Significance
In this study, we identify HKDC1 (hexokinase domain containing 1) as a direct target gene of TFEB (Transcription factor EB). The TFEB–HKDC1 axis plays an essential role in PINK1 (PTEN-induced kinase 1)/Parkin-dependent mitophagy by PINK1 stabilization. Additionally, HKDC1 and the VDACs (voltage-dependent anion channels) with which it interacts are important for the repair of damaged lysosomes and maintaining mitochondria–lysosome contact. Importantly, mitochondrial and lysosomal homeostasis maintained by HKDC1 counteracts cellular senescence. Our study reveals the mechanism through which the cells simultaneously maintain mitochondrial and lysosomal homeostasis and provides a potential therapeutic target for aging-related diseases.
Keywords: TFEB, HKDC1, mitophagy, mitochondria–lysosome contact, cellular senescence
Abstract
Mitochondrial and lysosomal functions are intimately linked and are critical for cellular homeostasis, as evidenced by the fact that cellular senescence, aging, and multiple prominent diseases are associated with concomitant dysfunction of both organelles. However, it is not well understood how the two important organelles are regulated. Transcription factor EB (TFEB) is the master regulator of lysosomal function and is also implicated in regulating mitochondrial function; however, the mechanism underlying the maintenance of both organelles remains to be fully elucidated. Here, by comprehensive transcriptome analysis and subsequent chromatin immunoprecipitation-qPCR, we identified hexokinase domain containing 1 (HKDC1), which is known to function in the glycolysis pathway as a direct TFEB target. Moreover, HKDC1 was upregulated in both mitochondrial and lysosomal stress in a TFEB-dependent manner, and its function was critical for the maintenance of both organelles under stress conditions. Mechanistically, the TFEB–HKDC1 axis was essential for PINK1 (PTEN-induced kinase 1)/Parkin-dependent mitophagy via its initial step, PINK1 stabilization. In addition, the functions of HKDC1 and voltage-dependent anion channels, with which HKDC1 interacts, were essential for the clearance of damaged lysosomes and maintaining mitochondria–lysosome contact. Interestingly, HKDC1 regulated mitophagy and lysosomal repair independently of its prospective function in glycolysis. Furthermore, loss function of HKDC1 accelerated DNA damage–induced cellular senescence with the accumulation of hyperfused mitochondria and damaged lysosomes. Our results show that HKDC1, a factor downstream of TFEB, maintains both mitochondrial and lysosomal homeostasis, which is critical to prevent cellular senescence.
Mitochondria and lysosomes are both essential organelles for cellular homeostasis. Mitochondria are double-membrane organelles that function as the main energy-producing center, while lysosomes are single-membrane acidic organelles that degrade a wide range of materials delivered from both inside and outside cells. Mitochondrial and lysosomal behaviors are intimately linked, and dysfunction of these organelles is associated with cellular senescence, aging, and multiple prominent diseases. For instance, mitochondrial dysfunction caused by oxidative stress or genetic deletion of mitochondrial proteins impairs lysosomal structure and function, leading to abnormal protein aggregation or storage of lipid (1–4). Macroautophagy (hereafter, autophagy) is a cytoplasmic degradation system in which double-membrane structures called autophagosomes randomly or selectively sequester cytoplasmic materials and deliver them to lysosomes for degradation. Acute mitochondrial stress stimulates autophagosome formation and thereby promotes lysosomal biogenesis as well as the degradation of mitochondria in lysosomes through selective autophagy termed mitophagy (1, 3). On the other hand, disturbed lysosomal function reduces mitochondrial metabolism and triggers mitochondrial proteome remodeling and mitochondrial functional decline (2, 5, 6). There has recently been increasing evidence that mitochondria and lysosomes communicate with each other through mitochondria–lysosome contact sites (7, 8). Mitochondria–lysosome crosstalk allows for bidirectional regulation of mitochondrial and lysosomal dynamics and metabolic exchange between the two organelles (7–11). However, it remains unclear how the coordination of mitochondrial and lysosomal homeostasis is achieved.
Mitochondrial dysfunction is induced by numerous endogenous and exogenous factors, such as nutrient deprivation, hypoxia, oxidative stress, and oxidative phosphorylation uncoupling. One protective process is the efficient elimination of damaged mitochondria through mitophagy. Among several mitophagy pathways identified thus far, one is that dependent on PTEN-induced kinase 1 (PINK1) and the E3 ubiquitin (Ub) ligase Parkin has been well characterized. Under basal conditions, newly synthesized PINK1 is imported to the mitochondrial inner membrane (MIM) and then sequentially cleaved by matrix-resident protease, mitochondrial processing peptidase after mitochondria targeting sequence and MIM-resident protease, presenilin-associated rhomboid-like protease (PARL) in the transmembrane domain (TMD). Cleaved PINK1 is subsequently degraded by proteasomes in the cytosol through the N-end rule pathway (12, 13). Upon mitochondrial depolarization, full-length (FL) PINK1 accumulates on the mitochondrial outer membrane (MOM), where it phosphorylates Ub and the Ub-like domain of Parkin. Parkin-dependent MOM ubiquitylation drives recruitment of autophagy receptors to damaged mitochondria; these mitochondria are subsequently engulfed by autophagosomes and delivered to lysosomes for degradation. On the other hand, lysosomes are also often damaged by internalized pathogens, crystals, or lysosomotropic agents (14–17). Harmful lysosomal leakage triggers multiple responses, collectively called the “lysosomal damage response,” which include repair of damaged lysosomes with relatively small ruptures by the endosomal sorting complexes required for transport (ESCRT) machinery (17–19), removal of damaged lysosomes by selective autophagy termed lysophagy (16, 20), and TFEB (Transcription factor EB)-mediated transcriptional upregulation of lysosomal biogenesis.
TFEB has been identified as the master transcription factor for autophagy and lysosomal biogenesis (21–23). During lysosomal damage, TFEB is dephosphorylated and then translocated to the nucleus, where it induces the expression of genes involved in lysosomal biogenesis (24, 25). It has recently been shown that TFEB is involved in mitochondrial biogenesis (26, 27). TFEB is also upregulated under mitochondrial stress, which in turn induces the expression of mitophagy receptors, including NDP52 and Optineurin (28, 29). This leads us to question whether there are factors downstream of TFEB that coordinate the homeostasis of both mitochondria and lysosomes.
Here, based on comprehensive transcriptome analysis followed by chromatin immunoprecipitation (ChIP)-qPCR, we demonstrated that hexokinase (HK) domain containing 1 (HKDC1), a member of the HK family, is the previously unidentified gene that is directly targeted by TFEB. During both mitochondrial depolarization and lysosomal damage, HKDC1 is significantly upregulated. HKDC1 is critical for maintaining mitochondrial homeostasis by regulating PINK1/Parkin-dependent mitophagy. Besides, HKDC1 is significant to maintain the lysosomal integrity and mitochondria–lysosome contact. In particular, the dual effect of HKDC1 physiologically prevents cellular senescence.
Results
HKDC1 Is a Direct Target of TFEB that Is Upregulated upon Mitochondrial Depolarization.
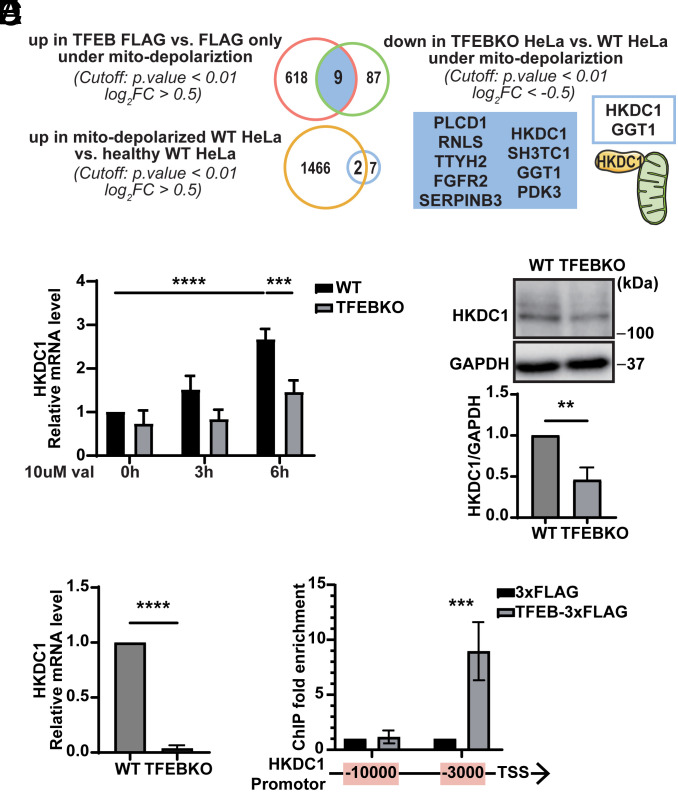
We considered it useful to uncover factors downstream of TFEB that coordinate the homeostasis of both mitochondria and lysosomes. To achieve this goal, we first sought to identify TFEB targets that are essential for mitochondrial homeostasis. Consistent with the previous report (28), we confirmed that mitochondrial depolarization promotes TFEB nuclear translocation in the HeLa cells stably expressing TFEB-mNeonGreen (mNG) (SI Appendix, Fig. S1A) after simultaneous treatment with antimycin A and oligomycin (A/O), which inhibit mitochondrial respiration. We compared transcriptome data obtained by RNA sequencing (RNA-seq) of differentially expressed genes (DEGs) between TFEB-3xFLAG-expressing and 3xFLAG-expressing HeLa cells under mitochondrial depolarization condition and identified 627 genes upregulated (Fig. 1A and SI Appendix, Fig. S1B). In parallel, we identified 96 genes downregulated in TFEB knockout (KO) HeLa cells (25) compared to wild-type (WT) HeLa cells under mitochondrial depolarization condition. The following 9 genes were overlapped between both sets and therefore possibly regulated by TFEB: PLCD1, RNLS, TTYH2, FGFR2, SERPINB3, HKDC1, SH3TC1, GGT1, and PDK3. To extract protective genes under mitochondrial depolarization condition, we next compared the transcriptomes of HeLa cells under mitochondrial depolarization condition and that under healthy condition. Comparison between them revealed that 1,468 genes were upregulated. Extracted from the intersection between the aforementioned 9 and 1,468 genes, HKDC1 and GGT1 are the most likely to be the target genes of TFEB and involved in maintenance of mitochondrial function (Fig. 1A). From the two candidates, the mitochondrially localized protein HKDC1 attracted our attention. Consistent with the RNA-seq analysis, HKDC1 transcripts were significantly upregulated in a TFEB-dependent manner during mitochondrial depolarization induced by valinomycin (Fig. 1B). TFEB KO cells consistently showed reduced HKDC1 protein levels compared to WT HeLa cells (SI Appendix, Fig. S1C).
Fig. 1.
HKDC1 is a direct target of TFEB that is upregulated upon mitochondrial depolarization. (A) RNA-seq analysis results are shown in Venn diagrams depicting the numbers of DEGs of these groups: upregulated genes in TFEB-3xFLAG expressing HeLa cells relative to 3xFLAG expressing HeLa cells under mitochondrial depolarization condition, downregulated genes in TFEB KO HeLa cells relative to WT HeLa cells under mitochondria depolarization condition, the overlap of two groups above, and upregulated genes in HeLa cells with depolarized mitochondria relative to healthy HeLa cells. N = 3. (B) Relative mRNA expression of HKDC1 in WT or TFEB KO HeLa cells treated with 10 μM val for 0 h, 3 h, or 6 h. Data were normalized by Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and are expressed relative to WT untreated with val. N = 3. (C and D) Representative western blot (WB) (C. Upper) and quantitated (C. Lower) and relative (D) mRNA expression of HKDC1 in immortalized kidney PTECs extracted from TFEBflox/flox (WT) and TFEBflox/flox; Kap-cre (KO) mice. N = 3. (E) ChIP-qPCR analyses of HeLa cells transiently expressing 3xFLAG or TFEB-3xFLAG in the regions 3,000 or 10,000 bp upstream of HKDC1 TSS. The bar chart shows the amount of immunoprecipitated DNA as detected by qPCR assay. N = 3. Values are represented as the mean ± SD, and P-values (**P < 0.01, ***P < 0.001, ****P < 0.0001) were determined by one-way ANOVA with Tukey’s multiple comparison test (B and E) or unpaired t test (C and D).
HKDC1 is a member of the HK family, which contains HK1–4 and HKDC1. To determine whether HKDC1 is specifically regulated by TFEB, we examined the expression of HK1–4 mRNA in WT HeLa cells and TFEB KO HeLa cells with or without valinomycin treatment. While the expression of both HK2 and HKDC1 was increased upon mitochondrial depolarization, only that of HKDC1 was dependent on TFEB (Fig. 1B and SI Appendix, Fig. S1D). Neither HK3 nor HK4 transcripts were detectable in HeLa cells, consistent with a previous report (30). Additionally, prompted by a recent report that kidney tubules are a major site of mammalian mitophagy (31), we examined HKDC1 expression in mouse immortalized kidney proximal tubular epithelial cells (PTECs) extracted from PTEC-specific TFEB-deficient (TFEBflox/flox; Kap-cre) and control (TFEBflox/flox) mice (25) and found that both mRNA and protein levels of HKDC1 were lower in TFEB KO PTECs than in control cells, even under steady-state conditions (Fig. 1 C and D). Therefore, HKDC1 expression is specifically regulated by TFEB in both human and mouse cell lines.
We next examined whether TFEB directly regulated HKDC1 expression. ChIP-Atlas integrating public ChIP-seq data (32) suggested that TFEB could bind to an approximately 3,000-bp region upstream of HKDC1 transcription start sites (TSS) (SI Appendix, Fig. S1E). ChIP-qPCR analysis indeed revealed that this 3,000-bp region upstream of TSS, but not a 10,000-bp region upstream of TSS, was strongly enriched in the TFEB-3xFLAG immunoprecipitated fraction, suggesting that HKDC1 is a direct target gene of TFEB (Fig. 1E).
HKDC1 Deficiency Impairs PINK1/Parkin-Mediated Mitophagy.
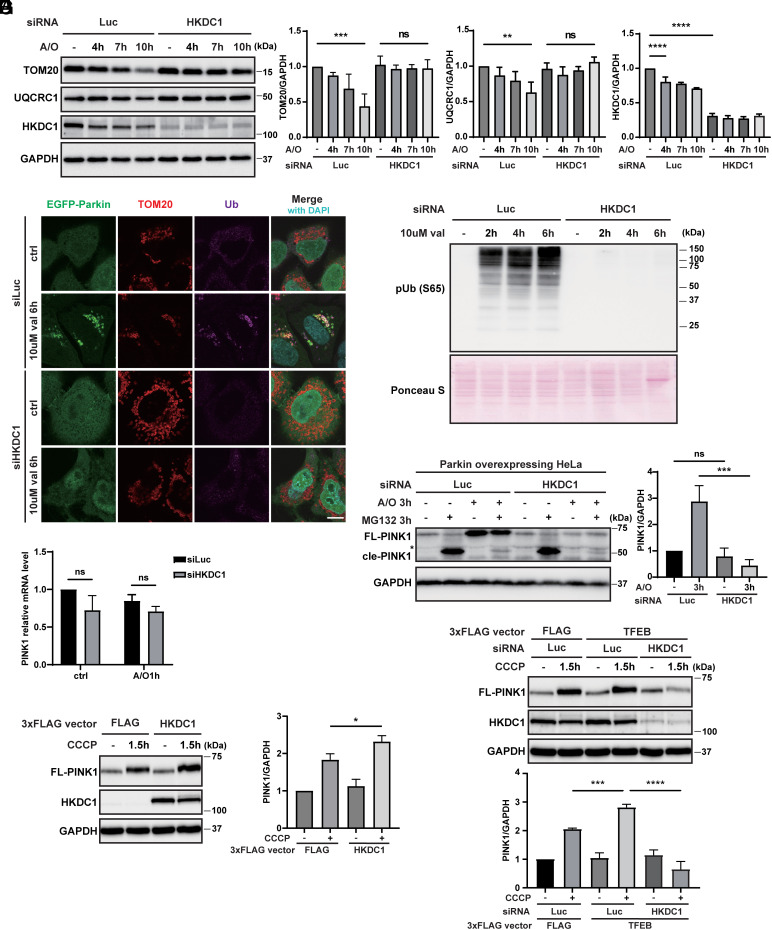
We confirmed that in agreement with a previous report (33), transiently expressed HKDC1-EGFP in HeLa cells was colocalized with a MOM protein, namely translocase of outer mitochondrial membrane 20 (TOM20) (SI Appendix, Fig. S1F). To address the possible role of HKDC1 in mitophagy, we first examined whether HKDC1 affected the well-characterized PINK1/Parkin-dependent mitophagy pathway. We knocked down its function using short interfering (si) RNA in stably EGFP-Parkin-expressing HeLa cells and examined the protein levels of TOM20 and the MIM/mitochondria matrix protein ubiquinol-cytochrome c reductase core protein 1 (UQCRC1) after A/O treatment. As the A/O treatment time was extended, TOM20 and UQCRC1 levels gradually decreased in HeLa cells transfected with siLuciferase (siLuc), while the reduction of these mitochondrial proteins was completely blocked in siHKDC1-transfected cells (Fig. 2A). We also found that Ub phosphorylation and EGFP-Parkin recruitment on mitochondria, both of which are critical steps during PINK1/Parkin-dependent mitophagy, were dramatically inhibited in HKDC1-deficient cells compared to siLuc-treated cells (Fig. 2 B and C and SI Appendix, Fig. S2A). Likewise, phosphorylated Ub and ubiquitinated EGFP-Parkin were manifest in the depolarized mitochondria extracted from siLuc-treated HeLa cells in comparison with those from siHKDC1-treated HeLa cells (SI Appendix, Fig. S2B). By contrast, HKDC1 depletion did not affect starvation-induced autophagy (SI Appendix, Fig. S2C), suggesting that HKDC1 is preferentially required for PINK1/Parkin-dependent mitophagy but not for non-selective bulk autophagy.
Fig. 2.
HKDC1 deficiency hampers PINK1/Parkin-mediated mitophagy. (A) Representative WB (Left) and quantification (Right) of TOM20, UQCRC1, and HKDC1 expression in siLuc- or siHKDC1-transfected HeLa cells overexpressing Myc-Parkin treated with A/O for 0, 4, 7, or 10 h. N = 3. (B) Representative images of EGFP-Parkin, TOM20, and Ub detected by immunofluorescence (IF) in siLuc- or siHKDC1-transfected HeLa cells overexpressing EGFP-Parkin treated with or without 10 μM val for 6 h. Nuclei were stained with DAPI. N = 3. (Scale bar, 10 μm.) (C) Expression of p-Ub in siLuc- or siHKDC1-transfected HeLa cells overexpressing EGFP-Parkin treated with 10 μM val for 0, 2, 4, or 6 h and detected by WB. N = 3. Ponceau S staining was performed for loading control. (D) Representative WB of full-length PINK1 (FL-PINK1) and cleaved PINK1 (cle-PINK1) in siLuc- or siHKDC1-transfected HeLa cells overexpressing Myc-Parkin treated with or without A/O and 10 μM MG132 for 3 h. Relative amounts of FL-PINK1 are quantified on the Right. N = 3. The asterisk denotes non-specific bands. (E) Relative mRNA expression of PINK1 in siLuc- or siHKDC1-transfected HeLa cells overexpressing Myc-Parkin treated with or without A/O for 1 h. N = 3. (F) Representative WB (Left) and quantification (Right) of FL-PINK1 in EGFP-Parkin-expressing HeLa cells with transient control 3xFLAG or HKDC1-3xFLAG overexpression treated with or without 7.5 μM CCCP for 1.5 h. N = 3. (G) Representative WB (Upper) and quantification (Lower) of FL-PINK1 in siLuc- or siHKDC1-transfected HeLa cells overexpressing EGFP-Parkin with transient control 3xFLAG or TFEB-3xFLAG overexpression treated with or without 7.5 μM CCCP for 1.5 h. N = 3. Values are represented as the mean ± SD, and P-values (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) were determined by one-way ANOVA with Tukey’s multiple comparison test.
Remarkably, we found that PINK1 accumulation on damaged mitochondria, which is the initial step of mitophagy, was severely compromised by HKDC1 knockdown (Fig. 2D and SI Appendix, Fig. S2 D and E). Similar phenotypes were observed in lung cancer cell line A549 (SI Appendix, Fig. S2F). Importantly, the PINK1 transcript level was not altered by HKDC1 knockdown under normal or mitochondrial depolarization conditions (Fig. 2E). Furthermore, under normal conditions, the proteasome inhibitor MG132 restored the cleaved form of PINK1 (52 kDa) in HKDC1-depleted cells as well as negative control knockdown cells (Fig. 2D), suggesting that HKDC1 is dispensable for mitochondrial targeting of PINK1 followed by PARL-mediated processing in healthy mitochondria. Intriguingly, neither treatment with MG132 nor another proteasome inhibitor, bortezomib, restored the accumulation of FL PINK1 upon mitochondrial depolarization in HKDC1 knockdown cells (Fig. 2D and SI Appendix, Fig. S2 D, E, and G), suggesting that the reduction of PINK1 in HKDC1-deficient cells is not mediated by proteasomes. We then investigated whether PINK1 is degraded in a lysosomal-dependent manner and found that the lysosomal inhibitor bafilomycin A1 (BafA1) did not restore PINK1 accumulation on depolarized mitochondria in HKDC1 knockdown cells (SI Appendix, Fig. S2H). We also examined whether mitochondrial proteases may be involved in PINK1 degradation following A/O treatment. OMA1 has been shown to inappropriately cleave PINK1 in depolarized mitochondria, and neither its depletion nor that of PARL suppressed the mitochondrial depolarization–dependent reduction of PINK1 in siHKDC1-treated cells (SI Appendix, Fig. S3A). Furthermore, knockdown of several mitochondrial proteases, as indicated in SI Appendix, Fig. S3A, did not inhibit the reduction of PINK1 in siHKDC1-treated cells. We also expressed several PINK1-YFP mutants in PINK1 KO cells, as shown in SI Appendix, Fig. S3B, but none of them accumulated in HKDC1-deficient cells upon mitochondrial depolarization, suggesting that the specific region tested is not responsible for the reduction of PINK1.
By contrast, HKDC1 overexpression sufficed to accelerate PINK1 accumulation upon mitochondrial depolarization (Fig. 2F). TFEB overexpression also consistently enhanced PINK1 stabilization on depolarized mitochondria (Fig. 2G). Importantly, HKDC1 silencing in the context of TFEB overexpression obviously inhibited PINK1 accumulation (Fig. 2G). Taken together, HKDC1 is required and sufficient for PINK1 accumulation under mitochondrial depolarization condition.
The Mitochondrial Localization of HKDC1 Is Required for PINK1 Accumulation and Parkin Recruitment.
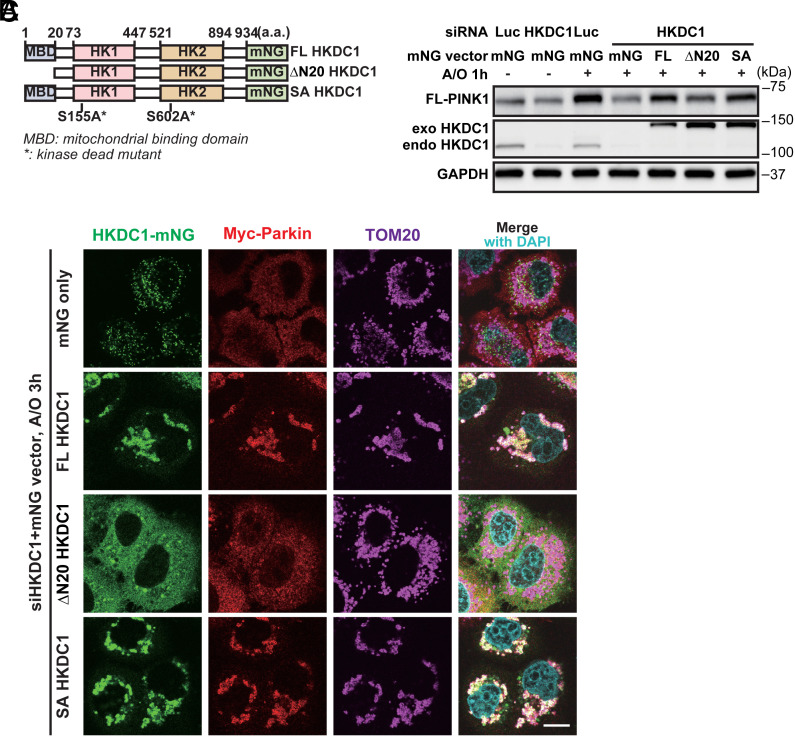
Next, we sought to determine which function of HKDC1 is required for PINK1 accumulation and Parkin recruitment. We overexpressed constructs containing a siHKDC1-resistant sequence and one of the following: C-terminal mNG-tagged HKDC1: full-length HKDC1 (FL HKDC1), an N-terminal 20-amino-acid deletion mutant [∆N20 HKDC1, which fails to localize on mitochondria due to lack of its N-terminal hydrophobic mitochondrial binding domain (MBD)], and the S155/602A mutant (SA HKDC1), which corresponds to the S155/603A mutant of HK2 that lost its HK activity (Fig. 3A and SI Appendix, Fig. S4 A and B) (34). We found that FL HKDC1 and SA HKDC1 rescued Parkin recruitment on mitochondria in HKDC1 knockdown cells, while ∆N20 HKDC1, which dispersed in the cytoplasm instead of remaining on the mitochondria, did not restore this recruitment (Fig. 3B and SI Appendix, Fig. S4 C–F). FL HKDC1 and SA HKDC1 partially rescued PINK1 accumulation on depolarized mitochondria in HKDC1 knockdown cells, whereas the effect of ∆N20 HKDC1 was less prominent (Fig. 3C). These results suggest that HKDC1 has a scaffolding function in mitochondria, and it is this role, rather than prospective HK activity, that is required for PINK1 accumulation and subsequent Parkin recruitment.
Fig. 3.
Localization of HKDC1 in mitochondria is required for PINK1 accumulation and Parkin recruitment. (A) Schematic domain structure of FL HKDC1, ∆N20 HKDC1, and SA HKDC1, showing the N-terminal MBD site, two HK domains, and the mNG tag. a.a. represents amino acids. (B) Representative images of mNG-HKDC1, Myc-Parkin, and TOM20 detected by IF in siHKDC1-transfected HeLa cells overexpressing mNG, FL HKDC1-mNG, ∆N20 HKDC1-mNG, or SA HKDC1-mNG, and treated with A/O for 3 h. Nuclei were stained with DAPI. N = 3. (Scale bar, 10 μm.) (C) Representative WB of FL-PINK1, exogenous (exo) and endogenous (endo) HKDC1 in siLuc- or siHKDC1-transfected HeLa cells overexpressing Myc-Parkin and either mNG, FL HKDC1–mNG, ∆N20 HKDC1–mNG, or SA HKDC1–mNG, and treated with A/O for 1 h. N = 3.
HKDC1 Interacts with the PINK1 Import Receptor TOM70 on Mitochondria.
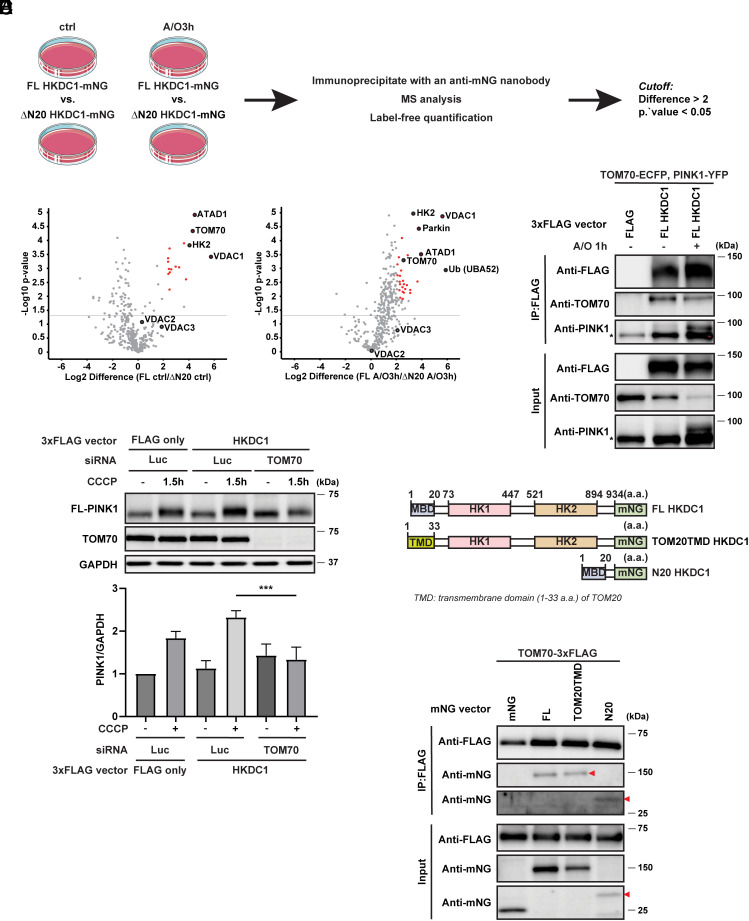
HKDC1 localizes to mitochondria via its N-terminal MBD. To gain insight into the mechanism through which HKDC1 regulates PINK1/Parkin-dependent mitophagy, we conducted an interactome analysis to find proteins that preferentially interact with HKDC1 on mitochondria. We immunoprecipitated the mNG-tagged FL HKDC1 or ∆N20 HKDC1 in HeLa cells stably expressing c-Myc–Parkin with or without A/O treatment, and performed mass spectrometry and label-free quantification (Fig. 4A). In comparison with the ∆N20 HKDC1-mNG-expressing samples, the FL HKDC1-mNG-expressing samples demonstrated significantly greater enrichment of a number of proteins (i.e., those with high abundance ratios) in both the untreated and A/O treated conditions (Fig. 4B). The proteins with the strongest interactions in the FL HKDC1-mNG-expressing samples under both conditions were the mitochondrial protein ATAD1 (outer mitochondrial transmembrane helix translocase) and HK family members HK2, VDAC1, and mitochondrial import receptor subunit TOM70. Under the A/O treated condition, Parkin and Ub (UBA52) showed the strongest interactions. The identification of these proteins demonstrated the effectiveness of our strategy to determine which proteins are in close proximity to mitochondria. Among these candidates, TOM70 attracted our attention since previous studies showed that TOM70 was an important protein for PINK1 import into mitochondria and that knockdown of TOM70 inhibited PINK1 accumulation in the PINK1–TOM complex in depolarized mitochondria (35, 36). Through co-immunoprecipitation, we confirmed that HKDC1 interacted with TOM70, and notably, PINK1 was also present in the HKDC1–TOM70 complex, especially under A/O treated conditions (Fig. 4C). In parallel, the physical interaction between PINK1-FLAG or HKDC1-FLAG and endogenous TOM70 was also detected by using a proximity ligation assay (PLA) (SI Appendix, Fig. S5 A and B). Moreover, to demonstrate that the interaction with TOM70 mediates HKDC1-dependent mitochondrial recruitment of PINK1, we assessed the effect of TOM70 knockdown on PINK1 accumulation. The silencing of TOM70 certainly blunts PINK1 recruitment in HKDC1-overexpressing cells (Fig. 4D). These data imply that HKDC1 functions as a scaffold-like protein in the initial step of mitophagy through interaction with TOM70.
Fig. 4.
HKDC1 interacts with the PINK1 import receptor TOM70 on mitochondria. (A) Experimental strategy for interactome analysis to identify proteins that interact with HKDC1 on mitochondria. (B) Volcano plots showing the distribution of quantified immunoprecipitated proteins from FL HKDC1-mNG-expressing cells vs. ∆N20 HKDC1-mNG-expressing cells under steady condition (Left) or mitochondrial depolarization condition (Right). Red puncta indicate candidates with a difference more than twofold and a Student’s t test P-value < 0.05. N = 3. (C) Representative immunoblots of FLAG, TOM70, and PINK1 in EGFP-Parkin-expressing HeLa cells transiently expressing TOM70-ECFP, PINK1-YFP, and 3xFLAG or FL HKDC1-3xFLAG treated with or without A/O for 1 h followed by immunoprecipitation with antibody against FLAG. N = 3. The asterisk denotes non-specific bands. (D) Representative WB (Upper) and quantification (Lower) of FL-PINK1 in siLuc- or siTOM70-treated HeLa cells overexpressing EGFP-Parkin with transient control 3xFLAG or HKDC1-3xFLAG overexpression treated with or without 7.5 μM CCCP for 1.5 h. N = 3. (E) Schematic domain structure of FL HKDC1, TOM20TMD HKDC1, and N20 HKDC1, showing the N-terminal MBD site or TMD site, two HK domains, and the mNG tag. a.a. represents amino acids. (F) Representative immunoblots of FLAG and mNG in EGFP-Parkin-expressing HeLa cells transiently expressing TOM70-3xFLAG and mNG, FL HKDC1-mNG, TOM20TMD HKDC1-mNG, or N20 HKDC1-mNG followed by immunoprecipitation with antibody against FLAG. N = 3. Red arrowheads denote TOM20TMD HKDC1-mNG or N20 HKDC1-mNG. Values are represented as the mean ± SD, and P-values (***P < 0.001) were determined by one-way ANOVA with Tukey’s multiple comparison test.
The MBD of HKDC1 enables its localization on mitochondria and interaction with mitochondrial TOM70. To further corroborate the role of its MBD, we utilized another two constructs: N20 HKDC1-mNG and TOM20TMD HKDC1-mNG (Fig. 4E). N20 HKDC1-mNG is the construct with only MBD of HKDC1. TOM20TMD HKDC1-mNG is the construct designed with the MBD of HKDC1 replaced by TOM20 N-terminal hydrophobic TMD regarding that TOM20 interacts with TOM70 via a C-terminal DDVE motif rather than its TMD (37). As expected, N-terminal-20-amino-acid of HKDC1 is sufficient to interact with TOM70-3xFLAG. Surprisingly, TOM20TMD HKDC1-mNG was also immunoprecipitated by TOM70-3xFLAG (Fig. 4F). Taken together, our results suggest that although the MBD of HKDC1 is sufficient to interact with TOM70, there might be another critical region other than its MBD for this interaction.
Complexes of HKDC1 with VDACs (Voltage-Dependent Anion Channels) Regulate Mitochondria–Lysosome Contact and Are Essential for Lysosomal Repair.
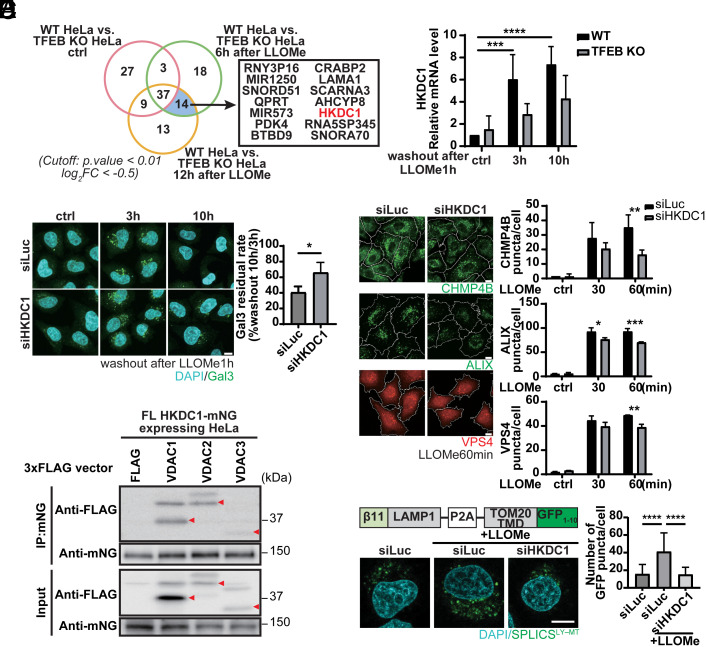
We recently showed that TFEB is activated upon lysosomal damage as part of the lysosomal damage response and is required for subsequent lysosomal recovery, presumably through induction of lysosomal biogenesis (25), although the actual TFEB targets during lysosomal damage remain elusive. To compare transcriptomes between WT and TFEB KO cells, we performed RNA-seq 6 h or 12 h after inducing lysosomal damage with the lysosomotropic compound Leu–Leu methyl ester (LLOMe), or after no treatment. Interestingly, HKDC1 was one of 14 genes with greater downregulation in TFEB KO cells compared to WT cells between 6 h and 12 h after LLOMe treatment (Fig. 5A). Furthermore, HKDC1 was significantly upregulated after LLOMe treatment in a TFEB-dependent manner (Fig. 5B). By contrast, HK1 and HK2 were not changed by LLOMe treatment (SI Appendix, Fig. S6A), suggesting that HKDC1 is specifically regulated by TFEB even under lysosomal damage conditions.
Fig. 5.
Complexes of HKDC1 with VDACs regulate mitochondria–lysosome contact and are essential for lysosomal repair. (A) RNA-seq analysis results are shown in Venn diagrams depicting the numbers of DEGs from WT vs. TFEB KO HeLa cells with or without treatment with LLOMe for 1 h followed by washout for 6 h or 12 h. N = 4. (B) Validation of qPCR-detected HKDC1 expression in WT or TFEB KO HeLa cells treated with LLOMe for 1 h followed by washout for 3 h or 10 h. N = 3. (C) Representative Gal3-antibody immunostaining images (Left) and quantification (Right) of damaged lysosomes in HKDC1 knockdown HeLa cells treated with LLOMe for 1 h followed by washout for 3 h or 10 h. Nuclei were stained with DAPI. N = 3. (Scale bar, 10 μm.) (D) Representative IF images (Left) and quantification (Right) of CHMP4B, ALIX, or VPS4 puncta in siLuc- or siHKDC1-transfected HeLa cells treated with 1 mM LLOMe for 0, 30, or 60 min. N = 3. (Scale bar, 10 μm.) (E) VDAC1-, VDAC2-, and VDAC3-3xFLAG were transiently expressed in HeLa cells stably expressing FL HKDC1–mNG. HKDC1-mNG was immunoprecipitated with mNG agarose beads. N = 3. Red arrowheads represent VDAC1, 2, and 3, respectively. (F) Schematic domain structure (Left Upper) of SPLICS reporter to detect lysosome–mitochondria (LY-MT) contact sites. Representative images (Left Lower) and quantification (Right) of SPLICS GFP punctate spots in siLuc- or siHKDC1-treated HeLa cells with or with LLOMe for 2 h. Nuclei were stained with DAPI. N (number of analyzed cells) ≥ 30. (Scale bar, 10 µm.) Values are represented as the mean ± SD, and P-values (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) were determined by one-way ANOVA with Tukey’s multiple comparison test (B, D, and F) or unpaired t test (C).
To determine whether HKDC1 plays a role in the lysosomal damage response, we knocked down HKDC1 and examined the turnover of damaged lysosomes after LLOMe treatment by detecting galectin-3 (Gal3) (16), a β-galactose–binding lectin and a marker of damaged endosomes and lysosomes. When lysosomes are damaged, its luminal glycochain is exposed to and then recruit cytosolic Gal3, forming puncta. Importantly, knockdown of HKDC1 impaired the clearance of Gal3 puncta within 10 h after LLOMe washout (Fig. 5C), suggesting that HKDC1 is also essential for the clearance of damaged lysosomes. To determine its role in this process, we first examined the possible involvement of HKDC1 in lysophagy. Upon lysosomal damage, damaged lysosomes are ubiquitinated and are thought to be recognized by the autophagic receptor p62/SQSTM1 (16, 20). We found that the numbers of Ub and p62 puncta in HKDC1-deficient cells were comparable to those in siLuc-treated cells (SI Appendix, Fig. S6B), suggesting that HKDC1 did not affect lysophagy. We then assessed whether HKDC1 participates in ESCRT-mediated repair, which is critical for lysosomal homeostasis after damage (18, 19). Intriguingly, knockdown of HKDC1 restrained the recruitment of ESCRT components on damaged lysosomes as shown by decreased numbers of puncta of the ESCRT-III component CHMP4B, the CHMP4A-interacting protein ALIX, and the ESCRT-III interactor VPS4 (Fig. 5D). Furthermore, we verified that HKDC1 mitochondrial localization is necessary for its role in lysosomal repair (SI Appendix, Fig. S6C). Because a previous report suggested that ESCRT recruitment requires calcium (19), we investigated intra-luminal calcium dynamics in lysosomes using Oregon Green 488 BAPTA-5N (OG-BAPTA-5N) and a calcium-insensitive probe, Texas Red–conjugated dextran. Of note, lysosomal calcium intensity was significantly reduced in HKDC1-deficient cells only under lysosomal damage conditions (SI Appendix, Fig. S6D). It was recently shown that mitochondria–lysosome contact mediated by mitochondrial VDAC1 and lysosomal TRPML1 is critical for calcium transport (7). Our interactome analysis confirmed that HKDC1 interacts with VDAC1, 2, and 3 (Fig. 5E). Notably, HKDC1 interacts with all the VDACs not through the MBD alone (SI Appendix, Fig. S6E), resembling TOM70. These results led us to hypothesize that HKDC1 contributes to lysosomal homeostasis through the mitochondria–lysosome contact site. We examined whether loss of HKDC1 function affected mitochondria–lysosome contact by using HeLa cells expressing split-GFP fragments—TOM20-GFP1-10 and nonfluorescent β11-LAMP1. The fluorescence signal of GFP exhibits when TOM20-GFP1-10 and β11-LAMP1 are located in close proximity (8 to 50 nm) (38). The number of GFP puncta was significantly increased upon lysosomal damage, in an HKDC1-dependent manner (Fig. 5F). Simultaneously, CLEM observations confirmed the GFP puncta locate between mitochondria and lysosomes (SI Appendix, Fig. S6F). We also verified the mitochondria–lysosome contact by using a PLA to evaluate the physical interaction between EGFP-TRPML1 and endogenous VDAC1 (SI Appendix, Fig. S6G). Similarly, knockdown of all VDACs (VDAC1, 2, and 3) showed a higher residual percentage of damaged lysosomes after LLOMe treatment (SI Appendix, Fig. S7A) and inhibition of VDAC1 decreased the number of ALIX puncta (SI Appendix, Fig. S7B). By contrast, depletion or overexpression of all three VDACs did not block PINK1 accumulation (SI Appendix, Fig. S7 C and D). These data suggest that HKDC1–VDACs is crucial for maintaining lysosomal homeostasis by affecting mitochondria–lysosome contact, and this ultimately affects lysosomal calcium levels that are essential for ESCRT-mediated repair.
Mitochondrial and Lysosomal Homeostasis Maintained by HKDC1 Counteracts Cellular Senescence.
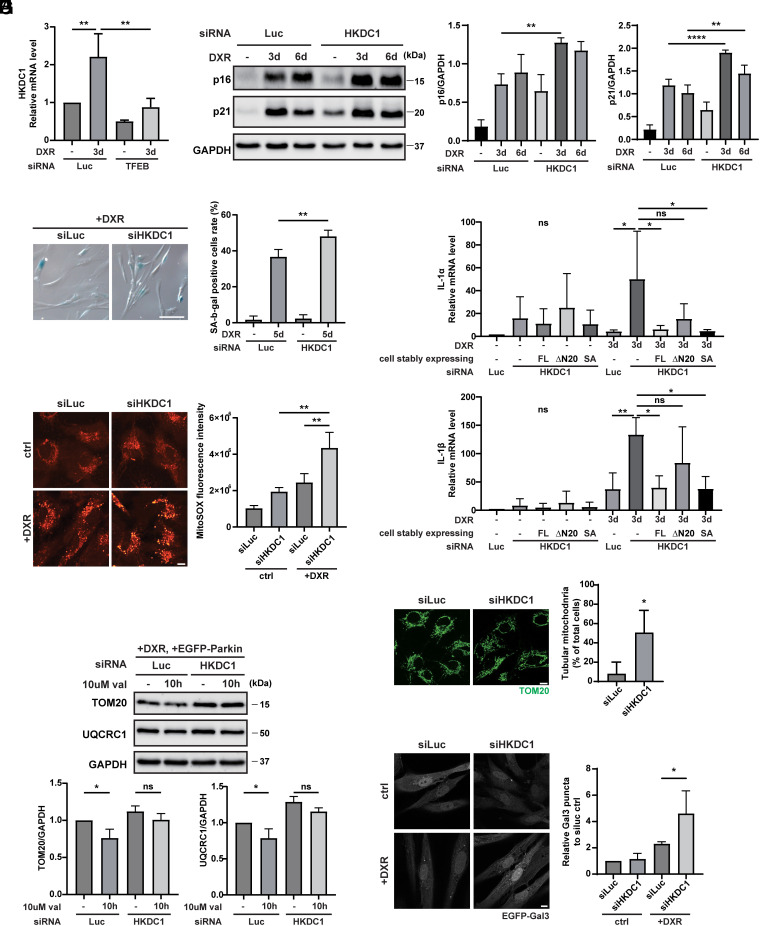
Mitochondria and lysosomes are critical organelles involved in cellular homeostasis, and abnormal mitochondria and lysosomes are implicated in cellular senescence (39–41). Since TFEB and HKDC1 are essential to maintain the homeostasis of both organelles, we wondered whether the TFEB–HKDC1 axis plays a pivotal role in preventing cellular senescence. We detected HKDC1 expression during DNA damage–induced senescence using human retinal pigment epithelial (RPE1) cell lines induced with the genotoxic agent doxorubicin (DXR) (42). Intriguingly, the HKDC1 expression level was upregulated under DXR treatment, and this upregulation was dependent on TFEB (Fig. 6A). We next examined DNA damage–induced senescence with knockdown of TFEB or HKDC1. In the siLuc-transfected group, DXR treatment for 3 d and 6 d induced cellular senescence, as shown by the increased expression of p16 and p21, whereas the siHKDC1-transfected group displayed further enhanced levels of p16 and p21 (Fig. 6B and SI Appendix, Fig. S8A). Similarly, knockdown of TFEB promoted elevation of p21 after DXR treatment for 3 d (SI Appendix, Fig. S8B). In addition, the number of senescence-associated β-galactosidase (SA-β-gal)–positive cells after DXR treatment for 5 d was markedly increased by HKDC1 knockdown (Fig. 6C). Senescent cells also showed the senescence-associated secretory phenotype (SASP), as demonstrated by increased levels of IL-1α and IL-1β (43). The lack of HKDC1 significantly stimulated IL-1α and IL-1β mRNA expression after DXR treatment for 3 d (Fig. 6D). To further clarify whether HKDC1's role in glycolysis or its scaffolding function in mitochondria is primarily responsible for this effect, we also performed rescue experiment utilizing the △N20 HKDC1 and SA HKDC1 mutant during cellular senescence. Compellingly, only ∆N20 HKDC1 mutant was unable to rescue the increased IL-1α and IL-1β, whereas SA HKDC1 mutant as well as FL HKDC1 showed restored IL-1α and IL-1β (Fig. 6D). It denotes that HKDC1’s responsibility for preventing cellular senescence lies in its scaffolding function on mitochondria.
Fig. 6.
Mitochondrial and lysosomal homeostasis maintained by HKDC1 counteracts cellular senescence. (A) qPCR-determined relative mRNA expression of HKDC1 in TFEB knockdown cells with or without DXR treatment for 3 d. N = 3. (B) Representative WB (Left) and quantification (Right) of p16 and p21 in siLuc- or siHKDC1-transfected RPE1 cells treated with 150 ng/mL DXR for 0, 3, or 6 d. N = 3. (C) Representative staining images (Left) and quantification (Right) of SA-β-gal–positive cells in siLuc- or siHKDC1-treated senescent RPE1 cells induced by DXR treatment for 5 d. N = 3. (Scale bar, 100 μm.) (D) qPCR-determined relative mRNA expression of IL-1α and IL-1β in siLuc- or siHKDC1-transfected RPE1 cells stably expressing FL HKDC1-mNG, ∆N20 HKDC1-mNG or SA HKDC1-mNG and treated with or without DXR for 3 d. N = 3. (E) Representative images (Left) and quantification (Right) of ROS with MitoSOX staining in siLuc- or siHKDC1-transfected RPE1 cells treated with or without 150 ng/mL DXR for 3 d. N = 3. (Scale bar, 10 μm.) (F) Representative images (Left) and quantification (Right) of mitochondrial morphology in siLuc- or siHKDC1-transfected RPE1 cells in the absence of DXR treatment detected by immunostaining with TOM20 antibody. N = 3. (Scale bar, 10 μm.) (G) Representative WB (Upper) and quantification (Lower) of TOM20 and UQCRC1 in siLuc- or siHKDC1-transfected RPE1 cells treated with 150 ng/mL DXR for 3 d, and with or without val for 10 h. N = 3. (H) Representative images (Left) and quantification (Right) of Gal3 puncta in siLuc- or siHKDC1-transfected RPE1 cells stably overexpressing GFP-Gal3 with or without DXR treatment. N = 3. (Scale bar, 10 μm.) Values are represented as the mean ± SD, and P-values (*P < 0.05, **P < 0.01, ****P < 0.0001) were determined by one-way ANOVA with Tukey’s multiple comparison test (A–E, G, and H) or unpaired t test (F).
Since HKDC1’s inhibitory effect on cellular senescence is dependent on its localization on mitochondria, we next detected how mitochondrial function is affected by suppression of HKDC1. We found knockdown of HKDC1 led to mitochondrial dysfunction by assessing mitochondrial reactive oxygen species (ROS) and oxygen consumption rate during cellular senescence (Fig. 6E and SI Appendix, Fig. S8C). Moreover, mitochondria in senescent cells always exhibited a hyperfused tubular structure (43, 44). HKDC1 knockdown resulted in an increased number of tubular mitochondria, as shown by immunostaining with TOM20, indicating a cell-cycle defect and accelerated cellular senescence (Fig. 6F). In addition to causing mitochondrial morphological changes, HKDC1 deficiency impaired mitophagy in EGFP-Parkin-expressing senescent RPE1 cells (Fig. 6G).
We then examined whether HKDC1 regulates lysosomal homeostasis in senescent cells as well. Knockdown of HKDC1 accelerated the accumulation of damaged lysosomes (identified by Gal3 puncta) during cellular senescence (Fig. 6H). Taken together, our results suggest that HKDC1 counteracts cellular senescence by maintaining both mitochondrial and lysosomal homeostasis.
Discussion
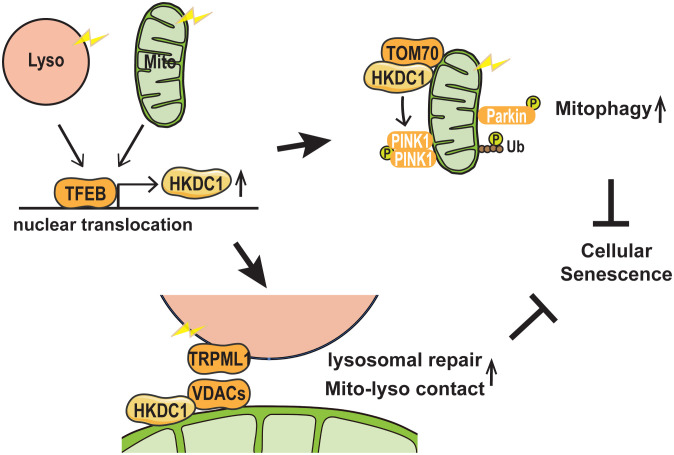
Our findings demonstrated that HKDC1, a direct target gene of TFEB, is upregulated by both mitochondrial and lysosomal stress. The TFEB–HKDC1 axis plays an essential role in PINK1/Parkin-dependent mitophagy by PINK1 stabilization, presumably through the interaction of HKDC1 with TOM70. Additionally, HKDC1 and the VDACs with which it interacts are important for the repair of damaged lysosomes, possibly as a result of regulating mitochondria–lysosome contact. We also showed that HKDC1 plays a key role in preventing DNA damage–induced cellular senescence in human cells through the maintenance of both mitochondrial and lysosomal homeostasis (Fig. 7).
Fig. 7.
Schematic model of HKDC1-mediated organelle maintenance. Both mitochondrial and lysosomal stress stimulate TFEB nuclear translocation, followed by increased HKDC1 expression. HKDC1 stabilizes PINK1 through interaction with TOM70, thereby facilitating PINK1/Parkin-dependent mitophagy. Additionally, HKDC1 and the VDAC proteins with which it interacts are important for repair of damaged lysosomes and maintaining mitochondria–lysosome contact. HKDC1 prevents DNA damage–induced cellular senescence by maintaining mitochondrial and lysosomal homeostasis.
Our work expanded upon the previously described physiological roles of HKDC1. HKDC1 is a putative fifth HK that mainly functions in glucose metabolism (45, 46). A recent report showed that mitochondrial localization of HKDC1 plays an essential role in liver cancer progression by regulating ATP levels (47). Here, we show that HKDC1 scaffolding in mitochondria is required for PINK1 accumulation and Parkin recruitment, the initial steps of PINK/Parkin-dependent mitophagy. Together with the previously demonstrated functions of TOM70 (36), HKDC1 stabilizes PINK1 by interacting with TOM70; this stabilization is important for PINK1 import into mitochondria under mitochondrial depolarization. Although we demonstrated that HKDC1 interacts with VDACs and it was previously reported that VDACs are required for Parkin recruitment into defective mitochondria (48), we proved that VDACs are not essential for PINK1 accumulation. Therefore, we exclude the possibility that HKDC1 stabilizes PINK1 via its interaction with VDACs. There is a broader range of possibilities as to why PINK1 becomes undetectable in HKDC1 KD cells after A/O treatment. On one hand, an efficient translation of mRNAs and trafficking of proteins across organelles allow cells to function exactly. Increasing evidence demonstrates that mitochondrial proteins are co-translated by mRNAs in its close proximity (49–51). Therefore, PINK1 translation could be achieved surrounding the mitochondria, which might be affected by HKDC1 deficiency. On the other hand, knockdown of HKDC1 might lead to PINK1 import into mitochondria and degraded by other peptidases in mitochondria, such as ATP23, IMMP1L, IMMP2L, HTRA2, and LACTB, etc., given that there have been various reports in which a single substrate can be cleaved by multiple mitochondria proteases (52–54). It would be interesting to examine why the level of PINK1 is reduced in HKDC1-deficient cells. It is also noteworthy that another HK, HK2, was highly expressed during mitochondrial depolarization and it was one of the prominent HKDC1-interacting proteins. Heo et al. recently found that the scaffolding function of HK2 promoted the assembly of a high-molecular-weight complex of PINK1, but the absence of HK2 had no obvious effect on PINK1 accumulation (55). Our findings demonstrate potential additional functions of mitochondrial HK family members in PINK1 stabilization on depolarized mitochondria.
We also revealed that HKDC1 was upregulated upon lysosomal stress and that lack of HKDC1 led to defective clearance of damaged lysosomes. HKDC1 deficiency impaired ESCRT recruitment on damaged lysosomes within 1 h after the damage, suggesting that HKDC1 is involved in a rapid response to lysosomal damage. Together with our experimental results that VDAC1 interacts with HKDC1, and they behave alike when forming mitochondria–lysosome contact and responding to lysosomal damage rather than accumulating PINK1, we assume that HKDC1 controls lysosome homeostasis through mitochondria–lysosome contact. The recruitment of ESCRT machinery is highly dependent on damage-related lysosomal calcium efflux that is sensed by the calcium-binding protein apoptosis-linked gene-2 (56). We found that knockdown of HKDC1 reduced the lysosomal luminal calcium level only during lysosomal damage, which could be due to decreased recruitment of ESCRT components. What then are the mechanisms by which HKDC1 maintains this calcium level during lysosomal damage? It was recently shown that at mitochondria–lysosome contact sites, calcium is transferred from lysosomes to mitochondria via the lysosomal channel TRPML1, mitochondrial VDAC1 (MOM protein), and the mitochondrial calcium uniporter (MIM protein) (7, 11). Importantly, we found that the number of mitochondria–lysosome contact sites was increased during lysosomal damage, and this increase was HKDC1-dependent. On the other hand, the calcium concentration in the lysosomal lumen is higher than that in the mitochondria; therefore, it is unclear whether the transfer of calcium from mitochondria to lysosomes via the increased number of mitochondria–lysosome contact sites actually occurs and is a mechanism by which HKDC1 maintains the calcium concentration in the lysosomal lumen. Interestingly, it has recently been reported that substance exchange takes place between the endoplasmic reticulum (ER), lysosomes, and mitochondria as a result of three-way contact between these structures (57). It has also been shown that calcium transfer from the ER to lysosomes is selectively mediated by ER–lysosomal contact sites when lysosomes are damaged (58). Considering that the ER is the primary site of intracellular calcium storage, the hypothesis that HKDC1 efficiently replenishes lysosomes with calcium by promoting the formation of three-way contact between ER, lysosomes, and mitochondria is attractive, although further examination is needed.
In addition to calcium, ions, ATP, and lipids are transferred at mitochondria–lysosome contact sites. The interaction between VDAC2 on mitochondria and PI3K on endolysosomes promotes endolysosomal maturation by contributing to ATP release from mitochondria and subsequent proton influx into endosomes (59). Furthermore, contact site–dependent lipid transfer via interaction between lysosomal Niemann–Pick type C1 protein and mitochondrial translocator protein is known to be involved in the maintenance of lysosomal and mitochondrial function (9, 10). In view of the above, future studies should verify in detail the specific HKDC1-regulated transfer of substances (other than calcium) between mitochondria and lysosomes.
Dysfunction of both mitochondria and lysosomes is closely associated with cellular senescence. The role of the TFEB–HKDC1 axis in maintaining mitochondria and lysosomes is essential for cellular senescence. A recent report has shown that the TFEB activation is a hallmark of senescence (40), which supports our observation that, as the target gene of TFEB, HKDC1 was increased in the DNA damage-induced senescent cells. Accumulating evidence shows that inhibition of glycolysis attenuates cellular senescence (60). However, deletion of HKDC1, which poorly impacts glycolytic activity (33), promoted cellular senescence. Moreover, our results demonstrated that expressing HK-deficient mutant in HKDC1 knockdown RPE1 cells was able to repress SASP expression, while the mutant without MBD was incapable. Thus, HKDC1 prevents cellular senescence in a mitochondrial anchor-dependent manner. Another point is worth mentioning that p16 and p21 levels are already higher in HKDC1 knockdown cells, even in the absence of DNA damage induction. We compared the fold increase in p16 and p21 levels. Due to increased level under no DXR condition in siHKDC1-transfected cells, the fold increase of DXR condition vs. Ctrl condition in siHKDC1-transfected cells was lower than that in siLuc-transfected cells. As it has been reported that the cell cycle inhibitors can be activated by depletion of HK2 (61) and HKDC1 structurally resembles HK2, we reasonably speculate that knockdown of HKDC1 also activates these cell cycle inhibitors, p16 and p21, the upregulation of which might simply reflect cell cycle inhibition without implicated in deleterious effects (62). Importantly, the SA-β-gal–positive cells and SASP in siHKDC1-transfected cells under no DXR condition did not show significant difference compared to siLuc-transfected cells, implying HKDC1’s key role in preventing cellular senescence. We, besides, confirmed that HKDC1 deficiency resulted in mitochondrial dysfunction, increased numbers of hyperfused mitochondria, impaired mitophagy, and the accumulation of damaged lysosomes, all of which are implicated in cellular senescence and multiple diseases exhibiting a senescence-like phenotype (41, 63–66). Therefore, our findings highlight that HKDC1 may be a potential therapeutic target for aging-related diseases.
In conclusion, HKDC1, which is the direct downstream target of TFEB, functions as a convergent factor regulating both mitochondrial and lysosomal homeostasis, and plays an important role in attenuating cellular senescence by improving mitochondrial and lysosomal function.
Materials and Methods
Cell Lines and Cell Culture.
HeLa Kyoto, A549, plat-E, and RPE1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, D6046, Sigma) supplemented with 10% fetal bovine serum (FBS, 10270106, Gibco) and 1% penicillin/streptomycin (P4333, Sigma). PTECs were cultured in low-glucose DMEM (08456-65, Nacalai) with 5% FBS and 1% penicillin/streptomycin (P4333, Sigma). All cells were cultured at 37 °C and 5% CO2. For transient expression studies, each plasmid was transfected by Lipofectamine 2000 (11668019, Invitrogen) and samples were used 24 h after transfection. For drug treatment experiments, cells were incubated in medium containing one or more of the following compounds: 10 μM valinomycin (V0627, Sigma), 7.5 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP, C2759, Sigma), 4 μM antimycin A (A8674, Sigma), 10 μM oligomycin (495455, Sigma), 10 μM MG132 (474790, Sigma), 1 mM LLOMe hydrobromide (LLOMe, L7393, Sigma), 150 ng/mL DXR (040-21521, FUJIFILM Wako Chemicals), 125 nM Bafilomycin A1 (BafA1, 11038, Cayman Chemical), 10 μM Bortezomib (504314, Sigma), or 10 μM MK6-83 (21944, Cayman Chemical), as indicated in the figure legends. For the nutrient starvation, the cells were cultured in Earle’s Balanced Salt Solution (EBSS, E2888, Sigma).
The remaining methods are described online in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the members of T. Yoshimori’s laboratory and N.M. laboratory for helpful discussions on this project. We also thank Osaka University’s Medical Innovation and Translational Research Center for assistance and access to experimental equipment and technical support. We are also grateful to Dr. Richard J. Youle (National Institute of Neurological Disorders and Stroke) and Dr. Eiji Hara (Osaka University) for kindly providing PARL KO, OMA1 KO, and PINK1 KO HeLa cells; and RPE1 cells, respectively. The CLEM experiment was performed in cooperation with Hiroko Omori (Research Institute for Microbial Diseases, Osaka University). M.C. is supported by the China Scholarship Council and Fujii Medical International Foundation. T. Yoshimori is supported by Japan Society for the Promotion of Science (JSPS) KAKENHI (22H04982), JST Core Research for Evolutional Science and Technology (CREST) (grant no. JPMJCR17H6), Japan Agency for Medical Research and Development (AMED) CREST (grant no. JP22gm1410014), and the Takeda Science Foundation. S.N. is supported by MEXT KAKENHI, a Grant-in-Aid for Transformative Research Area B (21H05145), JSPS KAKENHI (21H02428, 23K18140), AMED interstellar initiative beyond (23jm0610091h0001), the Astellas Foundation for Research on Metabolic Disorders, the Kao Research Council for the Study of Healthcare Science, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, Research grants in the Natural Sciences, the Mitsubishi Foundation, and the NOVARTIS Foundation (Japan) for the Promotion of Science. K. Yamano is supported by JSPS KAKENHI (22H02577 and 23H04923).
Author contributions
M.C., S. Minami, T.K., T. Yoshimori, and S.N. designed research; M.C., S. Minami, T. Yamamoto, and S. Matsui performed research; M.C., K. Yamano, M.O., B.T.L., Y.I., and N.M. contributed new reagents/analytic tools; M.C., K. Yamano, K. Yamamoto, H.Y.-I., T.S., M.T., K.N., H. Kato, H.O., S.O., Y.O., H. Kosako, N.M., and S.N. analyzed data; K. Yamano and N.M. provided us useful material and information on PINK1 and mitophagy; T.K. performed structural prediction; and M.C. and S.N. wrote the paper.
Competing interests
T. Yoshimori and S.N. are founders of AutoPhagyGO.
Footnotes
This article is a PNAS Direct Submission. M.R.P. is a guest editor invited by the Editorial Board.
Contributor Information
Tamotsu Yoshimori, Email: tamyoshi@fbs.osaka-u.ac.jp.
Shuhei Nakamura, Email: shuhei.nakamura@naramed-u.ac.jp.
Data, Materials, and Software Availability
RNA-seq data, Proteomic data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus, Proteome Xchange (jPOST) (GSE226743, PXD040970 for ProteomeXchange and JPST002097 for jPOST) (67, 68).
Supporting Information
References
- 1.Deus C. M., Yambire K. F., Oliveira P. J., Raimundo N., Mitochondria-lysosome crosstalk: From physiology to neurodegeneration. Trends Mol. Med. 26, 71–88 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Monteleon C. L., et al. , Lysosomes support the degradation, signaling, and mitochondrial metabolism necessary for human epidermal differentiation. J. Invest. Dermatol. 138, 1945–1954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X., et al. , MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 7, 12109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demers-Lamarche J., et al. , Loss of mitochondrial function impairs lysosomes. J. Biol. Chem. 291, 10263–10276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussi C., et al. , Lysosomal damage drives mitochondrial proteome remodelling and reprograms macrophage immunometabolism. Nat. Commun. 13, 7338 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clement D., et al. , The lysosomal calcium channel TRPML1 maintains mitochondrial fitness in NK cells through interorganelle cross-talk. J. Immunol. 211, 1348–1358 (2023), 10.4049/jimmunol.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng W., Wong Y. C., Krainc D., Mitochondria-lysosome contacts regulate mitochondrial Ca(2+) dynamics via lysosomal TRPML1. Proc. Natl. Acad. Sci. U.S.A. 117, 19266–19275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong Y. C., Ysselstein D., Krainc D., Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoglinger D., et al. , NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 10, 4276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J. X., et al. , Rab7a-mTORC1 signaling-mediated cholesterol trafficking from the lysosome to mitochondria ameliorates hepatic lipotoxicity induced by aflatoxin B1 exposure. Chemosphere 320, 138071 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Wong Y. C., Kim S., Peng W., Krainc D., Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 29, 500–513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S. M., et al. , Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamano K., Youle R. J., PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duewell P., et al. , NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464, 1357–1361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornung V., et al. , Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9, 847–856 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maejima I., et al. , Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thurston T. L., Wandel M. P., von Muhlinen N., Foeglein A., Randow F., Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radulovic M., et al. , ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, e99753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skowyra M. L., Schlesinger P. H., Naismith T. V., Hanson P. I., Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung Y. H., Chen L. M., Yang J. Y., Yang W. Y., Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 4, 2111 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Palmieri M., et al. , Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852–3866 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Sardiello M., et al. , A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Settembre C., et al. , TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia J., et al. , Galectins control mTOR in response to endomembrane damage. Mol. Cell 70, 120–135.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura S., et al. , LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat. Cell Biol. 22, 1252–1263 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Mansueto G., et al. , Transcription factor EB controls metabolic flexibility during exercise. Cell Metab. 25, 182–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastore N., et al. , TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol. Med. 9, 605–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nezich C. L., Wang C., Fogel A. I., Youle R. J., MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210, 435–450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park K., et al. , Lysosomal Ca(2+)-mediated TFEB activation modulates mitophagy and functional adaptation of pancreatic beta-cells to metabolic stress. Nat. Commun. 13, 1300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pusec C. M., et al. , Hepatic HKDC1 expression contributes to liver metabolism. Endocrinology 160, 313–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McWilliams T. G., et al. , mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oki S., et al. , ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zapater J. L., Lednovich K. R., Khan M. W., Pusec C. M., Layden B. T., Hexokinase domain-containing protein-1 in metabolic diseases and beyond. Trends Endocrinol. Metab. 33, 72–84 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaha C. S., et al. , A non-catalytic scaffolding activity of hexokinase 2 contributes to EMT and metastasis. Nat. Commun. 13, 899 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato H., Lu Q., Rapaport D., Kozjak-Pavlovic V., Tom70 is essential for PINK1 import into mitochondria. PLoS One 8, e58435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruszczak K. K., Jung M., Rasool S., Trempe J. F., Rapaport D., The role of the individual TOM subunits in the association of PINK1 with depolarized mitochondria. J. Mol. Med. (Berl.) 100, 747–762 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan A. C., et al. , Interaction between the human mitochondrial import receptors Tom20 and Tom70 in vitro suggests a chaperone displacement mechanism. J. Biol. Chem. 286, 32208–32219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cali T., Brini M., Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Nat. Protoc. 16, 5287–5308 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Cavinato M., et al. , Targeting cellular senescence based on interorganelle communication, multilevel proteostasis, and metabolic control. FEBS J. 288, 3834–3854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curnock R., et al. , TFEB-dependent lysosome biogenesis is required for senescence. EMBO J. 42, e111241 (2023), 10.15252/embj.2022111241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johmura Y., et al. , Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science 371, 265–270 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Gorgoulis V., et al. , Cellular senescence: Defining a path forward. Cell 179, 813–827 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto-Imoto H., et al. , Age-associated decline of MondoA drives cellular senescence through impaired autophagy and mitochondrial homeostasis. Cell Rep. 38, 110444 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J., A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U.S.A. 106, 11960–11965 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayes M. G., et al. , Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes 62, 3282–3291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irwin D. M., Tan H., Molecular evolution of the vertebrate hexokinase gene family: Identification of a conserved fifth vertebrate hexokinase gene. Comp. Biochem. Physiol. Part D Genomics Proteomics 3, 96–107 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Khan M. W., et al. , The hexokinase “HKDC1” interaction with the mitochondria is essential for liver cancer progression. Cell Death Dis. 13, 660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y., Vashisht A. A., Tchieu J., Wohlschlegel J. A., Dreier L., Voltage-dependent anion channels (VDACs) recruit Parkin to defective mitochondria to promote mitochondrial autophagy. J. Biol. Chem. 287, 40652–40660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fazal F. M., et al. , Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuboi T., et al. , Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. Elife 9, e57814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uszczynska-Ratajczak B., et al. , Profiling subcellular localization of nuclear-encoded mitochondrial gene products in zebrafish. Life Sci. Alliance 6, e202201514 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McBride H., Soubannier V., Mitochondrial function: OMA1 and OPA1, the grandmasters of mitochondrial health. Curr. Biol. 20, R274–R276 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Ohba Y., MacVicar T., Langer T., Regulation of mitochondrial plasticity by the i-AAA protease YME1L. Biol. Chem. 401, 877–890 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Potting C., Wilmes C., Engmann T., Osman C., Langer T., Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 29, 2888–2898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heo J. M., et al. , Integrated proteogenetic analysis reveals the landscape of a mitochondrial-autophagosome synapse during PARK2-dependent mitophagy. Sci. Adv. 5, eaay4624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheffer L. L., et al. , Mechanism of Ca(2)(+)-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 5, 5646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutry M., Kim P. K., ORP1L mediated PI(4)P signaling at ER-lysosome-mitochondrion three-way contact contributes to mitochondrial division. Nat. Commun. 12, 5354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atakpa P., Thillaiappan N. B., Mataragka S., Prole D. L., Taylor C. W., IP3 receptors preferentially associate with ER-lysosome contact sites and selectively deliver Ca2+ to lysosomes. Cell Rep. 25, 3180–3193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satoh A. O., et al. , Interaction between PI3K and the VDAC2 channel tethers Ras-PI3K-positive endosomes to mitochondria and promotes endosome maturation. Cell Rep. 42, 112229 (2023). [DOI] [PubMed] [Google Scholar]
- 60.Sabbatinelli J., et al. , Where metabolism meets senescence: Focus on endothelial cells. Front. Physiol. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf A., et al. , Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 208, 313–326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coppe J. P., et al. , Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J. Biol. Chem. 286, 36396–36403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carmona-Gutierrez D., Hughes A. L., Madeo F., Ruckenstuhl C., The crucial impact of lysosomes in aging and longevity. Ageing Res. Rev. 32, 2–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H., Tan J. X., Lysosomal quality control: Molecular mechanisms and therapeutic implications. Trends Cell Biol. 33, 749–764 (2023), 10.1016/j.tcb.2023.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cui M., Yoshimori T., Nakamura S., Autophagy system as a potential therapeutic target for neurodegenerative diseases. Neurochem. Int. 155, 105308 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Huang J., Meng P., Wang C., Zhang Y., Zhou L., The relevance of organelle interactions in cellular senescence. Theranostics 12, 2445–2464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui M., Nakamura S., Yoshimori T. HKDC1, a target of TFEB, is essential to maintain both mitochondrial and lysosomal homeostasis, preventing cellular senescence. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE226743. Deposited 06 March 2023.
- 68.Kosako H.. LFQ of mNeonGreen-Trap IP-MS from cells expressing HKDC1-mNeonGreen. jPOSTrepo. https://repository.jpostdb.org/preview/818775619643789bf442ac. Deposited 20 March 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA-seq data, Proteomic data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus, Proteome Xchange (jPOST) (GSE226743, PXD040970 for ProteomeXchange and JPST002097 for jPOST) (67, 68).