Abstract
Enhancing diversity and inclusion in clinical trial recruitment, especially for historically marginalized populations including Black, Indigenous, and People of Color individuals, is essential. This practice ensures that generalizable trial results are achieved to deliver safe, effective, and equitable health and healthcare. However, recruitment is limited by two inextricably linked barriers – the inability to recruit and retain enough trial participants, and the lack of diversity amongst trial populations whereby racial and ethnic groups are underrepresented when compared to national composition. To overcome these barriers, this study describes and evaluates a framework that combines 1) probabilistic and machine learning models to accurately impute missing race and ethnicity fields in real-world data including medical and pharmacy claims for the identification of eligible trial participants, 2) randomized controlled trial experimentation to deliver an optimal patient outreach strategy, and 3) stratified sampling techniques to effectively balance cohorts to continuously improve engagement and recruitment metrics.
Introduction
Clinical trial recruitment is limited by two inextricably linked barriers – the inability to recruit and retain enough trial participants, and the lack of diversity amongst trial populations whereby racial and ethnic groups are underrepresented when compared to national composition. Approximately 80% of clinical trials miss their enrollment timelines[1] and 55% of trials are terminated due to low accrual [2]. Moreover, despite Black, Indigenous, and People of Color (BIPOC) groups representing 42% of the United States (US) population [3], racial and ethnic diversity is limited. Of trials reporting ethnicity only 11% of participants were reported as Hispanic or Latino [4]. Further, of those reporting race, less than 1% were Native American, 6% were Asian, and 11% were Black [4]. The need for trial diversity extends beyond race and ethnicity (R/E), as many vulnerable groups including pregnant people and those with chronic conditions are often excluded in study designs and individuals outside of urban settings are grossly underrepresented. Inclusive trial design may increase heterogeneity of trial population and the generalizability of its results.
Inclusion practices to bolster trial diversity have been longstanding, but those efforts have done little to make substantial change. As far back as 1994, the National Institutes of Health (NIH) mandated the inclusion of women and other underrepresented populations in all NIH-sponsored clinical trials [5]. Over the subsequent decades, and continuing today, various stakeholders including the US Food and Drug Administration (FDA), sponsors, providers, academic centers, and patient advocacy groups have individually, and collaboratively, sought to address this disparity [6, 7]; however, review of clinical trial enrollment data has indicated little improvement in increasing diversity amongst clinical trial participants. A 2022 consensus study report by the National Academy of Sciences and the NIH found that, despite the priority of increasing diversity in clinical trials, the majority of participants continue to be White men [8].
Numerous initiatives have attempted to create infrastructures to enhance clinical trial diversity and the COVID-19 pandemic accelerated the digital transformation of clinical trial operations. Notably, the steadily decreasing prevalence of low accrual rates for trial termination is the result of informatics-enabled solutions to improve patient recruitment by contract research organizations [9]. While the expansion of decentralized trials has potential to begin to address access and recruitment, inclusive trial populations remain a significant barrier in their execution.
In addition to depriving historically marginalized participants of access to potentially lifesaving clinical research opportunities, the disparity of trial access also limits the generalizability of new drugs to the diverse and broad communities to which they will ultimately be prescribed. This disparity is particularly relevant in diseases which disproportionally impact BIPOC populations [10]. For example, cardiovascular disease disproportionally impacts Black adults, yet they remain under-represented in clinical trials designed to improve its prevention, diagnosis, and treatment. Systematic reviews demonstrated the proportion of non-White participants in cardiovascular-related clinical trials has not changed over several decades [11].
Recently, real-world data (RWD) and advanced analytics have shown great promise to inform clinical trial strategy, design, and execution [12, 13]. The COVID-19 pandemic accelerated the digital transformation of clinical research including the use of RWD sources such as electronic health data, patient-generated data, and claims databases. Additionally, RWD collaboration expanded more broadly to include insurers, retail, and pharmacies. The application of RWD can deliver innovative trial designs to address obstacles in clinical evaluations, including enrolling sufficient, and diverse, populations to improve trial inclusion [14]. Moreover, concomitant disruption and innovation associated with the application of advanced analytics including machine learning (ML) has been unprecedented, particularly in clinical research and trials, bringing more data-informed decision-making to its stakeholders.
Ensuring diversity, including racial and ethnic, representation is met in clinical trials is essential to advance science while concomitantly reducing bias, promoting social justice, and improving health equity. However, it’s estimated that only 43% of US trials reported R/E data [15]. This disparity of under enrolling racial and ethnic groups exacerbates their health inequities and creates bias in trial results. Moreover, structured health care data are plagued with missing, unreliable, or incomplete information including demographic fields such as individual designations for R/E. Despite temporal initiatives to expand the inclusion of BIPOC in trial enrollment and positive trends reporting R/E trial data following numerous mandates [16], their gaps in diversity and data remain.
Typically, people who do not volunteer identification data are historically marginalized, including BIPOC groups, and underrepresented in clinical trials [17]. Precedents and numerous negative experiences with the healthcare delivery system are correlated with mistrust, systemic racism, and stigma among these communities. Identity, including the consideration of one’s R/E, is a large ethical consideration and any attempt to mathematically impute, or infer, these fields to complete patient health information should be done with great scrutiny.
Imputing fields is long standing practice in clinical trials to correct for bias associated with missing data [18], but despite its limitations [19], it offers health equity many opportunities. With substantial evidence demonstrating health and healthcare disparities [20] are highly prevalent in BIPOC communities, the lack of R/E in trial data exacerbate those long–standing differences and promote further inequities. To overcome cyclical BIPOC health disparities, R/E data imputation can improve the trial design including patient outreach and enrollment strategies. Downstream impacts of these imputation provide the potential to investigate and address disparities in access to, utilization of, and outcomes of care.
This purpose of this study is to describe a framework and evaluate informatics techniques that aim to enhance equity in trial populations using enrollment and claims data to 1) operationalize Bayesian predictive and ML methods to impute missing R/E data and 2) dynamically adjust the outreach strategy based on patient engagement. The recruitment optimization framework utilizes a R/E imputation pipeline along with RCT experimentation and its associated patient cohort balancing techniques using stratified sampling. The real-world application and impact of this recruitment optimization framework and its model performance will be evaluated.
Methods
Framework Description of Techniques to Enhance Clinical Trial Equity
To overcome race-blindness within the data and the trial target population, the utilization of various probabilistic models, including Bayesian Improved Surname and Geocoding (BISG) [21] and Bayesian Improved First Name and Surname Geocoding (BIFSG) [22] methodologies (Figure 1) were applied to impute missing R/E within medical and pharmacy claims data. Both BISG and BIFSG are well-validated [21, 23-26] and widely used [27] R/E estimation methods to make inferences regarding missing demographic information, including R/E groups, based on proxy variables, such as their first name, surname, and address. First/surname analysis using US census data provides common names for racial and ethnic groups. When this information is combined with geocoding at the block group level using Bayesian estimation, R/E membership can be categorically classified. The BIFSG model is a direct expansion of BISG model that simultaneously includes first names to improve model robustness.
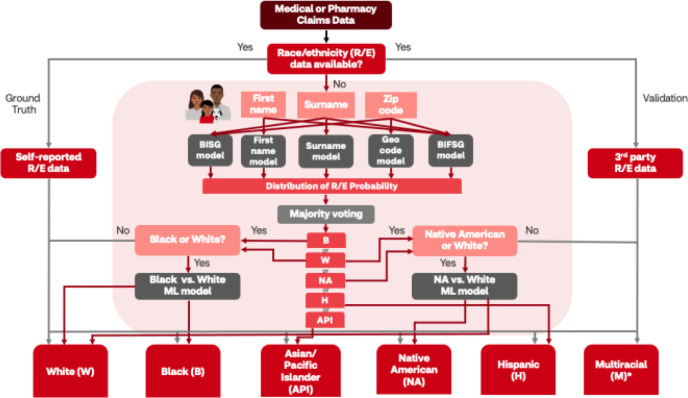
Figure 1.
Race and ethnicity (R/E) imputation pipeline. This decision tree illustrates the pathway by which three R/E data assets were utilized in this study: 1) self-reported R/E served as the ground truth for machine learning (ML) model development; 2) third-party R/E were used for model validation; and 3) any missing R/E data from medical and pharmacy retail data were imputed using a combination of BISG modified modeling (e.g., first name, surname, geocode, BIFSG derivations) that yielded a distribution of probabilities for R/E classification. Naïve model output was further post-processed using majority voting to select the primary R/E imputation. Additional post-processing was performed when White (W), Black (B), or Native American (NA) race were predicted. These classifications were further processed using race-specific ML models to improve the accuracy of the R/E imputation. No post-processing was performed on Hispanic (H) and Asian/Pacific Islander (API) classifications. Key: red arrows indicate R/E imputation and grey arrows indicate ground truth or validated R/E pathways. Abbreviations: BISG, Bayesian Improved Surname and Geocoding; BIFSG, Bayesian Improved First Name and Surname Geocoding
Race and Ethnicity Imputation Pipeline. When R/E data are unavailable, BISG/BIFSG imputation of the missing fields have demonstrated to reliably predict categories of explicit racial and ethnic membership including White, Black, Hispanic, and Asian individuals among medical health plan data [28]. However, identification of American Indian/Alaskan Native and multi-racial individuals was reported to be poor using this methodology [28]. Meanwhile, since BISG/BIFSG simultaneously requires surname (and first name for BIFSG) and geographical location to do the prediction, the algorithm will not work on members with missing name or geographical information.
In view of this, additional probabilistic models relying on only one of these fields (i.e., first name, surname, or geography only) were also used. To adequately capture a sample reflective of the target population, further post-processing occurred for the naïve algorithm to finalize race prediction and distinguish ethnicity from race using simple majority voting on the predictions of all the probabilistic models. Additionally, to address limitations of current model and boost predictive performance, two proprietary machine learning models were also employed to impute Black and Native American populations more accurately. Finally, to operationalize (Figure 1) Bayesian predictive and machine learning models to impute missing R/E from claims data, validation, including the use of self-reported and third-party data, were applied.
Randomized Controlled Trial (RCT) Experimentation. Data gaps and poor patient recruitment can potentially put clinical trials at risk; however, informatics approaches can be coupled with further experimentation to improve recruitment planning and its execution. Experimental design plays a critical role in recruitment enhancement; more specifically, campaign design, which includes the methods to identify, recruit, and retain eligible participants, is tailored to fit trial needs. Patient recruitment approaches can include campaigns to identify patients from imputation-adjusted medical or pharmacy claims data, retail visits, patient databases, community events, or existing partnerships.
To overcome recruitment challenges, a modularized and scalable randomized controlled experimentation design can enable clinical trial recruitment at scale. The methodology can be used to support experimental design, establish causality with high level of evidence, and conduct analysis to make data-informed decisions. Moreover, applying this methodology to recruitment interventions can enhance outreach strategies.
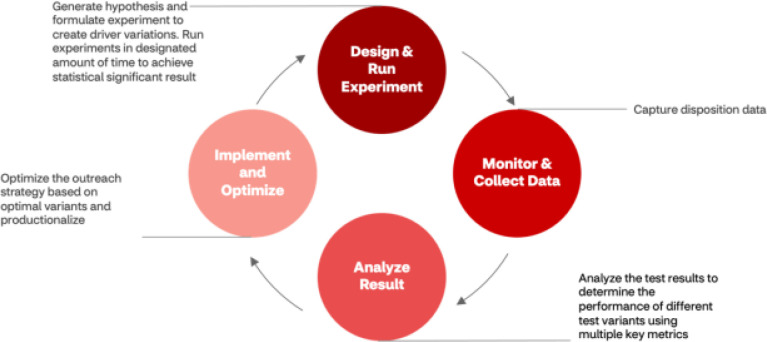
The goal of experimentation was to evaluate the impact of key campaign dimensions to operationalize outreach enhancements for recruitment improvement. The experimentation framework (Figure 2) included four components: 1) hypothesis generation and creation of driver variations as well as methodologies for patient eligibility and allocation, recruitment goal metrics, and statistical significance boundaries were established to plan and conduct experiment; 2) monitor and collect data; 3) analyze outreach drivers, whereby the intervention group was compared to control to test the key metrics (e.g., open rates, referrals, pre-screening, consents, randomization] of the outreach campaign for a specific period of time to achieve statistical significance; 4) implementation of levers demonstrating success into the production system and use data-informed decision-making to optimize outreach strategy for current and future states.
Figure 2.
Modularized and scalable RCT experimentation. This cyclical experimentation serves to inform current and future outreach strategy because of a priori hypothesis generation to execute quality improvement RCTs to evaluate the effectiveness of outreach strategies that serve to inform future decision-making.
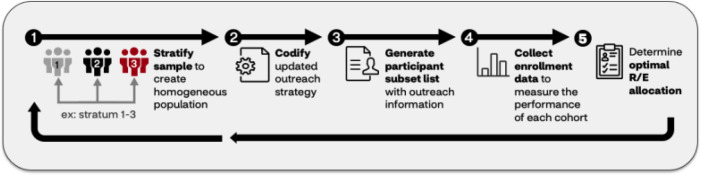
Patient Cohort Balancing Module Within RCT Experimentation. The opportunity to provide population-level estimates of clinical trial results can positively impact trial design when additional cohort balancing techniques were also applied. To consider the sample population representative to the target population, stratified sampling [29] was applied to improve the inclusion of trial recruitment design by balancing the patient cohort demographic (e.g., R/E) composition (Figure 3). To fill these needs, imputed R/E into claims data were leveraged to identify eligible trial participants. The population identified from trial enrollment data were divided into homogeneous strata according to multiple demographic factors (e.g., R/E, age, and gender), and a specific number of participants were chosen at random from each stratum. Upon applying stratified sampling to balance the outreach distribution of under-engaged patients, the strategy for enrollment outreach was dynamically adjusted. This process undergoes a feedback loop of iteration, measurement, and refinement until the trial engagement, enrollment distribution targets, and endpoints were satisfactorily met.
Figure 3.
An iterative loop to balance a patient cohort within experimentation. This feedback loop using stratified sampling, whereby enrollment data was split into homogeneous strata according to multiple demographic factors (e.g., race/ethnicity (R/E), age, and gender). A specific number of participants were chosen at random from each stratum and codified to generate a participant list. Next, enrollment data were collected to examine cohort performance, and if necessary, outreach was continued to meet desired R/E allocation.
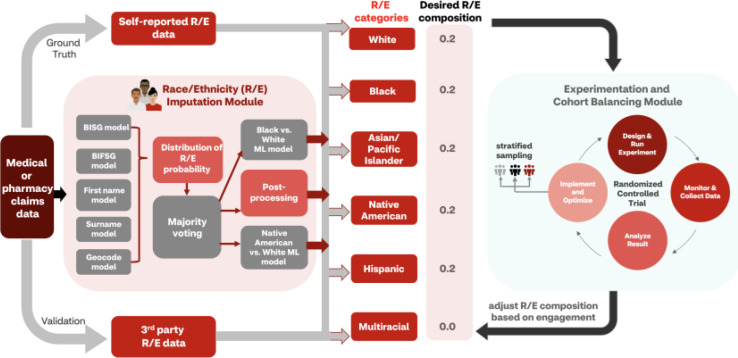
To overcome the limitations of clinical trials recruitment regarding R/E data gaps and race-related inequities, these modules (Figure 4), the R/E imputation pipeline along with RCT experimentation and its associated patient cohort balancing techniques using stratified sampling, were utilized.
Figure 4.
Schematic diagram of recruitment optimization framework. The R/E imputation module was applied to enhance cohort balancing techniques that utilized RCT experimentation for outreach strategy optimization with stratified sampling. Self-reported, third-party, or imputed race/ethnicity data were compared with trial-specific desired racial/demographic composition and incorporated into experimental design. Once enrollment data were collected, the composition was adjusted based on participant engagement.
Machine Learning Models to Improve Predictions of Black and Native American Groups
Model Features. Collected data were used for pre-modeling Exploratory Data Analysis to generate aggregate summary statistics and derive predictive model features (i.e., feature engineering). Features included binary, categorical, and continuous types, some of which were constructed to capture the dimension of time. Name embeddings, census data on zip code level including social determinants of health inferences, prescription fill/outreach patterns from pharmacy claims, previously imputed R/E with probabilistic models, and additional R/E information from third-party vendor were used as potential variables. As an example, total number of successful outreaches for retail pharmacy programs can be a variable to indicate member habit/patterns on effectiveness of previous outreach. Recursive feature elimination with 5-fold cross validation were used to select the most relevant features and reduce overfitting.
Model Training and Validation. After EDA, feature engineering, and feature selection, collected data were used to fit a binary classification extreme gradient boost (XGBoost) model. In view of the high misclassification rates using the BISG/BIFSG probabilistic models obtained from preliminary results, the two racial classes were Black or Native American versus (vs.) White. Two models were trained, one for Black vs. White and the other for Native American vs. White. The training and validation data for the first model were constructed using a 35% random sample of Blacks correctly classified with probabilistic model, a 25% sample of Black race misclassified to White previously, and a 10% sample of total White population to make the final Black:White = 1:2. The data were randomly partitioned into training and hold-out test sets with a ratio of 9:1, preserving the Black to White ratio. Similarly, a 5% sample of White race plus 50% of available Native American race were used to construct the training and validation set for the second model. Tenfold cross validation on the training set were employed for hyperparameter tuning and to optimize algorithm performance.
To incorporate the first name and surname information into the ML modeling and overcome the issue of high cardinality, a Name2Vec algorithm was employed to obtain distributed representations of first name and surname of an individual [30]. Name2Vec is a special implementation of Doc2Vec algorithm onto personal names, which effectively converts a document into vector embeddings by considering the internal structure and how topics are formed within the document [31]. Here, each name was considered a document while each letter in a name was equivalent to one word. Two separate Name2Vec models for first names and surnames were pretrained, using all first names and surnames within a proprietary database plus 730k first names and 983k surnames found publicly online [32], which contributed to the dataset containing ∼5.2M first names and ∼6.4M surnames in total. Both Doc2Vec models were pretrained with vector size = 30 and window = 3 for 50 epochs. After pretraining, 30-dimensional embeddings for all names were extracted in the database and stored for use in ML models as features.
Model Evaluation. Various algorithm performance metrics were considered (e.g., Precision, Recall, AUC/ROC, etc.) and examined on the hold-out test set.
Real-World Evaluation using the Recruitment Optimization Framework Study Design and its Data Analyses
Member month-level retrospective medical (e.g., commercial fully-insured, Medicare, and Medicaid) claims (N=6,348,500), pharmacy claims (N=127,407,048), personal (e.g., first name, surname), and demographic data (e.g., R/E, zip code) were used in this study. Member names and/or geographical location data were used as inputs to five probabilistic/Bayesian statistical models to obtain probabilities of an individual member falling into one of the six racial categories: White, Black, Asian American and Pacific Islander, Native American, Hispanic, or Multiracial. Next, all predicted probabilities were passed into the postprocessing module, which performed soft or hard majority voting to assign the most probable race to the member. At this step, necessary data normalizations and wrangling were performed to ensure the R/E imputed and its format were consistent with those in the dataset. This probabilistic pipeline was deployed to score every member in the database.
In view of the limitations of the probabilistic models, additional proprietary/in-house machine ML models were trained specifically to better identify Black and Native American populations with enhanced accuracy. The trained models were applied on members previously imputed as Black, Native American, or White receive a refined prediction/secondary confirmation upon final R/E assignment.
The two-step R/E prediction pipeline (probabilistic, and then ML) was validated using self-reported demographic data as the ground truth; inferential statistics were applied to larger claims set for testing and the model was validated using a subset of third-party claims data with self-reported R/E. In addition, to validate the predictions in real-world setting, A/B test/measurements were performed on collected participant response data between treatment (i.e., predicted R/E group) and control (i.e., self-reported R/E group) using the RCT experimentation framework. No difference observed between intervention and control groups indicated the success of using imputed R/E in actual recruitments.
To evaluate the performance of the model, the imputed R/E were incorporated to identify and outreach eligible patients in a clinical trial assessing the prevalence of valvular heart disease in older Americans. This study includes patients ≥ 65 years old and excludes patients with history of congenital heart disease. RCT experimentation was conducted to perform the stratified sampling method to meet the recruitment goal. Participants were divided into homogeneous strata based on R/E, age, and gender. Each stratum was randomly sampled and assigned to one unique vanity URL and one unique code that identified the demographic information of the group. After the recruitment campaign launch, the performance of each R/E subgroup engagement was examined by vanity URL clickthrough rate. These clickthrough rates across each R/E subgroup were compared to understand if stratified sampling improved engagement for cohorts with racial underrepresentation. Average R/E composition (i.e., percent improvement in coverage of cohorts) between stratified and simple random sampling was examined.
Results
Ensemble Probabilistic Imputation Module Accurately Fills in Missing R/E Information for Diversity Recruitment
To examine whether an ensemble of five proxy-based probabilistic models accurately fills in the missing R/E information, the model predictions were validated on internal self-reported R/E data from pharmacy claims data. Table 1 presents the performance of the predictive approach with an overall accuracy of 0.852. Predictions of White, Asian/Pacific Islander, and Hispanic race yielded an overall F1 score ≥ 0.8. However, Black, Native American, and Multiracial race predictions had a wide range of lower F1 scores (0.002 - 0.533). Both Black and Native American race predictions had higher precision (0.739 and 0.545, respectively), but both had low recall (0.417 and 0.013, respectively). Multiracial predictions showed unsatisfactory performance for both precision and recall (0.066 and 0.001, respectively).
Table 1.
Performance of ensemble method on imputing race/ethnicity for all pharmacy members with self-reported race/ethnicity data
| R/E Classification | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Overall | 0.852 | 0.936 | 0.695 | 0.798 |
| White | - | 0.814 | 0.966 | 0.884 |
| Black | - | 0.739 | 0.417 | 0.533 |
| Asian and Pacific Islander | - | 0.884 | 0.714 | 0.790 |
| Native American | - | 0.545 | 0.013 | 0.025 |
| Hispanic | - | 0.886 | 0.786 | 0.833 |
| Multiracial | - | 0.066 | 0.001 | 0.002 |
The predictive approach was further benchmarked using third-party R/E data for members with pharmacy claims, using the overlap population where both imputed values and third-party/external data were available. Table 2 shows a comparison of precision, recall, and F1 score by race. Since the Multiracial group was not available in the third-party data, it was excluded from the benchmark analysis to focus on the other five major racial categories. Overall, the R/E imputation approach achieves highly comparable, if not occasionally better, performance with RWD benchmarking.
Table 2.
Benchmark with third-party race/ethnicity data using overlap of pharmacy members
| R/E Classification | Precision imputed | Precision benchmarked | Recall imputed | Recall benchmarked | F1 imputed | F1 benchmarked |
|---|---|---|---|---|---|---|
| White | 0.816 | 0.871 | 0.967 | 0.915 | 0.885 | 0.892 |
| Black | 0.746 | 0.628 | 0.423 | 0.629 | 0.540 | 0.629 |
| Asian and Pacific Islander | 0.886 | 0.842 | 0.724 | 0.839 | 0.796 | 0.840 |
| Native American | 0.553 | 0.134 | 0.013 | 0.022 | 0.026 | 0.037 |
| Hispanic | 0.893 | 0.859 | 0.790 | 0.843 | 0.838 | 0.851 |
To validate the performance of this predictive approach in a real-world setting, a RCT was conducted to collect member response data in a live email campaign. The control group was derived from the self-reported R/E population while the intervention group was composed entirely from an imputed R/E population. The ratio of intervention:control was roughly 1:1. Table 3 shows recruitment engagement metrics observed during an email outreach campaign between the randomized intervention and control groups. There are no statistically significant differences between the groups for both the email open rate and clickthrough rate (Chi-squared, P=0.5575 and P=0.2408, respectively). This shows the overall success of leveraging imputed R/E information in real-world participant outreach when ground truth self-reported data is absent.
Table 3.
Email campaign statistics during a randomized controlled trial to validate the quality of imputed race/ethnicity (R/E)
| Metric | Control (Self-reported R/E) |
Intervention (Imputed R/E) |
P-value |
|---|---|---|---|
| Number emailed | 22,795 | 20,303 | - |
| Number emails opened (%) | 8,680 (38.08%) | 7,816 (38.5%) | 0.5575 |
| Number of clickthroughs (%) | 119 (1.37%) | 90 (1.15%) | 0.2408 |
Machine Learning Models Successfully Predict Minority Assignment for Targeted Races
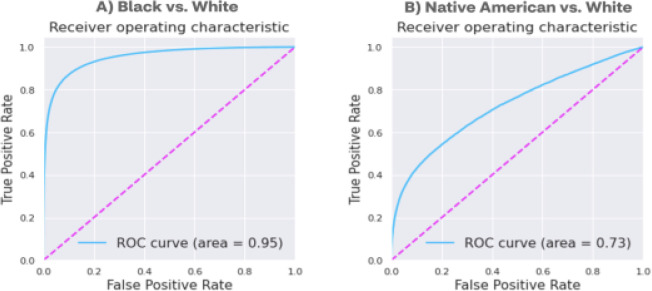
Upon further analysis, out of the total false negatives, 95.3% African Americans and 84.3% Native Americans were misclassified as Caucasians, which corresponded to the low precision of Caucasian prediction (0.814) relative to recall (0.966) (Table 1). In view of the relatively low predictive performance for Black and Native American racial identification, and that majority (>90%) of the misclassifications happen between when distinguishing those races from White, two additional XGBoost [33] models were built. These models, that differentiate Black and Native American races from White, were developed to further refine the identification of additional races from the initially predicted population. Both models show improved performance compared to the probabilistic approach when validated on the 10% hold-out test set (Table 4, Figure 5). In particular, the Black race predictive model displays superior performance with all metrics above 0.8 and an AUC score of 0.95, highlighting the success of ML-based approach to capture the multidimensional pattern difference and complex feature interactions that successfully classify these two racial groups. In contrast, the performance of predicting Native American race is still moderately low.
Table 4.
Benchmark with third-party race/ethnicity data using overlap of pharmacy members
| Model | Accuracy | Negative predictive value | Precision | Recall | F1-score | AUC | AUPR |
|---|---|---|---|---|---|---|---|
| Black vs. White | 0.903 | 0.913 | 0.878 | 0.802 | 0.838 | 0.953 | 0.921 |
| Native American vs. White | 0.889 | 0.934 | 0.390 | 0.353 | 0.371 | 0.726 | 0.351 |
Figure 5.
Receiver operating characteristic (ROC) curves for (A) Black and (B) Native American versus (vs) White race prediction models
Abbreviations: AUC, area under the curve; AUPR, area under the precision-recall curve.
When integrating the ML models into the ensemble probabilistic R/E imputation module, two new models were employed as an additional layer on top of the original predictions for Black, Native American, and White members. Essentially, the old race predictions were overwritten by the new ones primarily for White classifications as an attempt to identify additional minority members and boost the recall rate. An overall improvement of accuracy from 0.852 to 0.870 and F1-score from 0.798 to 0.828 was observed. Notably, the recall for Black race prediction was improved by an absolute 20.6% (or equivalently ∼1.5x of the original recall rate) with even slightly increased precision rate; however, significant change was not observed for Native American race predication, indicating limited generalizability of the specific-Native American prediction model (Table 5).
Table 5.
Performance of ensemble method plus ML models on imputing race/ethnicity for all pharmacy members with self-reported race/ethnicity data
| R/E Categories | Accuracy | Precision | Recall | F1-score |
|---|---|---|---|---|
| Overall | 0.870 | 0.932 | 0.745 | 0.828 |
| White | - | 0.839 | 0.961 | 0.896 |
| Black | - | 0.750 | 0.623 | 0.681 |
| Asian and Pacific Islanders |
- | 0.884 | 0.714 | 0.790 |
| Native American | - | 0.537 | 0.013 | 0.026 |
| Hispanic | - | 0.886 | 0.786 | 0.833 |
| Multiracial | - | 0.066 | 0.001 | 0.002 |
Race/Ethnicity Imputation Pipeline is Essential to Achieve Diversity in Recruitment Goals
After validating model performance, R/E imputation in a real-world clinical trial recruitment setting was examined, when the coverage of ground truth R/E data did not meet the targeting requirement. In the RCT utilized for the email campaign mentioned above, all potential participants meeting the eligibility criteria in self-reported R/E population only met 62.6% of the recruitment goal. Upon introducing the R/E imputation pipeline approach, an additional 193K potential and R/E diverse participants were identified and enriched the sample, which was equivalent to 48.4% of the targeting goal (Table 6).
Table 6.
Number of available participants that met eligibility criteria with and without R/E imputation pipeline and impact on percent target recruitment goal achieved
| R/E imputation impact | Number of total eligible diversity participants outreached (N) | Participant target goal achieved (% of 400K) |
|---|---|---|
| Self-reported race only (Ground Truth) | 250,471 | 62.6% |
| Self-reported race plus imputation pipeline (Experimentation) | 443,901 | 111.0% |
| Difference | 193,430 | 48.4% |
Recruitment Outreach to Minority Populations is Significantly Improved with Patient Cohort Balancing
After validation that R/E imputation techniques were effective, R/E imputations were combined with patient cohort balancing techniques to improve recruitment diversity in a real-world clinical trial. Since the goal of patient cohort balancing module is to represent the diversity of the target population and to promote clinical trial accessibility to underrepresented participants, random sampling methods were assessed to identify if stratification improved participant engagement when compared to simple traditional methods. Patient eligibility was defined based on the study protocol, and a subset of patients were selected from the participant pool based on eligibility criteria.
In this use case, Black and Hispanic R/E were two desired groups in this study, which was informed by imputed R/E methods. To demonstrate the effectiveness of these equity-based recruitment techniques, patient engagement data were collected via vanity URL clickthrough rates. Each clickthrough indicates a successful incidence of connecting/redirecting patients to the clinical trial who otherwise would typically not have access. For the baseline, the assumption was that clickthrough rates by race would remain the same regardless of intervention; however, to determine the expected response distribution those rates were multiplied by the outreach distribution without cohort balancing and normalized a sum of one. Then, this expected distribution of participant response (i.e., given access/connected to the trial) without cohort balancing was benchmarked with observed participant distribution collected with cohort balancing. Table 7 shows cohort balancing improves trial access/response distribution that for Black and Hispanic populations (513% and 312%, respectively). This equity-based enrichment, using that R/E imputation when combined with cohort balancing techniques, successfully connected 5-fold more Black and 3-fold more Hispanic participants to this particular trial.
Table 7.
Number of eligible Black and Hispanic participants connected to trial before and after equity-based enrichment utilizing race and ethnicity imputation combined with cohort balancing techniques.
| Targeted race and ethnicity recruitment demographic | Expected number of eligible and responded participants | Actual number of eligible and responded participants | % Improvement |
|---|---|---|---|
| Black | 338 | 2079 | 513% |
| Hispanic | 593 | 2444 | 312% |
Discussion
This study sought to characterize the development, implementation, and outcomes of an informatics-enabled framework designed to enhance equity in clinical trial populations. The framework, when applied in a real-world trial setting, demonstratively enriched the target Hispanic and Black composition 3- to 5-fold. This enrichment was achieved with the utilization of existing probabilistic and novel ML models that accurately imputed missing R/E fields in RWD from pharmacy claims to identify eligible target trial participants and effective RCT experimentation with stratified sampling techniques that delivered 48% improvement in participant availability. Imputed R/E techniques are essential to ensure recruitment of sufficient diverse population into clinical studies. The advantage of stratified sampling over simple random sampling is that with this proportionate sampling, participants were selected from each stratum in proportions observed in the general population, which allowed increased representation by members of R/E groups who are typically underrepresented in trial design. These results demonstrate the effectiveness of our equity-centered approach to improve access to trials and are directly aligned with research-sustaining goals set forth by the NIH Minority Health and Health Disparities Strategic Plan to “increase the overall proportion of participants from diverse populations included in NIH-funded clinical research to 40% by 2030 and within specific major disease categories” [34]. As the composition of the population continues to become more racially and ethnically diverse, combinatorial informatics-enabled techniques such as those described herein will become increasingly vital for ensuring that clinical trials are representative of the population for which their evidence base directly informs clinical practice and population health at large.
The application of informatics to advance clinical trial operations, including recruitment, have great potential mixed with inherent limitations. For example, to overcome missing values in big data (e.g., medical and pharmacy claims), BISG/BIFSG algorithms are widely used to supplement data sources with reported R/E. Most health plans lack R/E data on most of their enrollees, so these indirect estimations at the group level is a valuable methodology to leverage. However, despite its accuracy to predict membership for the four largest R/E groups at the population-level, BISG/BIFSG estimates have low concordance with reported values for BIPOC and Multiracial populations.
The ensemble Bayesian models employed in this analysis missed ∼60% of true Black and <90% of true Native American members due to low precision (0.814) of White prediction compared to its recall (0.966). This insufficiency illustrates the limits of using census-level name and geographical probabilities to infer race for Black and Native American members. To address this, two novel XGBoost algorithms were built to overcome the limited predictability of Black, White, and Native American races and demonstrated superior performance compared to the Bayesian-only algorithm approach. However, the model improving the predictability of Black from White race was superior to the model attempting to impute Native American race. The Native American-specific imputation model still had very low recall despite its moderate precision. This could indicate that Native Americans within the trial recruitment data asset were like Whites with respect to features including demographic- and behavior-level components that fail to distinguish between the two of them. Further, upon validation with third-party R/E data, poor performance was similarly observed to predict Native American race. Alternative explanations for this poor performance for Native American race imputation may include inconsistent definitions and/or low quality associated with self-reported race data for Native American members. There is a critical need for accurate data collection in the measurement of small populations including American Indian/Alaska Native communities. Undercounts of this population may be attributed to weighting issues in the sampling approach for national surveys and pervasive and long-standing social injustices.
In an increasingly diverse multiracial and multiethnic world, representation is critical and there is a moral imperative to ensure the widespread use of algorithms and their datasets are inclusive. A limitation of this study is that Multiracial imputation improvement was not addressed and remains a significant gap to be fulfilled, as the self-reported Multiracial classification increased 276% since the last US Census report [3]. Multiracial identification is intrinsically difficult for algorithmic modeling, as each racial and/or ethnic component may have low performance due to data inequality and data distribution mismatches between each group. Predictive modeling is dependent upon high-quality data, so further challenges to predict identity attributed to multiple groups stem from inconsistent data definitions, limited data collection processes, and socioracial considerations. For example, the term “multiracial” is seldomly used as opposed to “other race” categories when acquiring demographic information. Further, socioracial asymmetries persist, whereby individuals of mixed races and/or ethnicities do not self-report due to experienced social justice issues including racism.
Despite these R/E imputation limitations for Native Americans and Multiracial members, there is promise in its implementation to deliver high-impact, equity-centered clinical trial recruitment at scale. Trials are limited by participation and data gaps; however, the use of imputed R/E data derived from RWD sources can seemingly fulfill those of these gaps. Diverse and inclusive trial recruitment has been extremely limited to deliver information and decision-making related to R/E. In other words, in the absence of having complete and accurate data, it’s impossible to know the extent of the inequities and how to address them. The implementation of these ensemble BIFSG/BISG models with novel post-processing models augment participant recruitment and subsequently support the identification of R/E-related disparities within the trial participants, and more broadly, at the population level. Moreover, these novel ML methods that refine R/E classification of missing data fields in RWD will likely reveal the magnitude of disparities inherent in research today and necessitate a multitude of follow up trials to replicate, reproduce, and advance health equity over time.
Logical next steps to advance the utility of this equity-centered framework include the development and execution of structured qualitative studies to better understand research participant experiences and perspectives. Outcomes of particular interest include research participant preferences, motivation, perception, value proposition, and perceived benefit for various sub-groups and clinical populations of interest. Research participant archetypes and personas can be used to test and learn tailored campaigns and person-centered messaging tactics that aim to optimize recruitment identification, retention, and outcomes whereby durability of engagement persists across R/E groups.
Conclusion
Informatics-driven solutions are a promising transformational tool for stakeholders to enhance equity in clinical trial populations. Large-scale datasets with robust RWD and probabilistic modeling and methodologies to impute R/E directly enable the experimentation framework required for data-informed decision-making and clinical trial recruitment strategy implementation. Ongoing studies aim to further refine the technical performance of the model, optimize participant engagement, and highlight practical applications of this framework.
Figures & Tables
References
- 1.Desai M. Recruitment and retention of participants in clinical studies: critical issues and challenges. Perspect Clin Res. 2020;11(2):51–53. doi: 10.4103/picr.PICR_6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trial termination analysis unveils a silver lining for patient recruitment. Verdict Media Limited. Accessed March 8, 2023. https://www.clinicaltrialsarena.com/features/clinical-trial-terminations/
- 3.Jones N, Marks R, Ramirez R, Rios-Vargas M. Improved race and ethnicity measures reveal US population is much more multiracial. US Census Bureau. Accessed March 8, 2023 https://www.census.gov/library/stories/2021/08/ improved-race-ethnicity-measures-reveal-united-states-population-much-more-multiracial.html.
- 4.Flores LE, Frontera WR, Adrasik MP, et al. Assessment of the inclusion of racial/ethnic minority, female, and older individuals in vaccine clinical trials. JAMA Network Open. 2021;4(2):e2037640–e2037640. doi: 10.1001/jamanetworkopen.2020.37640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inclusion of women and minorities as participants in research involving human subjects, US Dept of Health and Human Services, Editor. 2022, National Institutes of Health: Bethesda, MD
- 6.Corneli A, Hanlen-Rosado E, McKenna K, et al. Enhancing diversity and inclusion in clinical trials. Clinical Pharmacology & Therapeutics. 2023;113(3):489–499. doi: 10.1002/cpt.2819. [DOI] [PubMed] [Google Scholar]
- 7.Ramamoorthy A, Araojo R, Vasish KP, Flenkeng M, Green D, Madabushi R. Promoting clinical trial diversity: a highlight of select US FDA initiatives. Clinical Pharmacology & Therapeutics. 2023;113(3):528–535. doi: 10.1002/cpt.2822. [DOI] [PubMed] [Google Scholar]
- 8.National Academies of Sciences and and Medicine . Improving representation in clinical trials and research: building research equity for women and underrepresented groups. Washington, DC: The National Academies Press; 2022. p. 280. [PubMed] [Google Scholar]
- 9.Inan OT, Tenaerts P, Prindiville SA, et al. Digitizing clinical trials. npj Digital Medicine. 2020;3(1):101. doi: 10.1038/s41746-020-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasanna A, Miller HN, Wu Y, et al. Recruitment of black adults into cardiovascular disease trials. J Am Heart Assoc. 2021;10(17):e021108. doi: 10.1161/JAHA.121.021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilcant V, Ceron C, Verma G, Zeltser R, Makaryus AN. Inclusion of under-represented racial and ethnic groups in cardiovascular clinical trials. Heart Lung Circ, 2022. 31(9):1263–1268. doi: 10.1016/j.hlc.2022.06.668. [DOI] [PubMed] [Google Scholar]
- 12.Rogers JR, Lee J, Zhou Z, Cheung YK, Hripscask G, Weng C. Contemporary use of real-world data for clinical trial conduct in the US: a scoping review. J Am Med Inform Assoc, 2021. 28(1):144–154. doi: 10.1093/jamia/ocaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shortreed SM, Rutter CM, Cook AJ, Simon GE. Improving pragmatic clinical trial design using real-world data. Clin Trials, 2019. 16(3):273–282. doi: 10.1177/1740774519833679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Designing sound clinical trials that incorporate real-world data . US FDA: Silver Spring, MD; 2022. [Google Scholar]
- 15.Turner BE, Steinberg JR, Weeks BT, Rodriguez F, Cullen MR. Race/ethnicity reporting and representation in US clinical trials: a cohort study. Lancet Reg Health Am. 2022. p. 11. [DOI] [PMC free article] [PubMed]
- 16.Adashi EY, Cohen IG. The FDA initiative to assure racial and ethnic diversity in clinical trials. The Journal of the American Board of Family Medicine. 2023. p. jabfm.2022.220290R1. [DOI] [PubMed]
- 17.George S, Duran N, Norris K. A systematic review of barriers and facilitators to minority research participation among African Americans, Latinos, Asian Americans, and Pacific Islanders. Am J Public Health. 2014;104(2):e16–31. doi: 10.2105/AJPH.2013.301706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobsen JC, Gluud C, Wetterslev J, Winkel P. When and how should multiple imputation be used for handling missing data in randomised clinical trials - a practical guide with flowcharts. BMC Med Res Methodol, 2017. 17(1):162. doi: 10.1186/s12874-017-0442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterne JA, White IR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:b2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Academies of Sciences and Medicine . Communities in action: pathways to health equity, in the state of health disparities in the United States. Washington, DC: The National Academies Press; 2017. p. 2. [PubMed] [Google Scholar]
- 21.Elliott MN, Fremont A, Morrison PA, Pantoja P, Lurie N. A new method for estimating race/ethnicity and associated disparities where administrative records lack self-reported race/ethnicity. Health Serv Res. 2008;43(5 Pt 1):1722–36. doi: 10.1111/j.1475-6773.2008.00854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voicu I. Using first name information to improve race and ethnicity classification. Statistics and Public Policy. 2018;5(1):1–13. [Google Scholar]
- 23.Brown DP, Knapp C, Baker K, Kaufmann M. Using bayesian imputation to assess racial and ethnic disparities in pediatric performance measures. Health Serv Res. 2016;51(3):1095–108. doi: 10.1111/1475-6773.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derose SF, Contreras R, Coleman KJ, Koebnick C, Jacobsen SJ. Race and ethnicity data quality and imputation using US Census data in an integrated health system: the Kaiser Permanente southern California experience. Med Care Res Rev, 2013. 70(3):330–45. doi: 10.1177/1077558712466293. [DOI] [PubMed] [Google Scholar]
- 25.Grundmeier RW, Song L, Ramos MJ, et al. Imputing missing race/ethnicity in pediatric electronic health records: reducing bias with use of US census location and surname data. Health Serv Res. 2015;50(4):946–60. doi: 10.1111/1475-6773.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elliott MN, Fremont A, Morrison PA, Pantoja P, Lurie N. Using the Census Bureau’s surname list to improve estimates of race/ethnicity and associated disparities. Health Serv Outcomes Res Method. 2009. pp. 69–83.
- 27.Haas A, Elliott MN, Dembosky JW, Adams JL, Wilson-Frederick SM, Mallett JS. Imputation of race/ethnicity to enable measurement of HEDIS performance by race/ethnicity. Health Serv Res, 2019. 54(1):13–23. doi: 10.1111/1475-6773.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahimi SE, Hallvik SE, Dameshghi N, Hildebran C, Fischer MA, Weiner SG. Enhancing race and ethnicity using bayesian imputation in an all payer claims database. Preprints. 2022. p. 2022010227.
- 29.Kernan WN, Viscoli CM, Makuch RW, Brass LM, Horwtiz RI. Stratified randomization for clinical trials. J Clin Epidemiol, 1999. 52(1):19–26. doi: 10.1016/s0895-4356(98)00138-3. [DOI] [PubMed] [Google Scholar]
- 30.Foxcroft JA, d’Alessandro A, Antonie ML. Name2Vec: personal names embeddings. Canadian Conference on AI. 2019.
- 31.Le Q, Mikolov T. Distributed representations of sentences and documents, in proceedings of the 31st international conference on machine learning. Proceedings of Machine Learning Research. 2014. pp. 1188–1196.
- 32.First and last names database. Accessed March 8, 2023. https://github.com/philipperemy/name-dataset.
- 33.Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Association for Computing Machinery. 2016. pp. 785–794.
- 34.National Institute on Minority Health and Health Disparities Diversity and inclusion in clinical trials. Accessed March 8, 2023. https://www.nimhd.nih.gov/resources/understanding-health-disparities/diversity-and-inclusion-in-clinical-trials.htm.