Abstract
We used competitive panning to select a panel of 10 different human antibodies from a large semisynthetic phage display library that distinguish between serum complement-resistant and complement-sensitive strains of the gram-negative diplococcus Moraxella (Branhamella) catarrhalis. Western blotting analyses and inhibition enzyme-linked immunosorbent assays showed that all phage antibodies were directed against the same or closely spaced epitopes on the target protein, which is the high-molecular-weight outer membrane protein (HMW-OMP) of M. catarrhalis. HMW-OMP was found in multiple isolates of complement-resistant but not complement-sensitive M. catarrhalis strains. Nucleotide sequence analysis demonstrated that the immunoglobulin heavy- and light-chain variable-region genes encoding the 10 phage antibodies were remarkably similar, with a strong preference for basic amino acid residues in the heavy-chain CDR3 regions. This is the first report showing that competitive panning is a successful procedure to obtain phage antibodies against differentially expressed structures on phenotypically dissimilar strains of prokaryotic cells.
The display of single-chain Fv (scFv) or Fab antibody fragments on the surface of filamentous phage particles and the selection of recombinant phages by binding to a target antigen provide a novel means of isolating antibodies with predetermined specificity (for reviews, see references 5 and 39). Libraries may be assembled from the variable (V) regions expressed by B lymphocytes from either an individual with a particular immune response or a nonimmunized individual in an attempt to recruit the diversity generated by the natural immune system. An alternative approach to create diverse libraries exploits the use of large collections of cloned V genes to which randomized CDR3 regions are fused in vitro by PCR. We recently constructed a large semisynthetic phage library of human scFv fragments with partially randomized heavy-chain CDR3 regions, designed to encode a high frequency of functional antibody molecules (8).
Selection of phage antibodies (PhAbs) of desired specificities is conventionally performed by panning of libraries on solid-phase-coated antigens and eluting bound phages with high- or low-pH buffers. Alternative strategies have also been successfully used. For example, PhAbs have been obtained by direct selection on structures expressed on the membranes of eukaryotic and bacterial cells expressing the target antigen as a recombinant fusion protein (3, 9, 25, 30). Furthermore, phage libraries may be preabsorbed to remove unwanted specificities, or alternatively, selections may be performed in the presence of a homologous competitor antigen to enrich for phages directed to nonhomologous regions of the target antigen (6, 8, 25). Finally, phages bound to the target structure may be eluted by competition with ligand or a conventional monoclonal antibody (MAb) (5, 26, 39).
In this study, we used our library in a competitive panning procedure to demonstrate the feasibility of obtaining antibodies specific for an unknown target structure predicted to be differentially expressed on two strains of the same bacterial species. Selections were performed on Moraxella (Branhamella) catarrhalis, a gram-negative bacterium that may cause upper respiratory tract disease in children and lower respiratory tract disease in elderly people and patients with chronic obstructive pulmonary disease (2, 7, 12, 28). Resistance against complement-mediated lysis is considered an important virulence factor of this bacterium (15, 17). Complement-resistant bacteria were coated onto a solid support, and phage selections were performed in the presence of a complement-sensitive strain as a particulate competitor antigen. After three rounds of selection, a collection of 10 different monoclonal PhAbs (MPhAbs) with binding specificity for complement-resistant but not complement-sensitive M. catarrhalis isolates was obtained. The molecule recognized was identified as the high-molecular-weight outer membrane protein (HMW-OMP) of M. catarrhalis (22).
MATERIALS AND METHODS
Phage display repertoire.
The semisynthetic phage display library used in this study has been described in detail elsewhere (8). In brief, PCR was applied to fuse synthetic CDR3 regions to a collection of 49 different germ line VH genes. The rearranged heavy-chain genes were combined to seven different light-chain genes in the pHEN1 vector (24), resulting in a repertoire of 3.6 × 108 different human scFvs displayed on filamentous bacteriophage particles.
M. catarrhalis strains and MAbs KV5 and 9E9.
All M. catarrhalis strains used in this study, except complement-resistant strain ATCC 25240, were kindly provided by C. Hol and C. Verduin (Eijkman-Winkler Institute, Utrecht, The Netherlands). Strain H2 is a complement-resistant M. catarrhalis isolate obtained from a patient in the Wilhelmina Children’s University Hospital. All other strains used in our panel were obtained from healthy carriers (16).
Mouse hybridoma KV5 (immunoglobulin G1 isotype) was raised against complement-resistant M. catarrhalis and obtained by screening for binding to complement-resistant strains and lack of binding to complement-sensitive strains. The target antigen on complement-resistant strains was identified as HMW-OMP (27a, 34).
HMW-OMP-specific MAb 9E9 (immunoglobulin M isotype) (22) was a kind gift of T. F. Murphy.
Purification of HMW-OMP.
M. catarrhalis 25240 was grown in brain heart infusion broth overnight at 37°C and centrifuged at 1,500 × g for 15 min, and the pellet was washed twice in 0.01 M HEPES buffer (pH 7.4). The pellet was resuspended in 5% Zwittergent 3-14–0.045 M Tris–0.001 M EDTA–0.25 M borate, sonicated four times at 100 W for 15 s on ice, and subsequently stirred on ice for 1 h to homogenize fully. Ethanol was added to 20% (final concentration), and the solution was incubated for 30 min at −20°C. DNA was precipitated by centrifugation at 17,000 × g for 20 min at 4°C, and the supernatant was dialyzed four times for 30 min against buffer Z (0.05 M Tris, 0.05% Zwittergent, 0.01 M EDTA [pH 8.0]). Saturated ammonium sulfate solution was added to a final concentration of 60%, and the solution was centrifuged at 17,000 × g for 30 min at 4°C. The resulting pellet was solubilized in 5% Zwittergent–0.05 M Tris–0.01 M EDTA (pH 8.0) and applied to a DEAE exchange column which had been equilibrated against buffer Z. HMW-OMP was eluted from the column at approximately 1 M NaCl. Fractions were pooled, concentrated, and dialyzed against buffer Z. Purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fractions that, upon silver staining, contained only a single band at the appropriate molecular weight were used. Immunoreactivity to 9E9 and KV5 was checked by immunoblotting and dot assay.
Complement resistance.
M. catarrhalis strains were subdivided into three groups according to their sensitivity to serum complement-mediated lysis as described previously (37): resistant (survival of more than 50% of the bacteria during a 3-h incubation in 50% human pooled serum), sensitive (less than 3% survival after incubation for 1 h in 50% pooled human serum), and with intermediate survival.
Panning procedure.
M. catarrhalis strains were grown overnight on blood agar plates at 37°C and suspended in phosphate-buffered saline (PBS). Strain 9.21R was adsorbed to MaxiSorp tubes (Nunc, Roskilde, Denmark) at a concentration of 108 CFU/ml by incubation for 1 h at 37°C followed by 16 h at 4°C. The tubes were washed twice with PBS and blocked for 1 h at 37°C with 2% low-fat dry milk powder in PBS. The phage display library (approximately 5 × 1012 phages) was preincubated for 15 min with 5 × 108 CFU of strain 3.21S in 3 ml of 2% low-fat dry milk powder in PBS at room temperature. The mixture was transferred to the 9.21R-coated immunotubes and incubated for a further 2 h at room temperature. Unbound phages and phages bound to 3.21S bacteria were removed by washing the tubes 10 times in 0.1% Tween 20–PBS followed by 20 times in PBS. Remaining phages were eluted in 0.1 M triethylamine and allowed to infect Escherichia coli XL1-Blue cells (Stratagene, La Jolla, Calif.). The E. coli cells were plated on agar containing the appropriate antibiotics and glucose and used to prepare phages for a next round of selection as described previously (24).
In selections in which no competition was used, all steps were essentially the same except for the omission of absorber strain 3.21S in the incubation mixture.
Enzyme-linked immunosorbent assay (ELISA).
Coating of MaxiSorp plates with M. catarrhalis strains and blocking of the plates were performed as described for the panning procedure. One hundred-microliter aliquots of 10-times-concentrated MPhAb preparation were added to each well and incubated for 1 h at 37°C. After washing of the plates with 0.05% Tween 20–PBS, binding of PhAbs was detected by using horseradish peroxidase-conjugated sheep anti-M13 antibody (Pharmacia, Uppsala, Sweden) according to the manufacturer’s recommendations.
For the HMW-OMP ELISA, wells were precoated with 2% low-fat dry milk powder for 30 min at room temperature, after which 5 μg of HMW-OMP was added to each well and incubation was continued for another 90 min. The rest of the procedure was performed exactly as described above.
Screening and nucleotide sequence analysis of clones.
After three rounds of selection, PhAbs were rescued from individual ampicillin-resistant colonies of infected XL1-Blue cells (24). Specific binding to a panel of antigens was verified by ELISA. The nucleotide sequences of selected clones were determined by the dideoxy-chain termination method (31), using primers LINKSEQ and PHENSEQ (18) to establish VH gene usage, heavy-chain CDR3 composition, and light-chain identity.
Inhibition ELISA.
ScFv fragments were produced in E. coli nonsuppressor strain SF110-F′ as described previously (24, 29). This E. coli strain lacks the periplasmic proteases OmpT and DegP, resulting in a higher yield of functional protein (11). Fifty-microliter aliquots of periplasmic scFv fragment preparations were mixed with 50 μl of 10-times-concentrated MPhAb preparation before being added to 9.21R-coated wells. Binding of PhAbs was detected by using horseradish peroxidase-conjugated sheep anti-M13 antibody (Pharmacia) according to the manufacturer’s recommendations.
Western blotting.
M. catarrhalis strains were grown as described above and adjusted to 109 CFU/ml in PBS. Ten-microliter aliquots of M. catarrhalis strains or 1 μg of purified HMW-OMP were run on a 10% reducing polyacrylamide gel in the presence of sodium dodecyl sulfate and transferred to nitrocellulose by using standard procedures. The nitrocellulose blots were blocked for 1 h in 5% low-fat milk powder in Tris-buffered saline (10 mM Tris, 150 mM NaCl [pH 7.4]) containing 0.5% Tween 20 (M-TTBS). For PhAb staining, the blots were subsequently incubated overnight in 1% M-TTBS containing approximately 5 × 1011 PhAbs, washed in TTBS, and incubated for 1 h in 1/1,500-diluted horseradish peroxidase-conjugated sheep anti-M13 in 1% M-TTBS. Blots were washed again in TTBS and developed with 3,3′-diaminobenzidine (Sigma Chemical Co., St. Louis, Mo.). The complete procedure was performed at room temperature.
Nitrocellulose blots used for staining with MAb were blocked as described above and subsequently incubated for 1 h with 1/5,000-diluted ascites fluid in 1% M-TTBS. After being washed in TTBS, the blots were incubated for 1 h in 1/2,000-diluted horseradish peroxidase-conjugated rabbit anti-mouse antibody (Dako, Glostrup, Denmark) in 1% M-TTBS and then washed and developed as described above.
Flow cytometry.
Bacterial suspensions of strain 9.21R were prepared as described above and blocked in 2% low-fat dry milk powder for 15 min. One hundred-microliter aliquots containing 5 × 108 bacteria were mixed with 100-μl periplasmic preparations containing scFv fragments. After being washed in 0.5% bovine serum albumin (BSA) in PBS, the scFvs were detected by using MAb 9E10, which specifically recognizes the Myc tag fused to the scFvs (24), followed by a fluorescein isothiocyanate-labeled goat anti-mouse antibody preparation (Becton Dickinson, San Jose, Calif.). Analyses were performed on a FACScan (Becton Dickinson).
RESULTS
Selection and binding specificity of PhAbs.
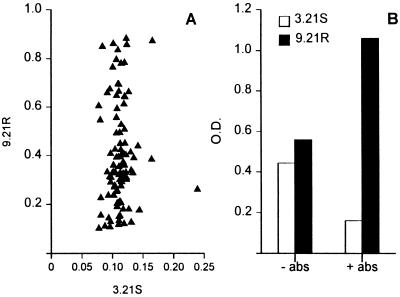
Phages from the semisynthetic scFv library were preincubated for 15 min with cells of the complement-sensitive strain 3.21S, and the mixture was added to immunotubes coated with complement-resistant target strain 9.21R. Nonbinding phages and the phages bound to 3.21S absorber bacteria were removed by washing, and the PhAbs bound to 9.21R bacteria were eluted and propagated. After three rounds of selection, 96 individual bacterial colonies were obtained and used to prepare MPhAbs. Binding of these MPhAbs to both strain 9.21R and strain 3.21S was assessed by ELISA. Twenty-one of 96 MPhAbs displayed significant (>0.6 OD [optical density unit]) binding to the target strain 9.21R, whereas none of the phage preparations displayed binding (<0.25 OD) to strain 3.21S (Fig. 1A). Specificity was checked with a panel of antigens consisting of ovalbumin, human immunoglobulin, hepatitis C virus polypeptide, high-mobility-group (HMG) box protein, FimD protein of Bordetella pertussis, E. coli O1K1, Salmonella paratyphi group B, Pseudomonas aeruginosa, Proteus mirabilis, Neisseria meningitidis groups A, B, and C, BSA, and low-fat dry milk, which was used for blocking. No binding to any of these antigens was observed.
FIG. 1.
Competitive selection and specificity of MPhAbs recognizing complement-resistant M. catarrhalis strains. (A) Screening of 96 individual MPhAbs for binding to strains 9.21R and 3.21S in ELISA. For each MPhAb, ODs are depicted on the x axis for binding to 3.21S and on the y axis for binding to 9.21R. (B) Comparison of selections performed in the presence (+ abs) or absence (− abs) of absorber strain 3.21S. Polyclonal phage preparations from entire selection rounds were tested for binding to 3.21S and 9.21R in ELISA.
We repeated phage selections on the target strain 9.21R in the absence of the absorber strain 3.21S. After three rounds of selection, a polyclonal phage preparation was generated and tested for binding to strains 9.21R and 3.21S in ELISA. No significant difference was observed in the binding to strain 9.21R or 3.21S. In contrast, the polyclonal phage preparation from selections performed in the presence of the absorber strain 3.21S bound significantly better to the target strain 9.21R (Fig. 1B).
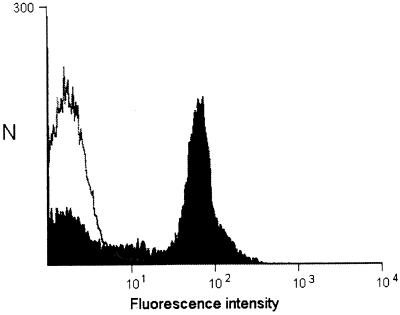
To establish whether the MPhAbs bound to cell surface determinants present on strain 9.21R, bacteria were incubated with scFv preparations from each clone. Binding of scFv fragments was detected by flow cytometry. The results show that the MPhAbs bind to intact bacterial cells; results of a representative experiment are shown in Fig. 2.
FIG. 2.
Flow cytometric detection of scFv binding to complement-resistant M. catarrhalis. Binding of scFv to whole 9.21R bacteria was visualized by the Myc tag-specific MAb 9E10, followed by a fluorescein isothiocyanate-labeled goat anti-mouse antibody. Black graph, scFv B12; white graph, negative control scFv HM1.
Expression of the target protein among a panel of M. catarrhalis strains.
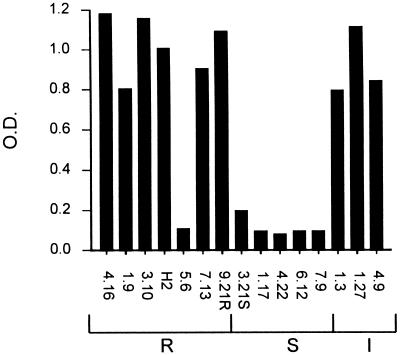
We tested the capacity of the 10 MPhAbs to bind in ELISA to a panel of 15 previously described isolates of serum complement-sensitive and -resistant strains of M. catarrhalis (16). None of the complement-sensitive strains were recognized by the 10 MPhAbs, whereas all but one (5.6) of the 10 complement-resistant and intermediately resistant strains were recognized by nine of the MPhAbs. Only MPhAb D11 bound to all complement-resistant and intermediate strains. A representative experiment with MPhAb F2 is shown in Fig. 3.
FIG. 3.
Selected MPhAbs recognize M. catarrhalis strains that are resistant (R) but not those that are sensitive (S) or intermediately sensitive (I) to complement-mediated lysis. ELISA results of MPhAb clone F2 are shown. Only 1 of 10 clones, D11, recognized strain 5.6.
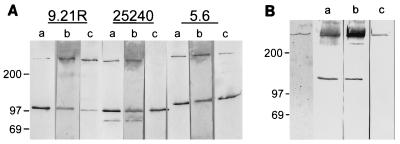
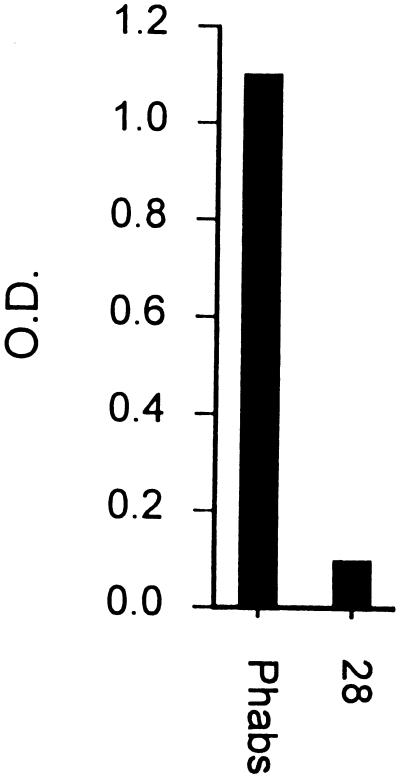
In Western blot analysis using a polyclonal PhAb preparation from the entire third round of competitive selection, HMW-OMP was detected in three complement-resistant M. catarrhalis strains as well as in a purified HMW-OMP preparation (Fig. 4). In addition to the reported oligomeric and monomeric bands of HMW-OMP of 350 to 720 kDa and 110 to 120 kDa, respectively (22), a band of 90 to 100 kDa was observed in strain ATCC 25240, which most probably represents a partial degradation product. Although the purified HMW-OMP preparation revealed only oligomeric HMW-OMP in Coomassie brilliant blue staining, an additional band of 120 to 140 kDa was detected in Western blot analysis by PhAbs and KV5 (Fig. 4B). No specific bands were observed in Western blots of lysates of complement-sensitive M. catarrhalis strains with either PhAbs or MAbs (not shown). An ELISA using purified HMW-OMP revealed strong binding by a polyclonal PhAb preparation directed against complement-resistant M. catarrhalis, while control MPhAb 28 showed no binding activity above background (Fig. 5).
FIG. 4.
Western blot analysis of whole-cell lysates of complement-resistant M. catarrhalis strains (A) and of purified HMW-OMP of strain 25240 (B). Adjacent lanes were loaded with lysates of the same strain and, after blotting, stained with either MAb KV5 (lanes a), polyclonal PhAbs (lanes b), or MAb 9E9 (lanes c). The first lane of panel B shows purified HMW-OMP stained with Coomassie brilliant blue. The strains used in panel A are indicated at the top. The positions of molecular size markers are shown in kilodaltons on the left.
FIG. 5.
Purified HMW-OMP is recognized in ELISA by polyclonal PhAbs but not by nonrelevant MPhAb 28.
Nucleotide sequence analysis of V regions encoding 9.21R-specific MPhAbs.
A total of 21 MPhAbs were selected for further study (Fig. 1A). Nucleotide sequence analysis showed that these 21 binders represented 10 different scFv antibodies, all encoded by members of the VH3 family and the Vλ3 light chain (Table 1). Remarkably, all 10 MPhAbs were encoded by only one of the seven randomized heavy-chain CDR3 primers used to construct the library (Table 1 and reference 8). Close inspection of the CDR3 regions of the 10 clones unveiled a strong, position-dependent preference for basic amino acid residues: 5 clones contained an arginine or lysine residue at position 96, 8 had an arginine at position 99, and 9 had an arginine or lysine at position 100 (numbering according to Kabat et al. [20]). Furthermore, the glycine residue at position 95 was invariant, and at position 98> a bias toward the small hydrophobic amino acids valine and alanine was observed in 7 of 10 clones.
TABLE 1.
Deduced amino acid sequences of VH chain CDR3 regions and VH and VL utilization of the 10 MPhAbs that selectively bind to complement-resistant M. catarrhalis strainsa
| Clone | Amino acid at position 94a | CDR3 sequence | VHb | VL |
|---|---|---|---|---|
| H3 | K | G L F S S R F D S | DP47 | Vλ3 |
| F3 | K | G F R A P K F D S | DP47 | Vλ3 |
| F2 | R | G W S P R R F D Y | DP47 | Vλ3 |
| E9 | K | G G T T R R F D S | DP49 | Vλ3 |
| A9 | R | G K M A R R F D Y | DP49 | Vλ3 |
| B12 | K | G R M V R R F D S | DP47 | Vλ3 |
| D9 | K | G R L V R R F D S | DP47 | Vλ3 |
| A11 | R | G T V V R R F D Y | DP47 | Vλ3 |
| D11 | R | G K V V R K F D Y | DP49 | Vλ3 |
| G1 | K | G R L V R P F D S | DP53 | Vλ3 |
| CR9A | K | A X X X X X F D S | ||
| R | D Y | |||
| G |
Framework position 94 is depicted because it is not fixed in the phage display library. Numbering of amino acids is according to Kabat et al. (20).
The MPhAbs bind to the same or closely spaced epitopes.
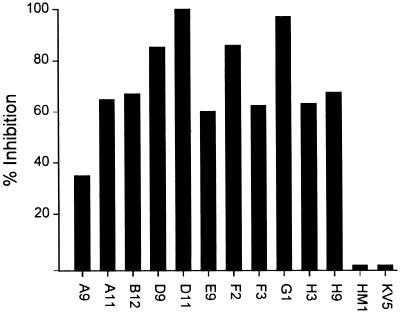
The uniformity of the VH and VL genes encoding the 10 different MPhAbs suggested that they were directed against the same or related epitopes on HMW-OMP. To explore this possibility, we produced scFv molecules from each of the 10 MPhAbs and assessed their ability to block the binding of each MPhAb to strain 9.21R in inhibition ELISA. The results show that each of the 10 scFvs was capable of inhibiting the interaction of each of the 10 MPhAbs with 9.21R, albeit to various degrees (Fig. 6; shown only for phage B12). The scFv fragment HM1, directed against HMG box protein and used as a control for the specificity of the inhibition assay, did not affect MPhAb binding to strain 9.21R. In addition, MAb KV5 also failed to block binding of the MPhAbs to strain 9.21R. Collectively, these experiments support the notion that all MPhAbs are directed against the same or closely spaced epitopes.
FIG. 6.
MPhAb binding to complement-resistant M. catarrhalis is inhibited by scFv antibody fragments. Plates were coated with serum-resistant strain 9.21R, and a mixture of PhAb and scFv was added. Bound phages were detected by anti-M13 antibody. Inhibition of MPhAb B12 binding by scFv fragments of each of the 10 clones was determined and presented as percentage inhibition of binding in absence of scFvs or KV5. Incubations with MAb KV5 (recognizing HMW-OMP on complement-resistant M. catarrhalis strains) and scFv HM1 (recognizing HMG box protein) served as inhibition controls.
DISCUSSION
We have used a large, synthetic phage antibody display library to search for membrane proteins differentially expressed on two types of strains of M. catarrhalis that differ in their sensitivity to complement-mediated lysis. Based on previous observations, we hypothesized that complement resistance in M. catarrhalis is dependent on the expression of one or multiple, trypsin-sensitive membrane-bound structures (38). The approach used here exploits the notion that phage libraries can be used in a subtractive procedure to obtain phages that specifically bind to target prokaryotic or eukaryotic cells of interest by using a closely related absorber cell to remove cross-reactive specificities binding to both target and absorber (6, 9, 25). By phage selection on a solid-phase-bound, complement-resistant strain in the presence of an excess of a complement-sensitive strain in the fluid phase (competitive panning), we selected a collection of 10 different MPhAbs which exclusively recognized the resistant M. catarrhalis strain used for selection. In addition, these MPhAbs displayed binding to a panel of nine independent isolates of complete or intermediate complement resistance, whereas none of the MPhAbs bound to five complement-sensitive isolates (Fig. 1A). The presence of the complement-sensitive strain during the selection procedure was essential, since selections performed in the absence of these absorbers (Fig. 1B) resulted in the isolation of phages that bound to both complement-resistant and complement-sensitive strains. These experiments confirm that competitive panning provides a powerful and generally applicable approach to unveil differences in membrane-protein expression patterns between prokaryotic cells established on a functional basis.
In Western blots of whole-cell lysates of three strains of M. catarrhalis and of purified HMW-OMP, the PhAbs selected recognized bands identical to those detected by the HMW-OMP-specific MAbs KV5 and 9E9 (Fig. 4). In addition to the oligomeric form of HMW-OMP of 350 to 720 kDa, the monomeric form of 110 to 120 kDa was observed in all strains. Using PhAbs and KV5, we detected in the purified HMW-OMP preparation an additional band which appeared to be slightly larger than the monomeric band observed in the whole-cell lysates (Fig. 4B). This band might represent a degradation product or, alternatively, could represent the monomeric form which appears larger than the monomeric band in the whole-cell lysates due to differences in preparation. In an ELISA, the PhAbs clearly bound to the purified HMW-OMP, while nonrelevant phages did not bind above background (Fig. 5). HMW-OMP was first described by Klingman and Murphy (21, 22), and is also known as ubiquitous surface protein A (UspA) (14). The protein shows strain-dependent variation for relative molecular mass and is expressed only in complement-resistant and intermediate strains. The variability in electrophoretic mobility of HMW-OMP is typical for OMPs of primary mucosal pathogens, e.g., Haemophilus influenzae and Neisseria gonorrhoeae. This variability, however, is not seen in other OMPs of M. catarrhalis (1, 27), which suggests that HMW-OMP might be an important target for the host immune response. This conclusion is supported by the following observations. Antibodies against HMW-OMP are protective (14), and carriership of M. catarrhalis among healthy children reverses with increasing age from more complement-resistant to mainly complement-sensitive strains (16, 17). Antibodies against HMW-OMP have been detected in convalescence but not in preimmune sera from patients with M. catarrhalis pneumonia (13). Moreover, the observation that HMW-OMP would be expressed by all M. catarrhalis strains was probably biased by the use of only clinical isolates in these analyses (14), which further supports the role of HMW-OMP as an important virulence factor.
The PhAbs and mouse MAb KV5 recognized different epitopes on HMW-OMP, based on inhibition ELISAs. In Western blots, both the monomeric form of HMW-OMP (110 to 120 kDa) and the oligomeric form (350 to 720 kDa) are detected by PhAbs and MAbs. The additional band of 90 to 100 kDa, observed in strain ATCC 25240, probably represents a partial degradation product of HMW-OMP.
The complexity of serum resistance in other, similar bacteria (e.g., Neisseria species) has led to the suggestion that serum resistance in M. catarrhalis is multifactorial (4, 19, 23). Indeed, the major OMP CopB of M. catarrhalis, with a molecular mass of approximately 81 kDa, has been shown to be involved in serum resistance (15), and antibodies against CopB are found in convalescent sera of patients with M. catarrhalis pneumonia as well (13). The strains described by Helminen et al. (15), including the serum-sensitive CopB mutants, all expressed HMW-OMP described in this report but were intermediately resistant in our assays (our unpublished observations), lending further support to the involvement of at least one additional membrane protein in serum complement resistance.
ScFv fragments derived from any of the selected MPhAbs inhibited binding of all other PhAbs, whereas nonrelevant scFv or MAb KV5 did not influence binding in inhibition ELISAs. This finding suggests that all MPhAbs bind to the same or closely spaced epitopes, including D11, which is the only clone recognizing strain 5.6. This notion is further supported by the remarkable similarity of the VH and VL regions encoding the 10 different MPhAbs. All were encoded by a single Vλ3 gene, one of seven light chains used to construct the library, and only 3 of 49 possible heavy-chain gene segments, all members of the VH3 gene family, with a strong preference for the DP47 (VH26) VH gene segment (6 of 10 MPhAbs). Strikingly, the CDR3 regions were uniform in length and showed an overwhelming, position-dependent preference for basic amino acids (5 of 10 at position 96, 8 of 10 at amino acid 99, and 9 of 10 at position 100). Additional homology was found at position 95 with glycine residues and 7 of 10 small hydrophobic amino acids at position 98. This restricted utilization in VH and VL gene segments and homology in CDR3 length and composition cannot be attributed to an artifact related to the construction of this antibody library since in a previous analysis of more than 100 different MPhAbs selected in various experiments, a broad variety of VH and VL genes as well as CDR3 lengths and compositions was noted (references 8 to 10 and unpublished results). This observation in combination with the strong preference for basic amino acids in VH CDR3 suggests that the phage selection have been biased toward a negatively charged epitope on HMW-OMP.
One mechanism used by M. catarrhalis to prevent lysis by human serum complement is the attachment of vitronectin to its surface (34). This serum protein, also known as S protein, interferes with the complement cascade at the level of membrane attack complex formation; it interferes with the insertion of C5b67 into target cell membranes and with the function of the C9 polymerase (33). HMW-OMP is the most likely candidate ligand for vitronectin at the membrane of M. catarrhalis, although published evidence is still circumstantial (35, 36). However, recent ELISAs clearly show that vitronectin binds to solid-phase-coated, purified HMW-OMP (34). Vitronectin contains a strongly positively charged region, the so-called heparin-binding site (amino acid sequence KKQRFRHRNRKGYR). Because this region resembles the VH CDR3 regions of the selected MPhAbs with respect to the high incidence of basic amino acids (Table 1), it is tempting to speculate that a negatively charged epitope recognized by the selected MPhAbs is also involved in vitronectin binding. Indeed, some MPhAbs did bind to heparin-conjugated BSA, while treatment of 9.21R with trypsin abolished the binding of both vitronectin and MPhAbs but not of KV5 (data not shown). However, we were not able to render resistant M. catarrhalis strains sensitive to complement-mediated lysis by preincubation with MPhAbs or scFvs. This might be due to a relatively low affinity of the MPhAbs compared with vitronectin or to the high concentration (0.25 to 0.45 mg/ml) of vitronectin in serum.
In conclusion, using competitive panning, we selected phages against an OMP differentially expressed on complement-resistant M. catarrhalis, which was identified to be HMW-OMP.
REFERENCES
- 1.Bartos L C, Murphy T F. Comparison of the outer membrane proteins of 50 strains of Branhamella catarrhalis. J Infect Dis. 1988;158:761–765. doi: 10.1093/infdis/158.4.761. [DOI] [PubMed] [Google Scholar]
- 2.Boyle F M, Georgiou P R, Tilse M H, McCormack J G. Branhamella (Moraxella) catarrhalis: pathogenic significance in respiratory infections. Med J Aust. 1991;154:592–596. doi: 10.5694/j.1326-5377.1991.tb121219.x. [DOI] [PubMed] [Google Scholar]
- 3.Bradbury A, Persic L, Werge T, Cattaneo A. Use of living columns to select specific phage antibodies. Bio/Technology. 1993;11:1565–1569. doi: 10.1038/nbt1293-1565. [DOI] [PubMed] [Google Scholar]
- 4.Brandtzaeg P, Mollnes T E, Kierulf P. Complement activation and endotoxin levels in systemic meningococcal disease. J Infect Dis. 1989;160:58–65. doi: 10.1093/infdis/160.1.58. [DOI] [PubMed] [Google Scholar]
- 5.Burton D R, Barbas C F. Human antibodies from combinatorial libraries. Adv Immunol. 1994;57:191–280. doi: 10.1016/s0065-2776(08)60674-4. [DOI] [PubMed] [Google Scholar]
- 6.Cai X, Garen A. Anti-melanoma antibodies from melanoma patients immunized with genetically modified autologous tumor cells: selection of specific antibodies from single-chain Fv fusion phage libraries. Proc Natl Acad Sci USA. 1995;92:6537–6541. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catlin B W. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin Microbiol Rev. 1990;3:293–330. doi: 10.1128/cmr.3.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Kruif J, Boel E, Logtenberg T. Selection and application of human single chain Fv antibody fragments from a semi-synthetic phage antibody display library with designed CDR3 regions. J Mol Biol. 1995;248:97–105. doi: 10.1006/jmbi.1995.0204. [DOI] [PubMed] [Google Scholar]
- 9.de Kruif J, Terstappen L, Boel E, Logtenberg T. Rapid selection of cell subpopulation-specific human monoclonal antibodies from a synthetic phage antibody library. Proc Natl Acad Sci USA. 1995;92:3938–3942. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kruif J, van der Vuurst de Vries A, Cilenti L, Boel E, van Ewijk W, Logtenberg T. New perspectives on recombinant human antibodies. Immunol Today. 1996;17:453–455. doi: 10.1016/0167-5699(96)30057-y. [DOI] [PubMed] [Google Scholar]
- 11.Georgiou G, Poetschke H L, Stathopoulos C, Francisco J A. Practical applications of engineering Gram-negative bacterial cell surfaces. Trends Biotechnol. 1993;11:6–10. doi: 10.1016/0167-7799(93)90068-K. [DOI] [PubMed] [Google Scholar]
- 12.Hager H, Verghese A, Alvarez S, Berk S L. Branhamella catarrhalis respiratory infections. Rev Infect Dis. 1987;9:1140–1149. doi: 10.1093/clinids/9.6.1140. [DOI] [PubMed] [Google Scholar]
- 13.Helminen M E, Beach R, Maciver I, Jarosik G, Hansen E J, Leinonen M. Human immune response against outer membrane proteins of Moraxella (Branhamella) catarrhalis determined by immunoblotting and enzyme immunoassay. Clin Diagn Lab Immunol. 1995;2:35–39. doi: 10.1128/cdli.2.1.35-39.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helminen M E, Maciver I, Latimer J L, Klesney-Tait J, Cope L D, Paris M, McCracken G H, Hansen E J. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J Infect Dis. 1994;170:867–872. doi: 10.1093/infdis/170.4.867. [DOI] [PubMed] [Google Scholar]
- 15.Helminen M E, Maciver I, Paris M, Latimer J L, Lumbley S L, Cope L D, McCracken G H, Hansen E J. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J Infect Dis. 1993;168:1194–1201. doi: 10.1093/infdis/168.5.1194. [DOI] [PubMed] [Google Scholar]
- 16.Hol C, Verduin C M, van Dijke E E M, Verhoef J, Fleer A, van Dijk H. Complement resistance is a virulence factor of Branhamella (Moraxella) catarrhalis. FEMS Immunol Med Microbiol. 1995;11:207–212. doi: 10.1111/j.1574-695X.1995.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 17.Hol C, Verduin C M, van Dijke E E M, Verhoef J, van Dijk H. Complement resistance in Branhamella (Moraxella) catarrhalis. Lancet. 1993;341:1281. doi: 10.1016/0140-6736(93)91185-o. [DOI] [PubMed] [Google Scholar]
- 18.Hoogenboom H R, Winter G. By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J Mol Biol. 1992;227:381–388. doi: 10.1016/0022-2836(92)90894-p. [DOI] [PubMed] [Google Scholar]
- 19.Joiner K A, Warren K A, Hammer C, Frank M M. Bactericidal but not nonbactericidal C5b-C9 is associated with distinctive outer membrane proteins in Neisseria gonorrhoeae. J Immunol. 1985;134:1920–1925. [PubMed] [Google Scholar]
- 20.Kabat E A, Wu T T, Oerry H M, Gottesman K S, Foeller C. Sequences of proteins of immunological interest. U.S. Bethesda, Md: Department of Health and Human Services, Public Health Service, National Institutes of Health; 1991. [Google Scholar]
- 21.Klingman K L, Murphy T F. Proceedings of the 1992 General Meeting of the American Society for Microbiology 1992. Washington, D.C: American Society for Microbiology; 1992. Identification and purification of the lipooligosaccharide-associated high-molecular-weight outer membrane protein (HMW-OMP) of Branhamella catarrhalis, abstr. B-388; p. 90. [Google Scholar]
- 22.Klingman K L, Murphy T F. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect Immun. 1994;62:1150–1155. doi: 10.1128/iai.62.4.1150-1155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandrell R E, Kim J J, John C M, Gibson B W, Sugai J V, Apicella M A, Griffiss J M, Yamasaki R. Endogenous sialylation of the lipooligosaccharides of Neisseria meningitidis. J Bacteriol. 1991;173:2823–2832. doi: 10.1128/jb.173.9.2823-2832.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty J, Griffiths A D, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 25.Marks J D, Ouwehand W H, Bye J M, Finnern R, Gorick B D, Voak D, Thorpe S J, Hughes-Jones N C, Winter G. Human antibody fragments specific for human blood group antigens from a phage display library. Bio/Technology. 1993;11:1145–1149. doi: 10.1038/nbt1093-1145. [DOI] [PubMed] [Google Scholar]
- 26.Meulemans E V, Slobbe R, Wasterval P, Ramaekers F C S, Van Eys G J J M. Selection of phage-displayed antibodies specific for a cytoskeletal antigen by competitive elution with a monoclonal antibody. J Mol Biol. 1994;244:353–360. doi: 10.1006/jmbi.1994.1735. [DOI] [PubMed] [Google Scholar]
- 27.Murphy T F. The surface of Branhamella catarrhalis: a systematic approach to the surface antigens of an emerging pathogen. Pediatr Infect Dis J. 1989;8:S75–S77. [PubMed] [Google Scholar]
- 27a.Murphy, T. F. Personal communication.
- 28.Murphy T F, Sethi S. Bacterial infections in chronic obstructive pulmonary disease. Am Rev Respir Dis. 1992;146:1067–1083. doi: 10.1164/ajrccm/146.4.1067. [DOI] [PubMed] [Google Scholar]
- 29.Plückthun A, Skerra A. Expression of functional antibody Fv and Fab fragments in Escherichia coli. Methods Enzymol. 1989;178:497–515. doi: 10.1016/0076-6879(89)78036-8. [DOI] [PubMed] [Google Scholar]
- 30.Portolano S, McLachlan S M, Rapoport B. High affinity, thyroid-specific human autoantibodies displayed on the surface of filamentous phage use V genes similar to other autoantibodies. J Immunol. 1993;151:2839–2851. [PubMed] [Google Scholar]
- 31.Sanger F, Nieklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 33.Tschopp J, Masson D, Schäfer S, Peitsch M, Preissner K T. The heparin binding domain of S-protein/vitronectin binds to complement components C7, C8 and C9 and perforin from cytolytic T-cells and inhibits their lytic activities. Biochemistry. 1988;27:41033–41039. doi: 10.1021/bi00411a029. [DOI] [PubMed] [Google Scholar]
- 34.Verduin C M. Complement resistance as a virulence factor of Moraxella (Branhamella) catarrhalis. Ph.D. thesis. Utrecht, The Netherlands: University of Utrecht; 1996. [Google Scholar]
- 35.Verduin C M, Bootsma H J, Hol C, Fleer A, Jansze M, Klingman K L, Murphy T F, van Dijk H. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Resistance of Moraxella (Branhamella) catarrhalis to killing by human serum is mediated by high-molecular-weight outer membrane protein (HMW-OMP) and vitronectin, abstr. B-137; p. 64. [Google Scholar]
- 36.Verduin C M, Hol C, Jansze M, van Dijke E E M, van Dijk H. Complement resistance in Moraxella (Branhamella) catarrhalis is species-restricted and mediated by the binding of human vitronectin. In: Verduin C M, Watson D A, van Dijk H, Verhoef J, editors. Constitutional resistance to infection. Austin, Tex: R. G. Landes Company (Medical Intelligence Unit); 1995. pp. 25–40. [Google Scholar]
- 37.Verduin C M, Hol C, van Dijke E E M, Faber J A J, Jansze M, Verhoef J, van Dijk H. Assessment of complement-mediated killing of Moraxella (Branhamella) catarrhalis isolates by a simple method. Clin Diagn Lab Immunol. 1995;2:365–368. doi: 10.1128/cdli.2.3.365-368.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verduin C M, Jansze M, Hol C, Mollnes T E, Verhoef J, van Dijk H. Differences in complement activation between complement-resistant and complement-sensitive Moraxella (Branhamella) catarrhalis strains occur at the level of membrane attack complex formation. Infect Immun. 1994;62:589–595. doi: 10.1128/iai.62.2.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]