/aging.202474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J., Lee K.S., Kim H., et al. The relationship between metabolic syndrome and the incidence of colorectal cancer. Environ. Health Prev. Med. 2020;25(1):6. doi: 10.1186/s12199-020-00845-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J., Park E.Y., Park E., Lim M.K., Oh J.K., Kim B. Metabolic syndrome and colorectal cancer risk: results of propensity score-based analyses in a community-based cohort study. Int. J. Environ. Res. Publ. Health. 2020;17(22) doi: 10.3390/ijerph17228687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu H.M., Lee Y.C., Tu C.H., et al. Effects of metabolic syndrome and findings from baseline colonoscopies on occurrence of colorectal neoplasms. Clin. Gastroenterol. Hepatol. : Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2015;13(6):1134–1142. doi: 10.1016/j.cgh.2014.10.022. e1138. [DOI] [PubMed] [Google Scholar]
- 16.Hong S.N., Kim J.H., Choe W.H., et al. Prevalence and risk of colorectal neoplasms in asymptomatic, average-risk screenees 40 to 49 years of age. Gastrointest. Endosc. 2010;72(3):480–489. doi: 10.1016/j.gie.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Chung K.C., Juang S.E., Chen H.H., et al. Association between metabolic syndrome and colorectal cancer incidence and all-cause mortality: a hospital-based observational study. BMC Gastroenterol. 2022;22(1):453. doi: 10.1186/s12876-022-02505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ajtajotms Siddiqui. Metabolic syndrome and its association with colorectal cancer: a review. 2011;341(3):227–231. doi: 10.1097/MAJ.0b013e3181df9055. [DOI] [PubMed] [Google Scholar]
- 19.Jinjuvadia R., Lohia P., Jinjuvadia C., Montoya S., Sjjocg Liangpunsakul. The association between metabolic syndrome and colorectal neoplasm: systemic review and meta-analysis. 2013;47(1):33–44. doi: 10.1097/MCG.0b013e3182688c15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinton S.K., Giovannucci E.L., Hursting S.D. The World cancer research Fund/American Institute for cancer research third expert report on diet, nutrition, physical activity, and cancer: impact and future directions. J. Nutr. 2020;150(4):663–671. doi: 10.1093/jn/nxz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith G.D., Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32(1) doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 22.Cornish A.J., Tomlinson I.P.M., Houlston R.S. Mendelian randomisation: a powerful and inexpensive method for identifying and excluding non-genetic risk factors for colorectal cancer. Mol. Aspect. Med. 2019;69:41–47. doi: 10.1016/j.mam.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler A., Mwambi H., König I.R. Mendelian randomization versus path models: making causal inferences in genetic epidemiology. Hum. Hered. 2015;79(3–4):194–204. doi: 10.1159/000381338. [DOI] [PubMed] [Google Scholar]
- 24.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porcu E., Rüeger S., Lepik K., Santoni F.A., Reymond A., Kutalik Z. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019;10(1):3300. doi: 10.1038/s41467-019-10936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alberti K.G.M.M., Eckel R.H., Grundy S.M., et al. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; national heart, lung, and blood Institute; American heart association; World heart federation; international atherosclerosis society; and international association for the study of obesity. Circulation. 2009;120(16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 27.Guerrero-Romero F., Rodríguez-Morán M. Concordance between the 2005 international diabetes federation definition for diagnosing metabolic syndrome with the national cholesterol education program adult treatment panel III and the World health organization definitions. Diabetes Care. 2005;28(10):2588–2589. doi: 10.2337/diacare.28.10.2588a. [DOI] [PubMed] [Google Scholar]
- 28.Liberopoulos E.N., Mikhailidis D.P., Elisaf M.S. Diagnosis and management of the metabolic syndrome in obesity. Obes. Rev. 2005;6(4):283–296. doi: 10.1111/j.1467-789X.2005.00221.x. [DOI] [PubMed] [Google Scholar]
- 29.Richmond R.C., Davey Smith G. Mendelian randomization: concepts and scope. Cold Spring Harb Perspect Med. 2022;12(1) doi: 10.1101/cshperspect.a040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lind L. Genome-wide association study of the metabolic syndrome in UK biobank. Metab. Syndr. Relat. Disord. 2019;17(10):505–511. doi: 10.1089/met.2019.0070. [DOI] [PubMed] [Google Scholar]
- 31.Elsworth B.L., Mitchell R.E., Raistrick C.A., Paternoster L., Hemani G., Gaunt T.R. 2017. MRC IEU UK Biobank GWAS Pipeline Version 1. [Google Scholar]
- 32.Chen J., Spracklen C.N., Marenne G., et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021;53(6):840–860. doi: 10.1038/s41588-021-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer C.J., Schmidt E.M., Sengupta S., et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.FinnGen I. FinnGen-tutkimushanke vie suomalaiset löytöretkelle genomitietoon. 2022. https://www.finngen.fi/fi/finngen_ [Internet]
- 35.Xu L., Borges M.C., Hemani G., Lawlor D.A. The role of glycaemic and lipid risk factors in mediating the effect of BMI on coronary heart disease: a two-step, two-sample Mendelian randomisation study. Diabetologia. 2017;60(11):2210–2220. doi: 10.1007/s00125-017-4396-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verbanck M., Chen C.-Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim N.H., Jung Y.S., Yang H.J., et al. Prevalence of and risk factors for colorectal neoplasia in asymptomatic young adults (20-39 Years old) Clin. Gastroenterol. Hepatol. : Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2019;17(1):115–122. doi: 10.1016/j.cgh.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Stocks T., Lukanova A., Johansson M., et al. Components of the metabolic syndrome and colorectal cancer risk; a prospective study. Int. J. Obes. 2008;32(2):304–314. doi: 10.1038/sj.ijo.0803713. [DOI] [PubMed] [Google Scholar]
- 39.Riondino S., Roselli M., Palmirotta R., Della-Morte D., Ferroni P., Guadagni FJWjog. Obesity and colorectal cancer: role of adipokines in tumor initiation and progression. 2014;20(18):5177–5190. doi: 10.3748/wjg.v20.i18.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uzunlulu M., Telci Caklili O., Oguz AJAon. metabolism. Assoc. between Metabol. Syndr. Cancer. 2016;68(3):173–179. doi: 10.1159/000443743. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe S., Hojo M., Ajjog Nagahara. Metabolic syndrome and gastrointestinal diseases. 2007;42(4):267–274. doi: 10.1007/s00535-007-2033-0. [DOI] [PubMed] [Google Scholar]
- 42.John B.J., Irukulla S., Abulafi A.M., Kumar D., Mendall M.A. Systematic review: adipose tissue, obesity and gastrointestinal diseases. Aliment Pharmacol. Therapeut. 2006;23(11):1511–1523. doi: 10.1111/j.1365-2036.2006.02915.x. [DOI] [PubMed] [Google Scholar]
- 43.Braun S., Bitton-Worms K., LeRoith D. The link between the metabolic syndrome and cancer. Int. J. Biol. Sci. 2011;7(7):1003–1015. doi: 10.7150/ijbs.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazawa-Hoshimoto S., Takahashi K., Bujo H., Hashimoto N., Saito Y. Elevated serum vascular endothelial growth factor is associated with visceral fat accumulation in human obese subjects. Diabetologia. 2003;46(11):1483–1488. doi: 10.1007/s00125-003-1221-6. [DOI] [PubMed] [Google Scholar]
- 45.Mendonça F.M., de Sousa F.R., Barbosa A.L., et al. Metabolic syndrome and risk of cancer: which link? Metab., Clin. Exp. 2015;64(2):182–189. doi: 10.1016/j.metabol.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Lega I.C., Lipscombe L.L. Review: diabetes, obesity, and cancer-pathophysiology and clinical implications. Endocr. Rev. 2020;41(1) doi: 10.1210/endrev/bnz014. [DOI] [PubMed] [Google Scholar]
- 47.Thrift A.P., Gong J., Peters U., et al. Mendelian randomization study of body mass index and colorectal cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American association for cancer research. Cosponsored by the Am. Soc. Preventive Oncol. 2015;24(7):1024–1031. doi: 10.1158/1055-9965.EPI-14-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki S., Goto A., Nakatochi M., et al. Body mass index and colorectal cancer risk: a Mendelian randomization study. Cancer Sci. 2021;112(4):1579–1588. doi: 10.1111/cas.14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loh N.Y., Wang W., Noordam R., Christodoulides C. Obesity, fat distribution and risk of cancer in women and men: a mendelian randomisation study. Nutrients. 2022;14(24) doi: 10.3390/nu14245259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jarvis D., Mitchell J.S., Law P.J., et al. Mendelian randomisation analysis strongly implicates adiposity with risk of developing colorectal cancer. Br. J. Cancer. 2016;115(2):266–272. doi: 10.1038/bjc.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao C., Patel C.J., Michailidou K., et al. Mendelian randomization study of adiposity-related traits and risk of breast, ovarian, prostate, lung and colorectal cancer. Int. J. Epidemiol. 2016;45(3):896–908. doi: 10.1093/ije/dyw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y., Tang H., Huang P., et al. Assessment of causal effects of visceral adipose tissue on risk of cancers: a Mendelian randomization study. Int. J. Epidemiol. 2022;51(4):1204–1218. doi: 10.1093/ije/dyac025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung S.Y., Papp J.C., Sobel E.M., Zhang Z.F. Mendelian randomization study: the association between metabolic pathways and colorectal cancer risk. Front. Oncol. 2020;10:1005. doi: 10.3389/fonc.2020.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H., Li D., Liu X., et al. Fasting insulin and risk of overall and 14 site-specific cancers: evidence from genetic data. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.863340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goto A., Yamaji T., Sawada N., et al. Diabetes and cancer risk: a Mendelian randomization study. Int. J. Cancer. 2020;146(3):712–719. doi: 10.1002/ijc.32310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwagami M., Goto A., Katagiri R., et al. Blood lipids and the risk of colorectal cancer: mendelian randomization analyses in the Japanese consortium of genetic epidemiology studies. Cancer Prev. Res. 2022;15(12):827–836. doi: 10.1158/1940-6207.CAPR-22-0146. [DOI] [PubMed] [Google Scholar]
- 57.Luo X., Tu Z., Chen H., Ding J. Blood lipids and risk of colon or rectal cancer: a Mendelian randomization study. J. Cancer Res. Clin. Oncol. 2021;147(12):3591–3599. doi: 10.1007/s00432-021-03790-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ibáñez-Sanz G., Díez-Villanueva A., Riera-Ponsati M., et al. Mendelian randomization analysis rules out disylipidaemia as colorectal cancer cause. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-49880-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Broadbent H., Law P.J., Sud A., et al. Mendelian randomisation implicates hyperlipidaemia as a risk factor for colorectal cancer. Int. J. Cancer. 2017;140(12):2701–2708. doi: 10.1002/ijc.30709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy N., Song M., Papadimitriou N., et al. Associations between glycemic traits and colorectal cancer: a mendelian randomization analysis. J. Natl. Cancer Inst. 2022;114(5):740–752. doi: 10.1093/jnci/djac011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Avgerinos K.I., Spyrou N., Mantzoros C.S., Dalamaga M. Obesity and cancer risk: emerging biological mechanisms and perspectives. Metab., Clin. Exp. 2019;92:121–135. doi: 10.1016/j.metabol.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Aleksandrova K., Mozaffarian D., Pischon T. Addressing the perfect storm: biomarkers in obesity and pathophysiology of cardiometabolic risk. Clin. Chem. 2018;64(1):142–153. doi: 10.1373/clinchem.2017.275172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Only publicly available data were used in this study; the data sources and processing of these data are described in the Materials and Methods. The UK Biobank is an open access resource. Bona fide researchers can apply to use the UK Biobank dataset by registering and applying at this website: http://ukbiobank.ac.uk/register-apply/.