Abstract
Immune evasive induced pluripotent stem cell (iPSC)-derived kidney organoids, known as “stealth” organoids, hold promise for clinical transplantation. To address immune rejection, we investigated the impact of genetically modifying human leukocyte antigen (HLA) class I in kidney organoids prior to transplantation. By using CRISPR-Cas9, we successfully knocked out beta-2-microglobulin (B2M), resulting in iPSCs devoid of HLA class I surface expression. In vitro, the B2M knockout protected kidney organoids derived from these iPSCs against T-cell rejection. To assess in vivo protection, unmodified (control) and B2M–/– kidney organoids were transplanted into humanized mice engrafted with human peripheral blood mononuclear cells (PBMCs). Successful engraftment of human PBMCs was confirmed, and after 4 weeks, we observed no discernible difference in the infiltration rate, proliferation, or cytotoxicity of CD4+ and CD8+ T cells between control and B2M–/– organoids. Both groups of organoids showed compromised tissue integrity, displaying tubulitis and loss of tubule integrity. Notably, while B2M–/– organoids failed to express HLA class I on their cell surface, there was preexisting expression of HLA class II in both control and B2M–/– organoids transplanted into mice with human PBMCs. HLA class II expression was not limited to antigen-presenting cells but also evident in epithelial cells of the kidney organoid, posing an additional immunological challenge to its transplantation. Consequently, we conclude that B2M knockout alone is insufficient to protect iPSC-derived kidney organoids from T-cell-mediated immune rejection. Additionally, our findings suggest that modulating HLA class II signaling will be necessary to prevent rejection following transplantation.
Keywords: induced pluripotent stem cells (iPSCs), differentiation, kidney, transplantation, CRISPR, immunogenicity, T cell
Graphical Abstract
Graphical Abstract.
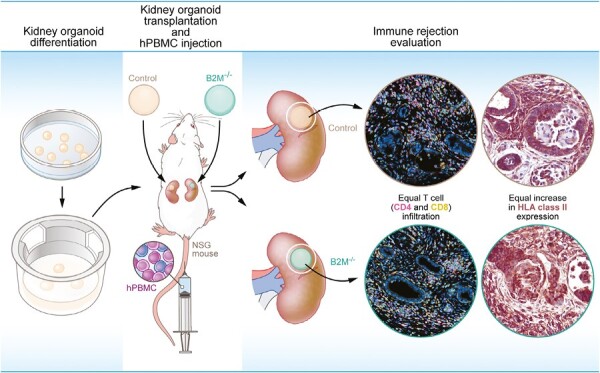
Significance Statement.
For patients with kidney failure, kidney transplantation is currently the only curative option. There are, however, not enough donor organs available to help all patients in need. Induced pluripotent stem cell (iPSC)-derived kidney organoids are a potential alternative treatment option. However, HLA expression on the transplanted tissues possibly elicits allorecognition and subsequent rejection. In this article, we show that knocking out beta-2-microglobulin (B2M) using CRISPR-Cas9 is not enough to prevent rejection. Interestingly we found preexisting expression of HLA class II in the kidney organoids, suggesting the need to modulate HLA class II to prevent rejection of kidney organoids.
Introduction
Organ transplantations are one of the most impactful medical treatments. Over time, advancements in organ matching and immune suppressive therapies have significantly enhanced the success rates of these procedures.1 However, the demand for donor organs has outpaced the available donors and extensive waiting lists have emerged, preventing timely assistance to all patients in need. Among the organs, the demand for kidney transplants is particularly high, highlighting an urgent need for alternative treatment options. Regenerative medicine has emerged as a promising field, aiming to restore tissue and organ function through innovative approaches. Induced pluripotent stem cells (iPSCs) can be differentiated into kidney organoids that contain multiple cell types of the nephron.2-4 Transplantation of kidney organoids in mice leads to vascularization, maturation, and tissue functionality shown by filtration,5-7 indicating their promising potential for regenerative medicine.
Successful transplantations have been performed with iPSC-derived cells for patients with age-related macular degeneration,8,9 and Parkinson’s disease.10 These studies showed successes in feasibility, safety, and effectivity but were limited to immune-privileged sites.11,12 In contrast the kidney is highly vascularized, linked to its function to regulate blood volume and composition,13 and subject to active immune surveillance.14-16 Therefore, transplantation of tissue or cells in the kidney can potentially elicit a strong immune response. With this study, we aim to investigate immune rejection of iPSC-derived kidney organoids.
The major histocompatibility complex molecules, in humans known as human leukocyte antigens (HLA), are a group of cell surface proteins capable of presenting processed peptides to T cells, allowing for immune surveillance. The highly polymorphic nature of HLA is a major factor in the development of an immune response to transplanted tissues or organs. A proportion of 1%-10% of circulating T cells is estimated to be able to be activated by mismatched HLA.17,18 HLA matching in organ transplantation showed significant benefits including better graft function, longer survival of graft and patient, and a lower requirement for immunosuppressive drugs.19 Newly developed techniques allow for a more precise comparison of HLA between donor and recipient,20 but further research is required to improve our understanding of varying mismatch effects and prove clinical value in rejection prediction and organ allocation.21-23
HLA genes can be divided into HLA class I and HLA class II. HLA class II expression is mostly limited to antigen-presenting cells such as dendritic cells, macrophages and B cells, while HLA class I is ubiquitously expressed. We have shown before that iPSC-derived kidney organoids express HLA class I with increased expression upon inflammatory stimulation,24 making these tissues vulnerable to T-cell-mediated rejection.
Previous studies have demonstrated the effectiveness of HLA modification in suppressing immune rejection for various iPSC-derived cell types, including endothelial cells and cardiomyocytes.25-27 In this study, we aimed to examine the significance of HLA expression in T-cell-mediated immune rejection of iPSC-derived kidney organoids. To achieve this, we used genetic modification to knockout the beta-2-microglobulin (B2M) gene expression in iPSCs. B2M is a crucial component of HLA class I surface molecules, and its absence destabilizes the HLA complex, preventing its transportation to the cell membrane for antigen presentation to T cells. To study immune evasion, we developed both an in vitro T-cell coculture model and an in vivo humanized mouse model.28 By comparing the T-cell response between B2M–/– kidney organoids and unmodified control organoids, we examined the susceptibility of iPSC-derived kidney organoids to T-cell-mediated rejection and the role of HLA expression in this process.
Materials and Methods
iPSC Culture and Genetic Modification by CRISPR-Cas9
Human iPSCs were generated by the Leiden University Medical Center (LUMC) iPSC Hotel using an RNA Simplicon reprogramming kit (Millipore) (LUMC0072iCTRL01, detailed information at Human Pluripotent Stem Cell Registry, https://hpscreg.eu/). iPSCs were cultured on recombinant human vitronectin (Thermo Fisher Scientific) coated culture plates in Essential 8 (E8) medium (Thermo Fisher Scientific) and passaged every 3-4 days using 0.5 mM UltraPure EDTA (Thermo Fisher Scientific). Genetic modification of iPSCs was performed using the P3 Primary Cell 4D-Nucleofector X Kit L (Lonza). 3 × 106 iPSCs and 8 µg total plasmid DNA were used per transfection in 100 µL volume. After completing program CA137, cells were transferred to pre-warmed culture plates containing E8 medium and RevitaCell (Thermo Fisher Scientific). RevitaCell was removed on day 1 after nucleofection. On day 4, iPSCs were dissociated into a single-cell suspension by TrypLE select (Thermo Fisher Scientific) and 2 × 105 cells/cm2 were transferred to new culture plates containing 0.2 µg mL–1 puromycin and RevitaCell. RevitaCell was kept for 1 day and puromycin selection was continued for 7 days. The remaining colonies were kept in culture until they had reached a size of approximately 2 mm in diameter. Single colonies were dissociated and transferred to new culture plates containing E8 medium. In total 12 iPSC clones were propagated, cryopreserved and material was obtained to validate the genetic modification. Three clones (B2M#2; B2M#7; B2M#10) were used for follow-up experiments.
Plasmid Production
In each nucleofection 3 different plasmids were used: A plasmid containing the sequence for Cas9 driven by a CAG promoter (Cas9 plasmid), a plasmid containing the B2M gRNA (B2M gRNA plasmid), and a plasmid containing the donor DNA for homologous recombination in the B2M locus (B2M donor DNA plasmid). The Cas9 plasmid (AV62_pCAG.Cas9.rBGpA) and gRNA scaffold backbone (S7 pUC.U6.sgRNA.Bvelstuffer) have been detailed elsewhere.29 The B2M gRNA plasmid, designed to target the DNA near the B2M start codon, was assembled by inserting annealed oligonucleotide pairs 5ʹ-ACCGGCCACGGAGCGAGACATCT-3ʹ and 5ʹ-AAACAGATGTCTCGCTCCGTGGC-3ʹ into the BveI digested gRNA scaffold backbone. The donor DNA plasmid (ordered at BaseClear B.V.) contained 1000 bp long homology arms of the sequence surrounding the start codon for B2M, with in between the ORF of eGFP followed by a PGK promoter and ORF for puromycin resistance gene in between loxP sites to allow for antibiotic selection of modified cells. Plasmids were transformed into chemo-competent Escherichia coli bacteria and isolated with maxiprep (Genomed). Correct plasmid sequences were validated by sanger sequencing by the Leiden Genome Technology Center.
DNA Isolation and Genomic PCR
Cell pellets of roughly 5 × 105 iPSCs were used for DNA isolation with the DNeasy kit (Qiagen) and DNA concentrations were measured with nanodrop (Thermo Fisher Scientific). PCR reactions were performed using GoTaq G2 Flexi DNA Polymerase (Promega) using the following conditions: 100-200 ng DNA, 1× Green GoTaq Flexi buffer, 1.25 units GoTaq G2 Flexi polymerase, 0.3 μM each primer, 0.3 mM each dNTP and 1.5 mM MgCl2 in a 30 μL volume for 30-40 cycles in a C1000 Touch Thermal Cycler (Bio-Rad). The 2 primer pairs used (Supplementary Table S1) span the homology arms at both sites of the inserted sequence to allow for validation of correct genomic insertion. PCR products and GeneRuler DNA Ladder mix (Thermo Fisher Scientific) were loaded on a 1% agarose gel for size separation and visualized by ethidium bromide in a Molecular Imager Gel Doc XR+ (Bio-Rad).
Kidney Organoid Differentiation
Differentiation of iPSCs to kidney organoids was performed following a previously described protocol.2,3 In short, iPSCs were cultured for 7 days as a monolayer after which they were dissociated and transferred as 3-dimensional clumps onto Transwell 0.4 µm pore polyester membranes (Corning). APEL2 medium containing rhFGF-9 and heparin was added to the culture well below the filter and refreshed on day 7 + 3. On day 7 + 5, growth factors were removed and the APEL2 medium was refreshed every 2 days for the remaining culture time.
RNA Analyses
Total RNA was isolated using nucleospin RNA/protein kit (Macherey-Nagel) at different time points during kidney organoid differentiation: day 0 (iPSCs, undifferentiated cells), day 7 (intermediate mesoderm), and day 7 + 20 (kidney organoids). RNA concentrations were measured with nanodrop. cDNA produced with SuperScript III Reverse Transcriptase (Invitrogen) was used as a template in real-time polymerase chain reaction (RT-qPCR). RT-qPCR was performed using SYBR Green Supermix (Bio-Rad) in a CFX Connect Real-Time System (Bio-Rad). Gene expression was normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and relative mRNA levels were determined based on the comparative Ct method (2–ΔΔCt) to the respective controls. Primers used in RT-qPCR are described in Supplementary Table S1.
RNA samples of all 3 differentiation stages were sent to GenomeScan B.V. for bulk RNA sequencing. Before sequencing, RNA quality was characterized using the Experion RNA StdSens 1K Analysis Kit (Bio-Rad, product number 7007103) on an Experion Automated Electrophoresis System (Bio-Rad) following the manufacturer’s protocol. Sequencing was performed with Illumina NovaSeq6000 (20M reads/sample). Raw read FASTQ files were processed using the opensource BIOWDL RNAseq pipeline v5.0.0 (https://zenodo.org/record/5109461#.Ya2yLFPMJhE) developed at the LUMC, adapter clipping was performed using Cutadapt (v2.10) with default settings and RNA-Seq read alignment was performed using STAR (v2.7.5a) on GRCh38 human reference genome. Read quantification was performed using HTSeq-count (v0.12.4) with the setting “–stranded = yes.” The gene annotation used for quantification was Ensembl version 104. Additional data quality controls were performed using fastqc (v0.11.9), Picard (v2.23.2) and MultiQC (v1.9). Principal component analysis (PCA) was performed with the counts per million and R-package stats (v3.6.2). The PCA plot, PCA plot with loadings, and expression heatmaps were visualized using R-package ggplot2 (v3.3.5 or v3.4.4) and NMF (v0.26.0).
HLA-II-Encoding Gene Expression
Processed scRNA-seq data from untransplanted and transplanted kidney organoids in chicken embryos (day 7 + 20, human cells) were obtained from,30 available in ArrayExpress repository under accession number E-MTAB-11429. Data were processed using R (v4.2.1) in Rstudio (v2022.02.3 build492 for Windows). The dataset was subset to the cell identities of interest (endothelial cells, podocytes, proximal tubules, loop of Henle-like, and distal tubule/collecting duct clusters). Scaled expression values and percentage of cells expressing each gene were obtained for each cell identity and organoid condition using the DotPlot () function from the Seurat package (v4.2.0) for HLA-II-encoding HLA-DRB1, HLA-DPB1, and HLA-DQB1 genes. The final dot plot was prepared with the ggplot2 package (v3.4.2) using the geom_point () function.
In Vitro T-Cell Coculture
CD8+ T cells had been transduced with a T-cell receptor (TCR) as described.31 Used CD8+ T cells were specific for either a peptide of human cytomegalovirus (CMV),32 not expressed in the kidney organoids (negative control), or specific for a peptide of household protein USP11 presented in HLA-A2 (Allo-A2).33 Control iPSCs are positive for HLA-A2, validated by the HLA laboratory at the LUMC, and therefore the Allo-A2 CD8+ T cells are expected to be activated upon coculture. CD8+ T cells were retrieved from liquid nitrogen storage 24 h before start of the coculture and cultured in T-cell medium consisting of Iscove’s Modified Dulbecco’s Medium (IMDM) (Lonza), 5% heat-inactivated fetal bovine serum (Gibco, Thermo Fisher Scientific), 5% human serum (Sanquin Reagents), 1.5% 200 mM l-glutamine (Lonza), 1% 1 × 105 U mL–1 penicillin/streptomycin (Lonza) and 100 IU mL–1 IL-2 (Novartis Pharma).
On days 7 + 18 of kidney organoid differentiation, T cells were added.34 APEL2 medium (1.5 mL) was added beneath the transwell membrane. 1 × 105 T cells were pipetted on top of each organoid in 10 µL volume. T cells were allowed to attach to the organoid by incubation for 15 min at 37 °C before adding 800 µL APEL2 medium on top of the filter. As a control for inflammatory stimulation, 500 IU mL–1 IFN-γ (R&D systems) was added to a separate culture well with organoids. T-cell coculture and IFN-γ stimulation were continued for 48 h, after which the supernatant on top of the filter was removed and stored for ELISA, and organoids with T cells were dissociated for flow cytometry.
Animals and Transplantation
All animal experimental procedures were approved by and performed in accordance with the guidelines of the animal welfare committee of the LUMC and the Dutch Animal Experiments Committee (AVD1160020198687). For all experiments, male NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG, Jackson 005557) mice between 6 and 8 weeks old were used. Transplantations were performed on day 0 using kidney organoids at differentiation day 7 + 11 or 7 + 12. Recipient mice were anesthetized with isoflurane and bupaq (buprenorphine) was added to the drinking water and subcutaneously injected as pre- and postoperative analgesic. Kidneys were exteriorized via flank incisions and bisected kidney organoids were transplanted under the renal capsule through a small incision. The experiment ended on day 35 at which the spleen and kidneys were collected.
PBMC Isolation, Injection, and Blood Collection
PBMCs were isolated from buffy coats ordered at the Netherlands’ central blood bank (Sanquin) of 3 separate healthy donors by ficoll density gradient separation (Pharmacy LUMC) and stored in liquid nitrogen until use. The HLA haplotype of the PBMC donors was examined and compared with that of the control iPSCs. Three PBMC donors were chosen with a similar high degree of mismatch in both HLA class I and HLA class II (Supplementary Table S2). Seven days after transplantation, 20 × 106 PBMCs were intravenously injected in 200 µL PBS per mouse. As a negative control, mice were injected with PBS instead of PBMCs. Blood was collected in K2E microvettes (Sarstedt) by performing a tail vein puncture on day 14 and a heart puncture on day 35 for flow cytometry analysis.
Flow Cytometry
Flow cytometry was performed on iPSCs, kidney organoids, and mouse peripheral blood. iPSCs were dissociated with TrypLE Select for 5 minutes at 37 °C. Kidney organoids with or without T cells were dissociated as described before.30 Upon dissociation, cell clusters were removed by passing the cells over a 30 μm filter. Single cells were resuspended in PBS + 0.1% BSA. Mouse peripheral blood was used directly and human PBMCs retrieved from liquid nitrogen storage were used as positive control for human leukocyte markers.
Single-cell suspension or blood was incubated for 30 minutes on ice with antibodies shown in Supplementary Table S3. Subsequently, iPSCs, kidney organoid cells and PBMCs were washed and resuspended in FACS buffer (0.1% BSA and 0.5 mM EDTA in PBS). For samples containing blood, red blood cells were lysed by the addition of a shock buffer consisting of 155 mM NH4Cl, 10 mM KHCO3 and 0.1 mM EDTA in distilled water. Samples were measured on an LSR-II (BD) and acquired results were analyzed using FlowJo (BD).
Immunohistochemistry
In vitro, kidney organoids were fixed in 2% PFA for 20 min at 4 °C and washed with PBS. Antibodies used for the detection of kidney structures (NPHS1, E-Cadherin, and LTL) are shown in Supplementary Table S3. Mouse kidneys with transplanted kidney organoids were fixed in 4% PFA overnight at 4 °C and washed with PBS. The kidneys with organoids were processed into paraffin and cut into sections of 4 µm thickness using a microtome. Evaluation of immune rejection was performed by incubation with antibodies shown in Supplementary Table S3. Sections incubated with HRP conjugated secondary antibody were subsequently processed by Vector NovaRED substrate following the manufacturer’s protocol (SK-4800, Vector Laboratories). In immunofluorescence stainings, nuclei were stained with Hoechst33342 (1:10 000, H3576, Invitrogen) and NovaRED stained slides were counterstained with Mayer’s hematoxylin (Sigma–Aldrich).
Periodic acid–Schiff (PAS) stainings were performed in periodic acid at 60 °C for 30 minutes, followed by washing in distilled water and incubation in Schiff’s reagent (Sigma–Aldrich) at room temperature for 45 minutes. Tissue slides were then washed in running water after which a counterstain was performed with Mayer’s hematoxylin.
Whole organoids were mounted using ProLong Gold Antifade Mountant (Invitrogen) in glass bottom dishes (MatTek). Stained paraffin sections were dehydrated and covered with Entellan mounting medium (Sigma–Aldrich) or ProLong Gold Antifade Mountant for fluorescent stainings. Images were acquired using a White Light Laser Confocal Microscope TCS SP8 and LAS-X Image software or using a PANNORAMIC 250 Flash III slide scanner (3DHISTECH) and Case Viewer (3DHISTECH). Positive cell detection was performed using QuPath.
ELISA
IFN-γ concentrations in the supernatant were measured by IFN-γ human ELISA (88-7316-88, Thermo Fisher Scientific) according to the manufacturer’s protocol. Light absorption was measured using an iMark microplate reader (Bio-Rad).
Statistical Analysis
Graphical results were acquired and statistical analysis was performed using GraphPad Prism software. Statistical tests performed were either a paired t test or one-way ANOVA with Dunnett’s or Šidák’s correction for multiple testing as indicated. Mean values are shown ±SD, and a P-value < .05 was considered statistically significant.
Results
Generation of B2M–/– iPSCs and Their Derived Kidney Organoids
iPSCs and iPSC-derived kidney organoids express HLA class I and this expression increases upon stimulation with inflammatory cytokine IFN-γ,24 making them vulnerable to T-cell-mediated immune rejection upon transplantation. Using CRISPR-Cas9-mediated homology recombination, we introduced a GFP/PGK-puromycin-containing cassette within exon 1 of the B2M gene to generate B2M–/– iPSCs (Fig. 1A and Supplementary Fig. S1). As expected, the resulting clones (B2M#2; B2M#7; B2M#10) showed no expression of B2M mRNA and HLA surface expression in normal conditions and after IFN-γ treatment, as determined by qPCR and flow cytometry respectively (Fig. 1B and 1C). Of note, the detection of GFP by flow cytometry showed low expression in the B2M-modified clones with an increase upon IFN-γ stimulation, indicating regulation by the IFN-γ responsive endogenous B2M promoter (Supplementary Fig. S2). These results confirm the successful generation of B2M–/– iPSC clones.
Figure 1.
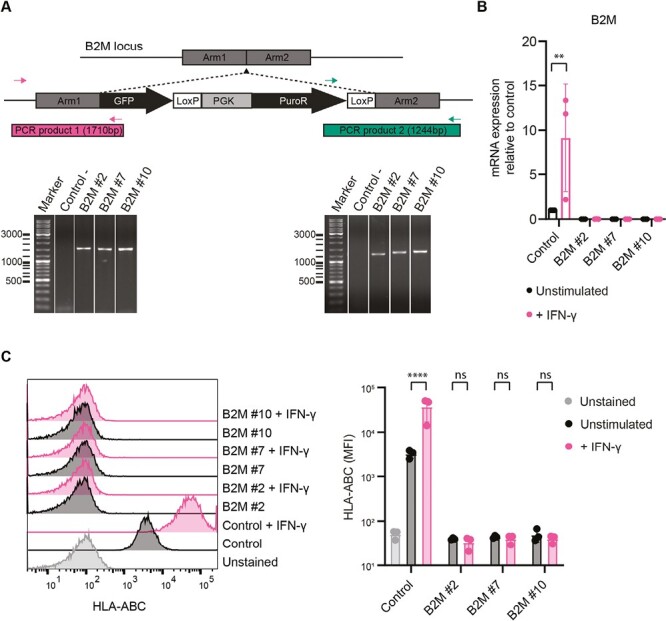
Genetic modification of iPSCs at the B2M locus. (A) Schematic of the genetic modification strategy at the B2M locus by CRISPR-Cas9 and genomic PCR for validation of correct insertion. Two primer pairs were used spanning the homology arms at both ends of the insert, producing PCR products of 1710 and 1244 bp in size. Unmodified control iPSCs were used as negative control (Control-) and the PCR results of 3 B2M–/– clones (#2, #7, and #10) are shown (see also Supplementary Fig. S1). (B) B2M mRNA expression measured by RT-qPCR for control iPSCs and B2M–/– clones in unstimulated and IFN-γ stimulated conditions (n = 3 independent experiments). (C) HLA-ABC surface expression measured by flow cytometry. Unstained control iPSCs were used as negative control and all cell lines were measured in unstimulated and IFN-γ stimulated conditions (n = 3 independent experiments). Results are shown as mean ± SD and significance was evaluated using one-way ANOVA with Šidák’s correction for multiple testing, comparing each sample to its own unstimulated control (ns = not significant, **P < .01, ****P < .0001).
To evaluate kidney organoid differentiation (Fig. 2A) upon genetic manipulation, we established the transcriptomic profile of B2M–/– cells at day 0 (undifferentiated), day 7 (intermediate mesoderm) and day 7 + 20 (kidney organoid) and compared this to unmodified control cells. The Principal Component Analysis (PCA) led to the identification of 3 principal clusters, illustrating the differentiation stages, with no sub-clustering of the B2M–/– lines (Fig. 2B). Top genes determining clustering of the samples include markers of kidney development like HOXB9, NPHS2, and WT135 (Supplementary Fig. S3A–C) with increased expression of kidney cell lineage markers along the differentiation process of B2M–/– and control iPSCs (Supplementary Fig. S3D). In addition, immunohistochemistry conducted on iPSC-derived kidney organoids in vitro confirmed the presence of glomeruli (NPHS1), proximal tubules (LTL), and distal tubules (ECAD) in control and B2M–/– tissues (Fig. 2C). Altogether, these results demonstrate that B2M disruption had no impact on the differentiation capacity of iPSCs toward kidney organoids.
Figure 2.
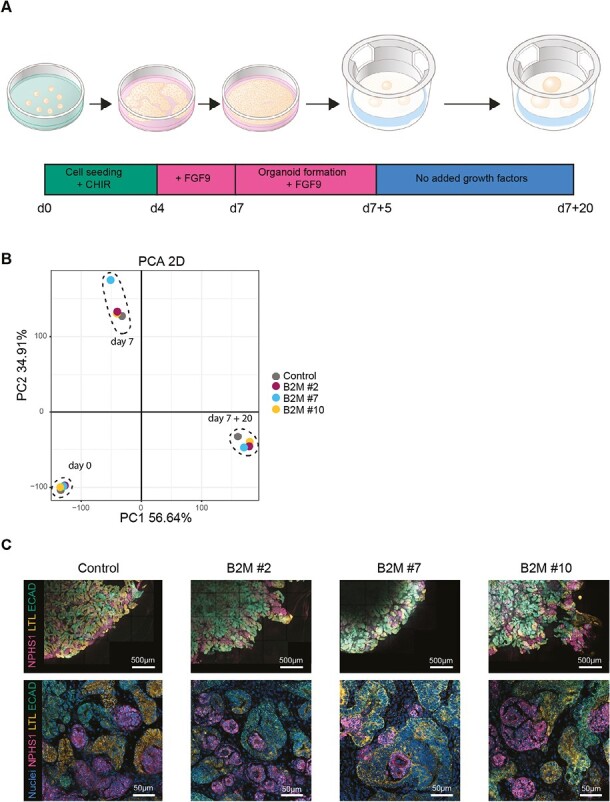
Kidney organoid differentiation of modified iPSCs. (A) Schematic representation of the differentiation protocol of iPSCs to kidney organoids. iPSCs are seeded on day 0 (d0), harvested on day 7 (d7) and cultured as cell clumps on top of a filter till day 7 + 20 (d7 + 20). The change of added growth factors to the culture medium is indicated in the timeline. (B) PCA plot of bulk RNA sequencing data of control and B2M–/– clones at 3 timepoints: undifferentiated iPSCs (day 0), intermediate mesoderm cells (day 7), and kidney organoids at the end of differentiation (day 7 + 20). Detailed analysis is displayed in Supplementary Fig. S3. (C) Representative fluorescent images of whole organoids with at the top a quarter of the organoid and below a higher magnification within the organoid. Specific units of the nephron were detected by NPHS1 (glomerulus), LTL (proximal tubule), and ECAD (distal tubule). Nuclei were stained with Hoechst.
B2M–/– Prevents CD8+ T-Cell Reactivity to Kidney Organoids In Vitro
To evaluate immunogenicity, we exposed control and B2M–/–organoids to HLA-A*02:01-allo-specific (Allo-A2) cytotoxic T cells or CMV-specific (CMV) cytotoxic T cells (negative control). After 2 days of coculture, organoids with T cells were dissociated and analyzed by flow cytometry for immune activation markers and the supernatant was collected for detection of IFN-γ using ELISA (Fig. 3A). The effect of the T-cell coculture on the organoid cells was evaluated by detection of HLA class I and CD54 (ICAM) expression. Kidney organoids treated with 500 IU mL–1 IFN-γ for 48 h were used as a positive control for HLA class I induction. As expected, IFN-γ treatment increased HLA class I expression in the control organoids but not in organoids of the B2M–/– clones (Fig. 3B). While coculture of control organoids with CMV T cells did not affect HLA class I surface expression in the kidney organoid cells, Allo-A2 T cells induced increased HLA class I surface expression to a similar degree as the IFN-γ stimulation, reflecting a possible response to IFN-γ endogenously produced by activated T cells during coculture (Fig. 3B). In B2M–/– organoids, HLA class I surface expression remained unchanged during coculture with either CMV or Allo-A2 T cells (Fig. 3B). Similar to HLA class I, CD54 expression in the control organoid was increased in coculture with Allo-A2 T cells, while the B2M–/– organoids were not affected (Fig. 3C).
Figure 3.
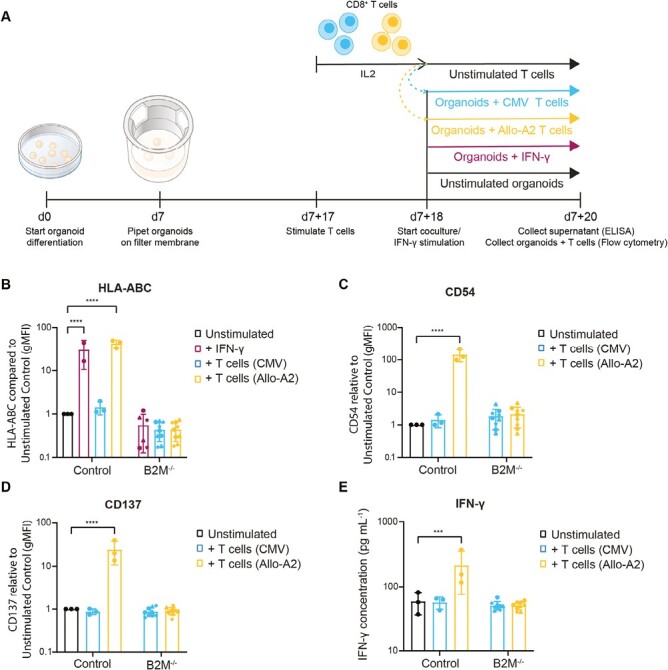
In vitro kidney organoid coculture assay with T cells. (A) Schematic of the in vitro coculture experiment and readouts. (B–C) Surface expression on organoid cells of (B) HLA-ABC and (C) CD54 (ICAM) measured by flow cytometry. Kidney organoid cells cultured without the addition of T cells (unstimulated) were used as a control and set to 1 for comparison. Results are shown of kidney organoids cultured with IFN-γ, CMV-reactive T cells (CMV), or HLA-A2-reactive T cells (Allo-A2) (n = 2 independent experiments for IFN-γ, n = 3 for others). (D) CD137 expression on CD8+ T cells cultured with control or B2M–/– kidney organoids measured by flow cytometry. T cells cultured alone (unstimulated) were used as a control and set to 1 for comparison (n = 3 independent experiments). (E) IFN-γ secretion in the supernatant by the T cells cultured with control or B2M–/– kidney organoids measured by ELISA. T cells cultured alone (unstimulated) were used as control for spontaneous IFN-γ secretion (n = 3 independent experiments). B2M–/– organoid clones are indicated with separate symbols; #2 (circle), #7 (triangle), and #10 (star). Results are shown as mean ± SD and significance was evaluated using one-way ANOVA with Dunnett’s correction for multiple testing, comparing each sample to the unstimulated control (***P < .001, ****P < .0001).
T-cell activation was evaluated by the detection of CD137 expression in CD8+ T cells by flow cytometry and IFN-γ secretion in the supernatant by ELISA. CD137 is a costimulatory molecule that is not expressed on unstimulated T cells and is upregulated upon activation.26 In these assays, Allo-A2 cells displayed an increased expression of activation marker CD137 (Fig. 3D) and increased secretion of effector cytokine IFN-γ (Fig. 3E) following coculture with control organoids. Thus, B2M–/– fully prevented T-cell activation in vitro.
B2M–/– Does Not Prevent T-Cell Mediated Immune Rejection of Transplanted Kidney Organoids In Vivo
To evaluate the capacity of B2M–/– organoids to escape T-cell-mediated immune rejection in vivo, we created a humanized mouse model with paired subcapsular transplantation (a control or a B2M–/– organoid on either the left or right kidney in the same mouse) by injecting NSG mice with HLA mismatched human PBMCs isolated from 3 healthy adult donors (Fig. 4A). This is an established mouse model which enables the engraftment of mainly human T cells (both CD4+ and CD8+) and can therefore be used to study T-cell-mediated rejection of human tissue.36 For all 3 PBMC donors there is an extensive mismatch in both HLA class I and HLA class II genes compared to the control iPSCs (Supplementary Table S2) potentially leading to immune rejection. We observed successful engraftment of human PBMCs as determined by the number of human leukocytes in the blood of these mice (0.2%-1.1% hCD45+ at day 14; 3%- 56% at day 35) (Fig. 4B and 4C). As expected, while the injected PBMCs contained all human leukocyte subtypes (CD4+, CD8+, CD14+, CD19+, and CD56+), hCD45+ cells in peripheral blood at day 14 contained predominantly CD4+ and CD8+ T cells, and at day 35 were restricted to CD4+ and CD8+ T cells (Fig. 4D). Of note, the ratio CD4:CD8 in the peripheral blood differed between PBMC donors with 20:80, 60:40, and 40:60 for donors 1, 2, and 3 respectively (Fig. 4D-4F).
Figure 4.
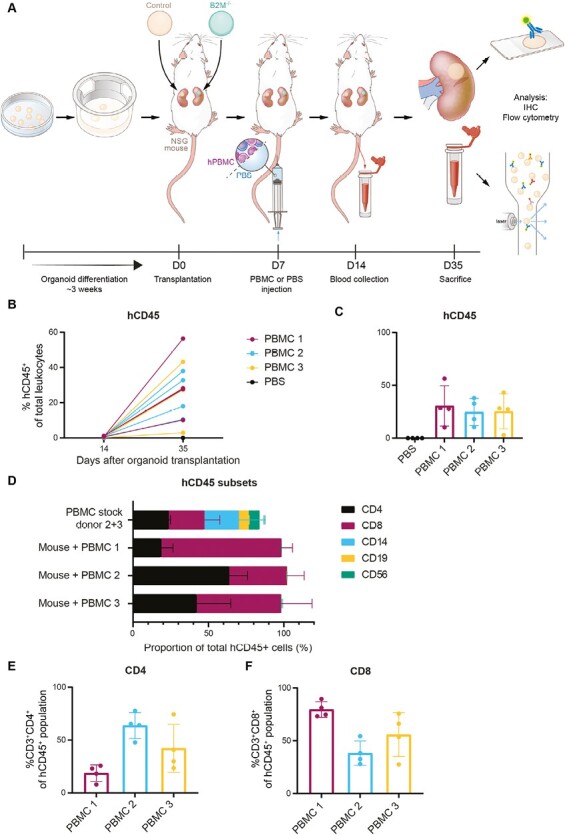
A humanized mouse model to study T-cell-mediated immune rejection of kidney organoids in vivo. (A) Schematic of the in vivo experimental setup. Control and B2M–/– organoids were transplanted in pairs in 16 mice. One week after transplantation, 4 mice were injected with PBS and 12 mice were injected with PBMCs of 3 different donors (4 mice per PBMC donor). (B-C) Proportion of human CD45+ cells of total leukocytes in the mouse peripheral blood measured by flow cytometry. (B) Results are shown for day 14 and day 35 after transplantation and are connected for each mouse (n = 16). (C) Results of day 35 are shown separately per group. Mice injected with PBS, negative control, are shown in black (n = 4 PBS injected mice). Mice injected with human PBMCs of 3 different donors, 4 mice per donor, are indicated in different colors (n = 12 PBMC injected mice). (D) Cell subsets in the hCD45+ population in the original PBMC stocks used for injection (positive control) and circulating in mouse peripheral blood at day 35 measured by flow cytometry. Subpopulations that were measured are T helper cells (CD4), cytotoxic T cells (CD8), monocytes/macrophages (CD14), B cells (CD19), and NK cells (CD56). Results for PBMC stock are shown for 2 donors combined (PBMC donors 2 and 3) and mouse peripheral blood results are shown per PBMC donor (n = 4 mice per donor). (E and F) Proportion of (E) T-helper cells (CD3+CD4+) and (F) cytotoxic T cells (CD3+CD8+) of the hCD45 + population at day 35 after transplantation measured by flow cytometry. Results are indicated per PBMC donor (n = 4 per PBMC donor) and are shown as mean ± SD.
Immunohistochemical staining performed on the transplanted tissues showed similar T-cell infiltration in B2M–/– and control kidney organoids (Fig. 5A). The presence of T cells in the epithelial layers of tubular structures, characteristic of tubulitis, was exclusively detected in the kidney organoid and not the mouse tissue, indicating a targeted inflammatory response toward the kidney organoid. Further characterization of the immune infiltrate showed the presence of both CD4+ and CD8+ T cells (Fig. 5B-5D). 40% of the infiltrated T cells were proliferative in both control and B2M–/– organoids, indicated by Ki-67 (Fig. 5E and Supplementary Fig. S4A). The cytotoxic potential of CD8+ T cells was evaluated by a co-staining with Granzyme B (GrB) and showed approximately 40% double-positive cytotoxic T cells in both control and B2M–/– organoids (Fig. 5F and Supplementary Fig. S4B).
Figure 5.
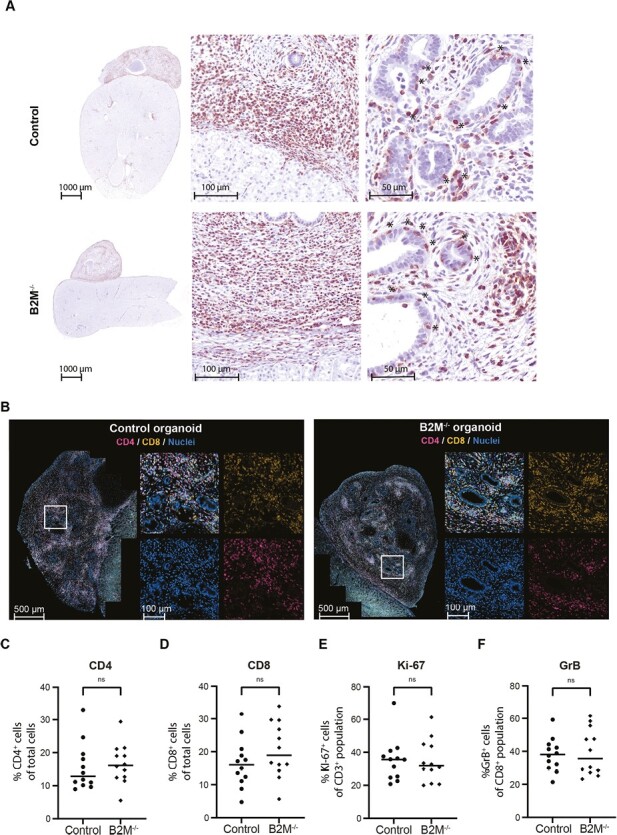
T-cell-mediated immune rejection in control and B2M–/– kidney organoids. (A) Images of transplanted kidney organoids on top of mouse kidneys stained for CD3 (NovaRED) to detect T cells. For both control and B2M–/– a representative organoid is shown. The left picture shows an overview of a section through the organoid on top of the kidney. The middle and right pictures show magnifications of respectively the border of the organoid with the mouse kidney and the center of the organoid. Locations where T cells have infiltrated the tubular wall (tubulitis) are indicated with *. (B) Representative fluorescent images of a control and B2M–/– kidney organoid transplanted in the same mouse. For each image a magnification is shown of the region indicated with a white square, including the separate fluorescent channels (CD4, CD8, Hoechst). (C-F) Quantifications in an organoid section of the proportion (C) CD4+ cells of the total cell population, (D) CD8+ cells of the total cell population (E) Ki-67+ cells in the CD3+ population, (F) granzyme B (GrB)+ cells in the CD8+ population analyzed by Qupath. For both control and B2M–/– individual results with the mean are shown of 12 organoids that were transplanted in pairs in 12 mice. Significance was evaluated using a paired t-test comparing organoids transplanted within the same mouse (ns = not significant).
The cytotoxicity of the T-cell infiltrate was further evaluated in PAS staining of the kidney organoids. Tissue morphology was similarly affected in T-cell infiltrated B2M–/– organoids compared to the paired control organoid (Supplementary Fig. S5). In both types of organoids, we noticed tubulitis, tubular dedifferentiation, and atrophy. These results indicate no difference in T-cell infiltration, proliferation, and cytotoxicity in transplanted B2M–/– organoids compared to control.
Since control and B2M–/– organoids have the same genetic background and are transplanted as a pair, an immune response toward the control organoid could induce rejection of the B2M–/– as a secondary response. To rule out this possibility, we repeated the experiment and transplanted either control or B2M–/– organoids in each mouse (Supplementary Fig. S6A). Similar to the previous transplantation experiment, the immune cell infiltrates of CD4+/CD8+ T cells, CD3+KI-67+ proportion and CD8+GrB+ proportion showed no differences between control and B2M–/– transplanted kidney organoids (Supplementary Fig. S6B-S6E). Thus, also in the separate transplantation model, no difference in T-cell-mediated immune rejection was detected in B2M–/– compared to control organoids. We conclude that B2M–/– is not sufficient to prevent T-cell-mediated immune rejection of iPSC-derived kidney organoids in vivo.
HLA Class II Upregulation in B2M–/– and Control Kidney Organoids
To further characterize the rejection response, transplanted kidney organoids were stained for HLA class II (HLA-DR/DP/DQ). Organoids transplanted in mice without human PBMCs showed faint coloring of the tubular structures, suggesting low expression of HLA class II in tubular epithelial cells (Fig. 6A and Supplementary Fig. S7). High expression of HLA class II in immune cells and in kidney organoids was detected in PBMC-injected mice. In particular, epithelial cells of nephron structures from the kidney organoid showed higher HLA class II expression in both the paired and the separate transplantation model (Fig. 6A and Supplementary Fig. S7). We verified our findings using our single-cell RNA sequencing dataset of untransplanted and transplanted kidney organoids in chicken embryos.30 Indeed, here endothelial and proximal tubular cells of the organoids express HLA class II both before and after transplantation (Fig. 6B). These results show that kidney organoid cells, including the tubular epithelial cells, are capable of expressing HLA class II. Moreover, HLA class II is already expressed before transplantation and in a transplantation model without a mature immune system. This suggests that preexisting HLA class II expression of the kidney organoid could be involved in the rejection in the humanized mouse model.
Figure 6.
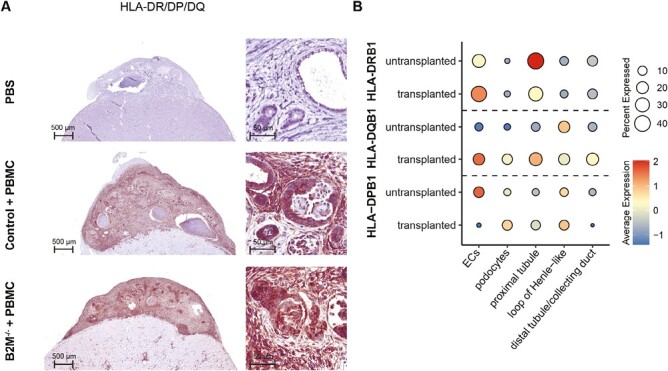
HLA class II expression in transplanted kidney organoids. (A) Representative images of transplanted kidney organoid sections stained for HLA-DR/DP/DQ with an overview of the organoid on the left and a magnification of nephron structures on the right. The upper panel shows a control organoid transplanted in a PBS-injected mouse. The middle and lower panels show a control and B2M–/– organoid transplanted in the same PBMC-injected mouse. (B) Dotplot displaying the scaled mRNA expression levels of HLA-DRB1, HLA-DPB1, and HLA-DQB1 in scRNA-sequencing dataset of untransplanted kidney organoids, and kidney organoids transplanted in chicken embryo for 8 days. Scaled expression levels are indicated for endothelial cells (ECs), podocytes, proximal tubular cells, loop of Henle-like cells, and distal tubular/ collecting duct cells. The color and size of the dot indicate the scaled expression level of the indicated gene and the percentage of cells expressing it, respectively.
Discussion
Here we show the successful development of HLA class I negative iPSCs and their derived kidney organoids by genetic modification of B2M using CRISPR-Cas9. We evaluated the immune-protective effect of B2M–/– in iPSC-derived kidney organoids compared to control organoids both in vitro and in vivo. Although we found complete protection of the B2M–/– organoids in vitro, this protection appeared to be insufficient to prevent T-cell-mediated immune rejection in vivo.
The in vitro system used a coculture of kidney organoids with CD8+ T cells with a TCR specific for a single antigen. Similar protection by B2M–/– in vitro was seen in other studies performing a CD8+ T-cell coculture.37,38 The use of CD8+ T cells specific for a single antigen allows for screening of T-cell reactivity in a short time with high sensitivity but does not consider the great variability of targets that could possibly initiate a T-cell-mediated response. The humanized mouse model used here resulted in the engraftment and proliferation of human T cells that allowed us to investigate T-cell-mediated alloimmune response toward the transplanted kidney organoids. Although humanized mice are routinely used to evaluate human cell engraftment and rejection processes, this in vivo model does not completely recapitulate all the components of the human immune system.39 While a full human PBMC mixture was injected in the mice, only T cells remained detectable in the blood at later time points. The presence of antigen-presenting cells at the reconstitution phase could be involved in indirect rejection but the main pathway for allogeneic immune rejection in this model will be driven by direct allorecognition. The lack of surviving human NK cells in the mouse peripheral blood makes this model not suitable for investigating NK cell reactivity. We cannot exclude that NK cells influenced the immune response early after reconstitution in our experiments, but to study the role of NK cells in more detail a model facilitating NK cell activity will be necessary.40 T cells have a lasting survival and are able to proliferate in the mouse model, and therefore we focused here on T-cell-mediated immune rejection. We used the quantification of CD4+ and CD8+ T cells within the organoid to assess cellular immune rejection. While the presence of T cells alone within a tissue does not prove rejection,41 we show a close correlation with injury to nephron structures, indicating the cytotoxic activity of the T cells within the tissue.
In our in vivo experiments we used 2 different approaches; paired transplantation of a control and B2M–/– organoid in each mouse, or separate transplantation of either control or B2M–/– organoids per mouse. The paired transplantation aimed to enable an adequate comparison between control and B2M–/–, since we have observed differences in the amount of circulating human immune cells per mouse. The paired model corrects for these differences and organoids can directly be compared within a mouse. However, the control and B2M–/– kidney organoids are interconnected through blood circulation, and an immunogenic response toward one organoid could impact the organoid in the second kidney. Therefore, we also performed separate transplantations and found that B2M–/– did not affect T-cell infiltration or the cytotoxic effect both in the paired and separate transplantation model.
While in our study the B2M–/– was not able to prevent T-cell-mediated immune rejection in vivo, other studies did observe a protective effect of B2M–/– on transplanted ESCs,38,42,43 including a decrease in T-cell infiltration.37,43 An important difference compared to these studies is the tissue specificity and maturation stage of the transplanted cells. The iPSC-derived kidney organoids transplanted in our study contained endothelial cells, epithelial cells of nephron structures, and interstitial cells,3 while the other studies examined ESC-derived teratomas.38,42,43 The immunogenic phenotype is dependent on cell type and differentiation stage.
Earlier studies have shown that human fetal kidney grafts are less immunogenic compared to adult kidneys, with fetal tissue showing decreased expression of HLA class I, HLA class II, adhesion molecules, and other immune mediators.44,45 Rossbach et al. have shown an increase in the immunogenic phenotype corresponding to the differentiation stage of stem cell-derived renal cells, with an increase in HLA class I and induction of HLA class II.46 We found increased expression of HLA class II in transplanted kidney organoids when infiltrated by T cells. The extent of the HLA class II expression matched the extent of T-cell infiltration. This is in contrast to the studies with fetal kidney grafts, which were unable to upregulate HLA class II in response to T-cell infiltration, and resembles more what is seen in rejection of a transplanted adult kidney.47 Also the cell type specificity of HLA class II corresponded to the situation in rejected adult kidneys, where an increase in HLA class II is seen mainly in the epithelial cells of the nephron tubules.47-49 Thus, considering HLA class II expression, the iPSC-derived kidney organoids are more mature in their immunogenic properties compared to fetal kidney tissue. In the adult human kidney, HLA class II is expressed in glomerular capillaries and peritubular microvasculature and is inducible in tubular epithelial cells by inflammatory stimulation.48 Single-cell RNA sequencing data of kidney organoids, however, shows that the HLA class II expression is already present in kidney organoids before transplantation, mainly in endothelial cells and proximal tubular cells. This expression can be upregulated after transplantation, even in a hypoimmunogenic environment such as the chicken embryo.30 Direct allorecognition is the earliest and most potent de novo graft-directed immune response since up to 10% of T cells can recognize a single HLA alloantigen while a much smaller proportion responds to a specific antigen.50 Therefore, it is possible that HLA class II expression is involved in direct allorecognition of the transplanted kidney organoids and is responsible for the absence of a protective affect by the B2M–/–.
Although the exact role of HLA class II is not demonstrated in this model, the available resources and the results of this study underscore the importance of preexisting class II regulation in the kidney organoids to prevent immune rejection. Additional disruption of HLA class II in addition to class I therefore seems crucial to prevent a T-cell alloimmune response. In addition to HLA knockout, other modifications are likely to benefit the immune protection of the iPSC-derived tissue. Importantly, the knockout of HLA class I has been shown to induce NK cell reactivity by the missing-self response.40 This can be prevented by selective maintenance of certain HLA subtypes, especially HLA-C,25 induced expression of minimally polymorphic HLA molecules,26,43,51 or induced expression of an inhibitory ligand such as CD47.27
In conclusion, our findings indicate that solely knocking out B2M is insufficient to counteract T-cell-mediated immune rejection of iPSC-derived kidney organoids in vivo. The expression of preexistent HLA class II molecules likely plays a significant role in this process. This study serves as a foundation for developing hypoimmunogenic kidney organoids and underscores the value of exploring diverse modification strategies to achieve immune protection for specific types of transplantable tissues.
Supplementary Material
Acknowledgments
We thank Christian Freund (hiPSC core facility, LUMC, Leiden, the Netherlands) for providing hiPSCs (LUMC0072iCTRL01), the HLA laboratory at the LUMC for the HLA typing, and Rosa van Amerongen and Mirjam Heemskerk for providing T cells. We acknowledge the technical support of Angela Koudijs and Anneloes Verwey and thank Manon Zuurmond for her illustrations. We thank the Light and Electron Microscopy Facility, the Flow Cytometry Core Facility and the Central Animal Facility for their technical support (all LUMC).
Contributor Information
Lonneke H Gaykema, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands; Department of Cell and Chemical Biology, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Rianne Y van Nieuwland, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Ellen Lievers, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Wessel B J Moerkerk, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Juliette A de Klerk, Department of Cell and Chemical Biology, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Sébastien J Dumas, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands; The Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW), Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Jesper Kers, Department of Pathology, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Arnaud Zaldumbide, Department of Cell and Chemical Biology, Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Cathelijne W van den Berg, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands; The Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW), Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Ton J Rabelink, Department of Internal Medicine (Nephrology) & Einthoven Laboratory of Vascular and Regenerative Medicine, Leiden University Medical Center (LUMC), Leiden, The Netherlands; The Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW), Leiden University Medical Center (LUMC), Leiden, The Netherlands.
Funding
This study was funded by LUF/Stichting Prof. Jaap de Graeff-Lingling Wiyadharma Fonds 2020-01 and the Novo Nordisk Foundation Center for Stem Cell Medicine (reNEW, supported by Novo Nordisk Foundation grants (NNF21CC0073729)).
Conflict of Interest
None declared.
Author Contributions
L.G. designed and performed experiments analyzed results and wrote the manuscript. R.N., E.L. and W.M. collected data. J.A.K. and S.D. performed analyses and created figures of RNAseq experiments. J.K. advised the project and aided in the interpretation of results. C.B. and A.Z. supervised the project and wrote the manuscript. T.R. arranged funding, supervised the project and wrote the manuscript. All authors have read and agreed to the submitted version of the manuscript.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Barker CF, Markmann JF.. Historical overview of transplantation. Cold Spring Harb Perspect Med. 2013;3(4):a014977. 10.1101/cshperspect.a014977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takasato M, Er PX, Chiu HS, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526(7574):564-568. 10.1038/nature15695 [DOI] [PubMed] [Google Scholar]
- 3. van den Berg CW, Ritsma L, Avramut MC, et al. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Rep. 2018;10(3):751-765. 10.1016/j.stemcr.2018.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morizane R, Bonventre JV.. Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nat Protoc. 2017;12(1):195-207. 10.1038/nprot.2016.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van den Berg CW, Koudijs A, Ritsma L, Rabelink TJ.. In vivo assessment of size-selective glomerular sieving in transplanted human induced pluripotent stem cell-derived kidney organoids. J Am Soc Nephrol. 2020;31(5):921-929. 10.1681/asn.2019060573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bantounas I, Ranjzad P, Tengku F, et al. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Rep. 2018;10(3):766-779. 10.1016/j.stemcr.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharmin S, Taguchi A, Kaku Y, et al. Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J Am Soc Nephrol. 2016;27(6):1778-1791. 10.1681/ASN.2015010096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mandai M, Watanabe A, Kurimoto Y, et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl J Med. 2017;376(11):1038-1046. 10.1056/NEJMc1706274 [DOI] [PubMed] [Google Scholar]
- 9. Sugita S, Mandai M, Hirami Y, et al. HLA-matched allogeneic iPS cells-derived RPE transplantation for macular degeneration. J Clin Med. 2020;9(7):2217. 10.3390/jcm9072217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schweitzer JS, Song B, Herrington TM, et al. Personalized iPSC-derived dopamine progenitor cells for Parkinson’s disease. N Engl J Med. 2020;382(20):1926-1932. 10.1056/nejmoa1915872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Streilein JW. Ocular immune privilege: therapeutic opportunities from an experiment of nature. Nat Rev Immunol. 2003;3(11):879-889. 10.1038/nri1224 [DOI] [PubMed] [Google Scholar]
- 12. Proulx ST, Engelhardt B.. Central nervous system zoning: how brain barriers establish subdivisions for CNS immune privilege and immune surveillance. J Intern Med. 2022;292(1):47-67. 10.1111/joim.13469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lopez MLSS, Gomez RA.. Development of the renal arterioles. J Am Soc Nephrol. 2011;22(12):2156-2165. 10.1681/ASN.2011080818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yatim KM, Gosto M, Humar R, Williams AL, Oberbarnscheidt MH.. Renal dendritic cells sample blood-borne antigen and guide T-cell migration to the kidney by means of intravascular processes. Kidney Int. 2016;90(4):818-827. 10.1016/j.kint.2016.05.030 [DOI] [PubMed] [Google Scholar]
- 15. Hughes AD, Lakkis FG, Oberbarnscheidt MH.. Four-dimensional imaging of T cells in kidney transplant rejection. J Am Soc Nephrol. 2018;29(6):1596-1600. 10.1681/asn.2017070800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park JG, Na M, Kim MG, et al. Immune cell composition in normal human kidneys. Sci Rep. 2020;10(1):15678. 10.1038/s41598-020-72821-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DeWolf S, Grinshpun B, Savage T, et al. Quantifying size and diversity of the human T cell alloresponse. JCI Insight. 2018;3(15):e121256. 10.1172/jci.insight.121256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suchin EJ, Langmuir PB, Palmer E, et al. Quantifying the frequency of alloreactive T cells in vivo: new answers to an old question. J Immunol. 2001;166(2):973-981. 10.4049/jimmunol.166.2.973 [DOI] [PubMed] [Google Scholar]
- 19. Zachary AA, Leffell MS.. HLA mismatching strategies for solid organ transplantation – a balancing act. Front Immunol. 2016;7:575. 10.3389/fimmu.2016.00575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Snanoudj R, Kamar N, Cassuto E, et al. Epitope load identifies kidney transplant recipients at risk of allosensitization following minimization of immunosuppression. Kidney Int. 2019;95(6):1471-1485. 10.1016/j.kint.2018.12.029 [DOI] [PubMed] [Google Scholar]
- 21. Sahin GK, Unterrainer C, Susal C.. Critical evaluation of a possible role of HLA epitope matching in kidney transplantation. Transplant Rev. 2020;34(2):100533. 10.1016/j.trre.2020.100533 [DOI] [PubMed] [Google Scholar]
- 22. Senev A, Emonds M-P, Van Sandt V, et al. Clinical importance of extended second field high-resolution HLA genotyping for kidney transplantation. Am J Transplant. 2020;20(12):3367-3378. 10.1111/ajt.15938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tambur AR, Das R.. Can we use Eplets (or molecular) mismatch load analysis to improve organ allocation? The hope and the hype. Transplantation. 2023;107(3):605-615. 10.1097/tp.0000000000004307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaykema LH, van Nieuwland RY, Dekkers MC, et al. Inhibition of complement activation by CD55 overexpression in human induced pluripotent stem cell derived kidney organoids. Front Immunol. 2022;13:1058763. 10.3389/fimmu.2022.1058763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu H, Wang B, Ono M, et al. Targeted disruption of HLA genes via CRISPR-Cas9 generates iPSCs with enhanced immune compatibility. Cell Stem Cell. 2019;24(4):566-578.e7. 10.1016/j.stem.2019.02.005 [DOI] [PubMed] [Google Scholar]
- 26. Han X, Wang M, Duan S, et al. Generation of hypoimmunogenic human pluripotent stem cells. Proc Natl Acad Sci USA. 2019;116(21):10441-10446. 10.1073/pnas.1902566116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deuse T, Hu X, Gravina A, et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol. 2019;37(3):252-258. 10.1038/s41587-019-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. King M, Pearson T, Shultz LD, et al. A new Hu-PBL model for the study of human islet alloreactivity based on NOD-scid mice bearing a targeted mutation in the IL-2 receptor gamma chain gene. Clin Immunol. 2008;126(3):303-314. 10.1016/j.clim.2007.11.001 [DOI] [PubMed] [Google Scholar]
- 29. Chen XY, Rinsma M, Janssen JM, et al. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44(13):6482-6492. 10.1093/nar/gkw524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koning M, Dumas SJ, Avramut MC, et al. Vasculogenesis in kidney organoids upon transplantation. NPJ Regen Med. 2022;7(1):40. 10.1038/s41536-022-00237-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Amerongen RA, Hagedoorn RS, Remst DFG, et al. WT1-specific TCRs directed against newly identified peptides install antitumor reactivity against acute myeloid leukemia and ovarian carcinoma. J Immunother Cancer. 2022;10(6):e004409. 10.1136/jitc-2021-004409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heemskerk MHM, Hagedoorn RS, van der Hoorn MAWG, et al. Efficiency of T-cell receptor expression in dual-specific T cells is controlled by the intrinsic qualities of the TCR chains within the TCR-CD3 complex. Blood. 2007;109(1):235-243. 10.1182/blood-2006-03-013318 [DOI] [PubMed] [Google Scholar]
- 33. Amir AL, van der Steen DM, Hagedoorn RS, et al. Allo-HLA-reactive T cells inducing graft-versus-host disease are single peptide specific. Blood. 2011;118(26):6733-6742. 10.1182/blood-2011-05-354787 [DOI] [PubMed] [Google Scholar]
- 34. van Amerongen RA, Morton LT, Chaudhari UG, et al. Human iPSC-derived preclinical models to identify toxicity of tumor-specific T cells with clinical potential. Mol Ther Methods Clin Dev. 2023;28:249-261. 10.1016/j.omtm.2023.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu HJ, Uchimura K, Donnelly EL, et al. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23(6):869-881.e8. 10.1016/j.stem.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ajith A, Mulloy LL, Musa MA, et al. Humanized mouse model as a novel approach in the assessment of human allogeneic responses in organ transplantation. Front Immunol. 2021;12. 10.3389/fimmu.2021.687715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riolobos L, Hirata RK, Turtle CJ, et al. HLA engineering of human pluripotent stem cells. Mol Ther. 2013;21(6):1232-1241. 10.1038/mt.2013.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang D, Quan Y, Yan Q, Morales JE, Wetsel RA.. Targeted disruption of the β2-microglobulin gene minimizes the immunogenicity of human embryonic stem cells. Stem Cells Trans Med. 2015;4(10):1234-1245. 10.5966/sctm.2015-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kooreman NG, de Almeida PE, Stack JP, et al. Alloimmune responses of humanized mice to human pluripotent stem cell therapeutics. Cell Rep. 2017;20(8):1978-1990. 10.1016/j.celrep.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flahou C, Morishima T, Takizawa H, Sugimoto N.. Fit-for-all iPSC-derived cell therapies and their evaluation in humanized mice with NK cell immunity. Front Immunol. 2021;12:662360. 10.3389/fimmu.2021.662360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scott EN, Gocher AM, Workman CJ, Vignali DAA.. Regulatory T cells: barriers of immune infiltration into the tumor microenvironment. Front Immunol. 2021;12:702726. 10.3389/fimmu.2021.702726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu P, Chen J, He L, et al. Generating hypoimmunogenic human embryonic stem cells by the disruption of beta 2-microglobulin. Stem Cell Rev Rep. 2013;9(6):806-813. 10.1007/s12015-013-9457-0 [DOI] [PubMed] [Google Scholar]
- 43. Gornalusse GG, Hirata RK, Funk SE, et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. 2017;35(8):765-772. 10.1038/nbt.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dekel B, Burakova T, Ben-Hur H, et al. Engraftment of human kidney tissue in rat radiation chimera II Human fetal kidneys display reduced immunogenicity to adoptively transferred human peripheral blood mononuclear cells and exhibit rapid growth and development. Transplantation. 1997;64(11):1550-1558. 10.1097/00007890-199712150-00008 [DOI] [PubMed] [Google Scholar]
- 45. Dekel B, Marcus H, Herzel B-H, et al. In vivo modulation of the allogeneic immune response by human fetal kidneys – the role of cytokines, chemokines, and cytolytic effector molecules. Transplantation. 2000;69(7):1470-1478. 10.1097/00007890-200004150-00044 [DOI] [PubMed] [Google Scholar]
- 46. Rossbach B, Hariharan K, Mah N, et al. Human iPSC-derived renal cells change their immunogenic properties during maturation: implications for regenerative therapies. Cells. 2022;11(8):1328. 10.3390/cells11081328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bishop GA, Hall BM, Duggin GG, et al. Immunopathology of renal-allograft rejection analyzed with monoclonal-antibodies to mononuclear cell markers. Kidney Int. 1986;29(3):708-717. 10.1038/ki.1986.56 [DOI] [PubMed] [Google Scholar]
- 48. Wuthrich RP, Glimcher LH, Yui MA, et al. Mhc Class-Ii, antigen presentation and tumor necrosis factor in renal tubular epithelial-cells. Kidney Int. 1990;37(2):783-792. 10.1038/ki.1990.46 [DOI] [PubMed] [Google Scholar]
- 49. Muczynski KA, Cotner T, Anderson SK.. Unusual expression of human lymphocyte antigen class II in normal renal microvascular endothelium. Kidney Int. 2001;59(2):488-497. 10.1046/j.1523-1755.2001.059002488.x [DOI] [PubMed] [Google Scholar]
- 50. Eleftheriadis T, Pissas G, Crespo M, Nikolaou E, Liakopoulos V, Stefanidis I., A role for human renal tubular epithelial cells in direct allo-recognition by CD4+ T-cells and the effect of ischemia–reperfusion. Int J Mol Sci. 2021;22(4):1733. 10.3390/ijms22041733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao L, Teklemariam T, Hantash BM.. Heterelogous expression of mutated HLA-G decreases immunogenicity of human embryonic stem cells and their epidermal derivatives. Stem Cell Res. 2014;13(2):342-354. 10.1016/j.scr.2014.08.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.


