Abstract
Background
Inability to identify the microbial etiology of lower respiratory tract infection leads to unnecessary antibiotic use. We evaluated the utility of the BioFire FilmArray Pneumonia Panel (BioFire PN) to inform microbiologic diagnosis.
Methods
Hospitalized adults with respiratory illness were recruited; sputa and clinical/laboratory data were collected. Sputa were cultured for bacteria and tested with BioFire PN. Microbial etiology was adjudicated by 4 physicians. Bacterial polymerase chain reaction (PCR) was compared with culture and clinical adjudication.
Results
Of 298 sputa tested, BioFire PN detected significantly more pathogens (350 bacteria, 16 atypicals, and 164 viruses) than sputum culture plus any standard-of-care testing (91% vs 60%, P < .0001). When compared with culture, the sensitivity of BioFire PN for individual bacteria was 46% to 100%; specificity, 61% to 100%; and negative predictive value, 92% to 100%. Cases were adjudicated as viral (n = 58) and bacterial (n = 100). PCR detected bacteria in 55% of viral cases and 95% of bacterial (P < .0001). High serum procalcitonin and bacterial adjudication were more often associated with sputa with 106 or 107 copies detected.
Conclusions
Multiplex PCR testing of sputa for bacteria is useful to rule out bacterial infection with added value to detect viruses and atypical bacteria.
Keywords: bacteria, diagnostic, respiratory illness, sputum, viruses
BioFire FilmArray Pneumonia Panel sputa testing is useful to rule out bacterial infection with increased yield of viruses and atypical bacteria. Combined with bacterial copy number and clinical variables, the assay could improve antibiotic management of respiratory infections.
Acute respiratory infections occur frequently throughout life and cause significant morbidity and mortality [1]. A range of viral and bacterial pathogens accounts for most acute respiratory infection cases, although establishing an etiology can be difficult. In a study of patients hospitalized with community-acquired pneumonia in the United States, a pathogen was identified in only 34% of cases [2]. Sensitive molecular diagnostic tests allow accurate and rapid diagnosis of many respiratory viruses, although impact on antibiotic management has been modest due to concern about bacterial coinfection [3, 4]. However, current bacterial diagnostics remain insensitive, and since many bacterial pathogens may colonize airways, sputum culture can be misleading [5]. Although the serum biomarker procalcitonin (PCT) has shown some promise to supplement clinical judgment, PCT has not been widely adopted by clinicians [6–8]. Diagnostic uncertainly has led to unnecessary use of broad-spectrum antibiotics, with resultant side effects and spread of antimicrobial resistance, which is currently one of the most urgent threats to global public health [9–13].
The BioFire FilmArray Pneumonia Panel (BioFire PN; BioMérieux) is a multiplex polymerase chain reaction (PCR) assay that analyzes sputum, tracheal aspirate, and bronchoalveolar lavage samples for the presence of bacteria, viruses, and genetic markers of antimicrobial resistance. Results are qualitative for viral and atypical bacterial targets and semiquantitative for 15 aerobic bacterial targets [14, 15]. Most studies using this assay have focused on the analytic features of BioFire PN as compared with either sputum cultures or tracheal aspirate or bronchoalveolar lavage samples in patients with pneumonia and respiratory failure [16–27]. In the latter setting, BioFire PN has been shown to have 96% positive and 98% negative agreement with cultures [16]. However, critically ill patients represent a small fraction of adults hospitalized with respiratory infections, and most respiratory samples submitted for microbial diagnosis are expectorated sputum samples. In addition, defining true pneumonia can be challenging, as patients often have ambiguous chest radiographs and complicating medical conditions, resulting in a significant proportion admitted with “possible” pneumonia who are then prescribed empiric antibiotics. Therefore, we evaluated BioFire PN testing of expectorated sputum in hospitalized patients with a variety of microbiologically adjudicated respiratory illnesses to determine the analytic characteristics and potential clinical utility of BioFire PN results.
METHODS
Full details of the clinical protocol, laboratory methods, and adjudication are available in the Supplementary Materials.
Study Period and Sites
The study was conducted in Rochester, New York, between March 2019 and March 2022. The present analysis represents a subgroup from a study evaluating whole blood gene expression for distinguishing bacterial from nonbacterial respiratory illnesses among those who had sputum samples collected.
Recruitment
Patients ≥18 years of age with signs and symptoms compatible with acute respiratory infection or acute cardiopulmonary illness were recruited within 24 hours of admission. Persons with significant immunosuppressive conditions or antibiotic use prior to admission were excluded. Written informed consent was obtained from participants or their authorized representatives.
Acute Illness Evaluation
Demographic, clinical, laboratory, and radiographic information was collected by medical record review and patient/family interview.
Laboratory Studies
All participants had a serum PCT measured by the VIDAS BRAHMS (bioMerieux) assay at enrollment. Other standard-of-care (SOC) testing was ordered by the treating physicians and could include a nasopharyngeal swab for viral PCR, blood cultures, sputum cultures, and Streptococcus pneumoniae and Legionella urine antigens (BinaxNow; Abbott).
Sputum Studies
Study personnel assisted in collection of expectorated sputum samples, but these were not induced. If BioFire PN testing could not be completed within 24 hours, samples were frozen and stored at −20 °C until testing could be performed.
Sputum quality was assessed by the clinical microbiology staff by examining the Gram stain and was classified as follows: good (>25 polymorphonuclear neutrophils [PMNs] and <10 epithelial cells per high-power field [hpf]), moderate (>25 PMN/hpf and > 10 epithelial cells/hpf), or poor quality (<25 PMN/hpf). Only sputa judged good or moderate quality underwent further analysis with BioFire PN.
BioFire PN Panel Testing
The BioFire PN assay reports semiquantitative results for 15 common bacteria (hereafter, “bacteria”), 3 atypical bacteria, and 9 viruses. The relative abundance of standard bacterial organisms was reported as 104, 105, 106, or ≥107 genomic copies per milliliter (see Supplementary Material for details).
Clinical and Microbiological Adjudication
Acute respiratory infection cases were adjudicated by a panel of 4 physicians and classified into 3 microbiologic categories: viral infection alone, bacterial infection alone, or bacterial-viral coinfection. Adjudicators were blinded to the BioFire PN bacterial PCR results but not to atypical bacterial and viral results. Microbiological classification required unanimous agreement by adjudicators, or the case was considered indeterminant.
Statistical Analysis
Patient characteristics were compared by Student t test, Mann-Whitney test, or chi-square test as appropriate. The analytic performance of BioFire PN bacterial results, as compared with culture and clinical adjudication as gold standards, was calculated via standard methods and included sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). P < .05 was considered significant.
RESULTS
Sample Characteristics
In total 737 participants were enrolled in the main study, of whom 428 (58%) were able to provide a sputum sample (Supplementary Figure 1). Of the 428 sputa, 203 were classified as good quality and 95 as moderate and underwent BioFire PN testing; 130 samples were poor quality and no further testing was done. Most samples were expectorated sputa (96%) and 4% were tracheal aspirates. Demographic and clinical data for the 298 patients in this analysis are shown in Table 1. The mean age was 63 years, 50% were female, and the majority were White and non-Hispanic. The leading discharge diagnoses were community-acquired pneumonia (32%) and acute exacerbation of chronic obstructive pulmonary disease (20%). Antibiotics were administered to 244 (81.9%) patients. Thirty-seven percent of sputum samples were collected prior to antibiotics administration, and of those collected afterward, the median time from administration to collection was 10 hours.
Table 1.
Demographic and Clinical Characteristics of Study Population (N = 298)
| No. (%) | |
|---|---|
| Age, ya | 63.1 ± 16.0 (19–98) |
| Female | 151 (50.1) |
| Race | |
| White | 209 (70.1) |
| Black | 76 (25.6) |
| Mixed/unknown | 11 (3.7) |
| Ethnicity | |
| Non-Hispanic | 269 (90.3) |
| Hispanic | 28 (9.4) |
| Unknown | 1 (0.3) |
| Underlying pulmonary conditions | |
| Asthma | 91 (30.5) |
| COPD | 140 (47.0) |
| Primary discharge diagnosis | |
| Respiratory failure | 24 (8.1) |
| Acute exacerbation of COPD | 60 (20.1) |
| Asthma | 22 (7.4) |
| Acute bronchitis | 9 (3.0) |
| Community-acquired pneumonia | 95 (31.9) |
| Congestive heart failure | 3 (1.0) |
| Viral syndrome | 24 (8.1) |
| Other | 61 (20.5) |
| Required ICU | 59 (19.8) |
| Mechanically ventilated | 13 (4.4) |
| Laboratoryb | |
| WBC, 103/μL | 10.9 (2.3–37.9) |
| Lactate, mmol/L | 1.4 (0.6–6.8) |
| PCT, ng/mL | 0.13 (0.04–100) |
Abbreviations: COPD, chronic obstructive pulmonary disease; ICU, intensive care unit; PCT, procalcitonin; WBC, white blood cell.
aMean ± SD (range).
bMedian (range).
Microbiologic Results
From the 298 good and moderate quality sputum samples, BioFire PN detected 350 bacteria and 16 atypical bacteria for an average of 1.23 potential bacterial pathogens per sample. Overall, 225 (75.5%) samples had typical bacteria detected. Of these, 58.2% were monomicrobial, 30.2% had 2 bacteria detected, and 11.6% had ≥3 bacteria detected with no significant difference between good and moderate quality sputa. The most commonly identified bacteria were Haemophilus influenzae, S pneumoniae, Staphylococcus aureus, and Moraxella catarrhalis (Supplementary Table 1).
In contrast, standard bacterial culture detected 144 organisms (143 bacteria and 1 atypical bacteria) for an average of 0.48 organisms per sample, significantly less (P < .0001) than what BioFire PN detected. An overall 126 samples (42.3%) grew a potential pathogen, of which most were monomicrobial (86.5%). H influenzae, S pneumoniae, S aureus, and low quantities of unspeciated Gram-negative rods (GNRs) were most frequently cultured from sputum.
Semiquantitative Bacterial Results
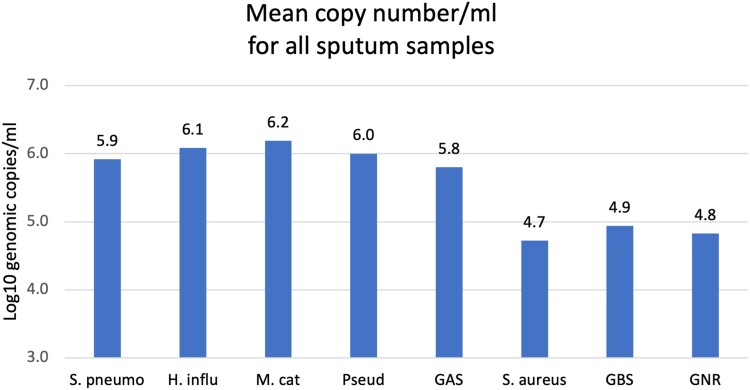
The specific organisms and semiquantitative values detected by BioFire PN are shown in Supplementary Table 1, as categorized by good and moderate quality sputum. The distribution of pathogens was similar between good and moderate quality samples, with the exception that pneumococcus was detected significantly more frequently in good quality samples (24% vs 13%, P = .02). Notably, the quantity of S pneumoniae, H influenzae, M catarrhalis, and Pseudomonas was significantly higher than S aureus, group B Streptococcus, and GNRs (P < .003 for all comparisons; Figure 1).
Figure 1.
Mean semiquantitative levels of bacteria in all 298 sputum samples tested by BioFire FilmArray Pneumonia Panel. Samples include sputa with good or moderate quality. Semiquantitative results are expressed a genomic copy per milliliter log10. Mean quantity was compared for S pneumoniae, H influenzae, M catarrhalis, P aeruginosa, group A streptococcus (GAS), S aureus, group B streptococcus (GBS), and other Gram-negative rods (GNR). For each comparison, values were significantly higher for S pneumoniae, H influenzae, M catarrhalis, and Pseudomonas aeruginosa vs S aureus, GBS, and other GNR. P < .003 for all comparisons.
Comparison of BioFire PN and Standard Sputum Culture
For good quality sputa, the overall concordance between the BioFire PN results and standard bacterial culture was modest, with only 37% of results fully concordant, 22% partially concordant (some but not all organisms match), and 41% discordant (no organism matched; Table 2A). Results for moderate quality sputum were similar: fully concordant (35%), partially concordant (17%) and discordant (46%; data not shown). BioFire PN detected more pathogens per sample than standard culture, with most discordant outcomes resulting from bacteria detected by BioFire PN and negative results in culture.
Table 2.
Concordance of BioFire PN With Sputum Culture According to Use of Antibiotics in Good Quality Sputum Samples
| Antibiotics | ||||
|---|---|---|---|---|
| Total (n = 203) | No (n = 85) | Yes (n = 118) | P Value | |
| A: Concordance of culture to BioFire PN | ||||
| Partially concordant | 43 (21) | 25 (29) | 18 (15) | .02 |
| Fully concordant | 75 (37) | 37 (44) | 38 (32) | .11 |
| Partially or fully concordant | 119 (59) | 62 (73) | 56 (47) | .0003 |
| Discordant | 85 (42) | 23 (27) | 62 (53) | .0003 |
| B: Culture | ||||
| Monomicrobial | 82 (40) | 48 (57) | 34 (29) | .0001 |
| Polymicrobial | 10 (5) | 5 (6) | 5 (4) | .74 |
| No growth/normal flora | 111 (55) | 32 (38) | 79 (67) | .0001 |
| C: BioFire PN | ||||
| Monomicrobial | 91 (45) | 33 (39) | 58 (49) | .16 |
| Polymicrobial | 64 (32) | 31 (37) | 33 (28) | .22 |
| No growth/normal flora | 48 (23) | 21 (25) | 27 (23) | .87 |
Data are presented as No. (%).
Abbreviation: BioFire PN, BioFire FilmArray Pneumonia Panel.
Concordance was significantly affected by antibiotic use, with discordant results increasing from 27% to 53% (P = .0003) for sputum collected prior to and after antibiotic administration, respectively (Table 2A). This finding was driven by decreased bacterial detection in sputum cultures collected after antibiotics (Table 2B), whereas BioFire PN results were not significantly different before and after antibiotic administration (Table 2C).
The sensitivity, specificity, PPV, and NPV of the BioFire PN panel were calculated for 7 commonly identified pathogens for good and moderate quality samples (Table 3). Because GNRs other than Pseudomonas were infrequently detected, their results were pooled as “other GNR.” With culture as the gold standard, sensitivity for good quality sputa (Table 3A) ranged from 46% for other GNR to 95% for S pneumoniae and H influenzae and 100% for group A Streptococcus, M catarrhalis, and Pseudomonas aeruginosa. Specificity was generally lower than sensitivity for most organisms, with H influenzae having the lowest value (75%). PPV was relatively low for all bacterial species, except group A Streptococcus (100%). In contrast, the NPV was very high (99% to 100%), except for other GNR at 92%. For the 46 samples with no bacteria detected by BioFire PN, 40 (87%) grew normal flora and 6 grew rare or 1+ nonspeciated GNR and 1 rare S aureus (see Supplementary Material for methods on reporting GNR).
Table 3.
Sensitivity, Specificity, and PPV and NPV of BioFire PN Panel Testing on Samples vs Standard Culture as Gold Standard
| Bacteria | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| A: Good quality (n = 203) | ||||
| S pneumoniae | 95 | 84 | 39 | 99 |
| S aureus | 77 | 84 | 24 | 99 |
| Group B streptococcus | 100 | 91 | 5 | 100 |
| Group A streptococcus | 100 | 100 | 100 | 100 |
| H influenzae | 95 | 75 | 29 | 99 |
| M catarrhalis | 100 | 91 | 39 | 100 |
| P aeruginosa | 100 | 98 | 64 | 100 |
| Other Gram-negative rod | 46 | 96 | 63 | 92 |
| B: Moderate quality (n = 95) | ||||
| S pneumoniae | —a | 87 | 0 | 100 |
| S aureus | 67 | 80 | 10 | 99 |
| Group B streptococcus | 100 | 90 | 18 | 100 |
| Group A streptococcus | 100 | 98 | 33 | 100 |
| H influenzae | 75 | 68 | 12 | 100 |
| M catarrhalis | —a | 91 | 0 | 100 |
| P aeruginosa | 100 | 98 | 50 | 100 |
| Other Gram-negative rod | 74 | 99 | 93 | 94 |
Data are presented as percentages.
Abbreviations: BioFire PN, BioFire FilmArray Pneumonia Panel; NPV, negative predictive value; PPV, positive predictive value.
aCannot be determined as the organism never grew in culture.
The results for moderate quality sputa were similar, although the sensitivity and specificity for several organisms were lower and PPV was substantially lower than good quality sputum samples (Table 3B). Notably, for moderate quality sputa, the NPV was also high, ranging from 94% to 100%.
We next determined if specificity and PPV could be improved by restricting analysis to good quality samples with high bacterial abundance (106 to ≥107 copies/mL; Supplementary Table 2). There was a decrease in sensitivity and an increase in PPV for S aureus and other GNR, with a smaller improvement in PPV for other organisms. Notably, the NPV remained high for all organisms.
Comparison of BioFire PN Results and Clinical Adjudication
Illnesses were adjudicated as viral infection alone, bacterial infection (with or without a virus identified), or indeterminate. For this analysis, atypical bacteria were excluded since adjudicators were given access to atypical bacterial results as part of the parent study.
Of the 158 cases with definitive microbiologic adjudication results, 58 were judged as viral alone and 100 bacterial. Of adjudicated bacterial and viral cases, 95% and 55% (P < .0001), respectively, were positive for bacteria by BioFire PN (Table 4). Restricting analysis to good quality samples provided similar results; however, moderate quality samples adjudicated as viral alone frequently (81%) had bacteria detected by BioFire PN. Of the 5 cases adjudicated as bacterial with no bacterial pathogen detected by BioFire PN, 4 were judged to have mixed anaerobic lung infections (necrotic pulmonary cavities, empyema), and the other grew Haemophilus parainfluenzae, which is an organism not in the BioFire PN panel. Considering clinical adjudication as the gold standard, the BioFire PN had a sensitivity of 95%, specificity of 45%, PPV of 75%, and NPV of 84%.
Table 4.
Sensitivity and Specificity of BioFire PN Panel Results Positive for Any Bacteria vs Clinical Adjudication as the Gold Standard
| No. BioFire Positive | Sensitivity, % | Specificity, % | PPV, % | NPV, % | |
|---|---|---|---|---|---|
| All sputum samples (n = 158) | 95 | 45 | 75 | 84 | |
| Viral (n = 58) | 32 | ||||
| Bacterial (n = 100) | 95 | ||||
| Good quality sputum (n = 122) | 94 | 59 | 84 | 81 | |
| Viral (n = 37) | 15a | ||||
| Bacterial (n = 85) | 80 | ||||
| Moderate quality sputum (n = 36) | 100 | 28 | 59 | 100 | |
| Viral (n = 21) | 17 | ||||
| Bacterial (n = 15) | 15 |
Standard bacteria were detected with or without a virus detected. Clinical adjudication resulted in definitive diagnosis of viral alone or bacterial. Bacterial included bacterial alone and bacterial + viral. Indeterminant cases are not included.
Abbreviations: BioFire PN, BioFire FilmArray Pneumonia Panel; NPV, negative predictive value; PPV, positive predictive value.
a P = .0001. Difference in percentage of BioFire PN–positive samples between bacterial and viral infection.
BioFire PN Results in the Context of Host Response Variables and Clinical Syndromes
Since neither BioFire PN nor culture can reliably distinguish colonization from invasive bacterial disease, we sought to determine if BioFire PN results correlated to host response variables and a diagnosis of pneumonia and thus might be indicative of invasive disease rather than colonization. Mean white blood cell and PCT values trended higher in those with bacteria identified by BioFire PN than those with viral alone, with good quality sputa providing the best discrimination (Supplementary Table 3). Of good quality sputa with bacteria detected, 38.6% had PCT values ≥0.25 ng/mL (a common threshold used to indicate bacterial infection), as opposed to 9.7% (P = .0001) of those with viruses only. Finally, when bacterial quantification was dichotomized into high (106–107) and low (104–105) copies per milliliter, there was a trend toward higher mean white blood cell and PCT values and significantly more patients with PCT values ≥0.25 ng/mL (41% vs 21%, P = .03) associated with high bacterial copy number (Table 5).
Table 5.
Comparison of Clinical Markers of Inflammation and Clinical Adjudication by Bacterial Abundance
| Quantity, Copies/mL | |||
|---|---|---|---|
| Variable | 104–105 (n = 40) | 106 to ≥107 (n = 109) | P Value |
| Median (IQR) | |||
| WBC, 103/μL | 12.35 (8.1–14.5) | 11.5 (9.4–15.3) | .23 |
| PCT, ng/mLa | 0.12 (0.06–0.21) | 0.16 (0.06–0.92) | .16 |
| No. (%) | |||
| PCT >0.25 ng/mLa | 8/39 (21) | 44/108 (41) | .03 |
| Adjudicated as bacterialb | 11/40 (28) | 69/109 (63) | <.0001 |
Results of good quality samples with quantifiable standard bacteria detected (n = 149) were analyzed by low and high genomic copy number. Samples with atypical bacteria detected were excluded because testing was qualitative.
Abbreviations: PCT, procalcitonin; WBC, white blood cell.
aPCT testing was missing in 1 participant in each group.
bBacterial = bacterial alone or bacteria + virus.
The clinical phenotype of pneumonia differed from nonpneumonia (Supplementary Table 4A and B) with a greater percentage of pneumonia cases with PCT ≥0.25 ng/mL and more frequent adjudication as bacterial. BioFire PN detected significantly more typical and atypical bacteria, including S pneumoniae and M catarrhalis, at higher copy numbers in pneumonia than nonpneumonia cases.
BioFire PN Samples With Negative or Low Bacterial Abundance
To assess the potential clinical utility of BioFire PN for avoiding or curtailing antibiotic treatment, we calculated the number of sputa with no bacteria detected (including atypical) or those with <106 copies/mL in conjunction with laboratory variables often used in clinical decision making (Supplementary Table 5). Of the 298 cases with sputa tested by BioFire PN, 134 (45%) had values <106 CFU/mL or no bacteria detected. Furthermore, 105 (35.2%) were associated with PCT <0.25 ng/mL, and 74 (24.8%) had chest radiographs with negative results. Antibiotic use was high in all 3 categories (64%–76%). Thus, potential opportunities for curtailing antibiotic use ranged from 16% to 34%, depending on the stringency of clinical criteria. Notably, 23% of pneumonia cases treated with antibiotics (Supplementary Table 4C) had low PCT values and BioFire PN values <106 CFU/mL or no bacteria detected, as compared with 44% of nonpneumonia cases.
Added Value of BioFire PN to Detect Atypical Bacteria and Viruses
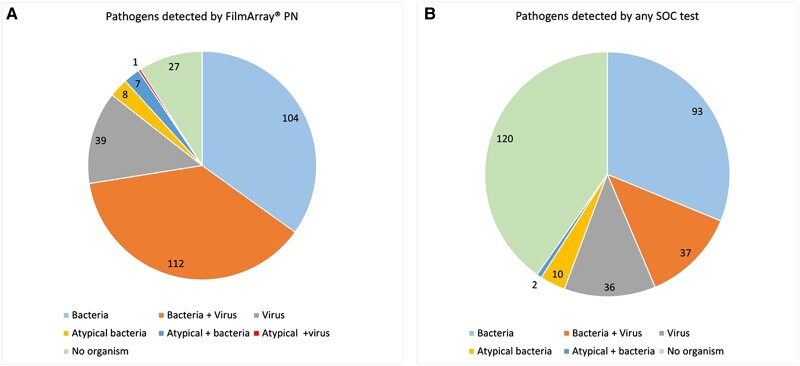
Atypical bacteria and viruses are generally not thought to colonize airways, and their presence suggests active infection. Notably, 164 viruses were detected in sputa by BioFire PN in 152 (51%) of the 298 samples. In addition, 8 cases each of legionella and mycoplasma were identified by BioFire PN. As compared with SOC testing, BioFire PN detected significantly more viral infections. In 2 cases, viruses were positive in SOC nasopharyngeal testing (1 RSV and 1 PIV-2) and negative in sputum; 53 samples were positive by SOC and BioFire PN; and 98 were positive by BioFire PN only. In 95% of cases positive by BioFire PN only, SOC viral testing for the pathogens identified was not performed. However, in 5 SOC cases, nasopharyngeal testing was negative with positive BioFire PN results (4 influenza and 1 RSV). Importantly, 2 legionella cases and 4 mycoplasma cases were missed by SOC testing. According to any diagnostic test (blood and sputum cultures, urine antigen testing, and nasal PCR), potential bacterial, atypical bacterial, or viral pathogens were detected in 178 (60%) SOC-tested cases, as compared with 272 (91%) of BioFire PN–tested cases (P < .0001). Overall, yield for SOC based on all available microbiologic techniques vs BioFire PN is shown in Figure 2.
Figure 2.
Diagnostic yield of BioFire FilmArray Pneumonia Panel (BioFire PN) vs any standard-of-care (SOC) test. A, Potential pathogens detected in 298 sputum samples of good and moderate quality by BioFire PN divided into microbiologic categories. Categories: bacteria alone, bacteria + virus, bacteria + atypical bacteria, virus alone, virus + atypical bacteria, atypical bacteria alone, no pathogen. B, Total microbiologic pathogens detected by any SOC test. Results include the sum of positive results from 126 sputum cultures, 5 blood cultures, and 19 urine antigen tests.
DISCUSSION
Historically, sputum Gram stain and culture have been used to diagnose the microbial etiology of lower respiratory tract infection (LRTI) [28]. However, recent guidelines have de-emphasized these traditional tools for a variety of reasons, including contamination by upper airway secretions, perceived urgency of antibiotic administration, and long turnaround times for culture results [29–31]. Molecular diagnostics, such as BioFire PN, that can rapidly identify bacterial and viral respiratory pathogens in sputa offers the possibility of improving the diagnostic capability and treatment of patients with LRTI [32]. Most studies using BioFire PN have described results of tracheal aspirates or bronchoalveolar lavage samples in patients with severe pneumonia [16, 19, 21–26]. Fewer studies address the majority of patients hospitalized with LRTI, who are treated outside intensive care settings, or utilize expectorated sputum samples [33–38]. In our study, we examined a range of patients hospitalized with respiratory complaints—including acute exacerbation of chronic obstructive pulmonary disease, bronchitis, and pneumonia—and the relationship of BioFire PN results with host response variables, clinical syndromes, and rigorous clinical adjudication of microbial etiology.
Overall, BioFire PN yielded a variety of potential pathogens, including typical and atypical bacteria and numerous viruses. Similar to other reports, we found that BioFire PN had high sensitivity for bacteria when compared with sputum culture as the gold standard [14, 15]. For most pathogens, specificity was lower than sensitivity, which was attributed to the greater number of identified potential pathogens vs standard culture [14, 34]. As noted by others, concordance between BioFire PN and culture results was affected by prior antibiotic use, resulting in negative cultures with positive BioFire PN results [14, 15, 21, 32]. Notably, in the study by Murphy et al, potentially “false positive” BioFire PN results with negative cultures were subsequently confirmed as accurate according to alternative molecular techniques [14]. Thus, BioFire PN results are unlikely to be false positive; rather, they are more likely detecting fastidious organisms or reflecting the effects of antibiotics on culture. Some experts suggest that new comparators are needed, particularly when a new test's accuracy exceeds that of the prior gold standard [39]. Solutions include clinically adjudicated reference standards, as in this study [40]. Importantly, and consistent with other reports, a negative BioFire PN result was highly predictive of normal flora on culture, with an NPV of 99% to 100% for most organisms [16, 18, 24, 25, 34].
Given these results, how does a clinician use the information provided by BioFire PN? With enhanced detection of bacteria, concern has arisen that inappropriate antibiotic use might actually increase with molecular testing [34, 41]. Previous reports have noted reasonable concordance of BioFire PN and culture results based on semiquantitative thresholds [14, 15, 20, 21]. However, since neither sputum culture nor molecular testing can accurately discriminate airway colonization from invasive bacterial infection, these data alone do not suffice. The relative abundance as well as the specific pathogens detected may help with interpretation. In our study, S pneumoniae, H influenzae, and M catarrhalis—common respiratory pathogens—were associated with higher genomic copies per milliliter than organisms often considered colonizers, such as S aureus, group B Streptococcus, and GNR. We found that high genomic copies were associated with a significantly greater percentage of patients with PCT values ≥0.25 ng/mL and more commonly adjudicated as bacterial illness. Notably, ≥106 copies of specific organisms (S pneumoniae and M catarrhalis) were more frequent in pneumonia, suggesting an association with invasive disease. As part of the PROGRESS trial, Kyriazopoulou et al also found that PCT values were higher in patients with bacteria detected at ≥105 copies/mL [42]. Thus, in the proper clinical setting, detection of a high quantity of a typical respiratory pathogen in conjunction with an elevated host biomarker may indicate a clinically important bacterial infection.
A potentially highly impactful aspect of the BioFire PN assay is the high NPV. It is noteworthy that a negative BioFire PN result in our study was very uncommon in patients with illness adjudicated as bacterial by a panel of experts. Thus, a negative result in patients outside the intensive care unit setting or in patients who are not immunocompromised—when unusual bacterial or fungal organisms not included in the BioFire PN panel are less of a concern—is very reassuring. This conclusion is supported by the finding that the BioFire PN results are not affected by recent prior antibiotics. Notably, patients with BioFire PN results yielding negative or low bacterial abundance with or without low PCT and normal chest radiograph results were frequently given antibiotics (64%–76%). It is possible that with sound clinical judgment and other clinical factors, negative BioFire PN results could provide clinicians with greater confidence to stop unneeded antibiotics.
Not to be overlooked is the additional value of BioFire PN to detect viruses and atypical organisms such as Mycoplasma and Legionella species. Non–Legionella pneumophila serotype 1 is not detected on urinary antigen assays and may be detectable by BioFire PN [43]. Viral detection doubled with BioFire PN when compared with SOC testing, consistent with a number of publications showing that viral testing of sputum offers increased diagnostic yield vs nasopharyngeal testing alone, particularly in hospitalized patients [44–46]. Sputum analysis has been severely limited by the difficulty of collecting adequate samples without oropharyngeal contamination from nonintubated patients. We observed that BioFire analytic performance was better in high quality samples than moderate quality samples, similar to a study by Andrews et al [34]. However, in the moderate quality samples, NPV remained excellent (>94%) and continued to demonstrate added value of detection of atypical bacterial and viruses. Gilbert et al assessed the value of BioFire PN vs a multitest bundle of SOC diagnostic testing in 274 patients with sputum available [33]. Investigators demonstrated that viral pathogens were more commonly detected with BioFire PN than SOC and that BioFire PN outperformed urinary antigen testing for pneumococcus and legionella. They also concluded that serum PCT was helpful to distinguish bacterial invasion from colonization. Our data align with these findings and suggest that use of BioFire PN in patients producing sputum could replace a number of SOC tests while offering the possibility of ruling out bacterial infection if negative.
Our study has several limitations. We were unable to collect sputum samples on all patients, and frequent administration of antibiotics prior to sputum collection hampered direct comparisons of BioFire PN and culture. In addition, our study population was limited to those who could provide informed consent. Finally, as BioFire PN results were not released to clinicians, clinical impact could not be directly assessed.
In summary, BioFire PN testing yielded significantly more potential pathogens than culture and was less affected by prior antibiotics in hospitalized patients with a variety of community-acquired LRTIs. The NPV for sputa of good and moderate quality was excellent, and with increased viral detections, BioFire PN offers a tool to reduce unnecessary antibiotic use. Randomized clinical trials are needed to further assess the clinical impact of BioFire PN testing.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Ann R Falsey, Infectious Diseases, Department of Medicine.
Angela R Branche, Infectious Diseases, Department of Medicine.
Daniel P Croft, Department of Pulmonary and Critical Medicine, University of Rochester.
Maria A Formica, Infectious Disease Unit, Rochester General Hospital, Rochester, New York, USA.
Michael R Peasley, Infectious Diseases, Department of Medicine.
Edward E Walsh, Infectious Diseases, Department of Medicine.
Notes
Acknowledgments. The authors thank Mary Criddle and Sharon Moorehead for patient recruitment.
Author contributions. A. R. F. and E. E. W. designed the study, collected and analyzed data, prepared the manuscript. A. R. B. and D. P. C. collected and analyzed data and edited the manuscript. M. A. F. and M. R. P. ran assays, analyzed data, and edited the manuscript.
Financial support. This work was supported by a grant from the National Institute of Allergy and Infectious Disease (R01AI137364) and a grant from BioFire Diagnostics.
References
- 1. GBD 2015 Disease and Injury Incidence and Prevalence Collaborators . Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016; 388:1545–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jain S, Self WH, Wunderink RG, Team CES. Community-acquired pneumonia requiring hospitalization. N Engl J Med 2015; 373:2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brendish NJ, Malachira AK, Armstrong L, et al. . Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): a pragmatic, open-label, randomised controlled trial. Lancet Respir Med 2017; 5:401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shiley KT, Lautenbach E, Lee I. The use of antimicrobial agents after diagnosis of viral respiratory tract infections in hospitalized adults: antibiotics or anxiolytics? Infect Control Hosp Epidemiol 2010; 31:1177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sethi S, Evans N, Grant BJ, Murphy TF. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 2002; 347:465–71. [DOI] [PubMed] [Google Scholar]
- 6. Branche A, Neeser O, Mueller B, Schuetz P. Procalcitonin to guide antibiotic decision making. Curr Opin Infect Dis 2019; 32:130–5. [DOI] [PubMed] [Google Scholar]
- 7. Schuetz P, Wirz Y, Mueller B. Procalcitonin testing to guide antibiotic therapy in acute upper and lower respiratory tract infections. JAMA 2018; 319:925–6. [DOI] [PubMed] [Google Scholar]
- 8. Ramilo O, Rodriguez-Fernandez R, Mejias A. Promise and limitations of procalcitonin to identify bacterial infections in the pediatric intensive care unit. J Pediatr 2016; 179:7–9. [DOI] [PubMed] [Google Scholar]
- 9. Paterson DL. “Collateral damage” from cephalosporin or quinolone antibiotic therapy. Clin Infect Dis 2004; 38(suppl 4):S341–5. [DOI] [PubMed] [Google Scholar]
- 10. Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf 2014; 5:229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Webb BJ, Sorensen J, Jephson A, Mecham I, Dean NC. Broad-spectrum antibiotic use and poor outcomes in community-onset pneumonia: a cohort study. Eur Respir J 2019; 54:1900057. [DOI] [PubMed] [Google Scholar]
- 12. Chambers HF, Bartlett JG, Bonomo RA, et al. . Antibacterial resistance leadership group: open for business. Clin Infect Dis 2014; 58:1571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larkin H. Increasing antimicrobial resistance poses global threat, WHO says. JAMA 2023; 329:200. [DOI] [PubMed] [Google Scholar]
- 14. Murphy CN, Fowler R, Balada-Llasat JM, et al. . Multicenter evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for detection and quantification of agents of lower respiratory tract infection. J Clin Microbiol 2020; 58:e00128-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginocchio CC, Garcia-Mondragon C, Mauerhofer B, Rindlisbacher C; EME Evaluation Program Collaborative . Multinational evaluation of the BioFire FilmArray Pneumonia Plus Panel as compared to standard of care testing. Eur J Clin Microbiol Infect Dis 2021; 40:1609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buchan BW, Windham S, Balada-Llasat JM, et al. . Practical comparison of the BioFire FilmArray Pneumonia Panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J Clin Microbiol 2020; 58:e00135-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Monard C, Pehlivan J, Auger G, et al. . Multicenter evaluation of a syndromic rapid multiplex PCR test for early adaptation of antimicrobial therapy in adult patients with pneumonia. Crit Care 2020; 24:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitton B, Rule R, Said M. Laboratory evaluation of the BioFire FilmArray Pneumonia Plus Panel compared to conventional methods for the identification of bacteria in lower respiratory tract specimens: a prospective cross-sectional study from South Africa. Diagn Microbiol Infect Dis 2021; 99:115236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rand KH, Beal SG, Cherabuddi K, et al. . Relationship of multiplex molecular pneumonia panel results with hospital outcomes and clinical variables. Open Forum Infect Dis 2021; 8:ofab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rand KH, Beal SG, Cherabuddi K, et al. . Performance of a semiquantitative multiplex bacterial and viral PCR panel compared with standard microbiological laboratory results: 396 patients studied with the BioFire Pneumonia Panel. Open Forum Infect Dis 2021; 8:ofaa560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee SH, Ruan SY, Pan SC, Lee TF, Chien JY, Hsueh PR. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J Microbiol Immunol Infect 2019; 52:920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kamel NA, Alshahrani MY, Aboshanab KM, El Borhamy MI. Evaluation of the BioFire FilmArray Pneumonia Panel Plus to the conventional diagnostic methods in determining the microbiological etiology of hospital-acquired pneumonia. Biology (Basel) 2022; 11:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karolyi M, Pawelka E, Hind J, et al. . Detection of bacteria via multiplex PCR in respiratory samples of critically ill COVID-19 patients with suspected HAP/VAP in the ICU. Wien Klin Wochenschr 2022; 134(9–10):385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foschi C, Zignoli A, Gaibani P, et al. . Respiratory bacterial co-infections in intensive care unit-hospitalized COVID-19 patients: conventional culture vs BioFire FilmArray Pneumonia Plus Panel. J Microbiol Methods 2021; 186:106259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen R, Babushkin F, Finn T, et al. . High rates of bacterial pulmonary co-infections and superinfections identified by multiplex PCR among critically ill COVID-19 patients. Microorganisms 2021; 9:2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Webber DM, Wallace MA, Burnham CA, Anderson NW. Evaluation of the BioFire FilmArray Pneumonia Panel for detection of viral and bacterial pathogens in lower respiratory tract specimens in the setting of a tertiary care academic medical center. J Clin Microbiol 2020; 58:e00343-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larry RC, Hoff BM, Bertram CM. Evaluation of microbiological concordance of a rapid molecular diagnostic pneumonia panel in a real-world population with pneumonia. J Appl Lab Med 2023; 8:514–22. [DOI] [PubMed] [Google Scholar]
- 28. Musher DM. The usefulness of sputum gram stain and culture. Arch Intern Med 2005; 165:470–1. [DOI] [PubMed] [Google Scholar]
- 29. Mandell LA, Wunderink RG, Anzueto A, et al. . Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007; 44(suppl 2):S27–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Houck PM, Bratzler DW, Nsa W, Ma A, Bartlett JG. Timing of antibiotic administration and outcomes for medicare patients hospitalized with community-acquired pneumonia. Arch Intern Med 2004; 164:637–44. [DOI] [PubMed] [Google Scholar]
- 31. Metlay JP, Waterer GW, Long AC, et al. . Diagnosis and treatment of adults with community-acquired pneumonia: an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 2019; 200:e45–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poole S, Clark TW. Rapid syndromic molecular testing in pneumonia: the current landscape and future potential. J Infect 2020; 80:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gilbert DN, Leggett JE, Wang L, et al. . Enhanced detection of community-acquired pneumonia pathogens with the BioFire Pneumonia FilmArray Panel. Diagn Microbiol Infect Dis 2021; 99:115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andrews V, Pinholt M, Schneider UV, Schonning K, Soes LM, Lisby G. Performance of PCR-based syndromic testing compared to bacterial culture in patients with suspected pneumonia applying microscopy for quality assessment. APMIS 2022; 130:417–26. [DOI] [PubMed] [Google Scholar]
- 35. Zacharioudakis IM, Zervou FN, Dubrovskaya Y, Inglima K, See B, Aguero-Rosenfeld M. Evaluation of a multiplex PCR panel for the microbiological diagnosis of pneumonia in hospitalized patients: experience from an academic medical center. Int J Infect Dis 2021; 104:354–60. [DOI] [PubMed] [Google Scholar]
- 36. Serigstad S, Markussen D, Grewal HMS, et al. . Rapid syndromic PCR testing in patients with respiratory tract infections reduces time to results and improves microbial yield. Sci Rep 2022; 12:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edin A, Eilers H, Allard A. Evaluation of the BioFire FilmArray Pneumonia Panel Plus for lower respiratory tract infections. Infect Dis (Lond) 2020; 52:479–88. [DOI] [PubMed] [Google Scholar]
- 38. Yoo IY, Huh K, Shim HJ, et al. . Evaluation of the BioFire FilmArray Pneumonia Panel for rapid detection of respiratory bacterial pathogens and antibiotic resistance genes in sputum and endotracheal aspirate specimens. Int J Infect Dis 2020; 95:326–31. [DOI] [PubMed] [Google Scholar]
- 39. Felker GM, North R, Mulder H, et al. . Clinical implications of negatively adjudicated heart failure events: data from the VICTORIA study. Circulation 2023; 147:694–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel R, Tsalik EL, Evans S, Fowler VG, Doernberg SB; Antibacterial Resistance Leadership Group . Clinically adjudicated reference standards for evaluation of infectious diseases diagnostics. Clin Infect Dis 2023; 76:938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanson KE, Azar MM, Banerjee R, et al. . Molecular testing for acute respiratory tract infections: clinical and diagnostic recommendations from the IDSA's Diagnostics Committee. Clin Infect Dis 2020; 71:2744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kyriazopoulou E, Karageorgos A, Lisskou-Antoniou L.. BioFire FilmArray pneumonia panel for severe lower respiratory tract infections: subgroup analysis of a randomized clinical trial. Infect Dis Ther 2021; 10:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Courboules C, Dournon N, Lawrence C, et al. . Non–Legionella pneumophila serogroup 1 pneumonia: diagnosis of a nosocomial legionellosis with the BioFire Pneumonia Plus Panel. IDCases 2022; 28:e01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Falsey AR, Formica MA, Walsh EE. Yield of sputum for viral detection by reverse transcriptase PCR in adults hospitalized with respiratory illness. J Clin Microbiol 2012; 50:21–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jeong JH, Kim KH, Jeong SH, Park JW, Lee SM, Seo YH. Comparison of sputum and nasopharyngeal swabs for detection of respiratory viruses. J Med Virol 2014; 86:2122–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lai T, Xiang F, Zeng J, et al. . Reliability of induced sputum test is greater than that of throat swab test for detecting SARS-CoV-2 in patients with COVID-19: a multi-center cross-sectional study. Virulence 2020; 11:1394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.