Abstract
Clinical trials are indispensable in developing new treatments, but they face obstacles in patient recruitment and retention, hindering the enrollment of necessary participants. To tackle these challenges, deep learning frameworks have been created to match patients to trials. These frameworks calculate the similarity between patients and clinical trial eligibility criteria, considering the discrepancy between inclusion and exclusion criteria. Recent studies have shown that these frameworks outperform earlier approaches. However, deep learning models may raise fairness issues in patient-trial matching when certain sensitive groups of individuals are underrepresented in clinical trials, leading to incomplete or inaccurate data and potential harm. To tackle the issue of fairness, this work proposes a fair patient-trial matching framework by generating a patient-criterion level fairness constraint. The proposed framework considers the inconsistency between the embedding of inclusion and exclusion criteria among patients of different sensitive groups. The experimental results on real-world patient-trial and patient-criterion matching tasks demonstrate that the proposed framework can successfully alleviate the predictions that tend to be biased.
1. Introduction
Clinical trials are an essential part of developing new treatments for diseases, as they provide rigorous scientific evidence for the safety and efficacy of new therapies. Despite their importance, clinical trials often face significant challenges in patient recruitment and retention due to the difficulty in obtaining the required number of participants. Several studies have examined the factors that affect patient participation in clinical trials. For instance, an existing study found that inadequate patient participation and recruitment can lead to delays and increased costs in clinical trials, ultimately hindering the development of new treatments [1]. A recent study also found that nearly one-third of publicly funded trials required time extensions due to low enrollment rates [2].
Recently, patient-trial matching has been the focus of research to accurately identify and recruit qualified patients [3]. The existing patient-trial matching methods could be divided into two categories: rule-based systems and machine learning approaches, which both have been proposed to accelerate the patient recruiting process. Rule-based systems rely on a vast number of human annotations to establish classification rules [4, 5]. However, they have limitations in terms of recall because of inadequate rule coverage, and require extensive manual efforts to set up rules. Alternatively, machine learning based models focus on extracting rules automatically. For instance, [6] adopts unsupervised clustering methods to automatically extract eligible rules. More recently, several studies also employ deep neural networks to further improve the model performance on patient-trial matching. For example, DeepEnroll [7] and COMPOSE [8] proposed utilizing deep embedding models to encode patient records and eligibility criteria of clinical trials for computing the similarity between patients and criteria in the embedding space. By considering the discrepancy between inclusion and exclusion criteria in clinical trials, these frameworks have shown promise in achieving more precise predictions and efficient matching.
However, there are disparity issues in patient-trial matching, which can be further amplified by machine learning models and lead to unfairness. Specifically, machine learning models trained on biased historical data can perpetuate disparities in patient-trial matching, resulting in underrepresented sensitive groups of individuals in clinical trials and limited treatment efficacy. Unfortunately, recent research has shed light on the potential for machine learning models to exhibit unfairness and bias [9], which may negatively impact the minority groups in the application fields. For example, a study found that a machine learning algorithm used to predict healthcare utilization showed bias against African-American patients, resulting in fewer healthcare resources allocated to them compared to white patients [10]. Current studies concentrate on developing bias mitigation methods to reduce discrimination in machine learning models. There are several existing fairness regularization [11, 12, 13, 14] and adversarial learning methods [15, 16, 17] are designed to ensure fairness by preventing discrimination based on sensitive attributes such as race, gender, or age. However, the existing fairness methods cannot be utilized to mitigate the fairness issue in patient-trial matching because of the complex inclusion and exclusion criteria, which require careful consideration to achieve accurate prediction.
The uniqueness of the patient-trial matching lies in its dual goals of matching inclusion criteria while mismatching exclusion criteria, which differs from other healthcare applications and adds extra complexity to the task. Specifically, the discrepancy between inclusion and exclusion criteria provides information about a clinical trial to better learn a patient-trial matching framework. To better characterize the uniqueness and tackle the fairness challenges, we propose FairPM, a fine-grained fairness framework for patient-trial matching tasks. Specifically, motivated by DeepEnroll [7] and COMPOSE [8], we develop a patient-trial matching framework by minimizing the distance between the embedding of qualified patients and inclusion criteria while maximizing the distance between the embedding of unqualified patients and exclusion criteria. To further mitigate the biased prediction behaviors, we propose a fine-grained fairness constraint to minimize the prediction differences among the inclusion and exclusion criteria and across different sensitive patient groups. We evaluate the proposed framework on a real-world EHR patient records dataset and six pivotal stroke clinical trials. The experimental results demonstrate that FairPM can improve two fairness metrics for both patient-criterion and patient-trial matching toward two sensitive attributes, albeit with a slight trade-off in prediction performance. The case study shows some eligibility criteria that may cause biased predictions for minority groups.
2. Background of Fairness in Patient-Trial Matching
In this section, we will first identify the fairness issue in the patient-trial matching from two levels, and then introduce the metrics to measure them from the computational perspective.
2.1. Fairness of patient-trial matching
We identified two critical fairness issues in matching patients with trials in previous matching systems, namely criteria-level and trial-level fairness. Criteria-level fairness indicates that criteria assessment should be consistent across all patient subgroups. For example, clinical trial eligibility criteria should be assessed the same way for patients of the majority and minority races. Conversely, trial-level fairness mean that different patient subgroups for the same clinical trial should be equal considered. For example, male and female patients in a clinical trial should have equal eligibility that is unrelated to gender. Although abundant of machine learning efforts have been made to predict patient eligibility for different clinical trials, they often ignore the fairness issues behind the clinical matching, as discussed before. Therefore, there is an urgent need to enable ML models product unbiased eligibility predictions from both the criteria and trial perspectives.
2.2. Fairness metrics
Fairness metrics have garnered considerable attention in recent years. For example, research in [18] delved into fairness definitions within political philosophy, attempting to establish connections with machine learning principles. Another study in [19] examined the evolution of fairness definitions over a period of five decades, focusing on the fields of education and machine learning. Additionally, comprehensive investigations have been conducted to enumerate and elucidate various definitions of fairness as they pertain to algorithmic classification challenges ([20, 21, 22]). In the subsequent section, we will reiterate and expound upon some of the most widely adopted definitions in our work.
Equal Opportunity (EO). It is a binary predictor Ŷ that adheres to the principle of equal opportunity with respect to protected attribute A and outcome Ŷ, if P(Ŷ = y|A = 0, Y = y) = P(Ŷ = y|A = 1, Y = y) [23]. This assertion implies that the likelihood of an individual belonging to the positive class being allocated a positive outcome should be equivalent for both protected and unprotected group members [20]. Thus, the equal opportunity definition stipulates that the true positive rates should be consistent across both protected and unprotected groups.
Demographic Parity (DP). Alternatively referred to as statistical parity, it is a predictor Y upholds demographic parity if P(Ŷ |A = 0) = P(Ŷ |A = 1) [24, 25]. This principle dictates that the probability of a positive outcome [20] should remain consistent irrespective of an individual’s membership in the protected group. In other words, demographic parity mandates that the likelihood of a positive outcome should be independent of the protected attribute.
3. Data and Problem Description
3.1. Data preparation
In the present investigation, we have undertaken a comprehensive analysis of data obtained from the renowned Texas Medical Center. This research focuses on six pivotal stroke trials, i.e., NCT03735979, NCT03805308, NCT03263117, NCT03496883, NCT03876457, and NCT03545607. The patient data are participants encompassed in these studies amounts to 825 individuals. The project was approved by the UTHealth Institutional Review Board (IRB) under HSC-SBMI-21-0529 - “Re-admission Risk Estimation for Stroke Patients”. For the purpose of this study, race and gender have been identified as sensitive demographic groups, warranting further examination. A meticulous breakdown of the demographic characteristics for these patients can be found in Table 1.
Table 1:
The demographic information pertaining to gender and race within the dataset.
| Dataset | Male / Female | White / Others | Total |
|---|---|---|---|
| Train | 308 / 217 | 185 /340 | 515 |
| Valid | 28 / 31 | 18 / 41 | 59 |
| Test | 135 / 116 | 83 / 168 | 251 |
3.2. Problem formulation
In this section, we will go over the notations and formulate the problems in this paper. We first define the notations for describing patient records and then introduce the two main tasks in this paper.
Definition 1: Patient records. We use P = [v1, v2, · · ·, vT] to represent a series of patient visit records within the longitudinal electronic health records (EHR). Every visit record contains three groups of observations: diagnosis D, medication M, and procedure P. These groups correspond to sets of diseases, medication types, and procedural operations, respectively. Given the three observation groups, each visit record of a patient can be represented by , where , , and . Since all the medical codes in D, M, and P are frequently utilized and can be considered as a single general concept, we represent them as gt for the sake of simplicity.
In this work, the sensitive attribute for each patient visit record is defined as Vs E S, where S is the sensitive attributes set, and the target sensitive groups place particular emphasis on the attributes of race and gender.
Definition 2: Clinical trials. Each clinical trial consists of two categories of eligibility criteria: inclusion criteria (CI) and exclusion criteria (CE). Therefore, we can denote each clinical trial as , where N and Q represent the number of inclusion and exclusion criteria, correspondingly. Note that each criterion is described in text.
Task 1: Patient-Criterion matching. When given the visit records of a patient P and an inclusion or exclusion criterion belonging to a clinical trial, we define the matching of patient-trial as a multi-class classification task. It means that a pair of patient P and criterion C can be classified into three possible categories: ”inclusion,” ”exclusion,” and ”unknown.” These three categories show whether the criteria include or exclude the patient. We can represent the patient-criterion matching task as ŷ(P, C) ∈ {inclusion, exclusion, unknown}.
Task 2: Patient-Trial matching. When given the visit records of a patient P and a clinical trial C, we define the patient-trial matching as a binary classification task, indicating whether a patient P is eligible for the clinical trial C. For a patient P to be eligible for the clinical trial C, all of the inclusion criteria CE ∈ C must apply to the patient and none of the exclusion criterion CE E C should apply to the patient. In other words, the patient-trial matching task is a 100% patient-criterion matching task where all the criteria belong to the same clinical trial C.
4. Fair Patient-Trial Matching (FairPM)
Inspired by DeepEnroll [7] and COMPOSE [8], we propose the FairPM framework that leverages deep embedding models for mapping input data to latent space and predict whether a patient is eligible for a criteria. To further tackle fairness issue, we introduce a specific fairness constraint that is tailored to the characteristics of patient-criterion and patient-trial matching tasks.
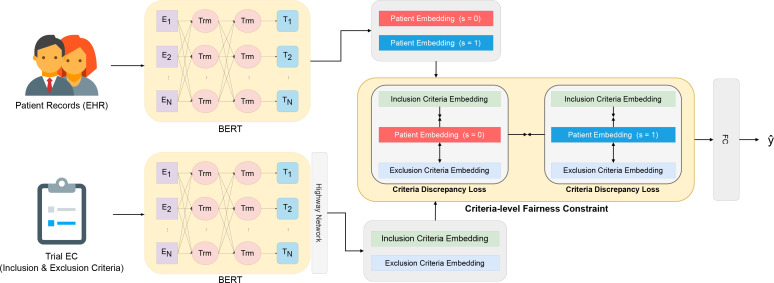
In this section, we present our FairPM to achieving fair patient-criterion and patient-trial matching. Figure 1 depicts an overview of the proposed FairPM, which comprises text encoder, criteria discrepancy loss, fair criteria matching loss, and a fully connected layer serving as the prediction head. First, we introduce the encoding model of patient records and eligibility criteria into embedding space (Section 4.1). Next, we present a joint learning approach to distinguish between inclusion and exclusion criteria for patient-criteria and patient-trial matching (Section 4.2). We then propose a fine-grained, criteria-level fairness constraint to achieve fair patient-trial matching (Section 4.3). Finally, we describe the training algorithm in its entirety (Section 4.4).
Figure 1:
An overview of the proposed framework FairPM, where BERTs refer to the encoding models for patient records and eligibility criteria (see Section 4.1), Criteria Discrepancy Loss represents the goal of optimizing the distance between patients and criteria (see Section 4.2), and Criteria-level Fairness Constraint refers to the fairness constraint considering the discrepancy among the eligibility criteria and sensitive groups (see Section 4.3).
4.1. Embedding of Patient Records and Eligibility Criteria
Patient Records Embedding. In a single patient visit record, the diagnosis results can include information on diagnosed diseases, medications, and procedures, all of which are expressed in natural language. Hence, latent representations of a patient’s visit record can be learned to use large language models (LLMs), such as BERT [26], RoBERTa [27], and more [28]. In this work, we leverage the pretrained BERT as the text encoder. Furthermore, since a patient is represented by a sequence of past diagnosis records, we employ a memory network [29] to process the patient records, which helps to effectively preserve the sequence of visit information in the embedding space. Formally, a patient record embedding, denoted by zP, is obtained through the encoding process gP(·), which can be represented as follows:
| (1) |
where d, m, and p denote the aggregated historical diagnosis, medications, and procedures embeddings, respectively. We incorporate a memory network Mmem(·) to preserve the sequence of visits information in the embedding space.
Eligibility Criteria Embedding. Each clinical trial is described by its eligibility criteria (ECs), including both inclusion and exclusion criteria, in unstructured textural description. Therefore, the embedding of each EC can also be learned by LLMs, and we use BERT, the same as for patient record embedding. However, one salient characteristic of ECs is the frequent appearance of concepts that express significant and detailed information, including numerical values and associated quantity units. To capture and encode these crucial features in the embedding space, we adopt a previous approach of using a convolutional neural network (CNN) [30] and a highway layer [31] to extract patterns across multiple levels for the semantic matching task [32]. Formally, the encoding process gc(•) is used to encode an EC embedding zc, which can be formulated as follows:
| (2) |
where, σ(•) represents the sigmoid activation function. Generally, we utilize the Highway layer Highway(•) to effectively capture the specific semantic concepts present in the ECs.
4.2. Joint Criteria Discrepancy Loss
Since the patient-trial matching task depends on the prediction results of patient-criteria matching, the objective of our learning workflow is to optimize the patient-criteria matching task. However, a crucial characteristic of the patient-criteria matching task is the discrepancy between the patient-inclusion and patient-exclusion criteria pairs. To train the framework taking the uniqueness of the task into account, we follow the objective loss proposed by the framework COMPOSE [8]. Specifically, the objective loss includes two parts as the following.
Cross-entropy Loss. Since the patient-criteria matching is formulated as a multi-class classification problem (Section. 3.2), we can develop a classification framework by optimizing a cross-entropy loss between the predicted labels ŷ and the ground truth y as follows:
| (3) |
Criteria Discrepancy Loss. Considering the characteristic of the patient-criteria matching task that the inclusion and exclusion criteria can have opposite effects, we adopt a loss that accounts for the difference between them. Specifically, the goal of the criteria discrepancy loss is to minimize the distance between the embedding of qualified patients and the inclusion criteria cI, while maximizing the distance between the embedding of unqualified patients and the exclusion criteria cE. Formally, the criteria discrepancy loss can be formulated as follows:
| (4) |
where d(•, •) is an arbitrary distance function in metric space, zP,CI represents the embedding of the patient who matches the inclusion criteria cI, zP,CE represents the embedding of the patient who doesn’t match the exclusion criteria cE, and K denotes the hyper-parameter of minimum distance between zP and .
Combination of the cross-entropy and criteria discrepancy loss, the joint objective loss for learning a patient-criteria matching framework is as follows:
| (5) |
4.3. Criteria-level Fairness Constraint
Despite the prediction efficacy of the LLM encoders and the designed objective function, the skewed demographic distribution in the training data inherently causes fairness issues for the trained deep models. However, directly adopting existing debiasing regularization without considering the unique characteristics of the training target may lead to the exacerbation of unfair predictions against protected groups. Therefore, it is necessary to carefully tailor the bias mitigating approach to the specific matching task.
To address the potential fairness issue resulting from the discrepancy between patient-inclusion and patient-exclusion criteria matching, we propose a novel approach that aims to minimize the prediction differences among the two types of criteria and across different sensitive patient groups. By doing so, we can mitigate the potential impact of biased model predictions on certain subgroups of patients. Formally, the proposed criteria-level fairness constraint can be formulated as follows:
| (6) |
where LCD,[vs=i] represents the criteria discrepancy loss LCD of a patient who belongs to the sensitive group i.
Finally, we can learn a fair patient-criteria and patient-trial matching framework by optimizing the joint criteria discrepancy loss LPC with the criteria-level fairness constraint LFC:
| (7) |
where λFC is the weighting hyper-parameter to balance the fairness constraint and the performance of predictions.
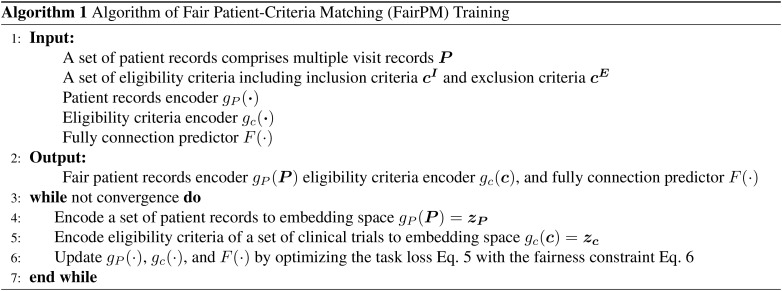
4.4. Algorithm of FairPM Training
The training outline of the proposed FairPM framework is given in Algorithm 1. The training aims to achieve the fair patient-criteria and patient-trial matching framework by optimizing the joint objective loss LPC with the proposed task-specific fairness constraint LFC. Specifically, FairPM first encode the patient records and eligibility criteria to embedding space (line 4-5), and then update the patient records encoder, eligibility criteria encoder, and predictor according to Eq. 5 and Eq. 6 (line 6) until it converges.
5. Experiment
In this section, we conduct experiments to evaluate the performance of FairPM framework, aiming to answer the following three research questions:
RQ1: How effective is the FairPM for improving different sensitive attributes (Section 5.4)?
RQ2: How does the hyper-parameter AFC impact the fairness performance of FairPM (Section 5.5)?
RQ3: What is the difference between the results of FairPM and the vanilla model (Section 5.6)?
5.1. Baseline methods
Baseline Model. COMPOSE [8] outperformed other baseline models, including LSTM+GloVE [33], LSTM+BERT [26], Criteria2Query [34], and DeepEnroll [7]. As our FairPM framework is inspired by and developed based on COMPOSE, we refer to the version of FairPM removing the proposed task-specific fairness constraint LFC (Eq. 6) as the baseline model for simplification.
Baseline Fairness Adversarial Learning Constraint (Baseline w/ ALC). In the seminal work of 2018, Zhang et al. introduced a novel approach utilizing adversarial networks as a means of mitigating model bias [15]. This innovative methodology was derived from the concept of generative adversarial networks [35]. The framework devised by Zhang et al. involved training the generator with a specific focus on a protected attribute, such as gender, ultimately leading to a structure in which the generator actively obstructs the discriminator’s ability to predict gender within a given overarching task. The proposed adversarial learning to mitigate bias successfully demonstrated an enhancement in the fairness of an income classification task. However, this improvement was accompanied by a slight reduction in overall accuracy. We implement this adversarial debiasing method as our baseline fairness constraint, named ALC.
5.2. Evaluation tasks and metrics
As mentioned in Section 3.2, there are two evaluation tasks:
Patient-Criterion matching. We begin by labeling all patient-criterion pairs, and then splitting them into training, validation, and testing sets. The training set is used for model training, the validation set for hyperparameter tuning, and the testing set for evaluating both the baselines and the proposed FairPM.
Patient-Trial matching: We define the patient-trial matching task as a binary classification problem that determines whether a patient P is qualified for a clinical trial C. A patient P is considered eligible for a clinical trial C only if they satisfy all the inclusion criteria CI and do not satisfy any of the exclusion criteria CE in the clinical trial C. The training, validation, and testing sets are split in the same way as for the patient-criterion matching task.
For both tasks, we evaluate the prediction performance using accuracy score (Acc.) and F1 score (F1), and assess the fairness of the models using demographic parity (DP) and equal opportunity (EO) as evaluation metrics.
5.3. Implementation details
We implement all the baseline models and FairPM in PyTorch. As for the LLM text encoders, we leverage the Clinical BERT [36] pretrained on PubMed and MIMIC-III [37] for increasing the medical knowledge of the text embedding.
5.4. Prediction performance
We conduct experiments to compare the prediction and fairness performance of the proposed FairPM with other baselines on patient-criterion and patient-trial matching tasks. Our target sensitive attributes are Race and Gender.
Results of patient-criterion matching. Table 2 summarizes the performance of FairPM and the two baseline methods on the testing set. Since the baseline model is only trained by the joint objective including two prediction task-oriented loss terms, cross-entropy LCE and criteria discrepancy loss LCD, it can achieve the best accuracy and F1 scores. However, the baseline model may learn the skew distribution of sensitive attributes in training data, which can be addressed by employing ALC [35] to the baseline model. Nevertheless, as shown in Table 2, adopting ALC would cause a huge performance drop. Comparing with the two baselines, FairPM improve the fairness metrics DP and EO, while maintaining the competitive performance for the patient-criterion matching task.
Table 2:
Performance comparison on patient-criteria matching task
| Model | Sensitive attribute: Race | Sensitive attribute: Gender | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc. | F1 | DP | EO | Acc. | F1 | DP | EO | |
| Baseline | 0.9595 | 0.9702 | 0.0301 | 0.0251 | 0.9595 | 0.9702 | 0.0216 | 0.0093 |
| Baseline w/ ALC | 0.8294 | 0.8839 | 0.0112 | 0.0248 | 0.8142 | 0.8642 | 0.0095 | 0.0010 |
| FairPM | 0.9130 | 0.9360 | 0.0001 | 0.0111 | 0.9299 | 0.9490 | 0.0093 | 0.0011 |
Results of patient-trial matching. Table 3 summarizes the performance of FairPM and the two baseline methods on the testing set. The prediction results of all three models are computed based on 100% matching patient-criterion pairs as mentioned in Section 3.2. Since the patient-trial matching results are derived from patient-criterion matching, we can observe a similar trend as in the patient-criterion matching task. Overall, FairPM shows promise in addressing fairness issues related to the two different sensitive attributes, race and gender, while causing a slight performance drop in patient-trial matching compared to the baseline model and the ALC debiasing method.
Table 3:
Performance comparison on patient-trial matching task
| Model | Sensitive attribute: Race | Sensitive attribute: Gender | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc. | F1 | DP | EO | Acc. | F1 | DP | EO | |
| Baseline | 0.8685 | 0.9296 | 0.0279 | 0.0279 | 0.8685 | 0.9296 | 0.0198 | 0.0198 |
| Baseline w/ ALC | 0.7088 | 0.7935 | 0.0125 | 0.0125 | 0.7106 | 0.7792 | 0.0106 | 0.0106 |
| FairPM | 0.8008 | 0.8894 | 0.0084 | 0.0084 | 0.8327 | 0.9087 | 0.0095 | 0.0095 |
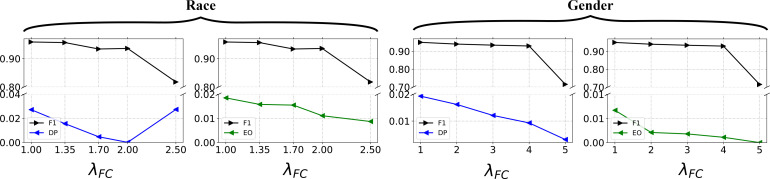
5.5. Analysis of sensitive hyper-parameter
In this section, we study the impact of the hyper-parameter λFC in Eq. 7 to answer the research question RQ2. We conduct the sensitive analysis on patient-criterion matching since the results of patient-trial matching are derived from it. As shown in Figure 2, the value of λFC does not significantly affect the patient-criterion matching performance up to a certain threshold, which is 2 for race and 4 for gender. After the certain value, the performance drop can be observed from Figure 2. Regarding the influence towards fairness, both the fairness metrics DP and EO can be improved when the value of λFC increases, except for the DP fairness metric of the sensitive attribute race. As the goal of FairPM is to mitigate the biased prediction outcomes against sensitive attributes while maintaining competitive performance in patient-criterion and patient-trial matching tasks, we can determine the appropriate value of λFC by identifying the threshold before performance drop occurs.
Figure 2:
Sensitive analysis of AFC for FairPM on patient-criterion matching task with respect to the sensitive attributes of race and gender.
5.6. Case study: Fairness-related criteria
To investigate the impact of the proposed FairPM framework on mitigating biased predictions, we compare the results between the baseline model and our FairPM for patients with different sensitive attributes vs. As shown in Table 4, the baseline model may provide biased predictions against the minority group for some eligibility criteria of clinical trials, which will exacerbate the underrepresentation problem. For example, for the inclusion criterion in NCT03545607 “Male or female subjects ≥ 18 years of age,” the baseline model predicts a patient-criterion matching pair incorrectly when a female patient is older than 18 years old. This unfair outcome may be attributed to the biased knowledge encoded in the model, which associates the term “Male” more strongly with the model’s predictions. In contrast, our FairPM can predict the criterion for different sensitive groups fairly.
Table 4:
Case study: different patient-criterion matching results between the baseline model and FairPM.
| Clinical trial | Criterion | Patient vs | Baseline | FairPM |
|---|---|---|---|---|
| NCT03735979 | (I) Acute ischemic stroke patients. | Male | ✔ | ✔ |
| Female | ✘ | ✔ | ||
| NCT03545607 | (I) Male or female subjects ≥ 18 years of age. | Male | ✔ | ✔ |
| Female | ✘ | ✔ | ||
| NCT03876457 | (I) Eligible for thrombectomy or medical management. | White | ✘ | ✔ |
| Others | ✔ | ✔ | ||
| NCT03496883 | (E) Patient suspected of not being able to comply with trial protocol (e.g., due to alcoholism, drug dependency, or psychological disorder). | White | ✘ | ✔ |
| Others | ✔ | ✔ |
6. Conclusion and future work
We present FairPM, an innovative framework designed to tackle the issue of AI fairness in clinical trial matching with deep learning. Our approach includes a novel patient-criterion level fairness constraint that can help mitigate this problem. One of the unique features of our proposed framework is that it focuses on the discrepancy between inclusion and exclusion criteria for both the model predicting and alleviating unfair predictions. In doing so, it helps ensure that the patient selection process is unbiased, transparent, and equitable, leading to fairer and more reliable clinical trials outcomes. To demonstrate the effectiveness of FairPM, we conducted several experiments using real-world patient records with six stroke clinical trials. Our results indicated that the framework significantly improved two fairness metrics while only marginally affecting overall model performance. As part of our future research directions, we plan to explore various perspectives on bias mitigation in patient-trial matching, specifically the different forms of skew distribution in training data. We aim to expand our concept of fairness and explore how to mitigate bias that may arise due to distribution shift, a key challenge in machine learning applications.
7. Acknowledgements
The authors thank the anonymous reviewers for their helpful comments. The work is in part supported by NSF grants NSF IIS-1900990, IIS-1939716, and IIS-2239257. The views and conclusions contained in this paper are those of the authors and should not be interpreted as representing any funding agencies. XJ is CPRIT Scholar in Cancer Research (RR180012), and he was supported in part by Christopher Sarofim Family Professorship, UT Stars award, UTHealth startup, the National Institute of Health (NIH) under award number R01AG066749, R01AG066749-03S1, R01LM013712, U24LM013755, U01TR002062, U01CA274576, U54HG012510 and the National Science Foundation (NSF) #2124789.
Figures & Tables
References
- 1.Ohmann C, Banzi R, Canham S, Battaglia S, Matei M, Ariyo C, et al. Sharing and reuse of individual participant data from clinical trials: principles and recommendations. BMJ open. 2017;7(12):e018647. doi: 10.1136/bmjopen-2017-018647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell et al. Recruitment to randomised trials: strategies for trial enrolment and participation study. The STEPS study. Health Technology Assessment. 2007. [DOI] [PubMed]
- 3.Yuan J, Tang R, Jiang X, Hu X. Large Language Models for Healthcare Data Augmentation: An Example on Patient-Trial Matching. arXiv preprint arXiv:230316756. 2023. [PMC free article] [PubMed]
- 4.Weng C, et al. EliXR: an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association. 2011;18(Supplement 1:i116–24. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang T, et al. EliIE: An open-source information extraction system for clinical trial eligibility criteria. Journal of the American Medical Informatics Association. 2017;24(6):1062–71. doi: 10.1093/jamia/ocx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alicante A, Corazza A, Isgro F, Silvestri S. Unsupervised entity and relation extraction from clinical records in Italian. Computers in biology and medicine. 2016;72:263–75. doi: 10.1016/j.compbiomed.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Xiao C, Glass LM, Sun J. DeepEnroll: patient-trial matching with deep embedding and entailment prediction. Proceedings of The Web Conference. 2020;2020:p. 1029–37. [Google Scholar]
- 8.Gao et al. COMPOSE: cross-modal pseudo-siamese network for patient trial matching. Proceedings of the 26th ACM SIGKDD international conference on knowledge discovery & data mining. 2020. pp. p. 803–12.
- 9.Ding S, Tang R, Zha D, Zou N, Zhang K, Jiang X, et al. Fairly Predicting Graft Failure in Liver Transplant for Organ Assigning. arXiv preprint arXiv:230209400. 2023. [PMC free article] [PubMed]
- 10.Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366(6464):447–53. doi: 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 11.Beutel A, et al. Putting fairness principles into practice: Challenges, metrics, and improvements. Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society. 2019. pp. p. 453–9.
- 12.Beutel A, et al. Fairness in recommendation ranking through pairwise comparisons. Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery & data mining. 2019. pp. p. 2212–20.
- 13.Jiang R, et al. Wasserstein fair classification. Uncertainty in artificial intelligence. PMLR. 2020. pp. p. 862–72.
- 14.Nam J, Cha H, Ahn S, Lee J, Shin J. Learning from failure: De-biasing classifier from biased classifier. Advances in Neural Information Processing Systems. 2020;33:20673–84. [Google Scholar]
- 15.Zhang BH, Lemoine B, Mitchell M. Mitigating unwanted biases with adversarial learning. Proceedings of the 2018 AAAI/ACM Conference on AI, Ethics, and Society. 2018. pp. p. 335–40.
- 16.Madras D, Creager E, Pitassi T, Zemel R. Learning adversarially fair and transferable representations. International Conference on Machine Learning. PMLR. 2018. pp. p. 3384–93.
- 17.Sweeney C, Najafian M. Reducing sentiment polarity for demographic attributes in word embeddings using adversarial learning. Proceedings of the 2020 Conference on Fairness, Accountability, and Transparency. 2020. pp. p. 359–68.
- 18.Binns R. Fairness in machine learning: Lessons from political philosophy. Conference on fairness, accountability and transparency. PMLR. 2018. pp. p. 149–59.
- 19.Hutchinson B, Mitchell M. 50 years of test (un) fairness: Lessons for machine learning. Proceedings of the conference on fairness, accountability, and transparency. 2019. pp. p. 49–58.
- 20.Verma S, Rubin J. Fairness definitions explained. Proceedings of the international workshop on software fairness. 2018. pp. p. 1–7.
- 21.Mehrabi N, Morstatter F, Saxena N, Lerman K, Galstyan A. A survey on bias and fairness in machine learning. ACM Computing Surveys (CSUR) 2021;54(6):1–35. [Google Scholar]
- 22.Chang CY, Chuang YN, Lai KH, Han X, Hu X, Zou N. Towards Assumption-free Bias Mitigation. arXiv preprint arXiv:230704105. 2023.
- 23.Hardt M, Price E, Srebro N. Equality of opportunity in supervised learning. Advances in neural information processing systems. 2016;29 [Google Scholar]
- 24.Dwork C, Hardt M, Pitassi T, Reingold O, Zemel R. Fairness through awareness. Proceedings of the 3rd innovations in theoretical computer science conference. 2012. pp. p. 214–26.
- 25.Kusner, et al. Counterfactual fairness. Advances in neural information processing systems. 2017;30 [Google Scholar]
- 26.Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805. 2018.
- 27.Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, et al. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:190711692. 2019.
- 28.Chuang YN, Tang R, Jiang X, Hu X. Spec: A soft prompt-based calibration on mitigating performance variability in clinical notes summarization. arXiv preprint arXiv:230313035. 2023. [DOI] [PMC free article] [PubMed]
- 29.Weston J, Chopra S, Bordes A. Memory networks. arXiv preprint arXiv:14103916. 2014.
- 30.Hu B, Lu Z, Li H, Chen Q. Convolutional neural network architectures for matching natural language sentences. Advances in neural information processing systems. 2014;27 [Google Scholar]
- 31.Srivastava RK, Greff K, Schmidhuber J. Highway networks. arXiv preprint arXiv:150500387. 2015.
- 32.You Q, Zhang Z, Luo J. End-to-end convolutional semantic embeddings. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. pp. p. 5735–44.
- 33.Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation. 1997;9(8):1735–80. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 34.Yuan C, Ryan PB, Ta C, Guo Y, Li Z, Hardin J, et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. Journal of the American Medical Informatics Association. 2019;26(4):294–305. doi: 10.1093/jamia/ocy178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodfellow et al. Generative adversarial networks. Communications of the ACM. 2020;63(11):139–44. [Google Scholar]
- 36.Alsentzer et al. Publicly available clinical BERT embeddings. arXiv preprint arXiv:190403323. 2019.
- 37.Johnson et al. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3(1):1–9. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]