Abstract
The Zinc finger and SCAN domain containing 4 (ZSCAN4) protein, expressed transiently in pluripotent stem cells, gametes, and early embryos, extends telomeres, enhances genome stability, and improves karyotypes in mouse embryonic stem (mES) cells. To gain insights into the mechanism of ZSCAN4 function, we identified genome-wide binding sites of endogenous ZSCAN4 protein using ChIP-seq technology in mouse and human ES cells, where the expression of endogenous ZSCAN4 was induced by treating cells with retinoic acids or by overexpressing DUX4. We revealed that both mouse and human ZSCAN4 bind to the TGCACAC motif located in CA/TG microsatellite repeats, which are known to form unstable left-handed duplexes called Z-DNA that can induce double-strand DNA breaks and mutations. These ZSCAN4 binding sites are mostly located in intergenic and intronic regions of the genomes. By generating ZSCAN4 knockout in human ES cells, we showed that ZSCAN4 does not seem to be involved in transcriptional regulation. We also found that ectopic expression of mouse ZSCAN4 enhances the suppression of chromatin at ZSCAN4-binding sites. These results together suggest that some of the ZSCAN4 functions are mediated by binding to the error-prone regions in mouse and human genomes.
Keywords: ZSCAN4, genome stability, CA/TG microsatellite, mouse ES cells, human ES cells
1. Introduction
Zinc finger and SCAN domain containing 4 (Zscan4 for mouse, ZSCAN4 for human gene symbols) gene expression was originally observed in two-cell embryos but can also be detected in 1–5% of mouse embryonic stem (mES) cells.1 The mouse genome contains six expressed genes (Zscan4a–Zscan4f, also referred to as the Zscan4 gene or mZscan4 gene and ZSCAN4 protein or mZSCAN4 protein as a group in this paper) and three pseudogenes (Zscan4-ps1–Zscan4-ps3), whereas the human genome contains only one gene (ZSCAN4, also referred to as the hZSCAN4 gene and hZSCAN4 protein in this paper to distinguish from mZscan4 gene mZSCAN4 protein).1 In mES cells, mZSCAN4 expression is transient and reversible, resulting in 1–5% ZSCAN4(+) cells and ZSCAN4(−) cells at a given time.2 Both normal and ectopic expression of mZSCAN4 decreases spontaneous sister chromatic exchange (SCE) but increases telomere SCE, thereby extending telomeres in mES cells.2 As elevated SCE indicates genome instability, the suppression of SCE by mZSCAN4 suggests that ZSCAN4 enhances genome stability.2 Furthermore, ectopic mZSCAN4 expression protects mES cells from DNA-damaging agents, such as mitomycin C.3 However, the mechanism by which mZSCAN4 contributes to genome stability and protection is not fully understood.
The regulation of mZscan4 is associated with global chromatin changes.4–6 It is, fittingly, upregulated by histone deacetylase inhibitor treatment,7 the deletion of Trim28 (aka Kap1) chromatin repressor,8 histone demethylase Kdm1a (aka Lsd1),9 and chromatin assembly factor Caf-1.10 Both mZSCAN4 and hZSCAN4 are also induced by mouse DUX and human DUX4, known for global transcriptional activation, including genes in heterochromatin regions11 and as the causative gene for facioscapulohumeral muscular dystrophy.12–14 mZSCAN4 expression is accompanied by heterochromatin decondensation, gene activation, and activation of retrotransposons that are selectively transcribed in mouse two-cell embryos.6,8,15 While the chromatins are in a highly active state, ZSCAN4 forms a complex with select chromatin repressors, including TRIM28, LSD1, and HDAC1,6,16 suggesting that mZSCAN4 enhances genome stability through repressing chromatin. However, it is not clear whether these mZSCAN4 and hZSCAN4 functions are mediated through direct binding to DNA, ZSCAN4’s association with other proteins involved in chromatin regulation, or both.
The evidence for direct binding of hZSCAN4 to DNA and binding motifs have been presented for the hZSCAN4 by the systematic evolution of ligands by exponential enrichment (SELEX).17 However, chromatin immunoprecipitation sequencing (ChIP-Seq) studies have not been performed for hZSCAN4, and therefore, hZSCAN4 binding sites on the human genome are not known. On the other hand, ChIP-Seq has been performed for the mZSCAN4 on mouse genomic DNA.18–20 These ChIP-Seq studies for mZSCAN4 used a peptide tag such as FLAG to do ChIP-seq to search for binding sequences, which requires the expression of a tag-fused mZSCAN4 protein.18–20 Although it is a common practice to use a tag-fused protein to do ChIP-seq, it is highly informative to perform ChIP-seq using an antibody against a protein itself so that the binding sites of endogenous protein can be detected. Interestingly, one study that analysed endogenous mZSCAN4 binding sites also revealed that mZSCAN4 protects the genome from DNA damage in mouse two-cell embryos by binding to subsets of CA/TG microsatellites.20
In this study, we performed ChIP-seq analyses of endogenous mZSCAN4 using an antibody against mZSCAN4. Furthermore, we also performed ChIP-seq analyses of endogenous hZSCAN4 using antibodies against hZSCAN4. To this end, we raised antibodies against hZSCAN4. One of the challenges was that the endogenous mZSCAN4 and hZSCAN4 are not constitutively expressed, and thus, we induced the expression of ZSCAN4 by treating cells with retinoic acid (RA)s in mES cells or by the overexpression of DUX4 in human embryonic stem (hES) cells. We revealed that mZSCAN4 and hZSCAN4 commonly bind to the TGCACAC motif enriched in CA/TG microsatellites.
2. Materials and methods
2.1. Culture of mES cells and mouse embryonic fibroblast
MC1 mES cells (129S6/SvEv)21 were cultured on gelatine-coated feeder-free plates in DMEM (Gibco) with 15% fetal bovine serum (FBS) (Atlanta Biologicals), 1,000 U/ml leukemia inhibitory factor (ESGRO, Chemicon), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 2 mM GlutaMAX, 0.1 mM beta-mercaptoethanol, and penicillin/streptomycin (50 U/50 µg/ml). The medium was changed daily, and cells were routinely split every 2–3 days. To increase mZSCAN4 positive cells, all-trans-RA was added to the medium at a final concentration of 1 μM. Mouse embryonic fibroblast (MEF) cells were cultured in DMEM (Gibco) with 15% FBS (Atlanta Biologicals), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 2 mM GlutaMAX, 0.1 mM beta-mercaptoethanol, and penicillin/streptomycin (50 U/50 µg/ml).
2.2. hES cell culture
SEES3 hES cells22 were obtained from the Center for Regenerative Medicine, National Research Institute for Child Health and Development, Japan. The cells were cultured in StemFit AK-02 medium (Ajinomoto) on iMatrix-511 (Nippi)-coated plates. The medium was changed daily, and cells were routinely split every 4–5 days. ES cells were transfected with synthetic mRNA encoding DUX4, as previously described,23–25 to induce hZSCAN4 expression. Briefly, DUX4 cDNA11 was subcloned into a plasmid containing a T7 promoter. DUX4 mRNA was synthesized following a previously described in vitro transcription protocol.26 The synthesized RNA was then transfected with Lipofectamine MessengerMAX (Invitrogen) according to the instructions.
2.3. Generation of hZSCAN4-Emerald knock-in hES cells
The targeting vector was designed to replace exon 3–5 of the hZSCAN4 genomic locus with Emerald-green-fluorescent-protein (Emerald)-polyA followed by a neomycin-resistant (Neo) gene cassette. Targeting arms of 1,295 bp (5') and 1,210 bp (3') fragments to the hZSCAN4 gene were generated by PCR from hES cell genomic DNA and directionally cloned in pKOII plasmid, flanking a pGK-Neo-polyA and a DT-A cassette. The homologous recombinant cells were isolated using hES cells after transfection of the targeting vector together with CRISPR/Cas9 pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene #42230) encoding specific guide RNA which directs 5'-cacagtcagttagagttgtc -3' at 3' downstream of exon 3 of the hZSCAN4 genomic locus. The G418-resistant hES clones were screened for homologous recombination in the hZSCAN4 locus by PCR using primers, hZ4-Wt-15257F (5'-gattcagggagtacatgtgcatgtttg-3') and hZscan4-EM-R (5'-cagctcctcgcccttgctcaccat-3') for 5'-arm (2036 bp); and hZscan4-Neo-F (5'-ACGGTATCGCCGCTCCCGATTCGC-3') and hZscan4-21853R (5'-ttgttcctcagcaggtaaagtgcc-3') for 3'-arm (1755 bp). The hZSCAN4-Emerald knock-in allele was verified to be heterozygous by Southern blotting after HindIII digestion using 5' probe.
2.4. Generation of hZSCAN4-knockout hES cells
In the hZSCAN4-Emerald knock-in ES cells, one allele of hZSCAN4 is replaced by Emerald, and the other allele is intact. To target the intact allele, the hZSCAN4-Emerald knock-in hES cells were transfected with CRISPR/Cas9 pX330-U6-Chimeric_BB-CBh-hSpCas9 vector encoding specific guide RNA, which directs 5'-gaaccatccgagaataatct-3' at 31 bp downstream from the start codon sequence (ATG) of the hZSCAN4 genomic locus. Fourteen colonies were picked and screened for indel mutations, which resulted in frameshifts leading to premature stop codons. PCR amplification and Sanger sequencing analysis were performed using primers, hZSCAN4-genomic-s2 (5'-gaagtgctgacctcagtaac-3') and hZSCAN4-genomic-a2 (5'-cagccatgagtgaaagatcc-3').
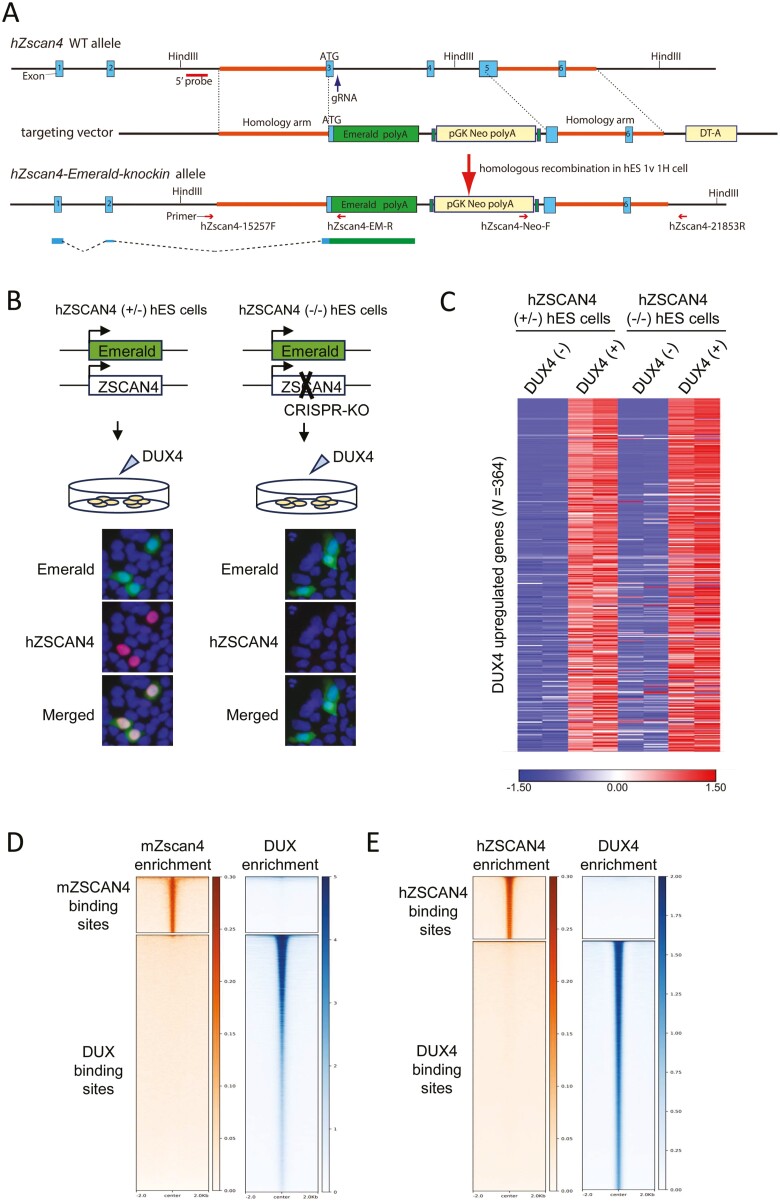
Deletion of the protein was confirmed by immunostaining using a specific antibody, as shown in Fig. 6.
Figure 6.
hZSCAN4 deletion does not affect gene expression changes induced by DUX4. (A) Schematic illustrations of the hZSCAN4 wildtype allele, the ZSCAN4-Emerald knock-in allele and the targeting vector. Coding exon 3 is followed by Emerald and polyA signal. 5' probe for Southern blotting is shown. (B) Experimental scheme for generating hZSCAN4 (+/−) and (−/−) hES cells, in which Emerald GFP replaces one allele of hZSCAN4, and the other allele is either intact (+/−) or disrupted by CRISPR-Cas9 (−/−). Those cells were transfected with synthetic mRNA of DUX4 to induce DUX4-regulated transcriptional burst, including hZSCAN4 expression. Immunostaining confirmed that hZSCAN4 was expressed in hZSCAN4 (+/−) cells but not in hZSCAN4 (−/−) cells. Emerald GFP was expressed in both cell lines. Nuclei were stained with DAPI (Blue). (C) RNA-sequencing analysis showed that transcriptional burst was similarly detected in hZSCAN4 (+/−) cells and hZSCAN4 (−/−) cells. Three hundred and sixty-four genes are upregulated by DUX4 induction in hZSCAN4 (+/−) cells (fold change > 2 compared with non-transfected cells). The data from two biological replicates are shown. The colour scale bar shows z-score values. (D) Density heatmaps showing enrichment levels of mZSCAN4 (this study) and DUX (GSE95517_mDUX-HA-rep1.bw)12 within a 4 kb window centred at mZSCAN4 and DUX binding sites. (E) Density heatmaps showing enrichment levels of hZSCAN4 (this study) and DUX4 (GSM2515762_DUX4-rep2.bw)12 within a 4 kb window centred at hZSCAN4 and DUX4 binding sites.
2.5. Generation of hZSCAN4 antibodies
The cDNA fragment encoding hZSCAN4 (a.a.1-432) protein was inserted in-frame into pET19b plasmid (Novagen), and the hZSCAN4 protein was produced in the Escherichia coli strain BL21-CodonPlus (DE3). His-tagged recombinant proteins were solubilized in a denaturing buffer (6 M HCl-Guanidine, 20 mM Tris–HCl [pH 7.5]) from the inclusion body and purified by Ni-NTA (QIAGEN) under denaturing conditions. After dialysing against phosphate-buffered saline (PBS), the purified protein was used to immunize mice, rats, and rabbits. The polyclonal antibodies were affinity-purified from the immunized crude serum with immobilized antigen on CNBr-activated Sepharose (GE Healthcare).
2.6. ChIP-seq for mZSCAN4 and hZSCAN4
ChIP experiments were performed with minor modifications according to a previously established protocol.27 Cells were cross-linked with 2 mM disuccinimidyl glutarate in PBS/1 mM MgCl2 for 40 min at room temperature, followed by 1% formaldehyde in PBS for 15 min at room temperature. The reaction was stopped by 125 mM glycine. The cells were washed with PBS and stored at –80°C prior to use. The cells were lysed in lysis buffer (10 mM Tris–HCl, pH8.0, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 0.1% Na-Deoxycholate, 0.5% N-lauroyl sarcosine) containing proteinase inhibitor cocktail (Roche). Sonication was conducted with the Handy Sonic UR-21P (Tomy) to generate DNA fragments of approximately 150–450 bp. The sonicated lysates from approximately 2 × 106 cells were diluted in ChIP dilution buffer (10 mM HEPES, pH 7.4, 50 mM NaCl, 1% IGEPAL-CA-630, 10% glycerol) containing proteinase inhibitor cocktail and incubated overnight at 4°C with 50 µl of protein G magnetic beads (Invitrogen) that were preincubated with ~2 µg of mZSCAN4 and hZSCAN4 antibodies. The precipitants were washed once with high salt wash buffer (20 mM Tris–HCl, pH 8.0, 400 mM NaCl, 2 mM EDTA, 0.1% SDS, 0.2% Triton-X), three times with LiCl wash buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 250 mM LiCl, 1% NP40, 1% Na-deoxycholate), and once with 10 mM Tris–HCl, pH 8.0, 5 mM EDTA, 10 mM NaCl. Bound chromatin was eluted in elution buffer (90 mM NaHCO3, 1% SDS), followed by RNase treatment at 37°C for 30 min and cross-link reversal with a decross-linking mixture (2 M NaCl, 0.1 M EDTA, 0.4 M Tris–HCl, pH 6.8) containing proteinase K at 65°C for 3 h. DNA was purified by phenol–chloroform–isoamyl alcohol extraction and ethanol precipitation.
2.7. Processing and analysis of ChIP-seq data
ChIP DNA libraries were prepared with the NEBNext ChIP-Seq Library Prep Kit for Illumina (New England BioLabs) and sequenced on an Illumina HiSeq 2500 using 50-nucleotide read length single-end sequencing at Macrogen. Fastq files from the Illumina pipeline were processed and analysed on the Biowardrobe platform.28 Sequence reads were aligned to the mouse (mm10) or human genome (hg19) using bowtie (version 1.2.0)29 with a maximum of one error in a sequence and one hit. MACS2 (version 2.1.1.20160309)30 was used to estimate fragment size and to find islands of enrichment with a q-value threshold less than 0.2. The data were uploaded to the UCSC genome browser for visualization, and fragment coverage was used with estimated fragment size from the MACS2 output. MAnorm31 was used to compare ChIP peaks between ZSCAN4 positive cells and control cells to identify ZSCAN4 specific peaks. Heat maps and average signal profiles were generated by Easeq32 or deepTools.33 BigWig, BAM, or BED files of sequence reads were uploaded as datasets. Doublet reads were excluded, and only one read at each position was allowed to map at each strand. Reads were normalized per million per 1 kbp. Heat maps show signals segmented in 200 bins. The X-axis represents regions surrounding the centre of the peaks. The regions were sorted on the Y-axis according to signals quantified within Easeq or deepTools. Signal profiles show average signals segmented into 400 bins with 1 bin smoothing at the regions surrounding the centre of the peaks. Motif discovery and enrichment analyses were performed by MEME-ChIP34 using Galaxy.35 FASTA files were created within Galaxy. All mZSCAN4 and hZSCAN4 binding regions were used as input for the analysis. The input was shuffled to create the negative set. Lists of mouse and human (CA/TG)n repeats were downloaded from RepeatMasker36 annotation track on the Mouse (mm10) and Human (hg19) UCSC genome browser. Overlap analysis of ZSCAN4 binding sites and simple repeats were performed using Galaxy. Identification of the ZSCAN4 binding motif (TGCACAC/GTGTGCA) in the (CA/TG)n repeats was performed using FIMO (version 5.1.1).37 The (CA/TG)n with more than 100 bp was used for this analysis.
2.8. RNA-sequencing analysis
Total RNA was extracted using TRIzol solution (Roche) according to the manufacturer’s instructions. cDNA libraries were prepared from 500 ng of each total RNA sample for massive parallel sequencing using the NEBNext Poly(A) mRNA Magnetic Isolation Module and an Ultra Directional RNA Library Prep Kit for Illumina (New England BioLabs). The cDNA library contained DNA ranging from 400 to 1000 bp, including the adaptor sequences. RNA-seq was performed on an Illumina HiSeq 2500 using 50-nucleotide read length single-end sequencing at Macrogen. The sequence reads were mapped to the human genome (hg19), and the expression values for genes were calculated as reads per kilobase of exon per million mapped reads (RPKM) using the Biowardrobe platform. Heatmaps were generated using the Morpheus software developed by the Broad Institute.
2.9. Datasets from publicly available sources
Previously published datasets were downloaded from the Gene Expression Omnibus (GEO) database: ATAC-seq (GSE85624),12 H3K27ac ChIP-seq (GSE51682),6 ZSCAN4-OE H3K27ac and H3K4me1 ChIP-seq data (GSE125238),18 DUX/DUX4 ChIP-seq (GSE85632).12
2.10. Data availability
All sequencing data have been deposited in the GEO database under series accession number GSE243628.
3. Results
3.1. Genome-wide analysis of endogenous mZSCAN4 binding sites in mES cells
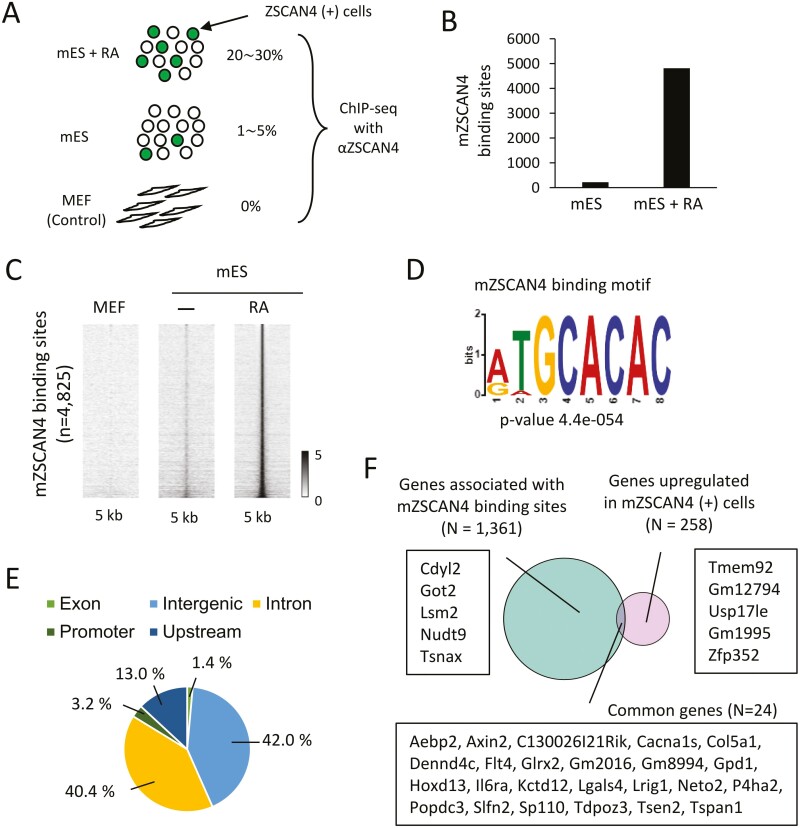
Due to the infrequent expression of endogenous mZSCAN4 in mES cells (1–5% of cells),1,2 we6 and others20 have FACS-sorted ZSCAN4(+) cells using mZscan4c promoter sequence identified previously.2 However, to take advantage of anti-mZSCAN4 antibodies to capture endogenous mZSCAN4, we first performed, without the enrichment of ZSCAN4(+) cells, chromatin immunoprecipitation with polyclonal antibodies against mZSCAN4,16,38 followed by sequencing analysis (ChIP-seq) and identified genome-wide binding sites of mZSCAN4 (Fig. 1A). We also performed ChIP-seq analyses of mES cells after treating with RA, because the RA treatment increased the percentage of ZSCAN4(+) cells to 20–30%.16,39 MEF, which does not express mZSCAN4, were used as negative controls. We identified 213 and 4,825 ChIP-seq peaks as mZSCAN4 binding sites in untreated mES cells and RA-treated mES cells, respectively (Fig. 1B). Binding sites were identified with weak signals in untreated mES cells but with strong signals in RA-treated mES cells (Fig. 1C). This suggests that RA treatment increased the number of ZSCAN4(+) cells and that ZSCAN4 binds to the same regions as untreated mES cells.
Figure 1.
Genome-wide profiles of mZSCAN4 binding sites. (A) Experimental scheme for mZSCAN4 ChIP-seq. mES + RA, mES cells after treatment with RA for 48 h. mES, mES cells without treatment of RA. MEF, mouse embryonic fibroblasts. (B) The number of mZSCAN4 bound sites identified in mES and mES + RA. (C) Heatmaps showing mZSCAN4 ChIP-seq signals at the mZSCAN4-bound sites in MEF, mES, and mES + RA cells. (D) Sequence motif enriched in mZSCAN4 binding regions. (E) Genomic distribution of mZSCAN4 binding sites (N = 4,825). (F) Analysis of gene expression and mZSCAN4 binding site relationships. Genes associated with mZSCAN4 binding sites are genes containing mZSCAN4 binding sites within 5,000 bp upstream, 1,000 bp downstream, and 20,000 bp max extension. GREAT tool (version 4.0.4)68 was used for this analysis. Upregulated genes are derived from previous RNA-seq data6 (Upregulation fold change > 2. P-value < 0.01). Example genes for each category are shown. For genes upregulated in Zscan4 (+) cells and associated with Zscan4 binding sites, all 24 gene symbols are shown. Among these 24 genes, only C130026I21Rik was upregulated, and only Gpd1 was downregulated when mZSCAN4 was overexpressed in mES cells.50
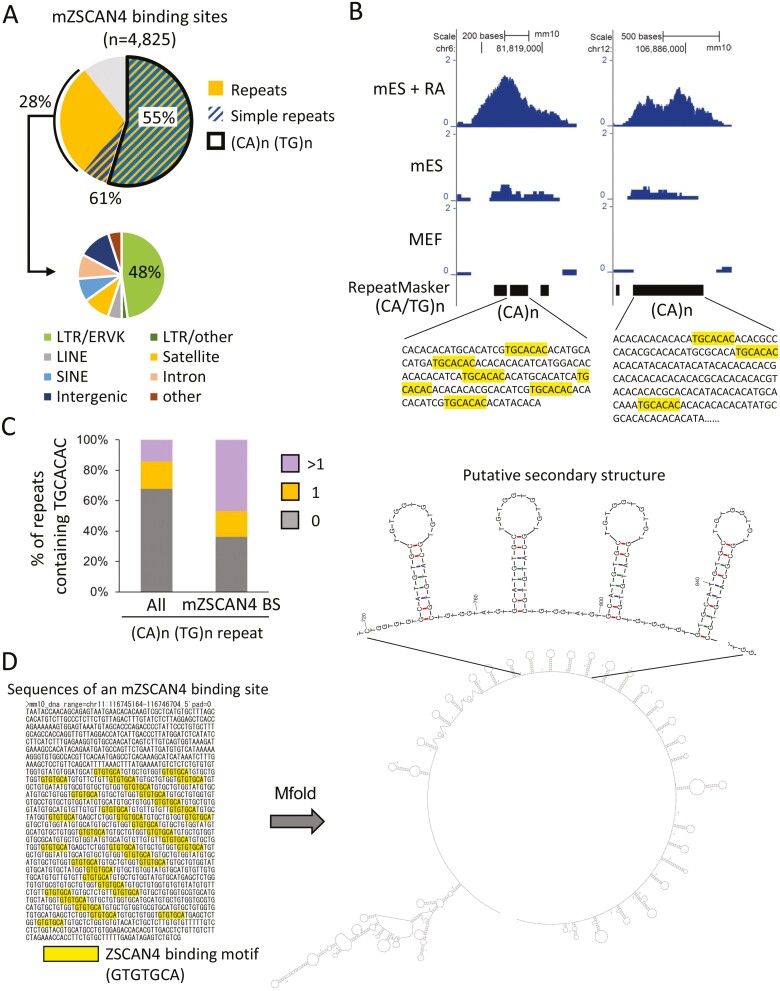
Motif discovery and enrichment analysis identified binding sites that were significantly enriched with a specific sequence motif: TGCACAC (Fig. 1D). This motif was essentially the same as the motifs identified by the SELEX for hZSCAN4 in vitro previously (JASPER ID, MA1155.1)17 and by the ChIP-seq for mZSCAN4.18,20 Genomic distribution analysis revealed that mZSCAN4 binding sites accumulate in intronic (40%) and intergenic (42%) regions of the genome (Fig. 1E), indicating that mZscan4 may bind in enhancer regions. The binding to the enhancer region suggests that mZSCAN4 increases the expression of downstream genes by binding these sites; however, we did not find significant correlations between mZSCAN4 binding sites and gene expression (Fig. 1F, Supplementary Tables S1–S3). Instead, we found that mZSCAN4 binding sites overlap with simple sequence repeats, especially repeats in (CA/TG)n repeating regions (Fig. 2A). Of 4,825 identified mZSCAN4 binding sites, 61% overlap with simple sequence repeats and 55% overlap with (CA/TG)n regions (Fig. 2A). The rest of mZSCAN4 binding sites are enriched with LTR/ERVK transposable elements (~13% [i.e. 48% of 28%]) (Fig. 2A). Among the ~300,000 regions that contain (CA/TG)n repeats in the mouse genome, mZSCAN4 binds to 4,825 specific regions within them. These results confirm the findings of Srinivasan et al.20 and suggest that mZSCAN4 is not primarily a transcription factor, as has been assumed previously, but rather regulates genome stability by binding to DNAs.
Figure 2.
Analysis of mZSCAN4 binding sites enriched with (CA/TG)n repeats. (A) Percentages of mZSCAN4 bound sites overlapping with the whole simple sequence repeats and (CA/TG)n repeats. mZSCAN4 bound repeats other than simple repeats (28%) are subclassified into transposable elements and satellite repeats. (B) Representative mZSCAN4 ChIP-seq signals detected at the simple repeat sequences containing the mZSCAN4 binding motif (TGCACAC). mES + RA, mES cells treated with RA; mES, untreated mES cells. MEF, mouse embryonic fibroblast cells. (C) Percentages of (CA/TG)n repeats containing mZSCAN4 binding motif (TGCACAC). All, whole (CA/TG)n repeats; mZSCAN4 BS, (CA/TG)n overlapped with mZSCAN4 bound sites; 0, repeats without the motif; 1, repeat with a single motif; >1, repeat with multiple motifs. (D) The putative secondary structure of mZSCAN4-binding regions. An example of mZSCAN4 binding sites is shown on the left (mm10, chr11:116745165-116746704). The DNA folding form was predicated by Mfold.44
We also wondered whether mZSCAN4 binds preferentially to (CA/TG)n repeats or to the mZSCAN4-binding motif, TGCACAC, which is buried in some (CA/TG)n repeats (Fig. 2B). Indeed, among all the (CA/TG)n repeats, ~30% contains more than one TGCACAC motif. However, among the subset of (CA/TG)n repeats where mZSCAN4 binds, more than half (55%) contain TGCACAC motif (Fig. 2C). These results indicate that mZSCAN4 binds primarily to TGCACAC motif, which is enriched in the (CA/TG)n repeats.
The (CA/TG)n repeats are known to form a left-handed helix structure that contributes to genome instability and human diseases.40–43 This prompted us to analyse the secondary structure of mZSCAN4-binding regions using the bioinformatics tool.44 We found that mZSCAN4-binding regions form tandemly repeated stem-loop structures (Fig. 2D). Such DNA structures contribute to genome instability and human diseases.45–47
3.2. Genome-wide analysis of endogenous hZSCAN4 binding sites in hES cells
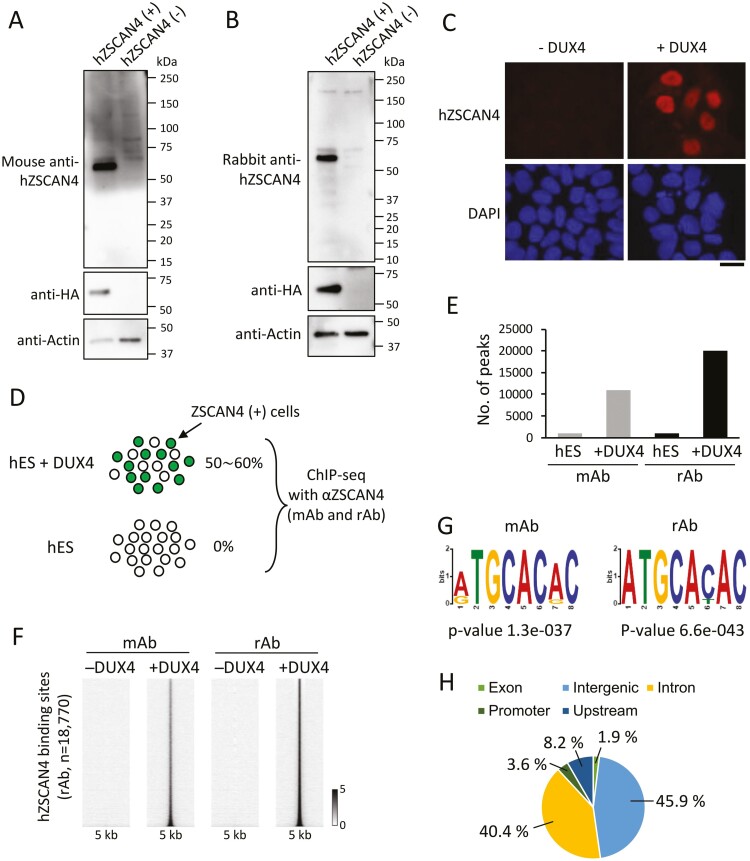
We next performed a ChIP-seq analysis of hZSCAN4 in the human genome. To this end, we generated and tested two antibodies against hZSCAN4: one produced in mice (mAb) (Fig. 3A, Supplementary Fig. S1) and another in rabbits (rAb) (Fig. 3B, Supplementary Fig. S1). To validate these antibodies, we transfected hES cells with or without a synthetic mRNA (synRNA) encoding for hZSCAN4 conjugated with the 3× FLAG and haemagglutinin peptide tags (ZSCAN4-3xFLAG-HA). Western blots showed that both mouse and rabbit polyclonal antibodies raised against hZSCAN4 protein recognized a unique protein of correct size in a synRNA-ZSCAN4-specific manner, successfully validating both antibodies.
Figure 3.
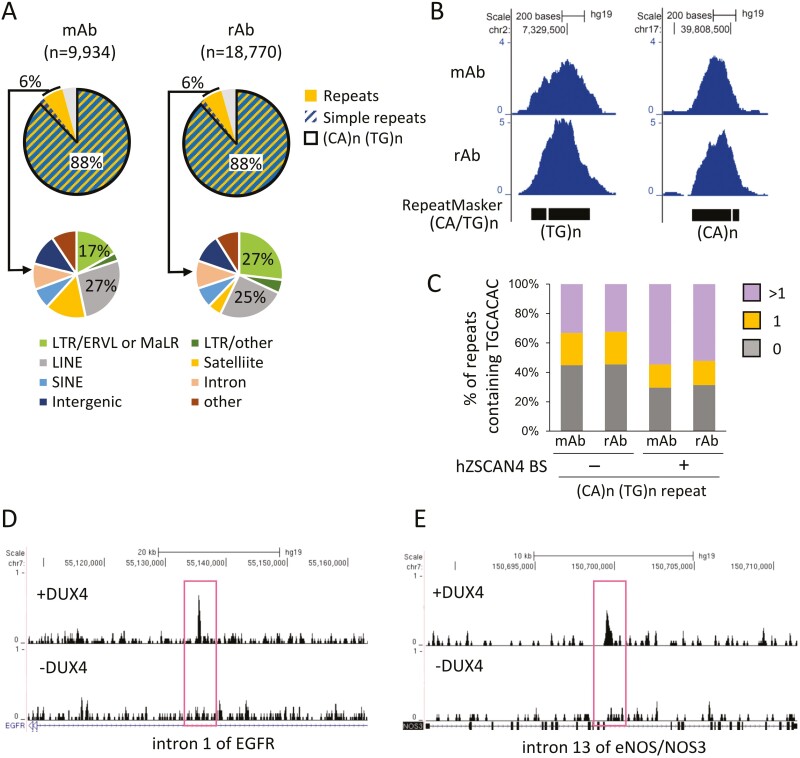
Genome-wide profiles of hZSCAN4 binding sites. (A) Specificity of mouse anti-human ZSCAN4 antibodies. Whole-cell lysates of hES cells transfected with (+) or without (−) synthetic RNA encoding hZSCAN4-3xFLAG-HA were probed with anti-hZSCAN4 antibodies raised in mice. Expression of ZSCAN4-3xFLAG-HA was probed by an anti-HA antibody as a positive control. (B) Rabbit anti-hZSCAN4 antibodies were examined, as in A. (C) hES cells were transfected with (+) or without (−) synthetic mRNA encoding for DUX4. Immunostaining analysis was performed using hZSCAN4 antibodies at 20 h post-transfection. DNA was counterstained with DAPI. Bar, 20 µm. (D) Experimental scheme for hZSCAN4 ChIP-seq. ChIP-seq experiments were conducted using mouse antibody (mAb) and rabbit antibody (rAb) against hZSCAN4 in hES cells overexpressed with DUX4 (hES + DUX) and non-treated hES cells. (E) The number of hZSCAN4 ChIP peaks in hES and hES +DUX identified by mAb and rAb ChIP-seq. (F) Heatmaps showing hZSCAN4 ChIP-seq signals at the hZSCAN4 binding sites identified by mAb and rAb in hES and hES+DUX cells. (G) Sequence motifs enriched in hZSCAN4 binding regions identified by mAb and rAb ChIP-seq. (H) Genomic distribution of hZSCAN4 binding sites (N = 18,770).
Unlike mZSCAN4 expression in mES cell cultures, hZSCAN4(+) cells are not usually observed in hES cell cultures. We, therefore, expressed DUX4 ectopically in hES cells (SEES3 line),22 as it is known that overexpression of DUX4 induces hZSCAN4 expression in hES cells.12–14 Following DUX4 induction, most cells expressed hZSCAN4 proteins (Fig. 3C).
ChIP-seq experiments used DUX4-overexpressed (+DUX4) or control wildtype (−DUX4) hES cells and the mAb and rAb hZSCAN4 antibodies to identify hZSCAN4 binding sites (Fig. 3D). The peak identification analysis demonstrated that both antibodies could detect a more than a 10-fold increased number of hZSCAN4 peaks in +DUX4 cells compared to −DUX4 cells (Fig. 3E). The number of peaks identified in +DUX4 cells using the rAb (18,770 peaks) was larger than the number identified by the mAb (9,934 peaks). Still, most of the mAb-detected signals with non-peaks were localized to sites also identified by rAb (Fig. 3F). This result indicates that both antibodies detect hZSCAN4 binding sites, but that rAb is more sensitive than mAb.
hZSCAN4-binding sites identified by both antibodies are enriched with the same DNA motif as the mZSCAN4 binding motif (TGCACAC; Fig. 3G). Several other characteristics of hZSCAN4 binding sites were also similar to mZSCAN4. For instance, hZSCAN4 occupies intronic (40%) and intergenic (45%) regions, which was similar to the proportions observed in mZSCAN4 (Fig. 3H). Additionally, hZSCAN4 binding sites correspond to (CA/TG)n repeats (Fig. 4A and B), and multiple hZSCAN4 motifs are contained in hZSCAN4-binding (CA/TG)n repeats (Fig. 4C). Nearly 60% of (CA/TG)n repeats bound by hZSCAN4 were abundant with hZSCAN4 consensus motif, whereas only 30% of hZSCAN4 unbound repeats are categorized in repeats with multiple motifs (Fig. 4C). Exemplary hZSCAN4-binding sites and sequences are shown for eNOS intron and EGFR intron (Fig. 4D and E). From these results, we concluded that both mZSCAN4 and hZSCAN4 bind to the TGCACAC motif in (CA/TG)n repeats. Interestingly, compared to mZSCAN4, hZSCAN4 binds more exclusively to (CA/TG)n repeats.
Figure 4.
Analysis of hZSCAN4 binding sites enriched with (CA/TG)n repeats. (A) Percentages of hZSCAN4 binding sites overlapping with the whole simple sequence repeats and (CA/TG)n repeats. hZSCAN4 bound repeats other than simple repeats (6%) are subclassified into transposable elements and satellite repeats. They are enriched with ERVL, MaLR, and LINE. (B) Representative hZSCAN4 ChIP-seq signals detected at the simple repeat sequences. (C) Percentages of (CA/TG)n repeats containing hZSCAN4 binding motif (TGCACAC) compared with percentages of repeats overlapping with hZSCAN4 binding sites (+) and without hZSCAN4 binding sites (−). 0, repeats without the motif; 1, repeat with a single motif; and >1, repeat with multiple motifs. (D and E). UCSC genome browser snapshots of hZSCAN4 ChIP-seq in DUX4 treated hES cells (+DUX4) and non-treated hES cells (−DUX4). hZSCAN4 peaks were detected in intron 1 of EGFR and intron 13 of eNOS (NOS3).
3.3. Comparison of ZSCAN4-binding sites between mouse and human
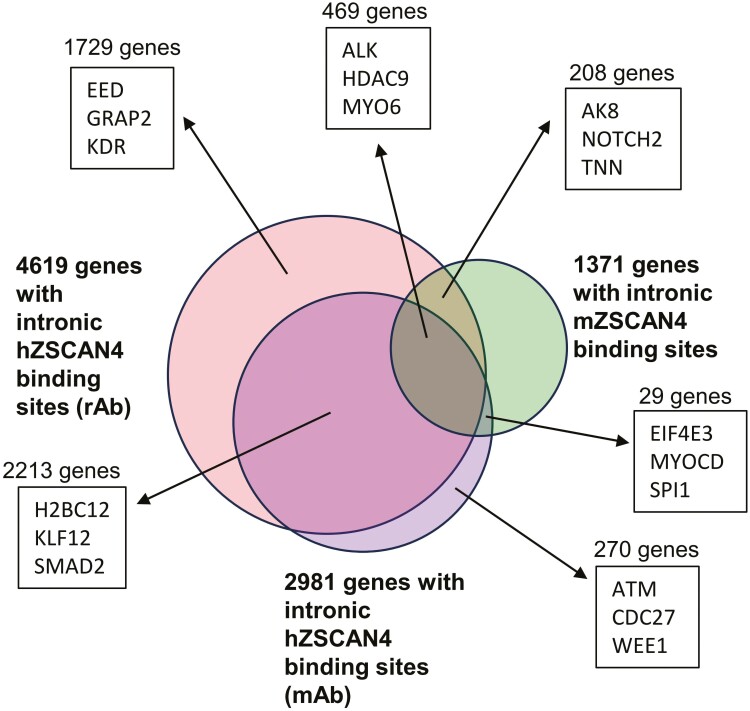
The results thus far suggest that mouse and hZSCAN4-binding sites are not directly linked to the function of genes but rather the structure of genomic DNAs. In general, the location of microsatellites in the genome is not evolutionary conserved among species, and only about 7% are conserved between human and mouse.48 If the locations of (CA/TG)n microsatellites are not well conserved between mouse and human, ZSCAN4-binding sites may also not be conserved. To test this notion, we first compared ZSCAN4-binding sites of introns that can be assigned to each gene between mouse and human genomes (Fig. 5, Supplementary Tables S4 and S5). For the human data set, ChIP-seq data from two hZSCAN4 antibodies (rabbit and mouse) were used: 4,619 genes were found by rabbit ab, and 2,981 genes were found by mouse ab. For the mouse data set, 1,371 genes were used. Among the intronic mZSCAN4-binding sites, 677 (49.4%) genes were overlapped with hZSCAN4 (rabbit Ab), and 498 (36.2%) genes were overlapped with hZSCAN4 (mouse Ab). However, a closer look at the aligned genomic regions of overlapped genes revealed that the microsatellites were not located in the same locations, and the binding sites were not overlapped between mouse and human genomes (Supplementary Fig. S2). Furthermore, the average size of the overlapped genes was much larger than the average size of all the genes: for human, 322,755 bp (the average size of the overlapped genes) versus 45,631 bp (the average size of all human genes); for mouse, 220,824 bp (the average size of the overlapped genes) versus 40,833 bp (the average size of all mouse genes). Thus, the overlapped genes identified in this analysis are most likely due to the higher chance of having at least one intronic ZSCAN4-binding microsatellite sequence in large genes. These results further support the notion that the function of both mouse and human ZSCAN4 is associated with genomic DNAs but not specific genes and their transcriptional regulation.
Figure 5.
The number of ZSCAN4-bound genes that overlap between mouse and human genomes. Mouse and human ChIP-seq peaks are analysed by HOMER’s annotatePeaks.pl to find the genes associated with ZSCAN4 binding. Genes with intron-annotated peaks were selected and compared between human and mouse genomes. ChIP-seq data from two hZSCAN4 antibodies (rabbit and mouse) were used for this analysis. Four thousand six hundred and nineteen genes were found by rabbit Ab, and 2,981 genes were found by mouse Ab. Thousand three hundred and seventy-one genes were used as mZSCAN4-binding genes. Venn diagram shows the overlapping and non-overlapping between mouse and human genomes for each gene set. The number of genes and the example gene names for each set are shown.
3.4. hZSCAN4 knockout does not affect transcriptome burst induced by DUX4 in hES cells
The results thus far suggest that ZSCAN4 does not function as a transcription factor but is involved in genome regulation. However, several reports, including ours, showed that mZSCAN4 is involved in the transcriptional regulation of two-cell embryos and germ cells in mice.18,49 Also, mZSCAN4(+) cells are accompanied by the activation of hundreds of genes and transposable elements, including MERVL.6,8,15,18 hZSCAN4 overexpression in hES cells upregulated 201 genes and downregulated 477 genes (FDR ≤ 0.05 and fold change ≤ 2).11,50 On the other hand, these are a relatively small number of genes, especially compared to DUX4, whose overexpression upregulated 11,733 genes and downregulated 1,517 genes (FDR ≤ 0.05 and fold change ≤ 2).11 DUX4 is a transcription factor, and its overexpression induces hZSCAN4 expression in hES cells.12–14
To investigate whether ZSCAN4 acts as a transcription factor in DUX4 transcriptional regulation, we generated hES cells, in which one ZSCAN4 allele was replaced by an emerald-green fluorescent-protein (Emerald), and the other ZSCAN4 allele was either intact (ZSCAN4+/−) or disrupted by CRISPR/Cas9 (ZSCAN4−/−) (Fig. 6A, Supplementary Fig. S3). We then expressed DUX4 ectopically and performed the transcriptome analyses (Fig. 6B and C). The results showed that hZSCAN4 knockout did not affect transcriptome changes induced by DUX4 in hES cells (Fig. 6C). Furthermore, DUX binding sites and mZSCAN4 binding sites did not overlap in the mouse (Fig. 6D) and human genomes (Fig. 6E).
Taken together, the massive transcriptome changes induced by DUX4 are not mediated by hZSCAN4, further suggesting that hZSCAN4 is not primarily a transcription factor but regulates genome functions by binding to microsatellite DNA.
3.5. Histone modifications and chromatin accessibility at the mZSCAN4-binding sites
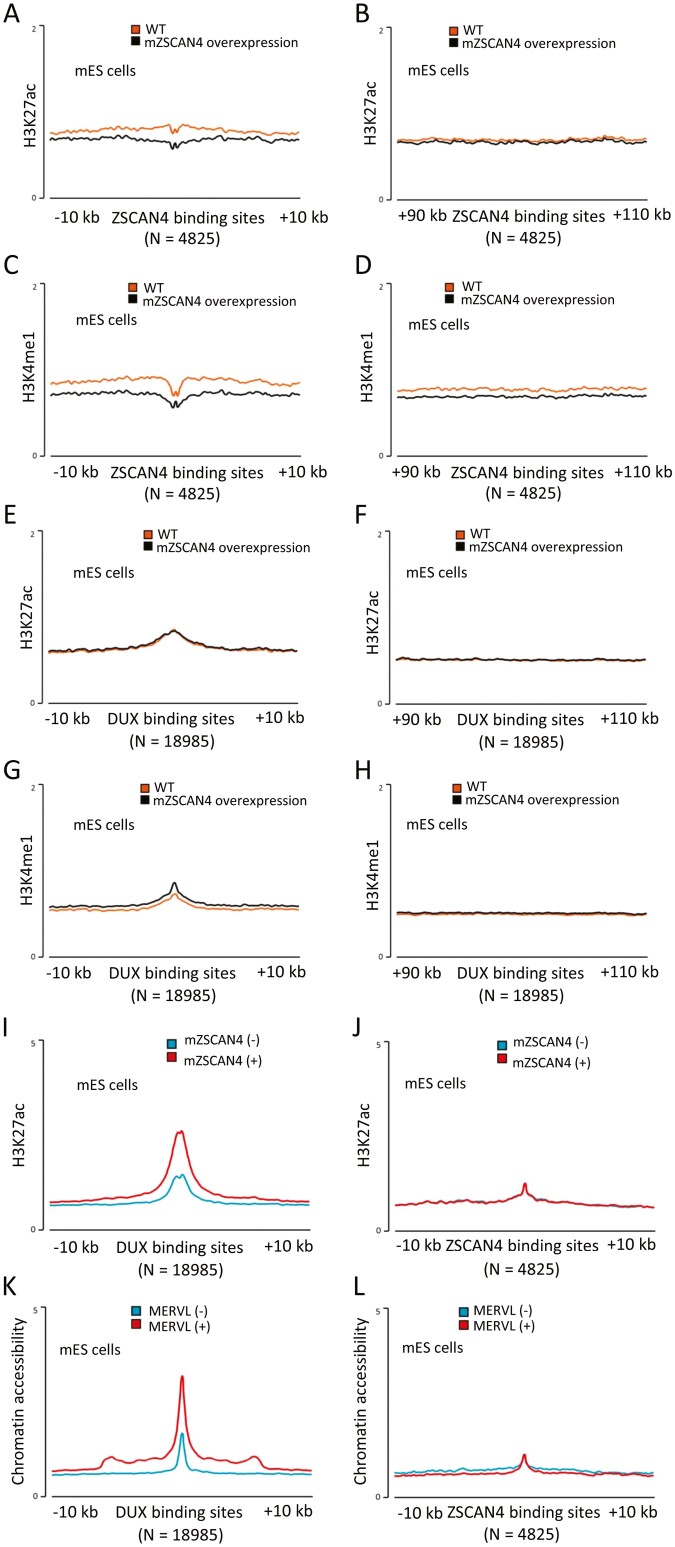
We have previously shown that mZSCAN4 complexes with chromatin repressive factors—KDM1A (aka LSD1), a demethylase of mono- or di-methylation of H3K4 (H3K4me1/2),51 and HDAC1, a deacetylase of histone H3/H4.6,16,52 To investigate the association of ZSCAN4-binding sites and chromatin regulation in mES cells, we examined the chromatin status of mZSCAN4-binding sites in previously published ChIP-seq data, in which histone modifications were analysed with or without the forced expression of mZSCAN4.18 The levels of active histone mark H3K27ac (acetylated histone H3 lysine 276) in mZSCAN4-binding sites were low even without mZSCAN4 overexpression (Fig. 6A). However, they were further reduced by mZSCAN4 overexpression (Fig. 7A), whereas those in non-mZSCAN4-binding sites (e.g. 100 kb away from the mZSCAN4-binding sites) were not changed by mZSCAN4 overexpression (Fig. 7B). Similarly, the levels of another active histone mark, H3K4me1 in mZSCAN4-binding sites, were slightly reduced by mZSCAN4 overexpression (Fig. 7C), whereas those in non-mZSCAN4-binding sites were not changed by mZSCAN4 overexpression (Fig. 7D). By contrast, both H3K27ac and H3K4me1 in DUX-binding sites were high, but not changed by mZSCAN4 overexpression (Fig. 7E and G), whereas those in non-DUX-bindings sites were low and not changed by mZSCAN4 overexpression (Fig. 7F and H). These results suggest that when mZSCAN4 is overexpressed, mZSCAN4 binds to mZSCAN4-binding sites and (CA/TG)n repeats, recruits chromatin repressors to the sites, and changes the chromatin to a further closed state. On the other hand, when mZSCAN4 is overexpressed, mZSCAN4 does not bind to the DUX binding site.
Figure 7.
Histone modifications and chromatin accessibility at the mZSCAN4-binding sites. (A) Average H3K27ac ChIP-seq profiles around mZSCAN4 binding sites (N = 4,825) in wildtype mES cells (WT) and mES cells overexpressing ZSCAN4 (ZSCAN4 overexpression). (B) Average H3K27ac ChIP-seq profiles around mZSCAN4 binding sites + 100 kb (N = 4,825) for controls. (C) Average H3K4me1 ChIP-seq profiles around mZSCAN4 binding sites (N = 4,825) in WT and mZSCAN4 overexpressing mES cells. (D) Average H3K4me1 ChIP-seq profiles around mZSCAN4 binding sites + 100 kb for controls. ChIP-seq data were reanalysed from previously published data.18 (E) Average H3K27ac ChIP-seq profiles around DUX binding sites (N = 18,985) in wildtype mES cells (WT) and mES cells overexpressing ZSCAN4 (ZSCAN4 overexpression). (F) Average H3K27ac ChIP-seq profiles around DUX binding sites + 100 kb (N = 18,985) for controls. (G) Average H3K4me1 ChIP-seq profiles around DUX binding sites (N = 18,985) in WT and mZSCAN4 overexpressing mES cells. (H) Average H3K4me1 ChIP-seq profiles around DUX binding sites + 100 kb (N =18,985) for controls. ChIP-seq data were reanalysed from previously published data.18 (I) Average H3K27ac ChIP-seq profiles around DUX binding sites (N = 18,990) in ZSCAN4 (−) and ZSCAN4 (+) cells. (J) Average H3K27ac ChIP-seq profiles around mZSCAN4 binding sites (N = 4,825) in ZSCAN4 (−) and ZSCAN4 (+) cells. Previously generated ATAC-seq data12 were reanalysed for A and B, and previously generated H3K27ac ChIP-seq data6 were reanalysed for C and D. (K) Average ATAC-seq profiles around DUX binding sites (N = 18,990) in MERVL (−) and MERVL (+) cells. (L) Average ATAC-seq profiles around mZSCAN4 binding sites (N = 4,825) in MERVL (−) and MERVL (+) cells.
Next, we investigated whether a naturally occurring transient expression of endogenous mZSCAN4 is associated with the changes in chromatin states in mES cells. ChIP-seq data of active histone mark H3K27ac showed that H3K27ac level of DUX binding site was high in ZSCAN4 (+) cells compared to ZSCAN4 (−) cells, whereas H3K27 level of mZSCAN4 binding site was low in both ZSCAN4 (+) cells and ZSCAN4 (−) cells (Fig. 7I and J). Similarly, chromatin accessibility examined by the previously published ATAC-seq analyses12 showed that DUX binding sites were high in MERVL (+) cells compared to MERVL (−) cells (Fig. 7K), as previously demonstrated.18 On the other hand, chromatin accessibility of the mZSCAN4 binding sites was low in both MERVL (+) and MERVL (−) cells (Fig. 7L). ZSCAN4 (+) cells and MERVL (+) cells are considered the same, as MERVL is usually co-expressed with mZSCAN4 in mES cells.4
These results indicate that in ZSCAN4 (+) mES cells (also MERVL (+) mES cells), chromatins of DUX binding sites were open, whereas chromatin of mZSCAN4 binding sites were closed. Unlike the mZSCAN4-overexpressing situation, the naturally occurring transient ZSCAN4 (+) state did not seem to further suppress the chromatin state of ZSCAN4-binding sites. This may be due to the transient nature of endogenous mZSCAN4 expression in mES cells. In mES cells, the ZSCAN4 (+) state is estimated to last for a short time,2 and thus, the suppression of chromatin may not be observed during the ZSCAN4 (+) state. Alternatively, the chromatin of the ZSCAN4-binding sites is already suppressed in the ZSCAN4 (−) state, and therefore, the further suppression of the chromatin state in ZSCAN4 (+) cells may not be observed.
4. Discussion
Since the initial identification of mZscan4 as a late two-cell embryo-specific gene,1 a variety of functions have been assigned to ZSCAN4, including telomere elongation,2 stabilization of genomes,2 karyotype corrections,15 DNA methylation inhibitors,5 enhanced generation of iPS cells with high quality.49,53 Because the ZSCAN4 protein has four zinc fingers and a SCAN domain, it was initially thought to be a transcription factor that binds to enhancer/promoter regions and is involved in the transcriptional regulation of two-cell embryos and germ cells in mice.18,49 On the other hand, it has also been demonstrated that ZSCAN4 forms protein complexes of various sizes, including chromatin remodellers and epigenetic regulators.6 Therefore, whether these ZSCAN4 functions are mediated primarily by direct binding to DNA, by association with other proteins involved in chromatin regulation, or both is unclear. Also, it was not clear whether ZSCAN4 indeed worked as a transcription factor. Our results show that the ZSCAN4 binding sites do not correlate with the expression of ZSCAN4-induced genes but are mostly located in intergenic and intronic regions of the genomes. Also, while there are many genomic sites that ZSCAN4 bind to, the ZSCAN4 overexpression alters the expression levels of a relatively small number of genes. We also showed that hZSCAN4 knockout does not affect transcriptome burst induced by DUX4 in hES cells.11 These results, together with previously published data, now support that ZSCAN4 indeed binds to the DNA but may not be directly involved in the transcriptional regulation.
The first evidence of ZSCAN4 binding to DNA was reported as a part of a large-scale identification of DNA binding motifs of transcription factors by SELEX.17 The SELEX in vitro method identified TGCACACACTGAAA as an hZSCAN4 binding motif but did not identify the location of ZSCAN4-binding sites in the genome. Zhang et al. expressed a mZSCAN4C-FLAG protein in mouse J1 ES cells, performed ChIP-seq using an anti-FLAG antibody, and identified GCACACACA as a mZSCAN4-binding motif.18 Srinivasan et al. expressed an mZSCAN4C-GFP fusion protein in mES cells, performed ChIP-seq using an anti-GFP antibody, and then identified TGCACACA as a mZSCAN4-binding motif.20 They also performed ChIP-seq using an anti-mZSCAN4 antibody and identified the same motif for an endogenous mZSCAN4.20 Cheng et al. expressed FLAG-mZSCAN4F protein in MEF cells during their conversion to iPS cells, performed ChIP-seq using an anti-FLAG antibody, and identified CCGCSGCB as mZSCAN4F-binding motif.19
In contrast to these earlier studies, we used an anti-ZSCAN4 antibody raised against mZSCAN4C to perform ChiP-seq for endogenous mZSCAN4 in mES cells. We also used two anti-hZSCAN4 antibodies (mouse and rabbit) raised against hZSCAN4 to perform ChIP-seq for endogenous hZSCAN4 in hES cells. In both mouse and human DNA, we identified TGCACAC as a ZSCAN4-binding motif, consistent with the motifs identified by several others17,18,20 but different from the motif identified by Cheng et al.19 It is unknown why the motif identified by Cheng et al. differs from all other studies, including ours, but it is the only study that used mouse MEF cells, whereas the others used mouse and hES cells. It is also possible that the use of MEF cells undergoing conversion from MEFs to iPS cells may have provided a different cellular environment for ZSCAN4 binding.
The study by Srinivasan et al. also revealed that mZSCAN4 binds to the mZSCAN4-binding motif enriched in CA/TG microsatellites.20 Our results confirmed the results of Srinivasan et al. and demonstrated that mZSCAN4 binds to mZSCAN4-binding motifs enriched in CA/TG microsatellites. Additionally, we show that hZSCAN4 binds to hZSCAN4-binding motifs enriched in CA/TG microsatellites. The CA/TG microsatellites are known for their formation of unstable left-handed duplexes called Z-DNA,43,54,55 which can induce double-strand DNA breaks and mutations56–58 and are also associated with chromosome breakpoints in human leukemias and lymphomas.59–61 The CA/TG microsatellites are frequently recombined and are a source of genome instability.40–43 Srinivasan et al. further demonstrated that in mice, mZSCAN4’s binding to CA/TG microsatellites is active in protecting the two-cell embryo genome from damage caused by zygotic genome activation (massive transcription upregulation).20 Using bioinformatic analyses, Burden et al. identified subsets of CA/TG microsatellites as a shared ‘vulnerability code’ for various types of DNA damage in mouse spermatocytes and early embryos, suggesting that mZSCAN4 binding protects those regions from DNA damage.62
This mode of action is consistent with the expression pattern of ZSCAN4, which is not expressed ubiquitously but expressed transiently during zygotic genome activation in mouse two-cell embryos,1 human 4–8-cell embryos,63 1–5% of mES cells,1,2,64 at pachytene-diplotene stage during mouse oogenesis and spermatogenesis,38 and in a rare population of human tissue stem cells, which is activated by inflammation and tissue damages.65 Thus, the timing of ZSCAN4 induction coincides with the openings of genome-wide chromatin, especially heterochromatin regions.6 The openings of heterochromatin may open the error-prone CA/TG microsatellites, which is prevented by the timely presence of ZSCAN4, as suggested by Srinivasan et al.20
Together, these findings are consistent with the earlier reports that normal and ectopic mZSCAN4 expression decreases SCE, resulting in the enhancement of genome stability in mES cells,2 and that ectopic mZSCAN4 expression protects mES cells from DNA-damaging agents, such as mitomycin C.3 Interestingly, a recent report shows that among patients with urothelial carcinoma of the upper urinary tract and urinary bladder, patients with high hZSCAN4 expression have a major survival advantage compared to patients with low hZSCAN4 expression.66 However, it remains to be seen whether the ZSCAN4 binding to the CA/TG microsatellites alone can also explain ZSCAN4’s other functions, such as repairing DNA damage and chromosome abnormalities,15,65,67 telomere elongation,2 DNA methylation inhibitor,5 and enhanced generation of iPS cells with high quality.49,53
Supplementary Material
Acknowledgements
This work was supported by JSPS KAKENHI grant number 17H01433 and 15K14453. The authors thank Allie Amick, PhD, for editing the manuscript and Hiromi Kimura for technical assistance.
Contributor Information
Tomohiko Akiyama, Department of Systems Medicine, Keio University School of Medicine, Tokyo 160-8582, Japan; Department of Molecular Biology, Yokohama City University, School of Medicine, Kanagawa 236-0027, Japan.
Kei-ichiro Ishiguro, Department of Systems Medicine, Keio University School of Medicine, Tokyo 160-8582, Japan; Department of Chromosome Biology, Institute of Molecular Embryology and Genetics (IMEG), Kumamoto University, Kumamoto 860-0811, Japan.
Nana Chikazawa, Department of Systems Medicine, Keio University School of Medicine, Tokyo 160-8582, Japan.
Shigeru B H Ko, Department of Systems Medicine, Keio University School of Medicine, Tokyo 160-8582, Japan.
Masashi Yukawa, Integrated Medical and Agricultural School of Public Health, Ehime University, Ehime 791-0295, Japan; Division of Allergy & Immunology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH 45229-3026, USA.
Minoru S H Ko, Department of Systems Medicine, Keio University School of Medicine, Tokyo 160-8582, Japan; Elixirgen Therapeutics, Inc., Baltimore, MD 21205, USA.
Conflict of Interest
MSHK is Professor Emeritus of Keio University and co-founder and Chief Scientific Officer of Elixirgen Therapeutics, Inc.
References
- 1. Falco, G., Lee, S.L., Stanghellini, I., Bassey, U.C., Hamatani, T., and Ko, M.S.. 2007, Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells, Dev. Biol., 307, 539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zalzman, M., Falco, G., Sharova, L.V., et al. 2010, Zscan4 regulates telomere elongation and genomic stability in ES cells, Nature, 464, 858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ko, M.S.H., Zalzman, M., and Sharova, L.V.. 2015, Methods for enhancing genome stability and telomere elongation in embryonic stem cells, US Patent, 9012223. [Google Scholar]
- 4. Eckersley-Maslin, M.A., Svensson, V., Krueger, C., et al. 2016, MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs, Cell Rep., 17, 179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dan, J., Rousseau, P., Hardikar, S., et al. 2017, Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells, Cell Rep., 20, 1936–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akiyama, T., Xin, L., Oda, M., et al. 2015, Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells, DNA Res., 22, 307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dan, J., Yang, J., Liu, Y., Xiao, A., and Liu, L.. 2015, Roles for histone acetylation in regulation of telomere elongation and two-cell state in mouse ES cells, J. Cell. Physiol., 230, 2337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macfarlan, T.S., Gifford, W.D., Driscoll, S., et al. 2012, Embryonic stem cell potency fluctuates with endogenous retrovirus activity, Nature, 487, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macfarlan, T.S., Gifford, W.D., Agarwal, S., et al. 2011, Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A, Genes Dev., 25, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ishiuchi, T., Enriquez-Gasca, R., Mizutani, E., et al. 2015, Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly, Nat. Struct. Mol. Biol., 22, 662–71. [DOI] [PubMed] [Google Scholar]
- 11. Nakatake, Y., Ko, S.B.H., Sharov, A.A., et al. 2020, Generation and profiling of 2,135 human ESC lines for the systematic analyses of cell states perturbed by inducing single transcription factors, Cell Rep., 31, 107655. [DOI] [PubMed] [Google Scholar]
- 12. Hendrickson, P.G., Dorais, J.A., Grow, E.J., et al. 2017, Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons, Nat. Genet., 49, 925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Iaco, A., Planet, E., Coluccio, A., Verp, S., Duc, J., and Trono, D.. 2017, DUX-family transcription factors regulate zygotic genome activation in placental mammals, Nat. Genet., 49, 941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whiddon, J.L., Langford, A.T., Wong, C.J., Zhong, J.W., and Tapscott, S.J.. 2017, Conservation and innovation in the DUX4-family gene network, Nat. Genet., 49, 935–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amano, T., Hirata, T., Falco, G., et al. 2013, Zscan4 restores the developmental potency of embryonic stem cells, Nat. Commun., 4, 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ishiguro, K.I., Nakatake, Y., Chikazawa-Nohtomi, N., et al. 2017, Expression analysis of the endogenous Zscan4 locus and its coding proteins in mouse ES cells and preimplantation embryos, In Vitro Cell. Dev. Biol. Anim., 53, 179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jolma, A., Yan, J., Whitington, T., et al. 2013, DNA-binding specificities of human transcription factors, Cell, 152, 327–39. [DOI] [PubMed] [Google Scholar]
- 18. Zhang, W., Chen, F., Chen, R., et al. 2019, Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes, Nucleic Acids Res., 47, 8485–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng, Z.L., Zhang, M.L., Lin, H.P., et al. 2020, The Zscan4-Tet2 transcription nexus regulates metabolic rewiring and enhances proteostasis to promote reprogramming, Cell Rep., 32, 107877. [DOI] [PubMed] [Google Scholar]
- 20. Srinivasan, R., Nady, N., Arora, N., et al. 2020, Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage, Sci. Adv., 6, eaaz9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olson, L.E., Bedja, D., Alvey, S.J., Cardounel, A.J., Gabrielson, K.L., and Reeves, R.H.. 2003, Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1, Cancer Res., 63, 6602–6. [PubMed] [Google Scholar]
- 22. Akutsu, H., Machida, M., Kanzaki, S., et al. 2015, Xenogeneic-free defined conditions for derivation and expansion of human embryonic stem cells with mesenchymal stem cells, Regen. Ther., 1, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goparaju, S.K., Kohda, K., Ibata, K., et al. 2017, Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors, Sci. Rep., 7, 42367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akiyama, T., Wakabayashi, S., Soma, A., et al. 2016, Transient ectopic expression of the histone demethylase JMJD3 accelerates the differentiation of human pluripotent stem cells, Development, 143, 3674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akiyama, T., Sato, S., Chikazawa-Nohtomi, N., et al. 2018, Efficient differentiation of human pluripotent stem cells into skeletal muscle cells by combining RNA-based MYOD1-expression and POU5F1-silencing, Sci. Rep., 8, 1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Warren, L., Manos, P.D., Ahfeldt, T., et al. 2010, Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA, Cell Stem Cell, 7, 618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian, B., Yang, J., and Brasier, A.R.. 2012, Two-step cross-linking for analysis of protein-chromatin interactions, Methods Mol. Biol., 809, 105–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kartashov, A.V. and Barski, A.. 2015, BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data, Genome Biol., 16, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langmead, B., Trapnell, C., Pop, M., and Salzberg, S.L.. 2009, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang, Y., Liu, T., Meyer, C.A., et al. 2008, Model-based analysis of ChIP-Seq (MACS), Genome Biol., 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shao, Z., Zhang, Y., Yuan, G.C., Orkin, S.H., and Waxman, D.J.. 2012, MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets, Genome Biol., 13, R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lerdrup, M., Johansen, J.V., Agrawal-Singh, S., and Hansen, K.. 2016, An interactive environment for agile analysis and visualization of ChIP-sequencing data, Nat. Struct. Mol. Biol., 23, 349–57. [DOI] [PubMed] [Google Scholar]
- 33. Ramirez, F., Dundar, F., Diehl, S., Gruning, B.A., and Manke, T.. 2014, deepTools: a flexible platform for exploring deep-sequencing data, Nucleic Acids Res., 42, W187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machanick, P. and Bailey, T.L.. 2011, MEME-ChIP: motif analysis of large DNA datasets, Bioinformatics, 27, 1696–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Afgan, E., Baker, D., van den Beek, M., et al. 2016, The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update, Nucleic Acids Res., 44, W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smit, A., Hubley, R., and Green, P.. 2013-2015, RepeatMasker Open-4.0. Available at: http://www.repeatmasker.org.
- 37. Grant, C.E., Bailey, T.L., and Noble, W.S.. 2011, FIMO: scanning for occurrences of a given motif, Bioinformatics, 27, 1017–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishiguro, K.I., Monti, M., Akiyama, T., et al. 2017, Zscan4 is expressed specifically during late meiotic prophase in both spermatogenesis and oogenesis, In Vitro Cell. Dev. Biol. Anim., 53, 167–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharova, L.V., Sharov, A.A., Piao, Y., et al. 2016, Emergence of undifferentiated colonies from mouse embryonic stem cells undergoing differentiation by retinoic acid treatment, In Vitro Cell. Dev. Biol. Anim., 52, 616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang, W., He, L., Liu, W., Sun, C., and Ratain, M.J.. 2009, Exploring the relationship between polymorphic (TG/CA)n repeats in intron 1 regions and gene expression, Hum. Genomics, 3, 236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma, V.K., Brahmachari, S.K., and Ramachandran, S.. 2005, (TG/CA)n repeats in human gene families: abundance and selective patterns of distribution according to function and gene length, BMC Genomics, 6, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bacolla, A. and Wells, R.D.. 2004, Non-B DNA conformations, genomic rearrangements, and human disease, J. Biol. Chem., 279, 47411–4. [DOI] [PubMed] [Google Scholar]
- 43. Zhao, J., Bacolla, A., Wang, G., and Vasquez, K.M.. 2010, Non-B DNA structure-induced genetic instability and evolution, Cell. Mol. Life Sci., 67, 43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zuker, M. 2003, Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Res., 31, 3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burns, J.A., Chowdhury, M.A., Cartularo, L., Berens, C., and Scicchitano, D.A.. 2018, Genetic instability associated with loop or stem-loop structures within transcription units can be independent of nucleotide excision repair, Nucleic Acids Res., 46, 3498–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McMurray, C.T. 1999, DNA secondary structure: a common and causative factor for expansion in human disease, Proc. Natl. Acad. Sci. U.S.A., 96, 1823–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McMurray, C.T., Goellner, G.M., Gacy, A.M., Spiro, C., Yao, J., and Dyer, R.D.. 1999, Improper DNA secondary causes trinucleotide repeat instability in human neurologic disease, Biophys. J., 76, A135–A135. [Google Scholar]
- 48. Buschiazzo, E. and Gemmell, N.J.. 2010, Conservation of human microsatellites across 450 million years of evolution, Genome Biol. Evol., 2, 153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hirata, T., Amano, T., Nakatake, Y., et al. 2012, Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells, Sci. Rep., 2, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Correa-Cerro, L.S., Piao, Y., Sharov, A.A., et al. 2011, Generation of mouse ES cell lines engineered for the forced induction of transcription factors, Sci. Rep., 1, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi, Y., Lan, F., Matson, C., et al. 2004, Histone demethylation mediated by the nuclear amine oxidase homolog LSD1, Cell, 119, 941–53. [DOI] [PubMed] [Google Scholar]
- 52. Taunton, J., Hassig, C.A., and Schreiber, S.L.. 1996, A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p, Science, 272, 408–11. [DOI] [PubMed] [Google Scholar]
- 53. Jiang, J., Lv, W., Ye, X., et al. 2013, Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation, Cell Res., 23, 92–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nordheim, A. and Rich, A.. 1983, The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids, Proc. Natl. Acad. Sci. U. S. A., 80, 1821–5. [Google Scholar]
- 55. Ho, P.S. 1994, The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA, Proc. Natl. Acad. Sci. U. S. A., 91, 9549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang, G., Christensen, L.A., and Vasquez, K.M.. 2006, Z-DNA-forming sequences generate large-scale deletions in mammalian cells, Proc. Natl. Acad. Sci. U. S. A., 103, 2677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Herbert, A. 2019, Z-DNA and Z-RNA in human disease, Commun. Biol., 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang, G. and Vasquez, K.M.. 2007, Z-DNA, an active element in the genome, Front Biosci., 12, 4424–38. [DOI] [PubMed] [Google Scholar]
- 59. Aplan, P.D., Raimondi, S.C., and Kirsch, I.R.. 1992, Disruption of the SCL gene by a t(1;3) translocation in a patient with T cell acute lymphoblastic leukemia, J. Exp. Med., 176, 1303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Adachi, M. and Tsujimoto, Y.. 1990, Potential Z-DNA elements surround the breakpoints of chromosome translocation within the 5' flanking region of bcl-2 gene, Oncogene, 5, 1653–7. [PubMed] [Google Scholar]
- 61. Wolfl, S., Wittig, B., and Rich, A.. 1995, Identification of transcriptionally induced Z-DNA segments in the human c-myc gene, Biochim. Biophys. Acta, 1264, 294–302. [DOI] [PubMed] [Google Scholar]
- 62. Burden, F., Ellis, P.J.I., and Farre, M.. 2023, A shared ‘vulnerability code’ underpins varying sources of DNA damage throughout paternal germline transmission in mouse, Nucleic Acids Res., 51, 2319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vassena, R., Boue, S., Gonzalez-Roca, E., et al. 2011, Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development, Development, 138, 3699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakai-Futatsugi, Y. and Niwa, H.. 2016, ZSCAN4 is activated after telomere shortening in mouse embryonic stem cells, Stem Cell Rep., 6, 483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ko, M.S. 2016, Zygotic genome activation revisited: looking through the expression and function of ZSCAN4, Curr. Top. Dev. Biol., 120, 103–24. [DOI] [PubMed] [Google Scholar]
- 66. He, H.L., Lai, H.Y., Chan, T.C., et al. 2023, Low expression of ZSCAN4 predicts unfavorable outcome in urothelial carcinoma of upper urinary tract and urinary bladder, World J. Surg. Oncol., 21, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Amano, T., Jeffries, E., Amano, M., Ko, A.C., Yu, H., and Ko, M.S.. 2015, Correction of Down syndrome and Edwards syndrome aneuploidies in human cell cultures, DNA Res., 22, 331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McLean, C.Y., Bristor, D., Hiller, M., et al. 2010, GREAT improves functional interpretation of cis-regulatory regions, Nat. Biotechnol., 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.