Abstract
Fecal pollution remains a significant challenge for recreational water quality management worldwide. In response, there is a growing interest in the use of real-time quantitative PCR (qPCR) methods to achieve same-day notification of recreational water quality and associated public health risk as well as to characterize fecal pollution sources for targeted mitigation. However, successful widespread implementation of these technologies requires the development of and access to a high-quality standard control material. Here, we report a single laboratory qPCR performance assessment of the National Institute of Standards and Technology Standard Reference Material 2917 (NIST SRM® 2917), a linearized plasmid DNA construct that functions with 13 recreational water quality qPCR assays. Performance experiments indicate the generation of standard curves with amplification efficiencies ranging from 0.95 ± 0.006 to 0.99 ± 0.008 and coefficient of determination values (R2) ≥ 0.980. Regardless of qPCR assay, variability in repeated measurements at each dilution level were very low (quantification threshold standard deviations ≤ 0.657) and exhibited a heteroscedastic trend characteristic of qPCR standard curves. The influence of a yeast carrier tRNA added to the standard control material buffer was also investigated. Findings demonstrated that NIST SRM® 2917 functions with all qPCR methods and suggests that the future use of this control material by scientists and water quality managers should help reduce variability in concentration estimates and make results more consistent between laboratories.
Keywords: qPCR, Microbial source tracking, Rapid fecal indicator, Standard control material
1. Introduction
Fecal pollution is the number one biological contaminant reported to cause water quality impairments in coastal, river, and stream environments across the United States (USEPA 2021). To help protect recreators, researchers have developed a series of real-time quantitative PCR (qPCR) methods that can characterize fecal waste in environmental water samples [for review see (Harwood et al. 2017)]. These methods have been employed to provide same day notification of recreational water public health risk by targeting fecal indicator bacteria such as enterococci and E. coli (Dorevitch et al. 2017, Shrestha and Dorevitch 2019), as well as characterize fecal pollution sources (i.e. sewage, wildlife, agricultural) (Li et al. 2021, Li et al. 2019, Shrestha et al. 2020). Many of these protocols have been subject to multiple laboratory validation studies (Aw et al. 2019, Ebentier et al. 2013, Shanks et al. 2016, Shanks et al. 2012) leading to the development of standardized procedures (USEPA 2013, 2019a, 2019b). qPCR is well suited for water quality testing applications due to high levels of precision, specificity, and sensitivity. However, implementation requires the generation of a standard curve to interpret results. Standard curves are constructed using a dilution series of a control material, usually five to six 10-fold dilution preparations, with a defined number of target sequences per dilution. However, reliance on standard curves is not without caveats, as they can strongly influence the precision of qPCR target concentration estimates. Variations in material type (e.g. plasmid DNA constructs, PCR amplicons, synthetic DNA, genomic DNA from biological samples), initial concentration determination method (e.g. droplet digital PCR, UV spectrophotometry, fluorometry), dilution preparation, and storage conditions can all introduce variability between different preparations, making it challenging to implement these methods on a broad scale in the absence of a readily available standard control material.
In response, the United States Environmental Protection Agency (USEPA) designed a DNA construct that functions with 13 recreational water quality testing qPCR protocols (Table 1) and partnered with the National Institute of Standards and Technology (NIST) to develop a large-scale preparation for mass distribution on a national scale. The result is Standard Reference Material 2917 (NIST SRM® 2917), a linearized plasmid preparation containing the 13 DNA target insert. Based on experiments conducted by NIST and reported elsewhere (Kralj et al. 2021), this material can be stored at 4 °C due to the addition of yeast carrier tRNA in the storage buffer and use of low-retention microtubes to maximize standard control material stability. Briefly, purity of the preparation was assessed using next generation sequencing to demonstrate the absence of any DNA contamination. Approximately 1000 sets were prepared for certification and distribution using an automated bottling and labeling system (Scinomix Sci-Print VXL, Earth City, MO). Each set is composed of five dilution levels allowing for the generation of qPCR calibration models with a range of quantification spanning approximately 10 to 105 copies per reaction. All NIST SRM® 2917 certification experiments were performed by NIST utilizing droplet digital PCR (Kralj et al. 2021). Certification included the determination of each dilution level mean concentration based on repeated measurements, demonstration of homogeneity between randomly selected dilution sets, and stability evaluation of the reference material under 4 °C and −20 °C storage conditions.
Table 1.
Summary of qPCR assay primer and probe sequences, targets, annealing temperatures, and references.
| qPCR Assay | Primer and Probe Sequences (5′ → 3′) | Target | Anneal Temp | Reference |
|---|---|---|---|---|
| Entero1a | F: GAGAAATTCCAAACGAACTTG R: CAGTGCTCTACCTCCATCATT P: [FAM]TGGTTCTCTCCGAAATAGCTTTAGGGCTA[TAMRA] |
Enterococci | 60°C | (Ludwig and Schleifer 2000, Siefring et al. 2008) |
| EC23S857 | F: GGTAGAGCACTGTTTTGGCA R: TGTCTCCCGTGATAACTTTCTC P: [FAM]TCATCCCGACTTACCAACCCG[TAMRA] |
E. coli | 56 °C | (Chern et al. 2011) |
| HF183/ BacR287 | F: ATCATGAGTTCACATGTCCG R: CTTCCTCTCAGAACCCCTATCC P: [FAM]CTGAGAGGAAGGTCCCCCACATTGGA[MGB] PIAC: [VIC]AACACGCCGTTGCTACA[MGB] |
Human fecal waste | 60°C | (Green et al. 2014a) |
| HumM2 | F: CGTCAGGTTTGTTTCGGTATTG R: TCATCACGTAACTTATTTATATGCATTAGC P: [FAM]TATCGAAAATCTCACGGATTAACTCTTGTGTACGC[TAMRA] PIAC: [VIC]CCTGCCGTCTCGTGCTCCTCA[TAMRA] |
60°C | (Shanks et al. 2009) | |
| CPQ_056 | F: CAGAAGTACAAACTCCTAAAAAACGTAGAG R: GATGACCAATAAACAAGCCATTAGC P: [FAM]AATAACGATTTACGTGATGTAAC[MGB] |
60°C | (Stachler et al. 2017) | |
| CPQ_064 | F:TGTATAGATGCTGCTGCAACTGTACTC R: CGTTGTTTTCATCTTTATCTTGTCCAT P: [FAM]CTGAAATTGTTCATAAGCAA[MGB] |
60°C | ||
| Rum2Bac | F:ACAGCCCGCGATTGATACTGGTAA R: CAATCGGAGTTCTTCGTGAT P: [FAM]ATGAGGTGGATGGAATTCGTGGTGT[BHQ-1] |
Ruminant fecal waste | 60°C | (Mieszkin et al. 2010) |
| CowM2 | F: CGGCCAAATACTCCTGATCGT R: GCTTGTTGCGTTCCTTGAGATAAT P: [FAM]AGGCACCTATGTCCTTTACCTCATCAACTACAGACA[TAMRA] PIAC: [VIC]TAGGAACAGGCGGCGACGA[TAMRA] |
Cattle fecal waste | 60°C | (Shanks et al. 2008) |
| CowM3 | F: CCTCTAATGGAAAATGGATGGTATCT R: CCATACTTCGCCTGCTAATACCTT P: [FAM]TTATGCATTGAGCATCGAGGCC[TAMRA] |
60°C | ||
| DG3 | F: TGAGCGGGCATGGTCATATT R: TTTTCAGCCCCGTTGTTTCG P: [FAM]AGTCTACGCGGGCGTACT[MGB] |
Canine fecal waste | 60°C | (Green et al. 2014b) |
| DG37 | F: CTTGGTTATGGGCGACATTG R: TTTTCTCCCACGGTCATCTG P: [FAM]TTGAACGTTTAAAGGAGCAGGTGGCAG[TAMRA] |
60°C | ||
| Pig2Bac | F: GCATGAATTTAGCTTGCTAAATTTGAT R: ACCTCATACGGTATTAATCCGC P: [FAM]TCCACGGGATAGCC[MGB] |
Swine fecal waste | 60°C | (Mieszkin et al. 2009) |
| GFD | F: TCGGCTGAGCACTCTAGGG R: GCGTCTCTTTGTACATCCCA |
Avian fecal waste | 57 °C | (Green et al. 2012) |
‘Anneal Temp’ indicates qPCR assay annealing temperature.
PIAC denotes the internal amplification control (IAC) probe in multiplex qPCR assay.
To evaluate NIST SRM® 2917 for qPCR standard curve generation, a series of experiments were conducted by USEPA. Study objectives include the investigation of qPCR performance in the presence and absence of yeast carrier tRNA, assessment of qPCR measurement variability at each standard control dilution level, and evaluation of the ability to generate high-quality standard curves with all qPCR assays. Findings demonstrate that the addition of yeast carrier tRNA does not interfere with qPCR assay performance, that repeated measures at each dilution level are highly reproducible, and that NIST SRM® 2917 functions with all qPCR assays.
2. Materials and methods
2.1. NIST SRM® 2917 construct design
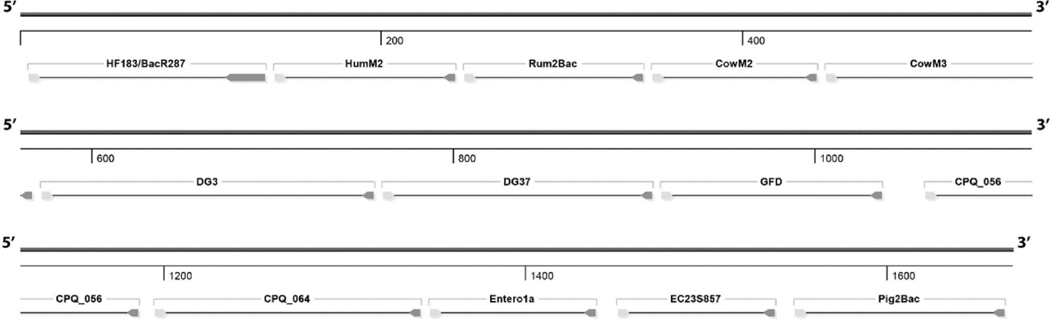
A single 1669 bp construct was designed to function with 13 recreational water quality monitoring qPCR methods including assays targeting host-associated genetic markers for human (HF183/BacR287, HumM2, CPQ_056, and CPQ_064), cattle (CowM2 and CowM3), canine (DG3 and DG37), ruminant (Rum2Bac), swine (Pig2Bac), and avian (GFD), as well as two general fecal indicator bacteria assays targeting enterococci (Entero1a) and E. coli (EC23S857) (Fig. 1). Primers, hydrolysis probes, source target, and citations for each qPCR assay are listed in Table 1.
Fig. 1.
Map of 13 recreational water quality qPCR assay target construct used to generate the pilot reference material and manufacture the NIST SRM® 2917. Each qPCR target location is defined with corresponding labels.
2.2. Reference material preparations
To investigate the potential influence of yeast carrier tRNA on qPCR assay performance, a pilot reference material was prepared by USEPA allowing for testing of the standard control in the presence and absence of yeast carrier tRNA. This was necessary because the NIST SRM® 2917 preparation contains 10 ng/μL yeast carrier tRNA (Thermo Fisher Scientific, Grand Island, NY) preventing a direct comparison (with and without yeast carrier tRNA) using the same reference material preparation. A plasmid-based internal amplification control (IAC) used to monitor for recreational water sample amplification inhibition was also prepared in accordance with standardized multiplex qPCR protocols for HF183/BacR287, HumM2, and CowM2 (Shanks et al. 2008, USEPA 2019a, b). Plasmid constructs (Integrated DNA Technologies, Coralville, IA) for the USEPA pilot reference material and IAC were linearized by either Not1 (pilot reference material) or Sca1 (IAC) restriction digestion (New England BioLabs, Beverly, MA), quantified with Quant-iT PicoGreen dsDNA assay kit (Thermo Fisher Scientific) on a Qubit 3 Fluorometer (Thermo Fisher Scientific) and diluted in 10 mM Tris and 0.1 mM EDTA (pH 8.0) to generate 10, 102, 103, 104, and 105 copies/2 μL for the pilot reference DNA material and 102 copies/2 μL for the IAC reference material. Pilot reference material and IAC preparations were stored in GeneMate Slick low-adhesion microcentrifuge tubes (ISC Bio-Express, Kaysville, UT) at −20 °C. NIST SRM® 2917 was supplied by NIST including five dilutions with mean concentrations as follows: Level 1 (10.3 copies/2 μL), Level 2 (1.11•102 copies/2 μL), Level 3 (1.06•103 copies/2 μL), Level 4 (1.06•104 copies/2 μL), and Level 5 (1.04•105 copies/2 μL). Details on NIST SRM® 2917 preparation are reported elsewhere (Kralj et al. 2021).
2.3. qPCR amplification
Thirteen qPCR assays (Table 1) were used in this study as previously reported with the following modifications. All reaction mixtures contained 1 × TaqMan Environmental Master Mix (version 2.0; Thermo Fisher Scientific, Grand Island, NY), 0.1 × SYBR Green I Dye (GFD assay only; Thermo Fisher Scientific), 0.2 mg/mL bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 1 μM each primer, and 80 nM 6-carboxyfluoroscein (FAM)-labeled probe (except GFD assay), and 80 nm VIC-labeled probe (multiplex reactions only). All reactions contained either 2 μL of reference material (pilot or NIST SRM® 2917) or laboratory grade water (no template controls) in a total reaction volume of 25 μL. HF183/BacR287, HumM2, and CowM2 also contained 102 copies of IAC template. Triplicate reactions were performed for all experiments. Amplifications were conducted on a QuantStudio 3 Real-Time PCR System in MicroAmp optical 96-well reaction plates with MicroAmp optical 96-well optical adhesive film (Thermo Fisher Scientific). The thermal cycling profile for all assays was 10 min at 95 °C followed by 40 cycles of 15 s at 95°C, and 1 min at 60°C (except GFD, 57°C and EC23S857, 56°C). The threshold was manually set to either 0.03 (Rum2Bac, Pig2Bac, DG3, DG37, CPQ_056, CPQ_064, Entero1a, and EC23S857) or 0.08 (HumM2, CowM2, CowM3, and GFD). Quantification cycle (Cq) values were exported to Microsoft Excel for further analysis.
2.4. Calculations and statistics
NIST SRM® 2917 master calibration models were generated for each qPCR assay from six independent standard curves using a Bayesian Markov Chain Monte Carlo approach (Sivaganesan et al. 2010, Sivaganesan et al. 2008). The lower limit of quantification (LLOQ) was defined as the 95% Bayesian credible interval (BCI) upper-bound from repeated measures (n = 18) of dilution Level 1 (10.3 copies per reaction). Amplification efficiency (E) for each master standard curve was calculated as follows: E = [10(−1/slope) −1] ± standard deviation. Outliers were defined as the absolute value of a Studentized residual of > 3. A Bayesian method was used to compare slope and intercept parameters generated from six instrument runs of four representative assays (HF183/BacR287, HumM2, Entero1a, and DG3) in the presence and absence of yeast tRNA (20 ng/reaction; Thermo Fisher Scientific) using the USEPA generated pilot reference material. If the 95% BCI for the difference in a respective parameter (e.g., slope or intercept) included zero, then no significant difference was observed. A two sample Z test (groups: with or without 20 ng/reaction of yeast carrier tRNA) was used to compare the mean Cq values at each dilution level for HF183/BacR287, HumM2, Entero1a, and DG3 assays. A Chi-square analysis was used to determine if there was a significant difference (α = 0.05) in the frequency of false positives in no template control reactions containing either laboratory grade water or yeast carrier tRNA (20 ng/reaction). All statistics were calculated with SAS software (Cary, NC) and WinBugs (https://www.mrc-bsu.cam.ac.uk/software/bugs/thebugs-project-winbugs). The 13 genetic target construct map was generated with DNASTAR Lasergene DNA SeqBuilder Pro™ Version 17.1.1 (Madison, WI).
3. Results
3.1. Yeast carrier tRNA does not introduce contamination
For each qPCR assay, no template controls with (n = 84) and without (n = 6) yeast carrier tRNA (20 ng/reaction) were tested to determine if this practice introduces contamination. All qPCR assays yielded no false positives (0 of 1080 reactions), except EC23S857. The EC23S857 assay indicated false positives in 16.7% (1 of 6) no template control reactions without the yeast carrier tRNA and 14.3% (12 of 84 reactions) containing yeast tRNA. To determine if there was a statistically significant difference (α = 0.05) in the occurrence of EC23S857 false positives between test conditions, paired no template control measurements were assessed with (n = 108) and without (n = 108) the yeast carrier tRNA. Fifteen false positives were observed with yeast carrier tRNA [13.9% (15 of 108)] and 10 false positives without yeast carrier tRNA [9.3% (10 of 108)] indicating no significant difference in occurrence (p = 0.288). EC23S857 false positive Cq values ranged from 36.9 to 38.8, all below the assay LLOQ (Table 2).
Table 2.
Summary of standard curve performance metrics based on six instrument runs for each qPCR assay using NIST SRM® 2917.
| Assay | Slope | Y-Intercept | E ± std | R2 | LLOQ |
|---|---|---|---|---|---|
| CowM2 | −3.36 ± 0.01 | 39.4 ± 0.06 | 0.98 ± 0.006 | 0.999 | 36.0 |
| CowM3 | −3.40 ± 0.01 | 37.7 ± 0.07 | 0.97 ± 0.005 | 0.998 | 34.3 |
| CPQ_056 | −3.35 ± 0.02 | 38.6 ± 0.13 | 0.99 ± 0.008 | 0.996 | 35.4 |
| CPQ_064 | −3.45 ± 0.01 | 39.9 ± 0.05 | 0.95 ± 0.006 | 0.998 | 36.4 |
| DG3 | −3.38 ± 0.04 | 35.9 ± 0.14 | 0.98 ± 0.014 | 0.998 | 32.7 |
| DG37 | −3.36 ± 0.02 | 36.1 ± 0.07 | 0.98 ± 0.008 | 0.998 | 32.8 |
| EC23S857 | −3.37 ± 0.02 | 36.7 ± 0.07 | 0.98 ± 0.008 | 0.998 | 33.3 |
| Entero1a | −3.37 ± 0.01 | 36.3 ± 0.05 | 0.98 ± 0.006 | 0.998 | 32.9 |
| GFD | −3.36 ± 0.02 | 34.6 ± 0.07 | 0.98 ± 0.008 | 0.998 | 31.2 |
| HF183/BacR287 | −3.37 ± 0.02 | 36.2 ± 0.08 | 0.98 ± 0.009 | 0.997 | 32.8 |
| HumM2 | −3.34 ± 0.03 | 38.7 ± 0.11 | 0.99 ± 0.011 | 0.997 | 35.4 |
| Pig2Bac | −3.36 ± 0.03 | 35.7 ± 0.12 | 0.98 ± 0.011 | 0.996 | 32.4 |
| Rum2Bac | −3.44 ± 0.02 | 39.3 ± 0.07 | 0.95 ± 0.009 | 0.998 | 35.9 |
E ± std denotes amplification efficiency [10(−1/slope) −1] ± standard deviation.
R2 represents linearity of the fitted curve.
LLOQ indicates lower limit of quantification reported as the quantitative threshold (Cq).
3.2. Yeast carrier tRNA does not influence calibration model performance
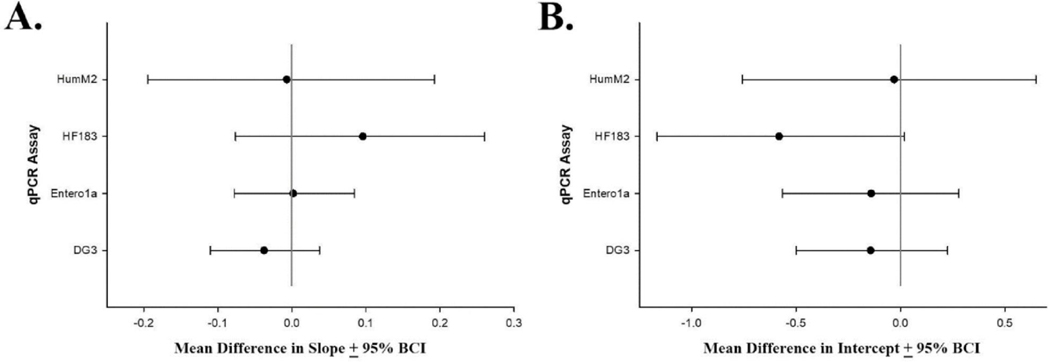
Standard curve slope, y-intercept, and Cq measurements at each dilution level were compared in the presence and absence of the yeast tRNA (20 ng/reaction) with HF183/BacR287, Entero1a, HumM2, and DG3 qPCR assays utilizing the USEPA generated pilot reference material. Using data generated from six independent instrument runs for each selected qPCR assay, no significant shift in slope (95% BCI includes zero; Fig. 2, Panel A) or y-intercept (95% BCI includes zero; Fig. 2, Panel B) were observed. No significant difference was also indicated for all qPCR assay and dilution level combinations (p ≥ 0.150) using a two sample Z test, except DG3 at 104 copies/reaction (p = 0.021). This exception was attributed to the small Cq standard deviations (0.019 and 0.020) in paired measurements and was considered negligible.
Fig. 2.
Scatter plots showing HF183/BacR287, HumM2, Entero1a, and DG3 qPCR assay mean difference (yeast carrier tRNA absent: yeast carrier tRNA present) for standard curve model slope (Panel A) and y-intercept (Panel B) with 95% Bayesian credible intervals (BCI). Shaded circles represent respective mean differences and error bars depict respective 95% BCI. The vertical line indicates a mean difference of zero. Calibration model parameter mean difference values where the 95% BCI intersects zero indicate no significant difference between the respective standard curve parameter with and without the yeast carrier tRNA stabilizer.
3.3. qPCR calibration model performance with NIST SRM® 2917
Six independent instrument run standard curves were generated for each qPCR assay DNA target using NIST SRM® 2917. Models indicated a range of quantification spanning 10.3 to 1.04•105 copies of target DNA per reaction (entire range tested in study) and R2 values of ≥ 0.996 regardless of qPCR assay. E ranged from 0.95 ± 0.006 (Rum2Bac) to 0.99 ± 0.008 (CPQ_056). A summary of standard curve performance parameters is shown in Table 2. A total of 31 outliers out of 1170 total measurements (2.6%) were identified and discarded when models were generated ranging from one (EC23S857, Pig2Bac, CPQ_064) to five (GFD) by assay with 93.5% (n = 29) occurring at the lowest standard dilution (Level 1 = 10.3 copies per reaction). HF183/BacR287, HumM2, and CowM2 multiplex IAC measurements (102 copies/2 μL) in no template controls yielded standard deviations ≤ 2.29 Cq (n = 18 measurements per assay). IAC measurements in NIST SRM® 2917 reactions performed as expected (data not shown).
3.4. NIST SRM® 2917 qPCR measurement variability at each dilution level
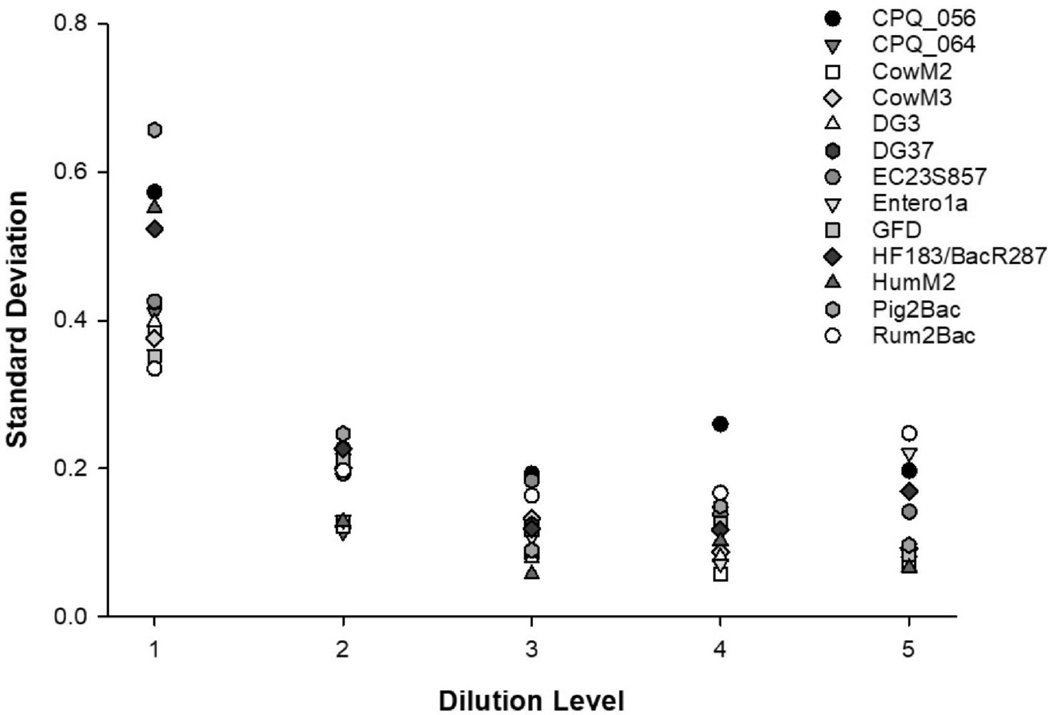
The standard deviation of repeated Cq measurements across six instrument runs for each qPCR assay were assessed at all dilution levels using NIST SRM® 2917 (Fig. 3). All qPCR assays exhibited a heteroscedastic trend with average Cq standard deviations of 0.443 at the 10.3 copies per reaction concentration (Level 1), 0.188 (1.11•102 copies per reaction; Level 2), 0.121 (1.06•103 copies per reaction; Level 3), 0.125 (1.06•104 copies per reaction; dilution Level 4), and 0.127 (1.05•105 copies per reaction; Level 5). For all qPCR assays, the highest Cq standard deviations were observed at the dilution Level 1 [range 0.657 (Pig2Bac) to 0.335 (Rum2Bac)].
Fig. 3.
Scatter plot depicting quantitative threshold (Cq) standard deviations in repeated measures (n = 18) generated across six instrument runs of NIST SRM® 2917 at each dilution (Levels 1–5) by qPCR assay.
3.5. Extraneous DNA controls
Across all experiments, no template control reactions indicated 99.75 % DNA-free (4 of 1620) excluding the EC23S857 qPCR assay. For EC23S857, a false positive rate of 22.2 % was observed (76 of 342 no template control reactions). All false positive Cq values were higher than respective LLOQ values.
4. Discussion
4.1. NIST SRM® 2917 performance for qPCR standard curve generation
NIST SRM® 2917 is designed to generate standard curves for 13 recreational water quality qPCR assays using a linearized plasmid DNA construct. Single laboratory performance assessment of this standard control material demonstrates the ability to generate high-quality calibration models for all qPCR assays with E values ranging from 0.95 ± 0.006 to 0.99 ± 0.008 and R2 ≥ 0.980, all within expert recommended guidelines for qPCR standard curves (Bustin et al. 2009). Regardless of qPCR assay, variability in repeated Cq measurements, generated from six independent instrument runs, at each dilution level were low (Cq standard deviations ≤ 0.657) and exhibited a heteroscedastic trend characteristic of qPCR standard curves (Bustin 2006), where measurement error is highest at the lowest concentration. Together these findings clearly demonstrate that NIST SRM® 2917 functions with all qPCR methods and suggest that the future use of this control material by scientists and water quality managers should help reduce variability in qPCR concentration estimates and make results more consistent between laboratories. Results also indicate that a multiple laboratory study is warranted to evaluate NIST SRM® 2917 performance across laboratories.
4.2. Use of yeast carrier tRNA
The addition of an RNA stabilizer such as yeast carrier tRNA to improve nucleic acid reference material storage stability is recommended for both qPCR and dPCR applications (Baoutina et al. 2019, dMIQE 2020, Jerome et al. 2002, Podivinsky et al. 2009). Yeast carrier tRNA was used in the preparation of NIST SRM® 2917 to maximize stability for storage at 4 °C (Kralj et al. 2021). Storage at 4 °C has been reported to be superior to −20 °C for DNA-based reference materials because this practice avoids unwanted degradation due to solution freezing and thawing after aliquot repeated use (Baoutina et al. 2019). To confirm that the presence of the yeast carrier tRNA did not introduce contamination or influence standard curve model performance (slope and y-intercept), a series of no template controls and standard curves were tested in the presence and absence of the yeast carrier tRNA additive. Findings indicate that the use of yeast carrier tRNA (10 ng/μL) as a stabilizer did not introduce contamination or alter qPCR performance suggesting that this practice may prove useful in future standard preparations for other nucleic acid-based control materials of interest.
4.3. The E. coli reagent contamination challenge
The presence of E. coli genomic fragments in commercially prepared molecular reagents has been well documented for more than 30 years (Corless et al. 2000, Hughes et al. 1994, Meier et al. 1993, Rand and Houck 1990, Schmidt et al. 1991, Silkie et al. 2008). These fragments are often identified in no template controls of PCR-based experiments designed to detect trace quantities of E. coli genetic material employing a high number of thermal cycles (> 35). The amount of E. coli contamination has been reported to vary across commercially prepared reagent lot preparations (Sivaganesan et al. 2019) and often occurs at levels, below the limit of quantification (< 10 copies per reaction). In this study, no template controls identified the presence of the E. coli multiple copy 23S ribosomal RNA gene in 22.2% of reactions (76 of 342) with all Cq values exceeding the EC23S857 LLOQ (33.3 Cq, Table 2) suggesting that the presence of contamination had a minimal influence on standard curves generated in this study. However, it is highly recommended for any PCR-based trace application targeting an E. coli genetic marker, such as recreational water quality monitoring, to conduct control experiments to characterize potential contamination in each reagent lot prior to sample testing.
4.4. Implications for water quality management
The lack of a standard control material is a key challenge for the successful widespread implementation of any qPCR-based recreational water quality testing method. The development of a high-quality material, readily available from a centralized source will have several important implications for water quality management. First, NIST SRM® 2917 allows for implementation of multiple qPCR water quality methods to assess recreational water public health risk as well as characterize human, dog, avian, ruminant, cattle, and pig fecal pollution sources, all from a single control material preparation. Because NIST SRM® 2917 functions with 13 qPCR water quality methods, practitioners can customize experiments to implement one or more of these methodologies using the same standard control material. Second, future use of this control material will help minimize concentration estimate variability and make results more comparable within and between laboratories. NIST SRM® 2917 will also benefit water quality management in less obvious ways. For example, standard curves generated with NIST SRM® 2917 may be used to establish performance benchmarks to help scientists, managers, reviewers, and the public evaluate the technical quality of future qPCR experiments against an established yardstick. These benchmarks could also potentially serve as performance metrics for future lab accreditation protocols. Finally, it is important to note that NIST SRM® 2917 qPCR targets were selected to augment routine recreational water quality testing, however, many of these assays are also commonly used in stormwater management (Ahmed et al. 2019, Staley et al. 2018), food production monitoring (Fu and Li 2014, Merino-Mascorro et al. 2018, Ravaliya et al. 2014), wastewater surveillance (Wilder et al. 2021), outbreak exposure route identification (Mattioli et al. 2021), or any other application seeking to characterize the amount and sources of fecal waste in a sample.
5. Conclusions
Single laboratory performance assessment of NIST SRM® 2917 demonstrates that this control material can consistently generate high-quality standard curves for all recreational water quality qPCR DNA targets. Key findings include:
NIST SRM® 2917 is the first standard control material produced for fecal pollution recreational water quality monitoring that functions with 13 qPCR assays that can assess recreational water public health risk, as well as characterize human, dog, avian, ruminant, cattle, and pig fecal pollution sources.
The addition of yeast carrier tRNA as a stabilizer does not compromise qPCR standard curve performance or introduce contamination.
All NIST SRM® 2917 dilution levels exhibited low variability across repeated measurements and instrument runs, regardless of qPCR assay with dilution Level 1 (10.3 copies per reaction) consistently exhibiting the highest variability.
Single laboratory performance assessment findings suggest that NIST SRM® 2917 is suitable for broad scale implementation for recreational water quality applications.
Genetic targets harbored on NIST SRM® 2917 could also serve as standard control material for stormwater, food production, wastewater surveillance, outbreak exposure route identification, or any other application seeking to characterize the amount and sources of fecal waste in a sample.
Although this study represents an important step towards the validation and widespread use of qPCR-based recreational water quality testing methods, it will be necessary to assess performance across multiple laboratories to confirm trends reported here.
Footnotes
Disclaimer
Information has been subjected to U.S. EPA peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use. These opinions, recommendations, findings, and conclusions do not necessarily reflect the views or policies of NIST or the United States Government. Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahmed A, Hamilton K, Toze S, Cook S, Page D, 2019. A review on microbial contaminants in stormwater runoff and outfalls: potential health risks and mitigation strategies. Sci. Total Environ 692, 1304–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw TG, Sivaganesan M, Briggs S, Dreelin E, Aslan A, Dorevitch S, Shrestha A, Isaacs N, Kinzelman J, Kleinheinz G, Noble R, Rediske R, Scull B, Rosenberg S, Weberman B, Sivy T, Southwell S, Siefring S, Oshima K, Haugland RA, 2019. Evaluation of multiple laboratory performance and variability in analysis of recreational freshwaters by a rapid Escherichia coli qPCR method (Draft Method C). Water Res. 156, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baoutina A, Bhat S, Partis L, Emslie KR, 2019. Storage stability of solutions of DNA standards. Anal. Chem 91, 12268–12274. [DOI] [PubMed] [Google Scholar]; Bustin SA, Bustin SA, 2006. A-Z of Quantitative PCR. In: International University Line. La Jolla, ca, pp. 3–29. [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, 2009. The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55 (4), 611–622. [DOI] [PubMed] [Google Scholar]
- Chern EC, Siefring S, Paar J, Doolittle M, Haugland R, 2011. Comparison of quantitative PCR Assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol 52, 298–306. [DOI] [PubMed] [Google Scholar]
- dMIQE G, 2020. The digital MIQE guidelines update: minimum information for publication of quantiative digital PCR experiments for 2020. Clin. Chem 66, 1012–1029. [DOI] [PubMed] [Google Scholar]
- Dorevitch S, Shrestha A, DeFlorio-Barker S, Breitenbach C, Heimler I, 2017. Monitoring urban beaches with qPCR vs. culture measures of fecal indicator bacteria: implications for public notification. Environ. Health 16, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebentier DL, Hanley KT, Cao Y, Badgley BD, Boehm AB, Ervin JS, Goodwin KD, Gourmelon M, Griffith JF, Holden PA, Kelty CA, Lozach S, McGee C, Peed LA, Raith M, Ryu H, Sadowsky MJ, Scott EA, Domingo JS, Schriewer A, Sinigalliano CD, Shanks OC, Van De Werfhorst LC, Wang D, Wuertz S, Jay JA, 2013. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 47 (18), 6839–6848. [DOI] [PubMed] [Google Scholar]
- Fu L, Li J, 2014. Microbial source tracking: a tool for identifying sources of microbial contamination in the food chain. Crit. Rev. Food Sci. Nutr 54, 699–707. [DOI] [PubMed] [Google Scholar]; Green HC, Dick LK, Gilpin B, Samadpour M, Field KG, 2012. Genetic markers for rapid PCR-based identificationof gull, Canada goose, duck, and chicken fecal contamination in water. Appl. Environ. Microbiol 78, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, Haugland R, Varma M, Millen HT, Borchardt MA, FIeld KG, Kelty CA, Sivaganesan M, Shanks OC, 2014a. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol 80 (10), 3086–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green HC, White KM, Kelty CA, Shanks OC, 2014b. Development of rapid canine fecal source identification PCR-based assays. Environ. Sci. Technol (48), 11453–11461. [DOI] [PubMed] [Google Scholar]
- Harwood VJ, Shanks OC, Korajkic A, Verbyla M, Ahmed A, Iriate M, 2017. General and Host-Associated Bacterial Indicators of Faecal Pollution. UNESCO, East Lansing, MI. [Google Scholar]
- Jerome KR, Huang M, Wald A, Selke S, Corey L, 2002. Quantitative stability of DNA after extended stroage of clinical specimes as determined by real-time PCR. J. Clin. Microbiol 40, 2609–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J, Servetas S, Hunter M, Toman B, Jackson S, 2021. Certification of standard reference material 2917 plasmid DNA for fecal indicator detection and identification. NIST Spec. Publ 1–41. [Google Scholar]
- Li X, Kelty CA, Sivaganesan M, Shanks OC, 2021. Variable fecal source prioritization in recreational waters routinely monitored with viral and bacterial general indicators. Water Res. 192, 116845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sivaganesan M, Kelty CA, Zimmer-Faust A, Clinton P, Reichman JR, Johnson Y, Matthews W, Bailey S, Shanks OC, 2019. Large-scale implementation of standardized quantitative real-time PCR fecal source identification procedures in the Tillamook Bay Watershed. PLoS One 14 (6), e0216827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Schleifer KH, 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol 23 (4), 556–562. [DOI] [PubMed] [Google Scholar]
- Mattioli MC, Benedict KM, Murphy J, Kahler A, Kline KE, Longenberger A, Mitchel PK, Watkins S, Berger P, Shanks OC, Barrett CE, Barclay L, Hall AJ, Hill V, Weltman A, 2021. Identifying septic pollution exposure routes during a waterborne norovirus outbreak - A new application for human-associated microbial source tracking qPCR. J. Microbiol. Methods 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino-Mascorro JA, Hernandez-Rangel LG, Heredia N, Garcia S, 2018. Bacteroidales as indicators and source trackers of fecal contamination in tomatoes and strawberries. J. Food Prot 81, 1439–1444. [DOI] [PubMed] [Google Scholar]
- Mieszkin S, Furet JP, Corthier G, Gourmelon M, 2009. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Appl. Environ. Microbiol 75, 3045–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieszkin S, Yala JF, Joubrel R, Gourmelon M, 2010. Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of rumaint faecal pollution by real-time PCR. J. Appl. Microbiol 108, 974–984. [DOI] [PubMed] [Google Scholar]
- Podivinsky E, Love JL, van der Colff L, Samuel L, 2009. Effect of storage regime on the stabiliyt of DNA used as a calibration standard for real-time polymerase chain reaction. Anal. Biochem 394, 132–134. [DOI] [PubMed] [Google Scholar]
- Ravaliya K, Gentry-Shields J, Garcia S, Heredia N, Fabiszewski de Aceituno A, Bartz FE, Leon JS, Jaykus L, 2014. Use of Bacteroidales microbial source tracking to monitor fecal contamination in fresh produce production. Appl. Environ. Microbiol 80, 612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Atikovic E, Blackwood AD, Lu J, Noble RT, Santo Domingo J, Siefring S, Sivaganesan M, Haugland RP, 2008. Quantitative PCR for Detection and Enumeration of Genetic Markers of Bovine Fecal Pollution. Appl. Environ. Microbiol 74 (3), 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, Field KG, Sivaganesan M, 2016. Data acceptance criteria for standardized human-associated fecal source identificationq quantitative real-time PCR methods. Appl. Environ. Microbiol 82 (9), 2773–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA, 2009. Quantitative PCR for genetic markers of human fecal pollution. Appl. Environ. Microbiol 75, 5507–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Sivaganesan M, Peed L, Kelty CA, Noble RT, Blackwood AD, Bushon RN, Stelzer EA, Kinzelman J, Anan’eva T, Sinagalliano CD, Wanless D, Griffith JF, Cao Y, Weisberg SB, Harwood VJ, Staley C, Oshima KH, Varma M, Haugland R, 2012. Inter-laboratory comparison of real-time PCR methods for quantification of general fecal indicator bacteria. Environ. Sci. Technol 46, 945–953. [DOI] [PubMed] [Google Scholar]
- Shrestha A, Dorevitch S, 2019. Evaluation of rapid qPCR method for quantification of E. coli at non-point source impacted Lake Michigan beaches. Water Res. 156, 395–403. [DOI] [PubMed] [Google Scholar]
- Shrestha A, Kelty CA, Sivaganesan M, Shanks OC, Dorevitch S, 2020. Fecal pollution source characterization at non-point source impacted beaches under dry and wet weather conditions. Water Res, 116014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefring SC, Varma M, Atikovic E, Wymer LJ, Haugland RA, 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health 6, 225–237. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Aw TG, Briggs S, Dreelin E, Aslan A, Dorevitch S, Shrestha A, Isaacs N, Kinzelman J, Kleinheinz JG, Noble R, Rediske R, Schull B, Rosenberg S, Weberman B, Sivy T, Southwell B, Siefring S, Oshima K, Haugland RA, 2019. Standardized data quality acceptance criteria for rapid E. coli qPCR method (Draft Method C) for water quality monitoring at recreational beaches. Water Res. 156, 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaganesan M, Haugland RA, Chern EC, Shanks OC, 2010. Improved strategies and optimization of calibration models for real-time PCR absolute quantification. Water Res. 44, 4726–4735. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Seifring S, Varma M, Haugland RA, Shanks OC, 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinf. 9, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler E, Kelty CA, Sivaganesan M, Li X, Bibby K, Shanks OC, 2017. Development of CrAssphage quantitative real-time PCR assays for human fecal pollution measurement. Environ. Sci. Technol 51, 9146–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley ZR, Boyd RJ, Shum P, Edge TA, 2018. Microbial source tracking using quantitative and digital PCR to identify sources of fecal contamination in stormwater, river water, and beach water in a Great Lakes area of concern. Appl. Environ. Microbiol 84, e01634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA, 2013. In: Water E.O.o. (Ed.), Method 1609: Enterococci in Water by TaqMan Quantitative Polymerase Chain Reaction (qPCR) with Internal Amplification Control (IAC) Assay. United States Environmental Protection Agency, Washington, DC. [Google Scholar]
- USEPA, 2019a. In: Water E.O.o. (Ed.), Method 1696: Characterization of Human Fecal Pollution in Water by HF183/BacR287 TaqMan quantitative Polymerase Chain Reaction (qPCR) Assay. United States Environmental Protection Agency, Washington DC. [Google Scholar]
- USEPA, 2019b. In: Water E.O.o. (Ed.), Method 1697: Characterization of Human Fecal Pollution in Water by HumM2 TaqMan Quantitative Polymerase Chain Reaction (qPCR) Assay. United States Environmental Protection Agency, Washington DC. [Google Scholar]
- USEPA (2021) ATTAINS: national summary of causes of impairment. [Google Scholar]
- Wilder ML, Middleton F, Larsen DA, Du Q, Fenty A, Zeng T, Insaf T, Kilaru P, Collins M, Kmush B, Green HC, 2021. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X 11, 100100. [DOI] [PMC free article] [PubMed] [Google Scholar]