Abstract
The types of clinical notes in electronic health records (EHRs) are diverse and it would be great to standardize them to ensure unified data retrieval, exchange, and integration. The LOINC Document Ontology (DO) is a subset of LOINC that is created specifically for naming and describing clinical documents. Despite the efforts of promoting and improving this ontology, how to efficiently deploy it in real-world clinical settings has yet to be explored. In this study we evaluated the utility of LOINC DO by mapping clinical note titles collected from five institutions to the LOINC DO and classifying the mapping into three classes based on semantic similarity between note titles and LOINC DO codes. Additionally, we developed a standardization pipeline that automatically maps clinical note titles from multiple sites to suitable LOINC DO codes, without accessing the content of clinical notes. The pipeline can be initialized with different large language models, and we compared the performances between them. The results showed that our automated pipeline achieved an accuracy of 0.90. By comparing the manual and automated mapping results, we analyzed the coverage of LOINC DO in describing multi-site clinical note titles and summarized the potential scope for extension.
Introduction
There are numerous types of clinical notes, including progress notes, discharge summaries, consultation notes, medication orders, and lab test results, that are commonly used in clinical settings1. Although these note types generally serve the purpose of documenting patient information and facilitating communication among healthcare professionals, they can vary widely as different healthcare providers may have different needs and preferences when it comes to the types of notes they use and the information that should be included in each type of notes. In addition, existing commercial electronic health record (EHR) systems used by different sites have their own technical requirements and local customizations, making it even more challenging to retrieve and share clinical notes across sites2. The use of standardized note types helps to ensure that information is consistently and accurately recorded, making it easier for healthcare providers to collaborate effectively and make rational decisions about patient care. Moreover, when using clinical notes in EHR for real-world studies across multiple sites, it is critical to standardize note types, so that the same types of clinical data can be collected from each site, to reduce potential inconsistency among participating sites. For this reason, the OHDSI (Observational Health Data Science and Informatics) consortium has proposed to formalize note types in the OMOP Common Data Model (CDM)3.
Over the last two decades, we have seen much progress in developing ontologies to provide standard representations of clinical note types. Logical Observation Identifiers Names and Codes (LOINC) is one of the most widely used universal standards for identifying medical laboratory results as well as other observations. The LOINC Document Ontology (DO) was created as a subset of LOINC that consistently describes the main characteristics of various types of clinical documents4. The LOINC DO defines and categorizes key attributes of clinical documents in five axes: Type of Service, Kind of Document, Setting, Role, and Subject Matter Domain. Each axis maintains a set of standardized terms in a poly-hierarchical structure. By assembling terms from multiple axes, a LOINC code from the Document Ontology can cover multiple aspects of one clinical document. For example, the LOINC DO code “100488-6” (Long Common Name: Neurology Outpatient Progress note) is a combination of terms from four of the DO axes: “Neurology” from Subject Matter Domain, “Outpatient” from Setting, “Progress” from Type of Service, and “note” from Kind of Document. The latest version of the LOINC DO has a total number of 3,894 codes. They are all indexed under the class “DOC.ONTOLOGY”. Since the release of LOINC DO, several studies have investigated the feasibility of standardizing clinical documents using this ontology5–7. Most research found that the LOINC DO has reasonable coverage on standalone EHR systems but lacks detailed representations on certain document types such as nursing notes. There are also studies that focused on extending the LOINC DO by aggregating it with terms or concepts from other ontologies8–11. The Role, Setting, and Subject Matter Domain are three axes that researchers believed should be extended.
Despite the efforts in assessing and extending LOINC Document Ontology and manually mapping from standalone EHR systems to LOINC DO codes, there is no existing application that can automatically map clinical note types from multiple health providers to the LOINC DO. In 2020 we conducted a preliminary study that evaluated the generalizability of LOINC DO axes across five institutions and developed an automated system that extracts information that aligns the definitions of LOINC DO axes from clinical note titles12. However, we did not map the note titles to the specific LOINC code. In this study, we would like to extend our research further and propose an automated approach that maps a clinical note title to a LOINC code in the Document Ontology, without accessing the content of the clinical note. For example, given a note title “Clinical Note - Infectious Disease”, our goal is to assign the most specific LOINC DO code (34782-3, with the Long Common Name - “Infectious disease Note” in this case) that best describes the note title. As we found that LOINC DO had limited coverage on certain note titles, we would also like to automatically detect note titles that cannot be mapped to any of the LOINC DO codes. This can be achieved by developing an entity linking pipeline that compares the semantic similarity between the note title and available LOINC DO codes and links the note title to its best match.
Having determined the main scope for our extended study, we decide to interpret this problem as an entity linking (or concept normalization) task: the input is an entire note title, and the output is a LOINC DO code. Conventional entity linking tasks can be divided into two steps: candidate retrieval and candidate ranking13. Previous entity linking studies often rely on lexical-based methods such as TF-IDF and BM25 to retrieve and rank relevant candidates from the knowledge base, then apply Learning to Rank algorithms, such as RankNet and LambudaRank, to re-rank the retrieved candidates14,15. Nevertheless, clinical note titles can be brief, confusing, and ambiguous without knowledge of note content and other metadata. More recently transformer-based large language models (LLM) have become the mainstream of natural language processing (NLP). We can easily load these pre-trained LLMs and then fine-tune them on a certain downstream task without spending substantial time and effort on feature engineering. The Bidirectional Encoder Representations from Transformers (BERT) is one of the most prominent pre-trained LLMs that can be fine-tuned on various downstream NLP tasks such as information retrieval and entity linking16. It uses a transformer encoder that can read text input sequentially from both directions and learns contextual word representations through two training tasks: masked language modeling and next-sentence prediction. Using BERT as the backbone, several domain-specific pre-trained LLMs, namely BioBERT, PubMedBERT, and ClinicalBERT have been released to overcome the challenges in clinical NLP tasks17–19, including concept normalization tasks20. They are pre-trained on large-scale biomedical text and show impressively higher performances on diverse clinical NLP tasks, such as named entity recognition, relation extraction, and concept normalization21–23. Given the prospects of such LLMs in previous clinical NLP studies, it is non-trivial to apply these LLMs and develop advanced transformer-based approaches for this entity linking problem.
Therefore, the objectives of this study are to (1) analyze the generalizability and limitations of the LOINC DO by mapping clinical note titles from five institutions to LOINC DO codes; (2) design and implement an automated pipeline with different transformer-based LLMs for mapping clinical note titles to LOINC DO codes. To achieve these goals, we used 18,075 clinical document types from five medical centers, implemented approaches to link note titles to LONIC DO codes, and quantitatively evaluated their performance across five institutions. Our best-performing LLM deployed in this standardization pipeline achieved a mapping accuracy of 0.90, outperforming the traditional algorithms by 17%. We believe this study will bring innovations for clinical note type standardization in real-world clinical settings and support information exchange for clinical research.
Methods
Dataset
Similar to our previous study12 we used clinical note titles retrieved from the following five institutions: Boston Children’s Hospital (BCH), Vanderbilt University Medical Center (VUMC), Stanford University Medical School (SUMS), The University of Texas Health Science Center at Houston (UTHealth), and Columbia University Medical Center (CUMC).
We manually mapped 800 note titles to LOINC DO codes and classified the mapping between the note title and LOINC codes into three categories with respect to semantic similarity. We randomly sampled 160 note titles from each institution, totaling 800 note titles. For each clinical note title, we aim to assign a unique LOINC code that best describes the note title semantically. In order to create our gold standard, we randomly sampled 800 note titles and manually mapped these note titles to LOINC DO codes by comparing them to the Long Common Name of the LOINC DO codes. Table 1 lists the three types of mapping categories we define in this study, along with their definitions and mapping examples. For mappings in the fuzzy mapping category, it is very likely that a clinical note title can be mapped to multiple LOINC DO codes. Given the complexity of fuzzy mapping, we developed fuzzy mapping with the following rules:
We try to map the note title to a LOINC code that covers the information without ambiguity. For example, for the note title “Emergency Department Patient Education note”, we map to its closest match “Emergency department Education note” (LOINC code:78273-0).
When a note title can be mapped to multiple LOINC DO codes, we map to the code that covers the greatest number of attributes. For example, for the note title “General Surgery Outpt H & P / Init Cons”, there are two options: “Surgery Outpatient History and physical note” (LOINC Code: 84019-9) and “Surgery Consult note” (LOINC Code: 34847-4). We choose “Surgery Outpatient History and physical note” in this case as it contains four attributes from four LOINC DO axes respectively (i.e., Surgery from Subject Matter Domain, Outpatient from Setting, History and physical from Type of Service, and note from Kind of Document), while the other code only has attributes from three axes (i.e., Surgery from Subject Matter Domain, Consult from Type of Service, and note from Kind of Document).
Table 1.
Examples and definitions of three types of mapping. “N/A” stands for “Not Applicable”.
| Mapping Category | Definition | Note Title | LOINC DO Code | Long Common Name | |
|---|---|---|---|---|---|
| Exact Mapping | The note title can be mapped to a LOINC DO code without any disambiguation. | NUTRITION DIETETICS EDUCATION NOTE | 78451-2 | Nutrition and dietetics | Education note |
| Fuzzy Mapping | The note title can be mapped to a LOINC DO code, but some information is missing or different. | Patient Result Letter Orthopedics | 68585-9 | Orthopaedic surgery Hospital Letter | |
| No Mapping | The note title cannot be mapped to any of the LOINC codes in the DO. | Master Problem List | N/A | N/A |
Pipeline Workflow
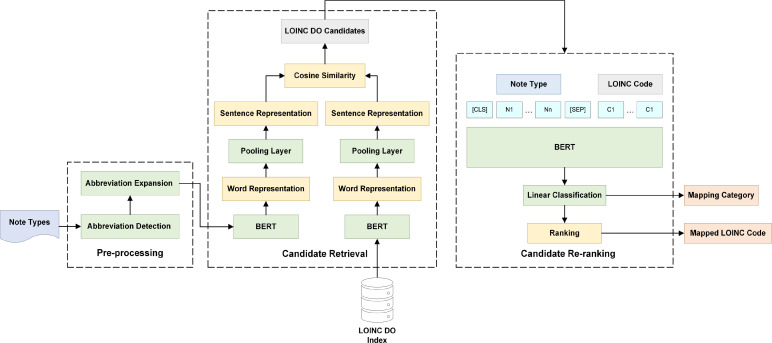
In this study, we follow the typical entity linking procedure and implement transformer-based architectures in both steps. Figure 1 below demonstrates the workflow of our mapping pipeline. It consists of three major modules: preprocessing, candidate retrieval, and candidate re-ranking. Given a note title, the candidate retrieval component compares the semantic similarity between the note title and all LOINC DO codes and generates a list of candidate LOINC DO codes that may be matched with the note title. During the candidate re-ranking process, we re-rank the list of candidates and sort the retrieved candidates by the ranking score, then classify the mapping into one of the three categories we defined in this study. If the note title-candidate pair was classified as “Exact Mapping” or “Fuzzy Mapping”, we return the LOINC code that has the highest-ranking score as the best match. If the note title-candidate pair is classified as “No Mapping”, we return only the note title. We describe the structure of each component in the following sub-sections.
Figure 1.
The workflow of the LOINC mapping pipeline.
Pre-processing: We used a popular biomedical abbreviation detection toolkit to identify and convert medical abbreviations in note titles24. When an abbreviation is detected, we replace the abbreviated word or phrase with its full-length text and use the de-abbreviated note title as the input for both candidate retrieval and candidate re-ranking steps.
Candidate retrieval: We adapt the model architectures from Sentence Transformers and construct a bi-encoder for the candidate retrieval component25. We first generate the word representations for both the note title and the Long Common Names of all LOINC DO codes. The word representations are fed into a pooling layer and converted into sentence representations. After this conversion, the sentence representations of the note title and the LOINC DO code will be two vectors of the same size. For each note title, we compare its sentence representations with representations of all LOINC DO codes by cosine similarity. In the end, we retrieve the top 10 LOINC DO codes that have the highest semantic similarity when compared with the clinical note title we wish to map.
Candidate re-ranking: We deploy a cross-encoder architecture for the candidate re-ranking component. Each time we form a note title-code sequence pair as the input. The first sequence is a clinical note title and the second sequence is the Long Common Name of a LOINC DO candidate. The two sequences are first tokenized and separated by the [SEP] token. We also add a [CLS] token at the beginning of the first sequence, indicating the start of the sentence pair. The sequence pair vector of the cross-encoder is forwarded to a linear classification layer to compute the ranking score as the class of this mapping.
Experiments and evaluation
Bi-encoder training: To optimize the accuracy in the candidate retrieval step, we fine-tune bi-encoders with several semantic textual similarity (STS) benchmark datasets. Prior to fine-tuning, we normalize the similarity scores of sentence pairs in the benchmark datasets from [0, 5] to [0, 1]. The bi-encoders are initialized with four types of LLMs: BERT-base, BioBERT-base, PubMedBERT-fulltext-uncased, and Bio_ClinicalBERT. The BERT-base model is developed by and pre-trained on BookCorpus and English Wikipedia17. It has 12 hidden layers with over 110 million parameters. Initialized with the embedding from BERT, the BioBERT model is pre-trained on PubMed abstracts and full-text articles18. On the other hand, the PubMedBERT model is a domain-specific model pre-trained from scratch with PubMed abstracts and full-text articles26. Unlike BioBERT which uses the same vocabulary list as the original BERT model, PubMedBERT uses a unique vocabulary list developed only from biomedical text. The Bio_ClinicalBERT model is initialized with the BioBERT embedding and pre-trained on over 2 million clinical notes from the MIMIC-III database27. We split the fine-tuning process into two phases. We first fine-tune bi-encoders on the open-domain STS Benchmark, which is a selection of English sentence pairs used in the SemEval Challenges from 2012 to 201728. It has 8,628 sentence pairs in total. We then further fine-tune three clinical STS benchmark datasets: ClinicalSTS 2018, ClinicalSTS 2019, and BIOSSES. The ClinicalSTS 2018 has 1,068 sentence pairs generated from EHRs at the Mayo Clinic’s data warehouse29. It was first released and used in the BioCreative/OHNLP Challenge in 2018. The ClinicalSTS 2019 dataset was first released in the n2c2/OHNLP Challenge in 2019 and added 1,006 new sentence pairs30. The BIOSSES contains 100 sentence pairs generated from PubMed Central Open Access Dataset31.
Cross-encoder training: We fine-tune cross-encoders with our gold standard mappings. Similar to the bi-encoder training process, we initialize the four types of word embedding. For each round of cross-validation, we use 8 folds for training, 1 fold for validation, and 1 fold for testing. For each fold, we randomly sample 16 note titles from each institution, making up 80 note titles in total.
Experimental setup and hyperparameters: For fine-tuning, most model hyperparameters were set as the default in the pre-trained model, except for the batch size and the number of training epochs. In this study, we tune the batch size with 16 and 32, the number of training epochs from 4 to 20, and select the model with the best performance. We also implement the Okapi BM25 model as our baseline throughout the experiments. In the baseline implementation, we retrieve and rank candidates based on BM25 scores without the re-ranking step.
Evaluation metrics: We evaluate the performance of different language models with 10-fold cross-validation. The evaluation metric is accuracy, which we define as the percentage of note titles that were correctly mapped. A mapping is considered correct if it fulfills the two following conditions at the same time:
The note title is correctly mapped to the LOINC DO code annotated in the gold standard.
The predicted mapping category is the same as in the gold standard.
For note titles that cannot be mapped, we consider the prediction is correct if its note title-code pair is classified into the “No Mapping” category.
Results
The total number of clinical note titles extracted from the EHR system of each institution, as well as the distribution of different types of gold standard mapping, are listed in Table 2. All clinical note titles are very short, as the mean lengths of note titles from all institutions are less than 10 tokens. We can see that only a small number of note titles can be mapped to an active LOINC DO code without any ambiguity, and the percentage of such exact mapping varies heavily between institutions. The note titles of Vanderbilt University Medical Center have a high percentage of exact mapping, while note titles from Stanford University Medical School are more difficult to map. Most mappings between the note title and the LOINC DO code are classified into the “Fuzzy Mapping” category.
Table 2.
The number of note titles retrieved and the number of note titles we manually mapped in the study.
| Institution Name | No. of Note Titles | Mean Value of No. of Tokens | Exact Mapping | Fuzzy Mapping | No Mapping |
|---|---|---|---|---|---|
| BCH | 7400 | 4.3 | 10 (6.3%) | 137 (85.6%) | 13 (8.1%) |
| Columbia | 881 | 5.7 | 11 (6.9%) | 133 (83.1%) | 16 (10%) |
| Stanford | 3232 | 6.2 | 5 (3.1%) | 131 (81.9%) | 24 (15%) |
| UTHealth | 3128 | 5.1 | 27 (16.9%) | 125 (78.1%) | 8 (5%) |
| Vanderbilt | 3434 | 4.6 | 38 (23.8%) | 120 (75%) | 2 (1.2%) |
Table 3 shows the evaluation results for all combinations of LLMs. The overall accuracy is calculated as a micro average of the accuracies of different mapping categories. We are unable to predict the mapping categories for BM25. We evaluate the overall accuracy by calculating the micro average of two categories that indicate valid mapping. From the results, we can imply that the overall accuracy is mainly determined by the results of fuzzy mapping. Surprisingly, the combination of PubMedBERT and BioBERT, which are two LLMs pre-trained on biomedical literature as opposed to clinical notes, shows the highest performance (Overall Accuracy = 0.90). However, using the Bio_ClinicalBERT model in the candidate retrieval step can bring high performance in the two minority classes: exact mapping and no mapping. We can also observe that using PubMedBERT in the candidate retrieval step can generally achieve high performance. However, if we consider only the exact mappings, applying the original BERT model achieves the highest accuracy of 0.95. Overall, the transformer-based pipeline with LLMs improves the mapping accuracy greatly, particularly in the fuzzy mapping category.
Table 3.
Mapping accuracy of different bi-encoder and cross-encoder LLM ensembles. The first model mention denotes the word embedding initialized for the bi-encoder in the candidate retrieval step, while the second model mention denotes the word embedding initialized for the cross-encoder in the candidate re-ranking step.
| Model Ensemble | Exact Mapping | Fuzzy Mapping | No Mapping | Overall |
|---|---|---|---|---|
| BM25 Only | 0.92 | 0.71 | N/A | 0.73 |
| BM25 + BERT | 0.95 | 0.80 | 0.65 | 0.80 |
| BERT + BERT | 0.95 | 0.83 | 0.78 | 0.84 |
| BERT + BioBERT | 0.93 | 0.85 | 0.81 | 0.85 |
| BERT + PubMedBERT | 0.95 | 0.84 | 0.83 | 0.85 |
| BERT + Bio_ClinicalBERT | 0.92 | 0.82 | 0.83 | 0.84 |
| BioBERT + BERT | 0.91 | 0.84 | 0.79 | 0.84 |
| BioBERT + BioBERT | 0.93 | 0.85 | 0.81 | 0.86 |
| BioBERT + PubMedBERT | 0.92 | 0.86 | 0.84 | 0.87 |
| BioBERT + Bio_ClinicalBERT | 0.93 | 0.85 | 0.79 | 0.86 |
| PubMedBERT + BERT | 0.95 | 0.88 | 0.86 | 0.89 |
| PubMedBERT + BioBERT | 0.93 | 0.90 | 0.84 | 0.90 |
| PubMedBERT + PubMedBERT | 0.93 | 0.87 | 0.86 | 0.88 |
| PubMedBERT + Bio_ClinicalBERT | 0.92 | 0.87 | 0.89 | 0.88 |
| Bio_ClinicalBERT + BERT | 0.95 | 0.85 | 0.83 | 0.86 |
| Bio_ClinicalBERT + BioBERT | 0.92 | 0.87 | 0.86 | 0.87 |
| Bio_ClinicalBERT + PubMedBERT | 0.93 | 0.87 | 0.90 | 0.88 |
| Bio_ClinicalBERT + Bio_ClinicalBERT | 0.92 | 0.86 | 0.89 | 0.87 |
We also evaluated the performances of different model ensembles at the institutional level. Table 4 lists the best model ensembles for each institution and the overall accuracy at the institutional level. By comparing the institutional level accuracy with the corresponding accuracy across all institutions in Table 3 above. We find the performances of note title mapping from UTHealth and VUMC are slightly higher than the overall results (UTHealth VS Overall: 0.87 > 0.86, VUMC VS Overall: 0.91 > 0.85), while the performance for the other three institutions are lower than the corresponding overall results. We can also notice that the BERT model achieves the highest performance for mapping note titles from VUMC, while mapping note titles from the rest of the institutions need domain-specific LLMs to achieve higher performances.
Table 4.
Best-performing models and corresponding mapping accuracy for clinical note titles from each institution.
| Institution Name | Best Model Ensemble | Accuracy |
|---|---|---|
| BCH | BioBERT + Bio_ClinicalBERT | 0.85 |
| CUMC | PubMedBERT + BioBERT | 0.85 |
| SUMC | PubMedBERT + Bio_ClinicalBERT | 0.86 |
| UTHealth | BioBERT + BioBERT | 0.87 |
| VUMC | BERT + BioBERT | 0.91 |
Discussions
In this study, we designed, implemented, and evaluated a transformer-based NLP pipeline that maps clinical document titles to the LOINC codes in the Document Ontology. This is the first attempt to automatically standardize clinical document types across institutions without accessing the document contents. From the analysis of manual mapping and automatic mapping results, we learn that the LOINC Document Ontology has a relatively high coverage over clinical note titles from multiple sites and potentially can be used as a universal standard for clinical note type normalization.
Overall, a transformer-based pipeline considerably improves the mapping accuracy, compared with traditional lexical-based methods. By assessing the mapping accuracy with different LLMs, we found that domain-specific LLMs, especially the PubMedBERT model, can generally achieve high performance for this note title mapping tasks, including mapping note titles to LOINC DO codes, and detecting note titles that cannot be mapped. Despite that the Bio_ClinialBERT model is pre-trained on clinical notes, it did not show better performance than other domain-specific language models. It may be because the notes for pre-training are all from the intensive care unit and the model doesn’t learn enough contextual diversity across different note titles. The best-performing model also slightly varies by institution. If the note title dataset has fewer note titles that can be mapped without any ambiguity, and more note titles that cannot be mapped, like note titles from Stanford University Medical School and Columbia University Medical Center, domain-specific LLMs will be helpful to improve the mapping accuracy. On the other hand, for those institutions that have more note titles that align better with the LOINC DO, like Vanderbilt University Medical Center and UTHealth, generic language models like the original BERT model can achieve relatively high accuracy. Although the findings validate the hypothesis that transformer-based LLMs can enhance automated mapping note titles with the LOINC DO, we are unable to recommend a universally effective LLM that functions well across sites. We will continuously strengthen generalizability of the pipeline as we try to scale this study up to distributed EHR systems that millions of clinical notes.
When developing our gold standard mapping, there are note titles that we can map very vaguely. For example, for the note title “Feet – Both, 3 Views or More-Lex”, its closest match is “Radiology Note” (LOINC Code: 75490-3). However, such LOINC codes can be difficult to retrieve based on semantic similarity only. More importantly, the portion of such in our dataset varies heavily across different sites. We did a string search and found that such note titles make up approximately 50% of the note titles retrieved from Stanford University Medical School. In contrast, there are less than 4% of note titles of similar patterns in the data from UTHealth and VUMC. This largely relates to the different documenting standards of multiple sites, as well as the range of clinical notes we can access in EHR systems. Therefore, we need a standardized way to retrieve clinical note titles from different EHR systems. Another issue that needs more investigation is to reduce the ambiguity in fuzzy mapping. Although we made specific rules for fuzzy mapping following our mapping definitions, there were note titles we found it difficult to decide on its mapping. For example, the note title “Cardiothoracic and Vascular Surgery Consult” can be mapped to either “Cardiothoracic surgery Consult note” (LOINC Code: 34849-0) or “Vascular surgery Consult note” (LOINC Code: 34853-2), as it did not specify the exact clinical specialty. Since a LOINC DO code only allows one attribute to be included for each of the Document Ontology axes, we are unable to find a perfect match that aggregates two attributes from the Subject Matter Domain axis. Such ambiguities in mapping are also reflected in our prediction errors. We believe that the number of exact mappings will increase considerably if a LOINC DO code is allowed to incorporate multiple attributes from the same DO axes. Moreover, it also indicates that an accurate mapping of a LOINC code needs to go beyond its title, e.g., to include the actual content in the note and structured fields in EHRs (e.g., specialty and roles of note writers).
Given the performance in fuzzy mapping, there is still space for improvement in our pipeline design. First, in the preprocessing step, we use only a tool for medical abbreviation detection. It is possible that the abbreviation detection tool cannot identify and convert some from certain healthcare providers, which will become a bottleneck for further performance improvement. Furthermore, we use a fully supervised approach in pipeline development, which requires a large amount of gold standard annotations. However, annotating and classifying the mapping is expensive in terms of time and labor. So far we only mapped 800 note titles, while we have more than 18,000 clinical note titles in total. We would like to investigate the possibility of a semi-supervised approach utilizing unlabeled note titles. One possible way is to create a silver corpus by using a fine-tuned cross-encoder and predicting the matched LOINC codes for unlabeled note titles32. Then we can add the silver corpus to the training process of the candidate retrieval component. Nevertheless, the success of this proposed semi-supervised method is under the assumption that the cross-encoder is a well pre-trained, high-performing model. Given the amount of data we used for training, our current cross-encoders still need to be improved before using it for silver corpus generation. Additionally, how to sample from the silver corpus is an important question. There is a risk that the pipeline performance will decrease when we add a large amount of silver data of low quality. We hope to release the pipeline for public use and continuously refine it by addressing the limitation mentioned above.
Conclusions
In this study, we developed and evaluated an NLP pipeline to map clinical note titles from multiple institutions to LOINC DO codes. Our results show that the transformer-based mapping pipeline, powered by multiple LLMs achieved good performance on clinical note title mapping across five institutions, compared with the baseline model. Additionally, the domain-specific LLMs pre-trained on large-scale biomedical text can further improve the mapping accuracy. We aim to release the pipeline for public use and hope it can facilitate textual data standardization in real world studies that use EHRs data (e.g., the All of Us initiative)33.
Acknowledgments
This study is partially supported by the following grants: NIA 1RF1AG072799, NIA 1R01AG080429, NIH U2C OD023196, and PCORI RI-MISSOURI-01-PS1.
Conflict of Interest
Dr. Hua Xu has research-related financial interests in Melax Technologies, Inc.
Figures & Tables
References
- 1.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012 Jun;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 2.Apathy NC, Vest JR, Adler-Milstein J, Blackburn J, Dixon BE, Harle CA. Practice and market factors associated with provider volume of health information exchange. Journal of the American Medical Informatics Association. 2021 Jul 14;28(7):1451–60. doi: 10.1093/jamia/ocab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Observational Health Data Sciences and Informatics (OHDSI)) Standardized Data: The OMOP Common Data Model [Internet] [cited 2023 Mar 19]. Available from: https://www.ohdsi.org/data-standardization/
- 4.Regenstrief Institute, Inc. LOINC Document Ontology [Internet] LOINC Document Ontology. 2023 [cited 2023 Mar 18]. Available from: https://loinc.org/document-ontology/
- 5.Li L, Morrey CP, Baorto D. Cross-mapping clinical notes between hospitals: an application of the LOINC Document Ontology. AMIA Annu Symp Proc. 2011;2011:777–83. [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ES, Melton GB, Engelstad ME, Sarkar IN. Standardizing Clinical Document Names Using the HL7/LOINC Document Ontology and LOINC Codes. AMIA Annu Symp Proc. 2010 Nov 13;2010:101–5. [PMC free article] [PubMed] [Google Scholar]
- 7.Hyun S, Bakken S. Toward the creation of an ontology for nursing document sections: mapping section names to the LOINC semantic model. AMIA Annu Symp Proc. 2006;2006:364–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Pakhomov S, Dale JL, Chen ES, Melton GB. Application of HL7/LOINC Document Ontology to a University-Affiliated Integrated Health System Research Clinical Data Repository. AMIA Jt Summits Transl Sci Proc. 2014;2014:230–4. [PMC free article] [PubMed] [Google Scholar]
- 9.Rajamani S, Chen ES, Akre ME, Wang Y, Melton GB. Assessing the adequacy of the HL7/LOINC Document Ontology Role axis. Journal of the American Medical Informatics Association. 2015 May 1;22(3):615–20. doi: 10.1136/amiajnl-2014-003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajamani S, Chen ES, Wang Y, Melton GB. Extending the HL7/LOINC Document Ontology Settings of Care. AMIA Annu Symp Proc. 2014;2014:994–1001. [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves RM, FitzHenry F, Brown SH, Kotter K, Gobbel GT, Montella D, et al. Who said it? Establishing professional attribution among authors of Veterans’ Electronic Health Records. AMIA Annu Symp Proc. 2012;2012:753–62. [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo X, Li J, Zhao B, Zhou Y, Dong X, Duke J, et al. Normalizing Clinical Document Titles to LOINC Document Ontology: an Initial Study. AMIA Annu Symp Proc. 2020;2020:1441–50. [PMC free article] [PubMed] [Google Scholar]
- 13.French E, McInnes BT. An overview of biomedical entity linking throughout the years. Journal of Biomedical Informatics. 2023 Jan;137:104252. doi: 10.1016/j.jbi.2022.104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson S, Zaragoza H. The Probabilistic Relevance Framework: BM25 and Beyond. FNT in Information Retrieval. 2009;3(4):333–89. [Google Scholar]
- 15.Liu T-Y. Learning to Rank for Information Retrieval. FNT in Information Retrieval. 2007;3(3):225–331. [Google Scholar]
- 16.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv:181004805 [cs] [Internet] 2019 May 24 [cited 2021 Dec 14]. Available from: http://arxiv.org/abs/1810.04805.
- 17.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. 2018 [cited 2023 Mar 18]. Available from: https://arxiv.org/abs/1810.04805.
- 18.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. Wren J, editor. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020 Feb 15;36(4):1234–40. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang K, Altosaar J, Ranganath R. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. 2019 [cited 2023 Mar 18]. Available from: https://arxiv.org/abs/1904.05342.
- 20.Luo Y-F, Henry S, Wang Y, Shen F, Uzuner O, Rumshisky A. The 2019 n2c2/UMass Lowell shared task on clinical concept normalization. Journal of the American Medical Informatics Association. 2020 Oct 1;27(10):1529–e1. doi: 10.1093/jamia/ocaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, et al. Deep learning in clinical natural language processing: a methodical review. Journal of the American Medical Informatics Association. 2020 Mar 1;27(3):457–70. doi: 10.1093/jamia/ocz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tutubalina E, Kadurin A, Miftahutdinov Z. Proceedings of the 28th International Conference on Computational Linguistics [Internet] Barcelona, Spain (Online): International Committee on Computational Linguistics; 2020 [cited 2023 Mar 19]. Fair Evaluation in Concept Normalization: a Large-scale Comparative Analysis for BERT-based Models; pp. p. 6710–6. Available from: https://aclanthology.org/2020.coling-main.588. [Google Scholar]
- 23.Ji Z, Wei Q, Xu H. BERT-based Ranking for Biomedical Entity Normalization. AMIA Jt Summits Transl Sci Proc. 2020 May 30;2020:269–77. [PMC free article] [PubMed] [Google Scholar]
- 24.Sohn S, Comeau DC, Kim W, Wilbur WJ. Abbreviation definition identification based on automatic precision estimates. BMC Bioinformatics. 2008 Dec;9(1):402. doi: 10.1186/1471-2105-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reimers N, Gurevych I. Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. 2019 [cited 2023 Mar 18]. Available from: https://arxiv.org/abs/1908.10084.
- 26.Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, et al. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans Comput Healthcare. 2022 Jan 31;3(1):1–23. [Google Scholar]
- 27.Alsentzer E, Murphy JR, Boag W, Weng W-H, Jin D, Naumann T, et al. Publicly Available Clinical BERT Embeddings. 2019 [cited 2023 Mar 18]. Available from: https://arxiv.org/abs/1904.03323.
- 28.Cer D, Diab M, Agirre E, Lopez-Gazpio I, Specia L. Proceedings of the 11th International Workshop on Semantic Evaluation (SemEval-2017) [Internet] Vancouver, Canada: Association for Computational Linguistics; 2017 [cited 2023 Mar 18]. SemEval-2017 Task 1: Semantic Textual Similarity Multilingual and Crosslingual Focused Evaluation; pp. p. 1–14. Available from: https://aclanthology.org/S17-2001. [Google Scholar]
- 29.Yanshan Wang, Afzal N, Sijia Liu, Rastegar-Mojarad M, Liwei Wang, Feichen Shen, et al. Overview of BioCreative/OHNLP Challenge 2018 Task 2: Clinical Semantic Textual Similarity. 2018 [cited 2023 Mar 18]. Available from: http://rgdoi.net/10.13140/RG.2.2.26682.24006.
- 30.Wang Y, Fu S, Shen F, Henry S, Uzuner O, Liu H. The 2019 n2c2/OHNLP Track on Clinical Semantic Textual Similarity: Overview. JMIR Med Inform. 2020 Nov 27;8(11):e23375. doi: 10.2196/23375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soğancıoğlu G, Öztürk H, Özgür A. BIOSSES: a semantic sentence similarity estimation system for the biomedical domain. Bioinformatics. 2017 Jul 15;33(14):i49–58. doi: 10.1093/bioinformatics/btx238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakur N, Reimers N, Daxenberger J, Gurevych I. Augmented SBERT: Data Augmentation Method for Improving Bi-Encoders for Pairwise Sentence Scoring Tasks. 2020 [cited 2023 Mar 18]. Available from: https://arxiv.org/abs/2010.08240.
- 33.All of Us Research Program Investigators. Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, et al. The “All of Us” Research Program. N Engl J Med. 2019 Aug 15;381(7):668–76. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]