Abstract
As Electronic Health Record (EHR) systems increase in usage, organizations struggle to maintain and categorize clinical documentation so it can be used for clinical care and research. While prior research has often employed natural language processing techniques to categorize free text documents, there are shortcomings relative to computational scalability and the lack of key metadata within notes’ text. This study presents a framework that can allow institutions to map their notes to the LOINC document ontology using a Bag of Words approach. After preliminary manual value- set mapping, an automated pipeline that leverages key dimensions of metadata from structured EHR fields aligns the notes with the dimensions of the document ontology. This framework resulted in 73.4% coverage of EHR documents, while also mapping 132 million notes in less than 2 hours; an order of magnitude more efficient than NLP based methods.
Introduction
As Electronic Health Record (EHR) systems are adopted, the amount of clinical information available greatly increases for both researchers and clinicians alike. While this is a great boon for clinicians, who can interpret text documents in relation to the patient’s chart, the ability to resolve clinical documentation semantic context is not as forthcoming for researchers. For a researcher, the first step is to isolate the notes they desire from millions of EHR documents, which remains a daunting task due to the lack of standardization and categorization. Ontologies and standards have been developed to overcome this challenge, such as the LOINC Document Ontology1 (LOINC DO), but these are rarely integrated into clinical workflows so that notes accurately categorized when initially authored. LOINC DO, a subset of the Clinical LOINC domain, enables institutes to label and organize their clinical documents with a wide range of well-defined codes and respective descriptors, ranging from generic “Hospital Notes” to more informative “Attending Physician Hospital Progress Notes.” Adopting this standard allows institutes to improve internal data collection and empowers cooperation and data sharing between organizations.
LOINC DO was created enable effective transfer of EHR data between institutes and organizations. Ideally, each clinical document will be assigned a specific LOINC Code based on the five LOINC Axis/Dimensions: Subject Matter Domain (SMD), Kind of Document (KOD), Type of Service (TOS), Setting and Role. Each of these dimensions consists of a unique part name and part number. For example, possible KOD values have the part names of Note and Report with their part numbers being LP173418-7 and LP173421-1, respectively. Similarly, the other axes all have a set of unique part numbers and corresponding part names. A LOINC Code is defined as a combination of two or more of these five axes, generally requiring at least a KOD and either TOS or SMD as a bare minimum.
While the LOINC DO provides consistent semantics for notes, the mapping is not a straightforward process. The task often requires vast amount of human time and/or computer resources, depending on how it is accomplished. Some of the earlier works by Hyun et al.2,3 involved a team of researchers manually mapping individual LOINC DO notes to corresponding LOINC Codes to validate the coverage and reliability of LOINC DO Standard. In more recent studies, an increasing amount of the mapping process is being automated but to-date there is poor adoption of the LOINC DO across large research networks supported by the Observational Medical Outcomes Partnership OMOP4 and PCORnet5 Common Data Models. While not exactly related to LOINC, Aronson et al.6 devised an early classifier that assigned ICD-9-CM codes to radiology reports specifically. Parr et al.7 used a noise-tolerant learning model pipeline to map laboratory tests to LOINC codes with promising results. Zou et al.8 leveraged the deep-learning model Bidirectional Encoder Representations from Transformer (BERT) to map a document to each of the five LOINC DO axes using the document title alone.
While several methods have attempted to solve the issue of LOINC Mapping, each of them has certain pitfalls that may make it undesirable in certain circumstances and warrants development of improved approaches. The manual mapping for example, used in the earlier days, is error prone, inefficient, and unscalable to the current EHR climate. On the other hand, the more recent efforts through techniques using machine learning, deep-learning and Natural Language Processing (NLP) show improved results but are computationally expensive, while also requiring manual consolidation and annotation of enormous amounts of training data. Even then, key axes of the ontology such as the type of service and role may not be contained within the document. Another issue with trained models lies the need to adapt to changes in LOINC DO documentation. Should LOINC add or change any of the LOINC Codes, the models would have to be retrained and many notes will potentially need to be reannotated every time.
For this study, we exploit the structured columns of data in the EHR that provide context regarding the note’s author of the note, patient location and type of encounter and then opted for a simpler design that relies on the bag-of-words matching approach and the concept of document distance using vector representations. Our framework does not build a model and therefore does not require any curated training data. The framework also does not aim to map each document to any of the five LOINC axes as is the common theme in many other mapping efforts, but instead assigns them to the closest valid LOINC Code directly. As this framework relies solely on associated metadata of the documents and not the specific contents of the documents themselves, there is a limit to the coverage of this framework based solely on the completeness of the metadata quality. However, due to this, the framework is also very generic, portable, and efficient. The pipeline was developed based on a preliminary investigation around value-set manual mapping, which will also be discussed to provide greater context for some of the decisions made during the designing phase.
Methods
Dataset
We used the EHR data from University of Missouri’s Cerner Millennium Database. It is in turn incorporated into the Patient-Centered Outcomes Research Network Common Data Model (PCORnet CDM) and consists of over 130 million decompressed notes. Each document has an EVENT_TITLE_TEXT, the title of the document, which holds essential information for our coding purposes. However often this information is not enough to fully map a document to a LOINC Code, so we use other columns in conjunction, such as ENC_TYPE, which broadly explains the type of encounter the document covers (inpatient, outpatient, emergency etc…). In our preliminary searches, we found seven such columns that contained helpful information mappable to LOINC dimensions9. These seven columns were selected from among the hundreds of columns in the millennium tables due to them containing possible clues for the LOINC Mapping. It should be noted that our columns names might not be an exact match for institutes using the Meditech, Epic, and other EHRs, but there should be suitable replacement columns that contain semantically similar information. The authors and additional collaborators are currently configuring the pipeline against Epic.
Preliminary Work
We first started by creating a value-set table for each of the LOINC dimensions using our metadata columns. Our goal was to study the initial coverage for the mapping and gain knowledge that could be used for automation. The look up tables were populated using distinct values of the following database columns:
- EVENT_TAG: A brief text string used to describe the event and displayed on the flowsheet.
- EVENT_TITLE_TEXT: The title used for document results related to the event.
- ENC_TYPE: Determines the type of encounter.
- EVENT_CD: The code that identifies the most basic unit of the storage, such as RBC (red blood cell), discharge summary, image, or other types of medical notes.
- PERFORMED_PERSONAL: The personnel ID of the provider who performed this result, meaning the healthcare professional who wrote or recorded the note.
- VERIFIED_PRSNL_ID: The personnel ID of the provider who verified the result, meaning the healthcare professional who reviewed and approved the note or event.
- NOTE_TYPE: A description of the note type, which initially matches the EVENT_CD display.
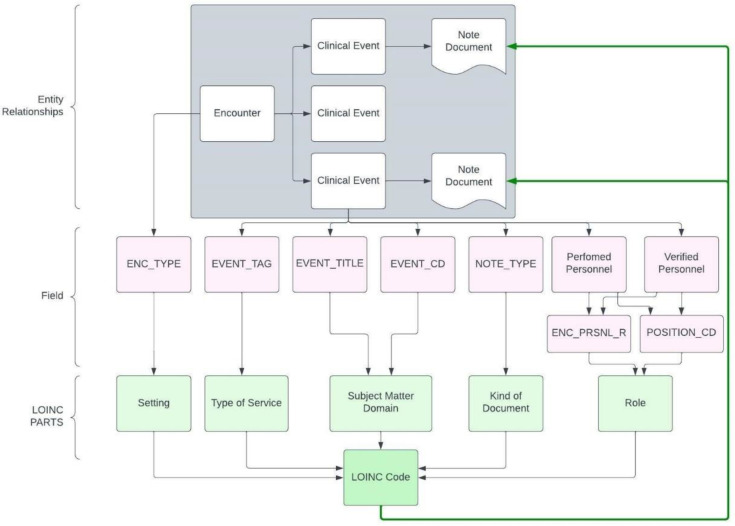
The tables were then mapped to the closest LOINC part name and number by the team. Once populated, the tables were validated for errors with the help of domain experts. Lastly, the notes were mapped to LOINC dimension values using the value-set tables and pattern matching SQL queries. This mapping work is visualized in Figure 1, which shows all the metadata columns used (in pink) as well as which of the LOINC dimensions they map to (in green). While the coverage results of this effort were less than encouraging, it offered a lot of insight for the next steps.
Figure 1:
The figure shows the process of going from a clinical document to LOINC Code. Each of the pink blocks are a metadata fields for each individual clinical document, mapping to a green block representing the LOINC dimensions. The distinct combination of the LOINC dimension points to a LOINC Code, which will connect back to the clinical document.
The key factor for the low coverage is inherent to the process of manual mapping itself. The EVENT_CD column, for example, had over 5,000 distinct values, so we had to choose the top 100 by frequency. This essentially limits our coverage far below the true value. To alleviate this issue, we looked to computational methods.
Bag of Words Implementation
The method we chose to employ here is called the Bag of Words (BOW) approach. While it is a traditional technique often found in various use cases of NLP11, it also has been shown to have viable application in classification problems12. In essence, BOW is a representation of text as an unordered set of words. The bag of words approach allows us to compare two different bags and find a match without the complexity of word order or frequency, and we will use this to facilitate the comparison of our clinical documents metadata to LOINC Codes and find sufficient matching. Additionally, we chose to implement the Bag of Words as vectors, using the Vector Space Model (VSM). VSM is widely used for document representation in information retrieval, such as search engines. Using cosine similarity, we can compare the similarity between two vectors.
The pipeline has three phases, and each may run separably from the other. The first is to convert all valid LOINC Codes into bags of words. Next, we will convert each of our document’s metadata into a bag of words. Lastly, we will compare each document to all the LOINC Codes and find the best match.
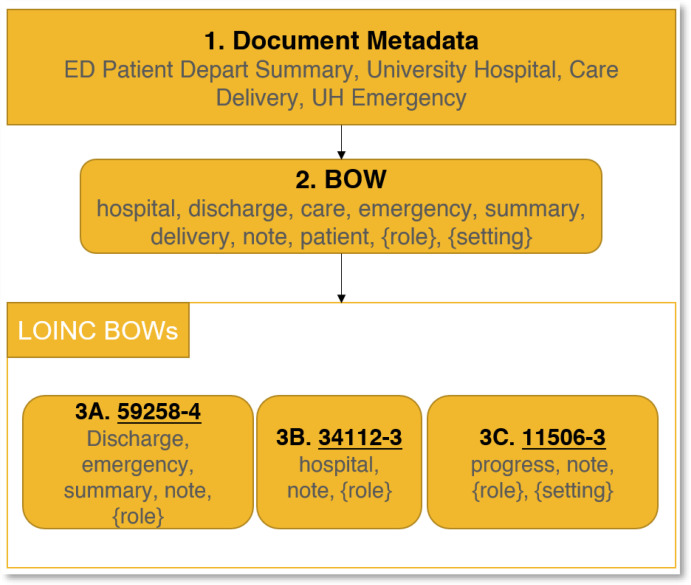
Figure 2 illustrates the step-by-step process. The first step is to take the Document’s Metadata and convert it into a Bag of Words (BOW) containing only relevant words. This is the reason for UH (University hospital) to be converted to simply hospital, getting rid of redundant information, and the keyword depart is converted to discharge as that is the word LOINC uses. We also add the two keywords {role} and {setting} as certain LOINC codes use these placeholders.
Figure 2:
Shows a example note’s metadata, its possible BOW as well as a few LOINC BOW’s. In this example, the LOINC Code that will be selected will be 3A due to it being a complete match to the note’s BOW. Note that while 3B is also a full match, 3A is given priority due to covering more LOINC Dimensions.
Box 1 shows the raw metadata, while Box 2 shows the BOW string. 3A, 3B and 3C are some possible LOINC Codes (underlined) and their respective bag of words (in gray). For this example, 3A will be chosen as every one of the words in its BOW is contained in the note’s BOW. While the same is true for 3B, 3A is given priority due to it covering more of the LOINC Dimensions.
LOINC Bag of Words
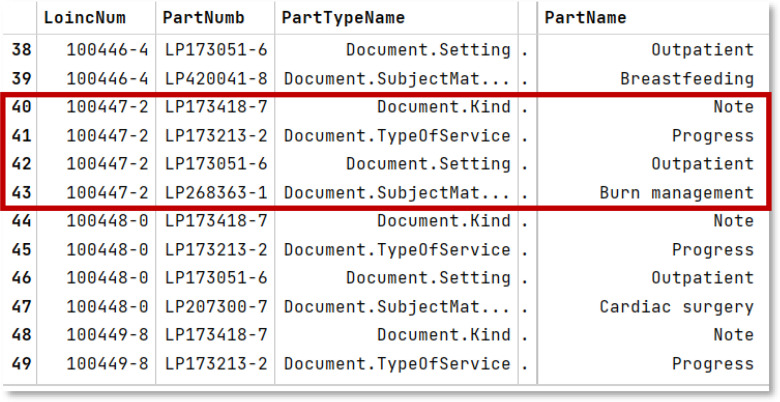
Our LOINC BOW is created using the DocumentOntology.csv file that ships with the official LOINC DO documentation. We used LOINC version 2.73, which was the latest at the time of writing this paper. The CSV file contains several lines per LOINC code, each line describing one of the various dimensions of the code. See Figure 3 for reference. Line 40-43 covers the LOINC code 100447-2 and shows the four related dimensions. We will be using the PartName field for each LOINC code to create our LOINC Bags of Words. LOINC code 110447-2, for instance, will have “Note Progress Outpatient Burn management”.
Figure 3:
The figure shows a few different LOINC Codes, each with their corresponding part numbers and part names defining their dimensions. The PartTypeName column specifies the exact dimension name for the given row.
Processing the string version of BOW is taxing and inefficient, which is why we now need to convert it into VSM compatible form. To define our vector space, we need a list of all the dimensions it can have. We will derive this list from the distinct values of PartName column of the DocumentOntology.csv file. This gives us a unique list of all the keywords used in LOINC DO, a dictionary of 520 terms. The vector will have 520 features, each representing a specific keyword from the LOINC dictionary. Following the VSM methodologies, the vector will contain a Boolean value of either 0 or 1 for any feature depending on weather the keyword appears in the LOINC PartName column or not. As an example, LOINC code 110447-2 from Figure 3 would have the value of 1 for the terms “Note”, “Progress”, “Outpatient”, “Burn”, “Management”, and a 0 for every other dimension of its vector. Once we repeat this method for every LOINC Code, we will have our LOINC BOW ready. This is a bit of a departure from standard practice where the value can be greater than 1 depending on the frequency of the term. However, using the frequency model introduced large amounts of incorrect matchings in our solution, and it was therefore left out. This error was caused by the use of multiple columns that sometimes contained the same information. This meant that the frequency was sometimes way higher than it should be due to the duplication of data in the database.
Before we create the LOINC BOW, we must follow some preprocessing steps to ensure maximum coverage. We followed some well-established preprocessing steps in the field13, such as case removal (making all text lower case), stop-word removal (removing words that don’t add semantical information such as ‘the’, ‘then’, ‘or’ etc…), and punctuation removal (for example, converting ‘wound,’ to ‘wound’) to improve our pipeline. We did not follow a few of lemmatization due to the added undue complexity without noticeable improvement in coverage. Lastly, there are some LOINC Codes that are now deprecated (such as 57140-6) or discouraged (80562-2) but still show up on the list; we took the time to identify these and removed them from our final list of BOW.
Notes’ Bag of Words
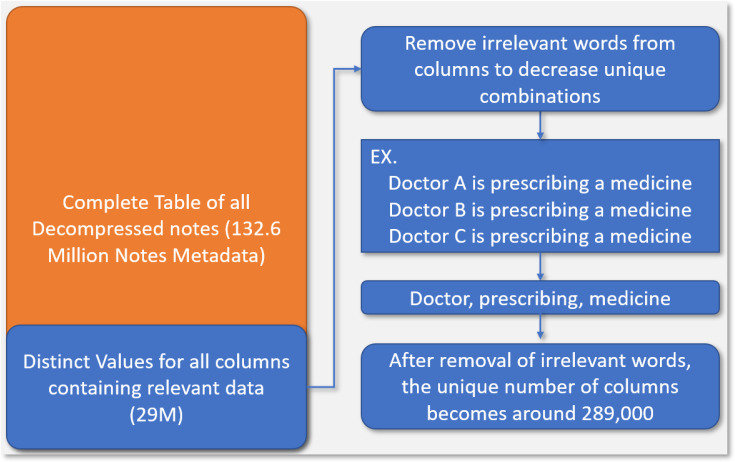
In order to create a BOW for individual notes, we utilized the seven columns that were identified earlier for the value- set mapping as these verifiably contained crucial information needed to match documents to LOINC codes. We could simply convert all of our documents to BOW, but this would be inefficient and unscalable for millions of notes. To optimize our pipeline, we decided to retrieve only the distinct combinations of the seven columns we needed, significantly reducing the problem space from over 130 million to around 29 million. However, there are still further steps we can take to prune the unique list substantially; There are many notes in our database that had very similar values, differing by a word or two, and this is the main cause of the 29 million unique rows. Consider for example the following two sentences: “Doctor A is prescribing a medicine to patient B,” and “Doctor C is prescribing a medicine to patient D”. The two statements are semantically similar and only differ in ways that do not affect its LOINC categorization. The two statements can be mapped to a single statement in the vein of “Doctor prescribing medicine patient”, while ensuring no loss of semantic information helpful for LOINC mapping. This cleanup step can be facilitated using our LOINC dictionary of 520 keywords, ensuring we do not remove any relevant word that may help match a clinical document to a LOINC code (We will see in the synonymy section that this is not entirely true). After the cleanup, we ended up with around 289,000 unique column combinations, which is computationally more tractable compared to the 130 million that we started out with. Finally, we also followed the same preprocessing steps here as in LOINC BOW section for coverage improvement. This complete process is illustrated in Figure 4.
Figure 4:
This figure visualizes the process of reducing the computational challenge of the pipeline. We start out with over 130 Million individual notes and get unique combinations of columns and remove irrelevant terms to decrease the computational complexity.
Synonymy List
To reduce complexity, the previous section described cleansing each document’s metadata using our generated LOINC dictionary. The intuition for this assumption is that if a metadata word is not in the LOINC dictionary, then it will offer no improvement to coverage by being included in a document’s BOW and can be safely removed. While this broadly makes sense for names and many words, it has many exceptions. For example, LOINC opts to use the term Diabetology instead of Diabetes. The issue stems from the fact that our metadata almost never mentions the word Diabetology, and instead uses Diabetes. Some other examples include discharge vs depart and procedure vs procedural where the former is used in LOINC while the latter is common in our Metadata. Due to words of similar ilk being missed by our above assumption, we created and maintained synonymy list that gets checked against each document’s metadata to maximize our coverage. The synonymy list has been built mainly from the knowledge base gained while doing the manual value-set mapping and has been pivotal in improving our pipeline’s Note-LOINC mapping coverage.
Match Condition
The implementation of BOW using VSM offers an elegant solution for matching different bags of words together. Vectors possess a property called Cosine Similarity (, where A and B are vectors and 𝜃 is the angle between them), which allows us to find how similar they are to each other using their dot products and magnitudes. We will be using a reduced version of this, specifically the dot-products of A and B. If we take two of our BOWS, which are internally vectors, and calculate their dot-products, the resulting value is equal to the count of words the two vectors share in common.
With that, we can now define a threshold for when a document can be acceptably matched to a LOINC code. Our pipeline uses an 85% threshold, meaning that a document must share at least 85% of the words in the LOINC BOW to be a valid match. If the result of the dot product between Note BOW and LOINC BOW is divided by the size of the LOINC BOW is greater than 0.85, then it will be noted down as a valid LOINC Code. This threshold means that the smaller the bag of words of a document, the stricter the matching algorithm is. For example, a bag with only five words, must match all five words before it can get an acceptable match, while a bag with 10 words can miss one word and still get accepted.
This threshold was selected after trial and error. We tested the program with a threshold value 0.75 and were getting many incorrect results. We tried 0.8 as well with little improvement in terms of accuracy. At 0.9, we saw more accurate results but in exchange we saw a severe drop in coverage. 0.85 was a good compromise between coverage and accuracy, resulting in far fewer incorrect mappings with minimal loss of coverage.
Results
We first undertook a manual mapping approach, where the team spent time mapping individual columns to individual LOINC dimensions and verifying the process. The initial results seemed pleasing, as we had mapped around 76% of our notes to at least two LOINC dimensions. Even with such a seemingly high coverage, however, mapping the notes to valid LOINC codes using the dimension values only gave us a coverage of around 23%. Table 1 summarizes the exact breakdown of these numbers. Given the primary task of evaluating the scalability of mapping millions of notes, sensitivity and specificity for a curated subset was not explored at this time.
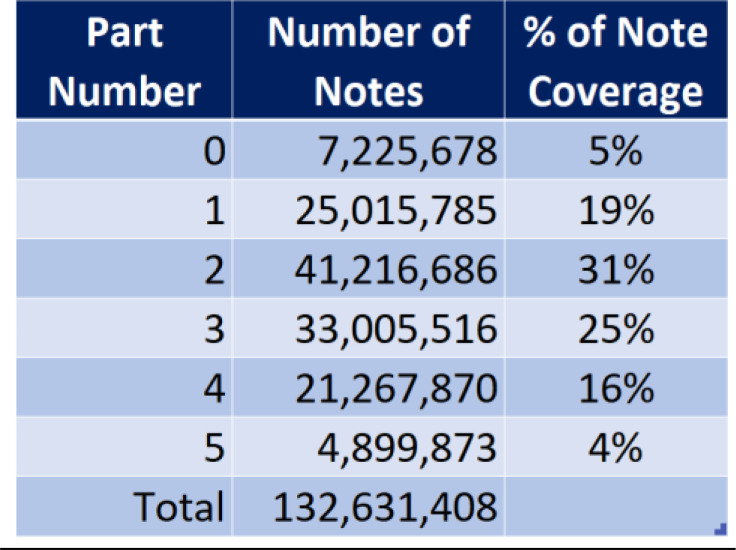
Table 1:
The table shows six levels of mapping to the LOINC DO axes, depending on the number of LOINC dimensions populated. 7 million notes which didn’t get mapped to any LOINC dimension, 25 million that mapped to exactly one LOINC dimension. Percentage coverage of notes mapped at each level are shown to the right.
We then looked to automation for improving the coverage and worked on the BOW approach. The initial result of the program came to around 38% coverage. The coverage decreased further with the removal of deprecated/discouraged LOINC codes, down to 27%.
Once we implemented the synonymy list, however, our coverage jumps substantially, first to 48% and then up to 73.4%. This occurred over a few weeks of analysis of the pipeline and careful accumulation of useful keywords.
Among the 26.6% of notes that did not have a LOINC code associated with them, nearly 10% is taken up by Phone messages and ‘Patient Portal Home Meds’ alone, followed by many other similarly low priority labels. In the Cerner EHR, fragments of text for when the patient updates their home medications list online are saved as independent text documents. There are also text fragments for portions of clinical documents (e.g. notes subjective, objective sections) that are saved redundantly along with the complete note and these form a bulk of the remaining 26.6%. There remains a gap of around 5-10% of our notes that are in need of mapping or exclusion but do not possess enough metadata for a valid categorization, and a still smaller percentage of our documents that do have enough data, but they are not caught by our synonymy list.
We further investigated the coverage of the documents by encounter types, focusing on outpatient, inpatient, and emergency notes specifically. For outpatient, there is 72% LOINC coverage. However, on further analysis we see that of the unmapped documents, over 40% are comprised of low priority Phone messages and “patient portal” notes, meaning our effective coverage is closer to around 85%. The same pattern follows with inpatient and emergency, but to a much lesser degree, implying that the main avenue of improvement likely lies in identifying patterns and gaps in these two categories and appropriately appending our synonymy list to close those gaps.
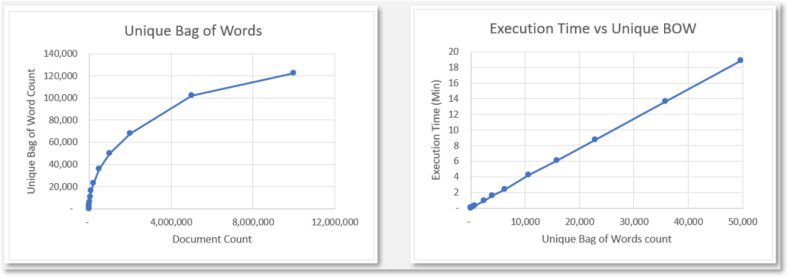
As an extension, some simple analysis was done for the complexity of the pipeline and to test its scalability for databases on a much larger scope. Figure 5 contains two graphs based on some test runs. The Unique Bag of Words graph has the count of Unique Bag of Words as its dependent variable and Document Count as the independent variable, and it shows the logarithmic growth of the Unique Bag of Words count in relation to the document count. The Execution Time vs Unique BOW graph shows the linear execution time, based solely on the number of Unique Bags of Words count.
Figure 5:
This figure shows two graphs: The left graph shows the logarithmic growth of the count of Unique BOW with respect to the actual document count and the right graph shows the linear runtime of the algorithm based on the count of Unique BOW.
Discussion
In this study, our goal was to create a generalized framework that can quickly and accurately map clinical documents to LOINC codes only through available metadata. The power of such a framework is limited based on the availability and quality of the metadata, but this also greatly reduces the complexity of the method, without sacrificing quality or speed.
Based on our results, we can say that our framework performs extremely well, given that it covers 73.4% of all our clinical notes. Looking at the remaining 26.6% of our notes in closer detail showed that a substantial number of them were phone messages, Portal notes, or some other type of outlier notes that are not the real focus of our initial mapping effort (These cases will be addressed in the future). While there are still many important notes/reports that are not getting mapped, it is only a matter of time before we identify them, gather their patterns, and expand our synonymy list to contain them.
The synonymy list has been quintessential in increasing our coverage for the LOINC Mapping. Over a matter of a few weeks, we identified a list of patterns that were not getting mapped and appended our synonym list with them, slowly improving our mapping as we went. However, this is also one of the main limitations of the program: the manual labor of maintaining a synonymy list is a non-trivial effort. While we attempted to find and use pre-existing synonymy lists such as the UMLS dictionary, we were unsuccessful in integrating it effectively into the program. However, while the synonymy list might seem like the manual annotation in ML methods, the synonymy list we created may be ported to other institutes, while the same is likely not true to the annotations in ML methodologies, where they’ll likely have to repeat the annotation steps. However, the goal will be to get rid of a local synonymy list in favor of something like the UMLS.
An aside on the validity of the program. At the time of writing, we were unable to perform formal validation for the program. However, a large part of the exercise to improve coverage using synonymy list involved repeatedly studying the results of the program, finding flawed mappings, and updating the synonymy lists to fix such errors. Validation is the next big step we are working towards and hopefully we will have plenty of results to share in a follow-up paper.
We made an important assumption in the creation of our framework, based on the LOINC documentation. LOINC mentions that while it makes a distinction between a report and a note, it will not create duplicate LOINC codes where the only difference is the note type. For example, consider LOINC code 11504-8 with the LOINC Name “Surgical Operation Note.” While the Name claims that it is a note, the description reads “note or report.” Our assumption dictates that to LOINC, notes and reports are the same, and this assumption is hard coded into our synonymy list. Should LOINC go back and amend its standing on this topic, the synonymy list will have to be updated.
The framework is also computationally scalable, running in logarithmic time based on note count. It takes under an hour to run on 100 million notes and will take no more than five hours on even a billion notes. While not an issue at UM Healthcare, many larger systems have billions of clinical documents where efficiency of this pipeline is desirable
This framework was created with simplicity and portability in mind so that health systems and academic medical centers can more easily make all their notes accessible via interoperable terminologies supported by research network’s common data models. That stands in contrast to other approaches of mapping a subset of notes for a predefined cohort. This framework can support the study feasibility use case by incorporating the document ontology into computable phenotyping tools like ATLAS and i2b2 where investigators often want to use whether a certain type of note exists as part of their search criteria.
Future directions for this effort include 1) current work to deploy the framework against sites using the Epic EHR in the Greater Plains Collaborative14 PCORnet Clinical Research Network and nationally; 2) align with the OMOP Common Data Model, and disseminate our software via GitHub; 3) revisit leveraging synonymy from sources available from the Unified Medical Language System supported by the National Library Of Medicine; 4) evaluating sensitivity and specificity with curated subsets of note types and clinical cohorts; 5) further refinement of detecting and blacklisting duplicative note fragments that may be impacting performance metrics; and 6) complementing this structured metadata approach with natural language processing or ChatGPT based methods that exploit the content of the document to resolve difficult classes of documents.
Conclusion
In this study, we developed a generalized framework, utilizing the Bag of Words approach, for rapidly mapping all clinical documents to LOINC DO codes using only the associated metadata. This framework has performed quite well at the University of Missouri and has mapped most of our important documents to a LOINC Code at 73.4% coverage, while also being general enough that it can be ported to other organizations through curation of the synonymy list.
Acknowledgements
This work is supported through a Patient-Centered Outcomes Research Institute (PCORI) Program Award RI-MISSOURI- 01-PS1.
Figures & Tables
References
- 1.Frazier P, Rossi-Mori A, Dolin RH, Alschuler L, Huff SM. The creation of an ontology of clinical document names . Studies in Health Technology and Informatics. 2001;84:94–8. [PubMed] [Google Scholar]
- 2.Hyun S, Ventura R, Johnson SB, Bakken S. Is the Health Level 7/LOINC document ontology adequate for representing nursing documents? Studies in health technology and informatics. 2006;122:527–31. [PubMed] [Google Scholar]
- 3.Hyun S, Shapiro JS, Melton G, Schlegel C, Stetson PD, Johnson SB, et al. Iterative evaluation of the health level 7-logical observation identifiers names and codes clinical document ontology for representing Clinical Document Names: A case report. Journal of the American Medical Informatics Association. 2009;16(3):395–9. doi: 10.1197/jamia.M2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong IC, Rijnbeek PR, Van Der Lei J. InMEDINFO 2015: eHealth-enabled Health. IOS Press; 2015. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers; pp. pp. 574–578. [PMC free article] [PubMed] [Google Scholar]
- 5.Qualls LG, Phillips TA, Hammill BG, Topping J, Louzao DM, Brown JS, Curtis LH, Marsolo K. Evaluating foundational data quality in the national patient-centered clinical research network (PCORnet®) Egems. 2018;6(1) doi: 10.5334/egems.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronson AR, Bodenreider O, Demner-Fushman D, Wah Fung K, Lee VK, Mork JG, et al. From indexing the biomedical literature to coding clinical text: experience with MTI and machine learning approaches. Proceedings of the Workshop on BioNLP 2007 Biological, Translational, and Clinical Language Processing. 2007. pp. 105–12.
- 7.Parr SK, Shotwell MS, Jeffery AD, Lasko TA, Matheny ME. Automated mapping of laboratory tests to LOINC codes using noisy labels in a National Electronic Health Record System Database. Journal of the American Medical Informatics Association. 2018;25(10):1292–300. doi: 10.1093/jamia/ocy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo X., Li J., Zhao B., Zhou Y., Dong X., Duke J., Natarajan K., Hripcsak G., Shah N., Banda J. M., Reeves R., Miller T., Xu H. Normalizing Clinical Document Titles to LOINC Document Ontology: an Initial Study. AMIA ... Annual Symposium proceedings. AMIA Symposium. 2021;2020:1441–1450. [PMC free article] [PubMed] [Google Scholar]
- 9.Shraboni Sarker, Md Kamruz Zaman Rana, Yahia Mohamed, Vasanti Mandhadi, Xing Song, Abu S, Mosa M., Waitman Russ, Rao Praveen. AMIA 2022 Annual Symposium. Washington, D.C.: 2022. “Mapping Clinical Notes to LOINC Document Ontology Using EHR Data.”. Poster. [Google Scholar]
- 10.Vreeman DJ, McDonald CJ. A Comparison of Intelligent Mapper and Document Similarity Scores for Mapping Local Radiology Terms to LOINC. AMIA Annual Symposium proceedings. 2006. pp. 809–13. [PMC free article] [PubMed]
- 11.Juluru K, Shih H-H, Keshava Murthy KN, Elnajjar P. Bag-of-words technique in natural language processing: A Primer for radiologists. RadioGraphics. 2021;41(5):1420–6. doi: 10.1148/rg.2021210025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pires R, Jelinek HF, Wainer J, Valle E, Rocha A. Advancing bag-of-visual-words representations for lesion classification in retinal images. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0096814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.HaCohen-Kerner Y, Miller D, Yigal Y. The influence of preprocessing on text classification using a bag-of-words representation. PLOS ONE. 2020;15(5) doi: 10.1371/journal.pone.0232525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waitman LR, Aaronson LS, Nadkarni PM, Connolly DW, Campbell JR. The Greater Plains Collaborative: a PCORnet Clinical Research Data Network. J Am Med Inform Assoc. 2014 Jul-Aug;21(4):637–41. doi: 10.1136/amiajnl-2014-002756. doi: 10.1136/amiajnl-2014-002756. Epub 2014 Apr 28. PMID: 24778202; PMCID: PMC4078294. [DOI] [PMC free article] [PubMed] [Google Scholar]