Abstract
PIWI-interacting RNAs (piRNAs) and their associated PIWI clade Argonaute proteins constitute the core of the piRNA pathway. In gonadal cells, this conserved pathway is crucial for genome defense, and its main function is to silence transposable elements. This is achieved through posttranscriptional and transcriptional gene silencing. Precursors that give rise to piRNAs require specialized transcription and transport machineries because piRNA biogenesis is a cytoplasmic process. The ping-pong cycle, a posttranscriptional silencing mechanism, combines the cleavage-dependent silencing of transposon RNAs with piRNA production. PIWI proteins also function in the nucleus, where they scan for nascent target transcripts with sequence complementarity, instructing transcriptional silencing and deposition of repressive chromatin marks at transposon loci. Although studies have revealed numerous factors that participate in each branch of the piRNA pathway, the precise molecular roles of these factors often remain unclear. In this review, we summarize our current understanding of the mechanisms involved in piRNA biogenesis and function.
Keywords: PIWI proteins, piRNA clusters, ping-pong loop, piRNA biogenesis, transposon control, transcriptional gene silencing, posttranscriptional gene silencing
INTRODUCTION
The discovery of RNA interference (RNAi) revealed broad roles for small noncoding RNAs, and the ribonucleoprotein complexes that they program, in plant and animal biology, with functions ranging from gene regulation to antiviral defense and transposon control (33). At the heart of RNAi-related pathways are small RNA-loaded Argonaute proteins that recognize and silence RNA targets. Argonaute proteins share characteristic structural features including an amino-terminal domain, a PAZ domain, a MID domain, and the PIWI domain (reviewed in 56, 135). Based upon their sequences, Argonautes are separated into three groups, the AGO clade, the WAGO clade, and the animal-specific PIWI clade (reviewed in 56). Argonautes from different clades associate with defined small RNA classes, with microRNAs and small interfering RNAs (siRNAs) acting through AGO and WAGO clade proteins and PIWI-interacting RNAs (piRNAs) associating with Argonautes of the PIWI family (reviewed in 56, 135). The piRNA pathway has many characteristics that distinguish it from canonical RNAi. In the canonical pathway, microRNAs and siRNAs are derived from double-stranded RNA (dsRNA) precursors and processed by the ribonuclease Dicer into 20- to 24-nucleotide (nt) species. In contrast, piRNAs, which range from 23 to 32 nt in length, originate from single-stranded precursors and are generated via a Dicer-independent mechanism (54, 149).
Unlike phenotypes observed for many AGO clade mutants, mutations in PIWI proteins typically result in sterility but are not lethal to the organism. These observations correlate with expression patterns, as PIWI proteins are most often restricted to gonadal tissues. Indeed, to date the function of piRNAs is best understood in animal germ cells, where they have been shown to silence the panoply of transposable elements (TEs) that threaten the integrity of germ cell genomes. The first indication that a small RNA-based regulatory mechanism could protect against transposon mobilization was the description of repeat-associated siRNAs (rasiRNAs), small RNAs found abundantly in germline tissue. These mapped primarily to repetitive regions and transposons (5, 6). In addition, early genetic studies in Drosophila had implicated the flamenco (flam) locus, now known to be a major piRNA source, in the regulation of gypsy family transposons (113, 117). RasiRNAs were later termed piRNAs when found to interact with PIWI proteins in mice, rats, flies, and fish (4, 10, 35, 39, 41, 54, 77, 130, 149, 164).
Contrary to the view that the piRNA pathway evolved with its activity centered on germ cells, new research on arthropods suggests that their last common ancestor contained pathways active in both the soma and germline and that several species lost that activity in all but gonadal tissues (81). piRNAs are also found in the somatic stem cells of several cnidarians and in Macrostomum lignano (38, 61, 85, 175). While transposon silencing is the primary function of piRNAs in fly germ cells, other roles for germline and somatic piRNAs have been described in a range of species, such as virus defense in the mosquito soma and sex determination in silkworm (70, 94, 97). The piRNA pathway can even be reactivated in somatic cells under certain conditions (31).
A decade of research has revealed the complexity of the piRNA pathway, involving a cadre of factors vastly exceeding those required for canonical RNAi pathways (23, 44, 99). Pioneering studies to uncover the framework of the ping-pong cycle provided the first concrete clues to piRNA biogenesis mechanisms (10, 41). It is now clear that piRNA precursors are transcribed from genomic clusters—loci harboring transposon fragments. These clusters provide a genetic memory of past transposition invasion. Cluster transcripts must be specifically selected for piRNA biogenesis, a poorly understood process referred to as licensing. Following nuclear export, piRNA precursors are either processed by the conserved endonuclease Zucchini (Zuc)/MitoPLD or alternatively consumed by the ping-pong cycle (7, 10, 41–43, 95). PIWI–piRNA complexes repress transposons via two alternative mechanisms depending on the PIWI protein involved. In Drosophila, the cytoplasmic proteins, Aubergine (Aub) and Argonaute3 (Ago3), participate in slicer-dependent posttranscriptional gene silencing (PTGS) via the ping-pong cycle, driven by Miwi and Mili in mouse and Siwi and Ago3 in silkworm (7, 10, 41, 64). In contrast, Drosophila Piwi and murine Miwi2 translocate to the nucleus when loaded with a piRNA (7, 10, 129). These proteins repress transposons through transcriptional gene silencing (TGS) (7, 78, 125, 141, 155). The piRNA pathway shows conceptual similarities to an immune system by distinguishing self from nonself. It adapts to new transposon challenges via the incorporation of sequences into piRNA clusters and acute transposon activation via ping-pong looping.
SOURCES AND LICENSING OF PIRNA PRECURSORS
Unlike precursors of siRNAs and microRNAs, which form dsRNA, piRNA precursors lack defined secondary structures and are consequently produced independently from Dicer proteins (54, 149). Moreover, universally recognized sequence tags that license precursors for small RNA biogenesis are absent. Thus, almost all cellular transcripts could theoretically be converted into piRNAs, including gene-coding mRNAs, depending on the species, tissue, and developmental stage. However, promiscuous processing operates at a comparatively low level, and only a few substrates are specifically selected for efficient piRNA biogenesis (licensing). Although this is poorly understood, several models have been advanced to explain piRNA precursor selection: One favors coupling the transcription of piRNA precursors to their delivery to biogenesis centers (fly dual-strand clusters); others propose that a sequence motif is sufficient to trigger processing (flam and traffic jam). The molecular mechanisms underlying these models remain obscure. Slicer targeting of transcripts was shown to result in the generation of piRNAs, suggesting an additional licensing mechanism based on sequence complementarity.
In most species studied, a discrete number of genomic loci stood out as hotspots responsible for producing the vast majority of piRNAs. These loci were dubbed piRNA clusters. piRNA clusters come in various configurations and are roughly divided into unistrand and dual-strand clusters based on their capacity to generate piRNAs from one or both genomic strands. In flies and mammals, piRNAs originating directly from piRNA clusters make up more than 90% of piRNA populations (4, 10, 35, 39, 77). Drosophila clusters (and mouse clusters at the prepachytene stage) are predominantly made up of transposon-derived content, thus constituting a genetic memory of past transposon invasion (7, 8, 10).
Unistrand Clusters Are the Most Prevalent Form of piRNA Source Loci
The defining feature of unistrand clusters is the production of piRNAs from only one genomic strand. These are the most widespread form of piRNA source loci and potentially the default type. Unistrand clusters appear indistinguishable from canonical genic transcription units, showing defined promoters that carry histone 3 lysine 4 dimethylation (H3K4me2) marks, transcription by RNA polymerase II (pol II), and canonical RNA processing (capping, splicing, and polyadenylation) (37, 96). Mouse pachytene piRNAs are produced from euchromatic regions and appear to require canonical transcriptional machineries and the transcription factor A-Myb (4, 35, 39, 83). Similarly, most silkworm piRNA clusters reside in euchromatin and are characterized by canonical mRNA features (63).
Only two major unistrand clusters are known in Drosophila, namely cluster 20A and flam (10). While cluster 20A is expressed in both somatic follicle cells and germ cells, flam is restricted to follicle cells (10, 76, 89, 117). The flam cluster is the master locus for piRNA production in these cells and is required for the repression of gypsy-like transposons (10, 113). Flam shows several remarkable features (Figure 1a). First, it is located at the boundary of euchromatin and heterochromatin, occupying at least 180 kb (10). Second, flam is populated with fragments of gypsy family TEs that are almost exclusively inserted on the minus strand, allowing the efficient production of piRNAs in antisense orientation to active TE mRNAs (10). Third, while the flam promoter is marked with H3K4me2, the cluster body is decorated with histone 3 lysine 9 dimethylation/trimethylation (H3K9me2/3) marks (37). Notably, production of flam piRNAs is impaired in Eggless (Egg)/dSETDB1 mutants, suggesting that Egg is the enzyme responsible for depositing its H3K9me3 marks (120). Flam is transcribed by RNA pol II and the resulting transcripts are capped and possibly spliced (37). Just as A-Myb is required by mouse unistrand clusters, flam expression in Drosophila requires the transcription factor Cubitus interruptus (37).
Figure 1.
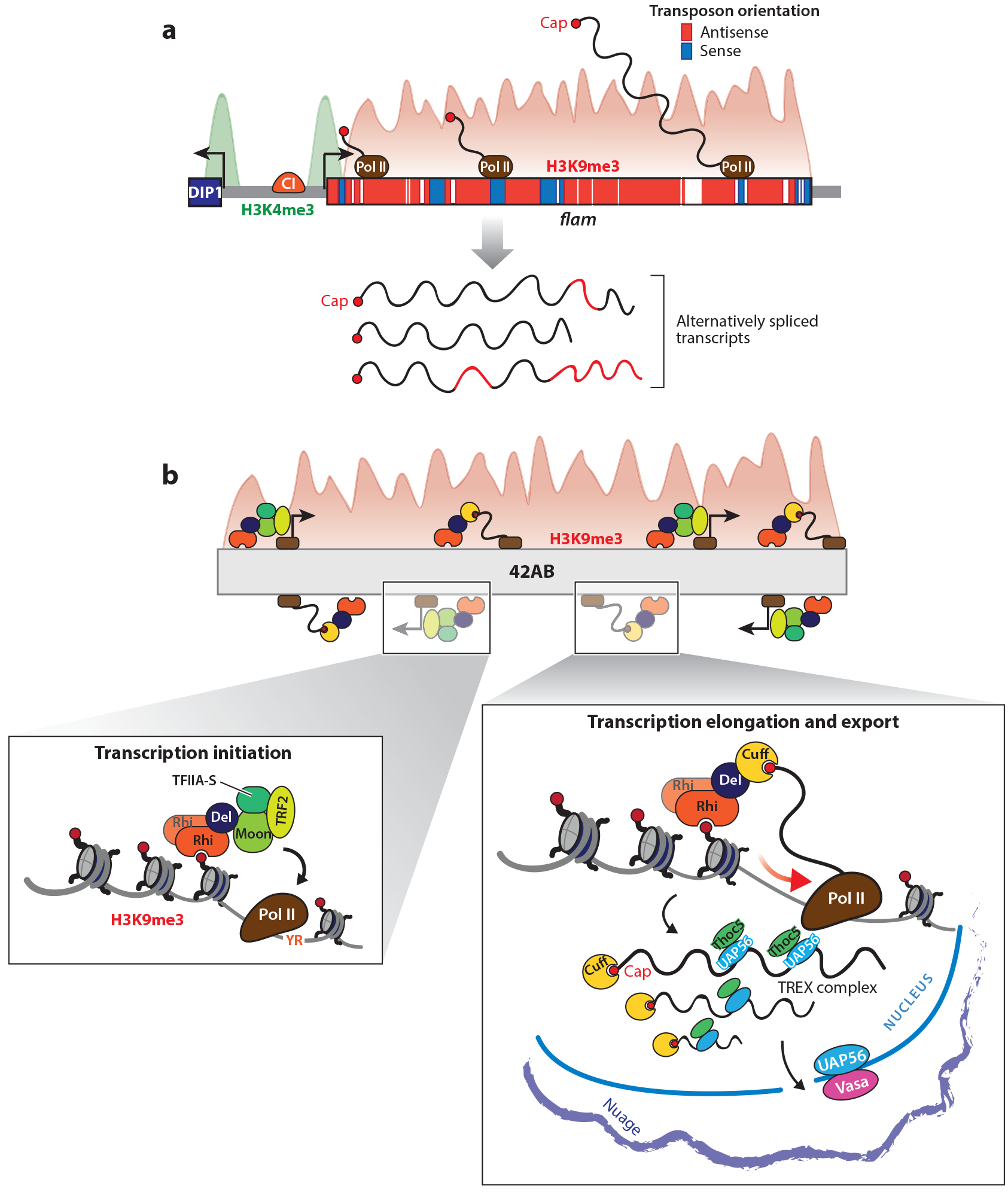
piRNA clusters are major piRNA source loci. (a) The flam locus is the major source of piRNAs in the follicle cells of the Drosophila ovary and a prime example for unistrand clusters. Like coding genes, flam features a promoter that is decorated with H3K4me3 marks and is controlled by the transcription factor CI. RNA pol II transcribes the flam locus; the resulting RNA is capped and shows splicing signatures. Unlike the promoter region, the body of flam is marked with H3K9me3. (b) Transcription from dual-strand clusters is noncanonical. Dual-strand clusters are decorated with H3K9me3 marks and, for unknown reasons, they are specifically bound by the HP1 homolog Rhi. Together with Del, Rhi serves as a binding platform for Moon, which recruits a specialized transcription initiation machinery and leads to RNA production by pol II. The accumulation of cluster-derived transcripts also depends on Cuff, which interacts with Rhi through Del. Cuff is thought to suppress transcription termination and to protect nascent RNAs from splicing and degradation. UAP56 and Thoc5 (the TREX complex) were proposed to transport cluster transcripts from their site of synthesis to their processing sites. Abbreviations: 42AB, the most prominent dual-strand piRNA cluster named for its cytological position; CI, Cubitus interruptus; Cuff, Cutoff; Del, Deadlock; DIP1, DISCO interacting protein 1; flam, flamenco; HP1, heterochromatin protein 1; Moon, Moonshiner; Nuage, perinuclear structures marked by the presence of PIWI proteins participating in the ping-pong loop; pol II, polymerase II; Rhi, Rhino; TFIIA, transcription factor II A; Thoc5, THO complex subunit 5; TRF2, TATA-box binding protein-related factor 2; UAP56, the conserved DEAD-box helicase U2AF65-associated protein also known as Hel25E; YR, dinucleotide signature of transcription initiation.
Though they are strictly speaking unistranded, source loci that produce worm piRNAs (also known as 21U-RNAs based on their length and 5′ nucleotide bias) have little in common with fly and mouse clusters. Most 21U-RNAs map to two clusters on chromosome IV that show low levels of histone occupancy (16, 126). Transcription by RNA pol II generates 28- to 29-nt-long precursor RNAs from individual promoters that carry a Ruby motif and are controlled by Forkhead family transcription factors as well as PRDE-1 (16, 36, 126, 159).
Specialized Dual-Strand Clusters Act in Drosophila Germ Cells
Dual-strand clusters are the predominant form of piRNA source loci in Drosophila germ cells. They produce piRNA precursors from both genomic strands by convergent transcription (3, 10, 71, 96, 171) (Figure 1b). The vast majority of these loci, presumably unique to Drosophilidae, are located at the boundaries between heterochromatin and euchromatin or in regions such as pericentromeric and subtelomeric heterochromatin (10).
While they are also transcribed by RNA pol II, dual-strand clusters do not follow canonical transcriptional rules. Not only do they lack H3K4me2 marks at a putative promoter but they seem to lack promoters altogether, requiring a different mechanism of transcription initiation. Transcripts from dual-strand clusters also escape splicing and polyadenylation (96, 171). Several studies have highlighted the importance of a multiprotein complex and an unusual expression mechanism. This complex is centered on the heterochromatin protein 1 (HP1) homolog, Rhino (Rhi), which specifically binds to H3K9me2/3 marks present at piRNA clusters (71, 96, 169, 171). The requirement of H3K9me3 for cluster transcription and its dependence on Egg was recognized early on (120). Rhi is now thought of as an anchor that brings other factors to the locus. Rhi is bound by Deadlock (Del), a mostly unstructured protein, which in turn serves as a platform for binding different adapter proteins that act at distinct steps (96). Recent structural work found that Rhi and Del interact in a one-to-one ratio through Rhi’s chromoshadow domain and the 60 amino-terminal residues of Del (164a). Moonshiner (Moon) recruits the TATA-box binding protein-related factor 2 and additional components of the transcription initiation complex. This complex was shown to bring in RNA pol II at YR sites, serving as an initiator analog to the TATA box (62, 118), and it was found to drive transcription straight from loci within heterochromatin that carry H3K9me3 and are bound by Rhi (3). Since this chromatin mark is not strand-specific, these findings also explain the transcription that occurs randomly from both genomic strands rather than precisely defined sites. Consequently, precursors of several sizes should be produced instead of, as previously hypothesized, a single, long transcript from read-through transcription (3, 96).
In addition to enabling transcription at cluster loci, the Rhi–Del platform also affects transcript processing and stability. Del is bound by Cutoff (Cuff), a protein with similarity to the Rai1 transcription termination factor (108). Yeast Rai1 has decapping activity; however, altered residues in the catalytic site of Cuff likely render it inactive. Instead, it is thought to bind the 5′ ends of precursor transcripts and shield nascent transcripts from degradation by the exonuclease Rat1 (18, 79, 96, 161, 171). Cuff was also reported to suppress RNA pol II termination by interfering with the recruitment of the cleavage and polyadenylation specificity factor (18). Additionally, Cuff was found to be essential for the recruitment of the THO–TREX complex to nascent piRNA precursors (55). This complex has an important role in the export of RNA from the nucleus, and it is necessary for dual-strand cluster transcription via the protein Thoc5 (55). Thus, in addition to transcription initiation, this Rhi–Del-centered complex also controls transcript processing. Intriguingly this was achieved through evolution of piRNA pathway-specific paralogs of factors involved in other pathways, including H3K9me3 binding (Rhi), Cap-binding (Cuff), and transcription initiation (Moon). Identification of this mechanism, while interesting, raises a further question of how the Rhi–Del platform is specifically guided to cluster loci and not to other regions of the genome decorated with H3K9me3. Why was it necessary to evolve this conceptually new cluster transcription machinery? It may have been driven by the need to produce transcripts from loci that share sequence homology with transcriptional silencing targets.
Export of piRNA Precursors
In cases where transcripts are licensed as piRNA precursors (cotranscriptionally) in the nucleus, particular mechanisms must exist that direct precursors to cytoplasmic processing centers. Indeed, transcripts from dual-strand clusters are specifically bound by the conserved DEAD-box helicase U2AF65-associated protein (UAP56, also known as Hel25E) (169). UAP56 localizes on the inner side of the nuclear pore opposite the RNA helicase Vasa, which sits on the outer side. It has therefore been suggested that piRNA cluster transcripts gathered by UAP56 are handed over to Vasa at perinuclear processing centers and are thereby destined for piRNA production (169). Interestingly, several RNA export factors were among the hits in screens for piRNA pathway components in both the germline and soma; however, their precise role in precursor selection is not yet understood (23, 44, 99).
Similarly, piRNA biogenesis from unistrand clusters seems to be linked to the RNA export machinery and might be coupled to upstream precursor specification events. Transport of the precursor transcript of the flam cluster in Drosophila follicle cells requires Nxf1/Nxt1 (27, 44, 99). Studies have identified a perinuclear granule right next to the Yb body, namely flam body or Dot COM, in which the majority of flam transcripts seem to reside in an Nxf1/Nxt1-dependent fashion (27, 28, 100). While mechanisms of specific recognition and recruitment of the flam precursor remain enigmatic, it has been shown that Yb binds to discrete flam sequences and that these are necessary and sufficient to trigger piRNA biogenesis from reporter constructs. Similar results were obtained with a fragment of the traffic jam 3′ UTR, a follicle cell-specific piRNA hotspot (51, 58, 100). However, the molecular mechanism underlying their recognition is unclear. There are no specific trigger sequences or secondary structures appearing as shared features of piRNA precursors. Proper localization of the precursor is indeed crucial to initiate piRNA production. In fact, tethering of artificial reporters to certain nuage/Yb body components seems sufficient for them to be parsed into bona fide piRNAs (107, 124). Nonetheless, this does not answer the question of which features of the endogenous substrates mark their fate. One possible feature is licensing through sequence-specific detection and the subsequent slicing of the precursors at nuage.
PIRNA BIOGENESIS
Processing of piRNA precursors into mature silencing triggers takes place within specialized subcellular compartments. Animal germ cells possess a perinuclear structure, termed nuage, that is marked by the presence of PIWI proteins participating in ping-pong looping (10, 32, 84, 89). In Drosophila follicle cells, which are of somatic origin, nuage is replaced by so-called Yb bodies, processing centers located on the cytoplasmic side of the nuclear envelope (100, 106, 119, 129, 145).
The biogenesis of piRNAs has historically been divided into two distinct branches: primary biogenesis, acting on cluster transcripts, and the ping-pong cycle, directed toward transposon mRNAs (also termed secondary biogenesis). Recent findings have challenged this separation, showing that the two routes are linked to one another with ping-pong triggering the generation of primary piRNAs. Therefore, instead of using these outdated terms, we suggest a separation based on the biogenesis machineries involved, those that involve Zuc and those that operate independently of Zuc. In the sections that follow, we discuss how piRNAs are released from their precursors and how the ping-pong cycle is connected to this (Figure 2). Further, we provide insights into alternative biogenesis routes that do not fall into the canonical categories.
Figure 2.
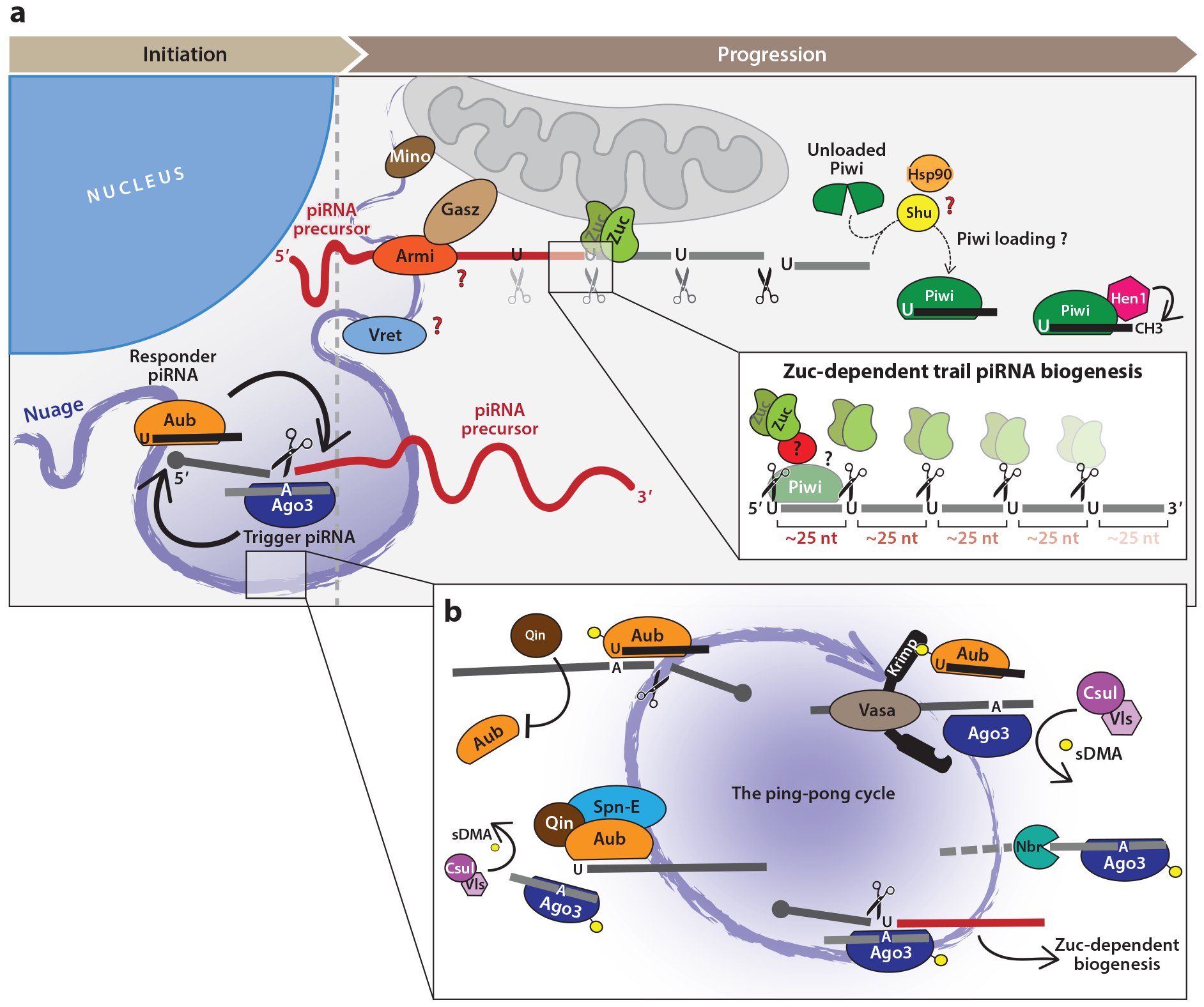
Biogenesis of piRNAs follows several routes. (a) Following export of precursor RNAs from the nucleus, piRNA biogenesis is initiated in nuage through ping-pong looping. There, precursor transcripts are recognized and cleaved by Ago3 in complex with a trigger piRNA. This slicing event gives rise to a responder piRNA, which is loaded into Aub. The remaining 3′ portion of the cleaved transcript (red) serves as substrate for further piRNA production. Subsequently, piRNA biogenesis proceeds in a 3′-oriented manner. The mitochondrial endonuclease Zuc generates waves of trail piRNAs approximately every 25 nt, guided by an as-yet-unidentified cofactor or by the footprint of Piwi itself (top inset). Several other factors participate in this process, including Vret, Mino, and Gasz, but their precise molecular functions remain to be clarified. The helicase activity of Armi is likely to unwind precursor RNAs before feeding them into Zuc. The loading of piRNAs relies on Shu and Hsp90 and causes a conformational change in Piwi, thereby allowing its nuclear translocation. As a final step, Hen1 methylates the 3′ end of mature, Piwi-loaded piRNAs prior to import to the nucleus. (b) The main steps of the ping-pong cycle. Aub bound to an antisense piRNA recognizes and cleaves a transposon mRNA. The resulting 3′ cleavage product is converted into a new sense piRNA that associates with Ago3; its 3′ end is generated through trimming by Nbr. Ago3 associated with a sense piRNA can in turn recognize and cleave cluster transcripts. The product of this slicing event reinitiates the cycle, becoming an Aub-bound piRNA, and the remaining 3′ slicing product (red) is processed into Piwi-loaded piRNAs via Zuc-mediated biogenesis. Specialized loading complexes ensure that products of slicing are loaded into the correct PIWI protein: Vasa and Krimp assist Ago3 loading, whereas Qin and Spn-E act on Aub. Furthermore, Qin prevents Aub cleavage products from being loaded into Aub, thus ensuring the directionality of the ping-pong cycle. This regulatory framework is based on posttranslational modifications of PIWI proteins in the form of sDMA residues, which are added by Csul and Vls. Abbreviations: Ago3, Argonaute3; Armi, Armitage; Aub, Aubergine; CH3, depicts a 2-O-methyl group; Csul, Capsuleen; Gasz, germ cell protein with ankyrin repeats/sterile-alpha motif/leucine zipper; Hen1, Hua enhancer 1; Hsp90, Heat shock protein 90; Krimp, Krimper; Mino, Minotaur; Nbr, Nibbler; nt, nucleotide; sDMA, symmetrical dimethylation of arginine; Shu, Shutdown; Spn-E, Spindle-E; Vls, Valois; Vret, Vreteno; Zuc, Zucchini.
Zuc-Mediated Processing of Precursors
Structural data and a combination of genetics and bioinformatic analyses of small RNAs have placed the conserved endonuclease Zuc/MitoPLD at the center of piRNA biogenesis (57, 103, 109, 153, 157). Originally, Zuc was identified as the enzyme responsible for forming piRNA 5′ ends, yet more recently it was shown also to directly or indirectly generate the 3′ ends of a subset of piRNAs (43, 95), further highlighting its essential position in the pathway. Zuc activity in Drosophila shows a remarkable degree of processivity, as piRNAs derived from reporter transcripts or the piRNA cluster flam precisely follow one another approximately every 25 nt. It is unknown what factors dictate the phasing of Zuc cleavage products. To some extent, the Piwi footprint on precursor molecules helps to determine the size of mature piRNAs (10, 33a). However, how tightly Piwi loading is connected to Zuc processing has not been fully investigated. Furthermore, all piRNAs derived from Zuc cleavage feature a pronounced bias for a uridine at their 5′ end (1U bias) (4, 10, 35, 64, 83). Crystal structures of Bombyx Siwi and of the MID domain from a mammalian PIWI protein suggest their ability to accommodate 5′ uridines (21, 90). However, Zuc processing of reporter constructs occurs selectively in correspondence to uridines (43, 47, 95), whereas in vitro assays with recombinant Zuc show no nucleotide bias (57, 103). Altogether, this hints at the existence of a specificity factor driving Zuc’s distinctive processing signature in vivo.
Besides Zuc, piRNA biogenesis requires numerous other factors that are organized into tissue-specific processing sites—Yb bodies in Drosophila follicle cells and nuage in animal germ cells. Intriguingly, several factors required for Zuc-mediated processing, including Zuc itself, show conserved localization to the mitochondria outer membrane, underscoring the importance of these organelles as platforms for the entire process. Other mitochondrial proteins required for Zuc-mediated piRNA biogenesis are the germ cell protein with ankyrin repeats, a sterile-alpha motif, and a leucine zipper (Gasz) and Minotaur (Mino)/glycerol-3-phosphate acyltransferase 2 (GPAT2) as well as Partner of PIWIs (Papi) and TDRKH (23, 44, 52, 86, 88, 133, 138, 151). Gasz appears involved in Armitage (Armi) recruitment because its depletion delocalizes Armi from nuage (23, 44). Mino is a GPAT, yet the catalytic activity of this domain is dispensable for its function in piRNA biogenesis (138, 151). Interestingly, Zuc and Mino both belong to protein families normally involved in lipid metabolism. Papi and TDRKH are Tudor proteins that are important for piRNA 3′ end formation in mice and silkworm but have little impact in flies, as discussed below. Among the cytosolic piRNA biogenesis factors, the helicase Armi, the mouse homolog of MOV10L1, has a pivotal role in both Yb bodies and nuage. It has been shown to associate with piRNA intermediates and likely unwinds them to facilitate processing (129, 154, 160, 173). Lastly, Shutdown in flies and Fkbp6 in mice colocalize with unloaded PIWI proteins in the cytosol and are thus probably involved in the loading process, which is also assisted by Heat shock protein 90 (105, 116, 162). Interestingly, murine TDRD9 also seems to be involved in the loading process, specifically for Miwi2 (139).
Tudor family proteins appear to be broadly implicated in Zuc-dependent piRNA biogenesis (45). These proteins typically detect and bind symmetrical dimethylation of arginine (sDMA) modifications, which are present on PIWI proteins (69, 122, 150). Yet, some of them might function independently of sDMA binding. Tudor proteins are believed to form a molecular scaffold interconnecting the various steps of the pathway. These proteins include female sterile (1) Yb (Yb, present in somatic follicle cells), its cognates Brother of Yb and Sister of Yb (both present in nuage), and Vreteno (Vret) (45, 106, 129, 167). Yb contains a helicase-like domain (similar to DEAD/DEAH-box RNA helicases) and a Tudor domain, and it accumulates in discrete perinuclear granules, namely, the Yb bodies (119). Yb bodies are located in close proximity to mitochondria and contain several other biogenesis factors, such as Armi, which directly interacts with Yb (45, 119, 129, 145). However, the correct localization of other biogenesis proteins depends on Yb itself. Upon Yb depletion, somatic piRNA production collapses, further supporting the relevance of Yb bodies in Zuc-dependent piRNA biogenesis (106, 119). Vret is another Tudor domain–containing protein, found in both Yb bodies and nuage, required for production of Piwi-bound piRNAs and the correct localization of all three PIWI proteins (45, 167).
In flies, the bulk of Zuc-dependent piRNAs are loaded into Piwi, and this step is essential for Piwi nuclear translocation (43, 95, 129, 134, 156). Unloaded Piwi appears to be extremely unstable. Its interaction with a mature piRNA was recently shown to induce a conformational change that exposes a nuclear localization signal and results in Importinα-mediated transport to the nucleus (163a). Upon their association with Piwi, the 3′ ends of piRNAs are methylated by Hen1 (53, 131). This modification is shared by other small RNAs and likely provides a protective function.
The Ping-Pong Cycle
The ping-pong amplification loop is a PTGS mechanism in which RNAs consumed by piRNA-guided transcript slicing provide substrates for additional piRNA production. Because it both represses transposons and increases the abundance of piRNAs capable of targeting active transposons, this germline-specific cycle was proposed to explain features of piRNAs associated with PIWI proteins purified from Drosophila ovaries (10, 41). While Aub- and Piwi-bound piRNAs tend to be antisense to transposons and display a preference for a 5′ terminal uridine, Ago3-bound piRNAs are primarily in the sense orientation and exhibit a bias for an adenosine at position 10. Moreover, the 5′ ends of sense–antisense piRNA pairs overlap by precisely 10 nt, a relationship termed the ping-pong signature (10, 41).
Analysis of piRNAs associated with Aub and Ago3 homologs has since identified ping-pong loops in other animals (8, 10, 38, 41, 54). The core mechanism of reciprocal cleavages, occurring in perinuclear nuage of germ cells, has proven to be well conserved. In Drosophila, maternally deposited or cluster-derived antisense piRNAs are loaded into Aub. These direct Aub to slice a transposon mRNA between the bases pairing with nucleotides 10 and 11 of the piRNA guide, a cleavage which defines the 5′ end of a complementary sense piRNA. Loading of the 3′ cleavage product into Ago3 followed by 3′ end processing produces a mature piRNA. Ago3-mediated slicing of cluster transcripts then specifies the 5′ end of an antisense piRNA, identical in sequence to the original piRNA that is funneled into Aub for continuation of the cycle (10, 41).
Within nuage, RNAs must be directed specifically into Aub or Ago3 in order to maintain their strand biases and drive continuous ping-pong looping. The regulatory framework to accomplish this is based on posttranslational modification of PIWI proteins in the form of sDMA residues. These modifications are added by Capsuleen (also known as dPRMT5), an enzyme conserved in silkworm and mouse, and its cofactor Valois (69, 102). sDMAs are recognized by a range of Tudor domain–containing cofactors, nuage-enriched proteins whose loss interrupts ping-pong looping (45). A key responsibility of these proteins is to ensure that products of Aub-mediated slicing are transferred to Ago3 and vice-versa, promoting Ago3–Aub heterotypic ping-pong, which is necessary for robust transposon repression (132, 172). For example, Krimper (Krimp) regulates Ago3 loading with sense piRNAs via a series of Tudor domain–dependent interactions (132, 158). Krimp interacts with unloaded and nonmethylated Ago3 in the cytoplasm, recruiting it to nuage. There, Krimp can form both homodimers and an sDMA-dependent interaction with Aub, bringing Aub and Ago3 together and ensuring that piRNA intermediates are efficiently passed from one to the other. Moreover, Krimp enforces PIWI strand biases by shielding Ago3 from associating with antisense piRNAs (132). Another Tudor domain protein, Qin, has an analogous function in maintaining the antisense bias of Aub-bound piRNAs via an sDMA-dependent interaction with Aub that prevents Aub–Aub homotypic ping-pong (156, 172). The role of additional Tudor domain proteins, namely, Spindle-E (Spn-E), Tejas, and Tapas in the ping-pong loop, is less well understood, but their loss typically leads to a disruption of the cycle (84, 89, 101, 111, 112).
Studies of the molecular features of ping-pong looping have greatly benefited from the availability of Bombyx ovary-derived cultured germ cells harboring an active ping-pong cycle (64). The RNA helicase Vasa, a key factor of germline piRNA pathways, was identified in silkworm as a cofactor in the release and transfer of cleaved RNAs from the Siwi complex, on which they are retained to prevent degradation, to unloaded Ago3 (101). In the same study, Spn-E and Qin silkworm homologs were found in a separate complex with Siwi and linked to the generation of Siwi-bound piRNAs (101). Recently it was shown that loss of l(3)mbt in ovarian somatic cells (OSCs) results in expression of core ping-pong components and the acquisition of a functional ping-pong cycle (144). Thus, these mutant OSCs should facilitate further biochemical probing of the ping-pong mechanism in a Drosophila cell culture system.
Links Between Ping-Pong Looping and Zuc-Mediated Processing
Generating piRNAs from the same transcripts targeted for silencing allows cells to refine the piRNA response against active transposons. However, ping-pong looping as described originally cannot generate piRNAs capable of targeting new sequences. Analysis of nucleotide distances between adjacent piRNAs revealed that in Drosophila the 5′ ends of antisense piRNAs mapping to the same genomic strand are most commonly separated by precisely one piRNA length (43, 95). Moreover, at the 5′ end of each wave of adjacent trail piRNAs lies a ping-pong pair of one sense trigger piRNA and one antisense responder piRNA, which are predominantly bound to Ago3 and Aub, respectively. This finding led to the idea that the ping-pong cycle could trigger a wave of downstream piRNA production. Indeed, Ago3-mediated slicing of a target mRNA triggers the production of Zuc-dependent, phased piRNAs from the downstream transcript, and this seems to be the predominant biogenesis mode for Piwi-bound piRNAs in the fly germline (134, 156).
In Drosophila nurse cells, it is likely that Aub and Ago3—in complex with piRNAs—reside in nuage and, there, scan all of the transcripts that exit nuclear pore complexes. Once they have identified a complementary target, a slicing event occurs, thereby generating the ping-pong responder piRNA together with the substrate for downstream trail piRNA production, which is remarkably processive (43, 95, 134, 156). This model also partially explains precursor recognition and licensing because target slicing generates a free 5′ end that is readily utilized by the biogenesis machinery. Phasing is processive; consecutive Zuc-mediated cleavage events define the 5′ and 3′ ends of newly produced piRNAs, which are then loaded into Piwi. Phased biogenesis also further diversifies the antisense piRNA pool produced against active transposons and represents an essential arm of the biogenesis pathway. It should be noted that the 3′-directed generation of antisense Piwi-bound piRNAs enables targeting of sense mRNAs closer to the 5′ end and eventually covers the entire transcript for robust transcriptional shutdown of a locus (134, 156).
Phased production of trail piRNAs is conceptually similar to RNA-dependent RNA polymerase (RdRP)-mediated sequence diversification in worms, though the machineries involved differ significantly. Upon target recognition, Caenorhabditis elegans piRNAs loaded into Piwi-related gene 1 (PRG-1) elicit local generation of complementary 22G-siRNAs, which in turn engage in nuclear silencing of nonself elements. PRG-1–piRNA complexes have the ability to recruit an RdRP that uses the target RNA as a template for the production of these secondary small RNAs, thus achieving sequence diversification of antisense RNAi triggers (80, 137). Although details might differ, phasing is conserved. In Bombyx cells, both piRNA pairs exhibiting ping-pong signatures and phased piRNAs are detectable (51). Mili-bound mouse precursor piRNAs are phased in a similar fashion, with MitoPLD-producing U-biased cleavages approximately every 40 nt (43, 95). Slicing events by Mili–piRNA complexes, in turn, give rise to trail piRNAs that populate Mili and Miwi2 (163).
While the 5′ ends of piRNAs are uniquely defined by endonucleolytic cleavage (via either Zuc or slicing), the 3′ ends can be formed by different mechanisms. In mouse and silkworm, Zuc-generated 3′ ends are extensively resected to their mature size by the Papi (TDRKH)/PNLDC1 pathway, whereas flies lack any PNLDC homolog (29, 43, 47, 60, 65, 95). Studies in Bombyx have found that the mitochondrial protein Papi, which lacks any recognizable nuclease domain, serves as a platform to recruit the 3′–5′ exonuclease PNLDC1, which trims piRNA precursors to their mature size. Similarly, mouse Mili associates with piRNA intermediates approximately 40 nt in length that are then resected to their mature size by PNLDC1 with the help of Papi/TDRKH (29, 133). Taking into consideration the 1U preference of Piwi/Siwi, this suggests that mouse and silkworm piRNAs are first incorporated into the RISC and then exonucleolytically trimmed to their mature length. Drosophila piRNAs, however, do not seem to rely on the same mechanism. Nibbler (Nbr) has been recently identified as the trimming enzyme in the piRNA pathway (47). However, in contrast with the other species, Nbr acts predominantly on Ago3-bound piRNAs. Piwi- and Aub-loaded piRNAs instead do not seem to require any trimming.
Interestingly, ping-pong competes with phased biogenesis for slicer-generated cleavage products; thus, the relative balance between Zuc and Nbr activities affects the ratio of Piwi-bound versus Aub- and Ago3-bound piRNAs and, in turn, of TGS versus PTGS (47). How is the destination of RNA cleavage products regulated? In particular, how are Ago3-generated precursors directed away from Nbr, localized to nuage, and specifically recognized by Zuc, a protein constrained to the mitochondrial membrane? Does this require an as-yet-unidentified cofactor? It is interesting to note that while most Piwi-bound piRNAs are initiated through Ago3 cleavage, there are some that are independent of Ago3, and it is unclear how their biogenesis occurs. Furthermore, somatic cells do not express Aub or Ago3 but have an active biogenesis pathway (82, 89). The proposed model therefore does not explain how somatic piRNA production is initiated, but it does imply key mechanistic differences between plain Zuc-mediated processing and that which is primed by ping-pong. Additional somatic piRNAs in mice and flies can also arise from selected mRNAs, especially from their 3′ UTRs (123, 128). This provides further evidence of different nonoverlap-ping entry points that eventually funnel a transcript into Zuc processing. It should be noted that several Tudor proteins that are essential for a functional ping-pong cycle are indirectly involved in Zuc-mediated piRNA production, likely connecting the initiation and progression steps of piRNA biogenesis.
Zuc-Independent piRNA Biogenesis
Biogenesis independent of Zuc is rare in mammals and Bombyx but does occur in fly ovaries. While ping-pong is usually linked to Zuc-mediated processing of 3′ ends (and trail piRNAs), it can also function independently. In the absence of Zuc, a piRNA 5′ end is generated normally via slicing, but its corresponding 3′ end becomes the product of a second slicing event followed by Nbr trimming (47). Novel ping-pong pairs can even arise to rescue impaired piRNA production upon combined depletion of Zuc and Nbr. In this case, piRNAs do not exhibit the canonical size profile, with some abnormally long or short species (47). Altogether, this shows that fly piRNA biogenesis is an extremely flexible and robust mechanism, capable of generating mature piRNAs through alternative routes, were one branch of the pathway to be disrupted. This intrinsic flexibility may be the reason why we have not observed transposon-derived inhibitors of the piRNA pathway in contrast to relatively common viral suppressors of RNAi.
The biogenesis of worm piRNAs is another example of Zuc-independent processing. C. elegans also does not have a ping-pong cycle; thus, piRNA biogenesis cannot be initiated via slicing and is remarkably different from what we have presented so far. 21U-RNAs are produced from 25 to 29 nt capped, small RNA precursors through removal of the first two nucleotides and trimming at the 3′ end (40). While the enzyme responsible for 5′ end generation is still unknown, PARN-1 is the 3′–5′ exonuclease that resects precursor 3′ ends (146). Interestingly, PARN-1 belongs to the same protein family as PNLDC1, which trims the 3′ ends of mouse and silkworm piRNAs.
PIRNA-GUIDED SILENCING
The mode of target regulation by piRNAs differs according to the involvement of PIWI proteins and their effectors. In general, these can be separated into the two major mechanisms, PTGS and TGS. PTGS mechanisms, such as the ping-pong cycle, rely on target transcript slicing and typically require cleavage-competent PIWI proteins (25, 121, 156). TGS, in contrast, can occur with catalytically inactive PIWI proteins (25, 141). Consequently, instead of regulating target transcripts directly through cleavage, TGS indirectly leads to reduced mRNA output via chromatin silencing through cooperation with histone modifying enzymes and, sometimes, with DNA methyltransferases. This additional layer of target control has coopted additional functions beyond transposon control, such as heterochromatin formation in general.
Concepts in Cotranscriptional Gene Silencing
The silencing responses of nuclear PIWI proteins are RNA-dependent and not thought to be mediated by recognition of DNA directly. Chromatin modifications that are usually regarded as repressive occur at PIWI-targeted loci; however, it remains to be investigated whether these are a cause or a consequence of TGS. The chromatin modifications include histone modifications such as removal of H3K4me2 and deposition of H3K9me2/3. In mammalian germ cells, PIWI-guided TGS also causes DNA methylation at CpG dinucleotides in addition to deposition of repressive histone marks (7). Conceptually, TGS can be divided into several stages. First, target RNAs engage the PIWI–piRNA complex (initiation); second, the chromatin silencing machinery recruits factors to the locus. This process ultimately leads to a transcriptionally inactive state (Figure 3).
Figure 3.
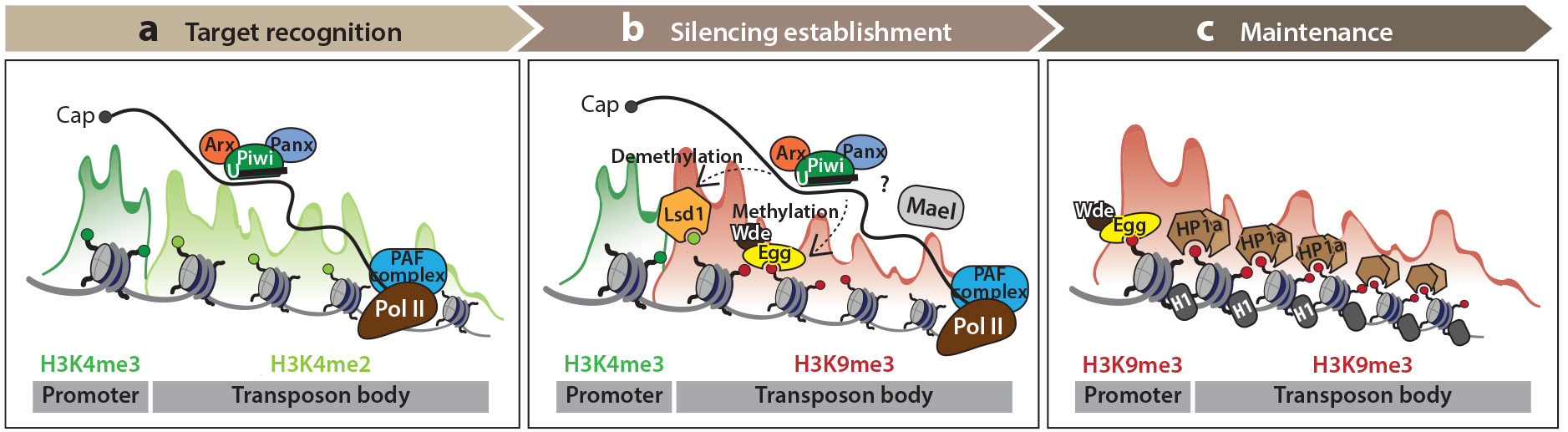
A stepwise model of piRNA-guided transcriptional silencing. (a) During target recognition, Piwi–piRNA complexes scan the nucleus and recognize nascent transcripts of TEs through sequence complementarity. This process is likely assisted by the cofactors Arx and Panx and is regulated by the PAF1 complex—the latter which is associated with elongating RNA pol II. (b) Silencing is established through interactions with several elements of the heterochromatin silencing machinery, yet the molecular details of these interactions remain to be resolved. The histone methyltransferase Egg and its cofactor Wde deposit H3K9me2/3 marks on transposon bodies. Mael appears to function independently of this histone mark, yet is crucial for proper silencing. Furthermore, silencing of some TEs requires Lsd1, which likely removes activating H3K4me2 modifications prior to Egg’s function. (c) Upon establishment of silencing, HP1a and histone H1 cover the entire locus and cooperate to maintain its repressed status. Across all three steps, histone modifications generally associated with transcriptional activity, namely H3K4me3 and H3K4me2, are represented in green, whereas the repressive H3K9me2/3 mark is shown in red. Abbreviations: Arx, Asterix; Egg, Eggless/dSETDB1; HP1a, heterochromatin protein 1a; Lsd1, lysine-specific demethylase 1 [also known as su(var)3–3]; Mael, Maelstrom; Panx, Panoramix; pol II, polymerase II; TEs, transposable elements; Wde, Windei.
Transcription-dependent heterochromatinization is a recurrent theme within the eukaryotic domain, and evolutionarily distant model organisms show surprising similarities. In Schizosaccharomyces pombe, for example, heterochromatin is formed at pericentromeric repeats. An siRNA-loaded Argonaute1 complex, termed RITS, is recruited to nascent long noncoding RNAs through sequence complementarity. RITS then recruits the H3K9 methyltransferase complex CLRC, which deploys H3K9me3 and recruits additional RITS in a positive-feedback loop (15, 20, 34, 152, 170). Additionally, RDRC, an RdRP complex, amplifies the transcript, which causes it to be processed into siRNAs in a role analogous to that of the Drosophila ping-pong machinery (98, 143). siRNA-dependent DNA methylation in Arabidopsis thaliana shows many similarities to the mechanism described in S. pombe (50, 174), as does small RNA-guided TGS in C. elegans, where 22G-RNAs are produced to repress TEs in the germline (24, 80). The Argonaute protein HRDE-1, loaded with 22G-RNAs, translocates to the nucleus and recognizes elongating mRNA precursors. The concerted action of several factors—including NRDE-1, NRDE-2, NRDE-4, and EMB-4—leads to RNA pol II stalling, H3K9me3 deposition, and the binding of the HP1a homolog HPL-2, to ultimately result in transcriptional silencing of the target locus (1, 9, 14, 87, 148). A recurring feature in all of these pathways is that recognition of nascent transcripts by an Argonaute family protein bound to a small RNA triggers recruitment of heterochromatin factors (histone methyltransferases and HP1 orthologs) that ultimately silence the targeted locus.
Among the three Drosophila PIWI clade proteins, Piwi is the only one to display a prevalent nuclear localization (10, 41, 46). Loss of Piwi protein causes sterility and severely compromises ovary development (22, 73), and deletions at its amino terminus or mutations of its nuclear localization signal are sufficient to compromise TGS (72, 73, 128, 141). While nuclear localization of Piwi is essential for its function in TE repression, its slicer activity is not required (128, 141). Harnessing catalytically inactive PIWI proteins for TGS is a recurring theme and has been described for mouse Miwi2 (25). In flies, association with a piRNA stabilizes Piwi and leads to its translocation from cytoplasmic processing sites to the nucleus (105, 106, 129, 163a). Consequently, compromised piRNA biogenesis results in cytoplasmic retention of Piwi (105, 106, 129). Moreover, piRNA loading-incompetent mutants result in full (129) or partial (78) delocalization of Piwi into the cytoplasm. Nuclear Piwi–piRNA complexes scan and interact with nascent transcripts through sequence complementarity. Transcription appears crucial for TGS, and nontranscribed loci escape deposition of repressive histone marks (114, 141, 147). Interestingly, Piwi-mediated silencing is favored when the PAF1 complex function is impaired (19). This function of PAF1 in antagonizing small-RNA mediated silencing is also conserved in S. pombe (74, 136). Following target engagement by Piwi, a conformational change is proposed to initiate TGS. Indeed, artificial Piwi recruitment to chromatin without piRNA–target interaction does not induce silencing of the reporter (78, 140, 165), whereas repression is conveyed when Piwi is recruited through the interaction between its loaded piRNA and the target transcript (115, 140). It is possible that the association of Piwi to chromatin follows a two-step mechanism: First, Piwi is recruited nonspecifically to chromatin; second, Piwi is guided to a specific locus through piRNA-mediated transcript recognition. In support of this model is the evidence that some Piwi protein still associates with the chromatin fraction even when it cannot bind piRNAs, although a large proportion of the protein fails to enter the nucleus (78). The factors loosely bringing Piwi to chromatin are possibly its interacting proteins histone H1 or HP1a (12, 59, 155).
Transcriptional Gene Silencing Relies on Several Factors and Machineries
The sequence-specific, cotranscriptional recognition of nascent mRNA precursors by Piwi–piRNA complexes requires additional factors that interact with Piwi, namely Asterix (Arx) and Panoramix (Panx) (30, 104, 140, 165). Arx is a small zinc-finger protein with a putative RNA binding function that is required for TGS (30, 99, 104). Its action likely stabilizes the piRNA-mediated Piwi interaction with the nascent transcript. Contrastingly, Panx, a protein with no known domain, is a key player in the establishment of repression (23, 44, 140, 165). Indeed, Panx is the only known TGS-specific factor capable of inducing repression when artificially tethered to chromatin or to an elongating transcript (140, 165). Nonetheless, other effectors must cooperate with Panx to achieve repression (165). The lack of either Arx or Panx results in derepression of TEs comparable to the absence of Piwi (23, 30, 44, 99, 104, 140, 165). Moreover, nuclear localization of Arx depends on Piwi and seems to overlap with a minor fraction of the total nuclear Piwi (30). Conversely, Piwi nuclear localization does not depend on the presence of either Arx or Panx (30, 140). Considered together, these data place Arx and Panx downstream of Piwi in the TGS pathway.
The repressive action of these components correlates with the deposition of H3K9me2/3 marks and with HP1a [also known as Su(var)205 in flies] recruitment. In Piwi mutants, these heterochromatic marks are lost on TEs (59, 72, 78, 125, 140, 141, 155, 165). The lack of HP1a induces transposon derepression both in vivo and in OSCs (104, 155). However, HP1a also represses the expression of some transposon families that are not regulated by piRNAs (104), suggesting a broader, Piwi-independent role in transposon transcriptional regulation. Intriguingly, HP1a and Piwi were shown to interact through HP1a dimerization of its chromoshadow domain and the PxVxL motif present at the amino terminus of Piwi (12, 93). However, mutations in this motif do not seem to affect TGS in the germline (155). It is noteworthy that upon Piwi recruitment H3K9me2/3 can spread for several kilobases around its target site with the subsequent recruitment of HP1a, and in some cases it can lead to lower expression of nearby protein-coding genes (59, 104, 141). However, opposite results were obtained by other studies, which found little to no change in the expression of protein-coding genes (72, 78). One possible explanation for these conflicting results could be the use of cultured cells versus in vivo assays.
The high mobility group protein Maelstrom (Mael) is another critical component of the TGS machinery (141). Early studies suggest shuttling of Mael between the nucleus and cytoplasm of the germline (32). Its RNase activity seems dispensable for transposon silencing (91), suggesting that it could act as an RNA binding protein instead (17). Although Mael mutants show impaired TGS similar to Piwi mutants, the impact of its impairment on H3K9me2/3 at target loci is moderate. Interestingly, H3K9me2/3 retention seems compatible with the derepression of TEs (141), indicating that this mark alone is insufficient to achieve silencing. Target repression may instead be achieved through chromatin compaction or even via HP1a-mediated organization of heterochromatic domains into a phase-separated liquid compartment (75, 142). These liquid droplet foci are thought to exclude other factors from interacting with heterochromatin; however, the underlying mechanistic details of this process are not well understood.
Recently, an involvement of histone H1 in TGS was reported; however, like HP1a, histone H1 has a broader role in the repression of TEs including those not regulated by Piwi (59). In addition, several general silencing effectors, including lysine-specific demethylase 1 [also known as Su(var)3–3], seem to function in the TGS pathway, as supported by in vivo and in vitro screens (23, 44, 99) and reporter assays (165). Understanding how the known TGS effectors cooperate with the downstream chromatin components continues to represent an important problem. In particular, one of the next big challenges for the field is to determine how recruitment of Piwi and its interacting partners can drive the surrounding chromatin to establish a silenced state.
TRANSMISSION OF EPIGENETIC INFORMATION THROUGH PIRNAS
In recent years it has become increasingly clear that small RNAs are passed from generation to generation through the female germline (reviewed in 48). Intergenerational inheritance of piRNAs is crucial for the fitness and fertility of offspring in flies and worms. These effects ultimately manifest in germ cells of the next generation as both reshaped piRNA populations and chromatin modifications across genomic TE insertions.
Strong evidence for a transgenerational component of the piRNA pathway came from C. elegans. Introduction of environmental dsRNAs or endogenous piRNAs was shown to be able to silence a predefined single-copy target sensor by triggering secondary siRNA production through RdRPs, and this ultimately led to repressive chromatin modifications inherited for several generations (87). It was further shown that maintenance of silencing is dependent on RNAi (exemplified by requirement of Argonaute proteins) and chromatin factors such as methyltransferases (9, 137). However, the epigenetic changes leading to transgenerational silencing did not depend on PRG-1–piRNA complexes. Thus, while the heritable silencing signal remains unknown, it is possibly mediated by deposition of secondary 22G-RNAs that are amplified in each subsequent generation.
Earlier evidence for a role of small RNAs in intergenerational silencing came from studies in Drosophila. In flies, the female deposits all material required for early embryonic development, including protein and mRNA gradients, into the egg. Male flies, in contrast, contribute their genome, but do not add cytoplasmic components. The maternal inheritance of transcripts and proteins controlling cell cycle and growth is required to direct DNA replication and rapid cell divisions in the period before zygotic transcription is active, and it also influences later development. Studies of the phenomenon hybrid dysgenesis also found a requirement for a maternal signal, termed the cytotype, for proper germ cell development in the offspring; lack of maternally deposited cytotypes resulted in sterility. Years later, this intergenerationally inherited signal was found to be piRNAs (11).
Hybrid Dysgenesis and Paramutation
Several distinct hybrid dysgenesis systems have been described in Drosophila, and they show very similar characteristics (Figure 4). Male, wild-caught flies crossed to laboratory strain females yield sterile offspring, whereas the reciprocal crosses are fertile (13, 68). The wild-caught flies contain genomic insertions of the underlying transposon, which is not carried by laboratory strains and differs from system to system. Sterility correlates with transposon expression in the germ cells of crosses between males harboring active TE insertions and females naïve to the transposon (13, 68). Although this phenomenon was first described in the 1970s, its underlying molecular mechanism was uncovered only much later. Maternal deposition of piRNAs targeting the specific transposon is sufficient to silence transcription in germ cells and provide lifelong protection (11). However, in some cases of hybrid dysgenesis, a lack of piRNA deposition can be compensated with increasing age of the offspring females (67). Recent work suggests that the failure of transposon silencing in hybrid dysgenesis systems is a result of the alternative splicing of transposons, which is normally regulated by the piRNA machinery (147).
Figure 4.
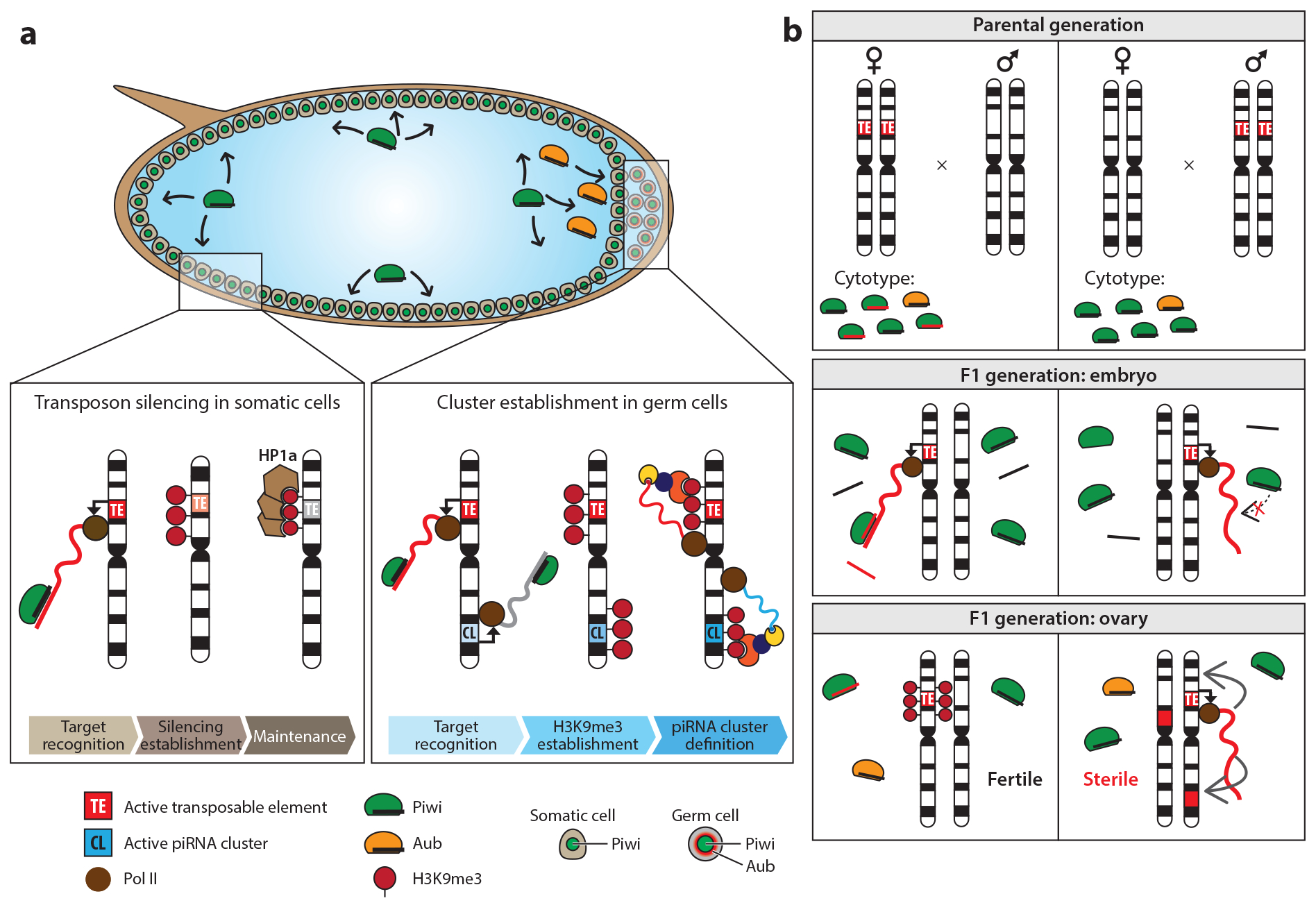
Intergenerationally inherited piRNAs function as epigenetic information carriers. (a) In addition to the maternal deposition of proteins and mRNAs, Drosophila embryos also inherit piRNAs associated with Aubergine (Aub) and Piwi. While Piwi is distributed in the nuclei of somatic and future germline cells throughout the entire embryo, Aub is specifically localized to germ cell progenitors known as pole cells. Upon activation of zygotic transcription, Piwi–piRNA complexes may engage with target transposon RNAs, resulting in recruitment of factors of the transcriptional silencing machinery and deposition of H3K9me2/3. Since Piwi expression in somatic cells is limited to the early stages of development, silencing of transposons must be maintained independently, likely through mechanisms involving heterochromatin protein 1a (HP1a). In developing germ cells, however, Piwi–piRNA complexes target nascent transcripts derived from transposon loci and piRNA clusters, and it is this mechanism that is thought to result in H3K9me2/3 establishment. Instead of recruiting HP1a, these loci are specifically bound by its homolog, Rhino (Rhi), through unknown mechanisms. Binding of Rhi and its cofactors Deadlock (Del), Moonshiner (Moon), and Cutoff (Cuff) ensures the production of piRNA cluster transcripts. Aub–piRNA complexes might serve to reinitiate the ping-pong cycle by generating new sense piRNAs for Ago3 loading, which can, in turn, produce a reshaped pool of trail piRNAs that associate with Piwi. This process might allow resetting of the piRNA pool and target diversification in each generation. (b) Maternally deposited piRNAs protect against hybrid dysgenesis. PIWI–piRNA complexes capable of targeting active transposon copies are maternally deposited. Males do not contribute piRNAs to their offspring. During embryonic development of the next generation, piRNAs scan for nascent transcripts with sequence complementarity. Targets are silenced through chromatin modifications such as H3K9me2/3. However, in cases where females lack piRNAs to protect against transposon insertions in the parental genome, these transposons can escape silencing and result in DNA damage and infertility.
Notably, maternal cytoplasmic inheritance of piRNAs is also responsible for the effects of paramutation (26, 49). Deposition of piRNAs targeting a specific gene locus can lead to it being embedded in heterochromatin and thus silence gene expression. This effect is genetically dominant and was shown to be propagated for over 50 generations in a Zuc-, Aub-, Rhi- and Cuff-dependent manner (26, 49).
Definition of piRNA Clusters by Intergenerationally Inherited piRNAs
Among the three PIWI proteins present in Drosophila germ cells, only Piwi and Aub were found to be maternally deposited, while little to no deposition was detected for Ago3 (Figure 4) (11, 41, 92). Piwi and Aub act through TGS and PTGS, respectively. Thus, epigenetic information encoded by piRNAs is transferred via both silencing mechanisms, though it has yet to be reported whether the other factors involved are also deposited. Prior to zygotic transcription, the genome of early embryos is thought to be naïve with respect to several chromatin marks, including H3K4me2 and H3K9me2/3 (127, 166), whereas other marks such as H3K27me3 are maternally transmitted (168). The precise mechanism involved in restoring histone marks at particular genomic locations is poorly understood.
Interestingly, a correlation found in adult ovaries from flies between deposited piRNAs and the occurrence of H3K9me2/3 marks at regions with homologous sequences suggested a role for intergenerationally inherited Piwi–piRNA complexes in piRNA cluster definition (79, 96). Furthermore, interspecies hybrid crosses between female Drosophila melanogaster and male Drosophila simulans flies produce piRNAs only from the D. melanogaster clusters, a finding that points toward a function of maternally inherited piRNAs in cluster definition (66, 79). Indeed, a recent study found that depletion of Piwi from developing germ cells during embryogenesis leads to decreased H3K9me3 levels as well as reduced Rhi binding at clusters in adult ovaries of D. melanogaster (2). The loss of Piwi in later stages of development had no impact on either H3K9me3 or Rhi binding, suggesting that dual-strand clusters are established and licensed during embryogenesis and maintained in later stages of development without the requirement of Piwi (2). Subsequent to chromatin modification and Rhi recruitment, other factors essential for noncanonical transcription and the production of precursor RNAs such as Del and Moon are thought to define these loci as piRNA clusters. However, why is Rhi specifically associated with clusters and not present at all loci decorated with H3K9me3? Some initial clues come from recent work on D. melanogaster and a related species, D. simulans, which demonstrated that the binding of Del to Rhi is required for accurate recruitment to piRNA cluster loci (110). It may be that the binding of Del to Rhi leads to a conformation change that enables this complex to specifically bind a combination of histone modifications that could be present at dual-strand clusters. However, in-depth analyses of the chromatin context at dual-strand clusters are required to test this option. Alternatively, it may be that as-yet-unknown proteins or other mechanisms function in defining dual-strand clusters.
In addition to defining dual-strand cluster loci, it is thought that maternally deposited piRNAs have a role in kick-starting the biogenesis machinery and in specifically amplifying piRNA species capable of targeting active transposons (79). We speculate that the lack of Ago3 deposition could necessitate the specific production and diversification of piRNA populations in each generation, which would enable the piRNA system to adapt and reshape its pool of antisense piRNAs. In this model, intergenerationally inherited Aub–piRNAs might be essential to detect and cleave transcripts derived from active TEs, thereby determining the piRNA content of Ago3 and, through ping-pong looping, of newly synthesized Aub protein.
SUMMARY POINTS.
The vast majority of piRNAs originate from genomic hotspots called piRNA clusters. RNAs derived from unistrand clusters appear similar to canonical transcripts, whereas RNAs originating from dual-strand clusters, which are likely specific to Drosophilidae, appear to be the products of noncanonical transcription that require specialized machineries.
Licensing of precursors for processing into piRNAs is not fully understood. A cascade of coupled transcription, export, and handover to biogenesis centers was proposed; however, others have suggested specific sequence tags or targeting by sequence complementarity as determinants for piRNA production.
piRNA biogenesis is a cytoplasmic event involving several nucleases. Zucchini (Zuc)-mediated processing requires a mitochondria-anchored machinery, while ping-pong looping depends on numerous Tudor domain proteins.
Ping-pong looping and Zuc-dependent biogenesis are interlinked and generate highly phased trail piRNAs that are predominantly Piwi-bound, thus enabling crosstalk between cytoplasmic target slicing and nuclear chromatin modifications.
Transcriptional gene silencing (TGS) requires piRNA-guided detection of complementary nascent RNAs and results in target silencing. This process requires interactions with factors of the heterochromatin silencing machinery and is accompanied by repressive chromatin marks.
Maternal piRNAs have a crucial role as carriers of epigenetic information that enable the offspring to distinguish self from nonself and consequently silence potentially detrimental transposon insertions while at the same time allowing the definition of piRNA source loci by as-yet-unknown mechanisms.
FUTURE ISSUES.
Which factors and processes are involved in the establishment and maintenance of piRNA source loci? How is the piRNA repertoire shaped across generations and how does cluster evolution take place? What determinants allow Rhino (Rhi) binding and the definition of piRNA clusters?
What molecular signals or processes license precursor transcripts from piRNA clusters for processing into mature piRNAs? How are these RNAs protected from degradation and flagged for transport to the processing centers? How is the transport regulated and what factors define the underlying protein machinery?
How precisely are piRNA processing and loading of piRNAs (or their precursors) into PIWI proteins happening? Are these processes linked? What causes the cleavage specificity of Zuc, which results in the 1U bias? While we have likely uncovered all of the relevant factors, their exact interplay is not yet understood.
What is the biochemical and molecular organization of the ping-pong machinery and how does it link to phased piRNA production? Although a rough framework is in place, we are far from understanding the molecular processes in detail.
How is TGS of transposable elements achieved at the mechanistic level? What is the hierarchy of events that lead to silencing? Is there a final silencing mark, or which chromatin alterations finally lead to transcriptional shutdown? How are piRNA pathway-specific factors linked to the general silencing machinery?
How, when, and where do maternally deposited piRNAs carry out their functions? What is Piwi’s role in somatic cells during early embryogenesis of Drosophila? Does cluster definition depend on maternally inherited Piwi or Aub, or the maternal inheritance of both proteins together?
ACKNOWLEDGMENTS
We thank the piRNA research community for their ongoing support and feedback. Work in the Hannon laboratory is supported by Cancer Research UK and by Wellcome Trust award 110161/Z/15/Z. M.M. is supported by a Boehringer Ingelheim Fonds PhD fellowship. F.C. is supported by a European Molecular Biology Organization Long-Term Fellowship. We apologize in advance to those investigators whose work was inadvertently overlooked or could not be included due to space restrictions.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Akay A, Di Domenico T, Suen KM, Nabih A, Parada GE, et al. 2017. The helicase Aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev. Cell 42:241–55.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkouche A, Mugat B, Barckmann B, Varela-Chavez C, Li B, et al. 2017. Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries. Mol. Cell 66:411–19.e4 [DOI] [PubMed] [Google Scholar]
- 3.Andersen PR, Tirian L, Vunjak M, Brennecke J. 2017. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549:54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207 [DOI] [PubMed] [Google Scholar]
- 5.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, et al. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337–50 [DOI] [PubMed] [Google Scholar]
- 6.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol 11:1017–27 [DOI] [PubMed] [Google Scholar]
- 7.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, et al. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31:785–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–47 [DOI] [PubMed] [Google Scholar]
- 9.Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150:88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–103 [DOI] [PubMed] [Google Scholar]
- 11.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322:1387–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, et al. 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21:2300–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucheton A, Paro R, Sang HM, Pelisson A, Finnegan DJ. 1984. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell 38:153–63 [DOI] [PubMed] [Google Scholar]
- 14.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, et al. 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489:447–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bühler M, Verdel A, Moazed D. 2006. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125:873–86 [DOI] [PubMed] [Google Scholar]
- 16.Cecere G, Zheng GX, Mansisidor AR, Klymko KE, Grishok A. 2012. Promoters recognized by Forkhead proteins exist for individual 21U-RNAs. Mol. Cell 47:734–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K-M, Campbell E, Pandey RR, Yang Z, McCarthy AA, Pillai RS. 2015. Metazoan Maelstrom is an RNA-binding protein that has evolved from an ancient nuclease active in protists. RNA 21:833–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y-CA, Stuwe E, Luo Y, Ninova M, Le Thomas A, et al. 2016. Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol. Cell 63:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark JP, Rahman R, Yang N, Yang LH, Lau NC. 2017. Drosophila PAF1 modulates PIWI/piRNA silencing capacity. Curr. Biol 27:2718–26.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colmenares SU, Buker SM, Buhler M, Dlakić M, Moazed D. 2007. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol. Cell 27:449–61 [DOI] [PubMed] [Google Scholar]
- 21.Cora E, Pandey RR, Xiol J, Taylor J, Sachidanandam R, et al. 2014. The MID-PIWI module of Piwi proteins specifies nucleotide- and strand-biases of piRNAs. RNA 20:773–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12:3715–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czech B, Preall JB, McGinn J, Hannon GJ. 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell 50:749–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, et al. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480:259–63 [DOI] [PubMed] [Google Scholar]
- 26.de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L, et al. 2012. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490:112–15 [DOI] [PubMed] [Google Scholar]
- 27.Dennis C, Brasset E, Sarkar A, Vaury C. 2016. Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila. Nat. Commun 7:13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis C, Zanni V, Brasset E, Eymery A, Zhang L, et al. 2013. “Dot COM”, a nuclear transit center for the primary piRNA pathway in Drosophila. PLOS ONE 8:e72752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding D, Liu J, Dong K, Midic U, Hess RA, et al. 2017. PNLDC1 is essential for piRNA 3′ end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun 8:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dönertas D, Sienski G, Brennecke J. 2013. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 27:1693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fagegaltier D, Falciatori I, Czech B, Castel S, Perrimon N, et al. 2016. Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev. 30:1623–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130:859–71 [DOI] [PubMed] [Google Scholar]
- 33.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–11 [DOI] [PubMed] [Google Scholar]; 33a. Gainetdinov I, Colpan C, Arif A, Cecchini K, Zamore PD. 2018. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol. Cell 71:775–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerace EL, Halic M, Moazed D. 2010. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol. Cell 39:360–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202 [DOI] [PubMed] [Google Scholar]
- 36.Goh W-SS, Seah JWE, Harrison EJ, Chen C, Hammell CM, Hannon GJ. 2014. A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans piRNA biogenesis. Genes Dev. 28:797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goriaux C, Desset S, Renaud Y, Vaury C, Brasset E. 2014. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 15:411–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, et al. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455:1193–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grivna ST, Beyret E, Wang Z, Lin H. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20:1709–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu W, Lee H-C, Chaves D, Youngman EM, Pazour GJ, et al. 2012. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151:1488–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, et al. 2007. A Slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315:1587–90 [DOI] [PubMed] [Google Scholar]
- 42.Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, et al. 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 24:2499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348:817–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handler D, Meixner K, Pizka M, Lauss K, Schmied C, et al. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell 50:762–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, et al. 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 30:3977–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris AN, Macdonald PM. 2001. aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128:2823–32 [DOI] [PubMed] [Google Scholar]
- 47.Hayashi R, Schnabl J, Handler D, Mohn F, Ameres SL, Brennecke J. 2016. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 539:588–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heard E, Martienssen RA. 2014. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157:95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermant C, Boivin A, Teysset L, Delmarre V, Asif-Laidin A, et al. 2015. Paramutation in Drosophila requires both nuclear and cytoplasmic actors of the piRNA pathway and induces cis-spreading of piRNA production. Genetics 201:1381–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. 2005. RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–20 [DOI] [PubMed] [Google Scholar]
- 51.Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, et al. 2015. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep. 12:418–28 [DOI] [PubMed] [Google Scholar]
- 52.Honda S, Kirino Y, Maragkakis M, Alexiou P, Ohtaki A, et al. 2013. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 19:1405–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horwich MD, Li C, Matranga C, Vagin V, Farley G, et al. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol 17:1265–72 [DOI] [PubMed] [Google Scholar]
- 54.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129:69–82 [DOI] [PubMed] [Google Scholar]
- 55.Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, et al. 2016. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev. 30:840–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hutvagner G, Simard MJ. 2008. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol 9:22–32 [DOI] [PubMed] [Google Scholar]
- 57.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. 2012. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491:279–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishizu H, Iwasaki YW, Hirakata S, Ozaki H, Iwasaki W, et al. 2015. Somatic primary piRNA biogenesis driven by cis-acting RNA elements and trans-acting Yb. Cell Rep. 12:429–40 [DOI] [PubMed] [Google Scholar]
- 59.Iwasaki YW, Murano K, Ishizu H, Shibuya A, Iyoda Y, et al. 2016. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol. Cell 63:408–19 [DOI] [PubMed] [Google Scholar]
- 60.Izumi N, Shoji K, Sakaguchi Y, Honda S, Kirino Y, et al. 2016. Identification and functional analysis of the pre-piRNA 3′ Trimmer in silkworms. Cell 164:962–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Juliano CE, Reich A, Liu N, Götzfried J, Zhong M, et al. 2014. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. PNAS 111:337–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaufmann J, Smale ST. 1994. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 8:821–29 [DOI] [PubMed] [Google Scholar]
- 63.Kawaoka S, Hara K, Shoji K, Kobayashi M, Shimada T, et al. 2013. The comprehensive epigenome map of piRNA clusters. Nucleic Acids Res. 41:1581–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, et al. 2009. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15:1258–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawaoka S, Izumi N, Katsuma S, Tomari Y. 2011. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell 43:1015–22 [DOI] [PubMed] [Google Scholar]
- 66.Kelleher ES, Edelman NB, Barbash DA. 2012. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLOS Biol. 10:e1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, et al. 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147:1551–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kidwell MG, Kidwell JF, Sved JA. 1977. Hybrid dysgenesis in Drosophila melanogaster: a syndrome of aberrant traits including mutation, sterility and male recombination. Genetics 86:813–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, et al. 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol 11:652–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, et al. 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509:633–36 [DOI] [PubMed] [Google Scholar]
- 71.Klattenhoff C, Xi H, Li C, Lee S, Xu J, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138:1137–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klenov MS, Lavrov SA, Korbut AP, Stolyarenko AD, Yakushev EY, et al. 2014. Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic Acids Res. 42:6208–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, et al. 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. PNAS 108:18760–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kowalik KM, Shimada Y, Flury V, Stadler MB, Batki J, Bühler M. 2015. The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature 520:248–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, et al. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547:236–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lau NC, Robine N, Martin R, Chung W-J, Niki Y, et al. 2009. Abundant primary piRNAs, endosiRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 19:1776–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, et al. 2006. Characterization of the piRNA complex from rat testes. Science 313:363–67 [DOI] [PubMed] [Google Scholar]
- 78.Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, et al. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27:390–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Thomas A, Stuwe E, Li S, Du J, Marinov G, et al. 2014. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 28:1667–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee H-C, Gu W, Shirayama M, Youngman E, Conte D Jr., Mello CC. 2012. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150:78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, et al. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol 2:174–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li C, Vagin VV, Lee S, Xu J, Ma S, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137:509–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, et al. 2013. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50:67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lim AK, Kai T. 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. PNAS 104:6714–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lim RSM, Anand A, Nishimiya-Fujisawa C, Kobayashi S, Kai T. 2014. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev. Biol 386:237–51 [DOI] [PubMed] [Google Scholar]
- 86.Liu L, Qi H, Wang J, Lin H. 2011. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138:1863–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luteijn MJ, van Bergeijk P, Kaaij LJT, Almeida MV, Roovers EF, et al. 2012. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31:3422–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, et al. 2009. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLOS Genet. 5:e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, et al. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137:522–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matsumoto N, Nishimasu H, Sakakibara K, Nishida KM, Hirano T, et al. 2016. Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA. Cell 167:484–97.e9 [DOI] [PubMed] [Google Scholar]
- 91.Matsumoto N, Sato K, Nishimasu H, Namba Y, Miyakubi K, et al. 2015. Crystal structure and activity of the endoribonuclease domain of the piRNA pathway factor Maelstrom. Cell Rep. 11:366–75 [DOI] [PubMed] [Google Scholar]
- 92.Megosh HB, Cox DN, Campbell C, Lin H. 2006. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol 16:1884–94 [DOI] [PubMed] [Google Scholar]
- 93.Mendez DL, Mandt RE, Elgin SC. 2013. Heterochromatin protein 1a (HP1a) partner specificity is determined by critical amino acids in the chromo shadow domain and C-terminal extension. J. Biol. Chem 288:22315–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miesen P, Girardi E, van Rij RP. 2015. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 43:6545–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mohn F, Handler D, Brennecke J. 2015. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348:812–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mohn F, Sienski G, Handler D, Brennecke J. 2014. The Rhino-Deadlock-Cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157:1364–79 [DOI] [PubMed] [Google Scholar]
- 97.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLOS Pathog. 8:e1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789–802 [DOI] [PubMed] [Google Scholar]
- 99.Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, et al. 2013. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell 50:736–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Murota Y, Ishizu H, Nakagawa S, Iwasaki YW, Shibata S, et al. 2014. Yb integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 8:103–13 [DOI] [PubMed] [Google Scholar]
- 101.Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, et al. 2015. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 10:193–203 [DOI] [PubMed] [Google Scholar]
- 102.Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, et al. 2009. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28:3820–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, et al. 2012. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491:284–87 [DOI] [PubMed] [Google Scholar]
- 104.Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. 2013. DmGTSF1 is necessary for Piwi–piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 27:1656–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Olivieri D, Senti K-A, Subramanian S, Sachidanandam R, Brennecke J. 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell 47:954–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 29:3301–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pandey RR, Homolka D, Chen K-M, Sachidanandam R, Fauvarque M-O, Pillai RS. 2017. Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLOS Genet. 13:e1006956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pane A, Jiang P, Zhao DY, Singh M, Schüpbach T. 2011. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 30:4601–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pane A, Wehr K, Schüpbach T. 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12:851–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parhad SS, Tu S, Weng Z, Theurkauf WE. 2017. Adaptive evolution leads to cross-species incompatibility in the piRNA transposon silencing machinery. Dev. Cell 43:60–70.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patil VS, Anand A, Chakrabarti A, Kai T. 2014. The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in the piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol. 12:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patil VS, Kai T. 2010. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol 20:724–30 [DOI] [PubMed] [Google Scholar]
- 113.Pélisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG. 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 13:4401–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pezic D, Manakov SA, Sachidanandam R, Aravin AA. 2014. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 28:1410–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Post C, Clark JP, Sytnikova YA, Chirn G-W, Lau NC. 2014. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 20:1977–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Preall JB, Czech B, Guzzardo PM, Muerdter F, Hannon GJ. 2012. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 18:1446–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A. 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139:697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Purnell BA, Emanuel PA, Gilmour DS. 1994. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 8:830–42 [DOI] [PubMed] [Google Scholar]
- 119.Qi H, Watanabe T, Ku H-Y, Liu N, Zhong M, Lin H. 2011. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem 286:3789–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, et al. 2011. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol 21:1373–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, et al. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480:264–67 [DOI] [PubMed] [Google Scholar]
- 122.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol 16:639–46 [DOI] [PubMed] [Google Scholar]
- 123.Robine N, Lau NC, Balla S, Jin Z, Okamura K, et al. 2009. A broadly conserved pathway generates 3′ UTR-directed primary piRNAs. Curr. Biol 19:2066–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rogers AK, Situ K, Perkins EM, Toth KF. 2017. Zucchini-dependent piRNA processing is triggered by recruitment to the cytoplasmic processing machinery. Genes Dev. 31:1858–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rozhkov NV, Hammell M, Hannon GJ. 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 27:400–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, et al. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127:1193–207 [DOI] [PubMed] [Google Scholar]
- 127.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, et al. 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3–3. Mol. Cell 26:103–15 [DOI] [PubMed] [Google Scholar]
- 128.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, et al. 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461:1296–99 [DOI] [PubMed] [Google Scholar]
- 129.Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, et al. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24:2493–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, et al. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20:2214–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev. 21:1603–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sato K, Iwasaki YW, Shibuya A, Carninci P, Tsuchizawa Y, et al. 2015. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol. Cell 59:553–63 [DOI] [PubMed] [Google Scholar]
- 133.Saxe JP, Chen M, Zhao H, Lin H. 2013. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 32:1869–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Senti K-A, Jurczak D, Sachidanandam R, Brennecke J. 2015. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 29:1747–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sheu-Gruttadauria J, MacRae IJ. 2017. Structural foundations of RNA silencing by Argonaute. J. Mol. Biol 429:2619–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shimada Y, Mohn F, Bühler M. 2016. The RNA-induced transcriptional silencing complex targets chromatin exclusively via interacting with nascent transcripts. Genes Dev. 30:2571–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, et al. 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150:65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shiromoto Y, Kuramochi-Miyagawa S, Daiba A, Chuma S, Katanaya A, et al. 2013. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA 19:803–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, et al. 2009. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell 17:775–87 [DOI] [PubMed] [Google Scholar]
- 140.Sienski G, Batki J, Senti K-A, Dönertas D, Tirian L, et al. 2015. Silencio/CG9754 connects the Piwi–piRNA complex to the cellular heterochromatin machinery. Genes Dev. 29:2258–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151:964–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547:241–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. PNAS 102:152–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sumiyoshi T, Sato K, Yamamoto H, Iwasaki YW, Siomi H, Siomi MC. 2016. Loss of l(3)mbt leads to acquisition of the ping-pong cycle in Drosophila ovarian somatic cells. Genes Dev. 30:1617–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Szakmary A, Reedy M, Qi H, Lin H. 2009. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J. Cell Biol. 185:613–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tang W, Tu S, Lee H-C, Weng Z, Mello CC. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164:974–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Teixeira FK, Okuniewska M, Malone CD, Coux R-X, Rio DC, Lehmann R. 2017. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552:268–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tyc KM, Nabih A, Wu MZ, Wedeles CJ, Sobotka JA, Claycomb JM. 2017. The conserved intron binding protein EMB-4 plays differential roles in germline small RNA pathways of C. elegans. Dev. Cell 42:256–70.e6 [DOI] [PubMed] [Google Scholar]
- 149.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313:320–24 [DOI] [PubMed] [Google Scholar]
- 150.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, et al. 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23:1749–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vagin VV, Yu Y, Jankowska A, Luo Y, Wasik KA, et al. 2013. Minotaur is critical for primary piRNA biogenesis. RNA 19:1064–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, et al. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O. 2012. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA 18:2128–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, et al. 2015. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 29:617–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang SH, Elgin SCR. 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. PNAS 108:21164–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang W, Han BW, Tipping C, Ge DT, Zhang Z, et al. 2015. Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol. Cell 59:819–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell 20:364–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Webster A, Li S, Hur JK, Wachsmuth M, Bois JS, et al. 2015. Aub and Ago3 are recruited to Nuage through two mechanisms to form a ping-pong complex assembled by Krimper. Mol. Cell 59:564–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Weick E-M, Sarkies P, Silva N, Chen RA, Moss SMM, et al. 2014. PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes Dev. 28:783–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wenda JM, Homolka D, Yang Z, Spinelli P, Sachidanandam R, et al. 2017. Distinct roles of RNA helicases MVH and TDRD9 in PIWI slicing-triggered mammalian piRNA biogenesis and function. Dev. Cell 41:623–37.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. 2009. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458:784–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, et al. 2012. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 47:970–79 [DOI] [PubMed] [Google Scholar]
- 163.Yang Z, Chen K-M, Pandey RR, Homolka D, Reuter M, et al. 2016. PIWI slicing and EXD1 drive biogenesis of nuclear piRNAs from cytosolic targets of the mouse piRNA pathway. Mol. Cell 61:138–52 [DOI] [PMC free article] [PubMed] [Google Scholar]; 163a. Yashiro R, Murota Y, Nishida KM, Yamashiro H, Fujii K, et al. 2018. Piwi nuclear localization and its regulatory mechanism in Drosophila ovarian somatic cells. Cell Rep. 23:3647–57 [DOI] [PubMed] [Google Scholar]
- 164.Yin H, Lin H. 2007. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450:304–8 [DOI] [PubMed] [Google Scholar]; 164a. Yu B, Lin YA, Parhad SS, Jin Z, Ma J, et al. 2018. Structural insights into Rhino–Deadlock complex for germline piRNA cluster specification. EMBO Rep. 19:e45418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yu Y, Gu J, Jin Y, Luo Y, Preall JB, et al. 2015. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350:339–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yuan K, O’Farrell PH. 2016. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev. 30:579–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, et al. 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138:4039–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zenk F, Loeser E, Schiavo R, Kilpert F, Bogdanović O, Iovino N. 2017. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357:212–16 [DOI] [PubMed] [Google Scholar]
- 169.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, et al. 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151:871–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang K, Mosch K, Fischle W, Grewal SIS. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol 15:381–88 [DOI] [PubMed] [Google Scholar]
- 171.Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, et al. 2014. The HP1 homolog Rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157:1353–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, et al. 2011. Heterotypic piRNA Ping-Pong requires Qin, a protein with both E3 ligase and Tudor domains. Mol. Cell 44:572–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, et al. 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. PNAS 107:11841–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, et al. 2014. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157:1050–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou X, Battistoni G, El Demerdash O, Gurtowski J, Wunderer J, et al. 2015. Dual functions of Macpiwi1 in transposon silencing and stem cell maintenance in the flatworm Macrostomum lignano. RNA 21:1885–97 [DOI] [PMC free article] [PubMed] [Google Scholar]


