Abstract
Background
In the ICU ward, septic cirrhotic patients are susceptible to suffering from sepsis-associated encephalopathy and/or hepatic encephalopathy, which are two common neurological complications in such patients. However, the mutual pathogenesis between sepsis-associated and hepatic encephalopathies remains unclear. We aimed to identify the mutual hub genes, explore effective diagnostic biomarkers and therapeutic targets for the two common encephalopathies and provide novel, promising insights into the clinical management of such septic cirrhotic patients.
Methods
The precious human post-mortem cerebral tissues were deprived of the GSE135838, GSE57193, and GSE41919 datasets, downloaded from the Gene Expression Omnibus database. Furthermore, we identified differentially expressed genes and screened hub genes with weighted gene co-expression network analysis. The hub genes were then subjected to Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway functional enrichment analyses, and protein-protein interaction networks were constructed. Receiver operating characteristic curves and correlation analyses were set up for the hub genes. Finally, we explored principal and common signaling pathways by using Gene Set Enrichment Analysis and the association between the hub genes and immune cell subtype distribution by using CIBERSORT algorithm.
Results
We identified seven hub genes—GPR4, SOCS3, BAG3, ZFP36, CDKN1A, ADAMTS9, and GADD45B—by using differentially expressed gene analysis and weighted gene co-expression network analysis method. The AUCs of these genes were all greater than 0.7 in the receiver operating characteristic curves analysis. The Gene Set Enrichment Analysis results demonstrated that mutual signaling pathways were mainly enriched in hypoxia and inflammatory response. CIBERSORT indicated that these seven hub genes were closely related to innate and adaptive immune cells.
Conclusions
We identified seven hub genes with promising diagnostic value and therapeutic targets in septic cirrhotic patients with sepsis-associated encephalopathy and/or hepatic encephalopathy. Hypoxia, inflammatory, and immunoreaction responses may share the common downstream pathways of the two common encephalopathies, for which earlier recognition and timely intervention are crucial for management of such septic cirrhotic patients in ICU.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-023-01774-7.
Keywords: ICU, Septic cirrhotic patients, Sepsis-associated encephalopathy, Hepatic encephalopathy, Bioinformatic analysis, Autopsy human brain tissue
Introduction
Sepsis—a major global healthcare concern—is one of the leading causes of morbidity and mortality in intensive care units (ICUs) [1, 2]. Cirrhosis is an independent risk factor for sepsis and sepsis-related mortality [3]. In ICU wards, cirrhotic patients are more susceptible to sepsis and severe complications than the general population because of their easier intestinal microbiota translocation, systemic immune dysregulation, and hyperdynamic circulation with high cardiac output and low systemic vascular resistance [4–9].
According to statistics, 30% of hospitalized patients with liver cirrhosis have a bacterial infection. If they develop sepsis, their mortality rate will reach 40–90% [10, 11].
Sepsis-associated encephalopathy (SAE) and hepatic encephalopathy (HE) are two common neurological complications in septic cirrhotic patients. HE is a severe reversible metabolic complication of sepsis-induced liver cirrhosis, demonstrating clinical manifestations of minor cognitive dysfunction such as personality changes, memory decline, attention deficit, and other neurological changes, including lethargy, delirium, and coma [12, 13]. ICU admission is usually required when patients with severe sepsis frequently develop multiple organ dysfunction syndrome (MODS), such as acute respiratory distress syndrome, acute kidney injury, and SAE [14–17]. Up to 70% of patients may develop SAE with increasing severity of disease, which is a diffuse brain dysfunction without direct intracranial infection, and whose clinical manifestations are extremely similar to those of HE, ranging from cognitive dysfunction to neurological changes [18, 19].
The pathogenesis of both encephalopathies, SAE and HE, is complex. Some authors have suggested that SAE may have similar metabolic pathogenesis to that of HE based on clinical similarities, including a wide range of neuropsychiatric alterations, ranging from confusion and delirium to coma [20, 21].
Few studies have reported on the common underling molecule mechanisms. Mizock et al. found elevated levels of phenylacetic acid in the blood and cerebrospinal fluid in patients with SAE and HE, suggesting that phenylethylamine metabolites may contribute to encephalopathy in systemic sepsis and hepatic failure [21]. Sari et al. demonstrated that hyperventilation decreases cerebral blood flow and CBF/CMRO2 (cerebral blood flow/cerebral metabolic rate for oxygen consumption), indicating that purposeful or unconscious hyperventilation may produce cerebral ischemia in the respiratory management of SAE or HE patients [22]. However, bioinformatic analysis of reciprocal genes in SAE and HE has never been exhaustively explored until now.
We speculated that SAE and HE—the two kinds of encephalopathy—might share a mutual pathophysiological molecule mechanism and cross-talk genes. Herein, the present study explored the common hub genes and potential biological mechanisms using a bioinformatics approach with precious autopsy brain tissue in SAE and HE for identification and treatment, thereby providing new promising insights into the clinical management of such septic cirrhotic illnesses.
Materials and methods
Data source
To explore the mutual hub genes between SAE and HE of septic cirrhotic patients, we selected three independent cohorts (GSE135838 [23], GSE57193 [24, 25], GSE41919 [24]) from the Gene Expression Omnibus database (GEO), a global public bioinformatic repository comprising high-throughput microarray and next-generation sequence functional genomic datasets [26]. The RNA-seq data of GSE135838 was generated from 24 post-mortem human brain tissue samples from 12 sepsis cases and 12 non-sepsis controls (These cases were deprived of the Internal Medicine Division of Pulmonary and Critical Care Medicine, University of Michigan, USA) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135838). The GSE57193 dataset comprises 12 post-mortem human brain tissue samples divided into 3 sample groups: 4 liver cirrhosis cases, 4 liver cirrhosis with HE cases, and 4 healthy controls (These cases were deprived of the Clinic for Gastroenterology, Hepatology and Infectiology, Germany) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57193). The GSE41919 cohort involves 19 post-mortem human brain samples from 8 cirrhotic patients with or 3 without HE and 8 non-cirrhotic controls (These cases were deprived of the Clinic for Gastroenterology, Hepatology and Infectiology, Germany). (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41919). All cohorts were used for microarray expression analysis.
Data extraction and management
All clinical, molecular information, and microarray datasets of these patients were publicly accessible at the GEO, and we used statistical analysis to investigate significantly differentially expressed genes (DEGs) on each microarray dataset. All aspects of experiment design, quality control, and data normalization conformed to standard Affymetrix protocols. The research was conducted in accordance with the International Conference and the Declaration of Helsinki.
Statistical analysis
Each dataset was first evaluated for normality of distribution by using the Kolmogorov-Smirnov test to decide whether a non-parametric rank-based or parametric analysis should be performed. The Fisher exact and Wilcoxon rank-sum tests were used for hypothesis testing with categorical and continuous variables, respectively. DEGs expression analysis was performed using the limma package [27]. P-value < 0.05 in unpaired t-test analysis and fold change (logFC) > 0.5 or < − 0.5 were used to determine the DEGs. The core modules and hub genes related to SAE and HE were identified using a weighted gene co-expression network analysis (WGCNA) [28]. An appropriate soft-thresholding power β = 8 was selected by using the integrated function (pick soft threshold) in the WGCNA package. Constructing a weighted gene network entails the choice of the soft thresholding power to which co-expression similarity is raised to calculate adjacency. We also used CIBERSORT [29, 30] and Gene Set Enrichment Analysis (GSEA) [31] of gene expression profiles to identify immune cell infiltration characteristics and related core genes sets. The Clusterprofiler [32] package was used to identify Gene Ontology (GO) enrichment terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways [33–35]. For all statistical analyses, a P-value < 0.05 was considered significant. The numerical variables and categorical variables were compared using the Wilcoxon rank-sum, Kruskal-Wallis, and Fisher exact tests. Co-expression analysis was conducted to detect correlated expression levels among hub genes by calculating Pearson’s correlation coefficient. The confidence interval was 95%. All statistical analyses were performed using R software 4.1.0 [36, 37].
Results
Identification of DEGs and screening of hub genes
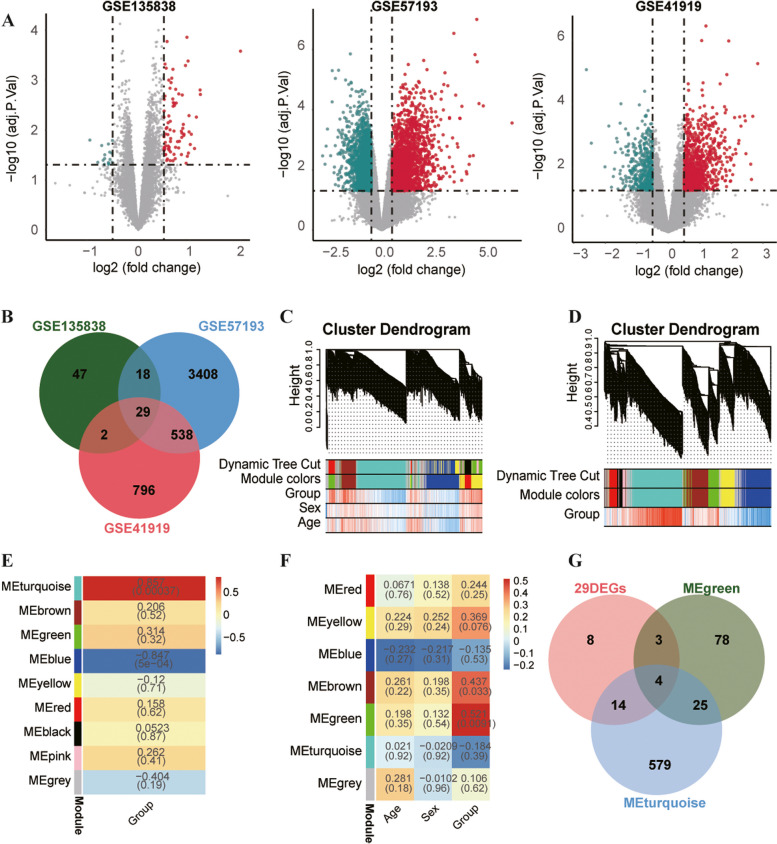
Based on the dataset GSE135838 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135838), we observed 96 DEGs between 12 sepsis samples and 12 non-sepsis samples, comprising 80 up-regulated DEGs and 16 down-regulated DEGs (Fig. 1A). The data were screened in another dataset GSE57193 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57193). There were 3993 DEGs between 4 liver cirrhosis patients with HE and 4 health controls, involving 2134 up-regulated DEGs and 1859 down-regulated DEGs (Fig. 1A). The third dataset was derived of GSE41919 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41919), which shows 1365 DEGs between 8 liver cirrhosis patients with HE and 8 non-cirrhotic controls, including 912 up-regulated DEGs and 453 down-regulated DEGs (Fig. 1A). The intersection of 29 DEGs from the aforementioned three datasets was determined using a Venn diagram (Fig. 1B). We further screened clusters or modules of highly correlated genes by using WGCNA analysis, which applies to small sample sizes. The results included Green module 110 genes and Turquoise module 622 genes (Fig. 1C-F); the gene list for these two modules are presented in Additional file 1: Table S1. Through the overlapping of the 29 DEGs, Green module genes, and Turquoise module genes, we obtained the following seven hub genes: GPR4, SOCS3, BAG3, ZFP36, CDKN1A, ADAMTS9, and GADD45B (Fig. 1G). The details of the seven hub genes are presented in Table 1.
Fig. 1.
Identification of DEGs and screening of hub genes. A The volcanic plots of DEGs analysis in GSE135838, GSE57193, and GSE41919 datasets. B The Venn diagram shows 29 DEGs at the intersection of three datasets. C, D Cluster dendrograms. In terms of dynamic tree cut, the genes from GSE135838 and GSE57193 were clustered into different modules through hierarchical clustering. Diverse colors indicate respective different modules. E, F The associations between clinical traits and modules, in which red represents a higher correlation coefficient. GSE57193 involves a Turquoise module containing 622 genes (r = 0.857, P<0.001), whereas GSE135838 involves a Green module containing 110 genes (r = 0.521, P<0.01). G The Venn diagram shows the seven hub genes that overlap 29 DEGs, Turquoise module genes, and Green module genes
Table 1.
Descriptions of the seven hub genes
| Gene | Full name | Protein family | Main function | Reference |
|---|---|---|---|---|
| GPR4 | G-protein-coupled receptors (GPR) 4 | GPCRs family | sensitive to both protons and lysolipids, expressed in multiple neuronal populations and vascular endothelial cells | [38–41] |
| SOCS3 | suppressor of cytokine signaling (SOC) 3 | SOCS family | positively and negatively regulate macrophage and dendritic cell activation and are involved in T-cell development/differentiation | [42] |
| BAG3 | B cell lymphoma-2-associated athanogene (BCL-2-associated athanogene) 3 | BAG family | development of both the neuronal and glial components of the central nervous system | [43, 44] |
| ZFP36 | Zinc finger protein (ZFP) 36 | ZFP family | encoding human tristetraprolin, which regulates TNF-α production by destabilizing TNF-α mRNA | [45, 46] |
| CDKN1A | cyclin-dependent kinase inhibitor p 21 (CDKN1A) | CDKN1 inhibitor family | negative regulation of the cell cycle | [47–49] |
| ADAMTS9 | a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) 9 | ADAMTS protein family | involved in neuroinflammation of the lesion core and neuroplasticity of surrounding tissues | [50, 51] |
| GADD45B | Growth arrest and DNA-damage-inducible protein (GADD) 45 beta | GADD45 family | anti-apoptotic factor in nonneuronal cells, an intrinsic neuroprotective molecule in neurons | [52] |
Expression levels of the seven hub genes
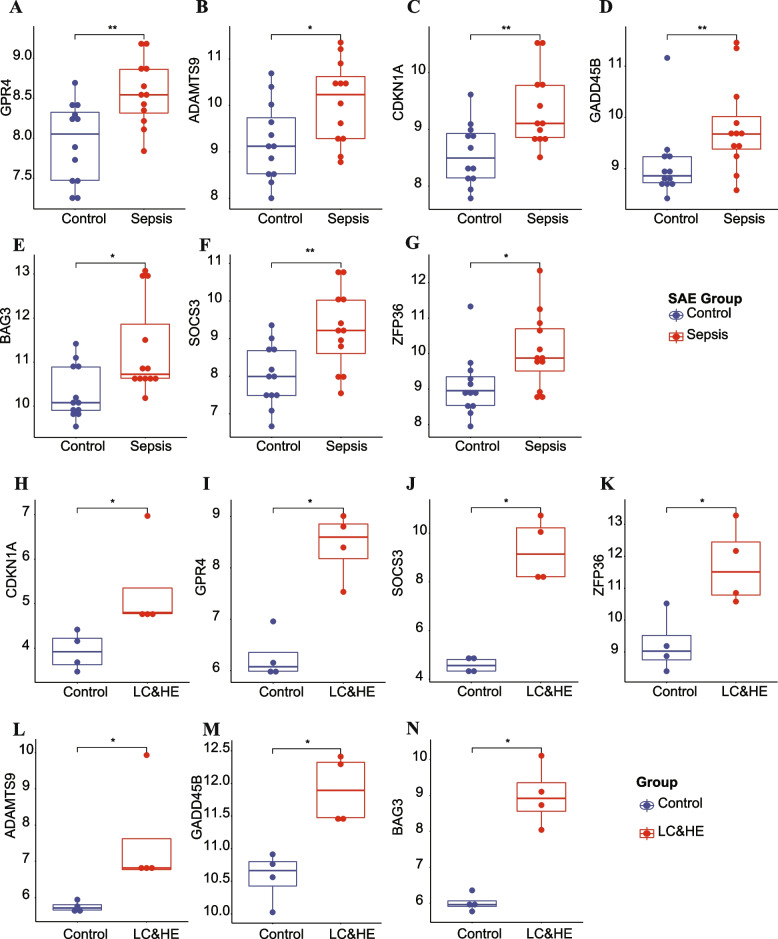
The expression levels of seven hub genes were significantly higher in SAE (Fig. 2A-G) or HE (Fig. 2H-N) than in control group.
Fig. 2.
All seven hub genes were significantly highly expressed in SAE/HE compared with that in controls. A-G The seven hub genes in SAE. H-N The seven hub genes in HE. SAE, sepsis-associated encephalopathy; LC, liver cirrhosis; HE, hepatic encephalopathy. *P <0.05, **P <0.01
Functional enrichment analysis and construction of protein-protein interaction (PPI) network
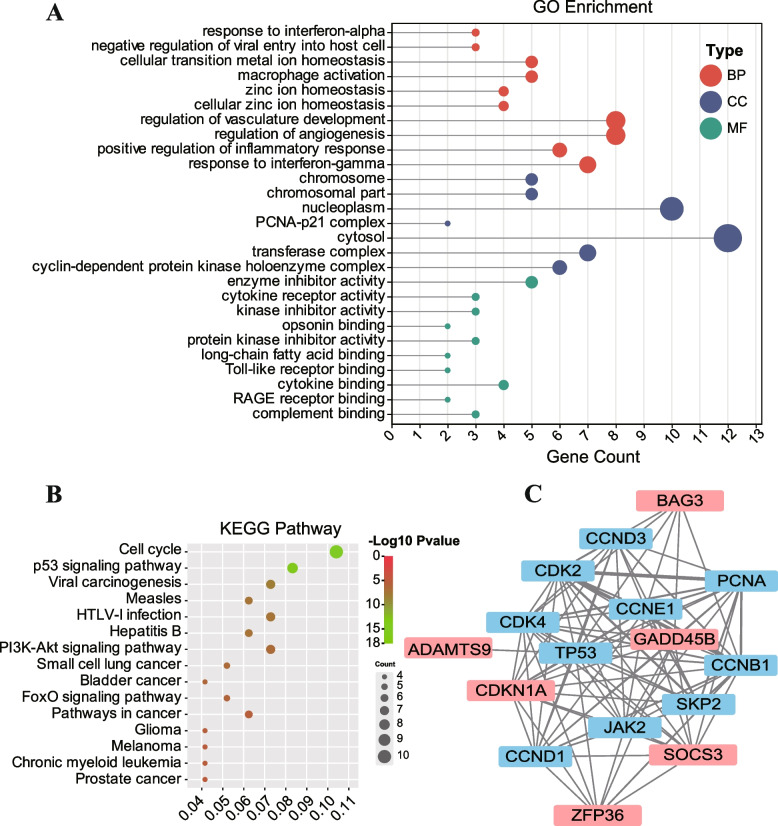
We performed GO and KEGG enrichment analyses [33–35] to explore the potential function of the seven hub genes (Fig. 3A, B). GO enrichment analysis comprises three parts: biological process (BP), cellular component (CC), and molecular function (MF) [53]. The BP indicated that these genes were associated with regulation of vasculature development, regulation of angiogenesis, response to interferon-gamma, positive regulation of inflammatory response, cellular transition, metal ion homeostasis, macrophage activation, zinc ion homeostasis, and cellular zinc ion homeostasis (Fig. 3A). Concerning the CC, these genes were involved in cytosol, nucleoplasm, transferase complex, cyclin-dependent protein kinase holoenzyme complex, chromosome, and PCNA-p21 complex (Fig. 3A). In terms of the MF, these seven hub genes were related with enzyme inhibitor activity, cytokine binding, cytokine receptor activity, kinase inhibitor activity, protein kinase inhibitor activity, complement binding, opsonin binding, long-chain fatty acid binding, Toll-like receptor binding, and RAGE receptor binding (Fig. 3A). The KEGG pathway analysis included the cell cycle, the p53 signaling pathway, the PI3K-Akt signaling pathway, and the FoxO signaling pathway (Fig. 3B). The PPI network results revealed that these seven hub genes interacted with each other (Fig. 3C). The detailed interaction relationship of PPI network are presented in Additional file 2: Table S2, which describes the weight of interaction relationship between all genes.
Fig. 3.
Function enrichment analysis and PPI network of the seven hub genes. A GO annotation. BP denotes the biological process; CC indicates the cellular component; MF represents the molecular function. B KEGG pathway enrichment analysis. C PPI network. The thickness of the lines represents the strength of interaction relationship between each hub gene
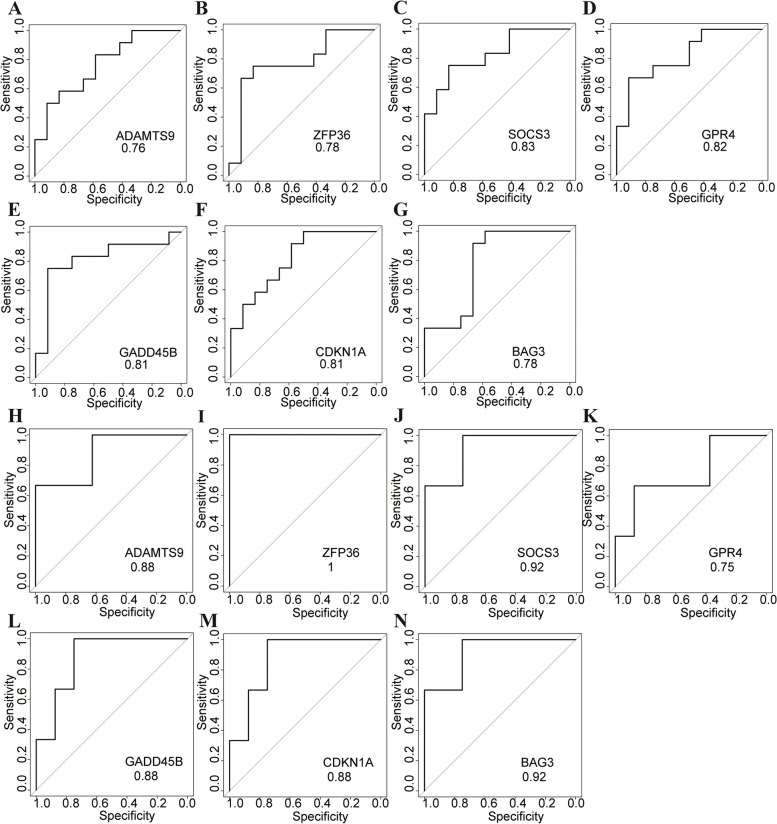
ROC curves analysis of the seven hub genes
The AUCs of the seven hub genes, ADAMTS9, ZFP36, SOCS3, GPR4, GADD45B, CDKN1A, and BAG3, were 0.76, 0.78, 0.83, 0.82, 0.81, 0.81, and 0.78, respectively, in SAE (Fig. 4A-G) and were 0.88, 1, 0.92, 0.75, 0.88, 0.88, and 0.92, respectively, in HE (Fig. 4H-N). These figures show that the AUCs of these seven hub genes are all greater than 0.7, indicating that these genes have preferable specificity, sensitivity, and excellent diagnostic properties in SAE and/or HE.
Fig. 4.
ROC diagnostic efficacy of the seven hub genes in SAE and/or HE. A-G The seven hub genes in SAE. H-N The seven hub genes in HE
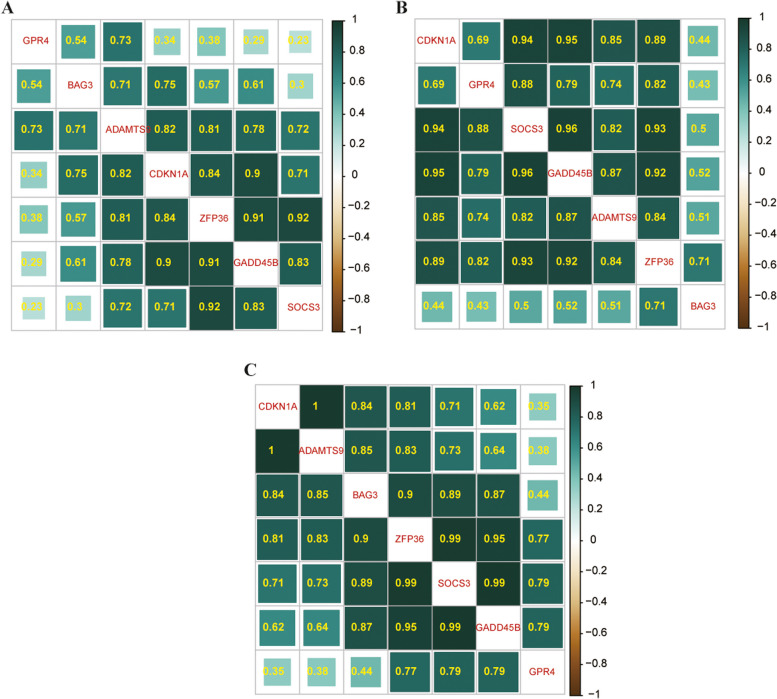
The correlation of the seven hub genes
Figure 5 shows the relationship between the seven DEGs. Figure 5A, B, and C represent three datasets, GSE135838, GSE57193, and GSE41919, respectively; the green color deeper, the stronger the positive correlation between each gene.
Fig. 5.
The heatmap of Spearman correlation analysis with the hub genes. A-C The dataset of GSE135838, GSE57193, and GSE41919, respectively. The greener the color, the greater the correlation coefficient
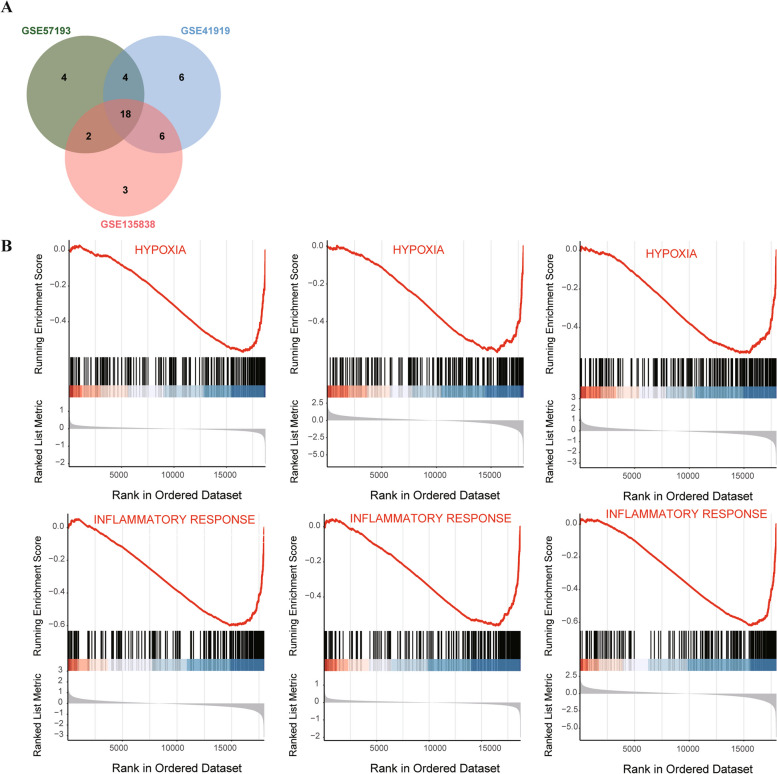
The GESA of the DEGs pathway
The Venn diagram depicts the intersection of 18 common pathways from the three datasets (Fig. 6A), of which GSEA was mainly enriched in hypoxia and inflammatory response (Fig. 6B). The detailed pathways for the three datasets are shown in Additional file 3: Table S3.
Fig. 6.
Venn diagram of GSEA results related to three datasets and the GSEA plot. A The intersection of signaling pathways. B Pathways mainly enriched in hypoxia and inflammatory response
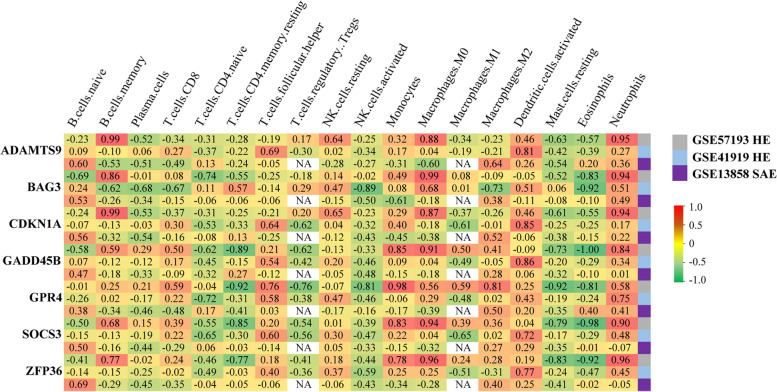
Immune infiltration analysis
CIBERSORT is an analytical tool for immune cell infiltration estimation analysis. Most of the seven genes are closely positively related to innate immune response, including neutrophils, dendritic cells, and macrophage, as well as negatively related with mast cells and NK cells, meanwhile, and also closely related to adaptive immune response comprising B cells and T cells, suggesting that they may be involved in the process of inflammatory and immunization response (Fig. 7).
Fig. 7.
Heatmap of the association between the seven hub genes and immune cell subtype distribution. Red represents a positive correlation, whereas green a negative correlation
Discussion
Cirrhotic patients are at an increased risk of developing sepsis and sepsis-related death and are predisposed to suffer critical complications requiring ICU admission [4, 54]. Septic cirrhotic patients have a significantly increased risk of short- and long-term mortality during their ICU hospital stay [55, 56]. Bacterial infection can act as a triggering factor development of HE [57]. Meanwhile, these extremely critical septic cirrhotic patients have a high risk of developing SAE. Subsequently, HE and SAE are common neurological complications that can co-occur and are difficult to differential diagnose in such patients.
Sepsis 3.0 is a life-threatening organ dysfunction that caused by a dysregulated host response to infection [58]. With an in-depth understanding of critical care medicine, studies have found that although critically ill patients are the most heterogeneous group in ICU, different critical illnesses often exhibit a similar clinical trajectory, in which the response to various acute critical conditions is relatively homogenous and may develop a critical illness [59]. PIRO model (predisposition, insult, response, organ dysfunction) is a well staging system for acute illness, which indicates the heterogeneity of critically ill patients and the identity of clinical manifestations that result from the body’s baseline physiology, the nature of the insult, and the way organ systems respond [60, 61]. Hence, the body’s response disorder usually induces hemodynamic disorders, hypoxia, and subsequent impairment of microcirculation, and mitochondria, resulting in lethal MODS, such as heart or brain, which may endanger the patients’ lives [62–64].
Cirrhotic patients are susceptible to bacterial infection insults, which share similar hemodynamic features to septic shock patients [65, 66]. The infection act as the “first hit”, triggering further dysregulated host response, and then the continuous dysregulated host response leads to a “second hit” to the body, causing MODS, such as SAE and/or HE [59, 67].
Therefore, we chose RNA-seq data from extremely precious autopsy brain tissue of HE and SAE deprived of GEO datasets to explore the possible underlying mutual genes, which maybe the molecular basis of mutual host-organ unregulated response [59] of the two kinds of encephalopathy, to provide new clues and perspectives for the diagnosis and treatment of the two common types of encephalopathy in such septic cirrhotic patients.
Based on the bioinformatics approach, we obtained seven hub genes: GPR4, SOCS3, BAG3, ZFP36, CDKN1A, ADAMTS9, and GADD45B. The expression levels of those genes were higher than that in the control group, and their AUCs were greater than 0.7, suggesting that their excellent diagnostic efficacy and performance in SAE and/or HE. As shown in Fig. 3, the GO and KEGG enrichment analysis suggested that these genes could be related to important biological processes, such as regulation of vasculature development and angiogenesis, cell cycle, cytokine receptor activity, macrophage activation, positive regulation of inflammatory response, and are associated with the p53 signaling pathway, PI3K-Akt signaling pathway, and FoxO signaling pathway. The detailed descriptions about the seven hub genes are shown in Table 1. CDKN1A—a member of the cyclin-dependent kinase inhibitor protein family—plays critical roles in the negative regulation of the cell cycle [47–49]. Sepsis, often complicated with hypoxia, which leads to cell cycle arrest, is linked to the transcription of CDKN1A [68]. Whereas hypoxia is usually accompanied by acidosis, in SAE or HE. There was a good linear relationship between cerebral blood flow and PaCO2, in which acidosis and hyperventilation may easily lead to brain ischemia [22]. GPR4 is a subfamily of G-protein-coupled receptors sensitive to both protons and lysolipids [38, 39]; hence, a GPR4 blocker—NE52-QQ57—can reduces CO2-induced hyperventilation that maybe the potential therapeutic target. GADD45B is an anti-apoptotic factor in nonneuronal cells and is an intrinsic neuroprotective molecule in neurons; its expression is up-regulated following focal and global cerebral ischemia, and it has a direct beneficial effect in cerebral ischemic injury [52, 69].
Furthermore, GSEA and immune infiltration analysis suggested that these genes could be involved in mutually intricate processes, such as hypoxia, inflammatory, and immunoreaction responses. As shown in Fig. 7, the seven hub genes are closely positively related to innate immune response, including neutrophils, dendritic cells, monocytes, and macrophages M0/M2, as well as negatively related to mast cells and activated NK cells.
After pathogenic microorganisms invade the human body, that is, after the body accepts the “first hit”, the host’s innate immune system is activated, comprising monocytes/macrophages, NK cells, neutrophils, dendritic cells, and other innate immune cells [70]. Infiltrating monocytes/macrophages and activation of microglia are closely related to excessive neuroinflammation in SAE [71]. Microglia is a type of macrophage distributed in all parts of the brain and critical immune cells in the nervous system, and microglia can be activated by inflammatory factors to release further proinflammatory cytokines [72], which is a considerable pathogenesis of SAE and/or HE [73, 74] and may be closely related to PI3K/AKT signaling pathway and FoxO signaling pathway [75–79] (Fig. 3).
In most cases, the host’s innate immune response can eliminate the invading pathogen. However, sometimes the host response gets overwhelmed and becomes further unbalanced and harmful [80], known as the “second hit”. Once the body has acquired a “second hit” and developed a critical sepsis illness, the central nervous system plays a vital role in responding to the “hit” and maintaining homeostasis. Subsequently, the body exhibits a series of stress-related decompensation syndrome, including the inflammatory immune system, vascular endothelial system, coagulation system, metabolism, and bioenergy. The inflammatory and immunoreaction responses may share the common downstream pathways, which are mainly characterized by the activation of several inflammatory cells and the release of proinflammatory mediators in the early stage; then, with the release of inflammatory mediators and immune cell apoptosis, the body enters a state of significant immunosuppression in both the innate and adaptive immune systems [59, 81–83], such as apoptosis of B cells and the decrease of CD4+/CD8+ T cell ratio [80, 84]. As shown in Fig. 7, the seven hub genes closely related to adaptive immune response were negatively correlated with plasma cells and CD4+ T cells and positively correlated with CD8+ T cells. Among them, SOCS3 can positively and negatively regulate macrophage and dendritic cell activation and are essential for T-cell development and differentiation [42], which may be involved in the highly complex neuroimmune response. To sum up, inflammation and the immune process interact closely, which is the underlying pathophysiological mechanism of the host response. The aforementioned seven hub genes are closely related and may be the mutual molecular mechanism of the host-organ dysregulated response in brain tissue.
The evolution of critical care medicine had two stages. The first stage is characterized by maintaining homeostasis and focusing on pathophysiology at the organ-level. In the current second stage, the main feature is transformed into a deeper understanding of the pathophysiological mechanism of the host response derived from various common ICU syndromes because of the complexity and heterogeneity of critical illness. Hence, identifying critical illness as a stress-related host response decompensation may assist us in improving current critical management [83]. However, excellent diagnostic biomarkers and molecular targets of host-organ dysregulated response for SAE and/or HE are unavailable in septic cirrhotic patients. The seven hub genes exhibit excellent diagnostic efficacy and have the potential to become promising central therapeutic targets blocked in host-organ dysregulated response, providing underlying molecular mechanisms for precise clinical treatment of such septic cirrhotic patients; earlier recognition and timely intervention are crucial for ICU management of such septic cirrhotic patients. Nevertheless, in the present study, the sample size was small because of the rarity and difficulty of obtaining autopsy brain tissue; therefore, we rely heavily on public autopsy brain sample data for analysis, with relevant conclusions and explicit molecular mechanisms to be experimentally verified in future work.
Conclusion
In summary, we identified the seven hub genes as potential diagnostic biomarkers and therapeutic targets in septic cirrhotic patients with HE and/or SAE. Hypoxia, inflammatory, and immunoreaction responses may share the common downstream pathways of the two common encephalopathies, SAE and HE, for which earlier recognition and timely intervention are crucial for management of such septic cirrhotic patients in ICU.
Supplementary Information
Additional file 1: Table S1. The genes list of Green and Turquoise modules.
Additional file 2: Table S2. The detailed interaction relationship of PPI network.
Additional file 3: Table S3. The detailed pathways deprived from the three datasets.
Acknowledgments
Sincere thanks to those patients who provided precious medical specimens in the global public database.
Abbreviations
- ICUs
Intensive Care Units
- SAE
Sepsis-associated encephalopathy
- HE
Hepatic encephalopathy
- MODS
Multiple Organ Dysfunction Syndrome
- CBF/CMRO2
Cerebral blood flow/cerebral metabolic rate of oxygen consumption
- GEO
Gene Expression Omnibus
- DEGs
Differentially expressed genes
- WGCNA
Weighted gene co-expression network analysis
- GSEA
Gene Set Enrichment Analysis
- GO
Gene Ontology enrichment terms
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- BP
Biological process
- CC
Cellular component
- MF
Molecular function
- PPI
Protein-protein interaction
Authors’ contributions
JL conceived idea of this research and finished the manuscript. DY, SG and YH performed analysis of data. LL and ZH revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Key Science and Technology Research Program of Health Commission of Hebei Province (No. 20181357).
Availability of data and materials
The data and materials in the current study are available from the corresponding author upon reasonable request. All raw and processed data are freely available from GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Declarations
Ethics approval and consent to participate
The research was conducted in accordance with the International Conference and the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shankar-Hari M, Phillips GS, Levy ML, Seymour CW, Liu VX, Deutschman CS, Angus DC, Rubenfeld GD, Singer M. Developing a new definition and assessing new clinical criteria for septic shock: for the third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315(8):775–787. doi: 10.1001/jama.2016.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, Machado FR, McIntyre L, Ostermann M, Prescott HC, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021;47(11):1181–1247. doi: 10.1007/s00134-021-06506-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun-Buisson C, Doyon F, Carlet J, Dellamonica P, Gouin F, Lepoutre A, Mercier JC, Offenstadt G, Régnier B. Incidence, risk factors, and outcome of severe sepsis and septic shock in adults. A multicenter prospective study in intensive care units. French ICU Group for Severe Sepsis. JAMA. 1995;274(12):968–974. doi: 10.1001/jama.1995.03530120060042. [DOI] [PubMed] [Google Scholar]
- 4.Foreman MG, Mannino DM, Moss M. Cirrhosis as a risk factor for sepsis and death: analysis of the National Hospital Discharge Survey. Chest. 2003;124(3):1016–1020. doi: 10.1378/chest.124.3.1016. [DOI] [PubMed] [Google Scholar]
- 5.Piano S, Tonon M, Angeli P. Changes in the epidemiology and management of bacterial infections in cirrhosis. Clin Mol Hepatol. 2021;27(3):437–445. doi: 10.3350/cmh.2020.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albillos A, Lario M, Álvarez-Mon M. Cirrhosis-associated immune dysfunction: distinctive features and clinical relevance. J Hepatol. 2014;61(6):1385–1396. doi: 10.1016/j.jhep.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Levesque E, Saliba F, Ichaï P, Samuel D. Outcome of patients with cirrhosis requiring mechanical ventilation in ICU. J Hepatol. 2014;60(3):570–578. doi: 10.1016/j.jhep.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Merli M, Lucidi C, Pentassuglio I, Giannelli V, Giusto M, Di Gregorio V, Pasquale C, Nardelli S, Lattanzi B, Venditti M, et al. Increased risk of cognitive impairment in cirrhotic patients with bacterial infections. J Hepatol. 2013;59(2):243–250. doi: 10.1016/j.jhep.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Arroyo V, Fernandez J, Ginès P. Pathogenesis and treatment of hepatorenal syndrome. Semin Liver Dis. 2008;28(1):81–95. doi: 10.1055/s-2008-1040323. [DOI] [PubMed] [Google Scholar]
- 10.Singal AK, Salameh H, Kamath PS. Prevalence and in-hospital mortality trends of infections among patients with cirrhosis: a nationwide study of hospitalised patients in the United States. Aliment Pharmacol Ther. 2014;40(1):105–112. doi: 10.1111/apt.12797. [DOI] [PubMed] [Google Scholar]
- 11.Aggarwal A, Ong JP, Younossi ZM, Nelson DR, Hoffman-Hogg L, Arroliga AC. Predictors of mortality and resource utilization in cirrhotic patients admitted to the medical ICU. Chest. 2001;119(5):1489–1497. doi: 10.1378/chest.119.5.1489. [DOI] [PubMed] [Google Scholar]
- 12.Vilstrup H, Amodio P, Bajaj J, Cordoba J, Ferenci P, Mullen KD, Weissenborn K, Wong P. Hepatic encephalopathy in chronic liver disease: 2014 practice guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the liver. Hepatology. 2014;60(2):715–735. doi: 10.1002/hep.27210. [DOI] [PubMed] [Google Scholar]
- 13.Hadjihambi A, Arias N, Sheikh M, Jalan R. Hepatic encephalopathy: a critical current review. Hepatol Int. 2018;12(Suppl 1):135–147. doi: 10.1007/s12072-017-9812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auriemma CL, Zhuo H, Delucchi K, Deiss T, Liu T, Jauregui A, Ke S, Vessel K, Lippi M, Seeley E, et al. Acute respiratory distress syndrome-attributable mortality in critically ill patients with sepsis. Intensive Care Med. 2020;46(6):1222–1231. doi: 10.1007/s00134-020-06010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarbock A, Nadim MK, Pickkers P, Gomez H, Bell S, Joannidis M, Kashani K, Koyner JL, Pannu N, Meersch M, et al. Sepsis-associated acute kidney injury: consensus report of the 28th acute disease quality initiative workgroup. Nat Rev Nephrol. 2023;19(6):401–417. doi: 10.1038/s41581-023-00683-3. [DOI] [PubMed] [Google Scholar]
- 16.Liu K, Chen X, Ren X, Wu Y, Ren S, Qin C. SARS-CoV-2 effects in the genitourinary system and prospects of sex hormone therapy. Asian J Urol. 2021;8(3):303–314. doi: 10.1016/j.ajur.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferlini L, Gaspard N. What's new on septic encephalopathy? Ten things you need to know. Minerva Anestesiol. 2023;89(3):217–225. doi: 10.23736/S0375-9393.22.16689-7. [DOI] [PubMed] [Google Scholar]
- 18.Mazeraud A, Pascal Q, Verdonk F, Heming N, Chrétien F, Sharshar T. Neuroanatomy and physiology of brain dysfunction in sepsis. Clin Chest Med. 2016;37(2):333–345. doi: 10.1016/j.ccm.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Mazeraud A, Righy C, Bouchereau E, Benghanem S, Bozza FA, Sharshar T. Septic-associated encephalopathy: a comprehensive review. Neurotherapeutics. 2020;17(2):392–403. doi: 10.1007/s13311-020-00862-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeppsson B, Freund HR, Gimmon Z, James JH, von Meyenfeldt MF, Fischer JE. Blood-brain barrier derangement in sepsis: cause of septic encephalopathy? Am J Surg. 1981;141(1):136–142. doi: 10.1016/0002-9610(81)90026-X. [DOI] [PubMed] [Google Scholar]
- 21.Mizock BA, Sabelli HC, Dubin A, Javaid JI, Poulos A, Rackow EC, Septic encephalopathy. Evidence for altered phenylalanine metabolism and comparison with hepatic encephalopathy. Arch Intern Med. 1990;150(2):443–449. doi: 10.1001/archinte.1990.00390140139029. [DOI] [PubMed] [Google Scholar]
- 22.Sari A, Yamashita S, Ohosita S, Ogasahara H, Yamada K, Yonei A, Yokota K. Cerebrovascular reactivity to CO2 in patients with hepatic or septic encephalopathy. Resuscitation. 1990;19(2):125–134. doi: 10.1016/0300-9572(90)90035-D. [DOI] [PubMed] [Google Scholar]
- 23.Bustamante AC, Opron K, Ehlenbach WJ, Larson EB, Crane PK, Keene CD, Standiford TJ, Singer BH. Transcriptomic profiles of sepsis in the human brain. Am J Respir Crit Care Med. 2020;201(7):861–863. doi: 10.1164/rccm.201909-1713LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sergeeva OA, Chepkova AN, Görg B, Rodrigues Almeida F, Bidmon HJ, Haas HL, Häussinger D. Histamine-induced plasticity and gene expression in corticostriatal pathway under hyperammonemia. CNS Neurosci Ther. 2020;26(3):355–366. doi: 10.1111/cns.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobczyk K, Jördens MS, Karababa A, Görg B, Häussinger D. Ephrin/Ephrin receptor expression in ammonia-treated rat astrocytes and in human cerebral cortex in hepatic encephalopathy. Neurochem Res. 2015;40(2):274–283. doi: 10.1007/s11064-014-1389-9. [DOI] [PubMed] [Google Scholar]
- 26.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Q, Zhang X, Ren X, Hang Z, Yin Y, Wang Z, Chen H, Sun L, Tao J, Han Z, et al. Biological characteristics and predictive model of biopsy-proven acute rejection (BPAR) after kidney transplantation: evidences of multi-omics analysis. Front Genet. 2022;13:844709. doi: 10.3389/fgene.2022.844709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung JH, Yang TH, Hu Z, Weng Z, DeLisi C. Gene set enrichment analysis: performance evaluation and usage guidelines. Brief Bioinform. 2012;13(3):281–291. doi: 10.1093/bib/bbr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2(3):100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu L, Shen H, Ren X, Wang A, Zhu S, Zheng Y, Wang X. Multi-omics analysis reveals the interaction between the complement system and the coagulation cascade in the development of endometriosis. Sci Rep. 2021;11(1):11926. doi: 10.1038/s41598-021-90112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwig MG, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, Hofstetter H, Wolf RM, Seuwen K. Proton-sensing G-protein-coupled receptors. Nature. 2003;425(6953):93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- 39.Tomura H, Mogi C, Sato K, Okajima F. Proton-sensing and lysolipid-sensitive G-protein-coupled receptors: a novel type of multi-functional receptors. Cell Signal. 2005;17(12):1466–1476. doi: 10.1016/j.cellsig.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Justus CR, Dong L, Yang LV. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front Physiol. 2013;4:354. doi: 10.3389/fphys.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong L, Li Z, Leffler NR, Asch AS, Chi JT, Yang LV. Acidosis activation of the proton-sensing GPR4 receptor stimulates vascular endothelial cell inflammatory responses revealed by transcriptome analysis. PLoS One. 2013;8(4):e61991. doi: 10.1371/journal.pone.0061991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7(6):454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 43.Gentilella A, Khalili K. BAG3 expression is sustained by FGF2 in neural progenitor cells and impacts cell proliferation. Cell Cycle. 2010;9(20):4245–4247. doi: 10.4161/cc.9.20.13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2(4):e141. doi: 10.1038/cddis.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor GA, Lai WS, Oakey RJ, Seldin MF, Shows TB, Eddy RL, Jr, Blackshear PJ. The human TTP protein: sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Res. 1991;19(12):3454. doi: 10.1093/nar/19.12.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26(24):9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 48.Nakayama K, Nakayama K. Cip/kip cyclin-dependent kinase inhibitors: brakes of the cell cycle engine during development. Bioessays. 1998;20(12):1020–1029. doi: 10.1002/(SICI)1521-1878(199812)20:12<1020::AID-BIES8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 49.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377(6549):552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 50.Reid MJ, Cross AK, Haddock G, Allan SM, Stock CJ, Woodroofe MN, Buttle DJ, Bunning RA. ADAMTS-9 expression is up-regulated following transient middle cerebral artery occlusion (tMCAo) in the rat. Neurosci Lett. 2009;452(3):252–257. doi: 10.1016/j.neulet.2009.01.058. [DOI] [PubMed] [Google Scholar]
- 51.Lemarchant S, Pruvost M, Montaner J, Emery E, Vivien D, Kanninen K, Koistinaho J. ADAMTS proteoglycanases in the physiological and pathological central nervous system. J Neuroinflammation. 2013;10:133. doi: 10.1186/1742-2094-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho CH, Byun HR, Jover-Mengual T, Pontarelli F, Dejesus C, Cho AR, Zukin RS, Hwang JY. Gadd45b acts as neuroprotective effector in global ischemia-induced neuronal death. Int Neurourol J. 2019;23(S1):11–21. doi: 10.5213/inj.1938040.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Wang J, Li L, Qin H, Wei Y, Zhang X, Ren X, Ding W, Shen X, Li G, et al. AC010973.2 promotes cell proliferation and is one of six stemness-related genes that predict overall survival of renal clear cell carcinoma. Sci Rep. 2022;12(1):4272. doi: 10.1038/s41598-022-07070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ginès P, Fernández J, Durand F, Saliba F. Management of critically-ill cirrhotic patients. J Hepatol. 2012;56(S1):13–24. doi: 10.1016/S0168-8278(12)60003-8. [DOI] [PubMed] [Google Scholar]
- 55.Chebl RB, Tamim H, Sadat M, Qahtani S, Dabbagh T, Arabi YM. Outcomes of septic cirrhosis patients admitted to the intensive care unit: a retrospective cohort study. Medicine (Baltimore). 2021;100(46):e27593. doi: 10.1097/MD.0000000000027593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arvaniti V, D'Amico G, Fede G, Manousou P, Tsochatzis E, Pleguezuelo M, Burroughs AK. Infections in patients with cirrhosis increase mortality four-fold and should be used in determining prognosis. Gastroenterology. 2010;139(4):1246–1256. doi: 10.1053/j.gastro.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 57.Jalan R, Fernandez J, Wiest R, Schnabl B, Moreau R, Angeli P, Stadlbauer V, Gustot T, Bernardi M, Canton R, et al. Bacterial infections in cirrhosis: a position statement based on the EASL special conference 2013. J Hepatol. 2014;60(6):1310–1324. doi: 10.1016/j.jhep.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 58.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang W, Liu D, Zhang H, Ding X, Wang X. Focus on host/organ unregulated response: a common cause of critical illness. Chin Med J. 2023;136(1):108–110. doi: 10.1097/CM9.0000000000002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall JC. The PIRO (predisposition, insult, response, organ dysfunction) model: toward a staging system for acute illness. Virulence. 2014;5(1):27–35. doi: 10.4161/viru.26908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maslove DM, Tang B, Shankar-Hari M, Lawler PR, Angus DC, Baillie JK, Baron RM, Bauer M, Buchman TG, Calfee CS, et al. Redefining critical illness. Nat Med. 2022;28(6):1141–1148. doi: 10.1038/s41591-022-01843-x. [DOI] [PubMed] [Google Scholar]
- 62.Hawchar F, Rao C, Akil A, Mehta Y, Rugg C, Scheier J, Adamson H, Deliargyris E, Molnar Z. The potential role of extracorporeal cytokine removal in hemodynamic stabilization in hyperinflammatory shock. Biomedicines. 2021;9(7):768. doi: 10.3390/biomedicines9070768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abraham E, Singer M. Mechanisms of sepsis-induced organ dysfunction. Crit Care Med. 2007;35(10):2408–2416. doi: 10.1097/01.CCM.0000282072.56245.91. [DOI] [PubMed] [Google Scholar]
- 64.Balestra GM, Legrand M, Ince C. Microcirculation and mitochondria in sepsis: getting out of breath. Curr Opin Anaesthesiol. 2009;22(2):184–190. doi: 10.1097/ACO.0b013e328328d31a. [DOI] [PubMed] [Google Scholar]
- 65.Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology. 2005;41(3):422–433. doi: 10.1002/hep.20632. [DOI] [PubMed] [Google Scholar]
- 66.Tsai MH, Peng YS, Chen YC, Liu NJ, Ho YP, Fang JT, Lien JM, Yang C, Chen PC, Wu CS. Adrenal insufficiency in patients with cirrhosis, severe sepsis and septic shock. Hepatology. 2006;43(4):673–681. doi: 10.1002/hep.21101. [DOI] [PubMed] [Google Scholar]
- 67.Marshall JC. Iatrogenesis, inflammation and organ injury: insights from a murine model. Crit Care. 2006;10(6):173. doi: 10.1186/cc5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23(9):1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu B, Li J, Li L, Yu L, Li C. Electrical stimulation of cerebellar fastigial nucleus promotes the expression of growth arrest and DNA damage inducible gene β and motor function recovery in cerebral ischemia/reperfusion rats. Neurosci Lett. 2012;520(1):110–114. doi: 10.1016/j.neulet.2012.05.044. [DOI] [PubMed] [Google Scholar]
- 70.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 71.Tian M, Qingzhen L, Zhiyang Y, Chunlong C, Jiao D, Zhang L, Li W. Attractylone attenuates sepsis-associated encephalopathy and cognitive dysfunction by inhibiting microglial activation and neuroinflammation. J Cell Biochem. 2019;120(5):7101–7108. doi: 10.1002/jcb.27983. [DOI] [PubMed] [Google Scholar]
- 72.Baaklini CS, Rawji KS, Duncan GJ, Ho MFS, Plemel JR. Central nervous system Remyelination: roles of glia and innate immune cells. Front Mol Neurosci. 2019;12:225. doi: 10.3389/fnmol.2019.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zemtsova I, Görg B, Keitel V, Bidmon HJ, Schrör K, Häussinger D. Microglia activation in hepatic encephalopathy in rats and humans. Hepatology. 2011;54(1):204–215. doi: 10.1002/hep.24326. [DOI] [PubMed] [Google Scholar]
- 74.Ye B, Tao T, Zhao A, Wen L, He X, Liu Y, Fu Q, Mi W, Lou J. Blockade of IL-17A/IL-17R pathway protected mice from sepsis-associated encephalopathy by inhibition of microglia activation. Mediat Inflamm. 2019;2019:8461725. doi: 10.1155/2019/8461725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Cheng X, Fu D, Liang Y, Chen C, Deng W, He L. Autophagy of spinal microglia affects the activation of microglia through the PI3K/AKT/mTOR signaling pathway. Neuroscience. 2022;482:77–86. doi: 10.1016/j.neuroscience.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 76.Cianciulli A, Calvello R, Porro C, Trotta T, Salvatore R, Panaro MA. PI3k/Akt signalling pathway plays a crucial role in the anti-inflammatory effects of curcumin in LPS-activated microglia. Int Immunopharmacol. 2016;36:282–290. doi: 10.1016/j.intimp.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 77.Tian Y, Liu B, Li Y, Zhang Y, Shao J, Wu P, Xu C, Chen G, Shi H. Activation of RARα receptor attenuates neuroinflammation after SAH via promoting M1-to-M2 phenotypic polarization of microglia and regulating Mafb/Msr1/PI3K-Akt/NF-κB pathway. Front Immunol. 2022;13:839796. doi: 10.3389/fimmu.2022.839796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang D, Pan N, Jiang C, Hao M. LncRNA SNHG8 sponges miR-449c-5p and regulates the SIRT1/FoxO1 pathway to affect microglia activation and blood-brain barrier permeability in ischemic stroke. J Leukoc Biol. 2022;111(5):953–966. doi: 10.1002/JLB.1A0421-217RR. [DOI] [PubMed] [Google Scholar]
- 79.Chen YN, Sha HH, Wang YW, Zhou Q, Bhuiyan P, Li NN, Qian YN, Dong HQ. Histamine 2/3 receptor agonists alleviate perioperative neurocognitive disorders by inhibiting microglia activation through the PI3K/AKT/FoxO1 pathway in aged rats. J Neuroinflammation. 2020;17(1):217. doi: 10.1186/s12974-020-01886-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017;17(7):407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- 81.Gotts JE, Matthay MA. Sepsis: pathophysiology and clinical management. BMJ. 2016;353:i1585. doi: 10.1136/bmj.i1585. [DOI] [PubMed] [Google Scholar]
- 82.Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD, Kreisel D, Krupnick AS, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 2011;306(23):2594–2605. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cuesta JM, Singer M. The stress response and critical illness: a review. Crit Care Med. 2012;40(12):3283–3289. doi: 10.1097/CCM.0b013e31826567eb. [DOI] [PubMed] [Google Scholar]
- 84.Markwart R, Condotta SA, Requardt RP, Borken F, Schubert K, Weigel C, Bauer M, Griffith TS, Förster M, Brunkhorst FM, et al. Immunosuppression after sepsis: systemic inflammation and sepsis induce a loss of naïve T-cells but no enduring cell-autonomous defects in T-cell function. PLoS One. 2014;9(12):e115094. doi: 10.1371/journal.pone.0115094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The genes list of Green and Turquoise modules.
Additional file 2: Table S2. The detailed interaction relationship of PPI network.
Additional file 3: Table S3. The detailed pathways deprived from the three datasets.
Data Availability Statement
The data and materials in the current study are available from the corresponding author upon reasonable request. All raw and processed data are freely available from GEO database (https://www.ncbi.nlm.nih.gov/geo/).