Abstract
Background
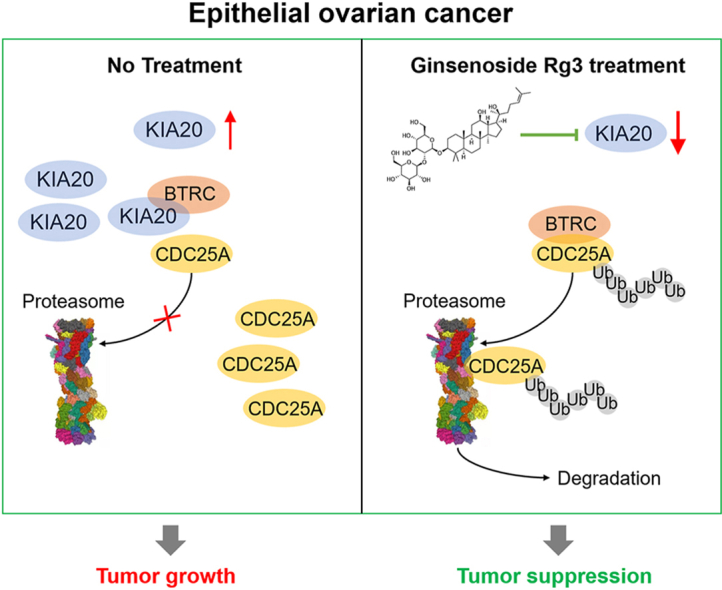
Ginsenoside 20(S)-Rg3 shows promising tumor-suppressive effects in ovarian cancer via inhibiting NF-κB signaling. This study aimed to explore the downstream tumor suppressive mechanisms of ginsenoside Rg3 via this signaling pathway.
Materials and methods
A systematical screening was applied to examine the expression profile of 41 kinesin family member genes in ovarian cancer. The regulatory effect of ginsenoside Rg3 on KIF20A expression was studied. In addition, we explored interacting proteins of KIF20A and their molecular regulations in ovarian cancer. RNA-seq data from The Cancer Genome Atlas (TCGA) was used for bioinformatic analysis. Epithelial ovarian cancer cell lines SKOV3 and A2780 were used as in vitro and in vivo cell models. Commercial human ovarian cancer tissue arrays were used for immunohistochemistry staining.
Results
KIF20A is a biomarker of poor prognosis among the kinesin genes. It promotes ovarian cancer cell growth in vitro and in vivo. Ginsenoside Rg3 can suppress the transcription of KIF20A. GST pull-down and co-immunoprecipitation (IP) assays confirmed that KIF20A physically interacts with BTRC (β-TrCP1), a substrate recognition subunit for SCFβ−TrCP E3 ubiquitin ligase. In vitro ubiquitination and cycloheximide (CHX) chase assays showed that via interacting with BTRC, KIF20A reduces BTRC-mediated CDC25A poly-ubiquitination and enhances its stability. Ginsenoside Rg3 treatment partly abrogates KIF20A overexpression-induced CDC25A upregulation.
Conclusion
This study revealed a novel anti-tumor mechanism of ginsenoside Rg3. It can inhibit KIF20A transcription and promote CDC25A proteasomal degradation in epithelial ovarian cancer.
Keywords: ginsenoside Rg3, ovarian cancer, KIF20A, CDC25A, ubiquitination
Graphical abstract
1. Introduction
Epithelial ovarian cancer (EOC) is a fatal malignancy in women. It acts as a "silent killer" since the symptoms are only noticeable at relatively late clinical stages (“silent”) and a high 5-year mortality rate ("killer") [1]. Most EOCs are highly sensitive to paclitaxel + platinum combination chemotherapy during initial treatment, but recurrence is inevitable due to the development of chemoresistance [2,3]. In the past decade, the application of Poly (ADP-ribose) polymerase (PARP) inhibitors significantly prolonged progression-free survival (PFS) in patients with Breast Cancer-Associated 1 and 2 (BRCA1/2) mutant ovarian cancer [4]. However, the BRCA1/2 mutation rate is around 16% to 24% in ovarian cancer patients in China [5,6]. Therefore, PARP inhibitors might not bring clinical benefits for more than 3/4 of ovarian cancer patients without BRCA mutations. The major mechanism of paclitaxel and platinum drugs is to promote microtubular aggregation and interfere with cell mitosis, transport and motility [7]. Therefore, an in-depth exploration of the biological characteristics of ovarian cancer cells in the process of mitosis is critical to overcome chemoresistance.
Ginsenoside 20(S)-Rg3 (ginsenoside Rg3) shows promising in vitro and in vivo tumor-suppressive effects in multiple cancers [[8], [9], [10]]. Ginsenoside Rg3 could induce cancer cell apoptosis [11,12], including ovarian cancer cells [13,14]. Besides, in ovarian cancer, it can inhibit hypoxia-induced epithelial-mesenchymal transition [15], indue autophagy [16], inhibit the Warburg effect [17], and suppress the expression of multiple tumor-promoting genes [17,18]. Therefore, it might be a potential candidate for adjuvant treatment. A clear understanding of the mechanisms underlying its anti-tumor effects would help promote its clinical application.
Mitosis is a highly regulated, highly ordered process. The precise progression of cytocentrosome isolation, spindle filament formation, chromosome collection, and cytoplasmic division requires the kinesin superfamily proteins (KIFs), a set of molecular motors [19]. The dysregulation of KIF proteins plays a key role in the development of human cancers [20,21]. In ovarian cancer, KIF14 expression is an independent biomarker of poor prognosis [22]. Inhibiting KIF5, an essential factor for spindle assembly, can suppress ovarian cancer growth [23]. KIF20A upregulation is associated with unfavorable clinical outcomes and tumor progression [24,25].
In this study, we systematically screened the KIF family member genes and confirmed KIF20A as a biomarker of poor prognosis and a functional tumor-promoting gene. Our bioinformatic analysis revealed that ginsenoside Rg3 might regulate KIF20A expression. Therefore, we further examined the effects of ginsenoside Rg3 on KIF20A transcription, the interacting proteins of KIF20A, and their molecular regulations in ovarian cancer.
2. Materials and methods
2.1. Bioinformatic analysis
RNA-seq data of 41 KIF family members from primary ovarian cancer cases in the cancer genome atlas (TCGA)-ovarian cancer (OV) (N = 419) and normal ovary and fallopian tube (N = 93) from the Genotype-Tissue Expression (GTEx) were extracted using the combined GTEx and TCGA dataset in UCSC Xena browser (https://xenabrowser.net/) [26]. Immunohistochemistry (IHC) staining of KIF20A expression was checked using data from the Human Protein Atlas (HPA) (https://www.proteinatlas.org/) [27]. In this database, the protein expression score is determined by manually scoring immunohistochemical data for staining intensity and the fraction of stained cells. The staining intensity is classified as negative, weak, moderate, or strong, while the fraction of stained cells is categorized as <25%, 25-75%, or >75%. Each combination of intensity and fraction is then automatically converted into a protein expression level score. A negative score indicates that the protein was not detected, while a weak score (<25%) also indicates that the protein was not detected. A weak score combined with 25-75% or 75% is classified as low. A moderate score (<25%) is classified as low, while a moderate score combined with either 25-75% or 75% is classified as medium. A strong score (<25%) is classified as medium, while a strong score combined with either 25-75% or 75% is classified as high (https://www.proteinatlas.org/about/help#4). The association between KIF20A expression and the survival outcomes of patients with primary ovarian cancer was assessed using the Kaplan-Meier plotter (http://kmplot.com/analysis/index.php?p=service&cancer=ovar), using the optimal cutoff [28].
2.2. Cell culture and treatment
Human ovarian surface epithelial (HOSE) cells and epithelial ovarian cancer cell lines (SKOV3, A2780, OV90, and OVCAR3) were cultured following the methods introduced previously [29]. Lentiviral KIF20A shRNAs (shKIF20A) were constructed by HanBio Technology, based on pLKO.1-puro plasmid. The following sequences were used: shKIF20A#1, 5′-CGTACACCATTCAAGGTACTA-3′, shKIF20A#2, 5′-GCTAGATGAAACAAGTCAATT-3′, shKIF20A#3, 5′- GCCACTCACAAATTTACCTTT-3’. Lentiviruses expressing myc-tagged BTRC (myc-BTRC), flag-tagged KIF20A (flag-KIF20A), and RELA (NF-κB p65) were generated based on the pLenti-puro backbone. pLenti puro HA-Ubiquitin (HA-Ub) (Plasmid #74218) was obtained from Addgene (Cambridge, MA, USA). Lentivirus for infection was produced by co-transfecting with the lentivirus packaging plasmids (pSPAX2 and pMD2.G, HanBio Technology) in 293T cells, as we previously described [30]. Cells were infected at a multiplicity of infection (MOI) of 15, with the presence of 6 μg/ml polybrene. Global protein translation inhibitor (cycloheximide, CHX) and the proteasome inhibitor MG132 were obtained from Selleck Chemicals. Ginsenoside Rg3 (purity>98%) was purchased from HerbSubstance (Chengdu, China).
2.3. Reverse transcription quantitative PCR (RT-qPCR)
In brief, cDNA was reversely transcribed using RNA samples extracted from cells and then used as the PCR assay template. Relative gene expression was normalized to GAPDH, by the 2−ΔΔCT method. The primers were as follows: KIF20A, forward: 5′-CAAGAGGCAGACTTTGCGGCTA-3′ and reverse: 5′-GCTCTGGTTCTTACGACCCACT-3’; RELA, forward: 5′-TGAACCGAAACTCTGGCAGCTG-3′ and reverse: 5′-CATCAGCTTGCGAAAAGGAGCC-3’; GAPDH, forward: 5′-GTCTCCTCTGACTTCAACAGCG-3′, and reverse, 5′-ACCACCCTGTTGCTGTAGCCAA-3’.
2.4. Western blotting
Cell samples were harvested, washed, and lysed using a lysis buffer containing protease inhibitors. After centrifugation, the supernatant was collected to determine protein concentration using a BCA kit (Beyotime). 30 μg protein samples were loaded into each lane. The following antibodies and dilutions were used, including anti-KIF20A (1:1000, 15911-1-AP, Proteintech, Wuhan, China), anti-BTRC (1:1000, 37-3400, ThermoFisher Scientific, Waltham, MA, USA), anti-Flag tag (1:5000, 66008-4-Ig, Proteintech), anti-CDC25A (1:1000, 55031-1-AP, Proteintech), anti-HA tag (1:5000, 51064-2-AP, Proteintech), anti-Myc tag (1:3000, 16286-1-AP, Proteintech), and anti-β-actin (sc-47778, Santa Cruz Biotechnology, Santa Cruz, CA, USA). HRP-conjugated secondary antibodies were used. Then, the signals were developed using the BeyoECL Star reagent (Beyotime) and a LAS-4000 imaging system (GE Healthcare, Piscataway, NJ, USA).
2.5. CCK-8 assays, colony formation, and cell cycle detection
48 h after lentiviral infections, SKOV3 and A2780 cells were seeded into 96-well plates at a density of 2,000 cells per well and were incubated for 24, 48, and 72 h. At each time point, the cell viability was measured using a CCK-8 Kit (Beyotime, Shanghai, China). 10 μl of CCK-8 reagent was added to each well and incubated for 2 h. Then, absorbance at 450 nm was measured using a microplate reader.
For colony formation assays, SKOV3 and A2780 cells with or without KIF20A knockdown were seeded in 6-well plates (1000 cells/per well) and cultured for 10-14 days. Then, the plates were washed, and the colonies were fixed with 4% formaldehyde and stained with 1.0% crystal violet.
Cell cycle distribution was assessed by Propidium Iodide (PI) staining. In brief, 48 h after lentiviral infection, cells were collected and washed twice with PBS and fixed with 70% ethanol at 4°C for 1h. Then, cells were resuspended in PBS with 50 μg/mL PI and 100 μg/mL RNase A. The cells were incubated at room temperature in the dark for 30 min. Then, cell cycle distribution was detected using flow cytometry (Novocyte Advanteon, Agilent, Santa Clara, CA, USA) and analyzed using NovoExpress software (version 1.5.4, Agilent).
2.6. Nude mouse xenograft model
Female Balb/c nude mice aged 4-6 weeks were purchased from Vital River Laboratory Animal Technology (Beijing, China). All animal protocols used were approved by the Institutional Animal Care and Use Committee of Shanxi Provincial People's Hospital, China (approval no. 2022-057). All animal housing and experiments were conducted in strict accordance with the institutional Guidelines for the Care and Use of Laboratory Animals. Mice were randomly divided into four groups (n = 6 per group). Approximately 2 × 106 of A2780 or SKOV3 cells with or without KIF20A knockdown or SKOV3 cells with KIF20A overexpression were inoculated subcutaneously in the left armpit of each mouse. For the ginsenoside Rg3 treatment group, Ginsenoside Rg3 was administered at 20 mg/ kg body weight to mice 3 times per week for 4 weeks via intraperitoneal injection. Tumors were measured every 3 days with a caliper. About 4 weeks after inoculation, xenograft tumors were removed, weighed, fixed, and sectioned. IHC staining was performed to check Ki-67 expression in tumor sections.
2.7. Glutathione-S-transferase (GST) pull-down and co-immunoprecipitation (IP) assays
The genes encoding BTRC, FBXW11, and FBXW7 were subcloned in pGEX-6p-1 between the EcoRI and XhoI restriction sites. GST-fusion proteins were expressed in E. coli BL21 (DE3). In brief, the ligation mixture was transformed into competent E. coli cells and plated on LB agar plates containing ampicillin. A single colony was picked and grown in LB broth containing ampicillin. When the OD600 of the LB medium reached 0.6-0.8, protein expression was induced by adding IPTG to a final concentration of 0.1 mM and continuing to incubate for 4 h at 37°C with shaking. The cells were harvested by centrifugation at 4,000 × g for 10 min at 4°C, resuspended in lysis buffer containing protease inhibitors, and lysed by sonication. The lysates were centrifugated at 12,000 × g for 15 min at 4°C to remove cell debris. The supernatant was collected and incubated with Glutathione-Sepharose beads for 1 h at 4°C with gentle shaking. Then, the beads were washed several times with lysis buffer to remove non-specifically bound proteins. The GST-tagged proteins were eluted from the beads by adding an elution buffer. Then, GST pull-down assays were performed using Pierce GST Protein Interaction Pull-Down Kit (ThermoFisher Scientific), following the protocol of the kit. The bait GST-tagged proteins were immobilized on the pierce spin column with equilibrated glutathione agarose. The supernatant from HEK293T cells overexpressing flag-tagged KIF20A (as prey protein) was prepared and added to the pierce spin column containing the immobilized GST-tagged proteins and then incubated at 4°C for 4 h with gentle rocking. Then, the column was washed several times and the bait-prey proteins were eluted with the glutathione elution buffer. The eluted samples were subjected to western blotting assays. Co-IP assay was performed as we previously described [30], using mouse anti-BTRC (37-3400, ThermoFisher Scientific), rabbit anti-KIF20A (15911-1-AP, Proteintech), and protein A agarose (sc-2001, Santa Cruz).
2.8. Immunofluorescent staining
Immunofluorescent staining was performed following the methods introduced previously [31]. In brief, cells were grown on coverslips. After about 50% confluence, cells were fixed and permeabilized. Then, the cells were washed and incubated with primary antibody against BTRC and KIF20A (mouse anti-BTRC, 1:200, 37-3400, ThermoFisher Scientific and rabbit anti-KIF20A, 1: 200, 15911-1-AP, Proteintech) at 4 °C overnight. The CoraLite594-conjugated Goat Anti-Mouse IgG(H + L) and CoraLite488-conjugated Goat Anti-Rabbit IgG(H + L) (Proteintech) were used as the secondary antibodies. Nuclei were stained with DAPI. Immunofluorescent images were acquired using IX83 (Olympus, Tokyo, Japan).
2.9. Statistical analysis
Welch's t-test was conducted to check the statistical difference between the two groups. One-way ANOVA with post-hoc Tukey test was performed for multiple-group comparisons. Log-rank test was performed in Kaplan-Meier (K-M) survival analysis. Pearson's r or Spearman's rho values were calculated to estimate correlations. p < 0.05 was considered statistically significant.
3. Results
3.1. Systematic screening identifies a link between KIF20A upregulation and unfavorable prognosis of ovarian cancer
To explore dysregulated KIF genes, we extracted RNA-seq data of 41 KIF family members from primary ovarian cancer cases in TCGA-OV (N = 419) and normal ovary and fallopian tube (N = 93) from GTEx (Fig. S1A). The dysregulated genes were identified by the following criteria: serous EOC (SEOC) vs. normal (log2 fold change ≥2) and adj. p < 0.05. After the screening, 14 candidates were identified, including KIF18B, KIF20A, KIF14, KIF11, KIF18A, KIF2C, KIF15, KIFC1, KIF1A, KIF23, KIF12, KIF26B, KIF6, and NEK6 (Fig. S1B). K-M survival analysis using data from the Kaplan-Meier plotter confirmed that high KIF20A expression was associated with significantly worse PFS (HR: 1.35, 95%CI: 1.12-1.63, p = 0.0015) and OS (HR: 1.25, 95%CI: 1.06-1.46, p = 0.006) (Fig. 1A–B). To explore KIF20A expression at the protein level, we conducted IHC staining using a commercial ovarian cancer tissue assay. Results confirmed medium to high KIF20A expression, mainly in the nucleus (Fig. 1C).
Fig. 1.
Systematic screening identifies a link between KIF20A upregulation and unfavorable prognosis of ovarian cancer, A-B. K-M plotter was generated to compare the difference in PFS (C) and OS (D) between patients with high and low KIF20A expression with survival data from the Kaplan-Meier plotter. C. Representative images of KIF20A expression in commercial ovarian cancer tissue arrays. Enlarged tissue areas were provided to show the subcellular distribution of KIF20A.
Then, we examined the IHC staining of KIF20A in the Human Protein Atlas (HPA) [27]. In the HPA, KIF20A was examined in 12 ovarian cancer tissues using two antibodies (HPA036909 and HPA036010) (Fig. S2). Medium to high expression of KIF20A was observed in most of the tumor tissues (Fig. S2). Subcellular analysis showed that KIF20A is mainly distributed in the nucleus, with minor distribution in the cytoplasm (Fig. S2, top panels, enlarged areas).
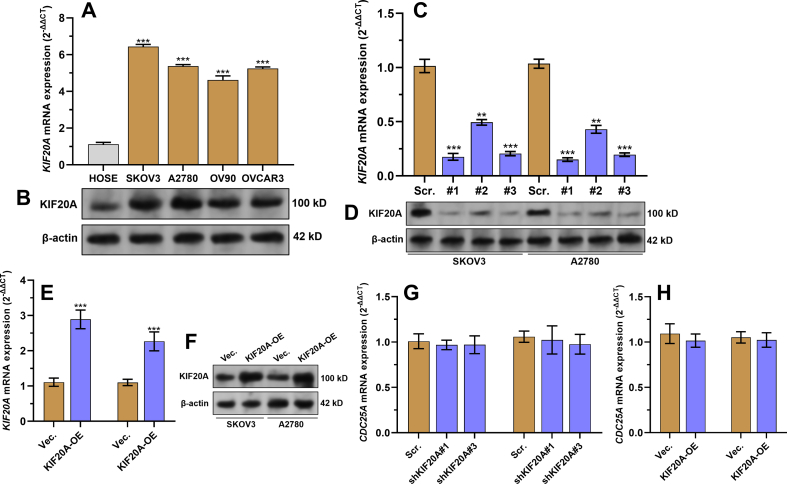
3.2. Knockdown of KIF20A impairs ovarian cancer cell proliferation in vitro
KIF20A expression in normal human ovarian surface epithelial (HOSE) cells and multiple ovarian cancer cell lines, including SKOV3, A2780, OV90, and OVCAR3, were examined. Results showed that the tumor cell lines had significantly elevated KIF20A expression at the mRNA and protein levels than HOSE cells (Fig. S3A-B). SKOV3 and A2780 were selected as representative cell lines for further studies. These two cell lines were subjected to KIF20A knockdown or overexpression using lentiviruses (Fig. S1C-F). CCK-8 (Fig. 2A–B), colony formation (Fig. 2E–F), and flow cytometric analysis (Fig. 2H–J) showed that KIF20A knockdown significantly reduced cell viability, colony formation, cells in the S phase, and increased G1 phase arrest of SKOV3 and A2780 cells (Fig. 2E–H). In comparison, KIF20A overexpression significantly enhanced proliferation and cell cycle progression (Fig. 2A–J).
Fig. 2.
Knockdown of KIF20A impairs ovarian cancer cell proliferation, A-D. CCK-8 assays showing the effects of KIF20A knockdown (A-B) or overexpression (C-D) on the viability of SKOV3 (A and C) and A2780 (B and D) cells. E-I. Colony formation (E-G) and flow cytometric analysis (H–I) were performed to show the effect of KIF20A knockdown or overexpression on colony formation and cell-cycle distribution of SKOV3 and A2780 cells. Data are expressed as mean ± SD. ∗, comparison between scramble (scr.) control and shKIF20A. # comparison between vector (vec.) and KIF20A-OE. ## and∗∗p < 0.01, ### and ∗∗∗p < 0.001.
3.3. Ginsenoside Rg3 suppresses KIF20A expression in ovarian cancer cells
By performing KIF20A promoter scanning, we observed high-potential binding sites of NF-κB p65/RELA (Figure S4, bold green nucleobases). Therefore, we further checked the expressional correlation between KIF20A and RELA in primary ovarian cancer cases in TCGA. Correlation analysis confirmed a moderate positive correlation (Pearson's r = 0.50) (Fig. 3A). Ginsenoside Rg3 has well-documented effects on inhibiting the activity of NF-κB p65 via suppressing its phosphorylation [[32], [33], [34]]. Therefore, we explored its effects on KIF20A expression in ovarian cancer cells. As expected, it suppressed KIF20A expression dose-dependently (Fig. 3B–C). To validate this finding, we overexpressed RELA in SKOV3 and A2780 cells (Fig. 3D–F). RELA overexpression was associated with increased NF-κB p65 phosphorylation (Fig. 3F–H) and increased KIF20A expression at the mRNA (Fig. 3D–E) and protein levels (Fig. 3F–H). Ginsenoside Rg3 treatment significantly weakened RELA overexpression-induced KIF20A upregulation (Fig. 3D–H).
Fig. 3.
Ginsenoside Rg3 suppresses KIF20A expression in ovarian cancer cells, A. A plot chart showing the expressional correlation between KIF20A and RELA in primary tumor cases in TCGA-OV. B–C. QRT-PCR (B) and western blotting assay (C) were performed to detect the expression of KIF20A in SKOV3 and A2780 cells 48 h after ginsenoside Rg3 treatment. D-H. QRT-PCR (D-E) and western blotting assay (F–H) were performed to detect the expression of RELA and KIF20A in SKOV3 and A2780 cells with RELA overexpression alone or in combination with ginsenoside Rg3 treatment (50 ng/ml for 48 h). ∗∗∗p < 0.001.
3.4. KIF20A directly interacts with BTRC
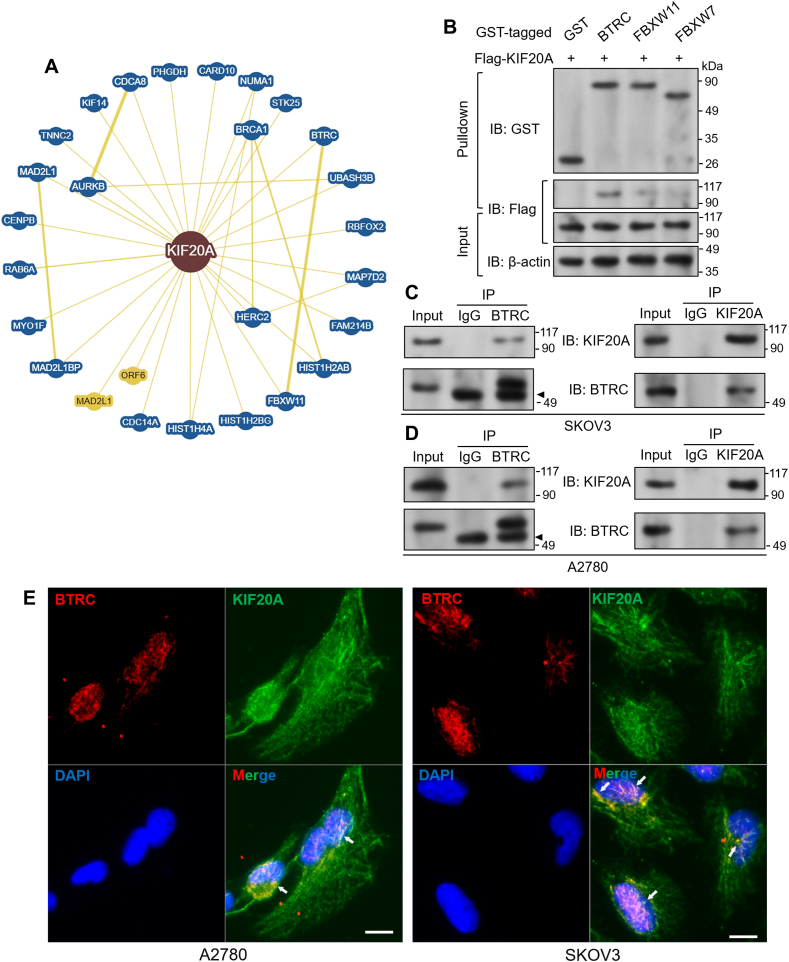
To explore the potential functional regulation of KIF20A, we checked its interacting proteins using BioGRID (https://thebiogrid.org/). Only the proteins with minimum evidence ≥2 (at least two previous experimental evidence) were identified (Fig. 4A). Among the candidates, BTRC (also known as β-TrCP1) and FBXW11 (β-TrCP2) were two potential candidates (Fig. 4A). BTRC serves as the substrate recognition subunit for SCFβ−TrCP E3 ubiquitin ligase, which specifically ubiquitinates phosphorylated substrates [35]. This E3 ligase exerts critical regulations on cell cycle progression and tumorigenesis by controlling the degradation of multiple essential cell cycle regulators [36].
Fig. 4.
KIF20A directly interacts with BTRC, A. Bioinformatic prediction of KIF20A interacting proteins. Prediction was performed using BioGRID. Only the proteins with minimum evidence ≥2 were identified as the candidates. B. Binding affinity of flag-KIF20A to the predicted E3 ligases. After various GST-fused proteins were expressed, a GST pull-down assay was performed. C-D. Co-IP was performed to show the interaction between KIF20A and BTRC at the endogenous level in SKOV3 (C) and A2780 (D) cells. IP was conducted with an anti-BTRC or anti-KIF20A antibody. Mouse IgG served as a negative control. Arrowheads: IgG heavy chain bands. E. Immunofluorescent staining was performed to visualize BTRC (red) and KIF20A (green) in SKOV3 and A2780 cells. Scale bar: 10 μm.
To validate the specificity of the potential interactions, we generated recombinant pGEX-GST vectors expressing BTRC, FBXW11, and, another well-characterized F-box protein FBXW7. GST-fusion proteins were expressed in E. coli BL21 (DE3). Flag-KIF20A was expressed in 293T cells. Then, a GST pull-down assay was performed. Results showed that KIF20A exhibited a weak binding to FBXW11 and FBXW7 and a strong affinity to BTRC (Fig. 4B). The interaction between KIF20A and BTRC was also validated at the endogenous level. In the cell lysates from SKOV3 and A2780 cells, BTRC was identified in the KIF20A enriched fraction, and KIF20A was co-isolated with BTRC reciprocally (Fig. 4C–D). Immunofluorescent staining confirmed the co-localization between endogenous BTRC and KIF20A in the nucleus (Fig. 4E).
3.5. Ginsenoside Rg3 promotes CDC25A proteasomal degradation via suppressing KIF20A expression
Since we validated the interaction between KIF20A and BTRC, we hypothesized that KIF20A might exert specific regulation on the SCFβ−TrCP−1 complex. To verify this hypothesis, we examined the expression of cell division cycle 25 A (CDC25A), a well-known SCFβ−TrCP−1 substrate [37] in SKOV3 cells. QRT-PCR assays confirmed that KIF20A knockdown or overexpression did not alter CDC25A transcription (S1 Fig. G-H). Western blotting data showed that KIF20A knockdown did not alter BTRC expression but reduced the expression of CDC25A (Fig. 5A, top panel). However, when MG132 was added, KIF20A knockdown could not induce CDC25A downregulation (Fig. 5A, bottom panel), suggesting that KIF20A might affect the proteasomal degradation of CDC25A. Therefore, we assessed the effect of KIF20A on SCFβ−TrCP−1 ubiquitin ligase activity for CDC25A. In brief, SKOV3 cells were infected with lentiviruses with selective overexpression of HA-ubiquitin (HA-Ub), Myc-BTRC, and/or flag-KIF20A (Fig. 5B). The ubiquitination of CDC25A was checked after IP using an anti-CDC25A antibody. The myc-BTRC group had a higher level of high molecular-weight smeared bands of CDC25A, indicating increased CDC25A poly-ubiquitination (Fig. 5B). KIF20A overexpression attenuated CDC25A poly-ubiquitination (Fig. 5B). In comparison, its knockdown increased CDC25A poly-ubiquitination (Fig. 5C). CHX chase assay showed that KIF20A overexpression increased the stability of CDC25A (Fig. 5D–E). KIF20A knockdown shortened the half-life of CDC25A (Fig. 5F–G).
Fig. 5.
Ginsenoside Rg3 promotes CDC25A proteasomal degradation via suppressing KIF20A expression, A. SKOV3 cells were subjected to lentiviral mediated KIF20A knockdown for 48 h, with or without the presence of MG132 (10 μM for 2 h before harvest). B–C. Western blot of in vitro ubiquitination assay. SKOV3 cells were infected with indicating selective combination of pLenti-puro-HA-Ubiquitin, myc-BTRC and/or flag-tagged KIF20A (B) or were infected with indicating selective combination of pLenti-puro-HA-Ubiquitin and shBTRC or shKIF20A (C). 48 h later, cells were lysed and the cell lysates were subjected to IP with an anti-CDC25A antibody. D-G. SKOV3 cells were infected for KIF20A overexpression (D-E) or knockdown (F-G). Then, the cells were incubated in the presence of 20 μM CHX for the indicated times (n = 3). Proteins were then analyzed by western blotting with the indicated antibodies. Relative levels of CDC25A (E and G) were determined from western blots using the ImageJ program based on the respective blots. H–I. Representative images (top panels) and quantitation (bottom panels) of western blotting analysis of KIF20A and CDC25A expression in SKOV3 and A2780 cells were subjected to ginsenoside Rg3 treatment for 48 h, with (I) or without (H) the presence of MG132 (10 μM for 2 h before harvest). J. SKOV3 and A2780 cells were subjected to lentiviral-mediated KIF20A knockdown. 48 h later, cells in indicated groups were treated with ginsenoside Rg3 for another 48 h. Then, cell samples were collected for western blotting analysis of KIF20A and CDC25A expression. Data are expressed as mean ± SD. Compared with the control group: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Ginsenoside Rg3 treatment reduced CDC25A protein levels in a dose-dependent manner (Fig. 5H), the effect of which was canceled by adding MG132 (Fig. 5I). In addition, it partly abrogated KIF20A overexpression-induced CDC25A upregulation (Fig. 5J). These findings imply that ginsenoside Rg3 can promote CDC25A proteasomal degradation by suppressing KIF20A expression.
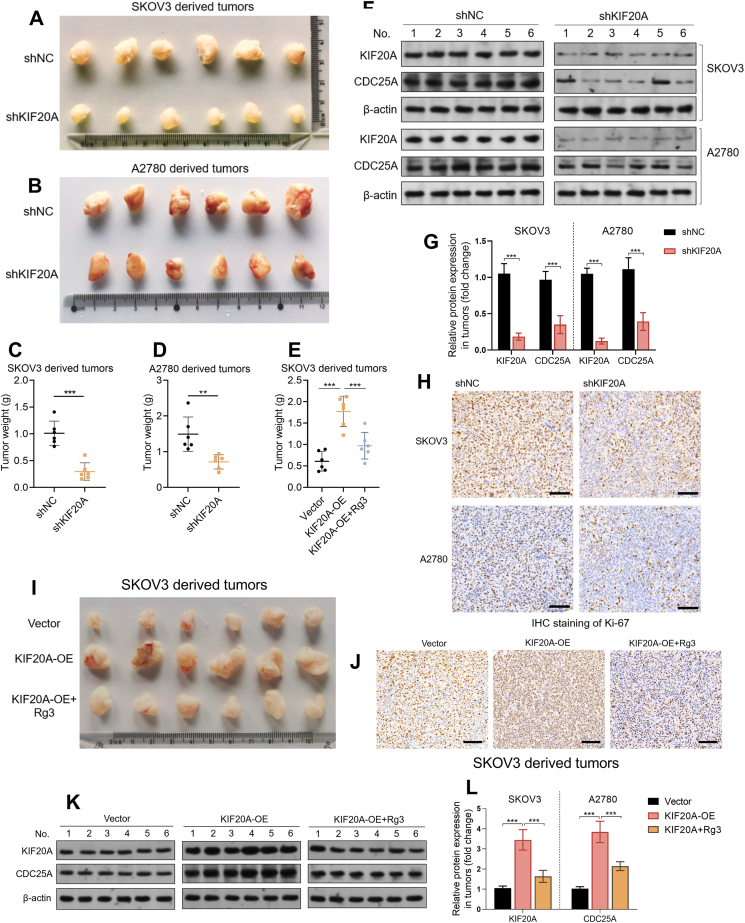
3.6. Ginsenoside Rg3 impairs ovarian tumor growth in vivo
To explore the regulatory effect of KIF20A on in vivo growth ovarian tumors, we generated xenograft tumor models in nude mice. Ovarian tumors derived from SKOV3 or A2780 cells with KIF20A knockdown grew significantly slower than those without KIF20A knockdown (Fig. 6A–D). CDC25A expression was also significantly lower in the group with KIF20A knockdown (Fig. 6F–G). IHC staining of Ki-67 confirmed that the tumors with KIF20A knockdown had fewer cells with positive Ki-67 staining (Fig. 6H). To validate the tumor suppressive effect of ginsenoside Rg3 in vivo, we generated xenograft tumor models in nude mice using SKOV3 cells with KIF20A overexpression. KIF20A overexpression significantly promoted tumor growth, accompanied by CDC25A upregulation (Fig. 6I–J). However, ginsenoside Rg3 treatment significantly retarded KIF20A overexpression-induced tumor growth and decreased CDC25A expression (Fig. 6I–J).
Fig. 6.
The in vivo regulatory effect of KIF20A and its correlation with CDC25A in human epithelial ovarian cancer tissues, A-D. Xenograft tumors (A-D) and their weight (C-D), derived from SKOV3 (A) and A2780 (B) cells with or without KIF20A knockdown (n = 6). F-G. Representative images (F) and quantitation (G) of KIF20A and CDC25A proteins in tumor tissues in panel A, detected by western blotting. β-actin was used as a loading control. H. Representative images of immunohistochemical staining for Ki-67 in xenograft tumor tissues in panel A. I and E. Xenograft tumors (I) and their weight (E), derived from SKOV3 cells with KIF20A overexpression alone or in combination with ginsenoside Rg3 treatment (n = 6). J. Representative images of immunohistochemical staining for Ki-67 in xenograft tumor tissues in panel I. K-L. Representative images (K) and quantitation (L) of KIF20A and CDC25A proteins in tumor tissues in panel I, detected by western blotting. β-actin was used as a loading control. Data are expressed as mean ± SD. Compared with the control group: ∗∗p < 0.01, ∗∗∗p < 0.001. Panels H and J, scale bar: 100 μm.
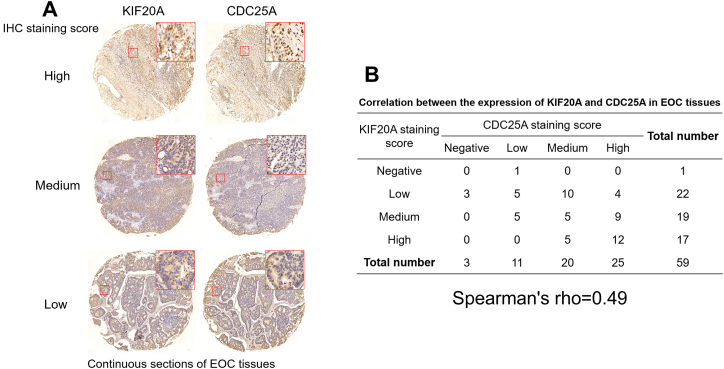
3.7. KIF20A expression was positively correlated with CDC25A expression in human epithelial ovarian cancer tissues
To validate the correlation between KIF20A and CDC25A expression, we used commercial tissue microarrays (continuous sections, Bioaitech, Xi'an, China), containing 59 human epithelial ovarian cancer cases. IHC staining and the following analysis of IHC scores by Spearman's correlation observed a moderate positive correlation between KIF20A and CDC25A in the 59 tumor tissue cases (Spearman's rho = 0.49, Fig. 7A–B).
Fig. 7.
The correlation between KIF20A and CDC25A expression in human epithelial ovarian cancer tissues, A. Representative IHC staining images of high, medium, and low KIF20A (left panel) and CDC25A (right panel) expression in two continuous ovarian cancer tissue microarrays (A). B. Correlation between KIF20A and CDC25A staining scores in 59 ovarian cancer tissues. Spearman's correlation analysis was conducted.
4. Discussion
Via bioinformatic analysis, we confirmed that KIF20A is significantly upregulated in ovarian cancer, and its upregulation is a biomarker of prognosis. The human KIF20A encodes a protein consisting of 890 amino acids. Aberrant KIF20A expression is associated with malignant behaviors and unfavorable clinical outcomes in multiple cancers [[38], [39], [40], [41]], including ovarian cancer [24,25]. KIF20A was transcriptionally activated by FOXM1 in ovarian cancer and can promote the proliferation and invasion of ovarian cancer cells [25]. In this study, using A2780 and SKOV3 cells as in vitro and in vivo cell models, we confirmed that the knockdown of KIF20A can significantly suppress tumor cell growth. These findings suggest that KIF20A might act as an important player in the pathological development of ovarian cancer.
KIF20A expression might be activated by NF-κB/P65, and ginsenoside Rg3 has confirmed effects on inhibiting the activity of P65 [[32], [33], [34]]. These clues suggest that ginsenoside Rg3 might suppress KIF20A expression. Our in vitro studies confirmed that ginsenoside Rg3 could inhibit the phosphorylation of P65 and transcription of KIF20A. Since ginsenoside Rg3 shows potent tumor-suppressive effects in ovarian cancer [[15], [16], [17], [18]], we infer that KIF20A might be a downstream effector of ginsenoside Rg3.
KIF20A exerts biological regulations mainly via interacting with other molecules [42,43]. It interacts with the chromosomal passenger complex (CPC) and mediates localization in anaphase [44]. It interacts with the Regulator of G Protein Signaling 3 (RGS3) and controls the division modes of neural progenitor cells during cortical neurogenesis [45]. Therefore, the current study focused on its interacting proteins in ovarian cancer. By applying GST pull-down and co-IP assays, we found that it physically interacts with BTRC, a substrate recognition subunit for SCFβ−TrCP E3 ubiquitin ligase. This protein can promote protein degradation by recognizing the phosphorylation of critical signaling molecules with crucial roles in cell cycle regulation, DNA damage response, and cancer progression [46,47]. SCFβ−TrCP E3 ubiquitin ligase can recognize phosphorylated AEBP2 and target it for ubiquitylation and proteasomal degradation, leading to cisplatin resistance in ovarian cancer [48].
Among the substrates of SCFβ−TrCP E3 ubiquitin ligase, CDC25A is a critical cell-cycle regulator, and its dysregulation is closely associated with chemoresistance in ovarian cancer [49]. CDC25A removes the inhibitory phosphorylation in cyclin-dependent kinase 4 (CDK4), CDK6, and CDK2 and promotes the cell-cycle progression during the G1/S and G2/M phases [50]. Inhibiting CDC25A using a small molecular inhibitor could sensitize ovarian cancer multicellular spheroids to chemotherapeutic agents [49]. The mechanisms underlying CDC25A upregulation are quite complex in cancer. Increased transcription, translation, and protein stability can elevate its expression in tumor cells. Some proteins can reduce CDC25A degradation via the ubiquitin-proteasome pathway via physical binding during cell cycle progression. For example, Rho-associated coiled-coil-containing protein kinase 2 (Rock2) can interact with CDC25A and reduce its ubiquitination and degradation in hepatocellular carcinoma cells [51]. CENPW can slow CDC25A degradation by interacting with and destabilizing the SCFβ−TrCP complex during the G2/M phase [52]. Therefore, we further explored whether KIF20A affects the stability of CDC25A. By performing in vitro ubiquitination and CHX chase assays, we confirmed that via interacting with BTRC, KIF20A could reduce BTRC-mediated CDC25A poly-ubiquitination and enhance its stability. In addition, we confirmed that ginsenoside Rg3 treatment could reduce CDC25A protein levels and abrogate KIF20A overexpression-induced CDC25A upregulation in SKOV3 and A2780 cells. Therefore, we infer that ginsenoside Rg3 can promote CDC25A proteasomal degradation by suppressing KIF20A expression.
5. Conclusion
In summary, KIF20A is a tumor-promoting gene in ovarian cancer. KIF20A physically interacts with BTRC, reduces BTRC-mediated CDC25A poly-ubiquitination, and enhances its stability. Ginsenoside Rg3 can inhibit KIF20A transcription, thereby promoting CDC25A proteasomal degradation. These findings revealed a novel anti-tumor mechanism of ginsenoside Rg3 in ovarian cancer.
Funding
This study was supported by Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-031A), Natural Science Foundation of Shanxi Province (grant no.202103021223422 and 202203021221261), and in part, by the National Natural Science Foundation of China (grant nos. 82073264).
Data availability statements
All data generated or analyzed during this study are included in this published article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2023.06.008.
Contributor Information
Ju Cui, Email: cuiju4366@bjhmoh.cn.
Yingmei Wang, Email: yingmeiwang@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
The promoter segment of KIF20A
Figure S1.
Systematic screening identifies KIF20A upregulation in ovarian cancer, A. A heatmap was generated to show the expression profile of 41 KIF family genes in ovarian cancer tissue (N = 419) in TCGA-OV and normal ovary and fallopian tubes (N = 93) in GTEx. B. A heatmap was generated to show the expression profile of 14 significantly dysregulated KIF family genes by using the following criteria: SEOC vs. normal (log2 fold change ≥2) and adj. p < 0.05.
Figure S2.
KIF20A expression in ovarian cancer tissues in the HPA, IHC staining images (top panels) and quantitation (bottom panels) of KIF20A expression in 12 ovarian cancer cases in the HPA. Enlarged tissue areas were provided in the right panel. Images were obtained from: v21, proteinatlas.org, https://www.proteinatlas.org/ENSG00000112984-KIF20A/pathology/ovarian+cancer#ihc.
Figure S3.
Relevant qRT-PCR and western blotting data, A-B. QRT-PCR (A) and western blotting assay (B) were performed to detect the expression of KIF20A in HOSE cells and multiple ovarian cancer cell lines (SKOV3, A2780, OV90 and OVCAR3). C–F. QRT-PCR (C and E) and western blotting assay (D and F) were performed to detect the expression of KIF20A in SKOV3 and A2780 cells 48 h after lentiviral shKIF20A or flag-KIF20A (KIF20A-OE) infection. G-H. QRT-PCR was performed to detect the expression of CDC25A in SKOV3 and A2780 cells 48 h after lentiviral shKIF20A (G) or flag-KIF20A (KIF20A-OE) (H) infection. ∗∗p < 0.01, ∗∗∗p < 0.001.
References
- 1.Shen L., Xia M., Zhang Y., Luo H., Dong D., Sun L. Mitochondrial integration and ovarian cancer chemotherapy resistance. Exp Cell Res. 2021;401(2) doi: 10.1016/j.yexcr.2021.112549. Epub 2021/03/01. PubMed PMID: 33640393. [DOI] [PubMed] [Google Scholar]
- 2.Coughlan A.Y., Testa G. Exploiting epigenetic dependencies in ovarian cancer therapy. Int J Cancer. 2021;149(10):1732–1743. doi: 10.1002/ijc.33727. Epub 2021/07/03. PubMed PMID: 34213777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crean-Tate K.K., Braley C., Dey G., Esakov E., Saygin C., Trestan A., et al. Pretreatment with LCK inhibitors chemosensitizes cisplatin-resistant endometrioid ovarian tumors. J Ovarian Res. 2021;14(1):55. doi: 10.1186/s13048-021-00797-x. Epub 2021/04/24. PubMed PMID: 33888137; PubMed Central PMCID: PMCPMC8063392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzese E., Diana A., Centonze S., Pignata S., De Vita F., Ciardiello F., et al. PARP Inhibitors in First-Line Therapy of Ovarian Cancer: Are There Any Doubts? Front Oncol. 2020;10:782. doi: 10.3389/fonc.2020.00782. Epub 2020/07/01. PubMed PMID: 32596142; PubMed Central PMCID: PMCPMC7303974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bu H., Chen J., Li Q., Hou J., Wei Y., Yang X., et al. BRCA mutation frequency and clinical features of ovarian cancer patients: A report from a Chinese study group. J Obstet Gynaecol Res. 2019;45(11):2267–2274. doi: 10.1111/jog.14090. Epub 2019/08/15. PubMed PMID: 31411802. [DOI] [PubMed] [Google Scholar]
- 6.Shi T., Wang P., Xie C., Yin S., Shi D., Wei C., et al. BRCA1 and BRCA2 mutations in ovarian cancer patients from China: ethnic-related mutations in BRCA1 associated with an increased risk of ovarian cancer. Int J Cancer. 2017;140(9):2051–2059. doi: 10.1002/ijc.30633. Epub 2017/02/09. PubMed PMID: 28176296. [DOI] [PubMed] [Google Scholar]
- 7.Mikula-Pietrasik J., Witucka A., Pakula M., Uruski P., Begier-Krasinska B., Niklas A., et al. Comprehensive review on how platinum- and taxane-based chemotherapy of ovarian cancer affects biology of normal cells. Cell Mol Life Sci. 2019;76(4):681–697. doi: 10.1007/s00018-018-2954-1. Epub 2018/11/02. PubMed PMID: 30382284; PubMed Central PMCID: PMCPMC6514066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang S.K., Jeong Y.J., Cho H.J., Park Y.Y., Song K.H., Chang Y.C. Rg3-enriched red ginseng extract promotes lung cancer cell apoptosis and mitophagy by ROS production. J Ginseng Res. 2022;46(1):138–146. doi: 10.1016/j.jgr.2021.05.005. Epub 2022/01/22. PubMed PMID: 35058730; PubMed Central PMCID: PMCPMC8753562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A., Yun E., Chang W., Kim J. Ginsenoside Rg3 protects against iE-DAP-induced endothelial-to-mesenchymal transition by regulating the miR-139-5p-NF-kappaB axis. J Ginseng Res. 2020;44(2):300–307. doi: 10.1016/j.jgr.2019.01.003. Epub 2020/03/10. PubMed PMID: 32148412; PubMed Central PMCID: PMCPMC7031736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan H., Yang L., Bai H., Luo J., Deng Y. Ginsenoside Rg3 increases gemcitabine sensitivity of pancreatic adenocarcinoma via reducing ZFP91 mediated TSPYL2 destabilization. J Ginseng Res. 2022;46(5):636–645. doi: 10.1016/j.jgr.2021.08.004. Epub 2022/09/13. PubMed PMID: 36090681; PubMed Central PMCID: PMCPMC9459078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park E.H., Kim Y.J., Yamabe N., Park S.H., Kim H.K., Jang H.J., et al. Stereospecific anticancer effects of ginsenoside Rg3 epimers isolated from heat-processed American ginseng on human gastric cancer cell. J Ginseng Res. 2014;38(1):22–27. doi: 10.1016/j.jgr.2013.11.007. Epub 2014/02/22. PubMed PMID: 24558306; PubMed Central PMCID: PMCPMC3915326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Z., Jiang H., Zhu X., Liu X., Li J. Ginsenoside Rg3 promotes cytotoxicity of Paclitaxel through inhibiting NF-kappaB signaling and regulating Bax/Bcl-2 expression on triple-negative breast cancer. Biomed Pharmacother. 2017;89:227–232. doi: 10.1016/j.biopha.2017.02.038. Epub 2017/02/24. PubMed PMID: 28231544. [DOI] [PubMed] [Google Scholar]
- 13.Zhao L., Shou H., Chen L., Gao W., Fang C., Zhang P. Effects of ginsenoside Rg3 on epigenetic modification in ovarian cancer cells. Oncol Rep. 2019;41(6):3209–3218. doi: 10.3892/or.2019.7115. Epub 2019/04/20. PubMed PMID: 31002353; PubMed Central PMCID: PMCPMC6489025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J.Y., Choi P., Lee D., Kim T., Jung E.B., Hwang B.S., et al. Effect of Amino Acids on the Generation of Ginsenoside Rg3 Epimers by Heat Processing and the Anticancer Activities of Epimers in A2780 Human Ovarian Cancer Cells. Evid Based Complement Alternat Med. 2016;2016 doi: 10.1155/2016/3146402. Epub 2016/04/07. PubMed PMID: 27051448; PubMed Central PMCID: PMCPMC4804038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T., Zhao L., Zhang Y., Chen W., Liu D., Hou H., et al. Ginsenoside 20(S)-Rg3 targets HIF-1alpha to block hypoxia-induced epithelial-mesenchymal transition in ovarian cancer cells. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0103887. Epub 2014/09/10. PubMed PMID: 25197976; PubMed Central PMCID: PMCPMC4157750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X., Chen W., Hou H., Li J., Li H., Sun X., et al. Ginsenoside 20(S)-Rg3 induced autophagy to inhibit migration and invasion of ovarian cancer. Biomed Pharmacother. 2017;85:620–626. doi: 10.1016/j.biopha.2016.11.072. Epub 2016/12/03. PubMed PMID: 27899249. [DOI] [PubMed] [Google Scholar]
- 17.Lu J., Chen H., He F., You Y., Feng Z., Chen W., et al. Ginsenoside 20(S)-Rg3 upregulates HIF-1alpha-targeting miR-519a-5p to inhibit the Warburg effect in ovarian cancer cells. Clin Exp Pharmacol Physiol. 2020;47(8):1455–1463. doi: 10.1111/1440-1681.13321. Epub 2020/04/10. PubMed PMID: 32271958. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L., Sun W., Zheng A., Zhang Y., Fang C., Zhang P. Ginsenoside Rg3 suppresses ovarian cancer cell proliferation and invasion by inhibiting the expression of lncRNA H19. Acta Biochim Pol. 2021;68(4):575–582. doi: 10.18388/abp.2020_5343. Epub 2021/05/27. PubMed PMID: 34038042. [DOI] [PubMed] [Google Scholar]
- 19.Zhong A., Tan F.Q., Yang W.X. Chromokinesin: kinesin superfamily regulating cell division through chromosome and spindle. Gene. 2016;589(1):43–48. doi: 10.1016/j.gene.2016.05.026. Epub 2016/05/20. PubMed PMID: 27196062. [DOI] [PubMed] [Google Scholar]
- 20.Liu X., Gong H., Huang K. Oncogenic role of kinesin proteins and targeting kinesin therapy. Cancer Sci. 2013;104(6):651–656. doi: 10.1111/cas.12138. Epub 2013/02/27. PubMed PMID: 23438337; PubMed Central PMCID: PMCPMC7657121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucanus A.J., Yip G.W. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37(7):833–838. doi: 10.1038/onc.2017.406. Epub 2017/10/24. PubMed PMID: 29059174. [DOI] [PubMed] [Google Scholar]
- 22.Theriault B.L., Pajovic S., Bernardini M.Q., Shaw P.A., Gallie B.L. Kinesin family member 14: an independent prognostic marker and potential therapeutic target for ovarian cancer. Int J Cancer. 2012;130(8):1844–1854. doi: 10.1002/ijc.26189. Epub 2011/05/28. PubMed PMID: 21618518. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto J., Amishiro N., Kato K., Ohta Y., Ino Y., Araki M., et al. Synthetic studies on mitotic kinesin Eg5 inhibitors: synthesis and structure-activity relationships of novel 2,4,5-substituted-1,3,4-thiadiazoline derivatives. Bioorg Med Chem Lett. 2014;24(16):3961–3963. doi: 10.1016/j.bmcl.2014.06.034. Epub 2014/07/09. PubMed PMID: 25001485. [DOI] [PubMed] [Google Scholar]
- 24.Li H., Zhang W., Sun X., Chen J., Li Y., Niu C., et al. Overexpression of kinesin family member 20A is associated with unfavorable clinical outcome and tumor progression in epithelial ovarian cancer. Cancer Manag Res. 2018;10:3433–3450. doi: 10.2147/CMAR.S169214. Epub 2018/09/27. PubMed PMID: 30254487; PubMed Central PMCID: PMCPMC6140728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Guo H., Wang Z., Bu H., Wang S., Wang H., et al. Cyclin F and KIF20A, FOXM1 target genes, increase proliferation and invasion of ovarian cancer cells. Exp Cell Res. 2020;395(2) doi: 10.1016/j.yexcr.2020.112212. Epub 2020/08/11. PubMed PMID: 32771525. [DOI] [PubMed] [Google Scholar]
- 26.Goldman M.J., Craft B., Hastie M., Repecka K., McDade F., Kamath A., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8. Epub 2020/05/24. PubMed PMID: 32444850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., et al. Towards a knowledge-based human protein atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. Epub 2010/12/09. PubMed PMID: 21139605. [DOI] [PubMed] [Google Scholar]
- 28.Gyorffy B., Lanczky A., Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. doi: 10.1530/ERC-11-0329. Epub 2012/01/27. PubMed PMID: 22277193. [DOI] [PubMed] [Google Scholar]
- 29.Garrido M.P., Hurtado I., Valenzuela-Valderrama M., Salvatierra R., Hernandez A., Vega M., et al. NGF-Enhanced Vasculogenic Properties of Epithelial Ovarian Cancer Cells Is Reduced by Inhibition of the COX-2/PGE2 Signaling Axis. Cancers (Basel) 2019;11(12) doi: 10.3390/cancers11121970. Epub 2019/12/11. PubMed PMID: 31817839; PubMed Central PMCID: PMCPMC6966471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R., Li L., Chen L., Suo Y., Fan J., Zhang S., et al. MAP7 interacts with RC3H1 and cooperatively regulate cell-cycle progression of cervical cancer cells via activating the NF-kappaB signaling. Biochem Biophys Res Commun. 2020;527(1):56–63. doi: 10.1016/j.bbrc.2020.04.008. Epub 2020/05/25. PubMed PMID: 32446391. [DOI] [PubMed] [Google Scholar]
- 31.Donaldson J.G. Immunofluorescence staining. Curr Protoc Cell Biol. 2015;69(3 7):4 3 1–4. doi: 10.1002/0471143030.cb0403s69. Epub 2015/12/02. PubMed PMID: 26621373. [DOI] [PubMed] [Google Scholar]
- 32.Kim B.M., Kim D.H., Park J.H., Surh Y.J., Na H.K. Ginsenoside Rg3 inhibits constitutive activation of NF-kappaB signaling in human breast cancer (MDA-MB-231) cells: ERK and akt as potential upstream targets. J Cancer Prev. 2014;19(1):23–30. doi: 10.15430/jcp.2014.19.1.23. Epub 2014/10/23. PubMed PMID: 25337569; PubMed Central PMCID: PMCPMC4189477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan X., Tian L.L., Zhang Y.M., Wang X.Q., Yan Q., Liu J.W. Ginsenoside Rg3 suppresses FUT4 expression through inhibiting NF-kappaB/p65 signaling pathway to promote melanoma cell death. Int J Oncol. 2015;47(2):701–709. doi: 10.3892/ijo.2015.3057. Epub 2015/06/23. PubMed PMID: 26094873; PubMed Central PMCID: PMCPMC6903900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu C., Wan B., Zeng Y. Ginsenoside Rg3 alleviates inflammation in a rat model of myocardial infarction via the SIRT1/NF-kappaB pathway. Exp Ther Med. 2020;20(6):238. doi: 10.3892/etm.2020.9368. Epub 2020/11/17. PubMed PMID: 33193843; PubMed Central PMCID: PMCPMC7646702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs S.Y., Spiegelman V.S., Kumar K.G. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23(11):2028–2036. doi: 10.1038/sj.onc.1207389. Epub 2004/03/17. PubMed PMID: 15021890. [DOI] [PubMed] [Google Scholar]
- 36.Bi Y., Cui D., Xiong X., Zhao Y. The characteristics and roles of beta-TrCP1/2 in carcinogenesis. FEBS J. 2021;288(11):3351–3374. doi: 10.1111/febs.15585. Epub 2020/10/07. PubMed PMID: 33021036. [DOI] [PubMed] [Google Scholar]
- 37.Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N.V., et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426(6962):87–91. doi: 10.1038/nature02082. Epub 2003/11/07. PubMed PMID: 14603323. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Z., Chai C., Shen T., Li X., Ji J., Li C., et al. Aberrant KIF20A Expression Is Associated with Adverse Clinical Outcome and Promotes Tumor Progression in Prostate Cancer. Dis Markers. 2019;2019 doi: 10.1155/2019/4782730. Epub 2019/10/01. PubMed PMID: 31565099; PubMed Central PMCID: PMCPMC6745134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Copello V.A., Burnstein K.L. The kinesin KIF20A promotes progression to castration-resistant prostate cancer through autocrine activation of the androgen receptor. Oncogene. 2022 doi: 10.1038/s41388-022-02307-9. Epub 2022/04/15. PubMed PMID: 35418689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong M., Zhuang K., Luo Y., Lai Q., Luo X., Fang Y., et al. KIF20A promotes cellular malignant behavior and enhances resistance to chemotherapy in colorectal cancer through regulation of the JAK/STAT3 signaling pathway. Aging. 2019;11(24):11905–11921. doi: 10.18632/aging.102505. Epub 2019/12/17. PubMed PMID: 31841120; PubMed Central PMCID: PMCPMC6949076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu M., Huang X., Chen Y., Fu Y., Xu C., Xiang W., et al. Aberrant KIF20A expression might independently predict poor overall survival and recurrence-free survival of hepatocellular carcinoma. IUBMB Life. 2018;70(4):328–335. doi: 10.1002/iub.1726. Epub 2018/03/04. PubMed PMID: 29500859. [DOI] [PubMed] [Google Scholar]
- 42.Qiu R., Runxiang Q., Geng A., Liu J., Xu C.W., Menon M.B., et al. SEPT7 Interacts with KIF20A and Regulates the Proliferative State of Neural Progenitor Cells During Cortical Development. Cereb Cortex. 2020;30(5):3030–3043. doi: 10.1093/cercor/bhz292. Epub 2019/12/10. PubMed PMID: 31813992; PubMed Central PMCID: PMCPMC7197076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu W.D., Yu K.W., Zhong N., Xiao Y., She Z.Y. Roles and mechanisms of Kinesin-6 KIF20A in spindle organization during cell division. Eur J Cell Biol. 2019;98(2–4):74–80. doi: 10.1016/j.ejcb.2018.12.002. Epub 2018/12/24. PubMed PMID: 30579662. [DOI] [PubMed] [Google Scholar]
- 44.Adriaans I.E., Hooikaas P.J., Aher A., Vromans M.J.M., van Es R.M., Grigoriev I., et al. MKLP2 Is a Motile Kinesin that Transports the Chromosomal Passenger Complex during Anaphase. Curr Biol. 2020;30(13):2628–2637. doi: 10.1016/j.cub.2020.04.081. Epub 2020/06/06. PubMed PMID: 32502404. [DOI] [PubMed] [Google Scholar]
- 45.Geng A., Qiu R., Murai K., Liu J., Wu X., Zhang H., et al. KIF20A/MKLP2 regulates the division modes of neural progenitor cells during cortical development. Nat Commun. 2018;9(1):2707. doi: 10.1038/s41467-018-05152-1. Epub 2018/07/15. PubMed PMID: 30006548; PubMed Central PMCID: PMCPMC6045631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaar J.R., Pagan J.K., Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13(12):889–903. doi: 10.1038/nrd4432. Epub 2014/11/15. PubMed PMID: 25394868; PubMed Central PMCID: PMCPMC4410837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee E.K., Diehl J.A. SCFs in the new millennium. Oncogene. 2014;33(16):2011–2018. doi: 10.1038/onc.2013.144. Epub 2013/04/30. PubMed PMID: 23624913. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Q., Wang W., Gao Q. beta-TRCP-mediated AEBP2 ubiquitination and destruction controls cisplatin resistance in ovarian cancer. Biochem Biophys Res Commun. 2020;523(1):274–279. doi: 10.1016/j.bbrc.2019.12.050. Epub 2019/12/23. PubMed PMID: 31864706. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y., Li S., Yang L., Zhang D., Zhao Z., Gao J., et al. CDC25A Facilitates Chemoresistance in Ovarian Cancer Multicellular Spheroids by Promoting E-cadherin Expression and Arresting Cell Cycles. J Cancer. 2019;10(13):2874–2884. doi: 10.7150/jca.31329. Epub 2019/07/10. PubMed PMID: 31281464; PubMed Central PMCID: PMCPMC6590049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen T., Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med Chem. 2012;12(6):631–639. doi: 10.2174/187152012800617678. Epub 2012/01/24. PubMed PMID: 22263797; PubMed Central PMCID: PMCPMC3544488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu T., Yu X., Li G., Yuan R., Wang Q., Tang P., et al. Rock2 regulates Cdc25A through ubiquitin proteasome system in hepatocellular carcinoma cells. Exp Cell Res. 2012;318(16):1994–2003. doi: 10.1016/j.yexcr.2012.04.017. Epub 2012/06/19. PubMed PMID: 22705122. [DOI] [PubMed] [Google Scholar]
- 52.Cheon Y., Lee S. CENP-W inhibits CDC25A degradation by destabilizing the SCF(beta-TrCP-1) complex at G2/M. FASEB J. 2018 doi: 10.1096/fj.201701358RRR. fj201701358RRR. Epub 2018/06/05. PubMed PMID: 29863914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The promoter segment of KIF20A
Data Availability Statement
All data generated or analyzed during this study are included in this published article.