Abstract
The structure and semantics of clinical notes vary considerably across different Electronic Health Record (EHR) systems, sites, and institutions. Such heterogeneity hampers the portability of natural language processing (NLP) models in extracting information from the text for clinical research or practice. In this study, we evaluate the contextual variation of clinical notes by measuring the semantic and syntactic similarity of the notes of two sets of physicians comprising four medical specialties across EHR migrations at two Mayo Clinic sites. We find significant semantic and syntactic variation imposed by the context of the EHR system and between medical specialties whereas only minor variation is caused by variation of spatial context across sites. Our findings suggest that clinical language models need to account for process differences at the specialty sublanguage level to be generalizable.
Introduction
The Health Information Technology for Economic and Clinical Health (HITECH) Act of 2009 has brought about widespread adoption of Electronic Health Record (EHR) usage as well as sweeping changes to healthcare data collection, storage, and retrieval1 with the intent to promote the “meaningful use” of EHRs by physicians as well as enable secondary research efforts, even among incentive-ineligible hospitals2. However, ample evidence exists that these facilitating changes have been accompanied by differences in information recording, productivity and sharing practices by clinicians, revealing considerable discrepancies across institutions and their respective EHR implementations3,4. For example, Edwards et al.5 reported that a significant portion of physician notes contained missing or mislabeled sections and subsections in a corpus of outpatient notes. Cohen et al.6 revealed considerable variation between the documentation of physicians within the same practice, clinical setting and note types in a large cohort of primary care physicians.
Since much of information about a patient narrative driving provider investigation is locked in unstructured clinical note text, understanding the differences across EHRs and institutions at the sublanguage level is critical for efforts toward standardization. Clinical natural language processing (NLP) applications which aim to classify arbitrary clinical text into common document ontologies require discernable patterns in language for the level of performance sufficient to translate trained language models into portable applications. Heterogeneity of language across EHRs and institutions dampers these efforts by confounding language model ability to conform outside medical records across institutions as well as to generalize across institutions and subspecialties. Several studies have confirmed that the considerable heterogeneity of EHR system usage across different EHR software implementations institutions has a significant effect on the performance of these models on an assortment of tasks and case studies7-9. Patterson and Hurdle10 illuminated significant differences in the vocabularies of medical specialty sublanguages through distinctly formed document clusters by medical specialty. Doing-Harris et al.11 extended that work by applying document clustering across subspecialties of different institutions, noting complications with documents that express multiple sublanguages, naming conventions, and document type incongruities. Further, national sublanguage trends have shown to drift and acutely change with trends over time12 as well as with clinical sublanguage semantics13,14.
While enforcement of universal documentation standards would alleviate the need for such generalizability of language models, reconciliation of the various types of heterogeneity across EHRs and institutions is currently pursued by building clinical NLP systems that are interoperable. Elkin et al.15 defined three levels of interoperability based on the previous work of Charles Morris: syntactic, semantic, and pragmatic interoperability. More recently, Sohn et al.16 and Fu et al.9 leveraged the definitions of these levels of interoperability to measure variation/similarity between EHRs, each finding significant variation between the compared EHR corpora for asthma and post-surgical complication terms, respectively. Moon et al.17 gauged the coverage of cardiovascular medicine concept terms received in outside medical records against Mayo Clinic concepts, finding that while there was overlap, a significant portion of the concepts were not recognized and therefore were not utilized once imported. Though these studies demonstrated differences using concepts specific to their case studies, further analysis is needed into the effect of differences in clinical documentation patterns caused by the each of the contextual dimensions of the clinical practice. An examination measuring more precisely the effects of context would facilitate pursuits toward portable NLP models needed for automated information sharing between institutions to build patient-level models for information extraction, information retrieval, and prediction applications, among many others. Furthermore, an understanding of contextual variation would contribute to broader theoretical NLP efforts by demonstrating how nuanced deviations across space, time, and institutional process influence language and should be accounted for in the processing of it.
The current study aims to better understand whether differences in spatial, temporal, and practical contexts significantly influence provider semantics and narrative structure of clinical documentation. We compared lexical, semantic, and process variations across Mayo Clinic EHR system migration from the Cerner EHR to Epic EHR as well as between two sites, Mayo Clinic Arizona (MCA) and Mayo Clinic Florida (MCF) for a selected subset of medical specialties. To our knowledge, this is the first attempt to compare sublanguages of a selected set of physicians over an EHR migration (i.e., temporal) or across specialities (i.e., practice) at multiple sites (i.e., spatial) of an institution sharing the same integrated data repository. This expands on the work of Fu et al.9 and Sohn et al.16 by comparing across Mayo Clinic sites, EHR systems, and specialties. While the current investigation lacks the granular analysis of those case studies, it instead highlights the effect that differences in context has on physicians.
Materials and Methods
Study design
Our examination uses several Mayo Clinic corpora to assess differences in sublanguage characteristics across the various practice settings in which physicians of Mayo Clinic function. We investigate variations in language use before and after the migration of EHR systems from Cerner EHR to the Epic EHR and across two sites, MCA and MCF. We define the criteria for physician selection and the bases for comparison below. Table 1 lists the timeframe of each EHR collection at each site as well the total number of physicians and in each specialty. To allow the comparison of corpora across sites as accurately as possible, we matched the number of physicians in each specialty. The study was considered exempted by Mayo Clinic IRB Board with the record number: 20-001137 “Corpus-based Natural Language Processing based on Big Data and Machine Learning“.
Table 1.
Corpus study period and physician count for each Mayo Clinic EHR, site, and medical specialty used.
| Arizona | Florida | |||
|---|---|---|---|---|
| Cerner | Epic | Cerner | Epic | |
| Study Period | 1/1/17 – 12/31/17 | 1/1/19 - 12/31/19 | 1/1/17 – 12/31/17 | 1/1/19 - 12/31/19 |
| Internal Medicine | 31 | 31 | ||
| Emergency Medicine | 13 | 13 | ||
| Gastroenterology and Hepatology | 20 | 20 | ||
| Neurology | 23 | 23 | ||
| Total Physicians | 87 | 87 | ||
Provider selection
We compared 87 attending physicians at each site practicing in one of four medical specialties: internal medicine, emergency medicine, gastroenterology and hepatology, or neurology. Providers included for sublanguage examination were selected only if they started serving as an attending physician prior to the start of the window for collection from the Cerner EHR, January 1, 2017, and remained in practice at the same Mayo Clinic site through the end of the Epic study period, December 31, 2019. Physicians who had too few notes (n < 200) for either period or had a three times difference between notes collected during either period were also excluded from consideration.
Corpora collection and preprocessing
The clinical notes for each set of selected physicians were retrieved from the Unified Data Platform (UDP), Mayo Clinic’s integrated enterprise data warehouse containing all Mayo Clinic sites’ data for each EHR. Word tokens, Unified Medical Language System (UMLS) concept semantic types and semantic group counts were collected for each section and aggregated to each document. Words and sentences were tokenized using the scikit-learn18 and medspacy19 python libraries, respectively. Punctuation, dates, and spaces were not counted as word tokens. UMLS concepts were matched using the QuickUMLS20 python library. An implementation of the ConText21 algorithm using a custom set of rules from the MedTagger Information Extraction pipeline22 was applied to tag concepts with their negation, hypothetical, and experiencer contextual modifiers. Sections were determined using the NLP-as-a- service toolkit according to the Health Level 7 section standard. Report header sections, containing physician and patient metadata, were removed before further calculations were performed once the section information was ascertained.
Dimensions of Clinical Context
In our attempt to quantify the degree of change across corpora that is influenced by context from the perspective of the physician, we consider the impact of three dimensions of context, formally defined in Table 2.
Table 2.
Definitions of contextual dimensions
| Context Dimension | Definition |
|---|---|
| Practical context | Factors from the set of note types, sections, templates used within the given institution, EHR implementation and medical specialty. |
| Spatial context | Factors dictated by the practice setting’s geographical locale such as patient population demographic variation, national and local laws, nearby competing healthcare institutions |
| Temporal context | Factors varying over time pertaining to the development and prognosis of a patient’s condition or the public perception about the condition at a given point in time. |
The confluence of these contextual dimensions in each physician’s practice setting determines the set of facts needed to interpret their clinical notes. In this analysis, the practical and temporal contexts are altered by the EHR migration from Cerner to Epic and the passage of time from 2017 to 2019, respectively, whereas the spatial context varies through Arizona and Florida regional differences. By comparing these corpora, we attempt to assess the degree to which physician documentation is influenced by these aspects of context.
Measurements of Clinical Corpus Variation
Process Variation
Variation of process here is adopted from previous work to refer to overall structural (i.e. syntactic) differences between physician documentation23. To measure the degree to which the physician practice process differs due the context of the EHR, we collected corpus statistics for each of the Cerner and Epic EHRs for the same set of physicians who met the inclusion criteria at the two Mayo Clinic sites. This included the total number of documents and sections as well as the unique number of note types. The total number of word tokens in each corpus was used to calculate the rates of words per document and words per concept, as noted below.
Semantic Variation
The source of variation by UMLS semantic group was of paramount interest. For this measurement, we aggregated concepts by UMLS semantic group and compared the rate of words per concept semantic group across sites. To limit the breadth of this analysis to terms most relevant to clinical practice, however, we focused on four of the thirteen UMLS semantic groups: Anatomy (ANAT), Chemicals & Drugs (CHEM), Disorders (DISO), and Procedures (PROC). A preliminary Kolmogorov-Smirnov normality test rejected the null hypothesis of normality for both the AZ (p<0.001) and FL (p<0.001) corpora. Hence, the rates of each practitioner’s words to these clinically-relevant concepts and groups were tested for statistical significance using the Wilcoxon signed-rank test. We also sought to examine this semantic variation at the specialty level at each site and EHR. For this comparison, we measured the rates of concepts per word for each semantic group across four selected medical specialties comprising both primary and secondary care. The areas included for this purpose were internal medicine, emergency medicine, gastroenterology and hepatology, and neurology.
In comparing the effect of the spatial context by measuring differences across MCA and MCF site corpora, we adapt the definition of semantic variation from Sohn et al.16 by implementing the class-based approach established by Grootendorst24 to assess lexical similarity using both standard tf-idf as well as a modified version accounting for the thematic medical specialty of the clinical note we call stf-idf. First the overall tf-idf was calculated using the sum of the tf (t) ∙ idf(t) for each term t: ∑𝑖 𝑡𝑓𝑖(𝑡) ∙ 𝑖𝑑𝑓(𝑡)/𝑁 , where
Next, we calculated the specialty term frequency-inverse document frequency (stf-idf), which weighs the term frequency by author physician specialty to adjust for variations in specialty lexicons. This overall stf-idf of each corpus was calculated using the sum of stf (t) ∙ idf(t) for each term t: ∑𝑠 𝑠𝑡𝑓𝑠(𝑡) ∙ 𝑖𝑑𝑓(𝑡)𝑁 , where
Here, the term frequency is calculated across each specialty s with respect to the total number of terms in the given medical specialty s, as opposed to the total number of terms in the document. As an example, a colon surgery such as intestinal anastomosis might be discussed several times in a clinical note but represent a small fraction of all patient procedures in the overall corpus. The stf-idf algorithm would more accurately capture the consistency of the surgery in the context of the site’s GI department as opposed to a general reflection of the number of times a procedure is repeated in a note. Thus, this scoring approach is intended to measure more precisely the salience of each term within each specialty instead of each document, since we suspect that the salience of a term to a document may depends excessively on the particularities of the patient case and specific encounter. The idf weighting in each of the equations is the same, as the log of the total number of documents in the corpus N is divided by the total number of documents with term t in the corpus, irrespective of specialty. To illustrate the validity of this method, we present the descending-ordered top ranked terms of each specialty across site and EHR, removing any provider- specific terms. The cosine similarity of each of these scores is calculated between the different EHR corpora at each site as well as the across MCA and MCF sites to assess their general likeness.
Results
Process Variation
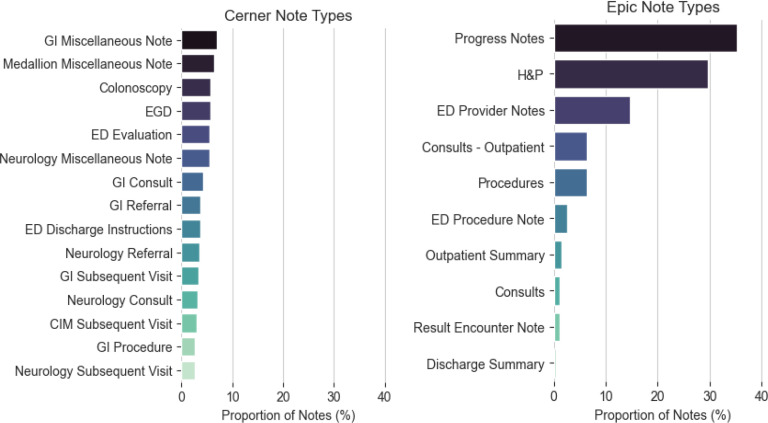
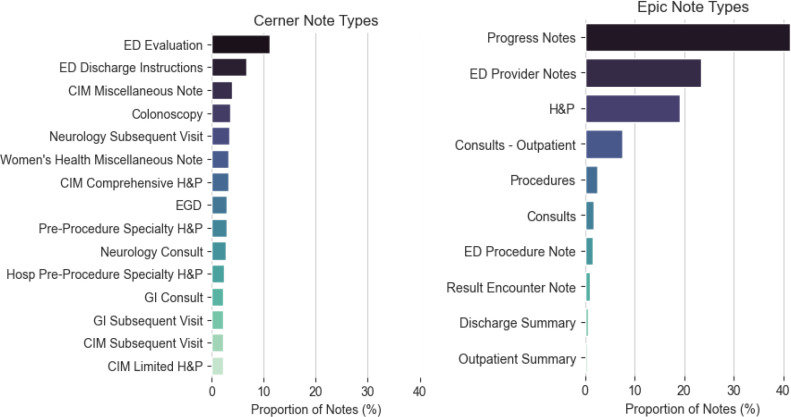
The overall corpus statistics, as well as the total UMLS concept and individual semantic group frequencies, are listed in Table 3. At both sites, more documents were found for the Cerner EHR than for the Epic EHR. The unique number of sections did not remarkably differ (Arizona: Cerner=43, Epic=44; Florida: Cerner=44, Epic=48), but the number of sections per document was consistently greater in Epic than Cerner at both sites (Arizona: Cerner=4, Epic=5; Florida: Cerner=2, Epic=5). The number of unique note types used in each EHR differed by about an order of magnitude (Arizona: Cerner=240, Epic=26; Florida: Cerner=270, Epic=27). This difference is attributed to the freedom given to providers to create note types in addition to catch-all categories as “Miscellaneous Note” and procedure specific categories such as “EGD”, as displayed in Figure 2 and Figure 3.
Table 3.
Corpus variation across EHR migration
| Metric | Mayo Clinic Arizona | Mayo Clinic Florida | ||||
|---|---|---|---|---|---|---|
| Cerner | Epic | p-value* | Cerner | Epic | p-value* | |
| Total Documents | 66781 | 56454 | 74874 | 56961 | ||
| Total Sections | 344063 | 288774 | 322802 | 266653 | ||
| Unique Note Types | 240 | 26 | 271 | 27 | ||
| Unique sections | 43 | 44 | 44 | 48 | ||
| Sections per document, median | 4 (7) | 5 (4) | 2 (5) | 5 (4) | ||
| Words per document, median (IQR) | 420 (628) | 515 (522) | 262 (508) | 423 (479) | ||
| Words per concept | 4.53 | 3.44 | <0.001 | 4.034 | 3.290 | <0.001 |
| Words per ANAT | 27.60 | 19.22 | <0.001 | 23.81 | 18.02 | <0.001 |
| Words per CHEM | 26.45 | 18.90 | 0.013 | 28.24 | 20.43 | 0.416 |
| Words per DISO | 10.08 | 7.80 | <0.001 | 8.54 | 7.14 | <0.001 |
| Words per PROC | 21.09 | 17.47 | <0.001 | 18.76 | 16.83 | 0.168 |
IQR: Interquartile range, ANAT: Anatomy, CHEM: Chemicals & Drugs, DISO: Disorders, PROC: Procedures
*Determined using Wilcoxon signed-rank test for every set of paired physician migration timeframe samples
Figure 2.
MCA Note Type distributions in descending order of relative frequency
Figure 3.
MCF Note Type distributions in descending order of relative frequency
Semantic Variation
The rate of total word tokens to total UMLS concepts as well as the rates of total word tokens to each (of the four pertinent) UMLS semantic groups is listed in Table 3. For each of the MCA and MCF sites, the p-values of each of the comparisons of Cerner EHR to Epic EHR, as measured by the Wilcoxon signed-rank of physician document collections before and after the migration, are listed in the column adjacent to the overall rates. Of the five measurements conducted at each site, we observed statistically significant differences in all paired sets except for words per chemicals and drugs concept (p=0.416) and words per procedure concept (p=0.168) for MCF using alpha=0.05. While words per chemicals and drugs concept (p=0.013) for MCA was not a statistically significant difference, the difference between that probability and other words per concept rates was notable.
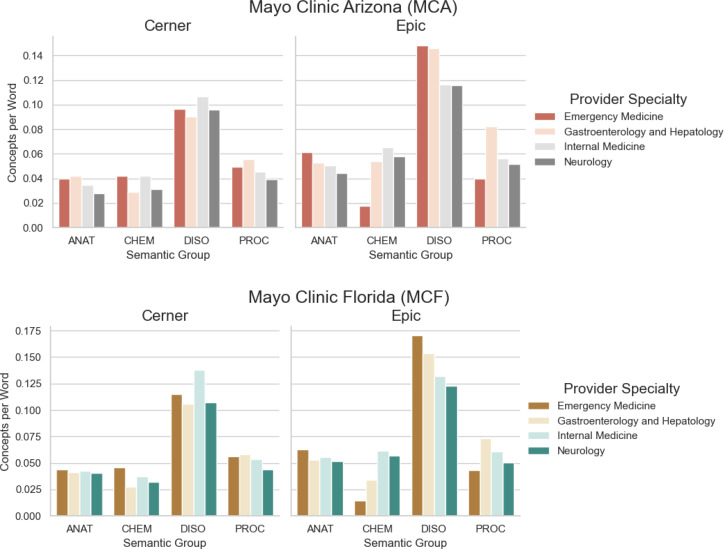
The rates of UMLS concepts per word for ANAT, CHEM, DISO, and PROC semantic groups for each of the four specialties are illustrated in Figure 4. These graphs illustrate the inverse of the words-per-concept rates of Table 2 but at the granularity of the specialty for each site; the higher the rate of semantic concepts per word, the greater the density of those concepts in the notes (i.e. the more frequently those concepts are mentioned in the notes). The graphs are shown this way to convey that a larger bar has a relatively higher density of concept term mentions in the notes. At each site, the figure shows a considerable relative increase in the rate of DISO concepts per word for Emergency Medicine and for Gastroenterology and Hepatology. A significant increase in the rate of PROC concepts per word is also observed for Gastroenterology and Hepatology across the MCA EHR transition.
Figure 4.
UMLS Semantic Group concept frequencies across the EHRs at each of Mayo Clinic Arizona and Florida sites.
As a way of quantifying the overall semantic variation between corpora, we also computed the cosine similarities between the sums of tf-idf and stf-idf values for each respective corpus pair, as presented in Table 4. While the tf-idf similarities indicate semantic deviation between EHRs (MCA Cerner to Epic = 0.779; MCF Cerner to Epic = 0.618), the semantic similarity across sites for the same Epic EHR was much higher (MCA Epic to MCF Epic = 0.965). The semantic similarity was much higher between EHRs at each site when adjusting for specialty using stf-idf (MCA Cerner to Epic = 0.941; MCF Cerner to Epic = 0.860) and even higher across sites for the same EHR (MCA Epic to MCF Epic = 0.987). These results demonstrate a high degree of semantic sensitivity to the specialty context.
Table 4.
Cosine similarities of MCA and MCF corpora
| Corpora comparison | tf-idf | stf-isf |
|---|---|---|
| MCA Cerner to Epic | 0.779 | 0.941 |
| MCF Cerner to Epic | 0.618 | 0.860 |
| MCA Epic to MCF Epic | 0.965 | 0.987 |
| MCA Cerner to MCF Cerner | 0.804 | 0.952 |
The top terms by specialty (determined by ranked descending order of stf-idf terms) are listed for each EHR and site in Table 5. The specificity of terms to the specialty under which they are listed demonstrates some validity to the approach of weighting terms by medical discipline. For example, in several of the stf-idf models, abbreviations for common terms specific to the area of practice (e.g. “eeg”, “mri” for Neurology and “eom”, “hent”, “hi” for Emergency Medicine) scored the highest.
Table 5.
Most informative words by specialty by stf-idf score
| Specialty | Site | EHR | Top stf-idf Terms+ |
|---|---|---|---|
| Internal Medicine | AZ | Cerner | healthy, age, good, died, regular, subject, pap, thyroid, issues, cholesterol, stress, sleep, mammogram, influenza, verified, exercise, hoc, ad, breast, vaccine |
| Epic | adult, ear, provided, bp, good, lesions, lungs, apply, hyperlipidemia, vitamin, thyroid, sleep, calcium, risk, stress, diet, healthy, breast, exercise | ||
| FL | Cerner | bilateral, surgical, surgery, mass, infection, hypertension, cancer, edema, weight, cuff, sleep, cardiac, kg, apnea, breast | |
| Epic | coronary, bang, glucose, index, perioperative, bedtime, hold, vitamin, breast, screening, calculated, stop, expected, ear, m², external, apnea, preoperative | ||
| Emergency Medicine | AZ | Cerner | operate, understand, medicines, rays, emergency, drink, important, received, indications, specialist, services, prescriptions, exitcare, seek, materials, document, caregiver, orally |
| Epic | eom, shortness, psychiatric, membranes, resp, nerve, temperature, spo2, fever, chills, motion, exhibits, hent, deficit, nursing, mucous, moist, ed, emergency, vitals |
||
| FL | Cerner | hi, neurological, tests, except, such, america, observed, regarding, prescriptions, exitcare, materials, account, indications, received, directed, seek, hpi, caregiver, emergency, orally | |
| Epic | hent, vomiting, temperature, spo2, resp, emergency, fever, shortness, dysuria, eom, moist, states, membranes, chills, mucous, personally, vitals, critical, ed, radiology |
||
| Gastroenterology and Hepatology | AZ | Cerner | forceps, colon, polyp, procedure, examined, mm, signed, preparation, mucosa, planned, monitored, duodenum, found, endoscopy, entire, procedural, diagnoses, op, scope, sedation |
| Epic | removal, sexual, consents, social, peripheral, pump, lumen, donor, wdl, anesthesia, connections, gastroenterology, catheter, overview, transplant, liver, implanted, clean, dressing, sedation | ||
| FL | Cerner | colonoscopy, advanced, examined, esophageal, introduced, biopsies, gi, les, forceps, reflux, found, esophagus, entire, duodenum, monitored, diagnoses, op, endoscopy, procedural, sedation |
|
| Epic | lumen, connections, line, ph, swallows, wdl, hepatology, gastroenterology, acid, balloon, implanted, reflux, les, sphincter, catheter, esophageal, sedation, endoscopy, clean, dressing | ||
| Neurology | AZ | Cerner | seizure, headaches, nerve, migraine, mental, visual, intact, reflexes, neurology, motor, eeg, tremor, stroke, headache, gait, ms, mri, brain, cognitive |
| Epic | movements, injection, neurology, facial, symmetric, headache, migraine, units, motor, eeg, bilaterally, seizure, stroke, seizures, gait, mri, intravenous, tremor, brain, cognitive |
||
| FL | Cerner | autonomic, head, cervical, mri, emg, headache, neurology, motor, brain, nerve, bilaterally, bp, migraine, headaches, nl | |
| Epic | midline, botox, flexion, muscle, symmetric, motor, mr, neu, tremor, touch, finger, gait, units, neurology, brain, facial, bilaterally, headache |
+In descending order of stf-idf score
Discussion
This study focused on assessing the degree of heterogeneity in clinical documentation. Toward that end, we sought to measure the variations brought on by the practice context (i.e., EHR), temporal context (i.e., time), and spatial context (i.e., regional characteristics and demographics of the site) by controlling as much as possible for the physician. We sampled a set of attending physicians at each of the Mayo Clinic Arizona and Florida sites who practiced for a full year prior to the start and after the completion of the transition from Cerner to Epic EHR systems and analyzed the lexical, semantic, and process variations of each associated clinical note corpus against each other using a combination of statistical and visual techniques. The results suggest that process variations are significant over different EHR systems and different subspecialties while matching as much as possible for physician. Moreover, the results also demonstrate a significant variation of semantics between subspecialties using the same EHR system at the same site.
Process Variation
The corpus statistics revealed a stark contrast in the amount of note types used by Cerner and Epic EHRs. While there is a direct mapping of many of the note types (e.g. Cerner “<Specialty> Subsequent Visit” maps to Epic “Progress Notes” and Cerner “<Specialty> Consult” maps to Epic’s “Consult – Outpatient”), Cerner contained an order of magnitude more note types as they were defined at the specialty level as well as for the type of operation being performed (e.g. “Colonoscopy” in Cerner as opposed to generic “Procedure” in Epic). Whereas almost all Epic notes conformed to one of a handful of pre-selected categories, Cerner allows providers to define new note types. Further, short communications between current providers and previous or referring providers often labelled as a short Miscellaneous Note instead of appending to Progress Notes. For Epic, these differences resulted in fewer notes that are shorter in length, set across a narrower distribution of note types. While the reduction in note types to consider has been shown in some cases to ease physician workflow and increase inter-provider communication of patient information due, in part, to the increased utilization of template structures25,26, this appears to come with the tradeoff of less contextual information conveyed in the note type name. For example, the type of procedure for which a note is written is not indicated by the note type “Procedure” in Epic but is readily apparent in “EGD” and “Colonoscopy” in Cerner. Thus, an interoperable system attempting to extract this information could establish which procedure was performed from the note type in a Cerner EHR but must rely on another source for Epic EHR, such as header metadata, unless the implementation in Epic is specifically tailored to include that level of information in the note types.
Semantic Variation
Although the overall word frequencies per UMLS semantic group concept and their associated Wilcoxon signed- rank test p-values listed in Table 3 exhibit statistically significant differences between EHRs at each site for most groups, the illustrated differences in Figure 4 at the specialty level reveal that these variations are attributed mostly to a subset, if not individual group. For example, the differences between the rates of DISO concepts per word between EHRs for both sites for Gastroenterology and Hepatology as well as Emergency Medicine were much greater than the differences in DISO concepts per word for Internal Medicine and Neurology. The overall statistically significant increase in semantic density (i.e. more UMLS semantic concepts per word for almost every semantic category) provides evidence that the Epic EHR system allowed communication of more clinically-relevant terms in the note text. However, it is unclear from our data whether that was due to an increase in redundant data, as duplicated information in clinical notes has been extensively documented27-30.
Limitations
One limitation of this work was the absence of diagnosis matching through ICD-code stratification, for instance, to account for differences in clinical cases between the various contexts examined in this study. Some of the observed differences in process and semantics, especially, could be attributed to geospatial or temporal shifts in the prevalence of disorders. Such differences could be confounding in our analyses and therefore should be controlled for as much as possible in similar future work.
Although we were able to compare medical specialty corpora across sites and EHRs, we acknowledge that this ignores the possible existence of more granular subspecialty sublanguages. For instance, within Gastroenterology and Hepatology, physicians subspecialize in physiological locales such as particular organs (e.g. liver, esophagus, small and large bowel) and often further by procedure and condition, such as Barrett’s Esophagus and eosinophilic esophagitis (also known as EOE) for esophageal subspecialists. Since this data on physician expertise within their specialty was not available and the stf-idf approach did not weigh terms at the subspecialty level but may influence which patient cases those providers are directed toward, we were not able to consider this in our study.
While it would be informative to consider each contextual dimension in isolation, the transition of EHR systems over the course of a year by the same physicians, as opposed to using both EHRs concurrently, prevents us from differentiating clinical and temporal contexts for the same set of physicians. A future study could hypothetically normalize clinical context changes to longitudinal changes in sublanguage trends, inspired by the previous trend analysis conducted by Shao et al.12, for instance, to sequester a more precise clinical context change measurement.
Conclusion
This study aimed to assess the effect of practical and temporal contexts of clinical practice settings by comparing the notes of a set of physicians at two Mayo Clinic sites across an EHR migration from Cerner to Epic. Additionally, we assessed the effect of the spatial context by comparing physicians within the same specialty and who utilized the same integrated EHR system (Epic) across the sites. We evaluated the lexical and semantic variations between these corpora by evaluating the word token, document, and concepts distributions to measure whether a statistically significant difference occurred between the timeframes and across the EHRs. We found a significant degree of variation in process and syntax between EHRs as well as semantic variation between EHRs and specialties at the same Mayo Clinic site. That physicians within the same speciality tend to converge onto a similar sublanguage when using the same EHR system is also worthy of remark. These findings have implications on future efforts toward building portable clinical language models.
Acknowledgements
This work was supported by the National Institutes of Health grant U01TR002062 and R01LM011934.
Figures & Tables
References
- 1.Buntin MB, Burke MF, Hoaglin MC, Blumenthal D. The benefits of health information technology: a review of the recent literature shows predominantly positive results. Health Aff (Millwood) 2011;30(3):464–71. doi: 10.1377/hlthaff.2011.0178. [DOI] [PubMed] [Google Scholar]
- 2.Adler-Milstein J, Jha AK. HITECH Act Drove Large Gains In Hospital Electronic Health Record Adoption. Health Affairs. 2017;36(8):1416–22. doi: 10.1377/hlthaff.2016.1651. [DOI] [PubMed] [Google Scholar]
- 3.Mennemeyer ST, Menachemi N, Rahurkar S, Ford EW. Impact of the HITECH Act on physicians’ adoption of electronic health records. J Am Med Inform Assoc. 2016;23(2):375–9. doi: 10.1093/jamia/ocv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gold M, Mc LC. Assessing HITECH Implementation and Lessons: 5 Years Later. Milbank Q. 2016;94(3):654–87. doi: 10.1111/1468-0009.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards ST, Neri PM, Volk LA, Schiff GD, Bates DW. Association of note quality and quality of care: a cross-sectional study. BMJ Quality & Safety. 2014;23(5):406–13. doi: 10.1136/bmjqs-2013-002194. [DOI] [PubMed] [Google Scholar]
- 6.Cohen GR, Friedman CP, Ryan AM, Richardson CR, Adler-Milstein J. Variation in Physicians’ Electronic Health Record Documentation and Potential Patient Harm from That Variation. Journal of General Internal Medicine. 2019;34(11):2355–67. doi: 10.1007/s11606-019-05025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan JW, Prasad R, Yabut RM, Loomis RM, Zisook DS, Mattison JE, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annu Symp Proc. 2011;2011:382–91. [PMC free article] [PubMed] [Google Scholar]
- 8.Glynn EF, Hoffman MA. Heterogeneity introduced by EHR system implementation in a de-identified data resource from 100 non-affiliated organizations. JAMIA Open. 2019;2(4):554–61. doi: 10.1093/jamiaopen/ooz035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu S, Wen A, Schaeferle GM, Wilson PM, Demuth G, Ruan X, et al. Assessment of Data Quality Variability across Two EHR Systems through a Case Study of Post-Surgical Complications. AMIA Annu Symp Proc. 2022;2022:196–205. [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson O, Hurdle JF. Document clustering of clinical narratives: a systematic study of clinical sublanguages. AMIA Annu Symp Proc. 2011;2011:1099–107. [PMC free article] [PubMed] [Google Scholar]
- 11.Doing-Harris K, Patterson O, Igo S, Hurdle J. Document Sublanguage Clustering to Detect Medical Specialty in Cross-institutional Clinical Texts. Proc ACM Int Workshop Data Text Min Biomed Inform. 2013;2013:9–12. doi: 10.1145/2512089.2512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao Y, Divita G, Workman TE, Redd D, Garvin J, Zeng-Treitler Q. Clinical Sublanguage Trend and Usage Analysis from a Large Clinical Corpus2020. pp. 3837–45 p.
- 13.Peterson KJ, Liu H. An Examination of the Statistical Laws of Semantic Change in Clinical Notes. AMIA Jt Summits Transl Sci Proc. 2021;2021:515–24. [PMC free article] [PubMed] [Google Scholar]
- 14.Moon S, He H, Liu H, editors. Sublanguage Characteristics of Clinical Documents. 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2022 6-8 Dec;2022 [Google Scholar]
- 15.Elkin PL, Froehling D, Bauer BA, Wahner-Roedler D, Rosenbloom ST, Bailey K, et al. Aequus communis sententia: defining levels of interoperability. Stud Health Technol Inform. 2007;129(Pt 1):725–9. [PubMed] [Google Scholar]
- 16.Sohn S, Wang Y, Wi C-I, Krusemark EA, Ryu E, Ali MH, et al. Clinical documentation variations and NLP system portability: a case study in asthma birth cohorts across institutions. Journal of the American Medical Informatics Association. 2017;25(3):353–9. doi: 10.1093/jamia/ocx138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon S, Liu S, Chen D, Wang Y, Wood DL, Chaudhry R, et al. Salience of Medical Concepts of Inside Clinical Texts and Outside Medical Records for Referred Cardiovascular Patients. J Healthc Inform Res. 2019;3(2):200–19. doi: 10.1007/s41666-019-00044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12(null):2825–30. [Google Scholar]
- 19.Eyre H, Chapman AB, Peterson KS, Shi J, Alba PR, Jones MM, et al. Launching into clinical space with medspaCy: a new clinical text processing toolkit in Python. AMIA Annu Symp Proc. 2021;2021:438–47. [PMC free article] [PubMed] [Google Scholar]
- 20.Soldaini L, editor. QuickUMLS: a Fast, Unsupervised Approach for Medical Concept Extraction. 2016.
- 21.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform. 2009;42(5):839–51. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, et al. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl Sci Proc. 2013;2013:149–53. [PMC free article] [PubMed] [Google Scholar]
- 23.Fu S. TRUST: clinical text retrieval and use towards scientific rigor and transparent process. University of Minnesota Digital Conservancy: University of Minnesota. 2021.
- 24.Grootendorst M. BERTopic: Neural topic modeling with a class-based TF-IDF procedure. arXiv preprint arXiv:220305794. 2022.
- 25.Koppel R, Lehmann CU. Implications of an emerging EHR monoculture for hospitals and healthcare systems. J Am Med Inform Assoc. 2015;22(2):465–71. doi: 10.1136/amiajnl-2014-003023. [DOI] [PubMed] [Google Scholar]
- 26.Kahn D, Stewart E, Duncan M, Lee E, Simon W, Lee C, et al. A Prescription for Note Bloat: An Effective Progress Note Template. Journal of Hospital Medicine. 2018;13(6):378–82. doi: 10.12788/jhm.2898. [DOI] [PubMed] [Google Scholar]
- 27.Cohen R, Elhadad M, Elhadad N. Redundancy in electronic health record corpora: analysis, impact on text mining performance and mitigation strategies. BMC Bioinformatics. 2013;14:10. doi: 10.1186/1471-2105-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, Pakhomov S, Melton GB. Longitudinal analysis of new information types in clinical notes. AMIA Jt Summits Transl Sci Proc. 2014;2014:232–7. [PMC free article] [PubMed] [Google Scholar]
- 29.Rule A, Bedrick S, Chiang MF, Hribar MR. Length and Redundancy of Outpatient Progress Notes Across a Decade at an Academic Medical Center. JAMA Network Open. 2021;4(7):e2115334–e. doi: 10.1001/jamanetworkopen.2021.15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinkamp J, Kantrowitz JJ, Airan-Javia S. Prevalence and Sources of Duplicate Information in the Electronic Medical Record. JAMA Network Open. 2022;5(9):e2233348–e. doi: 10.1001/jamanetworkopen.2022.33348. [DOI] [PMC free article] [PubMed] [Google Scholar]