Abstract
Oxysterol-binding Protein (OSBP) is a human lipid-transport protein required for the cellular replication of many types of viruses, including several human pathogens. The structurally-diverse small molecule compounds OSW-1, itraconazole (ITZ), T-00127-HEV2 (THEV) and TTP-8307 (TTP) inhibit viral replication through interaction with the OSBP protein. The OSW-1 compound reduces intracellular OSBP, and the reduction of OSBP protein levels persists multiple days after the OSW-1-compound treatment is stopped. The OSW-1-induced reduction of OSBP levels inhibited Enterovirus replication prophylactically in cells. In this report, the OSBP-interacting compounds ITZ, THEV, and TTP are shown not to reduce OSBP levels in cells, unlike the OSW-1-compound, and the OSW-1 compound is determined to be the only compound capable of providing prophylactic antiviral activity in cells. Furthermore, OSW-1 and THEV inhibit the binding of 25-hydroxycholesterol (25-OHC) to OSBP indicating that these compounds bind at the conserved sterol ligand binding site. The ITZ and TTP compounds do not inhibit 25-hydroxycholesterol binding to OSBP, and therefore ITZ and TTP interact with OSBP through other, unidentified binding sites. Co-administration of the THEV compound partially blocks the cellular activity of OSW-1, including the reduction of cellular OSBP protein levels; co-administration of the ITZ and TTP compounds have minimal effect on OSW-1 cellular activity further supporting different modes of interaction with these compounds to OSBP. OSW-1, ITZ, THEV, and TTP treatment alter OSBP cellular localization and levels, but in four distinct ways. Co-administration of OSW-1 and ITZ induced OSBP cellular localization patterns with features similar to the effects of ITZ and OSW-1 treatment alone. Based on these results, OSBP is capable of interacting with multiple structural classes of antiviral small molecule compounds at different binding sites, and the different compounds have distinct effects on OSBP cellular activity.
1. Introduction
Oxysterol-binding protein (OSBP) is a member of a lipid-binding protein family (i.e., OSBP and the OSBP-related proteins (ORPs)) conserved in all eukaryotes.1,2 Originally discovered based on its high affinity binding to 25-hydroxycholesterol and other oxysterols, the cellular function of OSBP has long been associated with the sensing or the transport of lipids, particularly sterols.1,2 All OSBP/ORP family members possess a C-terminal ~50 kDa OSBP-related ligand-binding domain (ORD).1,2 More recent work has identified OSBP as executing a critical lipid transport function at the membrane contact sites (MCS) between the ER and Golgi.3,4 OSBP transports cholesterol from the ER to the Golgi, while counter-transporting phosphosphoinositide-4-phosphate from the Golgi to the ER.3,4 In this role, OSBP mediates the necessary movement and regulation of the lipids between organelles, and OSBP is one of a growing roster of proteins, including multiple ORPs, involved in inter-organelle transport at MCS.5–7 ORP4 is the OSBP/ORP family member most closely related to OSBP based on conserved amino acid sequence (~64% overall sequence identity, ~67% in lipid binding domain).1,2 Despite their sequence similarity, OSBP and ORP4 perform different cellular functions.1,2 In immortalized cell lines8 and leukemia, ORP4 is highly expressed and drives proliferation.9–11 In leukemia stem cells, ORP4 is recently reported to transfer phosphoinositide-2-phosphate (PIP2) from the plasma membrane to PLCβ3 to trigger pro-growth cell signaling pathways.10,11 In contrast, reduction of OSBP in cancer cells lines by >90% has no observable effect on cellular proliferation or morphology.12,13
More recently, OSBP, but not ORP4, was determined to execute a critical role in the proliferation of RNA viruses.14 This unexpected discovery was made through identifying OSBP as the cellular protein through which the compound itraconazole prevented broad spectrum Enterovirus viral replication.15,16 OSBP executes a vital role in the replication of a growing number of clinically-important human pathogens including Enterovirus genus pathogens14,17,18, hepatitis C virus (HCV),19,20 encephalomyocarditis virus (EMCV)21, dengue virus22, and Zika virus.22 OSBP is important for the formation of the membrane bound viral replication organelles (RO), which form at the ER-Golgi interface.23–25
Four antiviral compounds have been identified as functioning through targeting OSBP: OSW-1 (2)13,17, itraconazole (ITZ) (3)15,16, T-00127-HEV-2 (THEV) (4)26, and TTP-8307 (TTP) (5)26 (Scheme 1). OSW-1 is an antiproliferative natural product compound that exerts its biological activity through interacting with OSBP and ORP4.12 The antiproliferative activity of the OSW-1 compound is likely due to interacting with ORP4, and the antiviral activity of OSW-1 is through its interaction with OSBP.13,14,17 ITZ, a clinically approved anti-fungal drug27, is a promiscuous binder reported to interact with VEGF228,29, Smoothened in the Hedgehog signaling pathway30, and both VDAC1 and NPC1 simultaneously.31 An additional report suggests that the ITZ antiviral activity is attributed to its interaction with the viral non-structural 3A protein in EV-A71, instead of OSBP.32
Scheme 1:
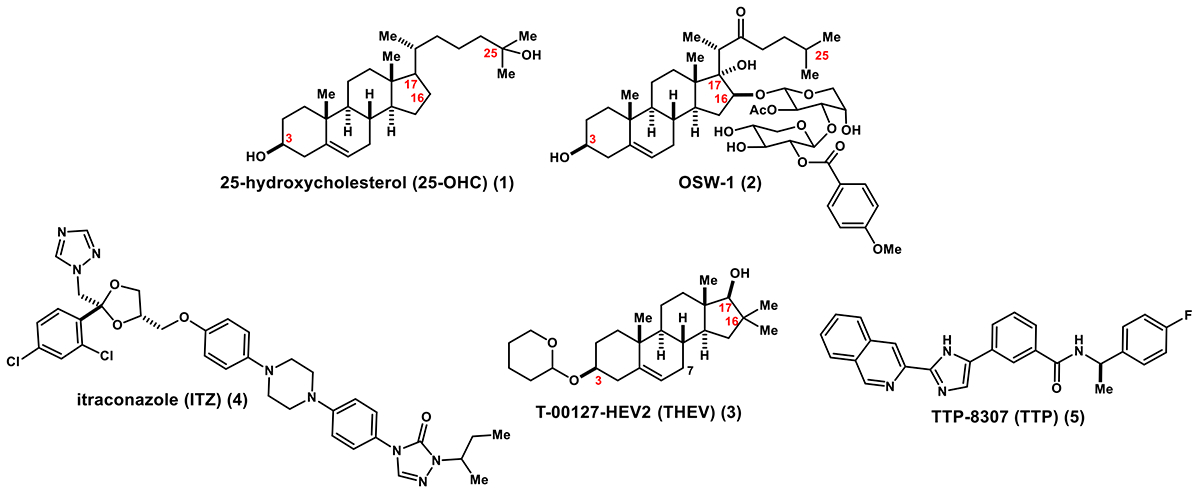
25-Hydroxycholesterol and OSBP Interacting Antiviral Compounds
There is no reported OSBP protein structure, but the conserved ORDs in related OSBP/ORP family members have been determined.1,2,6 The solved ORP protein structures provide evidence for two OSBP small molecule ligand binding sites on the ORD: 1) a well-established oxysterol binding site33; and 2) a separate phospholipid binding site, distinct from the N-terminal Pleckstrin Homology (PH) domain, which also has the capacity to bind phosphoinositides.6,34,35 Functional studies have also demonstrated that the residues required for OSBP ORD phospholipid binding are not required for oxysterol binding.3,4,36
Of the four OSBP-interacting compounds, only OSW-1 (2) is established to effect binding to one of the two established OSBP ligand binding sites (i.e., oxysterol or phospholipid binding sites). OSW-1 (2) competitively inhibits the binding of 25-hydroxycholesterol (25-OHC) (1) to OSBP and ORP4 with low nanomolar Kis.12,13 ITZ (3) is reported to have a KD of ~430 nM for full-length GFP-OSBP, as measured using microscale thermophoresis.15 This method has not been used to measure binding of any other potential ligand to OSBP, including 25-OHC (1) or OSW-1 (2).14 OSBP binding measurements for THEV (4) and TTP (5) have not been reported. The antiviral activity of the THEV compound (4) is potentiated by OSBP knockdown in cells, which supports the compound targeting OSBP in cells.26 The TTP compound (5) is shown to inhibit the ability of OSBP to transport cholesterol between vesicles, in vitro.26 13 Neither ITZ (3) or TTP (5) substantially inhibited the interaction of the N-terminal OSBP FFAT domain with the established OSBP-interacting protein VAP-A, as determined using bimolecular fluorescent complementation (BiMC) assay.37 Based on these results, ITZ (3) or TTP (5) are inferred not to bind to the N-terminal of region of OSBP which contains the FFAT domain, but instead likely binds to the C-terminal ORD.37 Lipid transfer assays using OSBP proteolyzed fragments suggested that ITZ interacted with the C-terminal ORD and blocked the ORD transfer activity of both sterols and PI4P.38 ITZ was computationally docked into a homology structure of the OSBP ORD overlapping with the putative oxysterol binding site.16 OSW-1 (2), ITZ (3), THEV (4), and TTP (5) have minimal overall structural similarity (Scheme 1), although THEV and OSW-1 share a steroidal core.
OSW-1 treatment induces a pronounced and lasting reduction in cellular OSBP levels.12,13 A short duration (1–6 h), non-cytotoxic dose of OSW-1 reduces OSBP levels in cells >90%, and the reduction of cellular OSBP levels persists multiple days after the compound treatment ceased.13 The cells with OSBP levels reduced >90% for multiple days show no effect on cellular proliferation, viability, or morphology.13 Cellular treatment with the OSBP high affinity ligands 25-OHC or the natural product schweinfurthin A compound do not reduce OSBP levels, indicating that a reduction of cellular OSBP levels is not a universal response to ligand binding.12 Furthermore, the OSW-1-induced reduction of OSBP levels in cells confers prophylactical antiviral activity against multiple Enterovirus strains 24 hours after the end of compound treatment.13
In this report, the other antiviral OSBP-interacting compounds ITZ, THEV, and TTP are shown not to reduce OSBP levels in cells or to provide prophylactic antiviral activity. Additionally, the THEV compound is shown to inhibit the binding of 25-OHC to OSBP and to ORP4, similar to OSW-1. ITZ and TTP do not inhibit 25-OHC binding to OSBP and ORP4, suggesting these compounds interact with OSBP through a different binding site. ITZ and THEV at high concentrations relative to OSW-1 can partially block the OSBP reduction caused by OSW-1. Interestingly, all four compounds—OSW-1, ITZ, THEV, and TTP—produce distinct changes on OSBP localization in cells. These results indicate that the structurally-diverse antiviral small molecule compounds are capable of interacting with and modulating the OSBP cellular activity through multiple different modes of action.
2. Materials and Methods
2.1. Cell lines and viruses
HCT-116, HEK293, RD, and HeLa cell lines were purchased from ATCC. HEK293 STF (CRL-3249), RD (CCL-136), and HeLa (CCL-2) were cultured in DMEM (Thermo 11995073) supplemented with 10% Hyclone (Fisher Sci SH3006603) and 1% penicillin streptomycin (Thermo 15140122). HCT-116 (ATCC CCL-247) was cultured in McCoys 5A media (Thermo 16600108) supplemented with 10% Hyclone and 1% penicillin streptomycin. Coxsackievirus A9 (strain CoxA9–01) and Echovirus 2 (strain Echo2–01) were obtained from the Oklahoma State Department of Health Laboratory.
2.2. Western blotting antibodies
Primary antibodies used for Western blot were OSBP A-5 (Santa Cruz sc-365771) and Beta-actin HRP (Santa Cruz sc-47778 HRP). Secondary antibodies used were goat anti-mouse IgG1-HRP (Santa Cruz sc-2060).
2.3. Immunofluorescent Imaging Antibodies
Primary antibodies used for immunofluorescent (IF) imaging were OSBP1 1F2 (Novus NBP2–00935) and TGN46 (Novus NBP1–49643). Secondary IF antibodies used were goat anti-mouse IgG H&L Alexa Fluor® 488 (Abcam ab150113) and donkey anti-rabbit IgG H&L Alexa Fluor® 594 (Abcam ab150076).
2.4. OSW-1, ITZ, THEV, and TTP Compounds
OSW-1 was generated either through chemical synthesis in our lab39 or isolation from the natural source40. ITZ was purchased from Sigma-Aldrich (I6657) as a 1:1:1:1 mixture of diastereomer. Both the T-00127-HEV2 (THEV) and TTP-8307 (TTP) compounds were synthesized in our group. TTP-8307 was made following the published routes.37 To our knowledge, a procedure for the synthesis of the THEV compound had not been reported. OSW-1, THEV, and TTP were subjected to HPLC purification to produce analytically pure compounds (>95% purity) and fully characterized using spectroscopic techniques (see Supporting Information for compound data).
2.5. OSBP Expression in Cells Upon Compound Treatments
HCT-116 or HEK293 cells were seeded out 5×105 into 6-well plates and incubated at 37 °C for 20 h. Compound stock solutions were diluted in complete media before treating the cells (1 nM OSW-1 and/or 10,000 nM THEV, TTP, ITZ). After compound treatment, cells were lysed according to the lysis procedure in the Supporting Information.
2.6. Viral Proliferation Assays
The CoxA9–01 or Echo2–01 clinically isolated viruses were passaged twice in RD cells and then stored at −80 °C until use.
Continual Compound Treatment Antiviral Experiment:
2.0 × 105 HeLa cells were plated in 1 mL of media in 24-well plates and incubated overnight. Then, the media was removed, and cells were infected with either CoxA9–01 or Echo2–01 viruses (estimated MOI = 1.0) in serum-free DMEM. The virus and cells were incubated for 30 min, at which point the virus inoculum was removed, and the culture was washed once with serum free media. Then, the infected cells were dosed with the indicated compound, 10,000 nM for THEV, TTP, and ITZ, or 10 nM for OSW-1 (4 treatments per virus for each compound) in 1 mL of complete media, and incubated with the compound for 10 h, and then snap-frozen and stored at −80 °C until processing. Viral titration was performed by thawing the plates, scraping the cells into microcentrifuge tubes, and then centrifuging the samples at 10,000g at 4 °C to produce the virus containing supernatant, which is assayed for TCID-50 titration on subconfluent RD cells (N=3). The graphs represent averages of three independent experiments.
Compound Washout Antiviral Experiment:
This experiment was performed identically to the antiviral assay described above, with the following changes. Cells were treated with the indicated compound-containing media for 6 h. Then, the cells were washed three times in compound-free complete media and incubated for 24 h in compound-free media. After 24 h washout recovery, the cells were then infected with CoxA9–01 or Echo2–01 viruses at an estimated MOI of 1.0 for 30 min, cultured for 10 h in complete media, and then subjected to analysis as described above. There were four treatments per virus for each compound, n=4. The graphs represent three independent experiments.
2.7. 96-well [3H]-25-OHC Competitive Binding Assay
The [3H]-25-OHC binding assay was conducted as previously described.12,13
2.8. Viability Assays
96-well Cytotoxicity Assay:
48 h cytotoxicity assays were performed in 96-well plates using standard CellTiter-Blue® (Promega) procedure. See supporting Information for a detailed protocol.
Trypan Blue Cell Viability Assay-
HCT-116 cells were seeded out 5 × 105 into 6 well plates incubated at 37 °C for 20 h. The cells were treated with DMSO 1 nM OSW-1, 10,000 nM Itraconazole, 10,000 nM TTP, 10,000 nM THEV, or a combination of treatments for 24 h. Cells were washed once with PBS, detached using TrypLE™ Express trypsin, and counted on a TC20™ Automated Cell Counter (BioRad) by combining 10 μL of cell solution with 10 μL Trypan Blue stain (Thermo 15250061).
2.9. Immunofluorescence Cell Imaging Experiments
HCT-116 immunofluorescent microscopy on endogenous OSBP was performed as described previously13, except with a 24 h compound treatment.
2.10. Statistical Analysis
All results are expressed as mean ± SD and are n ≥ 3 unless otherwise stated. All statistical tests were performed using GraphPad Prism 7.0. Comparison between groups was made by using a one-way ANOVA with a follow up Dunnett’s test. The p values are reported using GraphPad Prism: *p ≤ 0.05, **p ≤ 0.01 ***p ≤ 0.001, and ****p ≤ 0.0001.
2.11. Other Methods:
Any remaining experimental procedures and additional details of the methods outlined above are described in the Supporting Information.
3. Results
3.1. Only OSW-1, and not ITZ, THEV, or TTP, reduce OSBP levels in cells.
OSW-1 treatment is established to reduce OSBP cell levels in multiple cells lines, including HEK293, HCT-116, and HeLa.12,13 HEK293 and HCT-116 cells treated with 1 nM of the OSW-1 compound for 24 h show the pronounced reduction of OSBP cellular levels (Fig. 1A).12,13 10,000 nM treatment of the cells for 24 h with the ITZ, THEV, or TTP compounds do not reduce OSBP levels. Taxol, the tubulin binding anticancer drug, is included to show that OSBP levels are not reduced upon cellular treatment with a cytotoxic compound. 25-OHC also does not reduce OSBP levels in cells.12 OSW-1 is also the only compound to induce the reduction of OSBP in levels in HeLa cells after 6 h treatment and 24 h of compound-free recovery(Fig. 1B); ITZ, THEV, and TTP at 10,000 nM treatment did not affect OSBP levels after the 6 h treatment, 24 h recovery (Fig. 1B).
Figure 1: OSW-1 is the Only Compound That Reduces Cellular OSBP Levels.
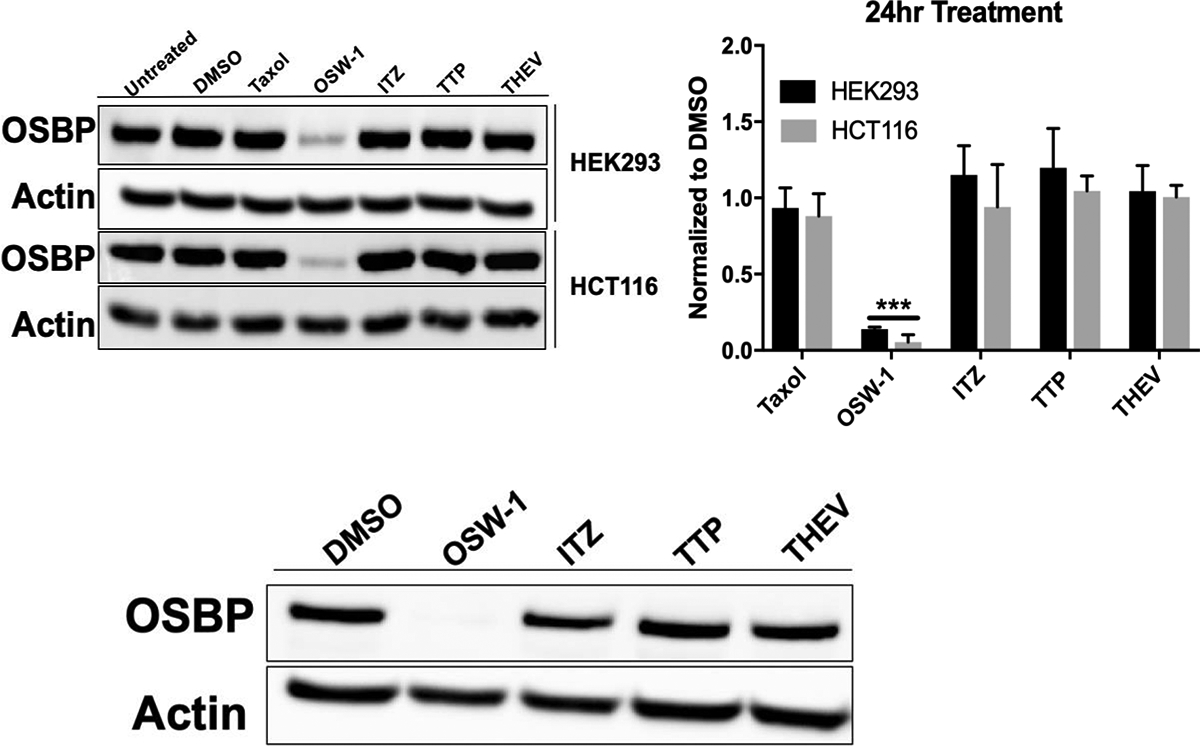
A) Effect of compound treatment on cellular OSBP expression in HCT-116 and HEK293 cells. Cells were treated with DMSO vehicle control, OSW-1 (1 nM), Taxol (10 nM), ITZ (10,000 nM), TTP (10,000 nM) or THEV (10,000 nM) for 24 h. Representative Western blot and average of multiple independent experiments shown. B) OSBP levels via Western blot in HeLa cells treated with compound (OSW-1 10 nM, THEV 10,000 nM, ITZ 10,000 nM, or TTP 10,000 nM) for 6 h followed by cell washing and incubation in compound-free media for 24 h. Western blot represents OSBP protein levels at the time of inoculation during washout treatment in anti-viral experiments (see Figure 2).
3.2. Only OSW-1, and not ITZ, THEV, or TTP, induces prophylactic antiviral activity in cells.
Treatment of cells with 1 nM OSW-1 for 6 h, followed by removing the compound from the cells triggers a repression of OSBP levels that last for 72 h after the compound treatment is stopped.13 Previously, a 1 nM OSW-1 treatment in HeLa cells conferred significant antiviral protection against the Enterovirus pathogens Cosackievirus 9a or Echovirus 2 24 h after OSW-1 treatment ended.13 The ITZ, THEV, and TTP compounds were tested for prophylactic antiviral activity similar to that of OSW-1 (Fig. 2). Cells infected with virus for 30 minutes, followed by a 10 h treatment with OSW-1 (10 nM), ITZ (10,000 nM), THEV (10,000 nM) or TTP (10,000 nM) (Fig. 2A) showed antiviral activity consistent with previous reports.13,15,17,26,37 However, OSW-1 is the only compound that displayed antiviral activity when viral infection occurred 24 h after compound treatment stopped (Fig. 2B). Cellular OSBP levels are reduced >90% after the 6 h OSW-1 treatment, 24 h recovery, which is just prior to viral infection (Fig. 1B). 6 h OSW-1 treatment of cells is not cytotoxic nor anti-proliferative.13
Figure 2: OSW-1 is the Only Compound That Provides Prophylactic Anti-Viral Activity.
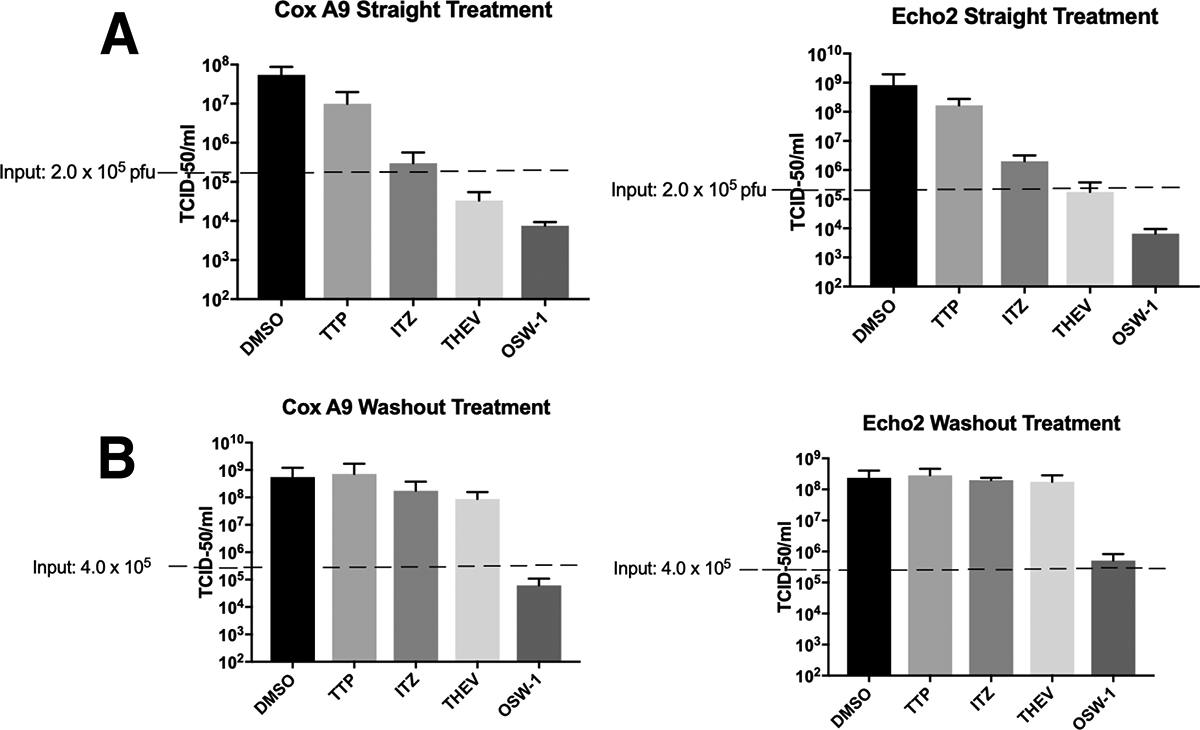
A) HeLa cells were infected with either Cosackievirus A9 or Echovirus 2 for 30 minutes, followed by compound treatment for 10 h OSW-1 (10 nM), THEV (10,000 nM), ITZ (10,000 nM), or TTP (10,000 nM). B) Compound Prophylactic Antiviral Activity. HeLa cells were treated with OSW-1 (10 nM), THEV (10,000 nM), ITZ (10,000 nM), or TTP (10,000 nM) for 6hr. Compound was removed from the cells and cell media, and the cells were incubated in compound-free media for 24 h. Then, cells were infected with Cosackievirus A9 or Echovirus 2 for 30 minutes, followed by 10 h incubation in compound-free media. Viral titers from three independent experiments shown.
3.3. Both OSW-1 and THEV inhibit oxysterol binding to OSBP and ORP4; ITZ and TTP do not.
OSW-1, ITZ, THEV, and TTP were tested in the established 96-well OSBP competition ligand binding assay, which measures compound binding to OSBP through inhibition of 25-OHC binding.12,13 KD of 25-OHC binding to human OSBP is 22+/−5 nM (Supp. Fig 2); human ORP4 25-OHC KD is reported as 54+/−23 nM.12 Compounds were measured for inhibition of binding of 20 nM of 3H-25-OHC to either OSBP or ORP4 overexpressed in HEK239T lysate. Consistent with published reports, OSW-1 produced a Ki in this binding assay of 16 +/− 4 nM for OSBP and 71 +/− 6 for ORP4 (Fig. 3).12,13 The THEV compound also showed strong competitive binding of 25-OHC, with Kis of 22 +/− 15 nM against OSBP and 98 +/− 15 nM for ORP4 (Fig. 3). ITZ and TTP showed no 25-OHC inhibition binding to OSBP or ORP4 at a concentration of 10,000 nM (Fig. 3).
Figure 3: OSW-1 and THEV Inhibit 25-OHC Binding to OSBP; ITZ and TTP Do Not.
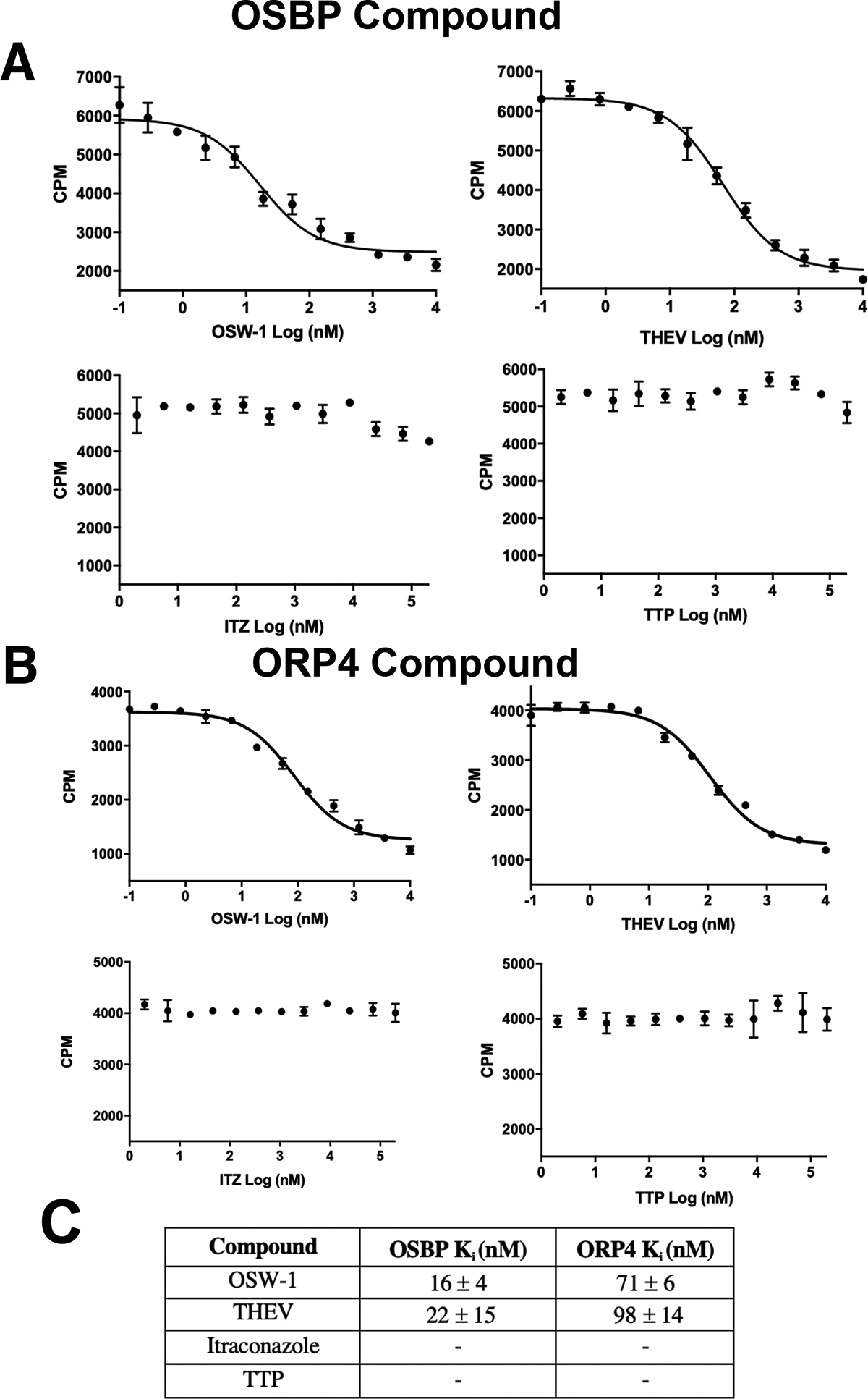
Compound binding to OSBP (A) or ORP4 (B) measured using a competitive 25-OHC binding assay. Representative inhibition binding curves are shown. C) Average Ki values from three independent binding experiments for OSW-1 and THEV to OSBP and ORP4. ITZ and TTP show no 25-OHC competitive binding.
3.4. THEV and ITZ, but not TTP, inhibits OSW-1 activity in cells supporting different modes of compound binding to OSBP.
Treatment of HCT-116 cells with 1 nM of OSW-1 and 10,000 nM ITZ shows a slight (~15–20%) effect on cell population after 24 h, assessed through cell counting after Trypan blue staining (Fig. 4A). 10,000 nM of THEV or TTP treatment does not affect the cell population (Fig. 4A). Co-administration of 1 nM OSW-1 with 10,000 nM of either ITZ, TTP, or THEV shows that OSW-1 and ITZ do not additively reduce the cell population (Fig. 4A). Co-administration of 10,000 nM of THEV with OSW-1 rescues cell population numbers (Fig. 4A). Extended cellular treatment with OSW-1 longer than 24 h is antiproliferative in immortalized cell lines, as determined using assays for cell metabolic activity.12,13 In a 48 h assay for cell metabolic activity, ITZ, THEV, and TTP showed no antiproliferative activity at <10,000 nM concentrations (Fig. 4B, Supp. Fig. 1). OSW-1 produced a growth inhibition 50% (GI50) value of 0.33 +/− 0.18 nM in HeLa cells and 0.22 +/− 0.09 nM in HEK293 cells after 48 h treatment. Co-administering either ITZ, THEV, or TTP at 10,000 nM does not significantly affect the antiproliferative OSW-1 activity in either HeLa or HEK293 cells (Fig. 4B). Co-administration of either 10,000 nM THEV or ITZ with 1 nM OSW-1 partially blocks the reduction of OSBP levels in both HEK293 and HCT116 cells (Fig. 4C). THEV rescues OSBP levels to a greater extent than ITZ (Fig. 4C). Co-administration of 10,000 nM of TTP with 1 nM OSW-1 has no effect on OSBP levels in cells (Fig. 4C).
Figure 4: THEV Co-administration blocks the cellular activity of OSW-1.
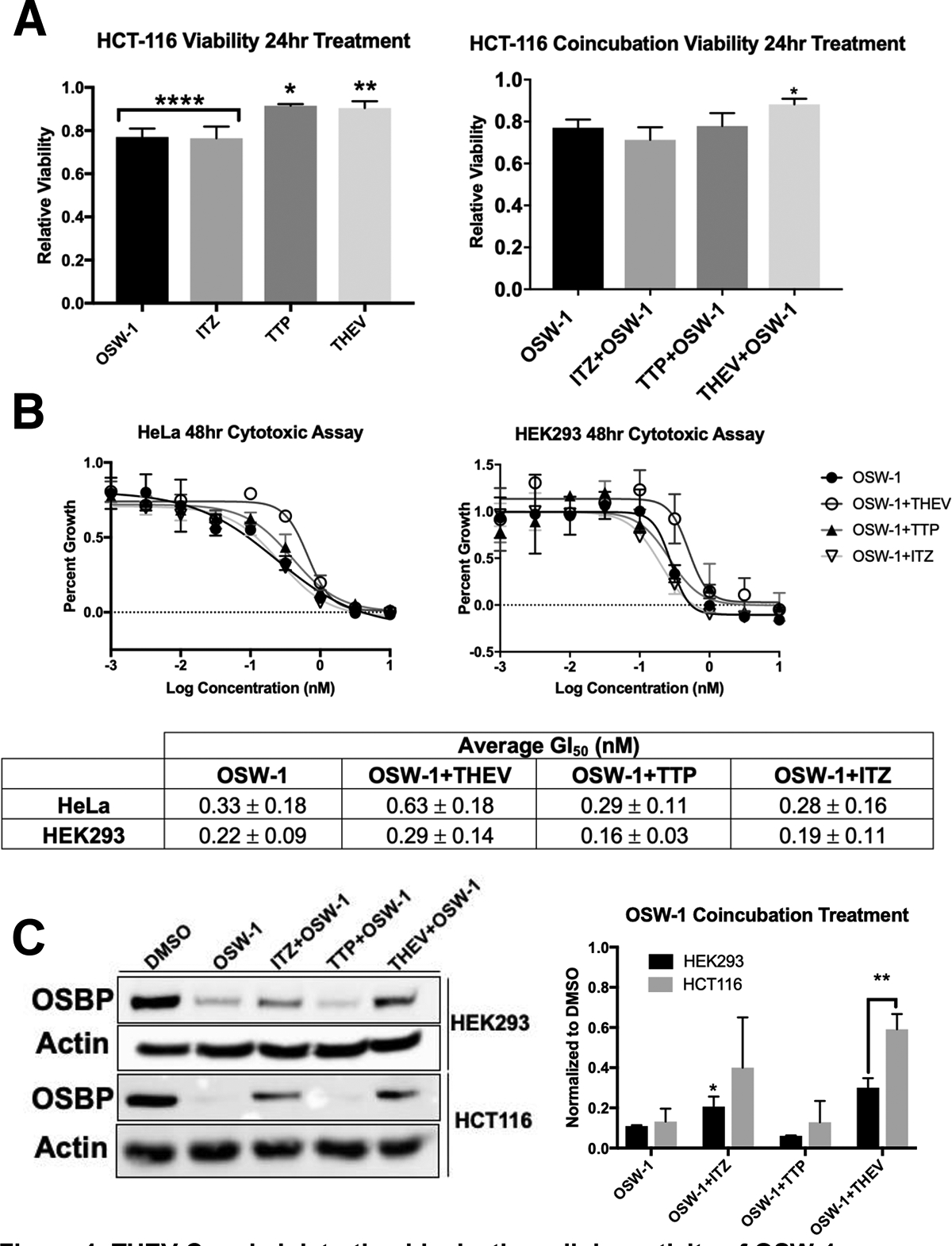
A) Viability of cells using Trypan Blue stain after 24 h of compound treatment or co-administration of OSW-1 (1 nM) with 10,000 nM of ITZ, THEV, or TTP. B) Cytotoxicity of 1 nM OSW-1 at 48 h co-administered with 10,000 nM THEV, TTP, or ITZ in HCT-116 and HEK293 cells. Representative cytotoxicity curves and average GI50 of three independent experiments shown. C) OSBP levels in HCT-116 and HEK293 cells of co-administration of 1 nM of OSW-1 with 10,000 nM of ITZ, THEV or TTP for 24 h. Representative Western blot and average of three independent experiments shown.
3.5. OSW-1, ITZ, THEV, and TTP treatments alter OSBP localization in cells in different ways.
OSBP localization patterns in cells are known to be altered by ligand binding, including through interacting with oxysterols3,12,41, OSW-112,13,37, ITZ14,37, THEV26, and TTP37. HCT-116 cells have been used for OSBP localization experiments previously.12,13 In HCT-116 cells, endogenous OSBP is largely localized with the Golgi marker in a diffused perinuclear pattern (Fig. 5A), which is consistent with OSBP localization in the ER/Golgi MCS.3 Treatment with 10,000 nM of 25-OHC for 24 h strongly colocalizes OSBP with the Golgi marker in a small area. 1 nM OSW-1 treatment for 24 h causes a significant reduction of cellular OSBP levels as expected based on the Western blot results13, and the remaining OSBP is tightly grouped around one location, possibly large vesicles. The Golgi marker in the OSW-1-treated cells is diffused and punctate in appearance (Fig. 5A). 10,000 nM of ITZ treatment causes OSBP to adopt a more punctate pattern closely associated with the Golgi marker, although the Golgi marker shows more diffused cytoplasm signal (Fig. 5A). 10,000 nM of TTP treatment induces a tight colocalization of OSBP with the Golgi marker with a perinuclear pattern close to the vehicle (DMSO) treated cell (Fig. 5A). 10,000 nM of THEV treatment induces OSBP localization to confined area tightly packed with the Golgi marker, very similar to 25-OHC treatment (Fig. 5A).
Figure 5: OSW-1, THEV, ITZ, and TTP Cellular Treatment All Have Distinct Effects on OSBP Cellular Localization.
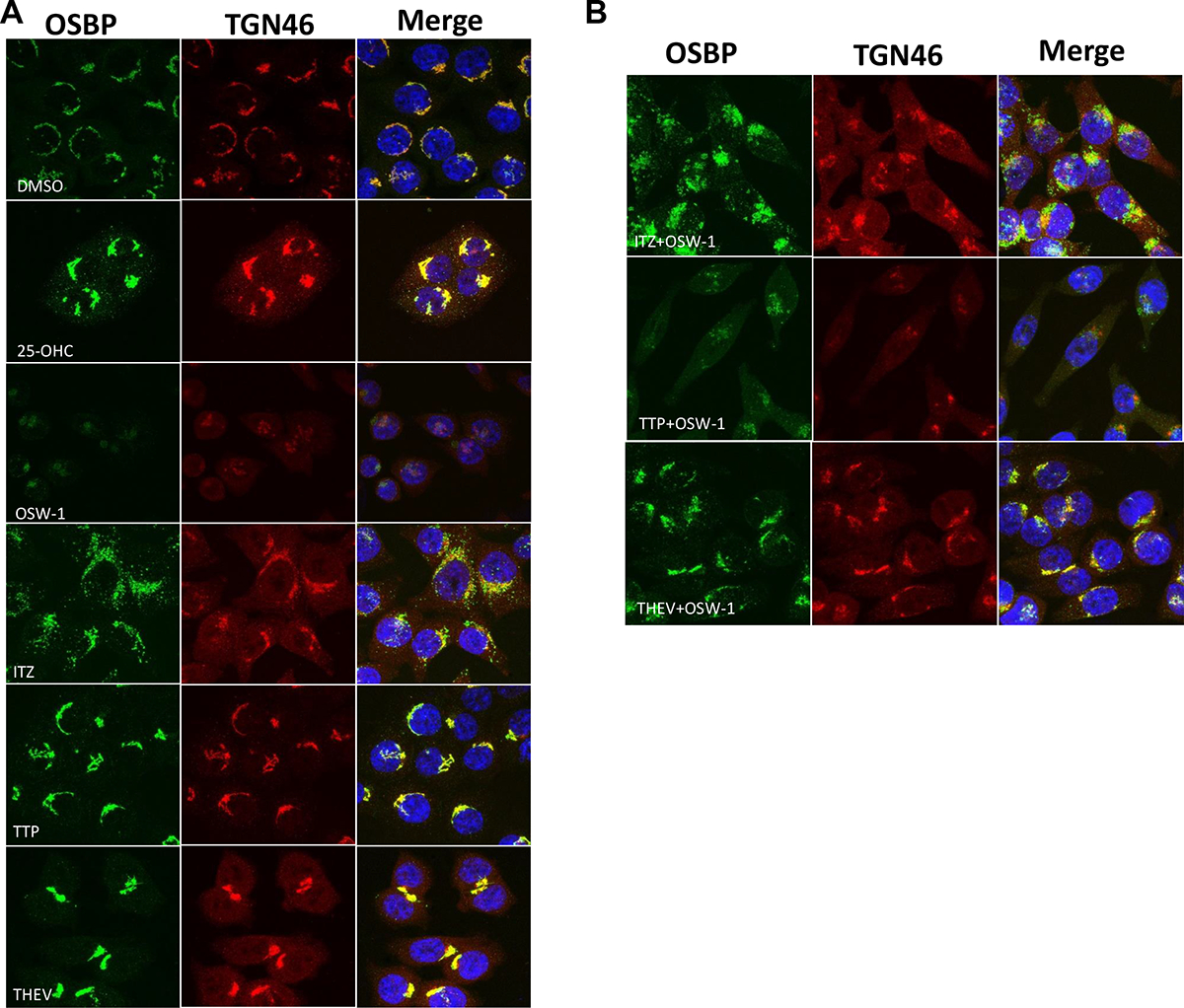
A) Immunofluorescent confocal microscopy of HCT-116 cells treated for 24 h with either DMSO (vehicle), 10,000 nM 25-OHC, 1 nM OSW-1, 10,000 nM ITZ, 10,000 nM TTP, or 10,000 nM THEV. OSBP shown in green; Golgi marker TGN46 shown in red; nucleus stain in blue. The merged images indicates colocalization of the OSBP and Golgi marker. B) Co-administration of 1 nM OSW-1 with 10,000 nM ITZ, 10,000 nM TTP, or 10,000 nM THEV.
The effects of co-administration of OSW-1 with the other compounds shows that ITZ and THEV alter the OSW-1-induced OSBP localization pattern (Fig. 5B). Co-administration of 1 nM of OSW-1 with 10,000 nM of ITZ produces a tight localization of OSBP with a stronger signal than OSW-1 alone (Fig. 5A and Fig. 5B). The Golgi marker in the OSW-1+ITZ treated cells is weakly diffused through the cytoplasm, similar to ITZ alone (Fig. 5A). Overall, the cellular effects of OSW-1+ITZ treatment is a hybrid of the effects of separate treatment of OSW-1 and ITZ, except for stronger OSBP signal (Fig. 5A and Fig. 5B). Similarly, co-administration of 1 nM of OSW-1 with 10,000 nM of THEV produces cellular localization effects with elements of both OSW-1 and THEV treatment alone (Fig. 5A and Fig. 5B). The OSW-1+THEV and OSW-1+ITZ treated cells possess more OSBP than the OSW-1 alone cells, verifying that THEV and ITZ can block the reduction in OSBP levels, as shown via Western blot (Fig. 4C). Similar to THEV treatment alone (Fig. 5A), the OSW-1+THEV treatment includes significant colocalization of OSBP with the Golgi marker, although the colocalization pattern is not as tight as with THEV alone (Fig. 5B). The OSW-1+TTP treated cells show low expression of OSBP with a pattern not consistent with the OSW-1 alone or TTP alone treatment.
Discussion
The results show that the identified OSBP small molecule ligands (i.e., OSW-1, ITZ, THEV, and TTP) do not interact with OSBP through a common binding site and do not have identical effects on OSBP in cells. OSW-1 is the only compound that: 1) causes a reduction in cellular OSBP levels (Fig. 1), and, 2) induces a prophylactic antiviral response in cells (Fig. 2B). These results suggest that it is the reduction of OSBP levels that confers the prophylactic antiviral activity. OSW-1 and THEV both compete with high affinity for 25-OHC binding to OSBP, suggesting that OSW-1 and THEV likely have the same or overlapping OSBP binding sites. Both OSW-1 and THEV possess a steroidal structure, although, importantly, THEV lacks the sterol side chain (Scheme 1). The ability of both OSW-1 and THEV to show high affinity interactions with OSBP and ORP4, as determined through Ki values, suggests the sterol component of OSW-1 and THEV is responsible for OSBP binding, and that the reduction of OSBP levels by OSW-1 is likely caused by the disaccharide portion of the compound.
Interestingly, neither ITZ or TTP inhibit 25-OHC binding to OSBP or ORP4. This suggests these two compounds, if they directly interact with OSBP, have binding sites separate from the sterol binding site. These results, in combination with previous reports, suggest that ITZ and TTP interact with C-terminal OSBP-related ligand-binding domain (i.e, ORD) at locations that do not effect oxysterol binding.
THEV substantially inhibits the activity of OSW-1 in cells, albeit at much higher relative concentrations (i.e.,1 nM OSW-1 vs. 10,000 nM THEV) (Fig. 4A, 4B, 4C and Fig. 5B), confirming the overlapping OSBP binding between OSW-1 and THEV. The requirement for much higher THEV concentration in cells to counter OSW-1 activity could be the result of: 1) THEV’s low specificity and multiple binding partners in cells; 2) poor cellular uptake of THEV in cells from the culture media, or 3) a high rate of cellular metabolism of the THEV compound. Similar to THEV, 25-OHC is commonly administered to cells at micromolar concentrations to affect OSBP, despite a low nanomolar 25-OHC OSBP KD (Fig. 5A).12,42 The tetrahydropyran acetal group on the 3-hydroxyl sterol position of THEV is expected to be labile under the acidic aqueous conditions likely encountered in cells, which could lead to degradation of the compound into an inactive form.
Co-administration of ITZ does somewhat prevent the reduction of OSBP levels induced by OSW-1 (Fig. 4C), and co-administration of ITZ and OSW-1 produces an OSBP cellular localization pattern that appears to have features in common with both OSW-1 and ITZ. These initial results suggest the possibility that OSBP might be capable of interacting with both OSW-1 and ITZ at different binding sites simultaneously. Interestingly, TTP did not alter the activity of OSW-1 in cells (Fig. 4A, 4B, 4C), although TTP co-administered with OSW-1 does induce a different OSBP localization pattern (Fig. 5B). These results suggest that TTP could have a modality of OSBP binding different than ITZ, indicating possibly a second, non-sterol binding site on the OSBP C-terminal domain. In aggregate, these results reveal OSBP to be able to bind multiple, structurally-diverse compounds through different binding sites to induce different biological activities. Therefore, the different OSBP targeting compounds can be used to potentially define different aspects of OSBP biology and present an array of modes for potential antiviral drug development through targeting OSBP.
Supplementary Material
Acknowledgements:
Thanks to the following funding sources: OCAST Health Grant HR17-116; NIH NCI: Innovation Molecular Analysis Technologies (IMAT) Program R21CA204706 (Burgett PI); NIH NIGMS R01GM116116; and the Barnes Family Foundation (Tulsa, Oklahoma). The laboratories of E. Zgurskaya and A. Duerfeldt are thanked for the use of laboratory equipment.
References
- (1).Kentala H; Weber-Boyvat M; Olkkonen VM OSBP-Related Protein Family: Mediators of Lipid Transport and Signaling at Membrane Contact Sites. In International Review of Cell and Molecular Biology; Elsevier Inc., 2016; Vol. 321, pp 299–340. [DOI] [PubMed] [Google Scholar]
- (2).Pietrangelo A; Ridgway ND Bridging the Molecular and Biological Functions of the Oxysterol-Binding Protein Family. Cell. Mol. Life Sci 2018, 75 (17), 3079–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Mesmin B; Bigay J; Moser von Filseck J; Lacas-Gervais S; Drin G; Antonny B A Four-Step Cycle Driven by PI(4)P Hydrolysis Directs Sterol/PI(4)P Exchange by the ER-Golgi Tether OSBP. Cell 2013, 155 (4), 830–843. [DOI] [PubMed] [Google Scholar]
- (4).Mesmin B; Bigay J; Polidori J; Jamecna D; Lacas-Gervais S; Antonny B Sterol Transfer, PI4P Consumption, and Control of Membrane Lipid Order by Endogenous OSBP. EMBO J. 2017, 36 (21), e201796687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wong LH; Gatta AT; Levine TP Lipid Transfer Proteins: The Lipid Commute via Shuttles, Bridges and Tubes. Nat. Rev. Mol. Cell Biol 2019, 20 (2), 85–101. [DOI] [PubMed] [Google Scholar]
- (6).Wang H; Ma Q; Qi Y; Dong J; Du X; Rae J; Wang J; Wu W-F; Brown AJ; Parton RG; et al. ORP2 Delivers Cholesterol to the Plasma Membrane in Exchange for Phosphatidylinositol 4, 5-Bisphosphate (PI(4,5)P2). Mol. Cell 2019, 73 (3), 458–473.e7. [DOI] [PubMed] [Google Scholar]
- (7).Chung J; Torta F; Masai K; Lucast L; Czapla H; Tanner LB; Narayanaswamy P; Wenk MR; Nakatsu F; De Camilli P PI4P/Phosphatidylserine Countertransport at ORP5- and ORP8-Mediated ER-Plasma Membrane Contacts. Science (80-. ). 2015, 349 (6246), 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Charman M; Colbourne TR; Pietrangelo A; Kreplak L; Ridgway ND Oxysterol-Binding Protein (OSBP)-Related Protein 4 (ORP4) Is Essential for Cell Proliferation and Survival. J. Biol. Chem 2014, 289 (22), 15705–15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhong W; Pan G; Wang L; Li S; Ou J; Xu M; Li J; Zhu B; Cao X; Ma H; et al. ORP4L Facilitates Macrophage Survival via G-Protein-Coupled Signaling. Circ. Res 2016, 119 (12), 1296–1312. [DOI] [PubMed] [Google Scholar]
- (10).Pan G; Cao X; Liu B; Li C; Li D; Zheng J; Lai C; Olkkonen VM; Zhong W; Yan D OSBP-Related Protein 4L Promotes Phospholipase Cβ3 Translocation from the Nucleus to the Plasma Membrane in Jurkat T-Cells. J. Biol. Chem 2018, 293 (45), 17430–17441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhong W; Xu M; Li C; Zhu B; Cao X; Li D; Chen H; Hu C; Li R; Luo C; et al. ORP4L Extracts and Presents PIP2 from Plasma Membrane for PLCβ3 Catalysis: Targeting It Eradicates Leukemia Stem Cells. Cell Rep. 2019, 26 (8), 2166–2177.e9. [DOI] [PubMed] [Google Scholar]
- (12).Burgett AWG; Poulsen TB; Wangkanont K; Anderson DR; Kikuchi C; Shimada K; Okubo S; Fortner KC; Mimaki Y; Kuroda M; et al. Natural Products Reveal Cancer Cell Dependence on Oxysterol-Binding Proteins. Nat. Chem. Biol 2011, 7 (9), 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Roberts BL; Severance ZC; Bensen RC; Le AT; Kothapalli NR; Nuñez JI; Ma H; Wu S; Standke SJ; Yang Z; et al. Transient Compound Treatment Induces a Multigenerational Reduction of Oxysterol-Binding Protein (OSBP) Levels and Prophylactic Antiviral Activity. ACS Chem. Biol 2019, acschembio.8b00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Strating JRPM; van der Linden L; Albulescu L; Bigay J; Arita M; Delang L; Leyssen P; van der Schaar HM; Lanke KHW; Thibaut HJ; et al. Itraconazole Inhibits Enterovirus Replication by Targeting the Oxysterol-Binding Protein. Cell Rep. 2015, 10 (4), 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Strating JRPMRPM; van der Linden L; Albulescu L; Bigay J; Arita M; Delang L; Leyssen P; van der Schaar HM; Lanke KHWHW; Thibaut HJJ; et al. Itraconazole Inhibits Enterovirus Replication by Targeting the Oxysterol-Binding Protein. Cell Rep. 2015, 10 (4), 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bauer L; Ferla S; Head SA; Bhat S; Pasunooti KK; Shi WQ; Albulescu L; Liu JO; Brancale A; Kuppeveld FJM Van; et al. Structure-Activity Relationship Study of Itraconazole , a Broad-Range Inhibitor of Picornavirus Replication That Targets Oxysterol-Binding Protein (OSBP). Antiviral Res. 2018, 156, 55–63. [DOI] [PubMed] [Google Scholar]
- (17).Albulescu L; Strating JRPM; Thibaut HJ; Van Der Linden L; Shair MD; Neyts J; Van Kuppeveld FJM Broad-Range Inhibition of Enterovirus Replication by OSW-1, a Natural Compound Targeting OSBP. Antiviral Res. 2015, 117, 110–114. [DOI] [PubMed] [Google Scholar]
- (18).Baggen J; Thibaut HJ; Strating JRPM; van Kuppeveld FJM The Life Cycle of Non-Polio Enteroviruses and How to Target It. Nat. Rev. Microbiol 2018, 16 (6), 368–381. [DOI] [PubMed] [Google Scholar]
- (19).Amako Y; Sarkeshik A; Hotta H; Yates J; Siddiqui A Role of Oxysterol Binding Protein in Hepatitis C Virus Infection. J. Virol 2009, 83 (18), 9237–9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Amako Y; Syed GH; Siddiqui A Protein Kinase D Negatively Regulates Hepatitis C Virus Secretion through Phosphorylation of Oxysterol-Binding Protein and Ceramide Transfer Protein. J. Biol. Chem 2011, 286 (13), 11265–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Dorobantu CM; Albulescu L; Lyoo H; van Kampen M; De Francesco R; Lohmann V; Harak C; van der Schaar HM; Strating JRPM; Gorbalenya AE; et al. Mutations in Encephalomyocarditis Virus 3A Protein Uncouple the Dependency of Genome Replication on Host Factors Phosphatidylinositol 4-Kinase IIIα and Oxysterol-Binding Protein. mSphere 2016, 1 (3), e00068–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Meutiawati F; Bezemer B; Strating JRPM; Overheul GJ; Žusinaite E; van Kuppeveld FJM; van Cleef KWR; van Rij RP Posaconazole Inhibits Dengue Virus Replication by Targeting Oxysterol-Binding Protein. Antiviral Res. 2018, 157 (June), 68–79. [DOI] [PubMed] [Google Scholar]
- (23).Nchoutmboube J; Ford-Siltz LA; Belov GA Enterovirus Replication: GO with the (Counter)Flow. Trends Microbiol. 2015, 23 (4), 183–184. [DOI] [PubMed] [Google Scholar]
- (24).Arita M Phosphatidylinositol-4 Kinase III Beta and Oxysterol-Binding Protein Accumulate Unesterified Cholesterol on Poliovirus-Induced Membrane Structure. Microbiol. Immunol 2014, 58 (4), 239–256. [DOI] [PubMed] [Google Scholar]
- (25).Dorobantu CM; Albulescu L; Harak C; Feng Q; van Kampen M; Strating JRPM; Gorbalenya AE; Lohmann V; van der Schaar HM; van Kuppeveld FJM Modulation of the Host Lipid Landscape to Promote RNA Virus Replication: The Picornavirus Encephalomyocarditis Virus Converges on the Pathway Used by Hepatitis C Virus. PLOS Pathog. 2015, 11 (9), e1005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Arita M; Kojima H; Nagano T; Okabe T; Wakita T; Shimizu H Oxysterol-Binding Protein Family I Is the Target of Minor Enviroxime-Like Compounds. J. Virol 2013, 87 (8), 4252–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hitchcock CA Resistance of Candida Albicans to Azole Antifungal Agents. Biochem. Soc. Trans 1993, 21 (4), 1039–1047. [DOI] [PubMed] [Google Scholar]
- (28).Chong CR; Xu J; Lu J; Bhat S; Sullivan DJ; Liu JO Inhibition of Angiogenesis by the Antifungal Drug Itraconazole. ACS Chem. Biol 2007, 2 (4), 263–270. [DOI] [PubMed] [Google Scholar]
- (29).Nacev BA; Grassi P; Dell A; Haslam SM; Liu JO The Antifungal Drug Itraconazole Inhibits Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) Glycosylation, Trafficking, and Signaling in Endothelial Cells. J. Biol. Chem 2011, 286 (51), 44045–44056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kim J; Tang JY; Gong R; Kim J; Lee JJ; Clemons KV; Chong CR; Chang KS; Fereshteh M; Gardner D; et al. Itraconazole, a Commonly Used Antifungal That Inhibits Hedgehog Pathway Activity and Cancer Growth. Cancer Cell 2010, 17 (4), 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Head SA; Shi WQ; Yang EJ; Nacev BA; Hong SY; Pasunooti KK; Li R-J; Shim JS; Liu JO Simultaneous Targeting of NPC1 and VDAC1 by Itraconazole Leads to Synergistic Inhibition of MTOR Signaling and Angiogenesis. ACS Chem. Biol 2017, 12 (1), 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gao Q; Yuan S; Zhang C; Wang Y; Wang Y; He G; Zhang S; Altmeyer R; Zou G Discovery of Itraconazole with Broad-Spectrum in Vitro Antienterovirus Activity That Targets Nonstructural Protein 3A. Antimicrob. Agents Chemother 2015, 59 (5), 2654–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Im YJ; Raychaudhuri S; Prinz W. a; Hurley JH Structural Mechanism for Sterol Sensing and Transport by OSBP-Related Proteins. Nature 2005, 437 (7055), 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tong J; Yang H; Yang H; Eom SH; Im YJ Structure of Osh3 Reveals a Conserved Mode of Phosphoinositide Binding in Oxysterol-Binding Proteins. Structure 2013, 21 (7), 1203–1213. [DOI] [PubMed] [Google Scholar]
- (35).Maeda K; Anand K; Chiapparino A; Kumar A; Poletto M; Kaksonen M; Gavin A Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins. Nature 2013, 501 (7466), 257–261. [DOI] [PubMed] [Google Scholar]
- (36).Tong J; Yang H; Yang H; Eom SH; Im YJ Structure of Osh3 Reveals a Conserved Mode of Phosphoinositide Binding in Oxysterol-Binding Proteins. Structure 2013, 21 (7), 1203–1213. [DOI] [PubMed] [Google Scholar]
- (37).Albulescu L; Bigay J; Biswas B; Weber-Boyvat M; Dorobantu CM; Delang L; van der Schaar HM; Jung Y-S; Neyts J; Olkkonen VM; et al. Uncovering Oxysterol-Binding Protein (OSBP) as a Target of the Anti-Enteroviral Compound TTP-8307. Antiviral Res. 2017, 140, 37–44. [DOI] [PubMed] [Google Scholar]
- (38).Albulescu L; Bigay J; Biswas B; Weber-Boyvat M; Dorobantu CM; Delang L; van der Schaar HM; Jung Y-S; Neyts J; Olkkonen VM; et al. Uncovering Oxysterol-Binding Protein (OSBP) as a Target of the Anti-Enteroviral Compound TTP-8307. Antiviral Res. 2017, 140, 37–44. [DOI] [PubMed] [Google Scholar]
- (39).Yu W; Jin Z Total Synthesis of the Anticancer Natural Product OSW-1. J. Am. Chem. Soc 2002, 124 (23), 6576–6583. [DOI] [PubMed] [Google Scholar]
- (40).Mimaki Y; Kuroda M; Kameyama A; Sashida Y; Hirano T; Oka K; Maekawa R; Wada T; Sugita K; Beutler JA Cholestane Glycosides with Potent Cytostatic Activities on Various Tumor Cells from Ornithogalum Saundersiae Bulbs. Bioorg. Med. Chem. Lett 1997, 7 (5), 633–636. [Google Scholar]
- (41).Ridgway ND; Dawson PA; Ho YK; Brown MS; Goldstein JL; Ridgway ND; Dawson PA; Ho YK; Brown MS; Goldstein JL Translocation of Oxysterol Binding Protein to Golgi Apparatus Triggered by Ligand Binding. 2008, 116 (2), 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ridgway ND; Dawson P. a; Ho YK; Brown MS; Goldstein JL Translocation of Oxysterol Binding Protein to Golgi Apparatus Triggered by Ligand Binding. J. Cell Biol 1992, 116 (2), 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


