Abstract
22q11.2 deletion syndrome (22q11DS) is the most frequently occurring microdeletion in humans. It is associated with a significant impact on brain structure, including prominent reductions in gray matter volume (GMV), and neuropsychiatric manifestations, including cognitive impairment and psychosis. It is unclear whether GMV alterations in 22q11DS occur according to distinct structural patterns. Then, 783 participants (470 with 22q11DS: 51% females, mean age [SD] 18.2 [9.2]; and 313 typically developing [TD] controls: 46% females, mean age 18.0 [8.6]) from 13 datasets were included in the present study. We segmented structural T1‐weighted brain MRI scans and extracted GMV images, which were then utilized in a novel source‐based morphometry (SBM) pipeline (SS‐Detect) to generate structural brain patterns (SBPs) that capture co‐varying GMV. We investigated the impact of the 22q11.2 deletion, deletion size, intelligence quotient, and psychosis on the SBPs. Seventeen GMV‐SBPs were derived, which provided spatial patterns of GMV covariance associated with a quantitative metric (i.e., loading score) for analysis. Patterns of topographically widespread differences in GMV covariance, including the cerebellum, discriminated individuals with 22q11DS from healthy controls. The spatial extents of the SBPs that revealed disparities between individuals with 22q11DS and controls were consistent with the findings of the univariate voxel‐based morphometry analysis. Larger deletion size was associated with significantly lower GMV in frontal and occipital SBPs; however, history of psychosis did not show a strong relationship with these covariance patterns. 22q11DS is associated with distinct structural abnormalities captured by topographical GMV covariance patterns that include the cerebellum. Findings indicate that structural anomalies in 22q11DS manifest in a nonrandom manner and in distinct covarying anatomical patterns, rather than a diffuse global process. These SBP abnormalities converge with previously reported cortical surface area abnormalities, suggesting disturbances of early neurodevelopment as the most likely underlying mechanism.
Keywords: 22q11 deletion syndrome, gray matter volume, magnetic resonnance imaging, source‐based morphometry
Using a novel source‐based morphometry method called SS‐Detect, we identified 12 structural brain patterns (SBPs) that discriminated individuals with 22q11.2 deletion syndrome from healthy controls. We further demonstrated that deletion size was related to structural covariance patterns; however, history of psychosis did not show a strong relationship with these covariance patterns.
1. INTRODUCTION
22q11.2 deletion syndrome (22q11DS) is the most commonly occurring microdeletion in humans, affecting an estimated 1 in 2000 to 4000 newborns (Blagojevic et al., 2021; McDonald‐McGinn et al., 2015). In addition to the congenital cardiovascular and craniofacial anomalies that most often lead to diagnosis, the vast majority of individuals with 22q11DS also exhibit anomalies related to the central nervous system (CNS) (Blagojevic et al., 2021; Hopkins et al., 2018; Linton et al., 2020; McDonald‐McGinn et al., 2015). The most prevalent CNS anomalies encompass a spectrum of intellectual and learning disabilities or cognitive deficits (>90%), psychiatric disorders (60%; estimated 25–30% with psychotic illness), and combinations thereof (Linton et al., 2020; Moberg et al., 2018).
A recent meta‐analysis showed that the neuropsychological profile in 22q11DS is characterized by a large effect on Full Scale intelligence quotient (FSIQ) (d = −2.5), with slightly worse performance IQ (PIQ) than verbal IQ (VIQ) impairments (d = −2.4 vs. d = −1.9, respectively), accompanied by significant impairments in domains of language, motor, executive function, and academic attainment as well (Moberg et al., 2018). Deletion size was recently shown to impact cognitive function, with the larger and more common (A‐D) deletion associated with 4 to 8‐point decreases in FSIQ, PIQ, and VIQ, relative to the smaller and less common (A‐B) deletion (Zhao et al., 2018). Furthermore, more severe baseline cognitive deficits—as well as early cognitive decline, particularly in the verbal domain—were associated with significantly increased risk of developing psychosis in 22q11DS (Pontillo et al., 2019; Vorstman et al., 2015). The broad range of cognitive impairment and diverse phenotypic manifestations involving the brain in individuals with 22q11DS suggests that the microdeletion has a widespread neuroanatomical impact, rather than a focal pattern of brain abnormalities.
Indeed, structural MRI (sMRI) studies have demonstrated that 22q11DS is associated with widespread brain abnormalities (Cheon et al., 2022; Ching et al., 2020; Lin et al., 2017; Schmitt et al., 2015; Simon, Ding, et al., 2005; Sun et al., 2020). Specifically, relative to TD controls, surface area is significantly lower across all brain lobes, while cortical thickness (CT) shows overall higher thickness, with localized thinning in the anterior cingulate and superior temporal gyrus. These neuroanatomic patterns led to correct classification of 22q11DS brain scans versus controls with an accuracy of 93.8% (Sun et al., 2020). In addition, 22q11DS is associated with subcortical alterations (Ching et al., 2020). There is also evidence that the magnitude of certain brain structural phenotypes (e.g., the extent of surface area reduction) depends on the size of the 22q11.2 deletion (Ching et al., 2020; Sun et al., 2020).
Gray matter volume (GMV) is a critical metric in sMRI. Voxel‐based morphometry (VBM) has been used in small to modest sample sizes in single‐site studies (N's ranging from 14 to 63) to characterize GMV alterations in 22q11DS (Baker et al., 2011; Campbell et al., 2006; Chow et al., 2011; Gothelf et al., 2011; Piervincenzi et al., 2022; Shashi et al., 2010; Simon, Ding, et al., 2005). Meta‐analyses have shown both a global deficit in gray matter as well as specific decreases predominantly in posterior brain regions (Tan et al., 2009). However, the cerebellum has typically not been included in these analyses.
VBM is a univariate approach that interrogates differences in single voxels between groups of interest. The underlying statistical assumption made in VBM is that the GMV value at each of the voxels is conditionally independent of the others, and thus independent statistics are computed for each of the thousands of voxels (e.g., using t tests). In contrast, source‐based morphometry (SBM) is a data‐driven multivariate method that takes into account the relationship of sMRI metrics amongst voxels. SBM originated from research that demonstrated the coordinated development and functioning of brain regions (Alexander‐Bloch et al., 2013; Zielinski et al., 2010). It was further supported by findings that communities of brain regions exhibit covariation in their morphological properties (Mechelli et al., 2005). SBM identifies spatially related imaging features (e.g., GMV) with common inter‐individual covariation to form spatial SBPs (Colloby et al., 2021; Gupta et al., 2019; Xu et al., 2009), and provides SBP‐specific loading scores for each subject on the sMRI metric of choice (e.g., GMV). SBM provides a means to assess whether GMV abnormalities in a disease involve a diffuse global process or occur according to distinct patterns. SBM has been primarily employed in single‐scanner studies to examine neuroanatomical distinctions between populations and explore the neuroanatomical associations with demographic or clinical characteristics (Ge et al., 2019; Ge, Liu, et al., 2021; Hafkemeijer et al., 2014; Park et al., 2022; Steenwijk et al., 2015; Xu et al., 2009). In recent years, there has been a growing trend toward collaborative studies that utilize SBM (Ge, Hassel, et al., 2021; Gupta et al., 2015; Luo et al., 2020; Mei et al., 2020), as the pooling of multisite MRI data from multiple research sites has gained momentum (Thompson et al., 2020). Our group has made a recent advance in SBM analysis with the development of a novel strategy called SS‐Detect (Ge, Ding, et al., 2021), which models scanner‐specific information and thereby infers more accurate estimates of subject‐specific loading parameters of each site in multisite studies.
In the present study, we assessed SBPs in 22q11DS and sex‐ and age‐matched TD controls (hereafter controls) from the ENIGMA 22q11.2 Deletion Syndrome Working Group using SS‐Detect. We addressed the following questions:
Are there specific covariation patterns of GMV abnormalities in 22q11DS? That is, do GMV in 22q11DS reflect a diffuse global process, or do they develop instead according to distinct anatomical patterns?
Is 22q11.2 deletion size associated with differences in 22q11DS SBPs?
Are SBP abnormalities associated with neuropsychiatric phenotypes in 22q11DS (namely, psychosis or IQ)?
2. MATERIALS AND METHODS
2.1. Participants and datasets
sMRI scan data from 783 participants (470 22q11DS individuals and 313 controls) from 10 research sites in the ENIGMA 22q11.2 Working Group were included in the present study. All research studies obtained approval from local institutional review boards and ethics committees. Written informed consent was obtained from all individual participants or their guardians.
Structural T1‐weighted brain MRI scans were acquired using 13 scanners at 10 participating international research sites. Sites with more than one scanner or acquisition protocol were treated as separate sites in the analysis. Details of the acquisition parameters for each contributing site can be found in previous ENIGMA studies using these data (Ching et al., 2020; Sun et al., 2020).
2.2. Image preprocessing
sMRI data preprocessing was performed with the Computational Anatomy Toolbox (CAT12; http://dbm.neuro.uni-jena.de/cat). For preprocessing, default settings were used as described in the toolbox manual (http://dbm.neuro.uni-jena.de/cat12/CAT12-Manual.pdf; see Supplementary Methods 1.1 for details). We evaluated data quality and did not exclude data because of poor quality as determined by CAT12 (see Supplementary Methods 1.2). We estimated the total intracranial volume (TIV) to be used as a covariate in the subsequent analyses.
2.3. Source‐based morphometry
SBM investigated the effect of 22q11DS diagnosis on variation between brain regions with covaried GMV, termed SBPs. The SBM protocol is outlined in detail in the Supplementary Material. Briefly, an SBM approach for multisite studies (Ge, Ding, et al., 2021) was implemented using the GMV images with independent component analysis (ICA; see supplementary material Figure S2 for details), with 17 components estimated by the minimum description length criterion (Li et al., 2007). In the literature of fMRI analysis using group ICA, there are commonly two principal component analysis (PCA) reduction steps, one at the individual subject level and a second at the group level. In this study, we adopted a similar strategy for SBM analysis. Specifically, we first reduced each single‐site dataset into 17 principal components, effectively preserving at least 95% of the variance for each dataset, followed by a reduction of the group‐level concatenated data into 17 principal components before utilizing them in ICA. As a supplement, we illustrated the effectiveness of this site‐specific PCA in mitigating site‐effect (see section S1.5 in supplement material). The site‐specific spatial brain patterns (SBPs) were subsequently derived through back‐reconstruction using dual regression (Beckmann et al., 2009). SBM obtains GMV covariation spatial maps (called SBPs) and their corresponding loading scores for each SBP; these SBPs capture GMV covariation patterns among participants. With SS‐Detect, correspondence of spatial SBPs across subjects acquired with different MRI scanners is robustly established; this is particularly important as SBM relies on the prerequisite hypothesis that the groups being compared exhibit similar spatial maps (Gupta et al., 2019; Xu et al., 2009). Furthermore, our previous study demonstrated that SS‐Detect enhances sensitivity and reduces false‐positive regions in detecting between‐group differences compared to the conventional SBM strategy. This improvement makes SS‐Detect particularly advantageous when utilized in multisite cross‐sectional clinical studies involving SBM (Ge, Ding, et al., 2021).
2.4. Statistical analyses
2.4.1. 22q11DS versus controls
Statistical analysis was performed on SBP loading scores: determination of group differences in SBPs was performed using a two‐sample t test on every column of the loading score matrix (supplementary material Figure S2); specifically, the effects of age, sex, TIV, and site were removed from the loading scores with a linear regression model with the “fitlm” function in MATLAB, then the residualized loading scores were submitted to two‐sample t tests. For each SBP the corresponding loading scores capture the covariation of this SBP among participants, and a group with larger loading scores is the group with larger GMV in this SBP (Gupta et al., 2019; Xu et al., 2009). The results were corrected for multiple comparisons using the false discovery rate (FDR) approach and the significance threshold was set at p < .05 (Benjamini et al., 2001). Post hoc analyses were carried out to evaluate SBP differences between 22q11DS and controls by calculating Hedges' g effect size for each SBP that exhibited a significant between‐group difference. In the present study, we computed each SBP's effect size in the pooled data across all 13 scanners (i.e., mega‐analysis). We also conducted meta‐analyses across all scanners that included both 22q11DS and control subjects by conducting the statistical comparisons of each study individually (11 studies were used, because 2 studies lacked controls), and combining the summary statistics across studies. To investigate the role of IQ on the results, we carried out secondary analyses with IQ as an additional independent variable that was removed from the loading scores of the SBPs (supplemental Figures S8 and S9). Post hoc analyses in all subjects were conducted on all 12 SBPs that exhibited significant difference between 22q11DS individuals and healthy controls. A general linear model was fit with IQ as the dependent variable, and SBP as an independent variable with TIV, age, sex, site, and group as covariates in the model.
We used the Neurosynth decoder function (https://neurosynth.org/; Yarkoni et al., 2011) to annotate the SBPs that showed significant between‐group differences to explore their putative functional role. We submitted the spatial maps (thresholded at Z > 2.5) of these SBPs to Neurosynth, and the top 10 functional roles assigned to each submitted SBP were used to infer its putative functionality. We calculated the energy associated with these Z > 2.5 voxels relative to the total energy (Wang et al., 2012) encompassing the entire brain's voxels. On average, these voxels accounted for 49.11% (standard deviation = 13.75%) of the total energy, and we deemed these voxels as robust representatives of the underlying patterns.
2.4.2. Effect of 22q11.2 deletion size on SBPs
Each subject with an A‐B deletion was matched with subjects with A‐D deletions, and healthy controls, based on same site and sex, and closest age following procedures according to our prior work (Sun et al., 2020). This comparison included 129 participants with 22q11DS who had deletion size information available (22 with a proximal nested A‐B deletion and 107 with a typical A‐D deletion). Deletion size was determined previously using multiplex ligation‐dependent probe amplification (MLPA) (see Sun et al., 2020 for details). Each subject with an A–B deletion was matched with four to five subjects with A–D deletions, and four to five healthy controls, based on same site and sex, and closest age. Determination of group differences in SBPs was performed using a two‐sample t test on the loading scores, after removing effects of age, sex, TIV, and site from the loading scores with a linear regression model. The results were corrected for multiple comparisons with FDR correction at the significance threshold q < 0.05. To investigate the effect of IQ on the results, we carried out sensitivity analyses by removing the effect of IQ from the loading scores.
2.4.3. Effects of psychosis on SBPs in 22q11DS
To compare SBPs between 22q11DS subjects with and without a psychotic disorder diagnosis, each 22q11DS + psychosis subject was matched to a 22q11DS‐no‐psychosis subject at the same site, with the same sex, and closest age following procedures according to prior work (Gur et al., 2017). This analysis included 117 demographically matched participants with 22q11DS who had consensus‐based diagnostic information about psychosis history (Sun et al., 2020), comparing two subgroups: n = 58 with confirmed diagnosis of psychotic disorder and n = 59 confirmed to have never had psychotic symptoms (Sun et al., 2020). A two‐sample t test on the loading scores was used to compare the two subgroups, after removing effects of age, sex, TIV, and site from the loading scores with a linear regression model with the “fitlm” function in MATLAB R2013b. The results were corrected for multiple comparisons, with the significance threshold q < 0.05 (FDR correction). To investigate the effect of IQ on the results, we carried out secondary analyses by removing the effect of IQ from the loading scores.
2.4.4. Conventional univariate analyses
Univariate VBM analyses of two‐sample t tests were conducted to compare participants with 22q11DS to controls, 22q11DS with the A‐B versus A‐D deletion, 22q11DS with and without psychosis, respectively (further details in Supplemental information 1.2). Before submitting the loading scores to t tests, the effects of age, sex, TIV, and site were removed from the loading scores with a linear regression model.
3. RESULTS
3.1. 22q11DS versus control differences in SBPs
A detailed description of the ascertainment procedures, inclusion/exclusion criteria and demographics of the current cohorts was published previously (Sun et al., 2020). Briefly, there were no significant group differences in sex (chi‐squared test, χ 2 = 2.01, p = .16) or age (two‐sample t test, t = 0.29, p = .77), but controls had significantly higher IQ (two‐sample t test, t = 31.95, p < .001) than 22q11DS participants (Table 1).
TABLE 1.
Participant demographics by site.
| Site | 22q11DS (n = 470) | Controls (n = 313) | p‐Value for sex | p‐Value for age | 22q11DS with IQ data (n = 442) | Controls with IQ data (n = 219) | p‐Value for sex | p‐Value for age | p‐Value for IQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex (F/M) | Mean age (SD) | Sex (F/M) | Mean age (SD) | Sex (F/M) | Mean age (SD) | IQ (SD) | Sex (F/M) | Mean age (SD) | IQ (SD) | ||||||
| All | 239/231 | 18.02 (9.20) | 143/170 | 18.21 (8.56) | .16 | .77 | 230/212 | 18.22 (8.66) | 75.13 (13.17) | 103/116 | 15.32 (6.28) | 111.67 (15.10) | .23 | .12 | <.001 |
| Cardiff | 8/7 | 15.07 (2.87) | 6/5 | 14.55 (1.63) | .95 | .59 | 8/6 | 14.79 (2.75) | 77.36 (19.12) | 5/5 | 14.50 (1.72) | 106.70 (7.45) | .73 | .77 | <.001 |
| UC Davis1 | 17/23 | 10.73 (2.06) | 18/19 | 10.43 (2.46) | .59 | .57 | 17/23 | 10.73 (2.06) | 72.65 (13.28) | 16/17 | 10.21 (2.48) | 114.70 (11.07) | .61 | .34 | <.001 |
| UC Davis2 | 30/34 | 11.34 (2.52) | 23/25 | 10.83 (2.39) | .91 | .28 | 28/34 | 11.35 (2.55) | 75.48 (13.15) | 22/23 | 10.78 (2.40) | 113.73 (14.64) | .70 | .24 | <.001 |
| IoP, London | 17/19 | 16.83 (7.19) | 15/10 | 19.64 (6.05) | .33 | .12 | 17/19 | 16.83 (7.19) | 84.50 (13.07) | 15/10 | 19.64 (6.05) | 115.88 (10.28) | .33 | .16 | <.001 |
| Maastricht | 13/14 | 29.22 (6.70) | 15/30 | 29.27 (9.62) | .21 | .98 | 13/14 | 29.22 (6.70) | 74.48 (11.30) | 5/18 | 24.43 (7.70) | 108.35 (15.82) | .05 | .02 | <.001 |
| Newcastle | 12/7 | 17.21 (2.92) | 14/12 | 16.77 (3.30) | .53 | .64 | 12/7 | 17.21 (2.92) | 73.74 (13.56) | 13/11 | 16.63 (3.37) | 108.67 (15.71) | .55 | .55 | <.001 |
| SUNY | 24/30 | 20.72 (2.33) | 8/12 | 20.50 (1.24) | .73 | .69 | 24/30 | 20.72 (2.33) | 73.65 (11.29) | 8/12 | 20.50 (1.24) | 105.20 (14.10) | .73 | .69 | <.001 |
| Toronto1 | 7/8 | 41.47 (7.58) | 4/8 | 42.42 (8.67) | .48 | .76 | 7/8 | 41.47 (7.58) | 72.00 (10.69) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| Toronto2 | 21/13 | 28.03 (10.26) | ‐ | ‐ | ‐ | ‐ | 20/13 | 28.09 (10.42) | 71.67 (9.85) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| UCLA1 | 12/14 | 14.69 (6.79) | 15/7 | 14.50 (5.79) | .13 | .92 | 11/13 | 13.50 (5.36) | 81.58 (14.15) | 14/7 | 13.95 (5.31) | 119.05 (20.58) | .16 | .78 | <.001 |
| UCLA2 | 21/17 | 15.84 (7.93) | 5/14 | 13.26 (3.60) | .04 | .18 | 21/15 | 15.47 (6.94) | 77.61 (12.15) | 5/13 | 12.89 (3.31) | 104.67 (17.83) | .03 | .14 | <.001 |
| UPenn | 19/30 | 17.29 (3.20) | 20/28 | 17.52 (3.22) | .77 | .72 | 14/15 | 17.62 (2.98) | 76.31 (18.58) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
| Utrecht | 38/15 | 17.57 (4.22) | ‐ | ‐ | ‐ | ‐ | 38/15 | 17.57 (4.22) | 69.74 (8.54) | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ |
Abbreviations: 22q11DS, 22q11 deletion syndrome; IoP, Institute of Psychiatry; IQ, intelligence quotient; SUNY, State University of New York; UC Davis, University of California Davis; UCLA, University of California, Los Angeles; Upenn, University of Pennsylvania.
The SBM procedure decomposed the GMV data into 17 independent SBPs. Overall, these SBPs exhibited symmetrically spatial distributions. SBPs were labeled and grouped according to their predominant similarity in topographical locations (supplementary material Figure S4, Figure S5, and Table S1). Group differences were observed in 12 of the 17 SBPs. Six SBPs showed lower loadings in 22q11DS compared to controls. The other six SBPs showed higher loadings in 22q11DS. Results of the meta‐analysis strategy (weighted summary statistics of 11 sites with both 22q11DS and controls) and mega‐analysis (pooling data from all 13 sites) were fully convergent. We proceeded with reporting in the main text the results of mega‐analysis only. Meta‐analytic results with effect sizes for each of the sites are presented in supplementary material Figures S6 and S7.
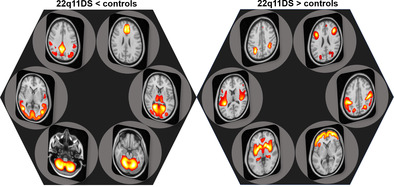
The spatial patterns of the six SBPs with significantly lower loading scores in 22q11DS relative to controls, along with the putative functionality of these SBPs according to the meta‐analytic decoder Neurosynth, are presented in Figure 1 (decoder labels are represented as word‐cloud plots on top of the violin plots). We observed highly significant differences (all p < .001) for SBP 1 and SBP 4 (both cerebellar SBPs), SBP 7 (parietal pattern), SBP 8 (lateral occipital pattern), SBP 9 (frontal pattern), and SBP 12 (medial occipital pattern), which had the largest effect size (−1.34 95% CI: [−1.5, −1.18]), indicating that 22q11DS had lower GMV than controls in those covariation patterns (supplementary material Table S2). Specifically, these results indicated lower GMV across almost the entire cerebellum, as well as middle temporal and occipital gyri, cuneus and precuneus, cingulate gyrus, anterior and posterior cingulate, and medial frontal gyrus in 22q11DS relative to controls (Supplementary Table S1 provides details of regions included in each of the SBPs). The spatial patterns of the six SBPs with higher loading scores in 22q11DS relative to controls are presented in Figure 2. These patterns included SBP 5 (temporal pattern), SBP 6 (subcortical pattern), SBP 10 (frontal pattern), SBP 14 (sensorimotor pattern), and SBP 13 and SBP 17 (two frontoparietal patterns). These results indicated higher GMV in these patterns in 22q11DS relative to controls (supplementary material Table S2). All of these results were robust to removing the effect of IQ from the loading scores in the analyses. Post hoc analyses with IQ as the dependent variable showed that the SBPs that demonstrated differences between 22q11DS individuals and controls did not show significant associations with IQ (FDR correction q < 0.05).
FIGURE 1.
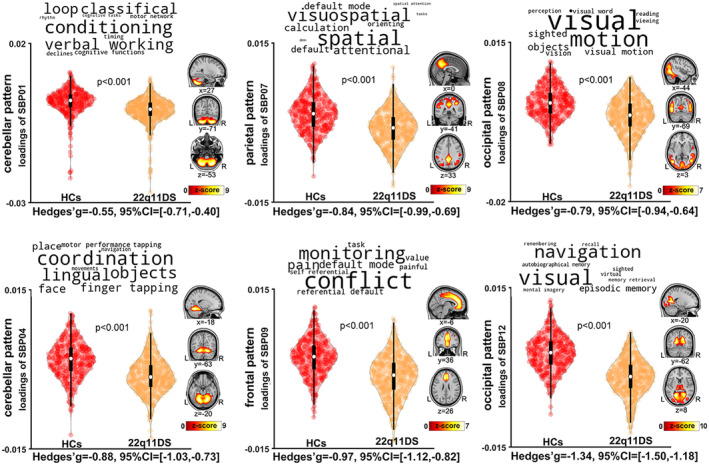
Six gray matter volume structural brain patterns (SBPs) exhibited lower loading in 22q11DS individuals relative to healthy controls (i.e., lower gray matter volume [GMV] in these SBPs). SBPs included two cerebellar patterns (SBP01 and SBP04, left column), one parietal pattern (SBP07) and one frontal pattern (SBP09, middle column), and two occipital patterns (SBP08 and SBP12, right column). Analyses included all participants (n = 783) across the 13 scanners, adjusting for effects of age, sex, total intracranial volume, and site. The top 10 terms associated with each SBP on Neurosynth decoding are shown in word‐cloud plots, and font size represents the relative correlation strength of each term to the SBP. L: left; R: right.
FIGURE 2.
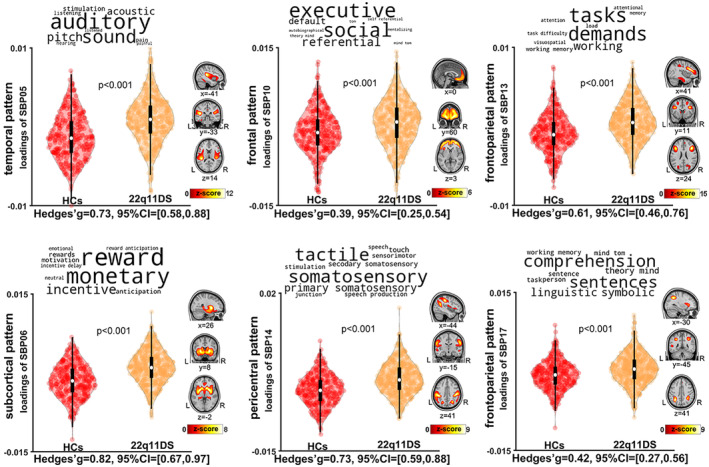
Six gray matter volume structural brain patterns (SBPs) exhibited higher loading in 22q11DS individuals relative to healthy controls (i.e., higher gray matter volume [GMV] in these SBPs). SBPs include one temporal pattern (SBP05) and one subcortical pattern (SBP06, on right column); one frontal pattern (SBP10), and one pericentral pattern (SBP14, on middle column); and two frontoparietal patterns (SBP13 and SBP17). Analyses included all participants (n = 783) across the 13 studies, adjusting for effects of age, sex, total intracranial volume, and research sites. The top 10 terms associated with each SBP on Neurosynth decoding are shown in word‐cloud plots, and font size represents the relative correlation strength of each term to the SBP. L: left; R: right.
3.2. Effect of deletion size on SBPs
There were no age or sex differences between 22q11DS subjects with the A‐B deletion (n = 22) and A‐D deletion (n = 107). However, consistent with previous reports (Zhao et al., 2018), IQ was significantly higher in those with A‐B deletions (n = 20; mean [SD] = 81.85 [9.48]) than those with A‐D deletions (n = 95; mean [SD] = 74.86 [14.58]) (p = .043) (supplementary material Table S3). For the SBPs, we observed three SBPs with higher loading scores in the A‐B deletion group than the A‐D deletion group (Figure 3). These patterns included SBP 9 (frontal pattern; p = .007), SBP 12 (medial occipital pattern; p = .004), and SBP 17 (frontoparietal pattern; p < .001). These results did not change after removing the effect of IQ from the loading scores.
FIGURE 3.
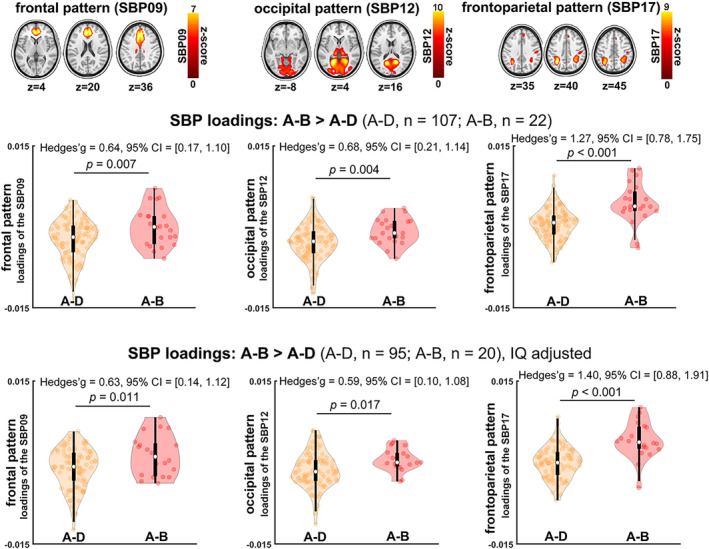
Three structural brain patterns (SBPs) exhibited higher loading in A‐B 22q11.2 deletion compared to A‐D deletion (i.e., higher gray matter volume [GMV] in these SBPs of A‐B). SBPs included one frontal pattern (SBP09), one occipital pattern (SBP12), and one frontoparietal pattern (SBP17). The frontal and the occipital SBPs had higher loading scores in healthy controls relative to A‐D deletion individuals, but had similar loading scores to A‐B deletion. The frontoparietal pattern had lower loading scores in controls than either A‐B and A‐D deletion. These results were similar after adjusting for intelligence quotient (IQ) (bottom panel).
3.3. SBPs abnormalities and psychosis
Demographics of participants included in this analysis (58 22q11DS individuals with psychosis, 59 without psychosis) are reported in Supplementary Table S3. We observed that the psychosis subgroup showed nominally higher loading scores in a medial occipital pattern (SBP 12) relative to the subgroup without psychosis (p = .031 uncorrected; Figure 4). However, these differences did not survive FDR correction.
FIGURE 4.
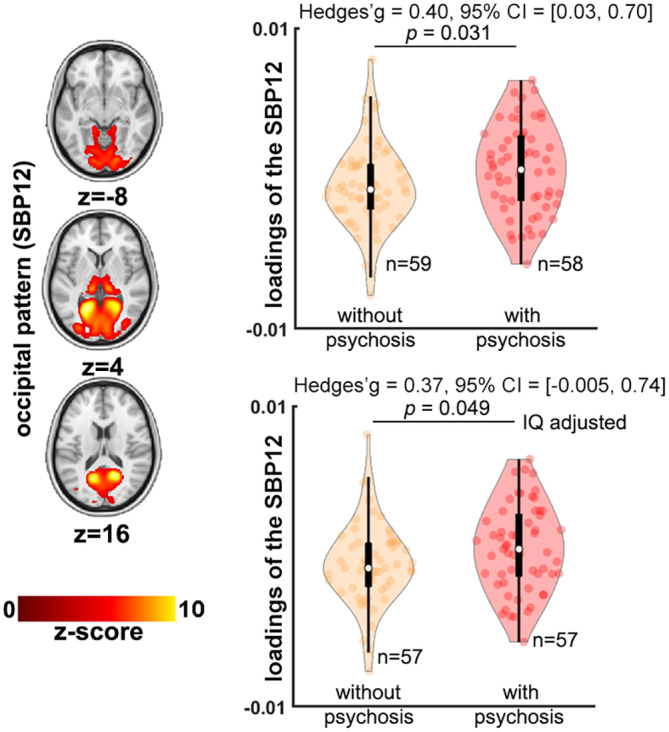
Gray matter volume‐based structural patterns comparison between 22q11DS individuals with and without psychosis. Only one structural brain pattern (SBP), occipital, showed higher loading scores (i.e., higher gray matter volume [GMV] in this SBP) in 22q11DS individuals with psychosis than those without psychosis (p < .05, uncorrected). Bottom panel shows results adjusted for intelligence quotient (IQ). Positive Hedges' g effect size value indicates 22q11DS individuals with psychosis have higher loadings than those without psychosis.
3.4. Univariate analysis of GMV
The VBM results showed a single global pattern of differences in GMV that was convergent with SBM results in regions where the differences were most pronounced (i.e., areas within SBNs with large effect sizes). Figure 5, supplementary Figure S11, and Table S4 show details of this analysis. These results indicated significantly reduced GMV in widespread cerebellar and medial regions and increased GMV in subcortical and lateral prefrontal regions in 22q11DS individuals, and results were not substantively altered by adjusting IQ of the participants. Site effects were minimal as evidence in supplementary Figure S10. Moreover, relative to the A‐D deletion group, the A‐B deletion group showed larger GMV in occipital regions including calcarine cortex, lingual cortex, and cuneus (Figure 5 and supplementary material Figure S11 and Table S5). No significant differences in VBM GMV results were detected between the 22q11DS‐psychosis and 22q11DS‐no‐psychosis groups.
FIGURE 5.
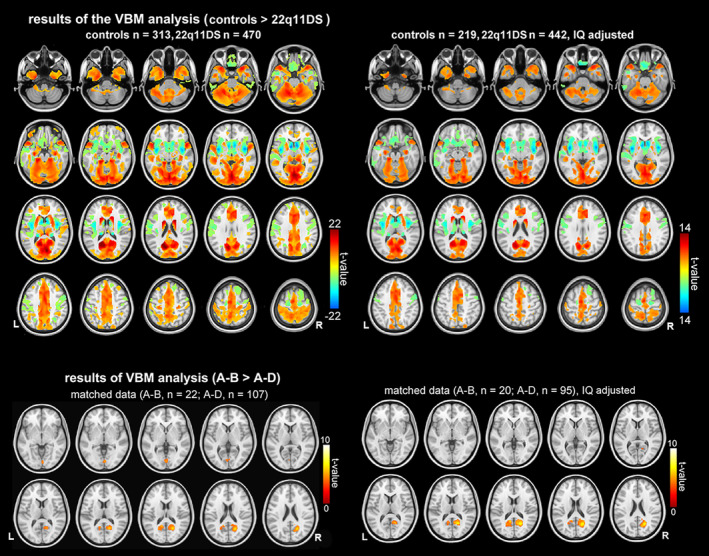
Voxel‐based morphometry (VBM) shows widespread cortical, subcortical, and cerebellar differences in gray matter volume (GMV) between 22q11DS and healthy volunteers (upper panel) and between individuals with A‐B deletion and individuals with A‐D deletion (lower panel). Upper panel: results of the VBM analysis showing regions where there are statistically significant differences between 22q11DS and controls. Warm colors (color spectrum range from red to yellow) indicate regions where gray matter volume is statistically larger in controls. Cool colors (color spectrum range from green to blue) indicate regions where gray matter volume is statistically larger in 22q11DS individuals. Lower panel: results of the VBM analysis where 22q11DS individuals with shorter (A‐B) deletions >22q11DS individuals with longer deletions (A‐D). Results shown in the left column were results with age, sex, total intracranial volume, and site included as covariates in the model, and results shown in the right column were results with age, sex, total intracranial volume, site, and intelligence quotient (IQ) included as covariates in the model. All results shown are corrected at p < .05 with false‐discovery rate (FDR). L, left; R, right.
4. DISCUSSION
This study represents the first investigation into patterns of GMV alterations in 22q11DS utilizing SBM methods optimized for multisite studies (Ge, Ding, et al., 2021). This approach yielded two major findings. First, 22q11DS was associated with widespread differences in GMV SBPs, displaying nonrandom patterns with symmetric distribution across various brain regions. These patterns encompassed sensorimotor areas, regions overlapping with the default mode system (Luo et al., 2012; Raichle, 2015), and cerebellar regions. Second, the SBM findings related to 22q11.2 deletion size; specifically, the smaller nested A‐B deletions compared to the typical larger A‐D deletions revealed distinct patterns involving higher loading in frontal, parietal, and occipital regions, which were robust to adjustment for IQ. These findings provide further evidence for distinct neuroimaging phenotypes in 22q11DS (Cheon et al., 2022; Ching et al., 2020; Sun et al., 2020; Villalón‐Reina et al., 2020), and extend on these findings by demonstrating that structural anomalies in 22q11DS manifest in a nonrandom manner and exhibit distinct anatomical patterns, rather than a diffuse global process.
Our method decomposed GMV covariance into 17 SBP patterns. Most SBPs (12/17; 75%) showed significant differences in 22q11DS relative to controls. Of these, half showed lower loadings, while the other half showed higher loadings in 22q11DS. The effect sizes were larger for SBPs that showed lower loadings in 22q11DS (supplementary Figure S6), and weak to moderate for most of the SBPs that showed higher loadings in 22q11DS (supplementary Figure S7), indicating that overall, lower GMV patterns were more pronounced than higher GMV patterns in 22q11DS. The lower GMV in 22q11DS formed SBP patterns comprising most of the cerebellum, frontoparietal, and thalamo‐occipital regions. These substantially overlap with cortical areas shown to have decreased surface area in 22q11DS (Sun et al., 2020). Unlike our previous ENIGMA 22q11DS cortical mapping study, the current investigation expands its scope to include analysis of the cerebellum. This analysis reveals significant deficits in GMV structural patterns within the cerebellum (Schmitt et al., 2022), indicating that the developmental processes governing cerebellar development in individuals with 22q11DS may share similarities with the molecular and cellular mechanisms involved in the formation of radial units in the cerebral cortex (Rakic, 1988; Rakic, 1995; Ten Donkelaar et al., 2003). Notably, our study revealed that the covariation of GMV exhibited distinct directions of effect between individuals with 22q11DS and controls across various SBPs. This finding aligns with recent research, particularly from the ENIGMA 22q11.2 Working Group, which has employed diverse methodologies to investigate cortical and subcortical structure (Ching et al., 2020; Sun et al., 2020), as well as white matter microstructure in 22q11DS (Seitz‐Holland et al., 2021; Villalón‐Reina et al., 2020). Specifically, Villalón‐Reina et al. found that fractional anisotropy (FA) of the white matter exhibited two distinct directions of effect in 22q11DS individuals relative to demographically matched control subjects: elevated FA in 22q11DS individuals relative to controls was observed in callosal and projection fibers (i.e., internal capsule and corona radiata), while decreased FA in 22q11DS individuals was observed in certain association fibers (Villalón‐Reina et al., 2020). Meanwhile, Ching et al. made a noteworthy discovery regarding subcortical structures, highlighting two distinct directions of effect between 22q11DS individuals and controls. Those with 22q11DS exhibited reduced volumes in the thalamus, putamen, hippocampus, and amygdala, alongside increased volumes of the lateral ventricles, caudate and accumbens, relative to controls (Ching et al., 2020). Furthermore, Sun et al. identified variations in CT and surface area, encompassing both thickening/expansion and thinning/reduction, in various brain regions among individuals with 22q11DS relative to controls (Sun et al., 2020).
In the present study, the outcomes derived from SBM and univariate VBM analyses exhibited substantial convergence. Specifically, regions within the SBPs that manifested group differences displayed a substantial overlap with regions showcasing group differences in the VBM analysis. Nonetheless, a noteworthy advancement offered by SBM over VBM was its ability to decompose the GMV data into discrete SBPs. This decomposition showcased that the structural anomalies associated with 22q11DS present themselves not in a random diffuse fashion, but rather as distinct, covarying anatomical patterns. Furthermore, SBM exhibited enhanced sensitivity compared to VBM in detecting group differences (Gupta et al., 2019; Xu et al., 2009). A notable illustration of this was the fact that while the univariate VBM did unveil differences in GMV within occipital regions between the A‐D deletion group and the A‐B deletion group, SBM further elucidated this contrast through the identification of three distinct SBPs that differentiate the two groups, with one of these SBPs was found to be specific to the occipital regions implicated in the VBM analysis.
We submitted the SBP maps that showed differences between 22q11DS and controls to Neurosynth to explore topographical overlap between the GMV SBPs and the putative functions attributed to those brain regions investigated using functional MRI. This qualitative hypothesis‐generating exercise showed that the attributed functions of these SBPs could be mapped onto cognitive domains such as visuo‐spatial deficits (SBP 7, 8, and 12); language (SBP 17); or verbal memory (SBP 1) that are impaired in 22q11DS (Bearden et al., 2001; Fiksinski et al., 2019; Glaser et al., 2002; Simon et al., 2005; Van Den Heuvel et al., 2018; Woodin et al., 2001). However, we did not find significant associations between IQ and these SBPs. It is plausible to consider that these SBPs identified in our study may be associated with particular cognitive deficits rather than a global cognitive deficit captured solely by IQ. Future studies are warranted to test the degree to which SBPs in 22q11DS are associated with specific cognitive deficits.
Our data showed that, when compared to the smaller nested A‐B deletion, the typical larger A‐D 22q11.2 deletion size was associated with lower loadings in SBPs encompassing mid‐frontal (SBP 9, Neurosynth “conflict monitoring”), occipito‐thalamic (SBP 12, Neurosynth “visuo‐spatial and verbal memory”) and frontoparietal (SBP 17, Neurosynth “language”) regions. Our findings that individuals with large A‐D deletions had lower GMV compared to individuals with A‐B deletions aligns with previous findings of greater surface area reductions in those with large A‐D deletions compared to those with A‐B deletions (Sun et al., 2020), consistent with studies showing similar genetic influences on surface area and GMV (Winkler et al., 2010). The convergence of reduced surface area and GMV SBPs points to the 22q11.2 deletion having an impact on early cortical development, as surface area is tightly linked to early cortical development and increased neuronal proliferation (Rakic, 1988). In addition, as the larger deletion size only impacts three SBPs rather than all 22q11DS relevant structural patterns, this finding could be a starting point for future research as it sheds light on SBPs that potentially reflect genetic vulnerability in 22q11DS. While three SBPs displayed higher loading scores in the A‐B deletion group compared to the A‐D deletion group, two of these (SBP 9 and 12) had lower loading scores in 22q11DS individuals than in healthy controls, whereas SBP 17 exhibited higher loading scores in 22q11DS individuals than controls. The variation in SBP 17 implies that the GMV within this parietal pattern/network may be influenced by factors not accounted for in the present study, such as modifying genes outside the locus. This divergence may also indicate that the deletion size does not always impact the brain in a consistent manner. Additional validation is necessary using an independent and larger sample of patients with atypical deletions.
Given the nominally significant difference between the 22q11DS‐psychosis and 22q11DS‐no‐psychosis groups and the smaller sample size for this comparison, this result should be interpreted with caution. It is of interest that our prior study using the same dataset uncovered significant differences in CT but not in surface area between these two groups (Sun et al., 2020), and a recent independent study revealed significant differences in structural covariance of CT between 22q11DS individuals with and without psychotic symptoms (Sandini et al., 2018). Considering that SBM serves as a multivariate alternative to univariate VBM, it offers enhanced sensitivity in detecting disease‐related structural anomalies (Xu et al., 2009), a future study using SBM to assess potential surface area correlates of psychosis in 22q11DS that cannot be captured with univariate analysis may be informative because surface area is tightly linked to cortical development in 22q11DS and in schizophrenia spectrum disorders (Cheon et al., 2022; Sun et al., 2020).
Limitations should be noted. First, while the methods applied here allow us to segment voxels into broad tissue categories (i.e., gray matter, white matter, CSF) based on intensities for each of the voxels in a scan, these intensities cannot inform us directly about what precise underlying biological processes may be changing intensity values. Second, our sample size is limited for the subgroup analyses of deletion size and history of psychosis, especially for the A‐B deletion type. Moreover, we cannot rule out that some of those classified as “no psychosis” may develop a psychotic illness in later life. Future studies that employ a more detailed and granular characterization of cognitive deficits in individuals with 22q11DS hold promise for exploring and addressing questions related to the underlying brain abnormalities in specific cognitive domains. The use of the “Neurosynth” decoder is an exploratory technique rather than an inference of putative function of the SBPs, and therefore the suggested functional labels of the SBPs should be interpreted as hypotheses. Third, while the present study utilized ICA to reveal spatially sparse structural patterns (Xu et al., 2009), alternative methods that could provide supplementary perspectives on the data can also be employed to quantify the SBPs and their associated loading scores. Among these methods is non‐negative matrix factorization (Dai et al., 2023; Neufeld et al., 2020; Sotiras et al., 2015; Sotiras et al., 2017) which has been demonstrated to be able to separate spatially more localized and independent components, in an analogous manner to ICA (Thompson et al., 2020). Forth, the influence of site or scanner effects can be efficiently mitigated by carefully designing new SBM protocols (Chen et al., 2014; Li et al., 2020), often necessitating the incorporation of a substantial number of components. In the present study, the relatively small to medium sample sizes at certain sites limited our ability to decompose the data into a large number of components. As an alternative, we employed SS‐Detect, which has shown its capacity to effectively model scanner‐specific SBPs and yield more precise estimates of subject‐specific loading parameters compared to conventional SBM analysis (Ge, Ding, et al., 2021), and demonstrated that the site‐specific PCA procedure successfully mitigated, if not entirely eliminated, the site effect. Even though we further accounted for inter‐scanner effects in our statistical analysis, it's possible that certain outcomes could still have been influenced by variations in MRI machines and protocols. This potential influence could be mitigated in future research by carefully balancing the inclusion of different scanning sites. This approach not only has the potential to eliminate this concern but also offers the added benefit of increased statistical power to reinforce methods aimed at correcting for any site‐related effects (Chen et al., 2022; Fortin et al., 2018; Pomponio et al., 2020; Sun et al., 2022). Finally, one limitation of utilizing SBM is its reliance on the prerequisite assumption that the compared groups have similar spatial maps.
5. CONCLUSION
In conclusion, our multivariate SBM analysis using novel methods for multisite data revealed new insights into the differential patterns of structural covariance associated with 22q11DS, involving SBPs that are distributed into distinct and nonrandom structural patterns which were not driven by differences in IQ between 22q11DS and TD individuals. This is so far the largest study implicating cerebellar deficits of 22q11DS, motivating future studies in this often‐ignored brain structure in the context of 22q11DS. We further demonstrated that while deletion size was related to structural covariance patterns, history of psychosis did not show a strong relationship with these patterns. Future translational studies linking these cortical patterns to in vitro and animal models of the 22q11.2 deletion are warranted.
Supporting information
FIGURE S1. t‐SNE visualization of GMV feature distribution.
FIGURE S2. Flowchart of data processing and analyses pipeline.
FIGURE S3. Neuroimaging quality control assessment.
FIGURE S4. Source‐based morphometry derives seventeen gray matter volume structural brain patterns.
FIGURE S5. Unthresholded gray matter volume structural brain patterns.
FIGURE S6. Effect size of meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with decreased gray matter in 22q11DS.
FIGURE S7. Effect size of meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with increased gray matter in 22q11DS.
FIGURE S8. Effect size with IQ adjusted meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with decreased gray matter in 22q11DS.
FIGURE S9. Effect size with IQ adjusted meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with increased gray matter in 22q11DS.
FIGURE S10. Site effects on VBM analysis.
FIGURE S11. Voxel‐based morphometry results of differences in gray matter volume between 22q11DS and healthy volunteers and between individuals with A‐B deletion and individuals with A‐D deletion.
TABLE S1. Regions included in each structural brain pattern.
TABLE S2. Effect sizes of the comparisons between 22q11DS individuals and healthy controls.
TABLE S3. Site Demographics of 22q11DS individuals with small (A‐B) vs. large (A‐D) deletions, and 22q11DS individuals with and without psychotic disorder diagnoses.
TABLE S4. Voxel‐based morphometry results: Differences between 22q11DS individuals and controls.
TABLE S5. Voxel‐based morphometry results: Differences between 22q11DS individuals with shorter deletion (A‐B) and longer deletion (A‐D).
ACKNOWLEDGMENTS
CC was reported support by U54EB020403 from the Big Data to Knowledge (BD2K) Program; R01MH116147‐04; R01AG058854‐02; R01MH129742‐01. GB was supported by European Commision grant: QLGU‐CT‐2001‐01081; NWO‐Veni grant 2006 ‐916.76.048; Dutch Brain Foundation: 15F07(2).55. ASB was supported by the Dalglish Family Chair in 22q11.2 Deletion Syndrome, Canadian Institutes of Health Research (CIHR) (MOP‐313331 and MOP‐111238), and NIMH grant number U01MH119741‐01. TvA was supported by the NIH via grants NIMH. ED reports support from Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 115300 (for EU‐AIMS) and No. 777394 (for AIMS‐2‐TRIALS). JLD was supported by a Wellcome Trust Clinical Research Training Fellowship (102003/Z/13/Z). MJ was supported by NIH grants K01MH112774 and R01MH129636 and a Next Generation Award from the Tommy Fuss Center for Neuropsychiatric Research. WRK was supported by NIH (R01 MH064824). DL reports support by Wellcome Trust Strategic Award “DEFINE”. DMM receives research support from NIH via grant 5U01MH119737‐04. DGM reports support by Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 115300 (for EU‐AIMS) and No. 777394 (for AIMS‐2‐TRIALS). MJO was supported by the NIH via grants NIMH U01MH119738 and U01MH101724 and the MRC via grant MR/T033045/1, MR/N022572/1, and MR/L011166/1. JVR was supported by the NIH via grants R01MH129742 and R01MH116147. GMR was supported by Fondecyt grants 1171014 and 1211411, and ACT 192064 (ANID‐Chile). KA was supported by NIH R01MH107018 and U54HD079125. MvdB was supported by the NIH via grants NIMH U01MH119758 and U01MH101724 and the MRC via grant MR/T033045/1, MR/N022572/1 and MR/L011166/1. JV was supported by The SickKids Psychiatry Associates Chair in Developmental Psychopathology and NIMH U01MH119741‐01. PMT was supported by the NIH via grants R01MH129742 and R01MH116147. CEB was supported by U54EB020403 from the Big Data to Knowledge (BD2K) Program, NIMH R01MH085953, NIMH U01MH101719. FVR receives research support from CIHR, Brain Canada, Michael Smith Foundation for Health Research, Vancouver Coastal Health Research Institute, and Weston Brain Institute for investigator‐initiated research. Philanthropic support from Seedlings Foundation. In‐kind equipment supports for this investigator‐initiated trial from MagVenture. He has received honoraria for participation in an advisory board for Allergan.
Ge, R. , Ching, C. R. K. , Bassett, A. S. , Kushan, L. , Antshel, K. M. , van Amelsvoort, T. , Bakker, G. , Butcher, N. J. , Campbell, L. E. , Chow, E. W. C. , Craig, M. , Crossley, N. A. , Cunningham, A. , Daly, E. , Doherty, J. L. , Durdle, C. A. , Emanuel, B. S. , Fiksinski, A. , Forsyth, J. K. , … Bearden, C. E. (2024). Source‐based morphometry reveals structural brain pattern abnormalities in 22q11.2 deletion syndrome. Human Brain Mapping, 45(1), e26553. 10.1002/hbm.26553
Fidel Vila‐Rodriguez and Carrie E. Bearden are joint senior‐authorship.
Contributor Information
Fidel Vila‐Rodriguez, Email: fidelvil@mail.ubc.ca.
Carrie E. Bearden, Email: cbearden@mednet.ucla.edu.
DATA AVAILABILITY STATEMENT
The data used for the analysis are made available upon request to the ENIGMA 22q11.2 Working Group.
REFERENCES
- Alexander‐Bloch, A. , Giedd, J. N. , & Bullmore, E. (2013). Imaging structural co‐variance between human brain regions. Nature Reviews Neuroscience, 14(5), 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, K. , Chaddock, C. A. , Baldeweg, T. , & Skuse, D. (2011). Neuroanatomy in adolescents and young adults with 22q11 deletion syndrome: Comparison to an IQ‐matched group. NeuroImage, 55(2), 491–499. [DOI] [PubMed] [Google Scholar]
- Bearden, C. E. ,Woodin, M. F., Wang, P., Moss, E., McGinn D.M., Zackai, E., Emannuel, B., & Tyrone, D. C. (2001). The neurocognitive phenotype of the 22q11.2 deletion syndrome: Selective deficit in visual‐spatial memory. Journal of Clinical Experimental Neuropsychology, 23(4), 447–464. [DOI] [PubMed] [Google Scholar]
- Beckmann, C. F. , Mackay, C. E. , Filippini, N. , & Smith, S. M. (2009). Group comparison of resting‐state FMRI data using multi‐subject ICA and dual regression: Organization for Human Brain Mapping 2009 Annual Meeting. NeuroImage, 47(suppl 1), S148. [Google Scholar]
- Benjamini, Y. , Drai, D. , Elmer, G. , Kafkafi, N. , & Golani, I. (2001). Controlling the false discovery rate in behavior genetics research. Behavioural Brain Research, 125(1–2), 279–284. [DOI] [PubMed] [Google Scholar]
- Blagojevic, C. , Heung, T. , Theriault, M. , Tomita‐Mitchell, A. , Chakraborty, P. , Kernohan, K. , Bulman, D. E. , & Bassett, A. S. (2021). Estimate of the contemporary live‐birth prevalence of recurrent 22q11.2 deletions: A cross‐sectional analysis from population‐based newborn screening. CMAJ Open, 9(3), E802–E809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, L. E. , Daly, E. , Toal, F. , Stevens, A. , Azuma, R. , Catani, M. , Ng, V. , van Amelsvoort, T. , Chitnis, X. , Cutter, W. , Murphy, D. G. M. , & Murphy, K. C. (2006). Brain and behaviour in children with 22q11.2 deletion syndrome: A volumetric and voxel‐based morphometry MRI study. Brain, 129(5), 1218–1228. [DOI] [PubMed] [Google Scholar]
- Chen, A. A. , Beer, J. C. , Tustison, N. J. , Cook, P. A. , Shinohara, R. T. , Shou, H. , & The Alzheimer's Disease Neuroimaging Initiative . (2022). Mitigating site effects in covariance for machine learning in neuroimaging data. Human Brain Mapping, 43(4), 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Liu, J. , Calhoun, V. D. , Arias‐Vasquez, A. , Zwiers, M. P. , Gupta, C. N. , Franke, B. , & Turner, J. A. (2014). Exploration of scanning effects in multi‐site structural MRI studies. Journal of Neuroscience Methods, 230, 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon, E. J. , Bearden, C. E. , Sun, D. , Ching, C. R. K. , Andreassen, O. A. , Schmaal, L. , Veltman, D. J. , Thomopoulos, S. I. , Kochunov, P. , Jahanshad, N. , Thompson, P. M. , Turner, J. A. , & van Erp, T. G. M. (2022). Cross disorder comparisons of brain structure in schizophrenia, bipolar disorder, major depressive disorder, and 22q11.2 deletion syndrome: A review of ENIGMA findings. Psychiatry and Clinical Neurosciences, 76(5), 140–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching, C. R. , Gutman, B. A., Sun, D., Villalon‐Reina, J., Ragothaman, A., Isaev, D., Zavaliangos‐Petropulu, A., Lin, A., Jonas, R. K., Kushan, L., Pacheco‐Hansen, L., Vajdi, A., Forsyth, J. K., Jalbrzikowski, M., Bakker, G., van Amelsvoort, T., Antshel, K. M., Fremont, W., Kates, W. R., … Bearden, C. E.. (2020). Mapping subcortical brain alterations in 22q11.2 deletion syndrome: Effects of deletion size and convergence with idiopathic neuropsychiatric illness. American Journal of Psychiatry, 177(7), 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, E. W. , Ho, A., Wei, C., Voormolen, E. H. J., Crawley, A. P., & Bassett, A. S. (2011). Association of schizophrenia in 22q11.2 deletion syndrome and gray matter volumetric deficits in the superior temporal gyrus. American Journal of Psychiatry, 168(5), 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colloby, S. J. , Nathan, P. J. , Bakker, G. , Lawson, R. A. , Yarnall, A. J. , Burn, D. J. , O'Brien, J. T. , & Taylor, J. P. (2021). Spatial covariance of cholinergic muscarinic M1/M4 receptors in Parkinson's disease. Movement Disorders, 36(8), 1879–1888. [DOI] [PubMed] [Google Scholar]
- Dai, R. , Herold, C. J. , Wang, X. , Kong, L. , & Schröder, J. (2023). Structural brain networks in schizophrenia based on nonnegative matrix factorization. Psychiatry Research: Neuroimaging, 334, 111690. [DOI] [PubMed] [Google Scholar]
- Fiksinski, A. M. , Breetvelt, E. J. , Lee, Y. J. , Boot, E. , Butcher, N. , Palmer, L. , Chow, E. W. C. , Kahn, R. S. , Vorstman, J. A. S. , & Bassett, A. S. (2019). Neurocognition and adaptive functioning in a genetic high‐risk model of schizophrenia. Psychological Medicine, 49(6), 1047–1054. [DOI] [PubMed] [Google Scholar]
- Fortin, J.‐P. , Cullen, N. , Sheline, Y. I. , Taylor, W. D. , Aselcioglu, I. , Cook, P. A. , Adams, P. , Cooper, C. , Fava, M. , McGrath, P. J. , McInnis, M. , Phillips, M. L. , Trivedi, M. H. , Weissman, M. M. , & Shinohara, R. T. (2018). Harmonization of cortical thickness measurements across scanners and sites. NeuroImage, 167, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, R. , Ding, S. , Keeling, T. , Honer, W. G. , Frangou, S. , & Vila‐Rodriguez, F. (2021). SS‐detect: Development and validation of a new strategy for source‐based morphometry in multiscanner studies. Journal of Neuroimaging, 31(2), 261–271. [DOI] [PubMed] [Google Scholar]
- Ge, R. , Downar, J. , Blumberger, D. M. , Daskalakis, Z. J. , Lam, R. W. , & Vila‐Rodriguez, F. (2019). Structural network integrity of the central executive network is associated with the therapeutic effect of rTMS in treatment resistant depression. Progress in Neuro‐Psychopharmacology and Biological Psychiatry, 92, 217–225. [DOI] [PubMed] [Google Scholar]
- Ge, R. , Hassel, S. , Arnott, S. R. , Davis, A. D. , Harris, J. K. , Zamyadi, M. , Milev, R. , Frey, B. N. , Strother, S. C. , Müller, D. J. , Rotzinger, S. , MacQueen, G. M. , Kennedy, S. H. , Lam, R. W. , & Vila‐Rodriguez, F. (2021). Structural covariance pattern abnormalities of insula in major depressive disorder: A CAN‐BIND study report. Progress in Neuro‐Psychopharmacology and Biological Psychiatry, 111, 110194. [DOI] [PubMed] [Google Scholar]
- Ge, R. , Liu, X. , Long, D. , Frangou, S. , & Vila‐Rodriguez, F. (2021). Sex effects on cortical morphological networks in healthy young adults. NeuroImage, 233, 117945. [DOI] [PubMed] [Google Scholar]
- Glaser, B. , Mumme, D. L. , Blasey, C. , Morris, M. A. , Dahoun, S. P. , Antonarakis, S. E. , Reiss, A. L. , & Eliez, S. (2002). Language skills in children with velocardiofacial syndrome (deletion 22q11.2). Journal of Pediatrics, 140(6), 753–758. [DOI] [PubMed] [Google Scholar]
- Gothelf, D. , Hoeft, F. , Ueno, T. , Sugiura, L. , Lee, A. D. , Thompson, P. , & Reiss, A. L. (2011). Developmental changes in multivariate neuroanatomical patterns that predict risk for psychosis in 22q11.2 deletion syndrome. Journal of Psychiatric Research, 45(3), 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, C. N. , Calhoun, V. D. , Rachakonda, S. , Chen, J. , Patel, V. , Liu, J. , Segall, J. , Franke, B. , Zwiers, M. P. , Arias‐Vasquez, A. , Buitelaar, J. , Fisher, S. E. , Fernandez, G. , van Erp, T. G. M. , Potkin, S. , Ford, J. , Mathalon, D. , McEwen, S. , Lee, H. J. , … Turner, J. A. (2015). Patterns of gray matter abnormalities in schizophrenia based on an international mega‐analysis. Schizophrenia Bulletin, 41(5), 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, C. N. , Turner, J. A. , & Calhoun, V. D. (2019). Source‐based morphometry: A decade of covarying structural brain patterns. Brain Structure and Function, 224(9), 3031–3044. [DOI] [PubMed] [Google Scholar]
- Gur, R. E. , Bassett, A. S. , McDonald‐McGinn, D. M. , Bearden, C. E. , Chow, E. , Emanuel, B. S. , Owen, M. , Swillen, A. , van den Bree, M. , Vermeesch, J. , Vorstman, J. A. S. , Warren, S. , Lehner, T. , Morrow, B. , & The International 22q11.2 Deletion Syndrome Brain Behavior Consortium . (2017). A neurogenetic model for the study of schizophrenia spectrum disorders: The international 22q11.2 deletion syndrome brain behavior consortium. Molecular Psychiatry, 22(12), 1664–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafkemeijer, A. , Altmann‐Schneider, I. , de Craen, A. J. M. , Slagboom, P. E. , van der Grond, J. , & Rombouts, S. A. R. B. (2014). Associations between age and gray matter volume in anatomical brain networks in middle‐aged to older adults. Aging Cell, 13(6), 1068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, S. E. , Chadehumbe, M. , Blaine Crowley, T. , Zackai, E. H. , Bilaniuk, L. T. , & McDonald‐McGinn, D. M. (2018). Neurologic challenges in 22q11.2 deletion syndrome. American Journal of Medical Genetics—Part A, 176(10), 2140–2145. [DOI] [PubMed] [Google Scholar]
- Li, H. , Smith, S. M. , Gruber, S. , Lukas, S. E. , Silveri, M. M. , Hill, K. P. , Killgore, W. D. S. , & Nickerson, L. D. (2020). Denoising scanner effects from multimodal MRI data using linked independent component analysis. NeuroImage, 208, 116388. [DOI] [PubMed] [Google Scholar]
- Li, Y. O. , Adalı, T. , & Calhoun, V. D. (2007). Estimating the number of independent components for functional magnetic resonance imaging data. Human Brain Mapping, 28(11), 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, A. , Ching, C. R. K. , Vajdi, A. , Sun, D. , Jonas, R. K. , Jalbrzikowski, M. , Kushan‐Wells, L. , Pacheco Hansen, L. , Krikorian, E. , Gutman, B. , Dokoru, D. , Helleman, G. , Thompson, P. M. , & Bearden, C. E. (2017). Mapping 22q11.2 gene dosage effects on brain morphometry. Journal of Neuroscience, 37(26), 6183–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton, S. R. , Popa, A. M. , Luck, S. J. , Bolden, K. , Carter, C. S. , Niendam, T. A. , & Simon, T. J. (2020). Neural and behavioral measures suggest that cognitive and affective functioning interactions mediate risk for psychosis‐proneness symptoms in youth with chromosome 22q11.2 deletion syndrome. American Journal of Medical Genetics—Part A, 182(7), 1615–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. , Xu, L. , Jung, R. , Pearlson, G. , Adali, T. , & Calhoun, V. D. (2012). Constrained source‐based morphometry identifies structural networks associated with default mode network. Brain Connectivity, 2(1), 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, N. , Sui, J. , Abrol, A. , Lin, D. , Chen, J. , Vergara, V. M. , Fu, Z. , du, Y. , Damaraju, E. , Xu, Y. , Turner, J. A. , & Calhoun, V. D. (2020). Age‐related structural and functional variations in 5,967 individuals across the adult lifespan. Human Brain Mapping, 41(7), 1725–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald‐McGinn, D. M. , Sullivan, K. E., Marino, B., Philip, N., Swillen, A., Vorstman, J. A. S., Zackai, E. H., Emanuel, B. S., Vermeesch, J. R., Morrow, B. E., Scambler, P. J., & Bassett, A. S. (2015). 22q11.2 deletion syndrome. Nature Reviews Disease Primers, 1(1), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechelli, A. , Friston, K. J. , Frackowiak, R. S. , & Price, C. J. (2005). Structural covariance in the human cortex. Journal of Neuroscience, 25(36), 8303–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, T. , Llera, A., Floris, D., Forde N., Tillmann, J., Durston, S., Moessnang, C., Banaschewski, T., Holt, R., Baron‐Cohen, S., Rausch, A., Loth, E., Dell'Acqua, F., Charman, T., Murphy, D., Ecker, C., Beckmann, C. F., & Buiterlaar, J. F. (2020). Gray matter covariations and core symptoms of autism: The EU‐AIMS Longitudinal European Autism Project. Molecular Autism, 11, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg, P. J. , Richman, M. J. , Roalf, D. R. , Morse, C. L. , Graefe, A. C. , Brennan, L. , Vickers, K. , Tsering, W. , Kamath, V. , Turetsky, B. I. , Gur, R. C. , & Gur, R. E. (2018). Neurocognitive functioning in patients with 22q11.2 deletion syndrome: A meta‐analytic review. Behavior Genetics, 48(4), 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, N. H. , Kaczkurkin, A. N. , Sotiras, A. , Mulsant, B. H. , Dickie, E. W. , Flint, A. J. , Meyers, B. S. , Alexopoulos, G. S. , Rothschild, A. J. , Whyte, E. M. , Mah, L. , Nierenberg, J. , Hoptman, M. J. , Davatzikos, C. , Satterthwaite, T. D. , & Voineskos, A. N. (2020). Structural brain networks in remitted psychotic depression. Neuropsychopharmacology, 45(7), 1223–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H. , Quide, Y., Schofield, P. R., Williams, L. M., & Gatt, J. M. (2022). Grey matter covariation and the role of emotion reappraisal in mental wellbeing and resilience after early life stress exposure. Translational Psychiatry, 12, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piervincenzi, C. , Fanella, M. , Petsas, N. , Frascarelli, M. , Morano, A. , Accinni, T. , di Fabio, F. , di Bonaventura, C. , Berardelli, A. , & Pantano, P. (2022). Structural cerebellar abnormalities and parkinsonism in patients with 22q11.2 deletion syndrome. Brain Sciences, 12(11), 1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponio, R. , Erus, G. , Habes, M. , Doshi, J. , Srinivasan, D. , Mamourian, E. , Bashyam, V. , Nasrallah, I. M. , Satterthwaite, T. D. , Fan, Y. , Launer, L. J. , Masters, C. L. , Maruff, P. , Zhuo, C. , Völzke, H. , Johnson, S. C. , Fripp, J. , Koutsouleris, N. , Wolf, D. H. , … Davatzikos, C. (2020). Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage, 208, 116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontillo, M. , Menghini, D. , & Vicari, S. (2019). Neurocognitive profile and onset of psychosis symptoms in children, adolescents and young adults with 22q11 deletion syndrome: A longitudinal study. Schizophrenia Research, 208, 76–81. [DOI] [PubMed] [Google Scholar]
- Raichle, M. E. (2015). The brain's default mode network. Annual Review of Neuroscience, 38(25), 433–447. [DOI] [PubMed] [Google Scholar]
- Rakic, P. (1988). Specification of cerebral cortical areas. Science, 241(4862), 170–176. [DOI] [PubMed] [Google Scholar]
- Rakic, P. (1995). Radial versus tangential migration of neuronal clones in the developing cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America, 92(25), 11323–11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandini, C. , Scariati, E. , Padula, M. C. , Schneider, M. , Schaer, M. , van de Ville, D. , & Eliez, S. (2018). Cortical dysconnectivity measured by structural covariance is associated with the presence of psychotic symptoms in 22q11.2 deletion syndrome. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 3(5), 433–442. [DOI] [PubMed] [Google Scholar]
- Schmitt, J. E. , Vandekar, S. , Yi, J. , Calkins, M. E. , Ruparel, K. , Roalf, D. R. , Whinna, D. , Souders, M. C. , Satterthwaite, T. D. , Prabhakaran, K. , McDonald‐McGinn, D. M. , Zackai, E. H. , Gur, R. C. , Emanuel, B. S. , & Gur, R. E. (2015). Aberrant cortical morphometry in the 22q11.2 deletion syndrome. Biological Psychiatry, 78(2), 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, J. E. , DeBevits, J., Roalf, D., Ruparel, K., Gallagher, R. S., Gur, R. C., Alexander‐Bloch, A., Emo, T., Alam, S., Steinberg, J., Akers, W., Khairy, K., Crowley, T. B., Emanuel, B., Zakharenko, S. S., McDonadl‐McGinn, D. M., & Gur, R. E. (2022). A comprehensive analysis of cerebellar volumes in the 22q11.2 deletion syndrome. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 8(1), 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz‐Holland, J. , Lyons, M., Kushan, L., Lin, A., Villalon‐Reina, J. E., Cho, K., Zhang, F., Billah, T., Bouix, S., Kubicki, M., Bearden, C. E., & Pasternak, O. (2021). Opposing white matter microstructure abnormalities in 22q11.2 deletion and duplication carriers. Translational Psychiatry, 11, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashi, V. , Kwapil, T. R. , Kaczorowski, J. , Berry, M. N. , Santos, C. S. , Howard, T. D. , Goradia, D. , Prasad, K. , Vaibhav, D. , Rajarethinam, R. , Spence, E. , & Keshavan, M. S. (2010). Evidence of gray matter reduction and dysfunction in chromosome 22q11.2 deletion syndrome. Psychiatry Research: Neuroimaging, 181(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, T. J. , Ding, L. , Bish, J. P. , McDonald‐McGinn, D. M. , Zackai, E. H. , & Gee, J. (2005). Volumetric, connective, and morphologic changes in the brains of children with chromosome 22q11.2 deletion syndrome: An integrative study. NeuroImage, 25(1), 169–180. [DOI] [PubMed] [Google Scholar]
- Simon, T. J. , Bearden, C. E., McDonald‐McGinn, D., & Zackai, E. (2005). Visuospatial and numerical cognitive deficits in children with chromosome 22q11.2 deletion syndrome. Cortex, 41(2), 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiras, A. , Resnick, S. M. , & Davatzikos, C. (2015). Finding imaging patterns of structural covariance via non‐negative matrix factorization. NeuroImage, 108, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiras, A. , Toledo, J. B. , Gur, R. E. , Gur, R. C. , Satterthwaite, T. D. , & Davatzikos, C. (2017). Patterns of coordinated cortical remodeling during adolescence and their associations with functional specialization and evolutionary expansion. Proceedings of the National Academy of Sciences of the United States of America, 114(13), 3527–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwijk, M. D. , Geurts, J. J. G. , Daams, M. , Tijms, B. M. , Wink, A. M. , Balk, L. J. , Tewarie, P. K. , Uitdehaag, B. M. J. , Barkhof, F. , Vrenken, H. , & Pouwels, P. J. W. (2015). Cortical atrophy patterns in multiple sclerosis are non‐random and clinically relevant. Brain, 139(1), 115–126. [DOI] [PubMed] [Google Scholar]
- Sun, D. , Ching, C. R. K. , Lin, A. , Forsyth, J. K. , Kushan, L. , Vajdi, A. , Jalbrzikowski, M. , Hansen, L. , Villalon‐Reina, J. E. , Qu, X. , Jonas, R. K. , van Amelsvoort, T. , Bakker, G. , Kates, W. R. , Antshel, K. M. , Fremont, W. , Campbell, L. E. , McCabe, K. L. , Daly, E. , … Bearden, C. E. (2020). Large‐scale mapping of cortical alterations in 22q11.2 deletion syndrome: Convergence with idiopathic psychosis and effects of deletion size. Molecular Psychiatry, 25, 1822–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D. , Rakesh, G. , Haswell, C. C. , Logue, M. , Baird, C. L. , O'Leary, E. N. , Cotton, A. S. , Xie, H. , Tamburrino, M. , Chen, T. , Dennis, E. L. , Jahanshad, N. , Salminen, L. E. , Thomopoulos, S. I. , Rashid, F. , Ching, C. R. K. , Koch, S. B. J. , Frijling, J. L. , Nawijn, L. , … Morey, R. A. (2022). A comparison of methods to harmonize cortical thickness measurements across scanners and sites. NeuroImage, 261, 119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, G. M. , Arnone, D. , McIntosh, A. M. , & Ebmeier, K. P. (2009). Meta‐analysis of magnetic resonance imaging studies in chromosome 22q11.2 deletion syndrome (velocardiofacial syndrome). Schizophrenia Research, 115(2–3), 173–181. [DOI] [PubMed] [Google Scholar]
- Ten Donkelaar, H. J. , den Dunnen, W., Lammens, M., Wesseling, P., Willemsen, M., & Hori, A. (2003). Development and developmental disorders of the human cerebellum. Journal of Neurology, 250(9), 1025–1036. [DOI] [PubMed] [Google Scholar]
- Thompson, E. , Mohammadi‐Nejad, A. R. , Robinson, E. C. , Andersson, J. L. R. , Jbabdi, S. , Glasser, M. F. , Bastiani, M. , & Sotiropoulos, S. N. (2020). Non‐negative data‐driven mapping of structural connections with application to the neonatal brain. NeuroImage, 222, 117273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Jahansha, N., Ching, C. R. K., Salminen, L., Thomopoulos, S., Bright, J., Baune, B., Bertolin, S., Bralten, J., Bruin, W., Bulow, R., Chen, J., Chye, Y., Dannlowski, U., de Kovel, C. G. F., Donohoe, G., Eyler, L., Faraone, S. V., Favre, P., … the ENIGMA Consortium. (2020). ENIGMA and global neuroscience: A decade of large‐scale studies of the brain in health and disease across more than 40 countries. Translational Psychiatry, 10, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Heuvel, E. , Manders, E., Swillen, A., & Zink, I. (2018). Atypical language characteristics and trajectories in children with 22q11.2 deletion syndrome. Journal of Communication Disorders, 75, 37–56. [DOI] [PubMed] [Google Scholar]
- Villalón‐Reina, J. E. , Martínez, K. , Qu, X. , Ching, C. R. K. , Nir, T. M. , Kothapalli, D. , Corbin, C. , Sun, D. , Lin, A. , Forsyth, J. K. , Kushan, L. , Vajdi, A. , Jalbrzikowski, M. , Hansen, L. , Jonas, R. K. , van Amelsvoort, T. , Bakker, G. , Kates, W. R. , Antshel, K. M. , … Bearden, C. E. (2020). Altered white matter microstructure in 22q11. 2 deletion syndrome: A multisite diffusion tensor imaging study. Molecular Psychiatry, 25(11), 2818–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstman, J. A. , Breetvelt, E., Duijff, S., Eliez, S., Schneider, M., Jalbrzikowski, M., Armando, M., Vicari, S., Shashi, V., Hooper, S. R., Chow, E. W. C., Fung, W. L. A., Butcher, N., Young, D. A., McDonald‐McGinn, D. M., Vogels, A., van Amelsvoort, T., Gothelf, D., Weinberger, R., … Bassett, A. S. (2015). Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry, 72(4), 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Zeng, W. , & Chen, L. (2012). A fast‐FENICA method on resting state fMRI data. Journal of Neuroscience Methods, 209(1), 1–12. [DOI] [PubMed] [Google Scholar]
- Winkler, A. M. , Kochunov, P. , Blangero, J. , Almasy, L. , Zilles, K. , Fox, P. T. , Duggirala, R. , & Glahn, D. C. (2010). Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. NeuroImage, 53(3), 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin, M. , Wang, P. P. , Aleman, D. , McDonald‐McGinn, D. , Zackai, E. , & Moss, E. (2001). Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genetics in Medicine, 3(1), 34–39. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Groth, K. M. , Pearlson, G. , Schretlen, D. J. , & Calhoun, V. D. (2009). Source‐based morphometry: The use of independent component analysis to identify gray matter differences with application to schizophrenia. Human Brain Mapping, 30(3), 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni, T. , Poldrack, R. A. , Nichols, T. E. , van Essen, D. C. , & Wager, T. D. (2011). Large‐scale automated synthesis of human functional neuroimaging data. Nature Methods, 8(8), 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Guo, T. , Fiksinski, A. , Breetvelt, E. , McDonald‐McGinn, D. M. , Crowley, T. B. , Diacou, A. , Schneider, M. , Eliez, S. , Swillen, A. , Breckpot, J. , Vermeesch, J. , Chow, E. W. C. , Gothelf, D. , Duijff, S. , Evers, R. , van Amelsvoort, T. A. , van den Bree, M. , Owen, M. , … International 22q11.2 Brain and Behavior Consortium . (2018). Variance of IQ is partially dependent on deletion type among 1,427 22q11.2 deletion syndrome subjects. American Journal of Medical Genetics—Part A, 176(10), 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, B. A. , Gennatas, E. D. , Zhou, J. , & Seeley, W. W. (2010). Network‐level structural covariance in the developing brain. Proceedings of the National Academy of Sciences of the United States of America, 107(42), 18191–18196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. t‐SNE visualization of GMV feature distribution.
FIGURE S2. Flowchart of data processing and analyses pipeline.
FIGURE S3. Neuroimaging quality control assessment.
FIGURE S4. Source‐based morphometry derives seventeen gray matter volume structural brain patterns.
FIGURE S5. Unthresholded gray matter volume structural brain patterns.
FIGURE S6. Effect size of meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with decreased gray matter in 22q11DS.
FIGURE S7. Effect size of meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with increased gray matter in 22q11DS.
FIGURE S8. Effect size with IQ adjusted meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with decreased gray matter in 22q11DS.
FIGURE S9. Effect size with IQ adjusted meta‐analysis, and mega‐analysis of gray matter volume structural brain patterns with increased gray matter in 22q11DS.
FIGURE S10. Site effects on VBM analysis.
FIGURE S11. Voxel‐based morphometry results of differences in gray matter volume between 22q11DS and healthy volunteers and between individuals with A‐B deletion and individuals with A‐D deletion.
TABLE S1. Regions included in each structural brain pattern.
TABLE S2. Effect sizes of the comparisons between 22q11DS individuals and healthy controls.
TABLE S3. Site Demographics of 22q11DS individuals with small (A‐B) vs. large (A‐D) deletions, and 22q11DS individuals with and without psychotic disorder diagnoses.
TABLE S4. Voxel‐based morphometry results: Differences between 22q11DS individuals and controls.
TABLE S5. Voxel‐based morphometry results: Differences between 22q11DS individuals with shorter deletion (A‐B) and longer deletion (A‐D).
Data Availability Statement
The data used for the analysis are made available upon request to the ENIGMA 22q11.2 Working Group.


