Abstract
Chenopodium quinoa possesses remarkable nutritional value and adaptability to various agroecological conditions. Panicle architecture influences the number of spikelets and grains in a panicle, ultimately leading to productivity and yield. Therefore, this study aimed to investigate the metabolites, nutrients, and minerals in Chenopodium quinoa accessions of varying panicle architecture. Metabolic profiling using liquid chromatography-mass spectrometry (LC–MS) analysis identified seventeen metabolites, including flavonoids, phenolics, fatty acids, terpenoids, phenylbutenoid dimers, amino acids, and saccharides. Eight metabolic compounds were reported in this study for the first time in quinoa. Some metabolites were detected as differentially expressed. The compound (Z)-1-(2,4,5-trimethoxyphenyl) butadiene and chrysin were found only in SPrecm. Sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl) methyl hydrogen phosphate and elenolic acid were detected only in CHEN-33, and quercetin, 3-hydroxyphloretin-3’-C-glucoside, kurarinone, and rosmarinic acid were identified only in D-12175. Variable importance in projection (VIP) scores annotated ten metabolites contributing to variability. Mineral analysis using atomic absorption spectrophotometry indicated that the quantity of magnesium and calcium is high in D-12175. In comparison, SPrecm showed a high quantity of magnesium compared to CHEN-33, while CHEN-33 showed a high quantity of calcium compared to SPrecm. However, the proximate composition showed no significant difference among quinoa accessions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-023-01398-2.
Keywords: Panicle architecture, LC–MS-MS, Metabolomic profiling, Novel compounds, Nutritional profiling
Introduction
Chenopodium quinoa, commonly known as quinoa, is an important pseudocereal originally cultivated in Andean regions of South America since the ancient era (Contreras-Jiménez et al. 2019). Quinoa is primarily cultivated in Peru, Chile, Bolivia, Ecuador, Colombia, and Argentina, but later on, it was familiarized in other states, such as North America, Europe, Asia, and Africa, with greater yields (Vilcacundo and Hernández-Ledesma 2017; Gomez-Pando et al. 2019). It belongs to the family Amaranthaceae and is considered a superfood because of its high nutritional value and ability to withstand various agroecological conditions (García-Parra et al. 2020). Quinoa exhibits high phenotypic variability that is easily distinguished by the plant’s pigmentation, seeds, inflorescence type, color, shape, density of the panicles, and variety of grain size and shape (Manjarres-Hernández et al. 2021).
Globally, it has gained more attention because of its gluten-free grains (Pathan and Siddiqui 2022) and exceptional nutritional properties, including high protein content and balanced essential amino acids, lipids, and vitamins (Carciochi et al. 2016; Vilcacundo and Hernández-Ledesma 2017). Additionally, quinoa has high mineral contents, including microelements (iron, copper, zinc, and manganese) and macronutrients (sodium, magnesium, calcium, potassium, and phosphorus) (Pathan et al. 2019; Adamczewska-Sowińska et al. 2021). Moreover, it also possesses terpenoids, flavonoids, phenolic acids, steroids, and nitrogen-containing compounds that have antidiabetic, antimicrobial, anti-inflammatory, anticancer, and immunoregulatory properties (Burrieza et al. 2019; Pereira et al. 2019, 2020). As an environment-resilient crop (Ruiz et al. 2014), it can thrive in high salinity, drought, frost, and extreme temperatures hence, can sustainably be cultivated in marginal environments, a considerable attribute due to the anticipated escalation of salinity and aridity in several parts of the world (FAO 2023). Thus, this adaptability makes quinoa a promising substitute for traditional crops in adverse climatic scenarios (Sosa‐Zuniga et al. 2017). Like quinoa grains, quinoa greens, including leaves, microgreens, and sprouts, also possess similar health-benefiting properties. These nutritional qualities make it a novel nutritious food with unique health benefits, and it is occasionally referred to as a “superfood” (Pathan and Siddiqui 2022).
Additionally, quinoa contains saponins (secondary metabolites) within its seed pericarp, conferring bitterness. Saponins are not restricted to seeds; they can also be found in other plant parts, such as leaves, fruits, and flowers (Kaur et al. 2022). The triterpenoid glycosides that make saponins in quinoa are primarily oleanolic acid (OA), hederagenin (HD), serjanic acid (SA), and phytolaccagenic acid (PA) derivatives (Pandya et al. 2021). Saponins have been extensively explored for their potential in agriculture due to their antifungal properties and possible use in the food industry as preservatives, food additives, or flavor modifiers (Rai et al. 2017). Furthermore, they are substantially involved in the pharmacology industry due to their contributions to anti-cholesterol activity and other valuable bioactive properties, such as antimicrobial, antioxidant, and anti-inflammatory activities (Pakbaz et al. 2021).
Metabolomics tools are extensively used to comprehend metabolism and analyze metabolic pathways in complex biological systems (Rai et al. 2017). Metabolomics studies are challenging ventures that heavily rely on instrumentation to precisely quantify and locate particular metabolites (Liu et al. 2020; Otterbach et al. 2021). Plant metabolome research analyzes a broader range of metabolites with diverse physical characteristics, such as ionic, organic and inorganic compounds, amino acids, hydrophilic carbohydrates, and hydrophobic lipid-derived compounds (Jorge et al. 2016). Liquid chromatography (LC), assisted with mass spectrometry (LC–MS-MS), is an efficient approach for metabolomic profiling (Otterbach et al. 2021). LC–MS is a robust tool that can distinguish and characterize differential polar metabolites such as lipids and secondary metabolites (Jorge et al. 2016; Cheong et al. 2019). It will help better understand quinoa grain’s potential as a functional food source (García-Parra et al. 2021). Several studies have emerged on quinoa’s nutritional constituents and therapeutic properties, representing the crop as a potential source for nutritious food development. Panicle architecture significantly affects grain yield by increasing the number of spikelets and grains per panicle, leading to higher productivity, while poor panicle architecture results in reduced yield due to fewer spikelets and grains (Li et al. 2021). So, this study aimed to identify the proximate, mineral, and metabolic compositions to highlight the impending benefits of selected quinoa accessions (SPrecm, CHEN-33, and D-12175) having diverse panicle architecture.
Materials and methods
Plant material
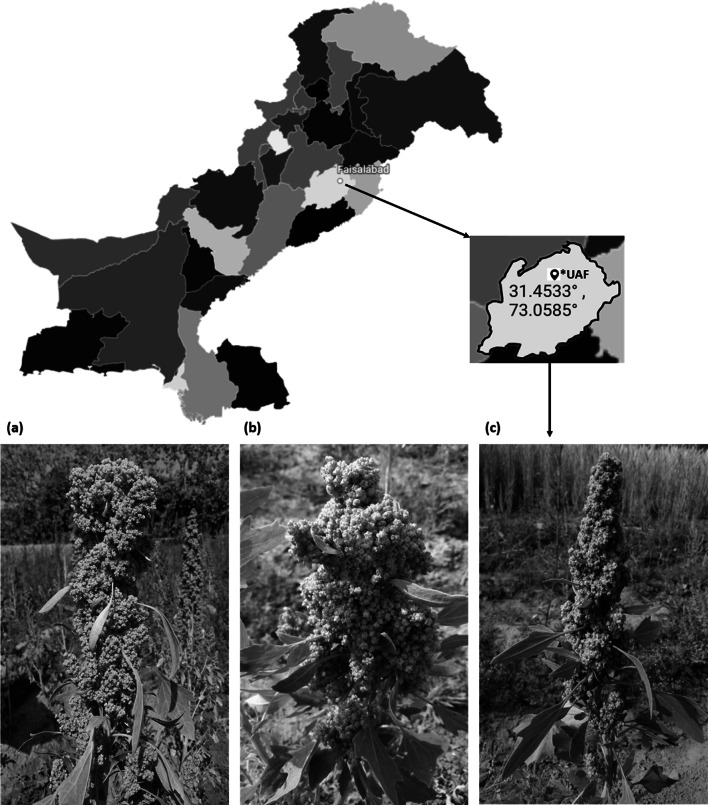
Three accessions of Chenopodium quinoa, well adapted to the agroecological zone of Pakistan located between geographical coordinates 31° 25ʹ 7.3740″ N (north) latitude and 73° 4ʹ 44.7924″ E (east) longitude, were used in this study and map was generated using datawrapper online tool (https://www.datawrapper.de/). These accessions (SPrecm, CHEN-33, and D-12175) were chosen based on excellent quality characteristics. However, with morphological discrepancies in panicle shape, leafiness, and panicle density (Fig. 1). The freshly harvested seeds of each accession were rinsed well with distilled water, dried with a paper towel with gentle tapping, and stored at − 20 °C for downstream analyses. Three biological and three analytical replications (n = 3) of each accession (SPrecm, CHEN-33, and D-12175) were used in this research study.
Fig. 1.
Chenopodium quinoa accessions a SPrecm**, b D-12175***, and c CHEN-33**** with morphological discrepancies in panicle architecture, grown in the agroecological zone of Pakistan. *University of Agriculture, Faisalabad. **Yellow, Intermediate, compact panicle with less leaves. ***Green, glomerulate, compact panicle with less leaves. ****Green, intermediate, intermediate panicles with less leaves (color figure online)
Metabolic profiling
Preparation of quinoa extract samples: The dried seed samples were ground in a mortar and pestle to make fine powder and then processed to prepare metabolite-rich extract, adopting the protocol described earlier (Sharma et al. (2008).
LC–MS analysis: The extraction samples were subjected to liquid chromatography-mass spectrometry (LC–MS) analysis using a linear ion trap mass spectrometer (Model; LTQ XL ThermoScientific, USA) equipped with an electrospray ionization (ESI) source. The LC–MS-MS analysis for metabolomic profiling analysis was conducted at a 10 μl/min flow rate under negative and positive scan ion modes with a scanning mass range of 50–2000 m/z. Peaks were selected for fragmentation (MS/MS) using collision-induced dissociation (CID) energy ranging from 2 to 20 and analyzed using Xcalibur2.0.7.
Data analysis: For metabolomic profiling, Metabolanalyst 5.0 (www.metaboanalyst.ca) was employed to perform principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA) (Xia et al. 2015) for demonstrating and comparing identified metabolites for contributing variations to three quinoa accessions (SPrecm, CHEN-33, and D-12175). Peak intensities of distinguishing ions and identified metabolites were loaded into MetaboAnalyst for exploratory statistical analysis. The datasets were then subjected to Pareto-scaling to reduce systematic variation within the features after the data had undergone normalization and log transformation (Xia et al. 2009). Further, a Heatmap was generated from TBtool for comparative metabolomic profiling. The Pearson correlation analysis was also performed to identify the significant correlation in metabolites. MetaboAnalyst 5.0 was used for Metabolite Set Enrichment Analysis (MSEA) and Metabolomic Pathway Analysis (MetPA).
Nutritional profiling
Mineral analysis
Sample preparation Sample preparation was performed using the digestion method. For that, 1 g of sample was weighed and shifted in the flask with a 10 ml mixture of HNO3 and HCLO3 (7:3 V/V). The solution was heated on hot plates at 180 °C until the solution became transparent (white fumes appeared). After that, the solution was cooled and filtered using Whatman filter paper No. 1. The filtrate was decanted into a volumetric flask and diluted with double distilled water to make a final volume of up to 100 ml for further analysis.
Mineral analysis Mineral analysis for micronutrients (copper, iron and zinc, Zn) and macronutrients (calcium and magnesium) was performed using an atomic absorption spectrophotometer (Hitachi Polarized Zeeman AAS, Z-8200, Japan). The concentration of minerals in the prepared samples was determined by following the conditions described in AOAC (1990).
Proximate analysis
The proximate analysis was conducted by adopting the methodology described by the Association of Official Analytic Chemists (AOAC 2006).
Determination of moisture content: Fresh quinoa seeds were taken and weighed on a dry petri plate (W1). The moisture content was determined by drying the samples at 60 °C for 48 h in an oven. After drying, the seeds were ground into a fine powder and weighed again (W2). The given formula was used to determine moisture content.
Determination of ash content: A blank, oven-dried crucible was weighed, followed by 1 g of sample in an already weighed blank crucible labeled W1. Each sample was charred on a heater and kept in a preheated muffle furnace at 350 °C for 24 h. After the burnt-through phase, the sample was removed from the furnace and cooled in a desiccator. The sample was weighed and labeled W2, and the ash content was calculated using the following formula.
Determination of crude protein content: The crude protein was determined using the Kjeldahl method. The seeds were ground into powder for sample digestion, and a 1 g sample was weighed and transferred to a digestion flask. Then, 5 g of the digestion mixture (CuSO4; MgSO4) was added to the digestion flask and heated for boiling under a fume hood. While boiling, 25 ml of concentrated H2SO4 was added to the flask. The digestion proceeded until the mixture turned light blue-green. After digestion, the sample mixture was cooled down and then subjected to the Kjeldahl apparatus for fractional distillation. Then, 10 ml of digested sample and 10 ml of NaOH were added into the distillation tube. Ammonia (NH3) was generated in the receiving flask and unruffled as ammonium hydroxide (NH4OH).
Furthermore, 10 ml of boric acid (H3BO4) and a few drops of methyl red as indicators were added to the receiving flask below the condenser. During distillation, the distillate was titrated against 0.1 N H2SO4 until a pink color appeared. As mentioned above, the process was repeated, and readings were noted. The crude protein content was determined using the following formula.
Determination of crude fat content: The crude fat was determined using a Soxhlet apparatus. One gram of sample was weighed, wrapped in filter paper, and placed in a Soxhlet apparatus with 400 ml of petroleum ether. The sample was heated for approximately 3 h until petroleum ether was circulated and reminisced via condensation in this apparatus. After the petroleum ether collection, the flask with extract was removed from the apparatus and cooled down. Then, the extract was poured into the already weighed petri plate, kept in the oven at 72 °C for 15 min, weighed again in the desiccator for 5 min, and weighed again. The crude fat was calculated as follows:
Determination of crude fiber content: After fat extraction, defatted samples were taken to detect crude fiber using the acid and base digestion method. For protein removal, 200 ml of H2SO4 (1.25%) was added to a beaker, stirred uninterruptedly, heated for 30 min, and filtered using a muslin cloth. The residues on muslin cloth were taken into the beaker and treated with 200 ml NaOH (1.25%) following the same procedure for carbohydrate removal. The residues were then shifted into an already-weighed crucible and dried for over 24 h. After that, the crucible was placed in a furnace for 24 h at 350 °C, kept in a desiccator, and weighed again. The crude fiber was calculated using the following formula.
Data analysis: For nutritional profiling, all measurements were taken in triplicate, and data was analyzed using HSD Tukey’s test at a significance level of p ≤ 0.05 in Statistix 8.0 software.
Results
Metabolomic profiling, nutritional profiling, and mineral analysis were performed to analyze the phytochemistry and nutritional chemistry of C. quinoa accessions.
Metabolic profiling: Metabolomic profiling with the LC–MS technique is proposed to be particularly significant in plants due to the highly complex biochemistry of plants, which includes a variety of semipolar molecules, including important secondary metabolite groups, which may be best separated and detected by this technique (El Sayed et al. 2020). This study performed metabolic profiling using liquid chromatography-mass spectrometry (LC–MS) analysis. The mass-to-charge ratio (m/z) of precursor ion peaks and MS/MS fragment ion peaks were consulted with the literature and mass spectrometry search tool (MASST) (Wang et al. 2020) for detecting metabolic compounds.
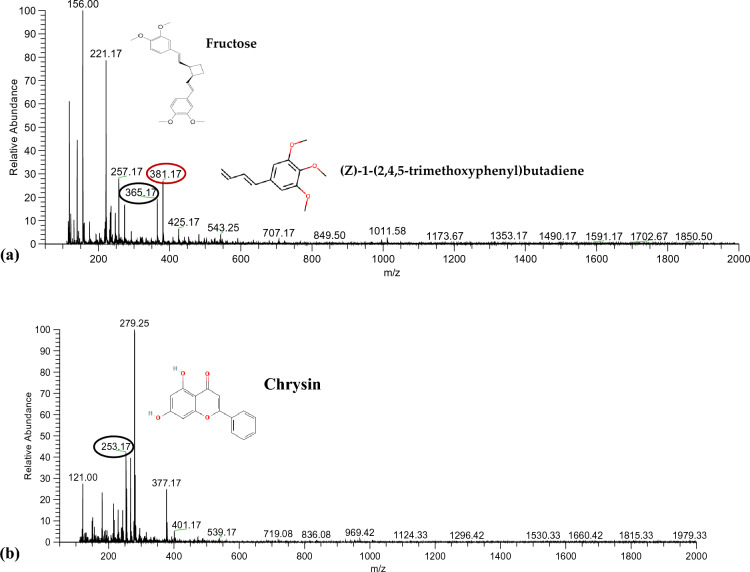
LC–ESI–MS analysis of SPrecm seed extract: LC–MS analysis of SPrecm was performed to assess the phytoconstituents, including phenolics, flavonoids, phenylbutenoids, and other organic compounds (Table 1). The full mass spectrum in positive ion mode indicated the five most abundant high precursor ion peaks at m/z 381.17, 365.17, 257.17, 221.17, and 156.00 (Fig. 2a). The MS–MS-ESI of the precursor ion peak at m/z 381.17 showed the presence of cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, a phenylbutenoid dimer (Fig. S1a). The peak at m/z 381.17 fragmented into the two most abundant fragment ion peaks at m/z 219.00 and 201.00, which could be due to the loss of 1,2-dimethoxy-4-vinylbenzene and 1,2-dimethoxybenzene, respectively. The MS–MS-ESI of the precursor ion peak at m/z 365.17 [2 M + Na–H2O]+ indicated the presence of fructose that gave two major fragment ion peaks at m/z 203.00 and 185.00 that could be a characteristic pattern of fructose (Fig. S1b). The MS–MS-ESI of the precursor ion peak at m/z 257.17 indicated the presence of pinocembrin. The peak at m/z 257.17 fragmented into one most abundant fragment ion peak at m/z 140.00, possibly due to the loss of the O-methyl group in the A ring (Fig. S1c). The MS–MS-ESI of the precursor ion peak at m/z 221.17 depicted the presence of (Z)-1-(2,4,5-trimethoxyphenyl) butadiene and phenylbutenoid dimers. The peak at m/z 221.17 fragmented into the two most abundant fragment ion peaks at m/z 104.17 and 60.00, which could be a loss of methoxyl or methyl radicals from the aromatic ring of the precursor ion (Fig. S1d). The MS–MS-ESI of the precursor ion peak at m/z 156.00 corresponds to the presence of alanine (an amino acid). The peak at m/z 156.00 resulted in the three most abundant fragment ion peaks at 156.00, 74.08, and 56.92, which could be due to the loss of pyrolysis products (3,6-dimethylpiperazine-2,5-dione, 1,3,6-trimethylpiperazine-2,5-dione, and O-methylalanylalanine dipeptide) of Alanine (Fig. S1e).
Table 1.
Identification of phytochemical constituents in seed extracts of three quinoa accessions (SPrecm, CHEN-33, and D-12175)
| Quinoa accessions | tR (min) |
Ion mode | Precursor ion peak (m/z) | ESI–MS-MS | Detected compounds | Type of compound | Molecular formula | Molecular weight (g/mol) | Abbreviations |
|---|---|---|---|---|---|---|---|---|---|
| SPrecm | 4.93 | + ve | 381.17 | 219.00, 201.00 | Cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane | Phenylbutenoid | C24H28O4 | 380.48 | 3,4-dmoCB |
| 4.59 | + ve | 365.17 | 203.00, 185.00 | Fructose | Saccharide | C6H12O6 | 180.16 | Fru | |
| 4.10 | + ve | 257.17 | 140.00 | Pinocembrin | Flavonoid | C15H12O4 | 256.25 | Pin | |
| 2.36 | + ve | 221.17 | 104.17, 60.00 | (z)-1-(2,4,5-trimethoxyphenyl)butadiene | Phenylbutenoid | C13H16O3 | 220.26 | Z-2,4-tmpBD | |
| 1.56 | + ve | 156.00 | 156.00, 74.08, 56.92 | Alanine | Amino acid | C3H7NO2 | 89.09 | Ala | |
| 17.27 | − ve | 377.17 | 179.00, 161.00 | Oleuropein aglycone | Phenolic | C19H22O8 | 378.4 | OA | |
| 15.13 | − ve | 279.25 | 261.17, 259.17, 243.00 | Paniculide C | Terpenoid | C15H18O5 | 278.30 | P_C | |
| 12.17 | − ve | 253.17 | 253.25, 193.00 | Chrysin | Flavonoid | C15H10O4 | 254.24 | Chr | |
| 11.27 | − ve | 121.00 | 120.92, 101.00 | 4-(1-Benzofuron-5-yl) benzaldehyde | Phenolic | C15H10O2 | 222.24 | 4,1-B-5-BA | |
| CHEN-33 | 3.32 | + ve | 381.17 | 219.00, 200.00 | Cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane | Phenylbutenoid | C24H28O4 | 380.48 | 3,4-dmoCB |
| 2.98 | + ve | 284.33 | 85.00, 268.25, 266.00 | Luteolin | Flavonoid | C15H10O6 | 286.24 | Lut | |
| 2.43 | + ve | 257.17 | 140.00 | Pinocembrin | Flavonoid | C15H12O4 | 256.25 | Pin | |
| 1.47 | + ve | 156.00 | 156.00, 74.00, 56.92 | Alanine | Amino acid | C3H7NO2 | 89.09 | Ala | |
| 9.64 | − ve | 393.33 | 393.33, 375.33, 313.00 240.92 | Sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl)methyl hydrogen phosphate | Flavonoid | C26H36O6 | 438.5 | S-dhy-mHP | |
| 8.30 | − ve | 377.17 | 178.92, 160.92 | Oleuropein aglycone | Phenolic | C19H22O8 | 378.4 | OA | |
| 7.73 | − ve | 279.33 | 261.17, 259.33, 235.08 | Paniculide C | Terpenoid | C15H18O5 | 278.30 | P–C | |
| 7.22 | − ve | 255.25 | 237.08, 211.00 | Palmitic acid | Fatty acid | C16H32O2 | 256.42 | PA | |
| 7.11 | − ve | 241.25 | 241.33, 223.08, 209.00, 197.08 | Elenolic acid | Fatty acid | C11H14O6 | 242.22 | EA | |
| 6.49 | − ve | 121.00 | 121.08, 101.00 | 4-(1-Benzofuron-5-yl) benzaldehyde | Phenolic | C15H10O2 | 222.24 | 4,1-B-5-BA | |
| D-12175 | 6.64 | + ve | 917.72 | 636.58, 622.58, 317.25 | Quercetin | Flavonoid | C15H10O7 | 302.23 | Qur |
| 5.10 | + ve | 453.42 | 435.25, 421.25, 392.42, 371.42, 355.17 | 3-hydroxyphloretin-3’-C-glucoside | Terpenoid | C21H25O11 | 452.4 | hyP-3-C-Glu | |
| 4.79 | + ve | 437.42 | 407.25 and 303.17 | Kurarinone | Flavonoid | C10H14N5NaO7P | 370.21 | Kur | |
| 3.68 | + ve | 284.42 | 268.33, 266.17 | Luteolin | Flavonoid | C15H10O6 | 286.24 | Lut | |
| 2.19 | + ve | 199.00 | 199.17, 181.00, 134.92, 109.00 | Rosmarinic acid | Phenolic | C18H16O8 | 360.3 | RA | |
| 10.06 | − ve | 279.33 | 261.17, 259.17, 243.25 | Paniculide C | Terpenoid | C15H18O5 | 278.30 | P–C | |
| 9.52 | − ve | 255.33 | 237.17, 211.08 | Palmitic acid | Fatty acid | C16H32O2 | 256.42 | PA |
Fig. 2.
LC‒MS analysis for metabolomic profiling of SPrecm: a Metabolomic profiling in positive-ion mode showing the presence of cis-1,2-bis-[(E)-3,4-dimethoxystyryl] cyclobutane, fructose, pinocembrin, (Z)-1-(2,4,5-trimethoxyphenyl)butadiene (E-12 or Z-12), and protocatechuic acid at precursor ion peaks m/z 381.17, 365.17, 257.17, 221.17, and 156.00, respectively. b Metabolomic profiling in negative-ion mode shows the presence of oleuropein aglycone, paniculide C, chrysin, and 4-(1-benzofuron-5-yl) benzaldehyde at precursor ion peaks m/z 377.17, 279.25, 253.17, and 121.00, respectively
The full mass spectrum in negative ion mode indicated the four most abundant high precursor ion peaks at m/z 377.17, 279.25, 253.17, and 121.00 (Fig. 2b). The MS–MS-ESI of the precursor ion peak at m/z 377.17 [C19H21O8]− showed the presence of deprotonated oleuropein aglycone (a phenolic compound). The peak at m/z 377.17 fragmented into one of the most abundant fragment ion peaks at m/z 179.00 and 161.00, which could result from 2‐butenal (C4H6O) loss from the precursor ion (Fig. S2a). The MS–MS-ESI of the precursor ion peak at m/z 279.25 corresponds to paniculide C; saponins belong to the class of terpenoids. The peak at m/z 279.25 presented the following most abundant fragmented ion peaks at m/z 261.17, 259.17, and 243.00, which could be due to [M + H − H2O]+, [M + H − H2O − CO]+, and [M + H − H2O − 2CO]+ ion adducts, respectively (Fig. S2b). The MS–MS-ESI of the precursor ion peak at m/z 253.17 indicated the presence of chrysin, a flavonoid. The peak at m/z 253.17 fragmented into the two most abundant fragment ion peaks at m/z 253.25 and 193.00, possibly due to the loss of rings A and B of dihydroxyflavones from the precursor compound (Fig. S2c). The MS–MS-ESI of the precursor ion peak at m/z 121.00 indicated the presence of 4-(1-benzofuron-5-yl) benzaldehyde, which belongs to the hydroxybenzaldehyde class. The peak at 121.00 fragmented into the two most abundant fragment ion peaks at 120.92 and 101.00, corresponding to 2,3-dihydrobenzofuran and 4-hydroxybenzaldehyde, which could be due to the removal of furan rings and H2O molecules (Fig. S2d).
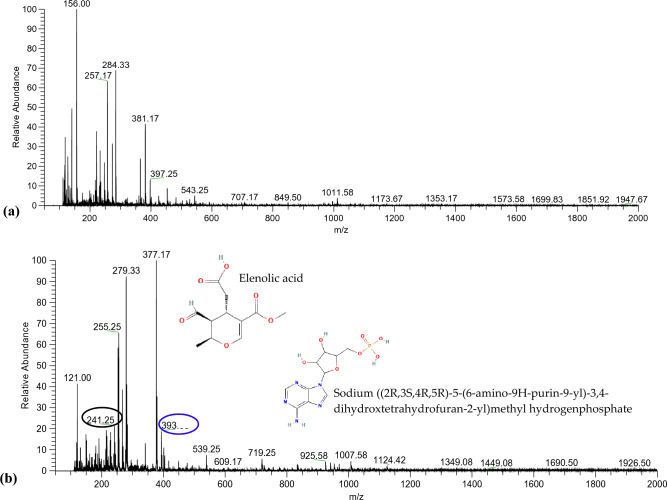
LC–ESI–MS analysis of CHEN-33 seed extract: LC–MS analysis of CHEN-33 was performed to assess the phytoconstituents, including phenolics, flavonoids, terpenoids, glycosides, and other organic compounds (Table 1). The full mass spectrum in positive ion mode indicated the four most abundant high precursor ion peaks at m/z 381.17, 284.33, 257.17, and 156.00 (Fig. 3a). The MS–MS-ESI of the precursor ion peak at m/z 381.17 showed the presence of cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, a phenylbutenoid dimer. The peak at m/z 381.17 fragmented into the two most abundant fragment ion peaks at m/z 219.00 and 200.00, which could be due to the loss of 1,2-dimethoxy-4-vinylbenzene and 1,2-dimethoxybenzene, respectively (Fig. S3a). The MS–MS-ESI of the precursor ion peak at m/z 284.33 depicted the presence of luteolin, which belongs to the flavonoid class. The peak at m/z 284.33 resulted in the two most abundant fragment ion peaks at m/z 85.00, 268.25, and 266.00, which could be due to the loss of deprotonated aglycone (Fig. S3b). The MS–MS-ESI of the precursor ion peak at m/z 257.17 indicated the presence of pinocembrin. The peak at m/z 257.17 fragmented into one most abundant fragment ion peak at m/z 140.00, possibly due to the presence of the O-methyl group in the A ring (Fig. S3c). The MS–MS-ESI of the precursor ion peak at m/z 156.00 showed the presence of alanine (an amino acid). The peak at m/z 156.00 resulted in the three most abundant fragment ion peaks at 156.00, 74.00, and 56.92, which could be due to the loss of pyrolysis products (3,6-dimethylpiperazine-2,5-dione, 1,3,6-trimethylpiperazine-2,5-dione, and O-methylalanylalanine dipeptide) of Alanine (Fig. S3d).
Fig. 3.
LC–MS analysis for metabolomic profiling of CHEN-33: a Metabolomic profiling in positive-ion mode showing the presence of cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, luteolin, pinocembrin, and alanine at the precursor ion peaks at m/z 381.17, 284.33, 257.17, and 156.00, respectively. b Metabolomic profiling in negative-ion mode showing the presence of sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl) methyl hydrogen phosphate, oleuropein aglycone, paniculide C, palmitic acid, deprotonated elenolic acid, and 4-(1-benzofuron-5-yl) benzaldehyde at precursor ion peaks m/z 393.33, 377.17, 279.33, 255.25, 241.25, and 121.00, respectively
The full mass spectrum in negative ion mode indicated the six most abundant high precursor ion peaks at m/z 393.33, 377.17, 279.00, 255.25, 241.25, and 121.00 (Fig. 3b). The MS–MS-ESI of the precursor ion peak at m/z 393.33 showed the presence of sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl)methyl hydrogen phosphate. The peak at m/z 393.33 fragmented into the four most abundant fragment ion peaks at m/z 393.33, 375.33, 313.00, and 240.92, which could be due to [M-Na]+ from the precursor compound (Fig. S4a). The MS–MS-ESI of the precursor ion peak at m/z 377.17 showed the presence of oleuropein aglycone, which belongs to phenolics. The peak at m/z 377.17 fragmented into the two most abundant fragment ion peaks at m/z 178.92 and 160.92, possibly due to the neutral loss of the C4H6O moiety (Fig. S4b). The MS–MS-ESI of the precursor ion peak at m/z 279.33 corresponds to paniculide C, a terpenoid. The peak at m/z 279.33 presented the following most abundant fragmented ion peaks at m/z 261.17, 259.33, and 235.08, which could be due to [M–H−H2O]−, [M−H−H2O−CO]−, and [M−H−H2O−2CO]− ion adducts, respectively (Fig. S4c). The MS–MS-ESI of the precursor ion peak at m/z 255.25 showed the presence of palmitic acid. The peak at m/z 255.25 resulted in the two most abundant fragment ion peaks at m/z 237.08 [M-H2O]− and 211.00 [M-H-CO2]− due to the neutral loss of H2O or CO2, which is a distinctive characteristic of acids (Fig. S4d). The MS–MS-ESI of the precursor ion peak at m/z 241.25 showed the presence of deprotonated elenolic acid. The peak at m/z 241.25 fragmented into the three most abundant fragment ion peaks at m/z 241.33, 223.08, 209.00, and 197.08 that could appear due to hydrolysis (Fig. S4e). The MS–MS-ESI of the precursor ion peak at m/z 121.00 depicted the presence of 4-(1-benzofuron-5-yl) benzaldehyde, which belongs to the hydroxybenzaldehyde class. The peak at 121.00 fragmented into the two most abundant fragment ion peaks at 121.08 and 101.00, corresponding to 2,3-dihydrobenzofuran and 4-hydroxybenzaldehyde, which could be due to the removal of furan rings and H2O molecules (Fig. S4f).
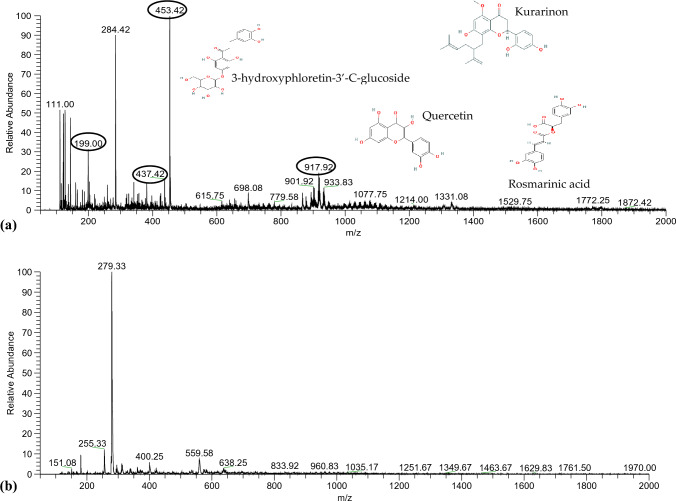
LC–ESI–MS analysis of D-12175 seed extract: LC–MS analysis of D-12175 accession was performed to assess the phytoconstituents, including phenolics, flavonoids, glucosides, and terpenoids (Table 1). The full mass spectrum in positive ion mode indicated the five most abundant high precursor ion peaks at m/z 917.92, 453.42, 437.42, 284.42, and 199.00 (Fig. 4a). The MS–MS-ESI of the precursor ion peak at m/z 917.92 showed the presence of quercetin, which belongs to the flavonoid class. The peak at m/z 917.92 fragmented into the three most abundant fragment ion peaks at m/z 636.58, 622.58, and 317.25, corresponding to 2-rhamnosyl–2-glucosyl quercetin and 3-glucosyl–2-rhamnosyl, which could be due to the loss of glycosyl (-C6H10O5) and rhamnosyl (-C6H10O4) groups from the precursor ion (Fig. S5a). The MS–MS-ESI of the precursor ion peak at m/z 453.42 showed the presence of 3-hydroxyphloretin-3’-C-glucoside (saponin from the terpenoid class). The peak at m/z 453.42 fragmented into the five most abundant fragment ion peaks at m/z 435.25, 421.25, 392.42, 371.42, and 355.17, which could be due to the loss of methyl (CH3) and carboxyl (-COOH) groups of water (H2O) from the precursor ion (Fig. S5b). The MS–MS-ESI of the precursor ion peak at m/z 437.42 showed the presence of kurarinone. The peak at m/z 437.42 fragmented into the two most abundant fragment ion peaks at m/z 407.25 and 303.17, possibly due to the [M-H]+ ion adduct (Fig. S5c). The MS–MS-ESI of the precursor ion peak at m/z 284.42 corresponds to luteolin, which belongs to the flavonoid class. The peak at m/z 284.42 resulted in the two most abundant fragment ion peaks at m/z 268.33 and 266.17 by the loss of deprotonated aglycone (Fig. S5d). The MS–MS-ESI of the precursor ion peak at m/z 199.00 showed the presence of rosmarinic acid, a phenolic compound. The peak at m/z 199.00 presented a fragmentation pattern of m/z 199.17, 181.00, 134.92, and 109.00, validating the presence of rosmarinic acid (Fig. S5e).
Fig. 4.
LC–MS analysis for metabolomic profiling of D-12175: a Metabolomic profiling in positive-ion mode showing the presence of quercetin, 3-hydroxyphloretin-3’-C-glucoside, kurarinone, luteolin, and rosmarinic acid at precursor ion peak m/z 917.92, 453.42, 437.42, 284.42, and 199.00, respectively. b Metabolomic profiling in negative-ion mode shows paniculide C and palmitic acid at the precursor ion peaks at m/z 279.33 and 255.33, respectively
The full mass spectrum of negative ion mode indicated the two most abundant high precursor ion peaks at m/z 279.33 and 255.33 (Fig. 4b). The MS–MS-ESI of the precursor ion peak at m/z 279.33 showed the presence of paniculide C. The peak at m/z 279.33 fragmented into three major fragment ion peaks at m/z 261.17, 259.17, and 243.25, which could be due to the removal of [M + H − H2O]+, [M + H − H2O − CO]+, and [M + H − H2O − 2CO]+ ion adducts, respectively (Fig. S6a). The MS–MS-ESI of the precursor ion peak at m/z 255.33 showed the presence of palmitic acid. The peak at m/z 255.33 resulted in the two most abundant fragment ion peaks at m/z 237.17 [M−H2O]− and 211.08 [M–H–CO2]− due to the neutral loss of H2O or CO2 (Fig. S6b).
Metabolomic profiling determined the presence of some common metabolites in two samples that were not detected in another sample. Similarly, some metabolites were found to be unique in one sample. LC–MS analysis in positive-ion mode indicated the presence of cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, pinocembrin, and Alanine in SPrecm and CHEN-33, and these compounds were not detected in D-12175. However, luteolin was found in CHEN-33 and D-12175 but was not detected in SPrecm. Moreover, two compounds, fructose and (Z)-1-(2,4,5-trimethoxyphenyl) butadiene, were unique to SPrecm and were not found in CHEN-33 and D-12175. Four compounds (quercetin, 3-hydroxyphloretin-3’-C-glucoside, kurarinone, and rosmarinic acid) were unique to the D-12175 accession and were not found in SPrecm and CHEN-33 (Table 1).
Similarly, metabolomic profiling in negative-ion mode indicated the presence of oleuropein aglycone and 4-(1-benzofuron-5-yl) benzaldehyde in SPrecm and CHEN-33 but not in D-12175, while palmitic acid was detected in SPrecm and D-12175 but not in CHEN-33. Similarly, chrysin was unique to SPrecm and was not found in CHEN-33 and D-12175. The two compounds sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl) methyl hydrogen phosphate and deprotonated elenolic acid were unique to CHEN-33 and were not detected in SPrecm and D-12175 (Table 1).
Metabolic profiling using liquid chromatography-mass spectrometry (LC–MS) analysis identified seventeen metabolites, including flavonoids, phenolics, fatty acids, terpenoids, phenylbutenoid dimers, amino acids, and saccharides. Of these, eight metabolic compounds were reported in this study for the first time in quinoa: cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, (Z)-1-(2,4,5-trimethoxyphenyl) butadiene, oleuropein aglycone, 4-(1-benzofuron-5-yl) benzaldehyde, sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydrox-tetrahydrofuran-2-yl) methyl hydrogen phosphate, elenolic acid, and kurarinone (Table 1).
Statistical analyses
Principal component analysis was performed to unveil the similarities and differences among three quinoa accessions (CHEN-33, D-12175, and SPrecm) based on their detected metabolites. A PCA plot was constructed to minimize the data dimensionality, thereby elucidating the variance within the data as orthogonal PCs (Fig. 5). The calculated model was based on components with 74.8% variation accounted for PC1 and PC2. PC1 explained 46.3% variations, whereas PC2 explained 28.5% of the total variance. Despite the differences observed in chromatograms, the PC scores plot grouped three accessions (CHEN-33, D-12175, and SPrecm) into distinct clusters based on their metabolites.
Fig. 5.
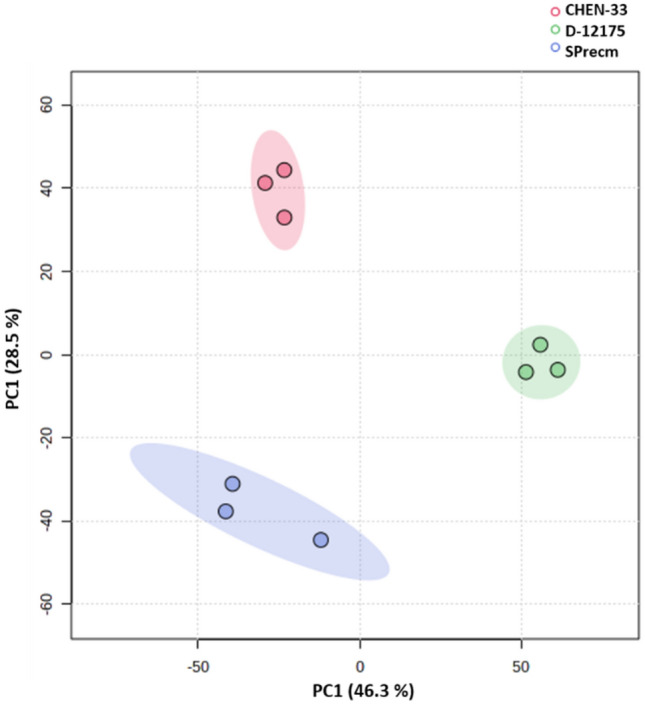
Principal components (PCs) scores plot based on the Pareto-scaled dataset. Principal component analysis (PCA) shows the differences among the three accessions and the distribution of samples in each accession: CHEN-33, D-12175, and SPrecm
The metabolomics composition in the three accessions was further investigated using the variable importance in projection (VIP) scores generated by PLS-DA to give more insights into the separation of accessions (CHEN-33, D-12175, and SPrecm) based on their metabolic profiles (Fig. 6). Metabolites that retain higher VIP scores contribute more significantly to the difference among groups. In contrast, those with lower VIP scores have less impact on the model in driving substantial differences among groups. VIP scores of metabolites elucidated ten metabolites (palmitic acid, chrysin, fructose, elenolic acid, luteolin, (z)-1-(2,4,5-trimethoxyphenyl)butadiene, sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl) methyl hydrogen phosphate, pinocembrin, Cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, and Paniculide) with VIP scores of more than 0.5 (VIP ≥ 0.5). They demonstrated that these metabolites contributed to three accessions, including CHEN-33, D-12175, and SPrecm (Fig. 6).
Fig. 6.
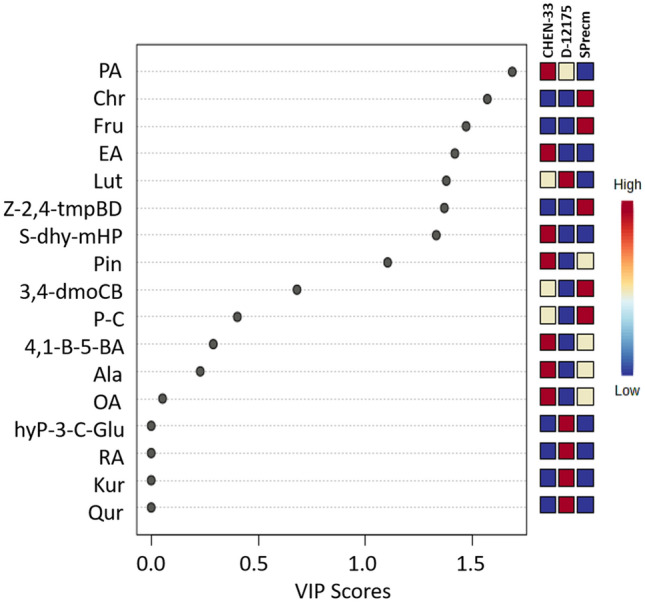
Variable importance in projection (VIP) scores plot: The VIP scores generated using partial least discriminant analysis (PLS-DA) show the metabolites at VIP ≥ 0.5 that contribute variation among accessions (CHEN-33, D-12175, and SPrecm)
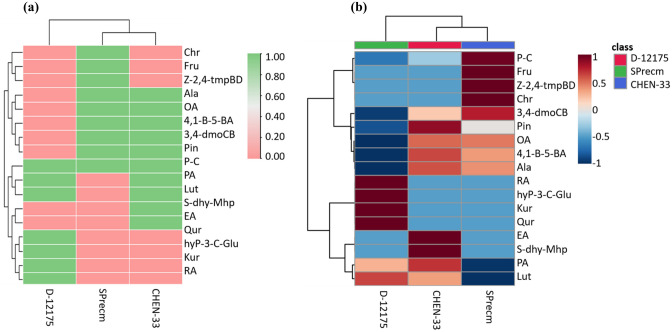
The hierarchical clustering heatmap of differentially expressed metabolites in three accessions (CHEN-33, D-12175, and SPrecm) grouped the metabolites into two major clusters (Fig. 7a&b). Cluster 1 comprised of nine metabolites. Among nine metabolites four metabolites, rosmarinic acid, kurarinone, 3-hydroxyphloretin-3’-C-glucoside, and quercetin, were unique to D-12175 and were absent in SPrecm and CHEN-33, while two metabolites, elenolic acid and sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl)methyl hydrogen phosphate, were unique to CHEN-33 and did not present in D-12175 and SPrecm. Palmitic acid and luteolin were two metabolites common in D-12175 and CHEN-33; however, paniculide C was found among all three accessions. Cluster II comprised of eight metabolites; among them three metabolites, chrysin, fructose, and (z)-1-(2,4,5-trimethoxyphenyl)butadiene, were unique to SPrecm whereas five metabolites, alanine, oleuropein aglycone, 4-(1-Benzofuron-5-yl) benzaldehyde, Cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobytane, and pinocembrin, were present in both accessions (CHEN-33 and SPrecm) and absent in D-12175 (Fig. 7a). Moreover, the cluster heatmap showed that SPrecm and CHEN-33 are closely related; however, D-12175 shows a distinct metabolic profile that contributes diversity to other accessions, including SPrecm and CHEN-33 (Fig. 7b).
Fig. 7.
Heat map clustering analysis: a showing Hierarchical cluster analysis of differentially expressed metabolites in three quinoa accessions (SPrecm, CHEN-33, and D-12175), where the green and pink color represents the presence and absence of a particular compound in each accession, respectively, b showing distinct metabolic profile contributing diversity among quinoa accessions
Correlation analysis Further, correlation analysis was performed among the differentially expressed potential metabolites depicted that rosmarinic acid was positively correlated with quercetin (r = 0.956; p < 0.001), kurarinone was positively correlated with quercetin (r = 0.934; p < 0.001) and rosmarinic acid (r = 0.986; p < 0.001), while chrysin and sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl)methyl hydrogenphosphate were found to be positively correlated with (z)-1-(2,4,5-trimethoxyphenyl)butadiene (r = 0.905; p < 0.001) and elenolic acid (r = 0.981; p < 0.001), respectively (Fig. 8). Further, quercetin was negatively correlated with both alanine (r = − 0.912; p < 0.001) and oleuropein aglycone (r = − 0.910; p < 0.001) and (z)-1-(2,4,5-trimethoxyphenyl)butadiene was negatively correlated with palmitic acid (r = − 0.940; p < 0.001) and luteolin (r = − 0.960; p < 0.001) (Table S1).
Fig. 8.
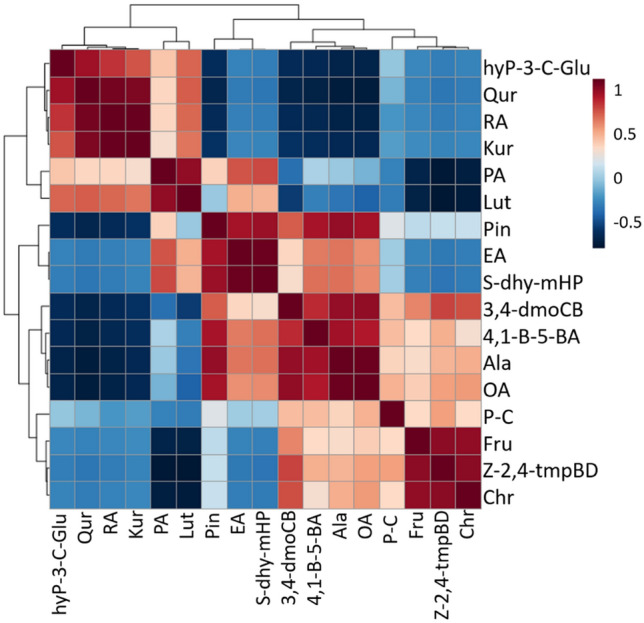
The Pearson correlation analysis unveiled the significant correlations among metabolites in three quinoa accessions (CHEN-33, D-12175, and SPrecm). The correlation strength is represented by blue (positive correlation) and red color (negative correlation). (p ≤ 0.001)
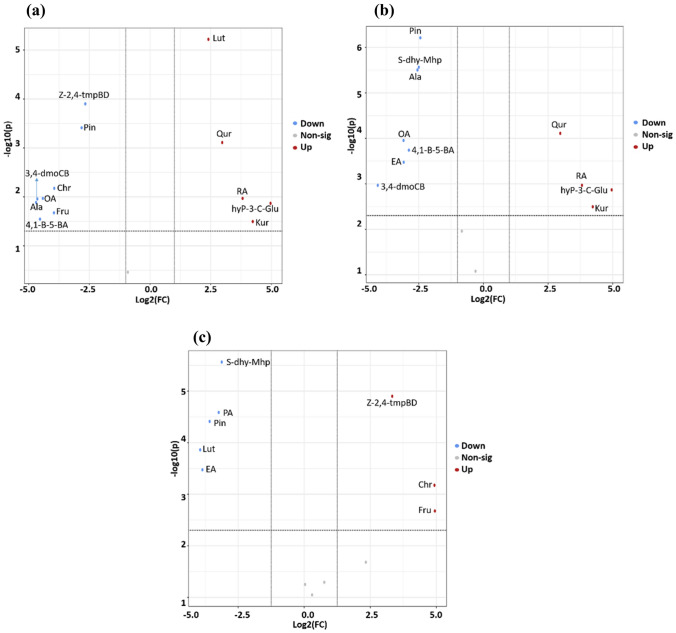
Furthermore, the differential pattern of metabolite accumulation in three quinoa accessions (SPrecm, CHEN-33, and D-12175) was analyzed using Volcano plot (Fig. 9). Metabolites with fold change ≥ 2 were referred to as differentially expressed metabolites. Results showed there were thirteen (five upregulated and eight downregulated) differentially expressed metabolites between D-12175 and SPrecm, eleven (four upregulated and seven downregulated) between D-12175 and CHEN-33, and eight (three upregulated and five downregulated) between SPrecm and CHEN-33.
Fig. 9.
Differentially expressed metabolites among three quinoa accessions: the volcano plots showing expression levels of differential metabolites between D-12175 and SPrecm (a), D-12175 and CHEN-33 (b) SPrecm and CHEN-33 (c). Red, blue, and grey dots indicate the up-, down-regulated, and non-significant metabolites, respectively (color figure online)
Moreover, the most significant metabolites were identified in three quinoa accessions. A comparison between D-12175 and SPrecm displayed a change factor ranging from 0.04 to 31.19 fold for thirteen metabolites, including 3-hydroxyphloretin-3’-C-glucoside (31.19-fold), kurarinone (18.77-fold), rosmarinic acid (14.05-fold), quercetin (7.85-fold), palmitic acid (7.39-fold), luteolin (5.3-fold) that were significantly upregulated, while (z)-1-(2,4,5-trimethoxyphenyl)butadiene (0.16-fold), pinocembrin (0.14-fold), chrysin (0.06-fold), fructose (0.06-fold), oleuropein aglycone (0.05-fold), Cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane (0.04-fold), alanine (0.04-fold), and 4-(1-Benzofuron-5-yl) benzaldehyde (0.04-fold) that were downregulated between D-12175 and SPrecm (Fig. 9a & Table S2a).
A comparison between D-12175 and CHEN-33 revealed a change factor ranging from 0.03 to 16.97 fold for eleven metabolites (Fig. 9b). Quercetin (7.85-fold), rosmarinic acid (14.05-fold), kurarinone (18.77-fold), and 3-hydroxyphloretin-3’-C-glucoside (31.19-fold) were significantly upregulated while, pinocembrin (0.18-fold), sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl) methyl hydrogenphosphate (0.17-fold), alanine (0.17-fold), oleuropein aglycone (0.12-fold), 4-(1-Benzofuron-5-yl) benzaldehyde (0.14), elenolic acid (0.12), and cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane (0.06) were identified as significantly downregulated metabolites between D-12175 and CHEN-33 (Fig. 9b & Table S2b). A comparison between SPrecm and CHEN-33 revealed a change factor ranging from 0.11 to 15.54 fold for eleven metabolites (Fig. 9c). Fructose (15.54-fold), chrysin (15.39-fold), and (z)-1-(2,4,5-trimethoxyphenyl) butadiene (6.34-fold) metabolites were found significantly upregulated while sodium ((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxtetrahydrofuran-2-yl)methyl hydrogenphosphate (0.18-fold), palmitic acid (0.17-fold), pinocembrin (0.14-fold), luteolin (0.11-fold), elenolic acid (0.12-fold) were downregulated between SPrecm and CHEN-33 (Fig. 9c & Table S2c).
Metabolic enrichment and metabolic pathway analyses
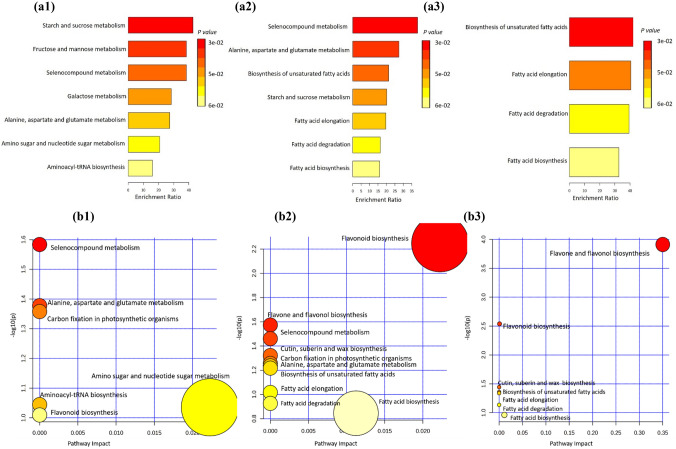
The metabolite enrichment and pathways analyses were performed using MetaboAnalyst 5.0 to identify the most significantly impacted metabolic pathways among three quinoa accessions (CHEN-33, D-12175, and SPrecm) (Fig. 10). Quantitative enrichment analysis (MESA) was employed to ascertain the key pathways with a score of more than 0.05 (p ≤ 0.05) that were significantly impacted across the three groups (Fig. 10a). Results showed starch and sucrose metabolism, fructose and mannose metabolism, selenocompound metabolism, and galactose metabolism were significantly enriched pathways in SPrecm (a1), selenocompound metabolism, alanine, aspartate, and glutamate metabolism, biosynthesis of unsaturated fatty acids, and fatty acid elongation were highly enriched in CHEN-3 (a2). In contrast, biosynthesis of unsaturated fatty acids and fatty acid elongation significantly enriched metabolic pathways in D-12175 (a3). Further, for metabolomic pathway analysis, the pathway impact value was computed using the topology pathway analysis, and pathways at a significance level of p ≤ 0.05 were considered significant metabolic pathways (Fig. 10b). Selenocompound metabolism was detected as a significant metabolic pathway in all accessions. In contrast, carbon fixation in photosynthetic organisms and alanine, aspartate, and glutamate metabolism were significant pathways in SPrecm (b1), flavonoid biosynthesis, flavone and flavonol biosynthesis, and cutin, suberin, and wax biosynthesis were significant metabolic pathways in CHEN-33 (b2) and D-12175 (b3).
Fig. 10.
Metabolic set enrichment and pathway analyses: a Metabolic set enrichment analysis a1 displaying starch and sucrose metabolism, fructose and mannose metabolism, selenocompound metabolism, and galactose metabolism as significantly enriched pathways in SPrecm; a2 C displays Selenocompound metabolism, alanine, aspartate, and glutamate metabolism, biosynthesis of unsaturated fatty acids, and starch and sucrose metabolism as highly enriched in CHEN-3; a3 E displays unsaturated fatty acids and fatty acid elongation as enriched metabolic pathways in D-12175, b Metabolic pathway analysis b1 displaying selenocompound metabolism, alanine, aspartate and glutamate metabolism, carbon fixation in photosynthetic organisms as significant pathways in SPrecm; b2 displays flavonoid biosynthesis, flavone and flavonol biosynthesis, selenocompound metabolism and cutin, suberin and wax biosynthesis as significant metabolic pathways in CHEN-33; b3 displays flavone and flavonol biosynthesis, flavonoid biosynthesis, and cutin, suberin and wax biosynthesis as significant metabolic pathways in D-12175
Nutritional profiling
Nutritional profiling approaches are developed to evaluate calorie content and nutritional composition, including macronutrients, micronutrients, and nutrient content (carbohydrates). Protein, fat, fiber, moisture, and ash) are present in a given food (Mondal et al. 2023). The quality and quantity of nutrients are essential for selecting plant species with nutraceutical significance, such as quinoa, for plant improvement programs (Contreras-Jiménez et al. 2019). This study assessed the nutritional value of seeds from three quinoa accessions (SPrecm, CHEN-33, and D-12175) through mineral and proximate analysis.
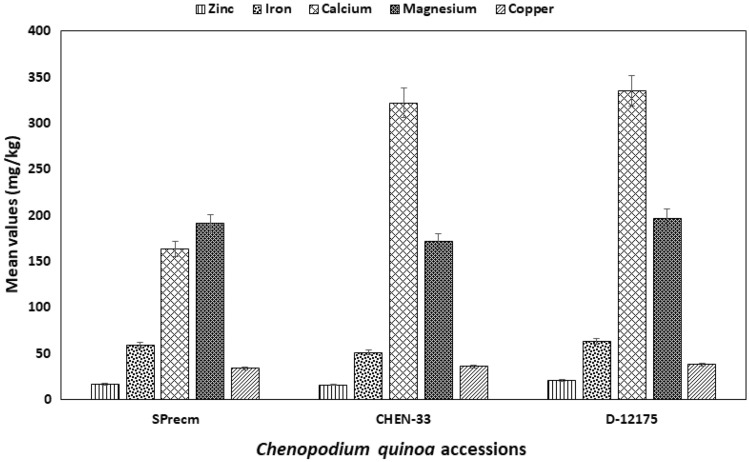
Mineral composition analysis was performed to identify the macronutrients and micronutrients in three quinoa accessions (SPrecm, CHEN-33, and D-12175) (Fig. 11). The current study evaluated three micronutrients (Cu, Zn, and Fe) and two macronutrients (Ca and Mg) in SPrecm, CHEN-33, and D-12175. The mineral contents varied among the studied accessions and are presented in Table 2. Zn was most abundant in D-12175 (21 ± 0.3) and less abundant in SPrecm (17 ± 0.5) and CHEN-33 (16 ± 0.4). Iron was found to be highest in D-12175 (63 ± 0.5) and SPrecm (59 ± 0.3) and lowest in CHEN-33 (51 ± 0.5). Similarly, Mg was also found to be highest in D-12175 (197 ± 0.2) and SPrecm (191 ± 0.4) and lowest in CHEN-33 (172 ± 0.8). Ca showed the highest concentration in D-12175 (335 ± 0.3) and CHEN-33 (322 ± 0.5) compared with SPrecm (164 ± 0.5). Cu was most abundant in D-12175 (38 ± 0.5), followed by CHEN-33 (36 ± 0.4) and SPrecm (34 ± 1.15). Further, comparative profiling using Tukey’s HSD test indicated that three minerals, including magnesium, calcium, and iron, significantly differed at a significance level of p ≤ 0.05 among quinoa accessions SPrecm, CHEN-33, and D-12175. In contrast, no significant differences at p ≤ 0.05 were observed for zinc and copper (Table 2).
Fig. 11.
A graphical illustration of comparative mineral profiling of Chenopodium quinoa accessions (SPrecm, CHEN-33, and D-12175)
Table 2.
Mean values of three micronutrients (iron, copper, and zinc) and two macronutrients (calcium and magnesium) (mg/kg) in the Chenopodium quinoa accessions (SPrecm, CHEN-33, and D-12175). Individual readings (n = 3) were averaged and given with ± standard error (SE)
| Minerals | SPrecm | CHEN-33 | D-12175 |
|---|---|---|---|
| Zinc (Zn) | 17 ± 2.3a | 16 ± 1.2a | 21 ± 1.7a |
| Iron (Fe) | 59 ± 1.7a | 51 ± 0.5b | 63 ± 0.5a |
| Magnesium (Mg) | 191 ± 1.2b | 172 ± 0.5c | 197 ± 0.5a |
| Calcium (Ca) | 164 ± 0.5c | 322 ± 2.3b | 335 ± 0.5a |
| Copper (Cu) | 34 ± 0.5a | 36 ± 1.2a | 38 ± 1.2a |
Different letters indicate significant differences (p ≤ 0.05) among treatments within the results taken at the same time interval according to Tukey’s HSD test
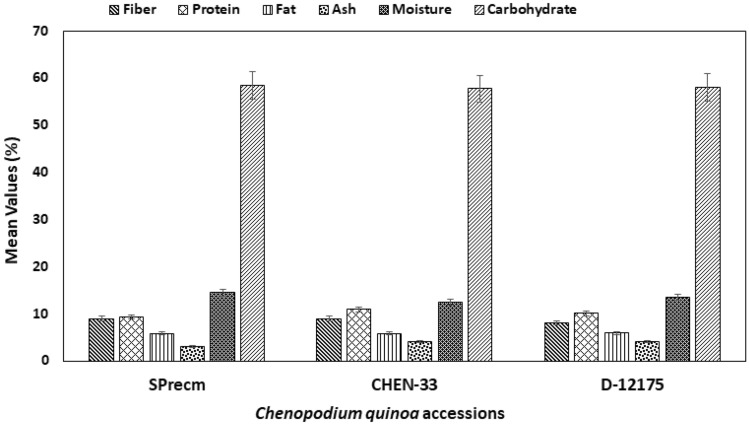
The nutrient contents (protein, fat, fiber, ash, and carbohydrate) were analyzed in three C. quinoa accessions (SPrecm, CHEN-33, and D-12175) (Fig. 12). The results showed that the moisture content was high in SPrecm (14.5%), followed by CHEN-33 (12.5%) and D-12175 (12%). Ash content was found to be higher in CHEN-33 (4%) and D-12175 (4%) and lower in SPrecm (3%). The crude fat was comparatively higher in D-12175 (5.92%) than CHEN-33 (5.85%) and lower in SPrecm (5.81%). The crude fiber was equally higher in SPrecm (9%) and CHEN-33 (9%) but lower in D-12175 (8%). The crude protein was found to be higher in CHEN-33 (10.93%) and lower in D-12175 (10.11%) and SPrecm (9.30%). The carbohydrate content was higher in SPrecm (58.39%) than in D-12175 (58.05%) and CHEN-33 (57.72%) (Table 3). Additionally, comparative profiling using Tukey’s HSD test showed no significant difference (at p ≤ 0.05) among quinoa accessions (SPrecm, CHEN-33, and D-12175) (Table 3).
Fig. 12.
A graphical illustration of proximate analysis of Chenopodium quinoa accessions (SPrecm, CHEN-33, and D-12175)
Table 3.
Proximate composition of three quinoa accessions (SPrecm, CHEN-33, and D-12175). Individual readings (n = 3) were averaged and given with ± standard error (SE)
| Nutrients | SPrecm | CHEN-33 | D-12175 |
|---|---|---|---|
| Fiber | 9 ± 0.5a | 9 ± 1.2a | 8 ± 0.6a |
| Protein | 9.30 ± 0.6a | 10.93 ± 0.6a | 10.11 ± 1.2a |
| Fat | 5.81 ± 0.2a | 5.85 ± 0.3a | 5.92 ± 0.2a |
| Ash | 3 ± 0.6a | 4 ± 0.6a | 4 ± 0.5a |
| Moisture | 14.5 ± 0.6a | 12.5 ± 0.3a | 12 ± 0.5a |
| Carbohydrate | 58.39 ± 2.3a | 57.72 ± 3.7a | 58.05 ± 0.8a |
Similar letters indicate no significant differences (p ≤ 0.05) among treatments within the results taken at the same time interval according to Tukey’s HSD test
Discussion
Metabolomic profiling using LC‒MS-based approaches is anticipated to be particularly significant in plants due to the highly complex biochemistry of plants, which includes a variety of semipolar molecules, including important secondary metabolite groups, which may be best separated and detected by LC–MS techniques (El Sayed et al. 2020). From an analytical stance, it is evident that all substances analyzed by HPLC require gradient elution and that particular compounds, such as saponins, cannot be detected by HPLC–UV analysis since they lack a potent chromophore (Hwang et al. 2019). Liquid chromatography with tandem mass spectrometry (LC–MS-MS) is sufficiently sensitive and selective to analyze specific phytochemical constituents. Electrospray ionization (ESI) in both positive and negative modes alongside the MS/MS technique has become an evident approach for more conclusive and accurate results (Göger et al. 2021). This approach has the benefit of requiring very small amounts of substance (< 1 mg) and enabling uninterrupted analysis of both mixtures and pure materials without the need for derivatization. The soft ionization method leads to the fragmentation of particular compounds in the plant extract. As a result, only the compounds with high concentrations were detectable (Jensen et al. 2022). The current study aimed to demonstrate the phytochemical, mineral, and proximate composition of selected quinoa accessions (SPrecm, CHEN-33, and D-12175) with diverse panicle architecture.
In the current study, a wide range of chemical constituents, including four flavonoids, three phenolic compounds, and their derivatives, 2 of each fatty acid, terpenoids, phenylbutenoid dimers, one of each amino acid, and monosaccharide, were detected among three quinoa accessions (SPrecm, CHEN-33, and D-12175). These phytochemical constituents effectively maintain human physiology and cure certain human ailments. The health benefits of phytochemicals result from extensive scientific research and ongoing studies. Three compounds, including cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane, pinocembrin, and alanine, were identified at precursor ion peaks m/z 381.17, 257.17, and 156.00, respectively, as reported by previous studies (Choi and Ko 2010; Yin et al. 2020; Kabkrathok et al. 2022). These compounds were common in SPrecm and CHEN-33 and not detected in D-12175. Cis-1,2-bis-[(E)-3,4-dimethoxystyryl]cyclobutane is a phenylbutenoid dimer that is potentially involved in anti-inflammatory, antimicrobial, antioxidant, anticancer, and neuroprotective activities (Han et al. 2021). However, this compound has not yet been documented in C. quinoa, so this is the first report on quinoa. Pinocembrin is a major flavonoid in many plant species with several therapeutic characteristics. It has been identified in Chenopodium graveolens (Mata et al. 1986), but no study has been reported yet for its identification in quinoa. Alanine is a nonessential amino acid documented in quinoa genotypes varying in seeds (Liu et al. 2020) and panicle color (Lujan et al. 2020).
Moreover, fructose and (Z)-1-(2,4,5-trimethoxyphenyl) butadiene were detected at precursor ion peaks m/z 221.17 and 257.17, respectively (Xia et al. 2012; Kabkrathok et al. 2022). These compounds are unique to SPrecm and were not detected in CHEN-33 and D-12175. Quinoa seeds typically contain only 3% total sugars, such as sucrose, glucose, and a minute level of fructose, conferring health-promoting qualities such as antidiabetic activity (Vilcacundo and Hernández-Ledesma 2017; Mohamed Ahmed et al. 2021). Further studies suggested that high levels of fructose in quinoa seeds exhibit hepatoprotective effects when experimented on animals; however, no study has been documented in humans yet (Al-Qabba et al. 2020; Ng and Wang 2021). Quercetin derivative (quercetin-3-O-glucosyl-rhamnosyl-(p-coumaroyl)hexoside) was detected at precursor ion peak m/z 917.92 as reported by (Ding et al. 2008), 3-hydroxyphloretin-3’-C-glucoside at m/z 453.42 (Leng et al. 2022; Zahnit et al. 2022), and kurarinone at m/z 437.42 (Liu et al. 2016). These compounds belong to the class flavonoid o-glycosides and are identified in D-12175 but not in the other two quinoa accessions (SPrecm and CHEN-33). It is the first report of these two compounds in quinoa; however, their derivatives have already been reported in previous studies in quinoa and other plant species (El-Sadek et al. 2017; Martínez-Villaluenga et al. 2020; Muller et al. 2021; Qian et al. 2023). These compounds and their derivatives are potentially involved in numerous biological activities, such as antioxidant, anticancer, and antibacterial activities (Martínez-Villaluenga et al. 2020). Another compound, rosmarinic acid, was identified at precursor ion peak m/z 199.00, validated by and not found in SPrecm and CHEN-33. The preceding study validated this identification; the precursor ion peak at m/z 199 was assigned to rosmarinic acid (Kashchenko and Olennikov 2022). This compound is a naturally occurring phenolic compound in quinoa leaves and seeds with potential therapeutic properties, making quinoa a promising nutraceutical cereal (Xu et al. 2018; Pereira et al. 2020).
Oleuropein aglycone is a strong phenolic compound detected and characterized in olives at m/z 377, conferring various bioactive characteristics (Ventura et al. 2019). However, this compound is reported for the first time in quinoa. The present study detected oleuropein aglycone at precursor ion peak m/z 377.17 in two accessions (SPrecm and CHEN-33) but not in D-12175. Furthermore, paniculide C (a triterpenoid) was identified at precursor ion peak m/z 279 among all studies of quinoa accessions (SPrecm, CHEN-33, and D-12175). This compound was also identified for the first time in this study. Previously, the precursor ion peak at m/z 279 was assigned to this compound in the leaves and seeds of Andrographis paniculata (Rafi et al. 2022). The precursor ion peak at m/z 121.00 corresponded to 4-(1-benzofuron-5-yl) benzaldehyde found in SPrecm and CHEN-33 but not in D-12175 and was also reported for the first time in quinoa. Moreover, chrysin was detected at the precursor ion peak m/z 253.17, validated by previous research (Simirgiotis et al. 2015). This compound was unique to SPrecm and not detected in CHEN-33 and D-12175. Chrysin is a flavonoid in quinoa seeds with potential health benefits and could be involved in treating cardiovascular and steatohepatitis diseases (Al-Okbi et al. 2020). Two fatty acids, palmitic acid and elenolic acid, were detected at precursor ion peaks m/z 255 (Yang et al. 2023) and 241 (Ventura et al. 2019), respectively. Palmitic acid was found in CHEN-33 and D-12175 but not in SPrecm, and elenolic acid was detected only in CHEN-33. Quinoa seeds are enriched in these major fatty acids and their derivatives, which are involved in metabolic and regulatory activities (Chen et al. 2019; Padmashree et al. 2019).
For metabolomic profiling, the detected metabolites from LC–MS-MS in the positive and negative ionization modes were further analyzed using various statistical analyses in MetaboAnalyst 5.0. Principal component analysis was performed to unveil the similarities and differences among three quinoa accessions (CHEN-33, D-12175, and SPrecm) based on their detected metabolites. The PC scores plot grouped three quinoa accessions (CHEN-33, D-12175, and SPrecm) into distinct clusters based on their metabolites. Previous studies demonstrated the clustering of three quinoa cultivars into distinct clusters varying in seed color that suggested obvious differences in metabolites of these cultivars via PCA and PLS-DA scores plot (Qian et al. 2023). Further, hierarchical cluster analysis was performed to elucidate the differences in metabolites among three quinoa accessions that classified metabolites into two major clusters. Results showed that two accessions (SPrecm and CHEN-33) are closely interrelated based on their metabolic profile, while D-12175 has a distinct metabolic profile, majorly contributing to diversity compared to SPrecm and CHEN-33. These results were validated by previous research studies that also demonstrated significant variations in chemical constituents of different species and genotypes’ varying morphological traits (Nurlela et al. 2022; Tabatabaei et al. 2022). The metabolic pathway analysis also showed significant differences in metabolic pathways among three quinoa accessions having distinct metabolic profiles. Our findings agreed with preceding studies that also displayed significant differences in metabolic pathways among different quinoa cultivars (Liu et al. 2022; Qian et al. 2023).
Nutritional profiling approaches are developed to evaluate calorie content and nutritional composition, including macronutrients, micronutrients, and nutrient content (carbohydrates). Protein, fat, fiber, moisture, and ash) are present in a given food (Mondal et al. 2023). The nutrients present in plants, such as minerals and carbohydrates, proteins, and lipids, are essential for improving human physiology (Radha et al. 2021). Quinoa is a promising “superfood” due to its exceptional nutritional qualities (Cañarejo-Antamba et al. 2021). Quinoa seeds vary significantly in genetic and agroecological conditions (Reguera et al. 2018). The current study evaluated the proximate composition (carbohydrate. protein, fat, fiber, moisture, and ash) of three quinoa accessions (SPrecm, CHEN-33, and D-12175). The crude fiber and moisture content was higher in SPrecm and CHEN-33 but lower in D-12175. The crude protein and ash contents were comparatively higher in CHEN-33 and lower in D-12175 and SPrecm. The crude fat was comparatively higher in D-12175 than CHEN-33 and lower in SPrecm. The carbohydrates were more abundant in SPrecm than D-12175 and CHEN-33.
However, results showed no statistically significant differences in proximate composition among three quinoa accessions. The crude fiber, protein, fat, and carbohydrate content (%) ranged from 7.0 to 14.1, 9.1 to 15.7, 4.0 to 7.6, and 48.5 to 69.8, respectively, as described by many authors (Marmouzi et al. 2015; Pathan and Siddiqui 2022) and our results correspond to these ranges. In this study, the moisture content was higher among all quinoa accessions (SPrecm, CHEN-33, and D-12175) than the previously reported moisture content (Marmouzi et al. 2015). Further, Nowak et al.’s extensive examination of different articles summarized infinitesimal variations in proximate composition among different quinoa genotypes (Nowak et al. 2016). Further preceding studies found no differences in carbohydrate, moisture (Rahimi and Bagheri 2020), protein, and fat (Präger et al. 2018) contents among different quinoa cultivars growing in similar agroecological regions. Conversely, Gonzalez et al. (2012) found differences in protein and other nutritional contents among ten quinoa cultivars originating from two agroecological states (Gonzalez et al. 2012).
For mineral profiling, two micronutrients, calcium and magnesium, were abundant, and three micronutrients were comparatively less abundant among the studied quinoa accessions (SPrecm, CHEN-33, and D-12175). Mineral content is an important factor in evaluating the nutritional quality of quinoa seeds. In the current study, calcium, copper, and zinc were found to be most abundant in D-12175, followed by CHEN-33 and SPrecm. A higher quantity of copper and calcium improves plants’s physiological and functional quality, enhancing crop yield (Saia et al. 2019). Quinoa seeds are enriched in calcium and are thus helpful in reducing hypertension by amplifying the parasympathetic nervous system (Agarwal et al. 2023). Copper is a critical component of various enzymes, particularly detoxifying enzymes. Copper-dependent enzymes are involved in several biochemical processes, such as antioxidant defense and neurotransmitter synthesis (Ibourki et al. 2022; Agarwal et al. 2023). Previous studies showed that zinc deficiency could lead to retarded plant growth (Hamzah Saleem et al. 2022). Further, it is an essential trace element for metalloenzymes and helps wound healing (Naqbi et al. 2022).
However, magnesium and iron were more abundant in D-12175 than SPrecm and CHEN-33. Magnesium is the second most abundant macronutrient, helping to improve quality and yield-related traits (Ishfaq et al. 2022), controlling blood sugar levels, and maintaining many biological activities in humans (Zamudio et al. 2022). Iron is an important metal ion that plays a vital role in oxygen transport, the production of red blood cells, and other health-promoting activities in humans (Ibourki et al. 2022). The mineral content level of quinoa seeds evaluated in this research study concurs with previous studies on different quinoa cultivars (Reguera et al. 2018; Pathan and Siddiqui 2022). It was validated by a previous research study documenting the mineral composition variation for three quinoa cultivars, Salcedo-INIA, Regalona, and Titicaca, from Chile, Spain, and Peru (Reguera et al. 2018). These findings revealed variations in several nutritional parameters, including minerals, amino acids, and protein contents, related to agroecological conditions and the genotype.
Further, it is anticipated that soil type and its composition, in particular, could be a potential contributing factor to these variations. Over the past few decades, quinoa has experienced a worldwide upsurge in popularity, potentially due to its exceptional nutritional content and its capacity to thrive in challenging environments (Langyan et al. 2023). Previous studies on different indica rice cultivars validated that changes in metabolic profiles can significantly affect their nutritional qualities (Yibo et al. 2022). It was further demonstrated that diversity in metabolites such as organic acids, flavonoids, and phenolic can certainly impact the nutritional quality of plants (Fang et al. 2019), thereby deciphering the metabolomic diversity is essential in metabolomics-assisted breeding (Martin and Li 2017).
Conclusion
The C. quinoa accessions with diverse panicle architecture were investigated for their metabolic and nutritional composition. Panicle architecture has a significant impact on crop yield and productivity. The quinoa accessions CHEN-33 and SPrecm were revealed as closely related based on their metabolic profile, while D-12175 exhibited a distinct metabolic profile that contributed diversity to it compared to others. The results elucidated that change in metabolic profiles has significantly affected the mineral composition of these accessions. D-12175, with a distinct metabolic profile, has high mineral levels, while SPrecm exhibited higher magnesium content than CHEN-33. Magnesium is an important nutrient for plant growth, thus substantially improving yield and quality in agriculture. However, no significant variation was observed for proximate composition among all three accessions. This research study provides a brief insight into diverse metabolic profiles that could be potentially helpful in upsurging the nutritional quality of quinoa, thereby imperative in metabolomics-assisted breeding for addressing global food scarcity.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the National Institute for Biotechnology and Genetics Engineering (NIBGE) for generously providing the facility for liquid chromatography‒mass spectrometry (LC‒MS) analysis for metabolomic profiling. Additionally, we are grateful to the Central Hi-Tech Laboratory and Animal Nutrition Laboratory, University of Agriculture Faisalabad, for their invaluable assistance in conducting the mineral and proximate analyses, respectively.
Funding
The research study is funded by Pakistan Agricultural Research Council (PARC), Agricultural Linkages Program (ALP) under the project "DNA barcoding of Quinoa (Chenopodium quinoa) germplasm and its associated pathogens for taxonomic identification to improve crop diversification (Project No. CS 444).
Declarations
Conflict of interest
The authors state they have no conflicts of interest.
Ethical approval
No ethical approval is required for this work.
Consent to participate
Informed consent was obtained from all individual participants included in this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adamczewska-Sowińska K, Sowiński J, Jama-Rodzeńska A. The effect of sowing date and harvest time on leafy greens of quinoa (Chenopodium quinoa willd.) yield and selected nutritional parameters. Agriculture. 2021;11:405–414. doi: 10.3390/agriculture11050405. [DOI] [Google Scholar]
- Agarwal A, Rizwana TAD, et al. Nutritional and functional new perspectives and potential health benefits of quinoa and chia seeds. Antioxidants. 2023;12:1413. doi: 10.3390/antiox12071413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Okbi SY, Hamed TE, Elewa TA, et al. Role of polar extracts from two quinoa varieties in prevention of steatohepatitis and cardiovascular diseases and improving glucose tolerance in rats. J Herbmed Pharmacol. 2020;10:93–101. doi: 10.34172/jhp.2021.09. [DOI] [Google Scholar]
- Al-Qabba MM, El-Mowafy MA, Althwab SA, et al. Phenolic profile, antioxidant activity, and ameliorating efficacy of chenopodium quinoa sprouts against CCl4-induced oxidative stress in rats. Nutrients. 2020;12:2904. doi: 10.3390/nu12102904v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AOAC (2006) International official methods of analysis of the AOAC, W. Horwitiz edn, 18th edn. AOAC International, Washington, DC, USA
- AOAC (1990) Official method of analysis 15th Edition. Association of Official Analytical Chemist, Washington, DC, USA
- Burrieza HP, Rizzo AJ, Vale EM, et al. Shotgun proteomic analysis of quinoa seeds reveals novel lysine-rich seed storage globulins. Food Chem. 2019;293:299–306. doi: 10.1016/j.foodchem.2019.04.098. [DOI] [PubMed] [Google Scholar]
- Cañarejo-Antamba MA, Bañuelos-Taváres O, Reyes-Trejo B, et al. Comparison of nutritional and nutraceutical properties of Chenopodium quinoa cultivated in Mexico and Ecuador. Chil J Agric Res. 2021;81:507–517. doi: 10.4067/S0718-58392021000400507. [DOI] [Google Scholar]
- Carciochi RA, Galván-D’Alessandro L, Vandendriessche P, Chollet S. Effect of germination and fermentation process on the antioxidant compounds of quinoa seeds. Plant Foods Hum Nutr. 2016;71:361–367. doi: 10.1007/s11130-016-0567-0. [DOI] [PubMed] [Google Scholar]
- Chen Y-S, Aluwi NA, Saunders SR, et al. Metabolic fingerprinting unveils quinoa oil as a source of bioactive phytochemicals. Food Chem. 2019;286:592–599. doi: 10.1016/j.foodchem.2019.02.016. [DOI] [PubMed] [Google Scholar]
- Cheong BE, Ho WWH, Biddulph B, et al. Phenotyping reproductive stage chilling and frost tolerance in wheat using targeted metabolome and lipidome profiling. Metabolomics. 2019;15:1–19. doi: 10.1007/s11306-019-1606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S-S, Ko J-E. Dimerization reactions of amino acids by pyrolysis. J Anal Appl Pyrolysis. 2010;89:74–86. doi: 10.1016/j.jaap.2010.05.009. [DOI] [Google Scholar]
- Contreras-Jiménez B, Torres-Vargas OL, Rodríguez-García ME. Physicochemical characterization of quinoa (Chenopodium quinoa) flour and isolated starch. Food Chem. 2019;298:124982. doi: 10.1016/j.foodchem.2019.124982. [DOI] [PubMed] [Google Scholar]
- Ding S, Dudley E, Plummer S, et al. Fingerprint profile of Ginkgo biloba nutritional supplements by LC/ESI-MS/MS. Phytochemistry. 2008;69:1555–1564. doi: 10.1016/j.phytochem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- El Sayed AM, Basam SM, El-Naggar E-MBA, et al. LC–MS/MS and GC–MS profiling as well as the antimicrobial effect of leaves of selected Yucca species introduced to Egypt. Sci Rep. 2020;10:17778. doi: 10.1038/s41598-020-74440-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sadek A, Balah M, Romani A, et al. Allelopathic potential of quinoa (Chenopodium quinoa Willd.) genotypes on the germination and initial development of some weeds and crops. Egypt J Desert Res. 2017;67:25–45. doi: 10.21608/ejdr.2017.5843. [DOI] [Google Scholar]
- Fang C, Luo J, Wang S. The diversity of nutritional metabolites: origin, dissection, and application in crop breeding. Front Plant Sci. 2019;10:1028–1037. doi: 10.3389/fpls.2019.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2023) The state of food and agriculture 2023 – revealing the true cost of food to transform agrifood systems. Rome. 10.4060/cc7724en
- García-Parra M, Zurita-Silva A, Stechauner-Rohringer R, et al. Quinoa (Chenopodium quinoa Willd.) and its relationship with agroclimatic characteristics: a Colombian perspective. Chil J Agric Res. 2020;80:290–302. doi: 10.4067/S0718-58392020000200290. [DOI] [Google Scholar]
- García-Parra M, Roa-Acosta D, García-Londoño V, et al. Structural characterization and antioxidant capacity of quinoa cultivars using techniques of FT-MIR and UHPLC/ESI-Orbitrap MS spectroscopy. Plants. 2021;10:2159. doi: 10.3390/plants10102159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göger G, Köse YB, Demirci F, Göger F. Phytochemical characterization of phenolic compounds by LC-MS/MS and biological activities of Ajuga reptans L., Ajuga salicifolia (L.) Schreber and Ajuga genevensis L. from Turkey. Turkish J Pharm Sci. 2021;18:616. doi: 10.4274/tjps.galenos.2021.33958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pando LR, Aguilar-Castellanos E, Ibañez-Tremolada M. Quinoa (Chenopodium quinoa Willd.) breeding. Adv Plant Breed Strateg Cereal. 2019;5:259–316. doi: 10.1007/978-3-030-23108-8_7. [DOI] [Google Scholar]
- Gonzalez JA, Konishi Y, Bruno M, et al. Interrelationships among seed yield, total proteinand amino acid composition of ten quinoa (Chenopodium quinoa) cultivars from twodifferent agroecological regions. J Sci Food Agric. 2012;92:1222–1229. doi: 10.1002/jsfa.4686. [DOI] [PubMed] [Google Scholar]
- Hamzah Saleem M, Usman K, Rizwan M, et al. Functions and strategies for enhancing zinc availability in plants for sustainable agriculture. Front Plant Sci. 2022;13:1033092. doi: 10.3389/fpls.2022.1033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A-R, Kim H, Piao D, et al. Phytochemicals and bioactivities of Zingiber cassumunarRoxb. Molecules. 2021;26:2377. doi: 10.3390/molecules26082377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JT, Park K-S, Ryuk JA, et al. Development of an oriental medicine discrimination method through analysis of steroidal saponins in Dioscorea nipponica Makino and their anti-osteosarcoma effects. Molecules. 2019;24:4022. doi: 10.3390/molecules24224022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibourki M, Ait Bouzid H, Bijla L, et al. Mineral profiling of twenty wild and cultivated aromatic and medicinal plants growing in Morocco. Biol Trace Elem Res. 2022;200:4880–4889. doi: 10.1007/s12011-021-03062-w. [DOI] [PubMed] [Google Scholar]
- Ishfaq M, Wang Y, Yan M, et al. Physiological essence of magnesium in plants and its widespread deficiency in the farming system of China. Front Plant Sci. 2022;13:802274–802291. doi: 10.3389/fpls.2022.802274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MB, Rød KE, Švarc PL, et al. Vitamin K (phylloquinone and menaquinones) in foods–cost-effective quantification by LC-ESI-MS/MS. Food Chem. 2022;385:132672. doi: 10.1016/j.foodchem.2022.132672. [DOI] [PubMed] [Google Scholar]
- Jorge TF, Rodrigues JA, Caldana C, et al. Mass spectrometry-based plant metabolomics: metabolite responses to abiotic stress. Mass Spectrom Rev. 2016;35:620–649. doi: 10.1002/mas.21449. [DOI] [PubMed] [Google Scholar]
- Kabkrathok P, Jarussophon S, Unger O, et al. Mass spectral analysis of secondary metabolites from Zingiber montanum rhizome extract using UHPLC-HR-ESI-QTOF-MS/MS. Phytochem Anal. 2022;33:57–71. doi: 10.1002/pca.3068. [DOI] [PubMed] [Google Scholar]
- Kashchenko NI, Olennikov DN. Glycosides of rosmarinic acid from Nepeta multifida. Chem Nat Compd. 2022;58:274–278. doi: 10.1007/s10600-022-03658-1. [DOI] [Google Scholar]
- Kaur H, Sunkaria B, Garg N. Quinoa: role and responses under abiotic stress. In: Kaur H, editor. Sustainable remedies for abiotic stress in cereals. Berlin: Springer; 2022. pp. 229–271. [Google Scholar]
- Langyan S, Khan FN, Kumar A. Advancement in nutritional value, processing methods, and potential applications of pseudocereals in dietary food: a review. Food Bioprocess Technol. 2023 doi: 10.1007/s11947-023-03109-x. [DOI] [Google Scholar]
- Leng Z, Zhong B, Wu H, et al. Identification of phenolic compounds in Australian-grown bell peppers by liquid chromatography coupled with electrospray ionization-quadrupole-time-of-flight-mass spectrometry and estimation of their antioxidant potential. ACS Omega. 2022;7:4563–4576. doi: 10.1021/acsomega.1c06532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhang H, Li J, et al. Genetic control of panicle architecture in rice. Crop J. 2021;9:590–597. doi: 10.1016/j.cj.2021.02.004. [DOI] [Google Scholar]
- Liu Y, Mo ZX, Wang CG, et al. Identification of metabolites of kurarinone from Sophora flavescens Ait in rat urine by ultra-performance liquid chromatography with linear ion trap orbitrap mass spectrometry. Trop J Pharm Res. 2016;15:1299–1305. doi: 10.4314/tjpr.v15i6.24. [DOI] [Google Scholar]
- Liu Y, Kong Z, Liu J, et al. Non-targeted metabolomics of quinoa seed filling period based on liquid chromatography-mass spectrometry. Food Res Int. 2020;137:109743. doi: 10.1016/j.foodres.2020.109743. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu J, Kong Z, et al. Transcriptomics and metabolomics analyses of the mechanism of flavonoid synthesis in seeds of differently colored quinoa strains. Genomics. 2022;114:138–148. doi: 10.1016/j.ygeno.2021.11.030. [DOI] [PubMed] [Google Scholar]
- Lujan AIB, Pardo FT, Carrión MLH, et al (2020) Flour amino acids and protein isolate of quinoa (ChenopodiumQuinoa) of the white and pink variety of Junín. EasyChair
- Manjarres-Hernández EH, Arias-Moreno DM, Morillo-Coronado AC, et al. Phenotypic characterization of quinoa (Chenopodium quinoa Willd.) for the selection of promising materials for breeding programs. Plants. 2021;10:1339. doi: 10.3390/plants10071339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmouzi I, El Madani N, Charrouf Z, et al. Proximate analysis, fatty acids and mineral composition of processed Moroccan Chenopodium quinoa Willd. and antioxidant properties according to the polarity. Phytothérapie. 2015;13:110–117. doi: 10.1007/s10298-015-0931-5. [DOI] [Google Scholar]
- Martin C, Li J. Medicine is not health care, food is health care: plant metabolic engineering, diet and human health. New Phytol. 2017;216:699–719. doi: 10.1111/nph.14730. [DOI] [PubMed] [Google Scholar]
- Martínez-Villaluenga C, Peñas E, Hernández-Ledesma B. Pseudocereal grains: nutritional value, health benefits and current applications for the development of gluten-free foods. Food Chem Toxicol. 2020;137:111178. doi: 10.1016/j.fct.2020.111178. [DOI] [PubMed] [Google Scholar]
- Mata R, Navarrete A, Alvarez L, et al. Flavonoids and terpenoids of Chenopodium graveolens. Phytochemistry. 1986;26:191–193. doi: 10.1016/S0031-9422(00)81509-5. [DOI] [Google Scholar]
- Mohamed Ahmed IA, Al Juhaimi F, Özcan MM. Insights into the nutritional value and bioactive properties of quinoa (Chenopodium quinoa): past, present and future prospective. Int J Food Sci Technol. 2021;56:3726–3741. doi: 10.1111/ijfs.15011. [DOI] [Google Scholar]
- Mondal DD, Chakraborty U, Bera M, et al. An overview of nutritional profiling in foods: bioanalytical techniques and useful protocols. Front Nutr. 2023;10:1124409. doi: 10.3389/fnut.2023.1124409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CJF, Joubert E, Chellan N, et al. New insights into the efficacy of aspalathin and other related phytochemicals in type 2 diabetes—a review. Int J Mol Sci. 2021;23:356. doi: 10.3390/ijms23010356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqbi NKMA, Karthishwaran K, Kurup SS, et al. Phytochemicals, proximate composition, mineral analysis and in vitro antioxidant activity of Calligonum crinitum Boiss. Horticulturae. 2022;8:156. doi: 10.3390/horticulturae8020156. [DOI] [Google Scholar]
- Ng CY, Wang M. The functional ingredients of quinoa (Chenopodium quinoa) and physiological effects of consuming quinoa: a review. Food Front. 2021;2:329–356. doi: 10.1002/fft2.109. [DOI] [Google Scholar]
- Nowak V, Du J, Charrondière UR. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.) Food Chem. 2016;193:47–54. doi: 10.1016/j.foodchem.2015.02.111. [DOI] [PubMed] [Google Scholar]
- Nurlela N, Nurfalah R, Ananda F, et al. Variation of morphological characteristics, total phenolic, and total flavonoid in Adenostemma lavenia, A. madurense, and A. platyphyllum. Biodiversitas J Biol Divers. 2022;23:1–7. doi: 10.13057/biodiv/d230818. [DOI] [Google Scholar]
- Otterbach SL, Khoury H, Rupasinghe T, et al. Characterization of epidermal bladder cells in Chenopodium quinoa. Plant Cell Environ. 2021;44:3836–3852. doi: 10.1111/pce.14181. [DOI] [PubMed] [Google Scholar]
- Padmashree A, Negi N, Handu S, Khan MA, Semwal AD, Sharma GK (2019) Effect of germination on nutritional, antinutritional and rheological characteristics of chenopodium quinoa. Energy 4:55–60. 10.14429/dlsj.4.12202
- Pakbaz N, Omidi H, Naghdi Badi H, Bostani A. Botanical, phytochemical and pharmacological properties of quinoa medicinal plant (Chenopodium quinoa Willd.): a review. J Med Herbs. 2021;12:1–11. doi: 10.22070/DANESHMED.2023.17462.1331. [DOI] [Google Scholar]
- Pandya A, Thiele B, Zurita-Silva A, et al. Determination and metabolite profiling of mixtures of triterpenoid saponins from seeds of chilean quinoa (Chenopodium quinoa) germplasm. Agronomy. 2021;11:1867. doi: 10.3390/agronomy11091867. [DOI] [Google Scholar]
- Pathan S, Siddiqui RA. Nutritional composition and bioactive components in quinoa (Chenopodium quinoa Willd.) greens: a review. Nutrients. 2022;14:558. doi: 10.5539/jfr.v8n6p55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan S, Eivazi F, Valliyodan B, et al. Nutritional composition of the green leaves of quinoa (Chenopodium quinoa Willd.) J Food Res. 2019;8:55–65. doi: 10.5539/jfr.v8n6p55. [DOI] [Google Scholar]
- Pereira E, Encina-Zelada C, Barros L, et al. Chemical and nutritional characterization of Chenopodium quinoa Willd (quinoa) grains: a good alternative to nutritious food. Food Chem. 2019;280:110–114. doi: 10.1016/j.foodchem.2018.12.068. [DOI] [PubMed] [Google Scholar]
- Pereira E, Cadavez V, Barros L, et al. Chenopodium quinoa Willd.(quinoa) grains: a good source of phenolic compounds. Food Res Int. 2020;137:109574. doi: 10.1016/j.foodres.2020.109574. [DOI] [PubMed] [Google Scholar]
- Präger A, Munz S, Nkebiwe PM, et al. Yield and quality characteristics of different quinoa (Chenopodium quinoa Willd.) cultivars grown under field conditions in Southwestern Germany. Agronomy. 2018;8:197–216. doi: 10.3390/agronomy8100197. [DOI] [Google Scholar]
- Qian G, Li X, Zhang H, et al. Metabolomics analysis reveals the accumulation patterns of flavonoids and phenolic acids in quinoa (Chenopodium quinoa Willd.) grains of different colors. Food Chem X. 2023;17:100594. doi: 10.1016/j.fochx.2023.100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radha KM, Puri S, et al. Evaluation of nutritional, phytochemical, and mineral composition of selected medicinal plants for therapeutic uses from cold desert of Western Himalaya. Plants. 2021;10:1429. doi: 10.3390/plants10071429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafi M, Karomah AH, Heryanto R, et al. Metabolite profiling of Andrographis paniculata leaves and stem extract using UHPLC-Orbitrap-MS/MS. Nat Prod Res. 2022;36:625–629. doi: 10.1080/14786419.2020.1789637. [DOI] [PubMed] [Google Scholar]
- Rahimi E, Bagheri M. Chemical, antioxidant, total phenolic and flavonoid components and antimicrobial effects of different species of quinoa seeds. Egypt J Vet Sci. 2020;51:43–54. doi: 10.21608/EJVS.2019.17122.1098. [DOI] [Google Scholar]
- Rai A, Saito K, Yamazaki M. Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 2017;90:764–787. doi: 10.1111/tpj.13485. [DOI] [PubMed] [Google Scholar]
- Reguera M, Conesa CM, Gil-Gómez A, et al. The impact of different agroecological conditions on the nutritional composition of quinoa seeds. PeerJ. 2018;6:e4442. doi: 10.7717/peerj.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz KB, Biondi S, Oses R, et al. Quinoa biodiversity and sustainability for food security under climate change. A review. Agron Sustain Dev. 2014;34:349–359. doi: 10.1007/s13593-013-0195-0. [DOI] [Google Scholar]
- Saia S, Colla G, Raimondi G, et al. An endophytic fungi-based biostimulant modulated lettuce yield, physiological and functional quality responses to both moderate and severe water limitation. Sci Hortic. 2019;256:108595–108612. doi: 10.1016/j.scienta.2019.108595. [DOI] [Google Scholar]
- Sharma B, Viswanath G, Salunke R, Roy P. Effects of flavonoid-rich extract from seeds of Eugenia jambolana (L.) on carbohydrate and lipid metabolism in diabetic mice. Food Chem. 2008;110:697–705. doi: 10.1016/j.foodchem.2008.02.068. [DOI] [Google Scholar]
- Simirgiotis MJ, Benites J, Areche C, Sepúlveda B. Antioxidant capacities and analysis of phenolic compounds in three endemic Nolana species by HPLC-PDA-ESI-MS. Molecules. 2015;20:11490–11507. doi: 10.3390/molecules200611490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Zuniga V, Brito V, Fuentes F, Steinfort U. Phenological growth stages of quinoa (Chenopodium quinoa) based on the BBCH scale. Ann Appl Biol. 2017;171:117–124. doi: 10.1111/aab.12358. [DOI] [Google Scholar]
- Tabatabaei I, Alseekh S, Shahid M, et al. The diversity of quinoa morphological traits and seed metabolic composition. Sci Data. 2022;9:323–330. doi: 10.1038/s41597-022-01399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura G, Calvano CD, Abbattista R, et al. Characterization of bioactive and nutraceutical compounds occurring in olive oil processing wastes. Rapid Commun Mass Spectrom. 2019;33:1670–1681. doi: 10.1002/rcm.8514. [DOI] [PubMed] [Google Scholar]
- Vilcacundo R, Hernández-Ledesma B. Nutritional and biological value of quinoa (Chenopodium quinoa Willd.) Curr Opin Food Sci. 2017;14:1–6. doi: 10.1016/j.cofs.2016.11.007. [DOI] [Google Scholar]
- Wang M, Jarmusch AK, Vargas F, et al. Mass spectrometry searches using MASST. Nat Biotechnol. 2020;38:23–26. doi: 10.1038/s41587-019-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:652–660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Zhou Y, Liu X, et al. Use of electrospray ionization ion-trap tandem mass spectrometry and principal component analysis to directly distinguish monosaccharides. Rapid Commun Mass Spectrom. 2012;26:1259–1264. doi: 10.1002/rcm.6219. [DOI] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43:251–257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang B, Pu Y, et al. Techniques for the analysis of pentacyclic triterpenoids in medicinal plants. J Sep Sci. 2018;41:6–19. doi: 10.1002/jssc.201700201. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhai Y, Zhang Z, et al. Liquid chromatography-mass spectrometry-based metabolomics reveals the comprehensive metabolites in Dioscorea opposita Thunb. peel. Sep Sci plus. 2023 doi: 10.1002/sscp.202300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yibo C, Zhidong W, Chongrong W, et al. Comparisons of metabolic profiles for carbohydrates, amino acids, lipids, fragrance and flavones during grain development in indica rice cultivars. Rice Sci. 2022;29:155–165. doi: 10.1016/j.rsci.2022.01.004. [DOI] [Google Scholar]
- Yin J, Ren W, Wei B, et al. Characterization of chemical composition and prebiotic effect of a dietary medicinal plant Penthorum chinense Pursh. Food Chem. 2020;319:126568. doi: 10.1016/j.foodchem.2020.126568. [DOI] [PubMed] [Google Scholar]
- Zahnit W, Smara O, Bechki L, et al. Phytochemical profiling, mineral elements, and biological activities of Artemisia campestris L. grown in Algeria. Horticulturae. 2022;8:914. doi: 10.3390/horticulturae8100914. [DOI] [Google Scholar]
- Zamudio FV, Hidalgo-Figueroa SN, Andrade RRO, et al. Identification of antidiabetic peptides derived from in silico hydrolysis of three ancient grains: Amaranth, Quinoa and Chia. Food Chem. 2022;394:133479. doi: 10.1016/j.foodchem.2022.133479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.