Abstract
The overall charge content and the patterning of charged residues have a profound impact on the conformational ensembles adopted by intrinsically disordered proteins. These parameters can be altered by charge regulation, which refers to the effects of post-translational modifications, pH dependent changes to charge, and conformational fluctuations that modify the values of ionizable residues. Although atomistic simulations have played a prominent role in uncovering the major sequence-ensemble relationships of IDPs, most simulations assume fixed charge states for ionizable residues. This may lead to erroneous estimates for conformational equilibria if they are linked to charge regulation. Here, we report the development of a new method we term -canonical Monte Carlo sampling for modeling the linkage between charge regulation and conformational equilibria. The method, which is designed to be interoperable with the ABSINTH implicit solvation model, operates as follows: For a protein sequence with ionizable residues, we start with all charge microstates and use a criterion based on model compound values to prune down to a subset of thermodynamically relevant charge microstates. This subset is then grouped into mesostates, where all microstates that belong to a mesostate have the same net charge. Conformational distributions, drawn from a canonical ensemble, are generated for each of the charge microstates that make up a mesostate using a method we designate as proton walk sampling. This method combines Metropolis Monte Carlo sampling in conformational space with an auxiliary Markov process that enables inter-conversions between charge microstates along a mesostate. Proton walk sampling helps identify the most likely charge microstate per mesostate. We then use thermodynamic integration aided by the multistate Bennett acceptance ratio method to estimate the free energies for converting between mesostates. These free energies are then combined with the per-microstate weights along each mesostate to estimate standard state free energies and pH dependent free energies for all thermodynamically relevant charge microstates. The results provide quantitative estimates of the probabilities and preferred conformations associated with every thermodynamically accessible charge microstate. We showcase the application of -canonical sampling using two model systems. The results establish the soundness of the method and the importance of charge regulation in systems characterized by conformational heterogeneity.
1. Introduction
Intrinsically disordered proteins (IDPs) highlight the functional importance of conformational heterogeneity 1, 2. Studies over the past decade have uncovered relationships between IDP sequences and global as well as local features of conformational ensembles 3–30. To first order, the sequence-ensemble relationships 31–33 of IDPs are governed by compositional biases such as the fraction of charged residues (FCR), net charge per residue (NCPR), and mean hydrophobicity 34. In addition, sequence patterning of oppositely charged residues 29 and the patterning of proline and charged residues vis-à-vis other residues 30 can have a profound impact on the overall dimensions, amplitudes of conformational fluctuations, and local conformational preferences of IDPs.
Atomistic simulations based on efficient and accurate implicit solvent models 7, 26, 28, 31–33, 35–40 as well as improved descriptions of protein-solvent interactions using explicit models for solvent molecules 41, 42 have been deployed to study a wide variety of IDPs 43–48. These simulations have yielded quantitative descriptions of sequence-ensemble relationships. The overall picture that has emerged may be summarized as follows: IDPs come in distinct sequence flavors; and the sequence-specific interplay between chain solvation vs. intramolecular interactions leads to distinct relationships between IDP sequences and conformational ensembles.
The knowledge base generated to date highlights the importance of charged residues, specifically parameters such as FCR, NCPR, and the patterning of charged residues, as determinants of sequence-ensemble relationships and as drivers of functional consequences of IDPs 29, 33, 34, 38, 49–52. The effects of charged residues can be altered through charge screening, charge renormalization, or charge regulation. Charge screening refers to the effects of solution ions and conformational fluctuations on the strengths of intra- and intermolecular electrostatic interactions among charged residues of IDPs 5, 26, 51, 53. In contrast, charge renormalization refers to alterations or even inversions of charge profiles 54, 55 that result from the accumulation of solution ions, specifically multivalent ions, around regions of high charge density. While the effects of charge renormalization have been well established for nucleic acids56, this has yet to be demonstrated for IDPs, although such effects might prevail for highly charged polyelectrolytes such as histone tails 57, 58, protamines 26, and acid-rich sequences such as prothymosin 53. It is also likely that charge renormalization contributes to phase separation via complex coacervation of complexing IDPs 59–61.
Charge regulation refers to changes in the FCR, NCPR, and charge patterning, and these changes may arise through post-translational modifications, due to pH effects such as intracellular pH gradients 62, 63, or through conformation-dependent changes to the protonation states of ionizable residues 34. Due to their conformational heterogeneity, solvent accessibility, and propensity for post-translational modifications, IDPs are likely to be much more susceptible to charge regulation than folded domains. The consequences of charge regulation through post-translational modifications such as Ser / Thr phosphorylation 8–10, 30, Lys acetylation 64–67, and Arg methylation / citrullination 68 are becoming topics of intense scrutiny for experimental studies of IDPs. Simulations of the impact of charge regulation via post-translational modifications are in their infancy, limited mainly by the absence of well-vetted forcefield parameters for modified amino acids.
Recent experiments indicate that charge regulation effects are likely to be important for disorder-to-order and order-to-disorder transitions in IDPs 69, the overall charge profiles of IDPs 6, and IDP-driven liquid-liquid phase separation 70. As an example, we consider data for the intrinsically disordered sidearms of neurofilaments 71, 72. These sequences are polymer brushes comprising of multiple repeats of the hexapeptide KSPAEA. The brush height is governed by the end-to-end distance distribution of these polymers. At extremes of pH, the sidearms are either cationic (low pH) or anionic polyelectrolytes (high pH) that form stiff brushes of maximal brush height 71, 72. Between these extremes, the polymer brushes should, in theory, be symmetric polyampholytes corresponding to lower brush heights. Interestingly, the pH dependence of measured brush heights is suggestive of upshifted values for some fraction of the Glu residues. An even more striking example of sequence-specific charge regulation in IDPs comes from single molecule nanoscale electrometry measurements on prothymosin 6. These measurements provide an estimate of the net charge of the system at very low salt concentrations. At pH 7.0, and very low salt concentrations, one would estimate the net charge to be –46 based on the sequence of prothymosin . In contrast, the electrometry measurements suggest an effective charge of –28.5 ± 1.2. This implies that at least a third of Glu / Asp residues in prothymosin have significantly upshifted values such that at pH 7.0 they are in their protonated (uncharged) states.
Despite their importance, the impacts of pH, sequence, and conformation dependent charge regulation are seldom accounted for in simulations of IDPs. Instead, it is common practice to fix the charges of ionizable groups using the intrinsic values of blocked amino acids as reference. Accordingly, the amine and guanido groups of Lys and Arg are assigned a charge of +1, the carboxyl groups of Asp and Glu are assigned charges of –1, and the imidazole group of His is typically set to be electroneutral. Critically these charge states are immutable throughout the course of a simulation, regardless of the local environment. The use of fixed charge models, which are based on the values of blocked amino acids, is a questionable approach 31. One way to work around this approximation is to fix the chemical potential of the proton, as in constant pH molecular dynamics 73–80 or Monte Carlo approaches 81–83. In these approaches, pH, sequence, and conformation dependent changes to the charge states of ionizable groups are realized via the uptake or release of protons to a proton bath. Successes have been achieved in the deployment of constant pH molecular dynamics methods 74–76 to calculate shifts of surface ionizable groups of globular proteins 84, 85 and for pH-dependent protein dissociation 86, 87. However for IDPs where over a third of the residues are ionizable, the combinatorics of proton uptake and release options becomes computationally unwieldy. This calls for different approaches that adapt advances in free energy calculations such as Wang-Landau sampling 88.
Here, we describe a two-stage method designated as -canonical Monte Carlo sampling that for a fixed temperature (a) samples conformational distributions for all thermodynamically relevant charge microstates (defined below) associated with an IDP sequence and (b) uses the multistate Bennett acceptance ratio (mBAR)89 estimator to calculate free energy changes associated with alchemic conversions between distinct pairs of charge microstates. This two-stage sampling allows us to compute pH-dependent weights of different charge microstates and their conformational ensembles. The current version of -canonical Monte Carlo sampling is designed to be interoperable with the ABSINTH implicit solvation paradigm 28, 90, 91, although the tenets of the model are interoperable with any implicit solvation model.
In what follows, we describe the overall methodology and the approaches we have incorporated to make the joint sampling of charge microstates and conformational distributions a tractable proposition. We illustrate the utility of the method by applying it to a pair of archetypal systems comprising of Asp, Glu, and Lys residues.
2. Theory and Computational Details
We introduce the different aspects of -canonical Monte Carlo sampling using peptide models of IDPs that contain multiple ionizable residues. For example, in a sequence such as ace-EEE-nme, where ace and nme respectively denote N-acetyl and N′-methylamide, each of the three Glu residues can either be protonated (E) or deprotonated (e). For a sequence with ionizable residues, there is a theoretical maximum of distinct charge microstates to consider. For ace-EEE-nme, these charge microstates are EEE, eEE, EeE, EEe, eeE, Eee, eEe, and eee, respectively. The overall “brute-force” strategy underlying -canonical Monte Carlo sampling is straightforward. For each of the conceivable charge microstates, we obtain conformational distributions sampled from the canonical ensemble at temperature . Conformational ensembles for each of the charge microstates are stitched together using free energy calculations. This allows us to compute pH-dependent relative populations for each of the charge microstates and the conformations for each charge microstate.
The overall computational complexity is vastly reduced by: a) Decreasing the number of charge microstates from its theoretical maximum and restricting considerations to the thermodynamically relevant microstates; b) Grouping microstates into mesostates and using a proton walk algorithm to reduce the computational cost of obtaining conformational ensembles to that of a single charge microstate; c) Minimizing the overall number of free energy calculations using maximum likelihood methods to identify the most representative charge microstates and associated conformational ensembles for each mesostate. The details are described in the following sections.
Eliminating forbidden charge microstates:
The effective number of thermodynamically relevant charge microstates can be considerably smaller than for sequences with ionizable residues. This pruning of charge microstates is readily achieved for sequences with a combination of acidic and basic groups. Consider the case of ace-EKEK-nme. Since = 4, there are, in theory, 16 distinct charge microstates. Based on the intrinsic values of Glu and Lys, we stipulate that the protonation of Glu and deprotonation of Lys are unlikely to occur simultaneously. This allows us to disregard eight of the charge microstates (ekek, ekeK, ekEK, ekEk, EKek, EkeK, eKEk, and eKek) and designate them as forbidden microstates. Accordingly, we reduce the effective number of charge microstates to 7 from 16. The ansatz of forbidden charge microstates is particularly effective for sequences that are symmetric polyampholytes, which refers to sequences where the number of groups that can be neutralized by uptake vs. release of a proton is essentially the same. We use a defined protocol to identify forbidden charge microstates, eliminate them, and prune the set of charge microstates to the thermodynamically relevant set of charge microstates per mesostate. The protocol uses model compound values to compute a reference free energy based on the assumption of additivity and calculates probabilities for each of the microstates along a mesostate. If this probability falls below 10–3, then the charge microstate in question is ignored. A particular advantage of the forbidden states ansatz is the well-documented observation that polyampholytic sequences make up roughly 70% of the sequence space of IDPs where FCR exceeds 0.25. Less than 5% of this sequence space comprises of polyelectrolytes. Therefore, the forbidden states ansatz leads to substantial reduction in the number of thermodynamically relevant charge microstates.
Grouping charge microstates into mesostates:
Charge microstates can be grouped into mesostates based on their net charge. In the example considered above, eEE, EeE, and EEe make up a mesostate where the charge per microstate is –2; similarly, eeE, Eee, and eEe constitute a mesostate where the charge per microstate is –1; finally, EEE and eee belong to the two mesostates where the charge per microstate is –3 and 0, respectively. Our grouping ensures that all charge microstates within a mesostate will have the same net charge. Accordingly, each mesostate is designated by the label where denotes the net charge per microstate within the mesostate and denotes the total number of charge microstates within the mesostate. Grouping of charge microstates into mesostates also allows us to reduce the cost of conformational sampling per charge microstate. If a pair of microstates is within the same mesostate then the free energy changes associated with converting between charge microstates will be independent of pH. We use this to our advantage in a proton walk sampling approach, as described next. This yields relative populations of charge microstates that belong to a specific mesostate.
Proton-walk Monte Carlo sampling:
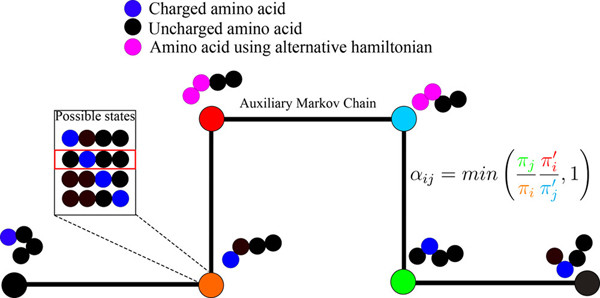
Consider a -mesostate of net charge that comprises of charge microstates. The goal is to obtain converged conformational ensembles for each of the charge microstates. We use Metropolis Monte Carlo methods92 to sample conformations from the canonical ensemble for each charge microstate. Sampling is initiated for a randomly chosen charge microstate designated as . In addition to switching between different conformations for a given charge microstate, we also propose switches to the identities of charge microstates. For example, for the –13 mesostate of EEE, a switch between charge microstates might involve a switch from microstate eeE to Eee or eEe. Such switches involve swapping the positions of protonated vs. unprotonated Glu residues in the sequence. One approach to switching of charge microstates involves a switch from charge microstate to charge microstate while keeping the conformation fixed. This can become computationally inefficient due to inevitable steric clashes involving the atom that is introduced at the site of protonation and the rest of the peptide. To solve this problem, we adapt the previously developed Hamiltonian Switch Metropolis Monte Carlo (HS-MMC or HS for short) 93 whereby we introduce an auxiliary Markov process that samples from an alternative potential function to alleviate problems associated with switching between charge microstates.
The HS method works as follows: we designate the current conformational state of the system of interest as ; an auxiliary Markov process is spliced in whereby sampling from the potential of interest is switched to sampling from an alternate, computationally inexpensive potential that also enhances conformational exploration; a series of conformational states are then generated using the alternate potential; these proposed conformations are accepted / rejected using the standard Metropolis criterion; after some number of moves, the system finds itself in a new conformation designated as ; the potential is now switched back from the alternative one to the actual potential and the new state is accepted / rejected according to the criterion 93:
| (1) |
In Equation (1), and are the Boltzmann weights associated with the potential of interest for conformations and whereas and are Boltzmann weights associated with the alternative potential for conformations and , respectively. Gelb has shown 94 that the structure of the acceptance ratio preserves detailed balance. The HS method allows us to make arbitrary choices for the alternative potential. In our case, we use HS aided proton walks to switch between charge microstates within a mesostate (Figure 1). To enable efficiency in proton walks, we choose an alternative potential – see below – to ensure that the sidechain to be protonated / deprotonated is solvent exposed thus reducing the rejection of proton walk moves because solvent exposed groups have minimal steric overlaps with surrounding peptide atoms. Our use of an implicit solvent model alleviates any concerns about overlaps with solvent atoms.
Figure 1: Schematic of the HS aided proton walk algorithm.
During the transformation, the potential is set such that the two interchanging residues are both uncharged, while having the high free energy of solvation of the charged state, and the Lennard-Jones are set to half of those of the fully grown atom. Spheres represent amino acids, colored in black for uncharged moieties, blue for charged moities, and pink for moieties that are in the alternative potential state used for charge transfer in the auxilary Markov chain.
For the standard ABSINTH potential 90, 91, the total potential energy for conformation is written as a sum of the direct mean field interaction (DMFI) with the implicit solvent, which is essentially the mean-field estimate of the conformation specific free energy of solvation, the electrostatic (el) interactions among polar and charged groups screened by a conformation specific inhomogeneous dielectric, the Lennard-Jones (LJ) potential to model van der Waals interactions, and any other terms that are incorporated to model stereoelectronic effects 28 and / or geometric constraints on the system. Accordingly, the potential energy for conformation takes the form:
| (2) |
In the alternative potential, we set the electrostatic term to zero and scale the LJ term by a factor of 0.5 using a soft-core potential 95 (see Equation (4) below) for residues whose charge microstates are to be altered.
The upshot of the HS aided proton walk algorithm is that the system, which is in conformation and charge microstate prior to the introduction of the auxiliary Markov process, can undergo a dual switch to conformation in charge microstate (Figure 1). Since all charge microstates along a mesostate have identical numbers of atoms, the switching between charge microstates does not require any special treatment, as would be the case in a grand or semi-grand canonical ensemble. Charge microstates and their associated conformational ensembles that belong to a specific mesostate make up what we refer to as a -canonical ensemble. From a single Markov process, aided by the auxiliary process wherein the switching of charge microstates is realized, one obtains a distribution of conformations associated with each of the charge microstates, and the sampling frequencies identify the most likely conformations and charge microstates for a given mesostate.
Overall, the HS aided proton walk algorithm requires the specification of three sets of parameters: (a) The frequency with which a HS aided proton walk move will be attempted; (b) the average number of steps to be taken using the alternative potential before accepting or rejecting charge / conformational states; and (c) the maximum number of protons that can change position in a single move. The choice of charge microstates for switching the positions of protonated vs. deprotonated residues in a proton walk move can either be chosen at random or from a pre-set list to restrict sampling to relevant states. If the assumptions made to designate forbidden states are correct, then the two methods should yield very similar results, although restricting the states to be used for sampling leads to substantially improved efficiency. For each mesostate, we pick the most likely charge microstate and use this – and its associated conformational ensemble – in free energy calculations as described next. When the number of charge microstates per mesostate is large, there is the formal possibility that several microstates will have similar probabilities that are equivalent and high. In this case, a charge microstate is chosen at random from the most likely set.
In the current setup, the ionizable residues are Asp, Glu, Lys, and His. Extant data suggest that Arg does not become deprotonated, even in seemingly hydrophobic environments such as the interiors of globular proteins 96 or well inside lipid bilayers 97. A recent potentiometric and NMR investigation sets the model compound of Arg to be 13.8 ± 0.1 98, which is 1.8 pH units higher than the value of ~12 that is quoted in textbooks. These data suggest that it is reasonable and appropriate to assume that Arg will always be protonated. Histidine has two neutral forms and it is treated as a special case in the proton walk algorithm where the proton is allowed to switch positions within a residue as well as between other His residues along the sequence. The fundamental structure of the algorithm does not change, but the frequency with which proton walks are attempted will increase in accord with the number of His residues in the sequence.
Setup of the free energy calculations:
We shall consider a pair of mesostates adjacent in charge space and designate them as and . The most likely charge microstate within mesostates and are designated as and , respectively. We calculate the free energy change associated with alchemic conversion between and . These calculations are performed using modules introduced into the CAMPARI modeling suite (http://campari.sourceforge.net). The transformation between charge microstates from adjacent mesostates is based on thermodynamic integration (TI) and uses three distinct Kirkwood coupling parameters99, 100 , and , respectively that are inserted as pre-factors into Equation (2). Each parameter λ takes values between 0 and 1. The transformation involves addition of a proton to a specific site on the peptide of interest. The new site is introduced using four distinct steps, with increasing systematically between 0 and 1 in steps of 0.25. Once = 1, the values for and , are set to unity in one step. Sampling along the alchemic path between charge microstates is aided by the use of Hamiltonian replica exchange 101, 102 between pairs of replicas corresponding to different potentials defined by the coupling parameters. Data gathered along the alchemic transformation are combined with the multistate Bennett Acceptance Ratio (mBAR) method 89 as implemented in the mBAR package (https://github.com/choderalab/pymbar) 103 to estimate the free energy changes associated with the transformation.
We use TI 100 to evaluate the free energy change associated with transforming between charge microstates from adjacent mesostates. In order to increase the reliability of the free energy changes assessed using TI, we calculate relative free energy changes referenced to model compounds for which the free energy change associated with protonation / deprotonation are known. Not using absolute free energies allows us to circumvent some of the limitations of standard free energy methods. For example, the use of fixed bond lengths and bond angles in the ABSINTH model 91 allows us to ignore the cost of changes to bond lengths and bond angles the in the free energy calculations 104, 105. This contribution remains constant and is independent of changes to the rest of the system properties and these contributions are dereferenced by subtraction. Accordingly, if the transformation is the protonation of a Glu / Asp or deprotonation of Lys, then we perform calculations of the free energy change associated with Protonation In a Reference Model-compound (PIRM) and dereference this against experimentally derived free energy changes for the same reference model compound. Accordingly, we estimate the free energy for the conversion between charge microstates and using the relationship:
| (3) |
In Equation (3), is the experimentally derived free energy change associated with protonation in the relevant reference model compound and is the estimate obtained from the simulations for the same model compound. This approach sets the reference energy scales using experimentally derived values for . The approach prescribed in Equation (3) allows us to reduce systematic errors in the free energy calculations that arise due to errors in the ABSINTH forcefield.
The ABSINTH model allows for simulations as a function of salt concentration using explicitly modeled solution ions 91, 106. Additionally, the model is versatile in allowing for the incorporation of temperature dependent reference free energies of solvation for model compounds that constitute the backbone and sidechain moieties 7. Therefore, care is taken to ensure that values for are derived by treating the temperature and salt concentration as part of the protein context that will influence changes in pH dependence of the protonation reaction. This is important given clear evidence for salt 107 and temperature dependence 108 of the deprotonation reaction of sidechains with ionizable groups. Salt and temperature dependent values used to calculate are obtained from the data of Platzer et al. 109.
Details of the paths chosen for TI:
Since the free energy of solvation is by far the most important change in energy among all transformation coordinates, we choose a path for alchemic transformation that allows a drastic reduction in the number of replicas while ensuring smooth transformations between replicas. To avoid singularities linked to having a charge assigned to a dummy atom, changes in LJ parameters are made after charges have been turned off. To further smooth the energy landscape across replicas, we use a soft core potential for the changes in LJ potential 110, using a dependent potential of the form:
| (4) |
Here, and are the well depth and hard sphere radii for the interaction pair in question and the overall functional form derives from previous calibrations 111, 112. We then change the free energy of solvation of the appropriate solvation groups, simultaneously, introducing an energy bath that compensates for the difference in reference free energy between the two end states. We arrived at an optimal schedule for changing the Kirkwood coupling parameters that result in relatively high overlaps and low variance for estimates to the changes in free energy 113, with just six steps (see Table 1). The creation of an atom ex nihilo is the single largest factor that influences rejection rates between adjacent steps along the transformation process. Accordingly, as noted above, we vary in increments of 0.25 whereas all other parameters are varied in one step from 0 to 1.
Table 1:
Values for the Kirkwood coupling parameters used in setting up efficient paths for TI calculations.
| Replicas | ||||
|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 0.25 | 0 | 0 |
| 2 | 1 | 0.5 | 0 | 0 |
| 3 | 1 | 0.75 | 0 | 0 |
| 4 | 1 | 1 | 0 | 0 |
| 5 | 1 | 1 | 1 | 1 |
Generation of pH dependent conformational ensembles:
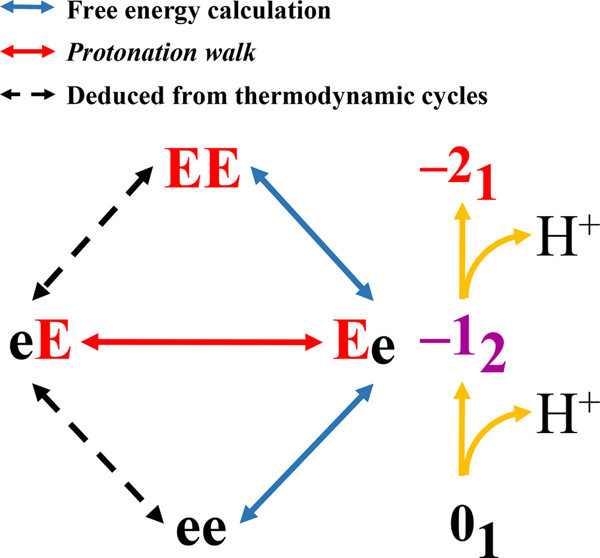
To illustrate the method, we introduce a simple system, ace-EE-nme, and the corresponding charge microstates as shown in Figure 2. This system calls for two free energy calculations between adjacent mesostates and one HS aided proton walk simulation along the –12 mesostate. For convenience, we set the free energy of the fully protonated state (ee) to be zero. In step (1) we perform HS aided proton walk simulations for the –12 mesostate that comprises of charge microstates eE and Ee, which identifies Ee as the most likely charge microstate. We also perform standard Metropolis Monte Carlo simulations for the charge microstates EE and ee. These simulations yield conformational distributions for each of the charge microstates. In step (2), we pick the most likely charge microstate for the –12 mesostate (Ee) and estimate the free energy changes associated with the transformation between Ee and EE as well as Ee and ee.
Figure 2: Illustration of -canonical sampling for the ace-EE-nme system.
The schematic lists the four charge microstates, depicts the grouping of charge microstates into mesostates, the use of proton walk sampling to extract weights for different charge microstates within a mesostate, and the use of free energy sampling for estimating the free energies for alchemic transformation between adjacent mesostates.
The standard state free energies associated with each of the relevant charge microstates viz., and changes are calculated using estimates based on mBAR by setting and using the following equations:
| (5) |
For an arbitrary pH, we have to also account for the chemical potential of the proton and accordingly, the relevant free energies, in terms of the thermal energy RT become:
| (6) |
For systems with ionizable residues, the standard state free energies and the pH dependent free energies for each charge microstate are calculated in direct analogy to the approach shown in Equations (5) and (6), respectively. The standard state and pH dependent free energies for each of the thermodynamically relevant charge microstates are calculated using the results obtained from the free energy estimator and the relative weights obtained for microstates along a mesostate using proton walk sampling. For a mesostate of net charge that comprises of charge microstates, the calculations analogous to those Equations (5) and (6) yield the standard state and pH-dependent free energies for charge microstate . If and are the weights obtained from visitation frequencies of charge microstates and from mesostate , then the relevant free energies for charge microstates are obtained using:
| (7) |
| (8) |
Given the information obtained in Equations (7) and (8), the free energies associated with each mesostate can be calculated using information regarding the free energies per microstate. For example, the standard state free energies and pH dependent free energies are calculated as:
| (9) |
In Equation (9), and are the standard state and pH dependent free energies, respectively for charge microstate from mesostate , while and are the standard-state and pH dependent mesostate free energies, respectively. For a system with thermodynamically relevant charge microstates distributed across all possible mesostates, we calculate the pH dependent, charge-microstate-specific Boltzmann probabilities as:
| (10) |
These calculations yield the pH dependent populations for the entire set of thermodynamically relevant charge microstates.
Obtaining estimates of errors:
The statistical error associated with free energy estimates based on mBAR are obtained using the method of Shirts et al. 89. Errors in estimates of visitation frequencies for distinct charge microstates along a mesostate are linked to the quality of the proton walk Monte Carlo simulations. These errors are estimated using bootstrapping and a non-parametric resampling of the state probabilities, using 103 distinct samples. Errors in estimates of standard state free energies for each of the charge microstates are obtained by propagation of the errors linked to each method along the path used for their determination. Because the probabilities are constructed relative to a reference charge microstate, the error grows as we consider charge microstates that are farther away from the reference microstate. To minimize the error for all charge microstates, we construct the free energies and their respective errors starting from both sides of the pH range (i.e., fully basic vs. fully acidic states). Since the charge microstates used for the construction are the same in both directions, the mean is independent of the direction used to retrieve the free energies, and only the estimates of errors will change. As a consequence, we set the error in the estimate of the free energy as the minimum error obtained for each microstate.
Archetypal systems used to demonstrate the -canonical method:
We demonstrate the applicability and working of -canonical sampling use two model peptides. The peptide ace--nme, designated hereafter as , is a model system that has been used to study the stabilities of so-called Charged Single Alpha Helices (CSAHs) 114. Sequences of CSAHs typically contain nearly perfect repeats of four Glu residues followed by four Lys residues 115. These repeats of blocks of four Glu and four Lys residues can range from being 25 to 200 residues long 116, and they are known to form long, stable alpha helices. The presence of alternating repeats of four Glu and four Lys residues lead to the postulate that alpha helicity in () repeats are stabilized by salt bridges between deprotonated Glu and protonated Lys residues that are four residues apart along the sequence 116. In this context, it is worth noting the () repeats are imperfect in that they are often interrupted by substitutions for Glu with Gln, Leu or other polar / non-polar residues 117. The second system NTL912–23 is a 12-residue peptide excised from the N-terminal domain of the ribosomal protein L9. Kuhlman et al.118 used this as a model system to quantify the pH dependence of alpha helicity in peptides that fold autonomously into structures they adopt in folded states. The sequence of NTL912–23 is: ace-KGKKGEIKNVAD-nme.
Details of the simulation setup for each of the systems:
The simulation temperature was set to 298 K. Four independent simulations were performed for the HS aided protonation walk and free energy calculations based on mBAR. Results from the four independent simulations were pooled for joint analysis and errors were estimated using bootstrap analysis methods as described above. The peptides were enclosed in spherical droplets (70 Å for and 75 Å for NTL912–23). Solution ions 106 including neutralizing counterions and ions to mimic NaCl concentrations of 10 mM and 100 mM, for and the NTL912–23 peptide, respectively were modeled explicitly as has been the case in all ABSINTH-based simulations reported to date.
Each HS aided protonation walk simulation comprises of 5×109 independent steps. On average, a HS step that switches to an auxiliary Markov chain was attempted once every 2.5×104 steps and each auxiliary process involved sampling for 10 steps using the alternative potential. This combination results in an acceptance of ca. 4% of the proposed transitions within the auxiliary process. The identities of charge microstates and associated conformations were recorded once every 104 steps. Alchemic transformation between charge microstates across adjacent mesostates uses 5×109 independent steps of Monte Carlo sampling along the TI path. Hamiltonian replica exchange was attempted once every 104 steps and the native and foreign energies of mBAR were evaluated once every 104 steps.
Results
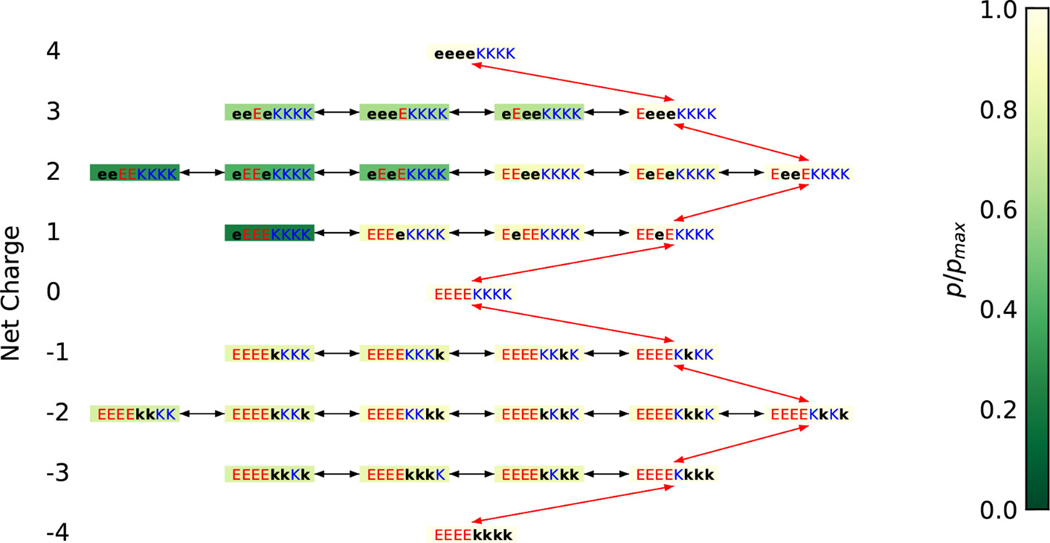
For the system, there are nine possible mesostates corresponding to net charges per microstate that range from –4 to +4 (Figure 3). All eight residues are ionizable; accordingly the theoretical maximum for the number of charge microstates is 28 = 256. However, once we deploy the forbidden microstates ansatz, only ~12% of the conceivable charge microstates are thermodynamically relevant. This reduces the number of relevant charge microstates to 31 from 256 (Figure 3). Similarly, for NTL912–23 a theoretical maximum of 26 = 64 charge microstates is reduced to 19 thermodynamically relevant charge microstates.
Figure 3: Demonstration of the maximum likelihood minimal transformation approach to select the free energy transformation path.
The color of the background of the boxes is representative of the ratio of the population of the corresponding charged microstate compared to that of the most populated microstate in the corresponding mesostate. Red arrows represent the free energy transformation chosen, and black arrows states that are on the same layer. Black lower-case letters represent the uncharged states of the corresponding amino acids.
For each of the 31 and 19 thermodynamically relevant charge microstates of and NTL912–23, respectively we perform HS aided proton walk simulations to obtain conformational ensembles for each of the charge microstates. We use a maximum likelihood approach to identify the charge microstate that is most representative of a particular mesostate as shown in Figure 3 for . Once the most likely charge microstates for each mesostate have been identified, we compute the free energy changes for alchemic transformations between mesostates. The path used for free energy calculations for is also shown in Figure 3.
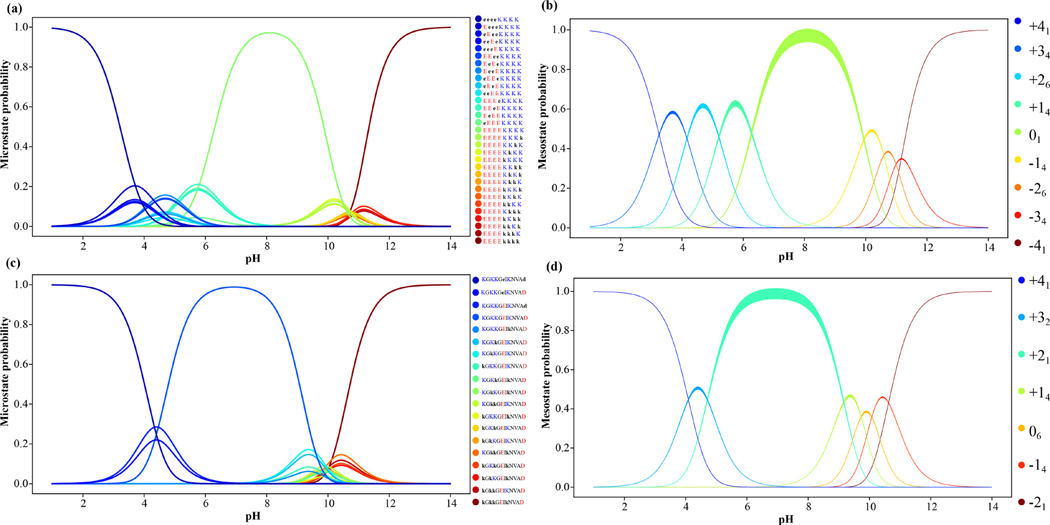
From the -canonical simulations that combine HS aided proton walks and free energy calculations we are able to compute the pH-dependent probabilities associated with each of the thermodynamically relevant charge microstates. These results, obtained for the and NTL912–23 systems are summarized in Figure 4. Panels (a) and (b) show the pH-dependence of charge microstate and mesostate probabilities for the whereas panels (c) and (d) shows the corresponding profiles for the NTL912–23 system.
Figure 4: Probability distributions for all charge microstates and mesostates obtained from -canonical sampling.
(a) Results for the 31 thermodynamically relevant charge microstates of ; (b) Results shown in panel (a) synthesized in terms of the mesostates for ; (c) Results for the 19 thermodynamically relevant charge microstates of NTL912–23; (d) Results shown in panel (c) synthesized in terms of the mesostates for NTL912–23. The envelopes for mesostate distributions quantify accumulated error in our estimates of the mesotstate statistics.
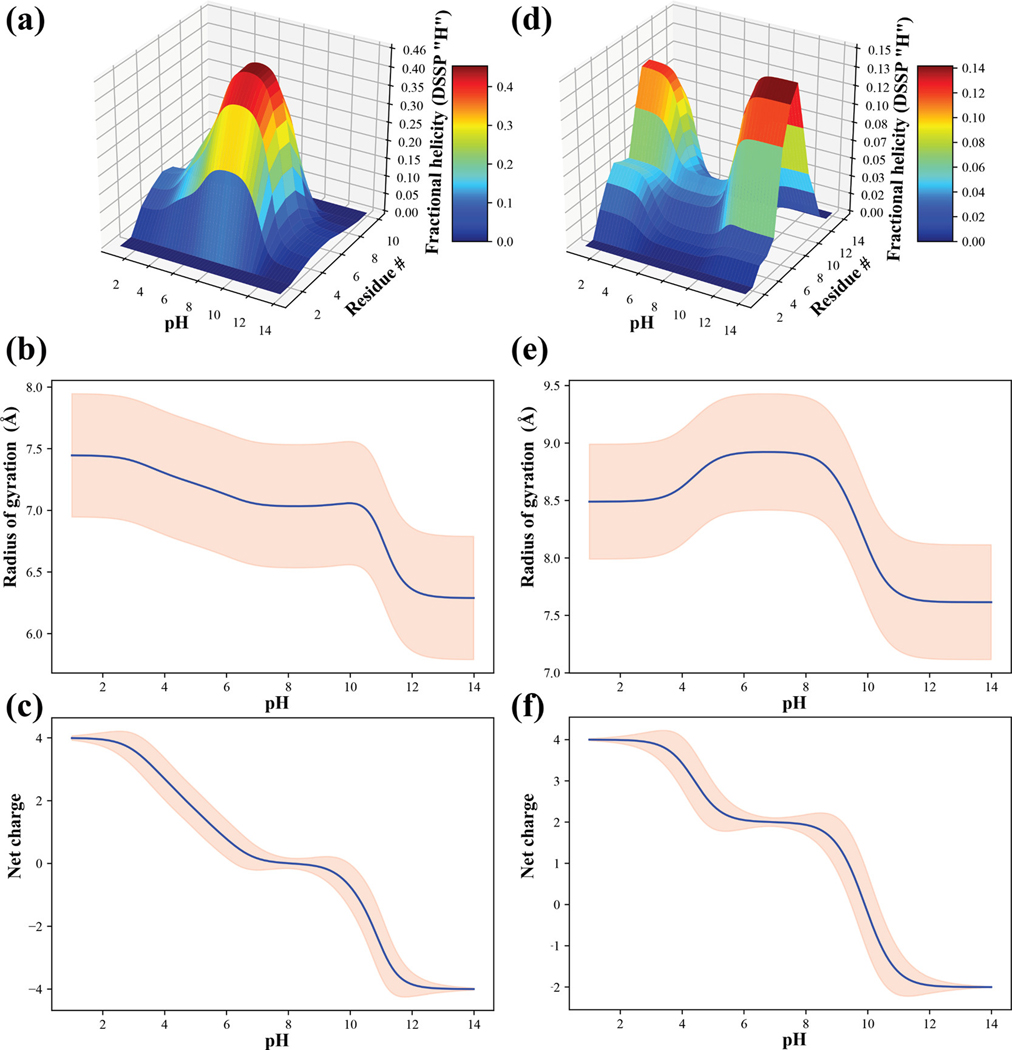
For , the single dominant charge microstate in the pH range of 7–9 is the one where all Glu residues are unprotonated and all Lys residues are protonated. Below a pH of 7.0, we record contributions from charge microstates corresponding to the +14 and +26 mesostates and likewise, above pH 9.0 the charge microstates from the -14 and −26 mesostates start to make significant contributions. The pH dependence of the per-residue fractional helicity profiles are shown in panel (a) in Figure 5 whereas panel (b) shows the pH dependence of the ensemble-averaged radii of gyration () and standard deviations. The average values change by ~0.5 Å between a pH range of 2.0 and 10.0 and decreases more substantially for pH values that are above 10.0. The net charge of the system, calculated as a weighted average over the contributions of the spectrum of charge microstates shows that the net charge is zero only within a narrow pH range of 7.0 – 9.0, falling below zero above 9.0 and becoming positive below a pH of 7.0.
Figure 5: Summary of results from -canonical sampling for the and NTL912–23 systems.
(a) Surface plot showing the per-residue alpha helicity, calculated using the DSSP algorithm 157, for each of the eight residues in as a function of pH. (b) Ensemble-averaged radius of gyration (blue curve) as a function of pH and standard deviations of the pH-dependent distributions for radii of gyration (pink envelope) for system. (c) Mean net charge (blue curve) as a function of pH and standard deviation for the pH-dependent net charge distributions for the system. Panels (d), (e), and (f) are results for the NTL912–23 system and are equivalent to panels (a), (b), and (c).
Results for the NTL912–23 system are also summarized in Figure 4 (panels (c) and (d)) and Figure 5 (panels (d) – (f)). The wild-type sequence belongs to the +21 mesostate. In the pH range between 5.0 and 8.5, the NTL912–23 system has a negligible preference for forming alpha helices or any other regular secondary structures. This is consistent with observations from the studies of Kuhlman et al.118 Below a pH of 6.0 there is a discernible increase in alpha helicity, especially within the C-terminal half of the peptide. A similar increase in helicity, mostly through the middle of the peptide is observed for pH values above 8.5. These preferences can be traced to the contributions of the +32 mesostate in the pH range between 3.0 and 6.0 and the +14, 06, and -14 mesostates in the pH range 8.0–11.0. These effects, which are indicative of smooth transitions in helical preferences of NTL923 well away from the model compound values of Asp, Glu, and Lys are suggestive of upshifted values for Glu 17 and Asp 23 combined with downshifted for Lys residues, although the magnitudes shifts are dependent on the specific Lys residue.
Apparent values of ionizable residues depend on sequence context:
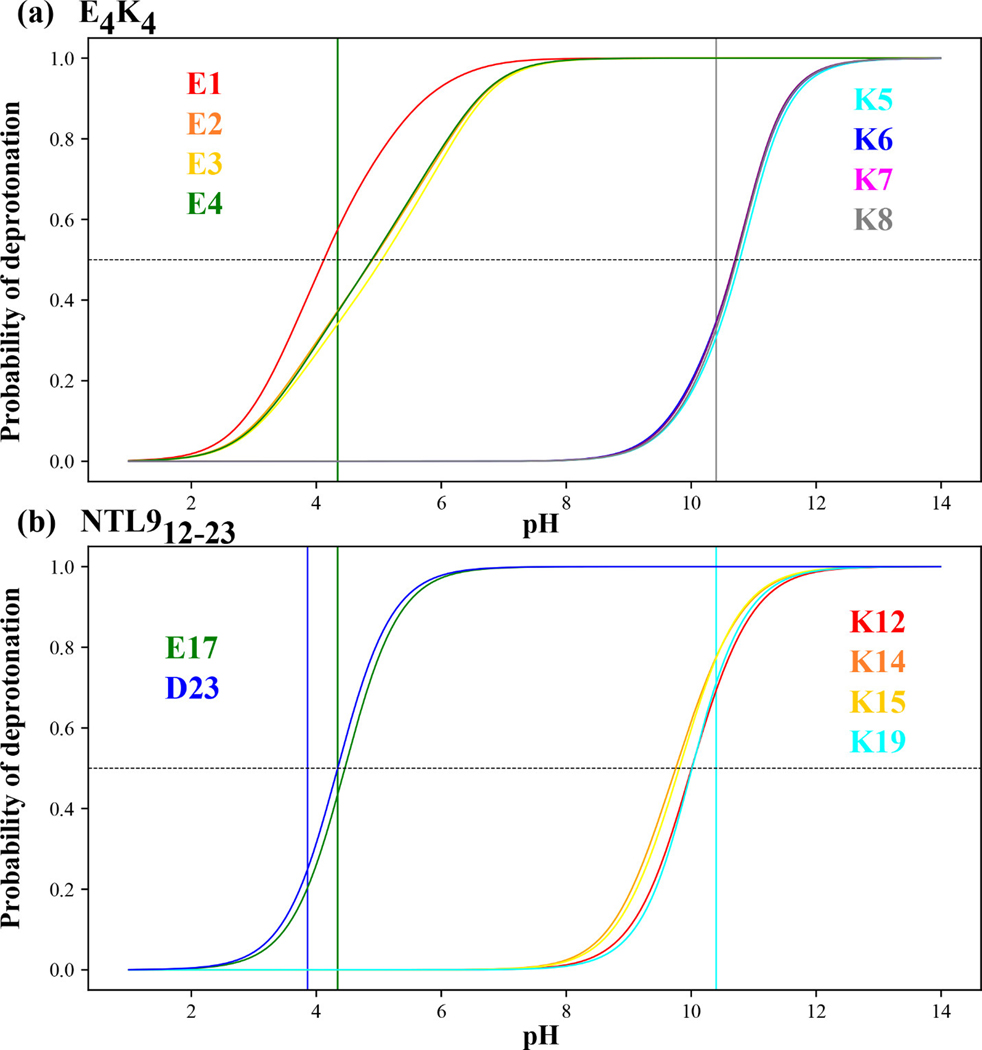
In Figure 6 we show our quantification of the probability of deprotonating different Asp, Glu, and Lys residues within the two systems. In panel (a) the vertical lines shown in green and black correspond, respectively to the model compound values of Glu and Lys. The apparent values of the different Glu and Lys residues within are estimated as the pH value at which the probability of observing the residue in a deprotonated state is precisely 0.5. Values for the apparent values calculated using the -canonical simulations are shown in Table 2. The apparent value of E1 is downshifted by 0.2 pH units with respect to the model compound value; conversely, when compared to the model compound value of Glu, the apparent values of E2, E3, and E4 are upshifted by 0.67, 0.73, and 0.57 pH units, respectively. The apparent values of the four Lys residues are upshifted by 0.4 pH units with respect to the model compound values.
Figure 6: Probability of deprotonating ionizable residues as a function of pH.
(a) Results for the eight ionizable residues within . The green and black vertical lines intersect the abscissa at model compound values for Glu and Lys, respectively. The horizontal dashed line intersects the ordinate at the value of 0.5. The intersection of this horizontal dashed line with the residue-specific “titration curves” is used to estimate the apparent value for the residue in question. The curves are colored according to the residues as shown in the legend. (b) Results for the six ionizable residues within NTL912–23. The vertical lines shown in blue, green, and cyan intersect the abscissa at pH values that correspond to the model compound values for Asp, Glu, and Lys, respectively.
Table 2:
Apparent values calculated using -canonical sampling for ionizable residues within and NTL912–23.
| E 4 K 4 | Residue | Apparent pK a | NTL9 12–23 | Residue | Apparent pK a |
| E1 | 4.11 | K12 | 10.00 | ||
| E2 | 4.98 | K14 | 9.74 | ||
| E3 | 5.04 | K15 | 9.80 | ||
| E4 | 4.88 | E17 | 4.45 | ||
| K5 | 10.77 | K19 | 10.00 | ||
| K6 | 10.72 | D23 | 4.34 | ||
| K7 | 10.70 | ||||
| K8 | 10.72 |
For NTL912–23 the apparent values for Glu17 and Asp23 are 4.45 and 4.35, respectively. Kuhlman et al.118 used pH titrations and chemical shift measurements to estimate the values of Glu17 and Asp23 to be 4.11 ± 0.17 and 4.11 ± 0.11, respectively. The values obtained using -canonical sampling (Table 2) are in accord with the estimates from experiments. The key point being that the apparent values of both residues are upshifted with respect to their model compound values, with the shift being more pronounced for Asp23, in accord with the experiments of Kuhlman et al.118 These results highlight the importance of local sequence contexts on the values of ionizable residues. The -canonical simulations also help quantify the apparent values for Lys residues as summarized in Table 2.
Comparison of q-canonical sampling to results obtained using unshifted values and fixed charge models:
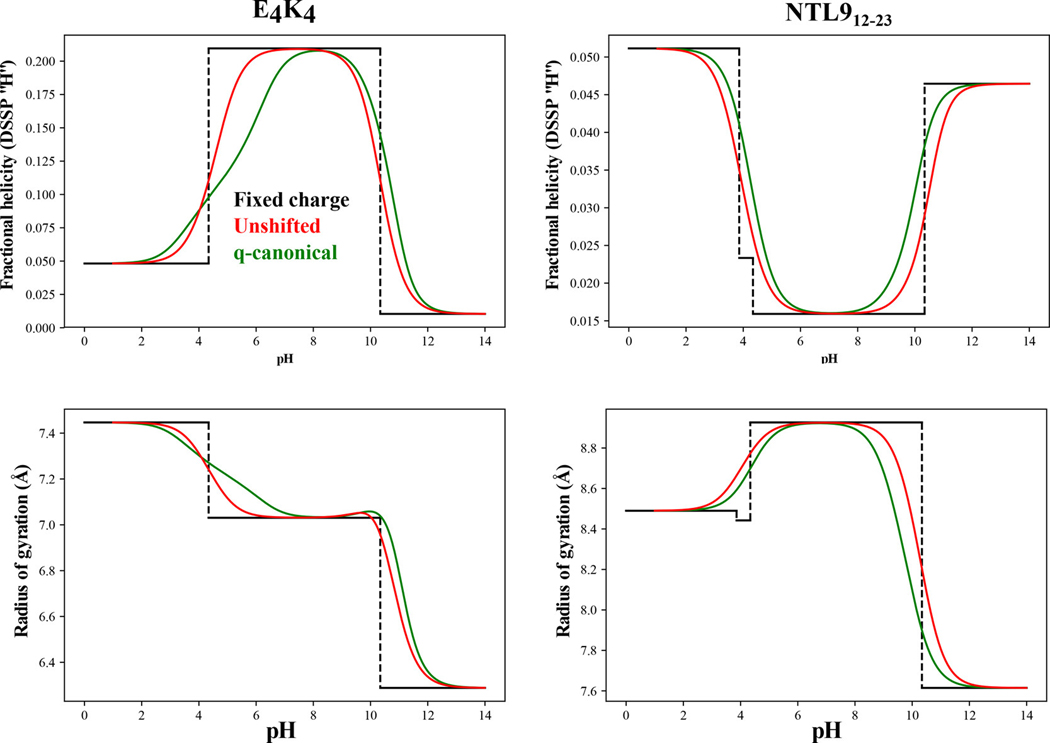
As noted in the introduction, fixed charge models specify charges for ionizable residues using the values of model compounds. We performed simulations using fixed charge models and compared the results to those obtained using the -canonical approach (Figure 7). In the fixed charge simulations, the charges of ionizable residues are immutable. For example, Glu is assumed to be deprotonated for all pH values above its model compound and protonated for all pH values below the model compound . Accordingly, for a sequence such as , only the , , and charge microstates contribute to the calculation of the pH dependent profiles. We can allow for charge state fluctuations while assuming unshifted values by weighting the contributions of all thermodynamically relevant charge microstates by model compound values. Comparisons are summarized in terms of the structural quantifies namely, the fractional helicity, calculated in terms of ensemble-averaged DSSP-H values – panels (a) and (b) – for and NTL912–23 and the ensemble-averaged values – panels (c) and (d). These comparisons show that pronounced deviations from the -canonical results come from the fixed charge simulations thus highlighting the errors associated with quenching charge state fluctuations. Accounting for these fluctuations vastly improves the calculated pH dependent profiles vis-à-vis the -canonical results and this is true even if we assume that the values are unshifted with respect to the model compounds. However, the impact of shifted values is made clear in the quantitative comparisons of the results from -canonical sampling and those obtained using unshifted values (Figure 7).
Figure 7: Comparison of results obtained using -canonical sampling (green), unshifted values that nevertheless use all of the thermodynamically relevant charge microstates (green), and fixed charge models (black).
The top row shows how the ensemble-averaged helical propensities, quantified as probabilities, vary with pH for the system (left) vs. the NTL912–23 system (right). The bottom row shows a similar comparative analysis for the ensemble-averaged radii of gyration.
Discussion
We have described a new method, which we designate as -canonical Monte Carlo sampling, to model the effects of pH, sequence, and conformation dependent charge regulation in peptides and IDPs. We applied -canonical Monte Carlo sampling to two systems and show how the method yields a complete pH dependent description of populations for charge microstates and their conformations. The results highlight the holistic picture one obtains for the diversity of charge microstates that contribute to conformational distributions. They also highlight the highly averaged descriptions one obtains using fixed charge models that do not allow for charge regulation. Strictly speaking, the results one obtains using fixed charge models will be accurate if and only if sequence context and conformational changes do not influence values of ionizable groups. Results obtained using fixed charge models are also likely to be reliable if a single mesostate dominates over a range of pH values. This is likely to be the case for proteins characterized by minimal conformational heterogeneity 119–121. In contrast, systems such as IDPs will require full consideration of contributions from all thermodynamically relevant charge microstates, and -canonical sampling enables this sort of sampling.
Although we have demonstrated the deployment of -canonical sampling using the ABSINTH implicit solvation and forcefield paradigm, there is nothing about its design that prevents its interoperability with other implicit solvation models. However, some of the distinct advantages of ABSINTH are likely to be lost in making -canonical sampling interoperable with other classes of implicit solvation models, especially those that come under the rubric of Poisson 122, 123 or generalized Born 82 approaches.
Ongoing work:
Effects of charge regulation are likely to become more pronounced in longer sequences where long-range interactions between non-nearest neighbor residues can alter charge states. Extant data already highlight the effects of charge regulation in sequences where the local charge density is high, as would be the case with the striking example of prothymosin 6. Long linear clusters of charged residues 34 and the modulating effects of long-range interactions 29 are rather common in intrinsically disordered proteins. Therefore, we expect that -canonical sampling applied to a host of recently studied systems will reveal the contributions from charge regulation.
Although we have focused our narrative on the application of -canonical sampling to the effects of charge regulation in IDPs, it is noteworthy that accounts of shifts have been well documented for folded proteins, especially variants of staphylococcal nuclease 96, 98, 119–121, 124–152. The design of -canonical sampling does not come with a formal restriction of being applicable to IDPs alone. The absence of this restriction is also true for ABSINTH / ABSINTH-C models. These are physics-based implicit solvation models that are interoperable with standard molecular mechanics forcefields. Accordingly, the model itself can be applied in conjunction with -canonical sampling to modeling the effects of shifts in folded proteins. This might require the use of hybrid Monte Carlo and torsional molecular dynamics methods that have been designed to work with the ABSINTH-style models 153. Estimators of enthalpies and entropies designed to be interoperable with the mBAR method 95 will help in uncovering the contributions of entropy to charge regulation effects. Grouping of microstates into mesostates, and the general approach of decoupling proton rearrangements and proton release should be usable in conjunction free energy methods for the study of charge regulation effects in folded proteins.
Conclusions
We have presented a detailed description of the -canonical sampling methodology and applied it to a set of short peptides, with the longest sequence being NTL912–23. Our demonstration of the initial version of -canonical sampling was based on the default ABSINTH implicit solvation model and forcefield paradigm with one essential distinction: we used the bond length and bond angle parameters derived from the CHARMM forcefield, which is different from the standard practice of using the Engh and Huber 154 values for bond lengths and bond angles. Importantly, recent efforts, based on the development of a quantitative touchstone for statistics of backbone dihedral angles for all twenty residues 155, Choi and Pappu developed an optimized version of ABSINTH, and referred to as ABSINTH-C 156, that vastly improves the description of local conformational equilibria. As a follow up study to the developments of ABSINTH-C and -canonical sampling for modeling charge regulation effects we are combining the results of the two efforts to perform a large-scale calibration of how the two major improvements to ABSINTH-based simulations alter our descriptions of conformational equilibria for a variety of IDP systems.
Acknowledgments
Grants from the National Institutes of Health (5R01NS056114) and the Human Frontier Science Program (RGP0034/2017) funded this work. We are immensely grateful to Ammon Posey and Tyler Harmon for helpful discussions and insights. We thank Alex Holehouse for comments on the manuscript and Bertrand Garcia-Moreno for inspiring discussions over the years about the importance of charge regulation in proteins.
References
- 1.Berlow RB; Dyson HJ; Wright PE, Expanding the Paradigm: Intrinsically Disordered Proteins and Allosteric Regulation. Journal of molecular biology 2018, 430 (16), 2309–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright PE; Dyson HJ, Intrinsically disordered proteins in cellular signalling and regulation. Nature Reviews Molecular and Cell Biology 2015, 16 (1), 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brucale M; Schuler B; Samori B, Single-Molecule Studies of Intrinsically Disordered Proteins. Chemical Reviews 2014, 114 (6), 3281–3317. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann H; Soranno A; Borgia A; Gast K; Nettels D; Schuler B, Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proceedings of the National Academy of Sciences of the United States of America 2012, 109 (40), 16155–16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller-Spaeth S; Soranno A; Hirschfeld V; Hofmann H; Rueegger S; Reymond L; Nettels D; Schuler B, Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (33), 14609–14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruggeri F; Zosel F; Mutter N; Rozycka M; Wojtas M; Ozyhar A; Schuler B; Krishnan M, Single-molecule electrometry. Nature Nanotechnology 2017, 12 (5), 488–495. [DOI] [PubMed] [Google Scholar]
- 7.Wuttke R; Hofmann H; Nettels D; Borgia MB; Mittal J; Best RB; Schuler B, Temperature-dependent solvation modulates the dimensions of disordered proteins. Proceedings of the National Academy of Sciences of the United States of America 2014, 111 (14), 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bah A; Forman-Kay JD, Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. Journal of Biological Chemistry 2016, 291 (13), 6696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bah A; Vernon RM; Siddiqui Z; Krzeminski M; Muhandiram R; Zhao C; Sonenberg N; Kay LE; Forman-Kay JD, Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519 (7541), 106–109. [DOI] [PubMed] [Google Scholar]
- 10.Borg M; Mittag T; Pawson T; Tyers M; Forman-Kay JD; Chan HS, Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proceedings of the National Academy of Sciences of the United States of America 2007, 104 (23), 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forman-Kay JD; Mittag T, From Sequence and Forces to Structure, Function, and Evolution of Intrinsically Disordered Proteins. Structure 2013, 21 (9), 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B; Chia D; Csizmok V; Farber P; Forman-Kay JD; Gradinaru CC, The Effect of Intrachain Electrostatic Repulsion on Conformational Disorder and Dynamics of the Sic1 Protein. Journal of Physical Chemistry B 2014, 118 (15), 4088–4097. [DOI] [PubMed] [Google Scholar]
- 13.Marsh JA; Dancheck B; Ragusa MJ; Allaire M; Forman-Kay JD; Peti W, Structural Diversity in Free and Bound States of Intrinsically Disordered Protein Phosphatase 1 Regulators. Structure 2010, 18 (9), 1094–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh JA; Forman-Kay JD, Sequence Determinants of Compaction in Intrinsically Disordered Proteins. Biophysical Journal 2010, 98 (10), 2383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh JA; Forman-Kay JD, Ensemble modeling of protein disordered states: Experimental restraint contributions and validation. Proteins-Structure Function and Bioinformatics 2012, 80 (2), 556–572. [DOI] [PubMed] [Google Scholar]
- 16.Mittag T; Kay LE; Forman-Kay JD, Protein dynamics and conformational disorder in molecular recognition. Journal of Molecular Recognition 2010, 23 (2), 105–116. [DOI] [PubMed] [Google Scholar]
- 17.Ozenne V; Schneider R; Yao M; Huang J -r.; Salmon, L.; Zweckstetter, M.; Jensen, M. R.; Blackledge, M., Mapping the Potential Energy Landscape of Intrinsically Disordered Proteins at Amino Acid Resolution. Journal of the American Chemical Society 2012, 134 (36), 15138–15148. [DOI] [PubMed] [Google Scholar]
- 18.Guerry P; Mollica L; Blackledge M, Mapping Protein Conformational Energy Landscapes Using NMR and Molecular Simulation. Chemphyschem 2013, 14 (13), 3046–3058. [DOI] [PubMed] [Google Scholar]
- 19.Jensen MR; Ruigrok RWH; Blackledge M, Describing intrinsically disordered proteins at atomic resolution by NMR. Current Opinion in Structural Biology 2013, 23 (3), 426–435. [DOI] [PubMed] [Google Scholar]
- 20.Kragelj J; Ozenne V; Blackledge M; Jensen MR, Conformational Propensities of Intrinsically Disordered Proteins from NMR Chemical Shifts. Chemphyschem 2013, 14 (13), 3034–3045. [DOI] [PubMed] [Google Scholar]
- 21.Communie G; Ruigrok RWH; Jensen MR; Blackledge M, Intrinsically disordered proteins implicated in paramyxoviral replication machinery. Current Opinion in Virology 2014, 5, 72–81. [DOI] [PubMed] [Google Scholar]
- 22.Huang J-R; Warner LR; Sanchez C; Gabel F; Madl T; Mackereth CD; Sattler M; Blackledge M, Transient Electrostatic Interactions Dominate the Conformational Equilibrium Sampled by Multidomain Splicing Factor U2AF65: A Combined NMR and SAXS Study. Journal of the American Chemical Society 2014, 136 (19), 7068–7076. [DOI] [PubMed] [Google Scholar]
- 23.Jensen MR; Blackledge M, Testing the validity of ensemble descriptions of intrinsically disordered proteins. Proceedings of the National Academy of Sciences of the United States of America 2014, 111 (16), E1557–E1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parigi G; Rezaei-Ghaleh N; Giachetti A; Becker S; Fernandez C; Blackledge M; Griesinger C; Zweckstetter M; Luchinat C, Long-Range Correlated Dynamics in Intrinsically Disordered Proteins. Journal of the American Chemical Society 2014, 136 (46), 16201–16209. [DOI] [PubMed] [Google Scholar]
- 25.Schwalbe M; Ozenne V; Bibow S; Jaremko M; Jaremko L; Gajda M; Jensen MR; Biernat J; Becker S; Mandelkow E; Zweckstetter M; Blackledge M, Predictive Atomic Resolution Descriptions of Intrinsically Disordered hTau40 and alpha-Synuclein in Solution from NMR and Small Angle Scattering. Structure 2014, 22 (2), 238–249. [DOI] [PubMed] [Google Scholar]
- 26.Mao AH; Crick SL; Vitalis A; Chicoine CL; Pappu RV, Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (18), 8183–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das RK; Crick SL; Pappu RV, N-Terminal Segments Modulate the alpha-Helical Propensities of the Intrinsically Disordered Basic Regions of bZIP Proteins. Journal of molecular biology 2012, 416 (2), 287–299. [DOI] [PubMed] [Google Scholar]
- 28.Radhakrishnan A; Vitalis A; Mao AH; Steffen AT; Pappu RV, Improved Atomistic Monte Carlo Simulations Demonstrate That Poly-L-Proline Adopts Heterogeneous Ensembles of Conformations of Semi-Rigid Segments Interrupted by Kinks. Journal of Physical Chemistry B 2012, 116 (23), 6862–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das RK; Pappu RV, Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proceedings of the National Academy of Sciences of the United States of America 2013, 110 (33), 13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin EW; Holehouse AS; Grace CR; Hughes A; Pappu RV; Mittag T, Sequence Determinants of the Conformational Properties of an Intrinsically Disordered Protein Prior to and upon Multisite Phosphorylation. Journal of the American Chemical Society 2016, 138 (47), 15323–15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao AH; Lyle N; Pappu RV, Describing sequence-ensemble relationships for intrinsically disordered proteins. Biochemical Journal 2013, 449, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruff KM; Pappu RV; Holehouse AS, Conformational preferences and phase behavior of intrinsically disordered low complexity sequences: insights from multiscale simulations. Current Opinion in Structural Biology 2019, 56, 1–10. [DOI] [PubMed] [Google Scholar]
- 33.Das RK; Ruff KM; Pappu RV, Relating sequence encoded information to form and function of intrinsically disordered proteins. Current Opinion in Structural Biology 2015, 32, 102–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holehouse AS; Das RK; Ahad JN; Richardson MOG; Pappu RV, CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophysical Journal 2017, 112 (1), 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuertes G; Banterle N; Ruff KM; Chowdhury A; Mercadante D; Koehler C; Kachala M; Estrada Girona G; Milles S; Mishra A; Onck PR; Grater F; Esteban-Martin S; Pappu RV; Svergun DI; Lemke EA, Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proceedings of the National Academy of Sciences of the United States of America 2017, 114 (31), E6342–e6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warner JB; Ruff KM; Tan PS; Lemke EA; Pappu RV; Lashuel HA, Monomeric Huntingtin Exon 1 Has Similar Overall Structural Features for Wild-Type and Pathological Polyglutamine Lengths. Journal of the American Chemical Society 2017, 139 (41), 14456–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newcombe EA; Ruff KM; Sethi A; Ormsby AR; Ramdzan YM; Fox A; Purcell AW; Gooley PR; Pappu RV; Hatters DM, Tadpole-like Conformations of Huntingtin Exon 1 Are Characterized by Conformational Heterogeneity that Persists regardless of Polyglutamine Length. Journal of molecular biology 2018, 430 (10), 1442–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staller MV; Holehouse AS; Swain-Lenz D; Das RK; Pappu RV; Cohen BA, A High-Throughput Mutational Scan of an Intrinsically Disordered Acidic Transcriptional Activation Domain. Cell systems 2018, 6 (4), 444–455.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KH; Chen J, Optimization of the GBMV2 implicit solvent force field for accurate simulation of protein conformational equilibria. Journal of computational chemistry 2017, 38 (16), 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X; Chen J; Chen J, Residual Structure Accelerates Binding of Intrinsically Disordered ACTR by Promoting Efficient Folding upon Encounter. Journal of molecular biology 2019, 431 (2), 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piana S; Donchev AG; Robustelli P; Shaw DE, Water dispersion interactions strongly influence simulated structural properties of disordered protein states. Journal of Physical Chemistry B 2015, 119 (16), 5113–23. [DOI] [PubMed] [Google Scholar]
- 42.Robustelli P; Piana S; Shaw DE, Developing a molecular dynamics force field for both folded and disordered protein states. Proceedings of the National Academy of Sciences of the United States of America 2018, 115 (21), E4758–e4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best RB; Zheng W; Mittal J, Balanced Protein-Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. Journal of chemical theory and computation 2014, 10 (11), 5113–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Best RB; Zheng W; Mittal J, Correction to Balanced Protein-Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. Journal of chemical theory and computation 2015, 11 (4), 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerze GH; Best RB; Mittal J, Sequence- and Temperature-Dependent Properties of Unfolded and Disordered Proteins from Atomistic Simulations. Journal of Physical Chemistry B 2015, 119 (46), 14622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Best RB, Computational and theoretical advances in studies of intrinsically disordered proteins. Current Opinion in Structural Biology 2017, 42, 147–154. [DOI] [PubMed] [Google Scholar]
- 47.Holmstrom ED; Holla A; Zheng W; Nettels D; Best RB; Schuler B, Accurate Transfer Efficiencies, Distance Distributions, and Ensembles of Unfolded and Intrinsically Disordered Proteins From Single-Molecule FRET. Methods in enzymology 2018, 611, 287–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerze GH; Zheng W; Best RB; Mittal J, Evolution of All-Atom Protein Force Fields to Improve Local and Global Properties. The journal of physical chemistry letters 2019, 10 (9), 2227–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyle N; Das RK; Pappu RV, A quantitative measure for protein conformational heterogeneity. Journal of Chemical Physics 2013, 139 (12), 121907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das RK; Huang Y; Phillips AH; Kriwacki RW; Pappu RV, Cryptic sequence features within the disordered protein p27Kip1 regulate cell cycle signaling. Proceedings of the National Academy of Sciences of the United States of America 2016, 113 (20), 5616–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherry KP; Das RK; Pappu RV; Barrick D, Control of transcriptional activity by design of charge patterning in the intrinsically disordered RAM region of the Notch receptor. Proceedings of the National Academy of Sciences of the United States of America 2017, 114 (44), E9243–e9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beveridge R; Migas LG; Das RK; Pappu RV; Kriwacki RW; Barran PE, Ion Mobility Mass Spectrometry Uncovers the Impact of the Patterning of Oppositely Charged Residues on the Conformational Distributions of Intrinsically Disordered Proteins. Journal of the American Chemical Society 2019, 141 (12), 4908–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borgia A; Borgia MB; Bugge K; Kissling VM; Heidarsson PO; Fernandes CB; Sottini A; Soranno A; Buholzer KJ; Nettels D; Kragelund BB; Best RB; Schuler B, Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555 (7694), 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen TT; Shklovskii BI, Model of inversion of DNA charge by a positive polymer: fractionalization of the polymer charge. Physical review letters 2002, 89 (1), 018101. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen TT; Grosberg AY; Shklovskii BI, Macroions in salty water with multivalent ions: giant inversion of charge. Physical review letters 2000, 85 (7), 1568–71. [DOI] [PubMed] [Google Scholar]
- 56.Shklovskii BI, Screening of a macroion by multivalent ions: correlation-induced inversion of charge. Physical Review E 1999, 60 (5 Pt B), 5802–5811. [DOI] [PubMed] [Google Scholar]
- 57.Watson M; Stott K, Disordered domains in chromatin-binding proteins. Essays in biochemistry 2019, 63 (1), 147–156. [DOI] [PubMed] [Google Scholar]
- 58.Greenberg RA, Histone tails: Directing the chromatin response to DNA damage. FEBS letters 2011, 585 (18), 2883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang LW; Lytle TK; Radhakrishna M; Madinya JJ; Velez J; Sing CE; Perry SL, Sequence and entropy-based control of complex coacervates. Nature communications 2017, 8 (1), 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pak CW; Kosno M; Holehouse AS; Padrick SB; Mittal A; Ali R; Yunus AA; Liu DR; Pappu RV; Rosen MK, Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Molecular cell 2016, 63 (1), 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barroso da Silva FL; Derreumaux P; Pasquali S, Protein-RNA complexation driven by the charge regulation mechanism. Biochemical and biophysical research communications 2018, 498 (2), 264–273. [DOI] [PubMed] [Google Scholar]
- 62.Munder MC; Midtvedt D; Franzmann T; Nüske E; Otto O; Herbig M; Ulbricht E; Müller P; Taubenberger A; Maharana S; Malinovska L; Richter D; Guck J; Zaburdaev V; Alberti S, A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 2016, 5, e09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Star WRL; Dijkema C; de Waard P; Picioreanu C; Strous M; van Loosdrecht MCM, An intracellular pH gradient in the anammox bacterium Kuenenia stuttgartiensis as evaluated by 31P NMR. Applied Microbiology and Biotechnology 2010, 86 (1), 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ortega E; Rengachari S; Ibrahim Z; Hoghoughi N; Gaucher J; Holehouse AS; Khochbin S; Panne D, Transcription factor dimerization activates the p300 acetyltransferase. Nature 2018, 562 (7728), 538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orren DK; Machwe A, Lysine Acetylation of Proteins and Its Characterization in Human Systems. Methods in Molecular Biology 2019, 1983, 107–130. [DOI] [PubMed] [Google Scholar]
- 66.Narita T; Weinert BT; Choudhary C, Functions and mechanisms of non-histone protein acetylation. Nature Reviews in Molecular and Cell Biology 2019, 20 (3), 156–174. [DOI] [PubMed] [Google Scholar]
- 67.Barnes CE; English DM; Cowley SM, Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays in biochemistry 2019, 63 (1), 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vassall KA; Jenkins AD; Bamm VV; Harauz G, Thermodynamic analysis of the disorder-to-alpha-helical transition of 18.5-kDa myelin basic protein reveals an equilibrium intermediate representing the most compact conformation. Journal of molecular biology 2015, 427 (10), 1977–92. [DOI] [PubMed] [Google Scholar]
- 69.Harmon TS; Pappu RV, Charge Patterned Sequences form Helical Structures through Charge Neutralization. Biophysical Journal 2016, 110 (3), 358a. [Google Scholar]
- 70.Franzmann TM; Jahnel M; Pozniakovsky A; Mahamid J; Holehouse AS; Nuske E; Richter D; Baumeister W; Grill SW; Pappu RV; Hyman AA; Alberti S, Phase separation of a yeast prion protein promotes cellular fitness. Science (New York, N.Y.) 2018, 359 (6371). [DOI] [PubMed] [Google Scholar]
- 71.Lei R; Lee JP; Francis MB; Kumar S, Structural Regulation of a Neurofilament-Inspired Intrinsically Disordered Protein Brush by Multisite Phosphorylation. Biochemistry 2018, 57 (27), 4019–4028. [DOI] [PubMed] [Google Scholar]
- 72.Srinivasan N; Bhagawati M; Ananthanarayanan B; Kumar S, Stimuli-sensitive intrinsically disordered protein brushes. Nature communications 2014, 5, 5145. [DOI] [PubMed] [Google Scholar]
- 73.Khandogin J; Chen J; Brooks CL 3rd, Exploring atomistic details of pH-dependent peptide folding. Proceedings of the National Academy of Sciences of the United States of America USA 2006, 103 (49), 18546–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khandogin J; Brooks CL 3rd, Toward the accurate first-principles prediction of ionization equilibria in proteins. Biochemistry 2006, 45 (31), 9363–9373. [DOI] [PubMed] [Google Scholar]
- 75.Khandogin J; Brooks CL 3rd, Constant pH molecular dynamics with proton tautomerism. Biophysical Journal 2005, 89 (1), 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee MS; Salsbury FR Jr.; Brooks CL 3rd, Constant-pH molecular dynamics using continuous titration coordinates. Proteins: Structure, Function, Bioinformatics 2004, 56 (4), 738–52. [DOI] [PubMed] [Google Scholar]
- 77.Radak BK; Chipot C; Suh D; Jo S; Jiang W; Phillips JC; Schulten K; Roux B, Constant-pH Molecular Dynamics Simulations for Large Biomolecular Systems. Journal of chemical theory and computation 2017, 13 (12), 5933–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y; Roux B, Constant-pH Hybrid Nonequilibrium Molecular Dynamics-Monte Carlo Simulation Method. Journal of chemical theory and computation 2015, 11 (8), 3919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vila-Vicosa D; Reis P; Baptista AM; Oostenbrink C; Machuqueiro M, A pH Replica Exchange Scheme in the Stochastic Titration Constant-pH MD Method. Journal of chemical theory and computation 2019, 15 (5), 3108–3116. [DOI] [PubMed] [Google Scholar]
- 80.Machuqueiro M; Baptista AM, Molecular dynamics at constant pH and reduction potential: application to cytochrome c(3). Journal of the American Chemical Society 2009, 131 (35), 12586–12594. [DOI] [PubMed] [Google Scholar]
- 81.Barroso daSilva FL; Dias LG, Development of constant-pH simulation methods in implicit solvent and applications in biomolecular systems. Biophysical Reviews 2017, 9 (5), 699–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Polydorides S; Simonson T, Monte Carlo simulations of proteins at constant pH with generalized Born solvent, flexible sidechains, and an effective dielectric boundary. Journal of computational chemistry 2013, 34 (31), 2742–56. [DOI] [PubMed] [Google Scholar]
- 83.Stern HA, Molecular simulation with variable protonation states at constant pH. Journal of Chemical Physics 2007, 126 (16), 164112. [DOI] [PubMed] [Google Scholar]
- 84.Permyakov SE; Makhatadze GI; Owenius R; Uversky VN; Brooks CL; Permyakov EA; Berliner LJ, How to improve nature: study of the electrostatic properties of the surface of alpha-lactalbumin. Protein Engineering Design and Selection 2005, 18 (9), 425–433. [DOI] [PubMed] [Google Scholar]
- 85.Chan CH; Wilbanks CC; Makhatadze GI; Wong KB, Electrostatic contribution of surface charge residues to the stability of a thermophilic protein: benchmarking experimental and predicted pKa values. PloS one 2012, 7 (1), e30296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang BW; Brunetti L; Brooks CL 3rd, Probing pH-dependent dissociation of HdeA dimers. Journal of the American Chemical Society 2011, 133 (48), 19393–19398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahlstrom LS; Law SM; Dickson A; Brooks CL 3rd, Multiscale modeling of a conditionally disordered pH-sensing chaperone. Journal of molecular biology 2015, 427 (8), 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Villa F; Simonson T, Protein p Ka's from Adaptive Landscape Flattening Instead of Constant-pH Simulations. Journal of chemical theory and computation 2018, 14 (12), 6714–6721. [DOI] [PubMed] [Google Scholar]
- 89.Shirts MR; Chodera JD, Statistically optimal analysis of samples from multiple equilibrium states. Journal of Chemical Physics 2008, 129 (12), 124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mittal A; Das R; Vitalis A; Pappu RV, ABSINTH Implicit Solvation Model and Force Field Paradigm for Use in Simulations of Intrinsically Disordered Proteins. In Computational Approaches to Protein Dynamics: From Quantum to Coarse-Grained Methods, Fuxreiter M, Ed. CRC Press (Taylor and Francis Group): Boca Raton, RL, 2015; pp 181–203. [Google Scholar]
- 91.Vitalis A; Pappu RV, ABSINTH: A New Continuum Solvation Model for Simulations of Polypeptides in Aqueous Solutions. Journal of computational chemistry 2009, 30 (5), 673–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Metropolis N; Rosenbluth AW; Rosenbluth MN; Teller AH; Teller E, Equation of State Calculations by Fast Computing Machines. The Journal of Chemical Physics 1953, 21 (6), 1087–1092. [Google Scholar]
- 93.Mittal A; Lyle N; Harmon TS; Pappu RV, Hamiltonian Switch Metropolis Monte Carlo Simulations for Improved Conformational Sampling of Intrinsically Disordered Regions Tethered to Ordered Domains of Proteins. Journal of chemical theory and computation 2014, 10 (8), 3550–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gelb LD, Monte Carlo simulations using sampling from an approximate potential. The Journal of Chemical Physics 2003, 118 (17), 7747–7750. [Google Scholar]
- 95.Wyczalkowski MA; Vitalis A; Pappu RV, New Estimators for Calculating Solvation Entropy and Enthalpy and Comparative Assessments of Their Accuracy and Precision. The Journal of Physical Chemistry B 2010, 114 (24), 8166–8180. [DOI] [PubMed] [Google Scholar]
- 96.Harms MJ; Schlessman JL; Sue GR; Garcia-Moreno B, Arginine residues at internal positions in a protein are always charged. Proceedings of the National Academy of Sciences of the United States of America USA 2011, 108 (47), 18954–18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoo J; Cui Q, Does arginine remain protonated in the lipid membrane? Insights from microscopic pKa calculations. Biophys J 2008, 94 (8), L61–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fitch CA; Platzer G; Okon M; Garcia-Moreno E, B.; McIntosh, L. P., Arginine: Its pKa value revisited. Protein science : a publication of the Protein Society 2015, 24 (5), 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watanabe M, Kirkwood coupling-parameter expansion method for the distribution function in a fluid. Physical Review A 1984, 29 (5), 2779–2786. [Google Scholar]
- 100.Kirkwood JG, Statistical Mechanics of Fluid Mixtures. The Journal of Chemical Physics 1935, 3 (5), 300–313. [Google Scholar]
- 101.Hritz J; Oostenbrink C, Hamiltonian replica exchange molecular dynamics using soft-core interactions. J Chem Phys 2008, 128 (14), 144121. [DOI] [PubMed] [Google Scholar]
- 102.Wyczalkowski MA; Pappu RV, Satisfying the fluctuation theorem in free-energy calculations with Hamiltonian replica exchange. Physical Review E 2008, 77 (2), 026104. [DOI] [PubMed] [Google Scholar]
- 103.Duarte Ramos Matos G; Kyu DY; Loeffler HH; Chodera JD; Shirts MR; Mobley D, Approaches for calculating solvation free energies and enthalpies demonstrated with an update of the FreeSolv database. Journal of Chemical Engineering Data 2017, 62 (5), 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boresch S; Karplus M, The Jacobian factor in free energy simulations. The Journal of Chemical Physics 1996, 105 (12), 5145–5154. [Google Scholar]
- 105.Wang L; Hermans J, Change of bond length in free‐energy simulations: Algorithmic improvements, but when is it necessary? The Journal of Chemical Physics 1994, 100 (12), 9129–9139. [Google Scholar]
- 106.Mao AH; Pappu RV, Crystal lattice properties fully determine short-range interaction parameters for alkali and halide ions. The Journal of Chemical Physics 2012, 137 (6), 064104. [DOI] [PubMed] [Google Scholar]
- 107.Neira JL; Rizzuti B; Iovanna JL, Determinants of the pKa values of ionizable residues in an intrinsically disordered protein. Arch Biochem Biophys 2016, 598, 18–27. [DOI] [PubMed] [Google Scholar]
- 108.Nagai H; Kuwabara K; Carta G, Temperature Dependence of the Dissociation Constants of Several Amino Acids. Journal of Chemical & Engineering Data 2008, 53 (3), 619–627. [Google Scholar]
- 109.Platzer G; Okon M; McIntosh LP, pH-dependent random coil (1)H, (13)C, and (15)N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. Journal of Biomolecular NMR 2014, 60 (2–3), 109–29. [DOI] [PubMed] [Google Scholar]
- 110.Beutler TC; Mark AE; van Schaik RC; Gerber PR; van Gunsteren WF, Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chemical Physics Letters 1994, 222, 529–539. [Google Scholar]
- 111.Shirts MR; Pande VS, Comparison of efficiency and bias of free energies computed by exponential averaging, the Bennett acceptance ratio, and thermodynamic integration. Journal of Chemical Physics 2005, 122 (14), 144107. [DOI] [PubMed] [Google Scholar]
- 112.Steinbrecher T; Mobley DL; Case DA, Nonlinear scaling schemes for Lennard-Jones interactions in free energy calculations. J Chem Phys 2007, 127 (21), 214108. [DOI] [PubMed] [Google Scholar]
- 113.Klimovich PV; Shirts MR; Mobley DL, Guidelines for the analysis of free energy calculations. Journal of Computer Aided Molecular Design 2015, 29 (5), 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baker EG; Bartlett GJ; Crump MP; Sessions RB; Linden N; Faul CF; Woolfson DN, Local and macroscopic electrostatic interactions in single alpha-helices. Nat Chem Biol 2015, 11 (3), 221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suveges D; Gaspari Z; Toth G; Nyitray L, Charged single alpha-helix: a versatile protein structural motif. Proteins: Structure, Function, Bioinformatics 2009, 74 (4), 905–916. [DOI] [PubMed] [Google Scholar]
- 116.Gaspari Z; Suveges D; Perczel A; Nyitray L; Toth G, Charged single alpha-helices in proteomes revealed by a consensus prediction approach. Biochim Biophys Acta 2012, 1824 (4), 637–46. [DOI] [PubMed] [Google Scholar]
- 117.Suveges D; Gaspari Z; Toth G; Nyitray L, Charged single alpha-helix: a versatile protein structural motif. Proteins 2009, 74 (4), 905–16. [DOI] [PubMed] [Google Scholar]
- 118.Kuhlman B; Luisi DL; Young P; Raleigh DP, pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry 1999, 38 (15), 4896–903. [DOI] [PubMed] [Google Scholar]
- 119.Peck MT; Ortega G; De Luca-Johnson JN; Schlessman JL; Robinson AC; Garcia-Moreno EB, Local Backbone Flexibility as a Determinant of the Apparent pKa Values of Buried Ionizable Groups in Proteins. Biochemistry 2017, 56 (40), 5338–5346. [DOI] [PubMed] [Google Scholar]
- 120.Richman DE; Majumdar A; Garcia-Moreno EB, pH dependence of conformational fluctuations of the protein backbone. Proteins 2014, 82 (11), 3132–43. [DOI] [PubMed] [Google Scholar]
- 121.Richman DE; Majumdar A; Garcia-Moreno EB, Conformational Reorganization Coupled to the Ionization of Internal Lys Residues in Proteins. Biochemistry 2015, 54 (38), 5888–97. [DOI] [PubMed] [Google Scholar]
- 122.Carstensen T; Farrell D; Huang Y; Baker NA; Nielsen JE, On the development of protein pKa calculation algorithms. Proteins: Structure, Function, Bioinformatics 2011, 79 (12), 3287–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gosink LJ; Hogan EA; Pulsipher TC; Baker NA, Bayesian model aggregation for ensemble-based estimates of protein pKa values. Proteins: Structure, Function, Bioinformatics 2014, 82 (3), 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee KK; Fitch CA; Lecomte JT; Garcia-Moreno EB, Electrostatic effects in highly charged proteins: salt sensitivity of pKa values of histidines in staphylococcal nuclease. Biochemistry 2002, 41 (17), 5656–67. [DOI] [PubMed] [Google Scholar]
- 125.Maldonado S; Irun MP; Campos LA; Rubio JA; Luquita A; Lostao A; Wang R; Garcia-Moreno EB; Sancho J, Salt-induced stabilization of apoflavodoxin at neutral pH is mediated through cation-specific effects. Protein Sci 2002, 11 (5), 1260–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mehler EL; Fuxreiter M; Simon I; Garcia-Moreno EB, The role of hydrophobic microenvironments in modulating pKa shifts in proteins. Proteins: Structure, Function, Bioinformatics 2002, 48 (2), 283–292. [DOI] [PubMed] [Google Scholar]
- 127.Schwehm JM; Fitch CA; Dang BN; Garcia-Moreno EB; Stites WE, Changes in stability upon charge reversal and neutralization substitution in staphylococcal nuclease are dominated by favorable electrostatic effects. Biochemistry 2003, 42 (4), 1118–28. [DOI] [PubMed] [Google Scholar]
- 128.Denisov VP; Schlessman JL; Garcia-Moreno EB; Halle B, Stabilization of internal charges in a protein: water penetration or conformational change? Biophysical Journal 2004, 87 (6), 3982–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garcia-Moreno EB; Fitch CA, Structural interpretation of pH and salt-dependent processes in proteins with computational methods. Methods in enzymology 2004, 380, 20–51. [DOI] [PubMed] [Google Scholar]
- 130.Whitten ST; Garcia-Moreno EB; Hilser VJ, Local conformational fluctuations can modulate the coupling between proton binding and global structural transitions in proteins. Proc Natl Acad Sci U S A 2005, 102 (12), 4282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fitch CA; Whitten ST; Hilser VJ; Garcia-Moreno EB, Molecular mechanisms of pH-driven conformational transitions of proteins: insights from continuum electrostatics calculations of acid unfolding. Proteins 2006, 63 (1), 113–26. [DOI] [PubMed] [Google Scholar]
- 132.Hilser VJ; Garcia-Moreno EB; Oas TG; Kapp G; Whitten ST, A statistical thermodynamic model of the protein ensemble. Chemical Reviews 2006, 106 (5), 1545–1558. [DOI] [PubMed] [Google Scholar]
- 133.Damjanovic A; Schlessman JL; Fitch CA; Garcia AE; Garcia-Moreno EB, Role of flexibility and polarity as determinants of the hydration of internal cavities and pockets in proteins. Biophysical Journal 2007, 93 (8), 2791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karp DA; Gittis AG; Stahley MR; Fitch CA; Stites WE; Garcia-Moreno EB, High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophysical Journal 2007, 92 (6), 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Damjanovic A; Miller BT; Wenaus TJ; Maksimovic P; Garcia-Moreno EB; Brooks BR, Open science grid study of the coupling between conformation and water content in the interior of a protein. Journal of chemical information and modeling 2008, 48 (10), 2021–9. [DOI] [PubMed] [Google Scholar]
- 136.Damjanovic A; Wu X; Garcia-Moreno EB; Brooks BR, Backbone relaxation coupled to the ionization of internal groups in proteins: a self-guided Langevin dynamics study. Biophys J 2008, 95 (9), 4091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fitzkee NC; Garcia-Moreno EB, Electrostatic effects in unfolded staphylococcal nuclease. Protein science : a publication of the Protein Society 2008, 17 (2), 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schlessman JL; Abe C; Gittis A; Karp DA; Dolan MA; Garcia-Moreno EB, Crystallographic study of hydration of an internal cavity in engineered proteins with buried polar or ionizable groups. Biophys J 2008, 94 (8), 3208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Castaneda CA; Fitch CA; Majumdar A; Khangulov V; Schlessman JL; Garcia-Moreno BE, Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins 2009, 77 (3), 570–88. [DOI] [PubMed] [Google Scholar]
- 140.Damjanovic A; Garcia-Moreno EB; Brooks BR, Self-guided Langevin dynamics study of regulatory interactions in NtrC. Proteins 2009, 76 (4), 1007–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harms MJ; Castaneda CA; Schlessman JL; Sue GR; Isom DG; Cannon BR; Garcia-Moreno EB, The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors. Journal of molecular biology 2009, 389 (1), 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Isom DG; Castaneda CA; Cannon BR; Velu PD; Garcia-Moreno EB, Charges in the hydrophobic interior of proteins. Proceedings of the National Academy of Sciences of the United States of America USA 2010, 107 (37), 16096–16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chimenti MS; Castaneda CA; Majumdar A; Garcia-Moreno EB, Structural origins of high apparent dielectric constants experienced by ionizable groups in the hydrophobic core of a protein. Journal of molecular biology 2011, 405 (2), 361–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goh GB; Garcia-Moreno EB; Brooks CL 3rd, The high dielectric constant of staphylococcal nuclease is encoded in its structural architecture. Journal of the American Chemical Society 2011, 133 (50), 20072–20075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kitahara R; Hata K; Maeno A; Akasaka K; Chimenti MS; Garcia-Moreno EB; Schroer MA; Jeworrek C; Tolan M; Winter R; Roche J; Roumestand C; Montet de Guillen K; Royer CA, Structural plasticity of staphylococcal nuclease probed by perturbation with pressure and pH. Proteins 2011, 79 (4), 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dellarole M; Kobayashi K; Rouget JB; Caro JA; Roche J; Islam MM; Garcia-Moreno EB; Kuroda Y; Royer CA, Probing the physical determinants of thermal expansion of folded proteins. The journal of physical chemistry. B 2013, 117 (42), 12742–9. [DOI] [PubMed] [Google Scholar]
- 147.Kukic P; Farrell D; McIntosh LP; Garcia-Moreno EB; Jensen KS; Toleikis Z; Teilum K; Nielsen JE, Protein dielectric constants determined from NMR chemical shift perturbations. J Am Chem Soc 2013, 135 (45), 16968–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Robinson AC; Castaneda CA; Schlessman JL; Garcia-Moreno EB, Structural and thermodynamic consequences of burial of an artificial ion pair in the hydrophobic interior of a protein. Proc Natl Acad Sci U S A 2014, 111 (32), 11685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dellarole M; Caro JA; Roche J; Fossat M; Barthe P; Garcia-Moreno EB; Royer CA; Roumestand C, Evolutionarily Conserved Pattern of Interactions in a Protein Revealed by Local Thermal Expansion Properties. J Am Chem Soc 2015, 137 (29), 9354–62. [DOI] [PubMed] [Google Scholar]
- 150.Robinson AC; Majumdar A; Schlessman JL; Garcia-Moreno EB, Charges in Hydrophobic Environments: A Strategy for Identifying Alternative States in Proteins. Biochemistry 2017, 56 (1), 212–218. [DOI] [PubMed] [Google Scholar]
- 151.Kougentakis CM; Grasso EM; Robinson AC; Caro JA; Schlessman JL; Majumdar A; Garcia-Moreno EB, Anomalous Properties of Lys Residues Buried in the Hydrophobic Interior of a Protein Revealed with (15)N-Detect NMR Spectroscopy. J Phys Chem Lett 2018, 9 (2), 383–387. [DOI] [PubMed] [Google Scholar]
- 152.Robinson AC; Schlessman JL; Garcia-Moreno EB, Dielectric Properties of a Protein Probed by Reversal of a Buried Ion Pair. The journal of physical chemistry. B 2018, 122 (9), 2516–2524. [DOI] [PubMed] [Google Scholar]
- 153.Vitalis A; Pappu RV, A simple molecular mechanics integrator in mixed rigid body and dihedral angle space. The Journal of Chemical Physics 2014, 141 (3), 034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Engh RA; Huber R, Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallographica Section A 1991, 47 (4), 392–400. [Google Scholar]
- 155.Choi J-M; Pappu RV, Experimentally Derived and Computationally Optimized Backbone Conformational Statistics for Blocked Amino Acids. Journal of chemical theory and computation 2019, 15 (2), 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Choi J-M; Pappu RV, Improvements to the ABSINTH Force Field for Proteins Based on Experimentally Derived Amino Acid Specific Backbone Conformational Statistics. Journal of chemical theory and computation 2019, 15 (2), 1367–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kabsch W; Sander C, Dictionary of Protein Secondary Structure - Pattern-Recognition of Hydrogen-bonded and Geometrical Features. Biopolymers 1983, 22 (12), 2577–2637. [DOI] [PubMed] [Google Scholar]