Abstract
Dry pea ( Pisum sativum ) seeds are valuable sources of plant protein, dietary fiber, and starch, but their uses in food products are restricted to some extent due to several off‐flavor compounds. Saponins are glycosylated triterpenoids and are a major source of bitter, astringent, and metallic off‐flavors in pea products. β‐amyrin synthase (BAS) is the entry point enzyme for saponin biosynthesis in pea and therefore is an ideal target for knock‐out using CRISPR/Cas9 genome editing to produce saponin deficient pea varieties. Here, in an elite yellow pea cultivar (CDC Inca), LC/MS analysis identified embryo tissue, not seed coat, as the main location of saponin storage in pea seeds. Differential expression analysis determined that PsBAS1 was preferentially expressed in embryo tissue relative to seed coat and was selected for CRISPR/Cas9 genome editing. The efficiency of CRISPR/Cas9 genome editing of PsBAS1 was systematically optimized in pea hairy roots. From these optimization procedures, the AtU6‐26 promoter was found to be superior to the CaMV35S promoter for gRNA expression, and the use of 37°C was determined to increase the efficiency of CRISPR/Cas9 genome editing. These promoter and culture conditions were then applied to stable transformations. As a result, a bi‐allelic mutation (deletion and inversion mutations) was generated in the PsBAS1 coding sequence in a T1 plant, and the segregated psbas1 plants from the T2 population showed a 99.8% reduction of saponins in their seeds. Interestingly, a small but statistically significant increase (~12%) in protein content with a slight decrease (~5%) in starch content was observed in the psbas1 mutants under phytotron growth conditions. This work demonstrated that flavor‐improved traits can be readily introduced in any pea cultivar of interest using CRISPR/Cas9. Further field trials and sensory tests for improved flavor are necessary to assess the practical implications of the saponin‐free pea seeds in food applications.
Keywords: CRISPR/Cas9, genome‐editing, pea, saponin, β‐Amyrin synthase
1. INTRODUCTION
Animal protein is currently an essential source of protein in the human diet. However, animal agriculture poses several risks to the environment in the form of climate change and land usage (Poore & Nemecek, 2018). The supplementation of some or all dietary animal protein with plant protein can mitigate these problems. Dry peas (Pisum sativum L.) are a member of the Fabaceae family characterized by their large edible cotyledons and ability to form symbiotic relationships with nitrogen‐fixing bacteria. Having an endogenous source of nitrogen lowers soil inputs needed for peas and is responsible for their high protein content (16–30% of dry weight), making peas have an exceptionally low land requirement per gram of protein (Burstin et al., 2007). Therefore, peas are an excellent source of plant proteins to address the issues posed by animal agriculture.
One hurdle preventing peas from supplanting animal proteins is the presence of off‐flavors in their seeds. The off‐flavors in pea seeds are partly caused by saponins, which have bitter and astringent flavors (Heng et al., 2006). Pea seeds accumulate two types of group B soyasaponins, soyasaponin Bb (saponin B), and 2,3‐dihydro‐2,5‐dihydroxy‐6‐methyl‐4H‐pyran‐4‐one (DDMP)‐conjugated soyasaponin, soyasaponin βg (DDMP saponin, Figure 1) (Tsurumi et al., 1992). DDMP saponin is significantly more bitter than saponin B and has been found to be the predominant saponin in pea seeds (Heng et al., 2006). Saponins interact with proteins and are retained during protein isolation (Lin et al., 2006). Therefore, foods derived from peas, or pea protein, require the addition of masking ingredients or chemical extraction to improve the food's palatability.
FIGURE 1.

Saponin biosynthesis in pea. BAS1, β‐amyrin synthase 1; BA22H, β‐amyrin C22‐hydroxylase; BA24H, β‐amyrin C24‐hydroxylase. Validated pea enzymes are shown in green, while pea orthologues to the saponin biosynthetic enzymes in soybean are shown in orange. DDMP saponin can be degraded to saponin B by heat and acid.
Many steps of DDMP saponin biosynthesis have been elucidated across several legumes, including pea. The first step of DDMP saponin biosynthesis is the formation of the triterpene backbone, β‐amyrin, from 2,3‐oxidosqualene by β‐amyrin synthase (BAS, Figure 1) (Morita et al., 2000). This backbone is then further decorated by a series of cytochrome P450s (CYPs) and UDP‐dependent glycosyltransferases (UGTs). In pea, β‐amyrin C‐24 hydroxylase (CYP93E8) has been the only experimentally proven enzyme for the oxygenation of saponins (Moses et al., 2014). However, in soybean, the biochemical identities of β‐amyrin C‐22 hydroxylase (CYP72A61), galactose‐, rhamnose‐, and DDMP‐transferase (UGT73P2, UGT91H4, and UGT73K5, respectively) have been fully elucidated (Yu et al., 2022, and references therein). The enzyme for the transfer of glucuronic acid to the C‐3 hydroxyl position has been a mystery for decades, but a cellulose synthase superfamily‐derived glycosyl transferase (CSyGT) was recently identified as a catalyst for this reaction (Chung et al., 2020).
Vernoud et al. (2021) utilized a TILLING population of spring pea cultivar Caméor to identify psbas1 mutants and demonstrated that pea BAS1 is responsible for the vast majority of saponin content in pea seeds. Caméor is the French pea cultivar used for whole genome sequencing (Kreplak et al., 2019). However, it is not currently cultivated for commercial purposes, owing to its low yield and poor resistance to lodging. The identified mutation in the Caméor background can be introgressed into elite breeding lines. However, despite the success of conventional breeding in producing elite pea varieties, introgression still requires significant resources and time, and the linkage drag can cause an unexpected problem during introgression.
Applying precision CRISPR/Cas9 genome‐editing directly in elite germplasms circumvents the problems of traditional breeding, allowing for transgene‐free mutant lines to be generated in two generations (Lyzenga et al., 2021). Additionally, many major importers of peas, China and the United States, have clear guidelines on genome‐edited crops with lower regulatory burdens than genetically modified (GM) ones (Hoffman, 2021; Mallapaty, 2022). CRISPR/Cas9 genome editing has been successfully used to edit phytoene desaturase (Li et al., 2023) and lipoxygenase 2 (Bhowmik et al., 2023) in peas. In this work, we optimized CRISPR/Cas9 using induced hairy roots from an elite yellow pea cultivar, CDC Inca (Warkentin et al., 2017). The developed system was then leveraged to produce two mutant alleles in stable mutant lines with a 99.8% reduction of saponin concentration in pea seeds. This work demonstrates that modern crop breeding can benefit from CRISPR/Cas9‐mediated mutations in peas for the creation of novel traits.
2. RESULTS
2.1. Saponins accumulate in pea embryos
DDMP saponin and saponin B accumulate in pea seeds, and saponin B is a biosynthetic intermediate as well as a degradation product of DDMP saponin (Figure 1). Yellow pea (cv. CDC Inca) seeds were separated into embryos and seed coats, and their saponin contents were measured by LC/MS (Table 1). A majority of saponins (>97.6%) in pea seeds were found in the embryo with only a negligible amount of saponins being detected in seed coats. Of the two types of saponins measured, DDMP saponin constitutes 99% of saponins in pea seeds. To determine whether the saponins are carried over to processed pea products, commercial pea flour and fiber were purchased for saponin analysis. The saponin content of pea flour was comparable to that from pea embryos, and pea fiber also contained ~1 mg saponin per g weight. These results showed that both pea seeds and processed pea products contain 0.1–0.3% (w/w) of saponins with DDMP saponin being the major component in pea embryos.
TABLE 1.
Saponin content in yellow pea seeds.
| Sample | DDMP saponin a (mg/g) | Saponin B a (μg/g) |
|---|---|---|
| 1 Embryo | 2.85 ± .32 | 31.7 ± 9.9 |
| Seed coat b | 0.07 ± .04 | 0.5 ± 0.3 |
| Flour c | 2.09 ± .07 | 53.0 ± 35.8 |
| Fiber c | 0.96 ± .29 | 26.1 ± 5.2 |
Data are mean ± SD (n = 4).
These tissues were from yellow pea, CDC Inca variety.
These commercial products were obtained from Parrheim Foods (Saskatoon, Canada).
2.2. Embryo‐specific expression of saponin biosynthetic genes in pea seeds
Transcriptome data in pea seeds can benefit trait developments through genome editing. Pea embryo and seed coat tissues were obtained from pea seeds 21 days after anthesis. Total RNA was separately isolated from the embryos and seed coats, followed by Illumina sequencing, resulting in 18–27 million reads from the two tissues in three individual plants. The cleaned reads were mapped to the P. sativum whole genome (Kreplak et al., 2019) to identify the genes differentially expressed in the embryo, relative to the seed coat. In differential expression (DE) analysis, 1,953 and 2,744 transcripts were identified as up‐regulated and down‐regulated genes, respectively, in pea embryos (Figure 2; all identified genes are listed in Dataset S1). Previously, PsBAS1 (Psat7g264880) was characterized as β‐amyrin synthase, while PsBAS2 (Psat4g188800) was determined to be a mixed α/β‐amyrin synthase producing five other minor triterpenes, in which β‐amyrin constituted ~30% of the total triterpenes (Morita et al., 2000). PsBAS2 was not included in our DE analysis due to its low expression, but PsBAS1 was the 218th most DE gene in the embryo. In addition, CYPs for β‐amyrin C‐22 and C‐24 hydroxylation, UGTs for galactose and DDMP transfer, and CSyGT for glucuronic acid transfer were identified. However, the UGT91H4 orthologue known to transfer rhamnose in soybean could not be found in the DE analysis, suggesting pea may use a different subclade of UGT for rhamnose conjugation. Two bHLH transcription factors (TSAR1/2) for saponin over‐production in soybean also showed significant DE in the embryo (Mertens et al., 2016). Other than saponins, lipoxygenases and protease inhibitors, known to have implications for aroma and nutritional quality of pea seeds, were identified in the DE genes in embryos (Robinson & Domoney, 2021). Overall, this DE analysis serves as a reference in selecting target genes for genome editing in pea seeds. Relevant to this work, these data showed that PsBAS2 plays a minor role in saponin biosynthesis in pea seeds. Hence, we decided to carry out CRISPR/Cas9‐induced mutations of a single gene, PsBAS1.
FIGURE 2.

Differential expression analysis (pea embryo vs. seed coat). Differentially expressed genes (embryo + and seed coat −) were identified using the DESeq2 method and plotted against fold change and statistical significance. The cut‐off values used were >1 transcript per million (TPM), log2 fold change >2, and p value <.05. See Dataset S1 for the entire list of differentially expressed genes and their expression levels in embryos and seed coats. Green dots are the two pea genes experimentally proven to catalyze the first two reactions in saponin biosynthesis in pea; red dots are pea saponin biosynthetic genes inferred by sequence homology to saponin biosynthetic genes in soybean or M. truncatula ; blue dots are possible target genes for genome‐editing to improve aroma and nutritional quality of pea seeds. The three most highly expressed lipoxygenases (TPM > 1,200) and trypsin inhibitors (TPM > 250) in the embryo are labeled in blue.
2.3. CRISPR/Cas9 vector design and in vitro test
Using the CRISPRater algorithm (Labuhn et al., 2018), five 20‐bp gRNA‐binding sites were identified from the first and second exon of PsBAS1 in the pea genome (Figure 3a). The cleavage efficacy of these sites was examined by an in vitro Cas9 cleavage assay. The 1.2‐kb cleavage template of PsBAS1 was amplified from pea genomic DNA and was incubated with recombinant Cas9 and each of the five in vitro transcribed gRNAs. Gel‐electrophoresis analysis of the in vitro Cas9 assays showed that gRNA3 exhibited the highest cleavage efficiency, followed by gRNA2 and 4; however, gRNA1 and 5 showed negligible cleavage activities (Figure 3b). Based on these results, gRNA2/3/4 were selected for CRISPR/Cas9 genome editing. In previous work, we developed a customizable CRISPR/Cas9 vector, in which multiple gRNAs linked by auto‐cleavable tRNAs can be expressed in a single transcript (Kwon et al., 2023; Xie et al., 2015). Using this system, two CRISPR/Cas9 vectors were constructed—Cas9 was expressed with the CaMV35 promoter in both constructs, but a transcript including three repeats of tRNA‐gRNA (gRNA2/3/4) sequences was expressed either by the CaMV35S or AtU6–26 promoter (Figures S1 and S2).
FIGURE 3.

Location of selected candidate gRNA‐binding sites on PsBAS1 and their in vitro cleavage analysis. (a) A map of the PsBAS1 gene and the location of candidate gRNA‐binding sites on the PsBAS1 cleavage template used for the in vitro cleavage assay. Exons are represented by black boxes; introns are represented by black lines; gRNA‐binding sites are represented by colored text; PAM sites of gRNAs are represented by black text. (b) In vitro test of the PsBAS1 candidate gRNAs cleavage abilities using recombinant Cas9, in vitro transcribed gRNAs, and a PCR generated PsBAS1 cleavage template. The negative control (−) does not contain any gRNA.
2.4. Evaluating CRISPR/Cas9 constructs in pea hairy roots
Pea is quite recalcitrant to transformation by A. tumefaciens. We reasoned that rapid assessment of CRISPR/Cas9 constructs in hairy roots can mitigate the risk of lengthy transformation processes in peas. Five different A. rhizogenes strains (AR10, AR1193, K599, R1000, and R1200) were tested for hairy root induction using the CDC Inca cultivar. Each strain carried a binary vector with a green fluorescent protein (GFP) reporter gene. Stems and stipules of 10‐ to 12‐day‐old pea seedlings were infected with each bacterial strain. The two best‐performing strains were AR1193 and AR10 with 88% and 92% efficiencies of hairy root induction, respectively (Table 2). Next, the induced hairy roots were examined for GFP signals by fluorescent microscopy. The hairy roots generated by AR10 showed the highest percentage of GFP signals (56%), while less than 25% of hairy roots from the other four strains showed GFP signals (Table 2). Based on these results, AR10 was chosen for hairy root assays in subsequent CRISPR/Cas9 experiments. Two CRISPR/Cas9 constructs were individually transformed to AR10, and hairy roots were induced at 24°C and at 37°C as improved Cas9 genome‐editing efficiency was reported at high temperatures (LeBlanc et al., 2018). For each construct, at either 24°C or 37°C, 10–12 hairy roots were generated, and the PsBAS1 locus was amplified and sequenced to assess the CRISPR/Cas9‐based mutations. The construct with the AtU6–26 promoter for gRNA expression at 37°C had the highest efficiency of mutations (36% major mutations; Table 3 and Figure S3). These results showed that the selection of the A. rhizogenes strain, promoter, and tissue culture temperature can influence genome‐editing efficiency in pea hairy roots.
TABLE 2.
Analysis of hairy roots produced from pea cv. CDC Inca using different A. rhizogenes strains.
| Strain | Percentage of explants, producing hairy roots | Average number of hairy roots per explant | Percentage of hairy roots with GFP signals |
|---|---|---|---|
| AR10 | 92% (36/39) | 2.9 (106/36) | 56% (9/16) |
| AR1193 | 88% (35/40) | 2.9 (100/35) | 24% (4/17) |
| K599 | 39% (20/51) | 1.7 (34/20) | 8% (1/12) |
| R1000 | 34% (19/56) | 2.4 (45/19) | 19% (3/16) |
| R1200 | 20% (10/49) | 1.5 (15/10) | 25% (3/12) |
TABLE 3.
Mutation efficiency of PsBAS1 CRISPR constructs in hairy roots.
| Promoter for gRNA | Growth conditions | Percentage of hairy roots with minor mutations | Percentage of hairy roots with major mutations |
|---|---|---|---|
| CaMV35S | 24°C | 9% (1/11) | 0% (0/11) |
| 37°C | 0% (0/10) | 20% (2/10) | |
| AtU6–26 | 24°C | 8% (1/12) | 25% (3/12) |
| 37°C | 9% (1/11) | 36% (4/11) |
2.5. Generating stable transgenic pea plants
Stable transformations of pea were performed using A. tumefaciens strain EHA105, carrying a CRISPR/Cas9 construct with the CaMV35S promoter for Cas9 expression and AtU6–26 promoter for gRNA expression. Slices of embryonic axis from pea seeds were incubated with A. tumefaciens, and the infected explants on callus‐inducing media were treated at 37°C for 24 h to promote Cas9 activity. Out of 396 explants infected, 45 plantlets were generated, and PCR‐genotyping of neomycin phosphotransferase II from these plantlets identified 8 transgenic plants, resulting in a 2% transformation efficiency. The PsBAS1 genomic locus of the 8 transgenic plants was amplified by PCR, followed by Sanger sequencing of the amplicons to examine the presence of mutations at double‐strand break (DSB) sites. Two transgenic pea lines, referred to as Lines 1 and 2, presented mixed sequencing chromatograms at DSB sites, implying the occurrence of CRISPR/Cas9‐induced mutations (Figure 4). Two overlapping chromatograms with equal area were detected in Line 1; however, the secondary sequencing signals in Line 2 were much weaker than those from Line 1. Seeds were set by self‐fertilization in these two lines, and multiple T2 plants from these two lines were further analyzed to examine the inheritability of the mutations.
FIGURE 4.
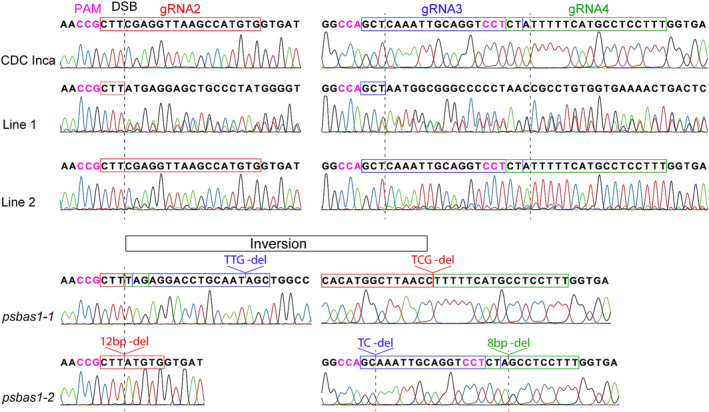
Sequencing chromatograms of PsBAS1 genomic loci in the CDC Inca parental line and psbas1 mutants. The mutant lines were produced using CRISPR constructs with the CaMV35S promoter for Cas9 expression, AtU6–26 promoter for gRNA expression, and 37°C heat treatment.
No sign of mutations at the DSB sites were detected in T2 plants from Line 2, indicating the mutations in the T1 plant were somatic. On the other hand, segregation of two mutant alleles was clearly observed in the T2 population from Line 1. The first mutant allele had an inversion of a 196‐bp DNA fragment between gRNA2 and gRNA4, together with two 3‐bp deletions at gRNA2 and gRNA3 sites, while the second allele includes a deletion in all three gRNA‐binding sites (Figure 4). The ratio of the two types of homozygous mutations and heterozygous mutation was 5:9:4, which approximately fits the Mendelian segregation ratio of 1:2:1 (Table S1). Therefore, the two distinct mutant alleles occurred at the early stage of callus formation in Line 1 and were inherited to the T2 generation. Among 18 T2 individual plants, two different homozygous mutants with no transgene were identified in Lines 1–5 and 1–9 (referred to as psbas1‐1 and psbas1‐2 henceforth). The seeds from these lines were used for further saponin analysis. In summary, using CRISPR/Cas9 to target the PsBAS1 locus, we acquired two psbas1 mutant alleles, and transgene‐free mutant lineages were established in T2 peas.
2.6. Characterization of β‐amyrin synthase mutants
Seeds were collected from the two homozygous T2 mutants, psbas1‐1 and psbas1‐2. The saponin content (DDMP saponin and saponin B) in the seeds from the parental pea cultivar (CDC Inca) and the two mutants were measured by LC/MS. The mutant pea seeds only contained ~0.1% of the saponin amount compared to that of the parental seeds (>99.8% reduction of saponins), indicating the mutation of PsBAS1 effectively eliminates saponins in pea seeds (Figure S4 and Table 4). Multiple individual plants from the two populations (lines psbas1‐1 and psbas1‐2) and control pea plants (CDC Inca) were grown under identical conditions in a phytotron chamber, and several key developmental phenotypes of peas were measured (Table 5 and Figure S5). In the T2 population of psbas1‐1, stem length, number of nodes, and number of seeds per plant were reduced with statistical significance compared to those from CDC Inca, suggesting that off‐target mutation(s) may be present in the psbas1‐1 lineage. On the other hand, no phenotypic differences were observed in the T2 population of line psbas1‐2. When protein and starch contents (dry weight basis) were measured by Near Infra‐Red (NIR) spectroscopy, a moderate increase in protein content (12.8–14.7% increase) and a slight decrease in starch content (4.2–5.3% decrease) was observed in both populations with statistical significance (p value < 0.01; Table 5). Although there is no obvious linkage between isoprenoid metabolism and protein/starch biosynthesis, abolition of saponin seems to indirectly alter protein/starch biosynthesis in pea seeds. Further metabolic studies are necessary to understand the underlying mechanism for the network of different metabolic pathways. In addition, field trials need to be performed to confirm that the saponin biosynthesis is truly controlled by a single gene, PsBAS1 (not by other hidden, inducible genes) and to re‐assess the influence of pabas1‐1 mutation in other agriculturally relevant traits, including those described in Table 5. Taken together, these results demonstrated that knockout of PsBAS1 effectively removed saponins (>99.8% removal in mutants) in yellow pea seeds.
TABLE 4.
Saponin content in CRISPR/Cas9‐induced psbas1 (β‐amyrin synthase) mutants.
| Sample | DDMP saponin a (mg/g) | Saponin B a (μg/g) |
|---|---|---|
| Wild type b | 2.96 ± 0.19 | 29.9 ± 9.2 |
| psbas1‐1 | 0.0020 ± 0.0009 | 0.009 ± 0.002 |
| psbas1‐2 | 0.0035 ± 0.0008 | 0.016 ± 0.002 |
Data are mean ± SD (n = 4). Replicates are from four individual plants grown under the same phytochamber conditions.
The parental variety used for genome‐editing was CDC Inca.
TABLE 5.
Developmental phenotypes of psbas1 mutants.
| Phenotypes | Control a | psbas1‐1 | psbas1‐2 |
|---|---|---|---|
| Germination rate b (%) | 100% ± 0 | 88% ± 5 | 100% ± 0 |
| Stem length c (cm) | 125.6 ± 5.8 | 98.9 ± 8.1*** | 120.2 ± 6.1 |
| Number of nodes c | 32.2 ± 2.2 | 26.7 ± 1.5*** | 32.0 ± 1.3 |
| Average of 10 seed weight d (g) | 3.1 ± 0.1 | 3.1 ± 0.2 | 3.0 ± 0.1 |
| Number of seeds per plant c | 139.3 ± 35.3 | 96.3 ± 23.8* | 140.8 ± 46.6 |
| Number of pods c | 46.7 ± 12.3 | 39.0 ± 8.8 | 48.0 ± 21.1 |
| Number of seeds per pod c | 3.0 ± 0.3 | 2.5 ± 0.2** | 3.0 ± 0.3 |
| Protein content c , e (DMB, %) | 21.1 ± 0.8 | 24.1 ± 0.5** | 23.8 ± 0.7** |
| Starch content c , e (DMB, %) | 60.0 ± 1.7 | 57.5 ± 1.1* | 56.8 ± 1.4** |
Control is the parental variety, CDC Inca, used for genome‐editing.
Data are mean ± SD (n = 3). Replicates are from three trials consisting of 10 individual seeds each.
Data are mean ± SD (n = 6). Replicates are from six individual plants grown under the same phytochamber conditions.
Data are mean ± SD (n = 5). Replicates are from five trials of 10 seeds randomly selected from six plants.
Protein and starch contents are measured by near infra‐red (NIR) on a dry matter basis (DMB).
p < .05 in comparison to the control.
p < .01 in comparison to the control.
p < .001 in comparison to the control.
3. DISCUSSION
3.1. Significance of CRISPR/Cas9 in creating new pea germplasm
A recent meta‐analysis showed that global food demand is expected to increase by 35–56% between 2010 and 2050 (van Dijk et al., 2021). Increased food demand is not a new issue, and scientific innovation has been able to meet the demand in the past. However, the contemporary need to increase the output of crop and animal agricultural products faces unique challenges that were not considered during the green revolution in the 1960s. Namely, increases in the productivity of global food must not be accompanied by increases in greenhouse gas (GHG) emissions. The greatest contributor to anthropogenic methane gas is enteric fermentation in ruminant animals, and methane is more than 20 times as effective at trapping heat as carbon dioxide. In addition, the nitrous oxide released from soils treated with synthetic N‐fertilizer is ~300 times more potent GHG than carbon dioxide (del Grosso et al., 2008). To mitigate global warming, decreasing the global dependency on animal proteins and synthetic N‐fertilizer is critically important. From this perspective, pea seeds are an excellent solution as they have high protein content from biological nitrogen fixation (i.e., without using synthetic N‐fertilizer) and thus are an ideal food source to partially replace animal protein and to reduce dependency on synthetic N‐fertilizer (Jensen et al., 2012). However, unfavorable off‐flavors, off‐aromas, and anti‐nutrients are associated with pea seeds, which significantly lowers consumer acceptance of pea products.
Novel favorable traits can be developed with traditional breeding, but this approach has intrinsic limitations as it relies on naturally occurring or randomly induced mutations. Additionally, trait development often requires mutations in multiple genes of the same function (i.e., functional redundancy), which is difficult to achieve through random mutations. CRISPR/Cas9 is an excellent complement to traditional breeding for targeted multigene mutations, as multiple gRNAs can be expressed to target multiple genes. CRISPR/Cas9 offers a practical solution for the development of elite varieties of peas and other legume crops with reduced levels of off‐flavors and off‐aromas as these traits are typically determined by simple biosynthetic pathways, which can be knocked‐out with single or multiple mutations of isoenzymes. This has been demonstrated with the generation of LOX mutants in both soybean (gmlox1, gmlox2, gmlox3, and combinations thereof; Wang et al., 2020) and pea (pslox2; Bhowmik et al., 2023). LOXs produce volatile compounds that cause off‐aromas. In these studies, mutation of a small number of LOX genes significantly reduced LOX activity in the seeds. As demonstrated in this work and others, well‐designed and executed CRISPR/Cas9 mutagenesis procedures can generate novel mutant alleles, which can be used to create standalone genome‐edited crops or to supply favorable new alleles to ongoing breeding programs.
3.2. Optimization of CRISPR/Cas9
Using hairy root induction in pea, the effectiveness of various promoters and tissue culture conditions on in planta CRISPR/Cas9 genome editing was assessed in 4 weeks compared to the several months typically required for transformation using A. tumefaciens and tissue culture. During the preparation of this study, Li et al. (2023) published work that also utilized hairy root transformation for the rapid assessment of CRISPR/Cas9 genome editing in pea. In their work, A. rhizogenes strain K599 was able to induce hairy roots with 66.6% efficiency, and 62.5% of these hairy roots were transgenic in the pea cultivar, Zhongwan 6. This is in stark contrast to the efficiency observed in this study for K599 (39% of explants produced hairy roots and only 8% of these expressed the GFP transgene) in the CDC Inca variety. In our work, five strains of A. rhizogenes were tested, resulting in AR10 being the best performing strain with 92% of explants inducing hairy roots and 56% of these expressing foreign genes (Table 2). Kaur et al. (2022) observed similar dramatic differences in transformation efficiencies between different pea cultivars using various A. tumefaciens strains. Taken together, these data highlight the importance of testing a variety of Agrobacteria strains, when optimizing transformation protocols for specific pea cultivars.
The AtU6–26 promoter for gRNA expression and 37°C heat treatments were the most effective conditions for genome editing that were tested here. Interestingly, for the mutation of PsLOX2, Bhowmik et al. (2023) employed a similar construct as the one in this study, which used the CaMV35S promoter for gRNA expression and did not expose the explants to 37°C heat treatments. Despite this, they were able to obtain four T1 mutant lines from 17 transgenic plants (24%). In the hairy root system used in our study, 9% of mutation rate (minor mutations) was observed when similar conditions to Bhowmik et al. (2023) were used. This difference in editing efficiency is likely due to differences in the chromatin structure at the PsLOX2 and PsBAS1 target loci, as well as difference in gRNA functionality (Jensen et al., 2017).
The CRISPR vectors deployed in this study shared some common features with those used by Li et al. (2023). Both used tRNA sequences in their gRNA expression cassettes. However, in our study, the tRNA sequences were used to produce multiple gRNAs from a single transcript but only a single gRNA was expressed in Li et al.'s work. Li et al. observed that constructs including the tRNA sequence had greater editing efficiencies than those without, possibly due to box A and B promoter elements present in tRNA sequences, known to help recruit the RNA polymerase III complex (Xie et al., 2015). The most effective CRISPR vectors used by Li et al. included a PsU6.3 promoter for gRNA expression and a CaMV35S promoter for Cas9 expression, which had 7.1–52.4% editing efficiencies in hairy roots. This was greater than the constructs that used the AtU6–26 promoter and had editing efficiencies of 1.9–1.1%. Similar improvements were observed in soybean when an endogenous GmU6 promoter was compared to the AtU6–26 promoter for CRISPR/Cas9 genome editing (Sun et al., 2015). Thus, we presume that the CRISPR vector used in our work can be further improved by using the PsU6.3 promoter. We are currently testing this possibility in pea.
3.3. Phenotyping of two mutant lines
As two mutant alleles of PsBAS1 were observed to have segregated in a standard Mendelian ratio (1:2:1) in the T2 population of line 1 (Table S1), we can conclude that the gRNA/Cas9 complex incurred two distinct mutations in primordial cells during the early stages of callus formation. These two alleles were then inherited to the T2 population. However, the two homozygous mutant lines (psbas1‐1 and psbas1‐2), selected for further phenotyping, showed different phenotypes (Table 5). The first mutant line (psbas1‐1) was shorter, with fewer nodes, fewer number of seeds per plant, and fewer seeds per pod than those from the control (CDC Inca variety). In contrast, the second mutant line (psbas1‐2) displayed identical patterns of growth and development as the control. As the two lines showed different phenotypes, the compromised growth and developmental patterns in psbas1‐1 are not the direct outcomes of psbas1 mutation but are likely to be caused by off‐target mutation(s). Indeed, in other studies involving the silencing or knock‐out of BAS in soybean (Takagi et al., 2011) and oat (Avena strigose; Haralampidis et al., 2001), respectively, no abnormalities in growth were detected, and EMS‐induced psbas1 mutants also displayed normal yields (Vernoud et al., 2021). This further supports the conclusion that the developmental phenotypes of psbas1‐1 are caused by off‐target mutation(s) and are not the result of the psbas1 mutant allele. The assumed off‐target mutation(s) could have been carried over to the T2 population of psbas1‐1, whereas the same off‐target mutation(s) was not inherited in the T2 population of psbas1‐2. Therefore, future breeding program to introgress saponin deficiency should use the psbas1‐2 line.
Besides the visible phenotypes, the protein and starch content were measured in seeds from control and mutant lines (Table 5). Unexpectedly, small (12–14%) yet statistically significant (p value < 0.01) increases of protein content were measured in the seeds of both mutant lines. On the contrary, small (~5%) decreases of starch content with statistical significance were observed in both mutant lines, compared to the control. It is difficult to link the isoprenoid metabolism to protein and starch biosynthesis, and thus, no straightforward explanation for the increased protein can be given at present. Saponins accumulate in the CDC Inca embryo at a relatively high concentration (2.7 mg/g). Although speculative, it is possible that saponins interfere with protein biosynthesis in the pea embryos. Having more protein in pea seeds is a serendipitous positive outcome. However, caution needs to be given to the facts that (i) high protein content can still be the result of unknown off‐target effects in both mutants, and (ii) the pea plants in this study were cultivated under controlled phytotron chamber conditions, which may not truly reflect the protein content in field grown CDC Inca seeds. CDC Inca has been a commercially popular cultivar in the Canadian prairies, and from 11 years of field tests in various locations, the protein content of field grown CDC Inca has been established to be an average of 22.4% (SaskSeed guide, 2023). This value is slightly higher than the phytotron chamber‐grown control in this study (21.1%) but still lower than the averages of the two mutant lines (23.8% and 24.2%). The protein contents of the control and two mutant lines fall in the low‐ and high‐end, respectively, of that from the field grown CDC Inca. It is imperative to carry out genetic and field studies to elucidate the cause and practical significance of the increased protein content in the mutant pea lines.
As reported previously, saponins have defensive roles against insects and pathogens. Increase in saponins was reported in pea when grown in soils naturally contaminated with Aphanomyces euteiches, and Rhizoctonia solani and Fusarium oxysporum (Oliete et al., 2022). Saponins in oat roots have been implicated in the prevention of zoospore cyst wall formation of Aphanomyces and fungal pathogens (Deacon & Mitchell, 1985), and saponin deficient oat mutants have been found to be more susceptible to fungal pathogens (Leveau et al., 2019; Papadopoulou et al., 1999). The application of exogenous saponins has also been utilized to prevent insect feeding on stored legume seeds (Applebaum et al., 1969; Taylor et al., 2004). Given that the saponin level is increased in pea plants during biotic stress and the role that saponins play in defense in other plant species, it is important to investigate the role of saponin in biotic interactions with insects and pathogens, particularly in field tests. Additionally, the over‐production of saponins in an M. truncatula line resulted in an increase in nodule formation (Confalonieri et al., 2009). Therefore, the nodule‐forming capabilities of psbas1 lines relative to control lines needs to be examined too.
4. MATERIALS AND METHODS
4.1. Pea growth conditions in soil, hairy root induction, and tissue culture
Yellow pea seeds (CDC Inca) were provided by the Crop Development Centre, University of Saskatchewan, Canada (Warkentin et al., 2017). Pea plants were grown in 4″ pots in Sunshine No. 4 potting mix (Sun Gro Horticulture, Vancouver, Canada) in phytotron chambers with 16/8 h day/night photoperiod and 20/18°C day/night temperatures. At 25 days old, the seedlings were transferred to 2 gal pots (two seedlings per pot). Plants were fertilized weekly with 20‐20‐20 fertilizer (Miracle‐Gro All Purpose Water Soluble Plant Food, The Scotts Company). Induction of pea hair roots and transformation by tissue culture are described in the Supplemental methods.
4.2. Cloning PsBAS1 CRISPR vectors
Appropriate gRNA‐binding sites were identified in the first two exons of PsBAS1 (AB034802). PsBAS1 was amplified with primers 1/2 (hereafter, all primers are listed in Table S2), and the resulting DNA fragment was cloned into the EcoRV site of pBluescript. Five candidate gRNA‐binding sites were identified and ranked, based on their CRISPRater (Labuhn et al., 2018) score using the web‐application CCTop (Stemmer et al., 2015). For in vitro cleavage assays, the PsBAS1 template was amplified with primers 3/4, and five gRNAs were in vitro synthesized with a HiScribe™ T7 Quick High Yield RNA Synthesis Kit (NEB). The DNA templates for in vitro transcription, comprised of the T7 promoter, spacer RNA, and tracrRNA, were prepared by amplifying the 76‐bp tracrRNA template using primers (5–9) and a common primer 10, and the synthesized gRNAs were purified using LiCl precipitation (Kwon et al., 2023). The PsBAS1 cleavage template, gRNAs, and recombinant Cas9 were incubated at 37°C for 1 h, and the cleavage efficiency of the gRNAs was evaluated using agarose gel electrophoresis. gRNAs 2/3/4 were cloned into the gRNA expression vector as described in Figure S2 using Gibson Assembly with primers 11–16. The synthesized AtU6–26 expression cassette (Figure S1) was used to assemble the gRNA cassette using primers 11–15 and 17.
4.3. RNA‐Seq library preparation and analysis
Libraries were prepared for strand‐specific RNA sequencing as described previously (Yan et al., 2020). Briefly, 2 μg of total RNA was used to enrich mRNA and underwent DNase‐digestion and fragmentation. Reverse transcription was employed to synthesize first‐strand cDNAs with fragmented mRNA. Second strand synthesis was performed with dNTPs, and double‐strand (ds) cDNAs were end‐repaired followed by dA‐tailing and adapter ligation. Solid‐phase reversible immobilization (SPRI)‐based size selection was conducted to isolate cDNA fragments of adapter‐coupled libraries. The second strand with dUTP was digested. PCR was employed to enrich and index RNA‐seq libraries. Libraries of three biological replicates were prepared for each sample and sequenced on an Illumina HiSeq 4000 system with paired‐end (PE) reads of 100 bp. Differential expression analysis was carried out by DESeq2 with cut‐off values of Log2 fold change >2 and adjusted p value of <.05 (Love et al., 2014).
4.4. Analysis of pea seed saponin, protein and starch content
A saponin B standard was purchased (Chromadex). A DDMP saponin standard was purified from commercial pea flour (Parrheim Foods) as described in the supplemental methods. For saponin analysis from pea seeds, pea flour, and fiber, saponins were extracted from 100 mg of samples for 2 h in 1 ml of 80% methanol under gentle agitation (100 rpm). Samples were centrifuged at 14,600g for 10 min, and the supernatants were collected. Supernatants were centrifuged again. Ten microliters of samples was loaded onto the LC system described in the supplemental methods. DDMP saponin and saponin B in samples were analyzed by the following LC gradient: 1 min hold at 90:10 (A/B; water/acetonitrile), 3 min change to 60:40, 4.8 min change to 22:78, 1.2 min change to 20:80, 1.5 min change to 0:100, hold for 1.5 min at 0:100. The solvents were passed through the column at 400 μl/min, and the column temperature was set to 30°C. Metabolites were ionized via H‐ESI (heated electrospray ionization), total ion scan was performed in positive ion mode, and specific ions were selected to visualize saponins. To increase detection sensitivity, saponins were also analyzed by multiple reaction monitoring (MRM) with the selected parent and daughter ions: m/z of 943.7 (423.3, 617.4, and 797.5) and 1,069.5 (423.3, 743.3, and 923.5). The protein and starch composition of the pea seeds were measured with a FOSS DS2500 Near Infrared Spectrophotometer (NIR; FOSS Analytics, Eden Prairie, MN, United StatesA) that collected spectra from whole pea samples. The percent protein and starch were predicted using an equation developed at the Crop Development Centre (University of Saskatchewan) following a procedure outlined in Arganosa et al. (2006).
AUTHOR CONTRIBUTIONS
Connor L. Hodgins performed the construction of the CRISPR/Cas9 vectors, in vitro Cas9 assays, stable pea transformation, and characterization of mutant peas and wrote the manuscript. Eman M. Salama and Susan A. Roth performed hairy root induction and characterization; Rahul Kumar and Connor L. Hodgins measured saponins in peas using LC/MS. Irene Z. Cheung performed the construction of CRISPR/Cas9 vector and in vitro Cas9 assays. Byung‐Kook Ham, Yang Zhao, and Jieyu Chen generated the Illumina sequencing libraries; Pankaj Bhowmik was consulted for the pea transformation; Gene C. Arganosa and Thomas D. Warkentin analyzed pea seed protein and starch content. Dae‐Kyun Ro designed the experiments, managed the project, and wrote the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors did not report any conflict of interest.
Supporting information
Data S1. Supporting Information
Figure S1. AtU6–26 driven gRNA expression cassette.
Figure S2. CRISPR construct cloning strategy A) Cloning of the CaMV35S gRNA expression cassette is represented here. To express multiple gRNAs in a single transcript each gRNA fragment was flanked by an auto‐cleavable tRNA sequence. To assemble this gRNA‐tRNA array, each gRNA fragment was produced using a tracrRNA‐pre‐tRNAGly template. gRNA fragment 1 underwent a second amplification step to add a tRNA sequence to the 5′ end. Note that the reverse primer for gRNA fragment 3 in the AtU6 (AtU6–26) vector did not contain a 5′ overhang. The CaMV35S and AtU6 gRNA expression cassettes were synthesized and added to the NotI/AscI site of the pENTR™/D‐TOPO™ vector. The three gRNA fragments were cloned into the BsaI site of the gRNA expression cassettes in a single step using Gibson assembly. The gRNA expression cassette was then transferred to a modified pK7WG2D binary vector containing a CaMV35S driven Cas9 expression cassette using LR Clonase™ II Enzyme mix. B) The final CRISPR constructs.
Figure S3. Analysis of mutations in PsBAS1 from hairy roots transformed with CRISPR constructs. A) Hairy roots growing from a pea explant. B) Hairy roots growing on selection media after being removed from the explant. C) Agarose gel images of PsBAS1 PCR products amplified from transgenic hairy roots. Some PCR products vary in size from the wild‐type (WT) and therefore represent mutations. PCR products were also Sanger sequenced to identify lines with more subtle mutations. WT samples were generated using the native AR10 strain, lacking the CRISPR construct. (+) samples are from CRISPR construct plasmid templates. D) An example Sanger sequencing chromatogram from line 3 produced with the AtU6–26 promoter for gRNA expression and grown at 24 °C. This is an example of a chromatogram signal which would be interpreted as a mutation. This line was determined to be mutated because a secondary peak appeared near the DSB (represented by a dashed line) of gRNA4 indicating a mixture of alleles.
Figure S4. LC–MS analysis of saponins extracted from wild‐type and psbas1 seeds. A) MRM chromatograms for the saponin B standard and metabolite extracts from CDC Inca and psbas1 mutant seeds. The m/z values for parental and daughter ions were 943.7, and 423.3, 617.4, and 797.5, respectively. B) MRM chromatograms for the DDMP saponin standard and metabolite extracts from CDC Inca and psbas1 mutant seeds. The m/z values for parental and daughter ions were 1,069.5, and 423.3, 743.3, and 923.5, respectively.
Figure S5. Wild‐type (CDC Inca) and psbas1 mutant lines at different developmental stages.
A) 4‐day‐old germinated seeds. B) 15‐day‐old seedlings. C) 66‐day‐old plants.
Table S1. Genotype of line 1 T2 progeny.
Table S2. Primers used in this article. Underlined sequences represent overhanging sequences used for Gibson assembly which will not bind to the desired PCR template.
ACKNOWLEDGMENTS
We thank Christina Yanta (University of British Columbia, Canada) for the differential expression analysis and Parrheim Foods (Saskatoon, Canada) for donating pea flour and fiber samples. We thank the funding supporters from Alberta Innovates (grant number: 2018F160R) to DKR, Discovery Grant program from Natural Sciences and Engineering Research Council of Canada (NSERC) to BKH and DKR, and Aquatic and Crop Resource Development Research Centre of the National Research Council Canada (NRC) as part of the Sustainable Protein Production (SPP) program to PB.
Hodgins, C. L. , Salama, E. M. , Kumar, R. , Zhao, Y. , Roth, S. A. , Cheung, I. Z. , Chen, J. , Arganosa, G. C. , Warkentin, T. D. , Bhowmik, P. , Ham, B.‐K. , & Ro, D.‐K. (2024). Creating saponin‐free yellow pea seeds by CRISPR/Cas9‐enabled mutagenesis on β‐amyrin synthase. Plant Direct, 8(1), e563. 10.1002/pld3.563
DATA AVAILABILITY STATEMENT
The transcriptome data generated and analyzed in this study are available through NCBI under BioProject PRJNA1020263 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1020263).
REFERENCES
- Applebaum, S. W. , Marco, S. , & Birk, Y. (1969). Saponins as possible factors of resistance of legume seeds to the attack of insects. Journal of Agricultural and Food Chemistry, 17, 618–622. 10.1021/jf60163a020 [DOI] [Google Scholar]
- Arganosa, G. C. , Warkentin, T. D. , Racz, V. J. , Blade, S. , Phillips, C. , & Hsu, H. (2006). Prediction of crude protein content in field peas using near infrared reflectance spectroscopy. Canadian Journal of Plant Science, 86, 157–159. 10.4141/P04-195 [DOI] [Google Scholar]
- Bhowmik, P. , Yan, W. , Hodgins, C. , Polley, B. , Warkentin, T. , Nickerson, M. , Ro, D.‐K. , Marsolais, F. , Domoney, C. , Shariati‐Ievari, S. , & Aliani, M. (2023). CRISPR/Cas9‐mediated lipoxygenase gene‐editing in yellow pea leads to major changes in fatty acid and flavor profiles. Frontiers in Plant Science, 14, 1246905. 10.3389/fpls.2023.1246905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstin, J. , Marget, P. , Huart, M. , Moessner, A. , Mangin, B. , Duchene, C. , Desprez, B. , Munier‐Jolain, N. , & Duc, G. (2007). Developmental genes have pleiotropic effects on plant morphology and source capacity, eventually impacting on seed protein content and productivity in pea. Plant Physiology, 144, 768–781. 10.1104/pp.107.096966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S. Y. , Seki, H. , Fujisawa, Y. , Shimoda, Y. , Hiraga, S. , Nomura, Y. , Saito, K. , Ishimoto, M. , & Muranaka, T. (2020). A cellulose synthase‐derived enzyme catalyses 3‐O‐glucuronosylation in saponin biosynthesis. Nature Communications, 11, 5664. 10.1038/s41467-020-19399-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri, M. , Cammareri, M. , Biazzi, E. , Pecchia, P. , Fevereiro, M. P. S. , Balestrazzi, A. , Tava, A. , & Conicella, C. (2009). Enhanced triterpene saponin biosynthesis and root nodulation in transgenic barrel medic (Medicago truncatula Gaertn.) expressing a novel β‐amyrin synthase (AsOXA1) gene. Plant Biotechnology Journal, 7, 172–182. 10.1111/j.1467-7652.2008.00385.x [DOI] [PubMed] [Google Scholar]
- Deacon, J. W. , & Mitchell, R. T. (1985). Toxicity of oat roots, oat root extracts, and saponins to zoospores of Pythium spp. and other fungi. Transactions of the British Mycology Society, 84, 479–487. 10.1016/S0007-1536(85)80010-3 [DOI] [Google Scholar]
- del Grosso, S. J. , Wirth, T. , Ogle, S. M. , & Parton, W. J. (2008). Estimating agricultural nitrous oxide emissions. Eos, Transactions, American Geophysical Union, 89, 529–529. 10.1029/2008EO510001 [DOI] [Google Scholar]
- Haralampidis, K. , Bryan, G. , Qi, X. , Papadopoulou, K. , Bakht, S. , Melton, R. , & Osbourn, A. (2001). A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proceedings of the National Academy of Sciences of the United States of America, 98, 13431–13436. 10.1073/pnas.231324698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, L. , Vincken, J. , van Koningsveld, G. , Legger, A. , Gruppen, H. , van Boekel, T. , Roozen, J. , & Voragen, F. (2006). Bitterness of saponins and their content in dry peas. Journal of the Science of Food and Agriculture, 86, 1225–1231. 10.1002/jsfa.2473 [DOI] [Google Scholar]
- Hoffman, N. E. (2021). Revisions to USDA biotechnology regulations: The SECURE rule. Proceedings of the National Academy of Sciences of the United States of America, 118, e2004841118. 10.1073/pnas.2004841118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, E. S. , Peoples, M. B. , Boddey, R. M. , Gresshoff, P. M. , Hauggaard‐Nielsen, H. , Alves, B. J. R. , & Morrison, M. J. (2012). Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agronomy for Sustainable Development, 32, 329–364. 10.1007/s13593-011-0056-7 [DOI] [Google Scholar]
- Jensen, K. T. , Fløe, L. , Petersen, T. S. , Huang, J. , Xu, F. , Bolund, L. , Luo, Y. , & Lin, L. (2017). Chromatin accessibility and guide sequence secondary structure affect CRISPR‐Cas9 gene editing efficiency. FEBS Letter, 591, 1892–1901. 10.1002/1873-3468.12707 [DOI] [PubMed] [Google Scholar]
- Kaur, R. , Donoso, T. , Scheske, C. , Lefsrud, M. , & Singh, J. (2022). Highly efficient and reproducible genetic transformation in pea for targeted trait improvement. ACS Agricultural Science and Technology, 2, 780–787. 10.1021/acsagscitech.2c00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreplak, J. , Madoui, M.‐A. , Cápal, P. , Novák, P. , Labadie, K. , Aubert, G. , Bayer, P. E. , Gali, K. K. , Syme, R. A. , Main, D. , Klein, A. , Bérard, A. , Vrbová, I. , Fournier, C. , d'Agata, L. , Belser, C. , Berrabah, W. , Toegelová, H. , Milec, Z. , … Burstin, J. (2019). A reference genome for pea provides insight into legume genome evolution. Nature Genet, 51, 1411–1422. 10.1038/s41588-019-0480-1 [DOI] [PubMed] [Google Scholar]
- Kwon, M. , Hodgins, C. , Salama, E. , Dias, K. , Parikh, A. , Mackey, A. , Catenza, K. , Vederas, J. , & Ro, D.‐K. (2023). New insights into natural rubber biosynthesis from rubber‐deficient lettuce mutants expressing goldenrod or guayule cis‐prenyltransferase. New Phytologist, 239, 1098–1111. 10.1111/nph.18994 [DOI] [PubMed] [Google Scholar]
- Labuhn, M. , Adams, F. F. , Ng, M. , Knoess, S. , Schambach, A. , Charpentier, E. M. , Schwarzer, A. , Mateo, J. L. , Klusmann, J.‐H. , & Heckl, D. (2018). Refined sgRNA efficacy prediction improves large‐and small‐scale CRISPR–Cas9 applications. Nucleic Acids Research, 46, 1375–1385. 10.1093/nar/gkx1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc, C. , Zhang, F. , Mendez, J. , Lozano, Y. , Chatpar, K. , Irish, V. F. , & Jacob, Y. (2018). Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. The Plant Journal, 93, 377–386. 10.1111/tpj.13782 [DOI] [PubMed] [Google Scholar]
- Leveau, A. , Reed, J. , Qiao, X. , Stephenson, M. J. , Mugford, S. T. , Melton, R. E. , Rant, J. C. , Vickerstaff, R. , Langdon, T. , & Osbourn, A. (2019). Towards take‐all control: A C‐21β oxidase required for acylation of triterpene defence compounds in oat. New Phytologist, 221, 1544–1555. 10.1111/nph.15456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Liu, R. , Xu, R. , Varshney, R. K. , Ding, H. , Li, M. , Yan, X. , Huang, S. , Li, J. , & Wang, D. (2023). Development of an agrobacterium‐mediated CRISPR/Cas9 system in pea (Pisum sativum L.). The Crop Journal, 11, 132–139. 10.1016/j.cj.2022.04.011 [DOI] [Google Scholar]
- Lin, J. , Krishnan, P. G. , & Wang, C. (2006). Retention of isoflavones and saponins during the processing of soy protein isolates. Journal of the American Oil Chemists' Society, 83, 59–63. 10.1007/s11746-006-1176-0 [DOI] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga, W. J. , Pozniak, C. J. , & Kagale, S. (2021). Advanced domestication: Harnessing the precision of gene editing in crop breeding. Plant Biotechnology Journal, 19, 660–670. 10.1111/pbi.13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty, S. (2022). China's approval of gene‐edited crops energizes researchers. Nature, 602, 559–560. 10.1038/d41586-022-00395-x [DOI] [PubMed] [Google Scholar]
- Mertens, J. , Pollier, J. , van den Bossche, R. , Lopez‐Vidriero, I. , Franco‐Zorrilla, J. M. , & Goossens, A. (2016). The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula . Plant Physiology, 170, 194–210. 10.1104/pp.15.01645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, M. , Shibuya, M. , Kushiro, T. , Masuda, K. , & Ebizuka, Y. (2000). Molecular cloning and functional expression of triterpene synthases from pea (Pisum sativum). European Journal of Biochemistry, 267, 3453–3460. 10.1046/j.1432-1327.2000.01357.x [DOI] [PubMed] [Google Scholar]
- Moses, T. , Thevelein, J. M. , Goossens, A. , & Pollier, J. (2014). Comparative analysis of CYP93E proteins for improved microbial synthesis of plant triterpenoids. Phytochemistry, 108, 47–56. 10.1016/j.phytochem.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Oliete, B. , Lubbers, S. , Fournier, C. , Jeandroz, S. , & Saurel, R. (2022). Effect of biotic stress on the presence of secondary metabolites in field pea grains. Journal of the Science of Food and Agriculture, 102, 4942–4948. 10.1002/jsfa.11861 [DOI] [PubMed] [Google Scholar]
- Papadopoulou, K. , Melton, R. E. , Leggett, M. , Daniels, M. J. , & Osbourn, A. E. (1999). Compromised disease resistance in saponin‐deficient plants. Proceedings of the National Academy of Sciences of the United States of America, 96, 12923–12928. 10.1073/pnas.96.22.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore, J. , & Nemecek, T. (2018). Reducing food's environmental impacts through producers and consumers. Science, 360, 987–992. 10.1126/science.aaq0216 [DOI] [PubMed] [Google Scholar]
- Robinson, G. H. J. , & Domoney, C. (2021). Perspectives on the genetic improvement of health‐ and nutrition‐related traits in pea. Plant Physiology and Biochemistry, 158, 353–362. 10.1016/j.plaphy.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer, M. , Thumberger, T. , del Sol Keyer, M. , Wittbrodt, J. , & Mateo, J. L. (2015). CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE, 10, e0124633. 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Hu, Z. , Chen, R. , Jiang, Q. , Song, G. , Zhang, H. , & Xi, Y. (2015). Targeted mutagenesis in soybean using the CRISPR‐Cas9 system. Scientific Reports, 5, 10342. 10.1038/srep10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, K. , Nishizawa, K. , Hirose, A. , Kita, A. , & Ishimoto, M. (2011). Manipulation of saponin biosynthesis by RNA interference‐mediated silencing of β‐amyrin synthase gene expression in soybean. Plant Cell Reports, 30, 1835–1846. 10.1007/s00299-011-1091-1 [DOI] [PubMed] [Google Scholar]
- Taylor, W. G. , Fields, P. G. , & Sutherland, D. H. (2004). Insecticidal components from field pea extracts: Soyasaponins and lysolecithins. Journal of Agricultural and Food Chemistry, 52, 7484–7490. 10.1021/jf0308051 [DOI] [PubMed] [Google Scholar]
- Tsurumi, S. , Takagi, T. , & Hashimoto, T. (1992). A γ‐pyronyl‐triterpenoid saponin from Pisum sativum . Phytochemistry, 31, 2435–2438. 10.1016/0031-9422(92)83294-9 [DOI] [PubMed] [Google Scholar]
- van Dijk, M. , Morley, T. , Rau, M. L. , & Saghai, Y. (2021). A meta‐analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nature Food, 2, 494–501. 10.1038/s43016-021-00322-9 [DOI] [PubMed] [Google Scholar]
- Vernoud, V. , Lebeigle, L. , Munier, J. , Marais, J. , Sanchez, M. , Pertuit, D. , Rossin, N. , Darchy, B. , Aubert, G. , & le Signor, C. (2021). β‐amyrin synthase1 controls the accumulation of the major saponins present in pea (Pisum sativum). Plant & Cell Physiology, 62, 784–797. 10.1093/pcp/pcab049 [DOI] [PubMed] [Google Scholar]
- Wang, J. , Kuang, H. , Zhang, Z. , Yang, Y. , Yan, L. , Zhang, M. , Song, S. , & Guan, Y. (2020). Generation of seed lipoxygenase‐free soybean using CRISPR‐Cas9. The Crop Journal, 8, 432–439. 10.1016/j.cj.2019.08.008 [DOI] [Google Scholar]
- Warkentin, T. , Tar'an, B. , Banniza, S. , Vandenberg, A. , Bett, K. , Arganosa, G. , Barlow, B. , Ife, S. , Horner, J. , de Silva, D. , Wagenhoffer, S. , Liu, Y. , Prado, T. , & Mikituk, K. (2017). CDC Inca yellow field pea. Canadian Journal of Plant Science, 98, CJPS‐2017‐0141. 10.1139/cjps-2017-0141 [DOI] [Google Scholar]
- Xie, K. , Minkenberg, B. , & Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proceedings of the National Academy of Sciences of the United States of America, 112, 3570–3575. 10.1073/pnas.142029411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y. , Ham, B.‐K. , Chong, Y. H. , Yeh, S.‐D. , & Lucas, W. J. (2020). A plant SMALL RNA‐BINDING PROTEIN 1 family mediates cell‐to‐cell trafficking of RNAi signals. Molecular Plant, 13, 321–335. 10.1016/j.molp.2019.12.00 [DOI] [PubMed] [Google Scholar]
- Yu, B. , Patterson, N. , & Zaharia, L. I. (2022). Saponin biosynthesis in pulses. Plants, 11, 3505. 10.3390/plants11243505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information
Figure S1. AtU6–26 driven gRNA expression cassette.
Figure S2. CRISPR construct cloning strategy A) Cloning of the CaMV35S gRNA expression cassette is represented here. To express multiple gRNAs in a single transcript each gRNA fragment was flanked by an auto‐cleavable tRNA sequence. To assemble this gRNA‐tRNA array, each gRNA fragment was produced using a tracrRNA‐pre‐tRNAGly template. gRNA fragment 1 underwent a second amplification step to add a tRNA sequence to the 5′ end. Note that the reverse primer for gRNA fragment 3 in the AtU6 (AtU6–26) vector did not contain a 5′ overhang. The CaMV35S and AtU6 gRNA expression cassettes were synthesized and added to the NotI/AscI site of the pENTR™/D‐TOPO™ vector. The three gRNA fragments were cloned into the BsaI site of the gRNA expression cassettes in a single step using Gibson assembly. The gRNA expression cassette was then transferred to a modified pK7WG2D binary vector containing a CaMV35S driven Cas9 expression cassette using LR Clonase™ II Enzyme mix. B) The final CRISPR constructs.
Figure S3. Analysis of mutations in PsBAS1 from hairy roots transformed with CRISPR constructs. A) Hairy roots growing from a pea explant. B) Hairy roots growing on selection media after being removed from the explant. C) Agarose gel images of PsBAS1 PCR products amplified from transgenic hairy roots. Some PCR products vary in size from the wild‐type (WT) and therefore represent mutations. PCR products were also Sanger sequenced to identify lines with more subtle mutations. WT samples were generated using the native AR10 strain, lacking the CRISPR construct. (+) samples are from CRISPR construct plasmid templates. D) An example Sanger sequencing chromatogram from line 3 produced with the AtU6–26 promoter for gRNA expression and grown at 24 °C. This is an example of a chromatogram signal which would be interpreted as a mutation. This line was determined to be mutated because a secondary peak appeared near the DSB (represented by a dashed line) of gRNA4 indicating a mixture of alleles.
Figure S4. LC–MS analysis of saponins extracted from wild‐type and psbas1 seeds. A) MRM chromatograms for the saponin B standard and metabolite extracts from CDC Inca and psbas1 mutant seeds. The m/z values for parental and daughter ions were 943.7, and 423.3, 617.4, and 797.5, respectively. B) MRM chromatograms for the DDMP saponin standard and metabolite extracts from CDC Inca and psbas1 mutant seeds. The m/z values for parental and daughter ions were 1,069.5, and 423.3, 743.3, and 923.5, respectively.
Figure S5. Wild‐type (CDC Inca) and psbas1 mutant lines at different developmental stages.
A) 4‐day‐old germinated seeds. B) 15‐day‐old seedlings. C) 66‐day‐old plants.
Table S1. Genotype of line 1 T2 progeny.
Table S2. Primers used in this article. Underlined sequences represent overhanging sequences used for Gibson assembly which will not bind to the desired PCR template.
Data Availability Statement
The transcriptome data generated and analyzed in this study are available through NCBI under BioProject PRJNA1020263 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1020263).


