Abstract
Cryptococcus neoformans is a major fungal pathogen for patients with debilitated immune systems. However, no information is available on the stability of virulence or of phenotypes associated with virulence for C. neoformans laboratory strains. A serendipitous observation in our laboratory that one isolate of C. neoformans ATCC 24067 (strain 52D) became attenuated after continuous in vitro culture prompted us to perform a comparative study of nine strain 24067 isolates obtained from six different research laboratories. Each isolate was characterized by DNA typing, virulence for mice, proteinase production, extracellular protein synthesis, melanin synthesis, carbon assimilation pattern, antifungal drug susceptibility, colony morphology, growth rate, agglutination titers, phagocytosis by murine macrophages, capsule size, and capsular polysaccharide structure. All isolates had similar DNA typing patterns consistent with their assignment to the same strain, although minor chromosome size polymorphisms were observed in the electrophoretic karyotypes of two isolates. Several isolates had major differences in phenotypes that may be associated with virulence, including growth rate, capsule size, proteinase production, and melanization. These findings imply that C. neoformans is able to undergo rapid changes in vitro, probably as a result of adaptation to laboratory conditions, and suggest the need for careful attention to storage and maintenance conditions. In summary, our results indicate that C. neoformans (i) can become attenuated by in vitro culture and (ii) is capable of microevolution in vitro with the emergence of variants exhibiting new genotypic and phenotypic characteristics.
Cryptococcus neoformans is a frequent cause of life-threatening meningoencephalitis in immunocompromised patients, such as those with human immunodeficiency virus infection or lymphoproliferative disorders or patients who have undergone organ transplantation or immunosupressive therapies (30). One of the striking characteristics of this fungal pathogen is its ability to cause persistent infections. The virulence characteristics that allow C. neoformans to persist in the host are poorly understood, but there is accumulating evidence that cryptococcal strains can undergo genetic changes in vitro and in vivo (16, 45). This phenomenon may contribute to survival in the host by providing a means to evade host defenses.
Previous studies have documented genetic and phenotypic instability among C. neoformans strains from clinical and environmental sources. Genetic instability has been demonstrated by electrophoretic karyotype changes after murine passage (16) and in serial isolates from individual patients (2, 44, 45). Another indication that C. neoformans can undergo rapid genetic change was provided by Block et al. (1), who reported that the development of 5-fluorocytosine resistance in vitro was associated with a high rate of mutation, i.e., 1.2 × 10−7 to 4.8 × 10−7 mutations per cell division. In addition, structural variation in the capsular polysaccharide has been observed among serial isolates from patients with recurrent meningitis and in one isolate of a serotype C strain in vitro (7, 8). Passage of environmental isolates in mice has been shown to alter the sterol content and composition and antifungal drug susceptibility (9). More recently, it was shown that one C. neoformans strain also displays differences in colony morphology suggestive of the phenomenon of phenotypic switching described for Candida albicans (17). Hence, there is strong circumstantial evidence from multiple studies that under certain conditions, C. neoformans strains can undergo genetic and phenotypic changes.
During the course of studying the murine immune response to C. neoformans, we noted that one isolate of strain ATCC 24067 became attenuated with respect to virulence following continuous passage in vitro. This observation raised the question of whether strain ATCC 24067 could change with time under laboratory conditions. ATCC 24067 (also known as 52D) is one of the best characterized C. neoformans strains and has been the subject of intensive study for several years by various laboratories (Table 1). We hypothesized that the attenuation of our ATCC 24067 stock was the result of unknown selection pressures during in vitro maintenance, and we proceeded to investigate this phenomenon further. Our approach was to compare strains from multiple laboratories, with the premise that minor differences in laboratory handling could result in different selection pressures that may lead to emergence of new variants. Consistent with this hypothesis, we found that some isolates from the various laboratories had different phenotypes, including virulence, implying that strain ATCC 24067 can change with time. These results have important implications for pathogenesis, comparison of results obtained in different laboratories, and maintenance of C. neoformans strains.
TABLE 1.
Summary of studies that have employed C. neoformans ATCC 24067 (or 52D)
| Study subject | Reference(s) |
|---|---|
| rRNA genes | 12, 13 |
| URA5 gene | 4 |
| Protease production | 5, 6 |
| Melanin production | 47 |
| Mitochondrial DNA RFLPs | 46 |
| Electrophoretic karyotype | 48 |
| Pulmonary infection | 11, 14, 19, 20, 29 |
| Cytokine inflammatory response | 18 |
| Phagocytosis | 23, 24, 26, 32 |
| Complement activation | 25, 38 |
| Polysaccharide binding | 21 |
| Cell charge and hydrophobicity | 23, 35 |
| Antibody protection | 31 |
MATERIALS AND METHODS
Strains and maintenance conditions.
Nine C. neoformans var. neoformans ATCC 24067 (serotype D) isolates were obtained from six different laboratories, including our own (A. Casadevall) and those of M. Scharff, Albert Einstein College of Medicine, New York, N.Y.; R. Cherniak, Georgia State University, Atlanta; G. Huffnagle, University of Michigan, Ann Arbor; M. Lipscomb, University of New Mexico, Albuquerque; and J. Murphy, University of Oklahoma, Oklahoma City. Table 2 describes the origin and maintenance conditions of each isolate as provided to us by the various laboratories.
TABLE 2.
Sources of the strains
| Strain code | Laboratory | Origin | Yr obtained | Maintenance
|
|
|---|---|---|---|---|---|
| Conditions | Frequency of subcloning | ||||
| 24067-A | A. Casadevall | 24067-B | 1992 | 25–30°C, Saba brothb | Frequent |
| 24067-B | M. Scharff | ATCC | 1991 | 4°C, Sab slants | ∼12 mo |
| 24067-C | M. Scharff | 24067-Bc | 1993 | 4°C, Sab slants | Infrequent |
| 24067-D1 | R. Cherniak | ATCC | 1993 | Lyophilized, 10% skim milk | None |
| 24067-D2 | R. Cherniak | 24067-D1 | 1993 | 4°C, Sab slants | 3–4 mo |
| 24067-E | G. Huffnagle | ATCC | 1991 | −70°C, Sab slants | 6–12 mo |
| 24067-F1 | M. Lipscomb | ATCC | 1993 | −70°C, Sab slants | 6 mo |
| 24067-F2 | M. Lipscomb | ATCC | 1996 | −70°C, Sab slants | 6 mo |
| 24067-G | J. Murphy | K. J. Kwon-Chungd | 1987 | 4°C, Sab slants | ∼12 mo |
Sab, Sabouraud dextrose.
This is not a standard storage condition in our laboratory.
24067-C was derived from a single colony recovered from a mouse infected with 24067-B.
National Institutes of Health.
Electrophoretic karyotyping.
C. neoformans cultures were grown overnight at 30°C in Sabouraud dextrose broth, and chromosomal DNA plugs were prepared from protoplasts as described previously (15, 37, 40). Protoplasts were obtained by treating cells with 10 mg of NovoZym per ml for 3 h at 30°C. Chromosomes were resolved in a 1% agarose gel with a contour-clamped homogeneous electric field (CHEF) DRIII variable-angle pulsed-field electrophoresis system (Bio-Rad) (15).
CNRE-1 restriction fragment length polymorphism (RFLP) analysis.
Genomic DNA was extracted from each isolate by using a modification of an existing protocol (4, 15) and examined by Southern blot hybridization analysis with the CNRE-1 probe as described previously (15). CNRE-1 is a highly discriminatory DNA probe which hybridizes to a repetitive sequence found in all C. neoformans chromosomes (43).
Antifungal susceptibility assays.
Antifungal susceptibility assays were performed by the broth macrodilution method proposed by the National Committee for Clinical Laboratory Standards (33) to determine the MICs of amphotericin B (Boehringer Mannheim), fluconazole (Roerig-Pfizer, New York, N.Y.), and 5-fluorocytosine (Sigma Chemical Co., St. Louis, Mo.). Assays were performed in RPMI 1640 medium (Sigma) supplemented with l-glutamine, without bicarbonate, and buffered to pH 7.0 with 0.165 M MOPS (morpholinepropanesulfonic acid) (Sigma). Inocula of 500 to 2,500 cells/ml in 0.9 ml of RPMI 1640 medium were added to polystyrene plastic tubes containing 0.1-ml aliquots of each drug at 10 times the final concentration. Final drug concentrations ranged from 0.03 to 128 μg/ml for fluconazole and 5-flucytosine and from 0.03 to 2 μg/ml for amphotericin B. The MICs were recorded after incubation at 35°C for 72 h. Fluconazole and 5-flucytosine MICs were defined as the lowest concentration which achieved 80% growth inhibition compared to the drug-free control. Amphotericin B MICs were defined as the lowest drug concentration at which there was an absence of growth.
Growth studies.
For growth studies, 50-ml cultures in Sabouraud dextrose broth were inoculated with ∼100,000 organisms from an overnight starter culture. Isolates were incubated with moderate shaking (150 rpm) at 37°C, and absorbance (600 nm) was measured periodically with an Ultrospec 2000 UV/visible-light spectrophotometer (Pharmacia Biotech, Cambridge, United Kingdom). For isolates A, B, D2, and F2, growth curves were also performed by hemacytometer and CFU counts. Isolates were grown as described above, and at 0, 24, 30, 36, 48, and 54 h, aliquots were used for hemacytometer counting and plated on Sabouraud dextrose agar for CFU determination.
Carbohydrate assimilation.
The ID 32C system (bioMerieux Vitek, Hazelwood, Mo.) was used to analyze the carbon assimilation pattern of each isolate with 32 carbohydrate substrates. Isolates were streaked to single colonies on Sabouraud agar plates and analyzed according to the manufacturer’s instructions.
Colony phenotypes and capsule size.
Colony phenotypes were determined by plating cells from a single colony of each isolate onto fresh Sabouraud agar plates. The plates were incubated at 30°C for 7 days, and the colonies were examined for size, shape, and texture. To measure capsule size, isolates were grown for 48 h in Sabouraud dextrose broth at 30°C with agitation (150 rpm). Capsule size was measured in India ink preparations under oil immersion (×100), using a grid with a resolution of 0.1 μm. Measurements of the capsule size (distance from the edge of the capsule to the cell wall) and cell diameter (including the capsule) were averaged (n = 20).
Melanization.
Isolates were grown on 1.5% agar plates made of defined minimal medium (15 mM glucose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine, 3.0 μM thiamine) with 1.0 mM l-dopa (Sigma) for 21 days at 30°C. Colony color was examined daily. In medium containing l-dopa, cultured cells are usually gray-brown (moderately melanized) after 3 to 5 days and black (heavily melanized) after 6 to 8 days. Isolates which differed in time to melanization (faster melanization as opposed to slower melanization) were chosen for further quantitative determination of melanin. Four isolates were selected for melanin content determination: 24067-A, 24067-D1, 24067-E, and 24067-G. Melanin quantification was performed as described previously (47). Briefly, melanized cells were collected at day 14 of culture and washed, and protoplasts were obtained as described above. Protoplasts were then suspended in 4 M guanidinium isothiocyanate for 30 min at room temperature. Cell debris was collected by centrifugation and digested in 6 M HCl for 2 h at 100°C. The HCl-insoluble material is melanin (47). The melanin was then dialyzed against distilled water for 10 days, lyophilized, weighed, and expressed as the amount of melanin (picograms) per cell.
Proteinase activity.
Each isolate was examined for protein agar clearance according to the protocol described by Chen et al. (5). Isolates were grown on plates containing defined minimal medium (as described above) supplemented with 0.1% azoalbumin (pH 4.5) at 30°C until clearance halos appeared.
Extracellular protein profile.
The extracellular protein profiles of the nine isolates of ATCC 24067 were determined as described by Chen et al. (6). Cells were incubated in defined minimal medium (as described above) supplemented with 20 to 25 μCi of 35S per ml for 10 days. Cells were pelleted, and the supernatants were filtered and subjected to scintillation counting. The supernatants were analyzed for protein content by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12% acrylamide gels and visualized by autoradiography. Band intensities were measured by exposure to a phosphorimaging screen, scanned, and then analyzed by Image Quant (Molecular Dynamics, Sunnyvalle, Calif.). Each band within a lane was corrected for background intensity by subtracting the signal from the regions without apparent bands. The protein patterns observed by autoradiography and phosphorimaging analysis were very similar if not identical.
Isolation of GXM from culture supernatant and NMR.
Each isolate was grown in 100 ml of defined minimal medium for 14 days at 30°C and then autoclaved, and the cells were removed by centrifugation. Hexadecyltrimethylammonium bromide (0.75 g total) was slowly added to 25 ml of culture supernatant with stirring at 23°C. The precipitated glucuronoxylomannan (GXM)-hexadecyltrimethylammonium bromide complex was recovered by centrifugation (5,000 × g, 15 min, 23°C), the pellet was triturated with 10% ethanol, and the suspension was again centrifuged as described above. The resulting pellet was dissolved in 1 M NaCl (5 ml) by stirring overnight at room temperature or until the precipitate was completely dissolved, and the GXM was precipitated by the slow addition of 3 volumes of 95% ethanol. The flask was placed at 4°C, and the GXM was recovered by centrifugation (5,000 × g, 15 min, 4°C). This material was dissolved in 2 M NaCl (∼10 ml) by stirring at room temperature and then was treated by ultrasonic irradiation (Branson sonifier, model 450; 3-in. cup horn, 2 h, 80% power, 40% pulse). The sample was O deacetylated by incubation at pH 11 (NH4OH) for 24 h at 23°C, followed by dialysis and lyophilization.
For nuclear magnetic resonance (NMR) spectroscopy, each O-deacetylated GXM sample (2 to 10 mg) was dissolved in 0.70 ml of 99.66% D2O and transferred into a 5-mm NMR tube. Data from all 1H-NMR experiments were recorded at 80°C with a Varian UnityPlus 500 spectrometer equipped with a 5-mm triple-resonance probe. 1H chemical shifts were measured relative to the methyl groups of sodium 4,4-dimethyl-4-silapentane-1-sulfonate taken as 0.00 ppm, and NMR data were processed off-line by using the FELIX 95.0 software package (Biosym/Molecular Simulations, San Diego, Calif.) with a Silicon Graphics Indy Workstation. Spectra were resolution enhanced by applying a sine bell window function over all real datum points. The portion of each proton spectrum between 5.00 and 5.40 ppm, where only the mannosyl residues resonated, was analyzed separately. This is the structure reporter group region used to assign the structure of each GXM (8).
Agglutination assays.
Monoclonal antibody (MAb)-mediated agglutination of the ATCC 24067 isolates was determined in a 96-well microtiter plate (Corning Glass Works, Corning, N.Y.) containing serial twofold dilutions of the immunoglobulin G1 MAb 2H1 (ranging from 0.049 to 100 μg/ml) and 106 cells/well in 1% bovine serum albumin in phosphate-buffered saline (PBS), pH 7.2. Agglutination was determined by microscopic examination after the plates were incubated at 4°C overnight. The agglutination end point was defined as the highest dilution of antibody that agglutinated C. neoformans relative to control wells containing no MAb 2H1.
In vitro murine macrophage assay.
The murine macrophage-like cell line J774.16 was grown in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (Harlan Bioproducts for Science, Indianapolis, Ind.), 10% NCTC-109 (Life Technologies Inc., Grand Island, N.Y.), and 1% nonessential amino acids (Mediatech, Washington, D.C.). Cells were plated at 4 × 105 cells per well on 96-well tissue culture plates, stimulated with 500 U of gamma interferon (Genzyme, Cambridge, Mass.) per ml, and incubated at 37°C overnight. The medium in each well was then replaced with fresh medium containing 500 U of gamma interferon per ml, 3 μg of lipopolysaccharide (from Escherichia coli serotype O127:B8 [Sigma]) per ml, 1.6 × 105 cryptococci, and 10 μg of protein G affinity-purified MAb 2H1 per ml. The plates were incubated for 2 h at 37°C, the medium was removed, and the monolayer was fixed with ice-cold methanol, stained with a 1:20 dilution of Giemsa stain, and examined with a light microscope. Phagocytosis was measured by determining the phagocytic index, defined here as the number of ingested and attached yeast cells divided by the number of macrophages per field. Five fields of approximately 200 macrophages were counted for each measurement.
Virulence studies.
A murine model of intravenous cryptococcal infection was used to study the relative virulences of isolates A, B, D2, and E. C. neoformans cells, selected from a single colony, were grown in Sabouraud dextrose broth at 30°C with shaking (150 rpm) for approximately 20 h. Organisms were harvested, washed three times with sterile PBS, and counted multiple times with a hemacytometer. Adult female A/J mice (10 per group) were infected intravenously with 5 × 105 organisms via the lateral tail vein. The inoculum size was confirmed by plating a dilution of each inoculum solution on Sabouraud dextrose agar. Mice were observed twice daily. Seventy days postinfection, mice infected with isolate A were sacrificed by cervical dislocation. The lungs were harvested and homogenized in 10 ml of sterile PBS, and 0.1 ml of the homogenate was plated on Sabouraud dextrose agar. Plates were incubated at room temperature for 3 days, and colonies were counted to determine CFU per lung.
Statistical analysis.
The results obtained for capsule size and colony diameter were compared among each group of isolates. Statistical significance was evaluated by analysis of variance followed by the Bonferroni t test. Pairwise comparisons of the melanin contents and phagocytic indices were performed by the Student t test. In vivo virulence data were analyzed by using True Epistat (standard version, 1991). Survival data were analyzed by a Wilcoxon test for censored data. Pairwise comparison between groups was performed by log-rank analysis. P values of ≤0.05 were considered significant.
RESULTS
DNA typing studies.
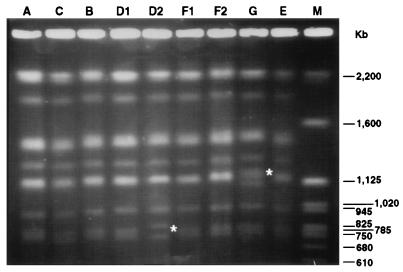
The first step in the comparison of ATCC 24067 isolates was to establish that all were in fact the same strain. Two highly discriminatory DNA typing methods were used: electrophoretic karyotyping by CHEF analysis (39) and RFLP analysis with the CNRE-1 probe (10, 15, 44). All nine isolates had the same number of chromosomes with similar if not identical molecular weights, consistent with their historical classification within one strain. Of the nine isolates, only 24067-B had been passed in vivo prior to assay for karyotype polymorphisms (Table 2). Two isolates, D2 and G, had minor differences in the electrophoretic mobilities of small chromosomes (Fig. 1). These differences were reproducible in two independent CHEF analyses and appeared to be true karyotype polymorphisms. All nine isolates had the same CNRE-1 RFLP pattern, consistent with identity within one strain (not shown). Hence, the DNA typing studies are consistent with and support the notion that all isolates were the same strain.
FIG. 1.
Electrophoretic karyotypes of the nine ATCC 24067 isolates as obtained by CHEF analysis. The molecular weight marker (Saccharomyces cerevisiae chromosomal DNA; Bio-Rad) is shown in lane M, and the corresponding sizes in are indicated on the right. The asterisks in lanes D2 and G indicate the differences in the electrophoretic karyotypes of these isolates.
Phenotypic studies.
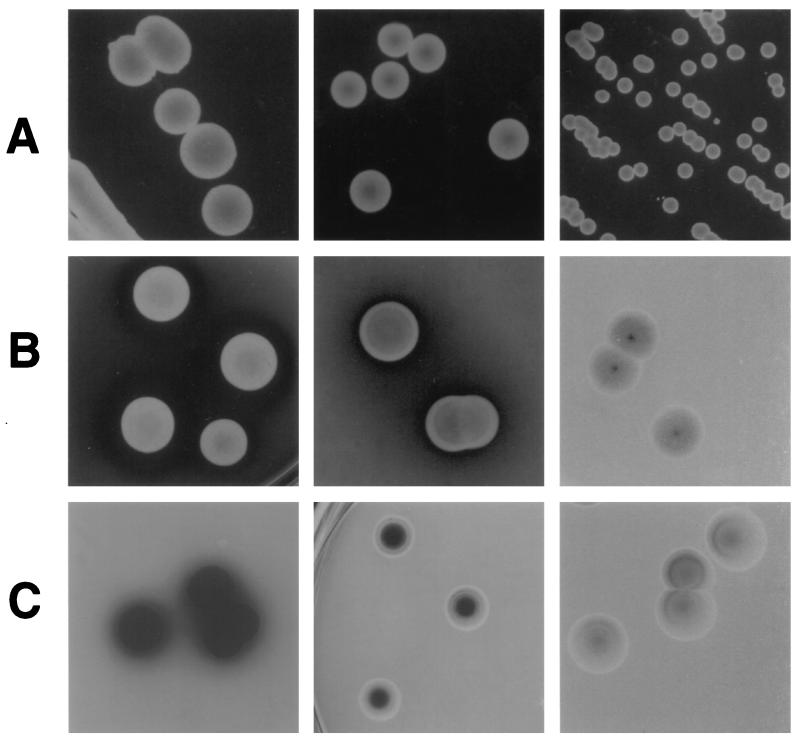
Colony morphology was assessed as described in Materials and Methods. Phenotypes were distinguished by differences in colony size and texture (Table 3; Fig. 2A). Colony types were grouped by size. Colonies of isolates C and F1 were larger than all others, with average diameters of 5.5 ± 0.7 and 5.4 ± 0.7 mm, respectively. Isolates D1, D2, E, and F2 produced colonies ranging from 4.2 ± 0.3 to 4.8 ± 0.3 mm in diameter, which were typically smooth and mucoid in texture. Isolates B and G produced smaller colonies, with average diameters of 3.6 ± 0.4 and 3.8 ± 0.3 mm, respectively. Isolate A produced the smallest colonies, being only 2 ± 0.5 mm in diameter and having a drier texture. Statistical analysis of colony size differences among these four groups revealed significant differences among each of them (P < 0.05).
TABLE 3.
Summary of phenotypic characterization of the nine ATCC 24067 isolates
| Strain code | MIC (μg/ml) ofa:
|
Melanin content (pg/cell)b | Proteolytic activity | Clearance halo size (mm)c | Agglutination titer (μg/ml) | Phagocytic indexd | Capsule size (μm)e | Cell diam (μm)f | Colony diam (mm)g | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | FLU | 5FC | |||||||||
| 24067-A | 0.25 | 1 | 2 | 2.60 ± 0.07 | Yes | 2.6 | 1.56 | 2.0 ± 0.1 | 0.6 ± 0.3 | 5.7 ± 1.1 | 2.0 ± 0.5 |
| 24067-B | 0.25 | 2 | 2 | Yes | 2.3 | 3.12 | 2.2 ± 0.3 | 3.3 ± 1.0 | 11.0 ± 1.8 | 3.6 ± 0.4 | |
| 24067-C | 0.25 | 1 | 2 | Yes | 5.0 | 6.25 | 2.2 ± 0.2 | 2.3 ± 0.7 | 9.8 ± 1.0 | 5.5 ± 0.7 | |
| 24067-D1 | 0.25 | 1 | 2 | 2.50 ± 0.07 | Yes | 4.3 | 6.25 | 2.5 ± 0.4 | 4.2 ± 0.8 | 13.4 ± 1.9 | 4.8 ± 0.3 |
| 24067-D2 | 0.25 | 2 | 0.5 | Yes | 4.1 | 12.5 | 2.4 ± 0.2 | 3.9 ± 1.3 | 13.8 ± 3.3 | 4.8 ± 0.3 | |
| 24067-E | 0.25 | 1 | 1 | 3.20 ± 0.14 | Yes | 4.2 | 3.12 | 2.2 ± 0.3 | 1.4 ± 0.4 | 7.6 ± 1.4 | 4.2 ± 0.3 |
| 24067-F1 | 0.25 | 0.5 | 2 | Yes | 2.0 | 6.25 | 2.3 ± 0.5 | 2.8 ± 0.9 | 10.0 ± 0.9 | 5.4 ± 0.7 | |
| 24067-F2 | 0.25 | 2 | 2 | Yes | 4.8 | 3.12 | 2.7 ± 0.1 | 1.3 ± 0.3 | 8.5 ± 0.7 | 4.4 ± 0.3 | |
| 24067-G | 0.25 | 0.5 | 1 | 2.00 ± 0.07 | No | 0.78 | 2.2 ± 0.4 | 0.7 ± 0.2 | 5.3 ± 0.5 | 3.8 ± 0.3 | |
AMB, amphotericin B; FLU, fluconazole; 5FC, 5-fluorocytosine.
Means ± standard deviations.
Halo refers to clearance of turbidity in protein agar plates and is a measure of proteinase activity.
Average number (± standard deviation) of C. neoformans cells per macrophage.
Capsule size was defined as the distance from the cell wall to the outer capsular border. Results are means ± standard deviations.
Cell diameter includes the capsule size. Results are means ± standard deviations.
Colony diameter measured at 7 days of growth in Sabouraud agar. Results are means ± standard deviations.
FIG. 2.
Representative phenotypes of ATCC 24067 isolates distinguished by colony size (A), extracellular proteinase production (B), and melanin production (C). (A) From left to right, colonies of isolates F1, D2, and A after 7 days of growth in Sabouraud dextrose agar plates. (B) From left to right, colonies of isolates C, F1, and G in minimal medium agar plates supplemented with 0.1% azoalbumin after 12 days of growth. Note the difference in halo size for isolates C and F1, as well as the absence of a clearance halo for isolate G. Halo sizes are listed in Table 3. (C) From left to right, colonies of isolates E, C, and G after 15 days of growth in minimal medium agar plates containing l-dopa. Note the different levels of melanization among the isolates. All photographs were taken at the same magnification (×1.5 to ×2). Actual colony sizes are listed in Table 3. Note that the isolates shown and times of growth are different for rows A, B, and C and that comparisons between rows should not be made.
Proteolytic activity and melanin production were studied by plating the isolates on minimal medium agar plates supplemented with 0.1% azoalbumin (5) and l-dopa (47), respectively. Proteolytic activity was observed as clearance of the turbid yellow azoalbumin agar surrounding the colonies (Fig. 2B). All isolates except 24067-G exhibited proteolytic activity, with halo sizes ranging from 2 to 5 mm after 16 days of growth (Table 3). Melanin production was indicated by development of black pigmentation on colonies grown in minimal medium agar plates containing l-dopa (Fig. 2C). When the nine isolates were plated on l-dopa agar plates, differences in the degree of melanization as a function of time were noted. Isolate E became completely melanized after 9 days of growth, whereas isolates B, C, D1, D2, F1, and F2 became fully melanized only after 18 days of growth. Remarkably, isolates A and G were only slightly melanized, with colonies being light brown after 21 days of growth. These differences were confirmed by measuring the melanin content per cell for isolates A, D1, E, and G at day 14 (Table 3). Statistical analysis of the melanin content per cell for these isolates showed significant differences when isolates E and G were compared to the others (P < 0.028 and P < 0.019, respectively [t test]). No significant differences in melanin content were found between isolates A and D1 (P = 0.293 [t test]). No differences were observed among the nine isolates in their ability to assimilate 32 different carbon sources.
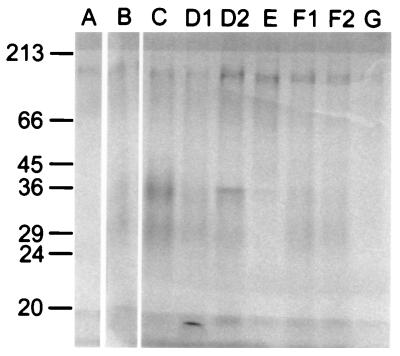
There were differences in the extracellular protein profiles of the nine isolates of ATCC 24067. Although all isolates exhibited a protein band at approximately 100 kDa, differences were apparent in the bands at 29 and 36 kDa. Visual differences for protein bands were confirmed by pixel intensity measurements through phosphorimaging analysis. With the exception of isolates A and G, all isolates exhibited a protein band between 29 and 36 kDa; however, when corrected for total counts per minute loaded, the relative protein contents associated with these bands differed as follows: for 36 kDa, C > D2 > D1 > B, F1 > E > F2 > A, G; for 29 kDa, C > D1 > B > D2 > F1 > F2 > A > E, G (Fig. 3).
FIG. 3.
Extracellular proteins of ATCC 24067 isolates on a sodium dodecyl sulfate–12% polyacrylamide gel. Each lane contained ∼400,000 total cpm from the indicated isolate. Molecular mass markers are indicated to the left in kilodaltons. A 6-day exposure is shown.
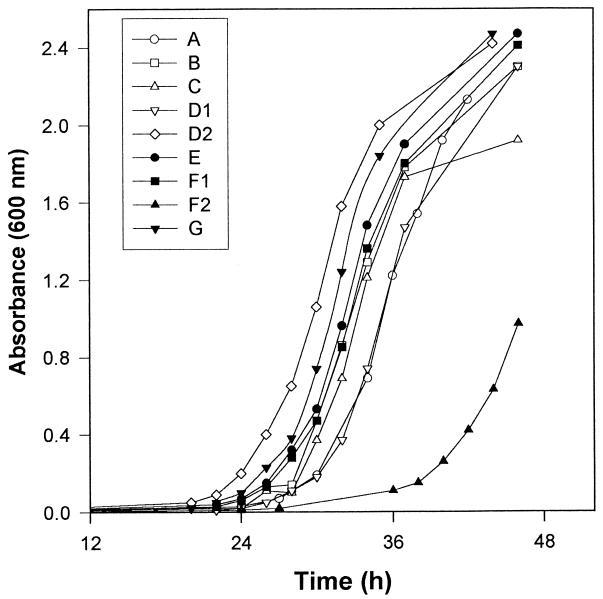
In two separate experiments, we found that the relative rates of growth of the isolates as measured by absorbance at 600 nm were as follows: D2 > G > E > B, C, F1 > A, D1 >> F2 (Fig. 4). For isolates A, B, D2, and F2, differences in growth were confirmed by a third experiment which measured cell numbers by hemacytometer and CFU counts. All isolates grew in defined minimal medium, indicating that none were auxotroph mutants.
FIG. 4.
Growth curves for the nine ATCC 24067 isolates in Sabouraud dextrose broth. This experiment was performed twice with similar results. For isolates A, B, D2, and F2 the results were confirmed by an additional experiment in which growth was measured by hemacytometer and CFU counts (not shown).
Isolates showed marked differences in capsule size and cell diameter (Table 3). The capsule size for isolates grown under identical conditions ranged from 0.6 to 4.2 μm, while the total cell diameter (including the capsule) ranged from 5.3 to 13.8 μm. Based on the capsule sizes, the isolates were categorized into four groups: those with the smallest capsule size (from 0.6 ± 0.3 to 0.7 ± 0.2 μm), represented by isolates A and G; those with the second smallest capsule size (from 1.3 ± 0.3 to 1.4 ± 0.4 μm), represented by isolates E and F2; those with an intermediate capsule size (from 2.3 ± 0.7 to 3.3 ± 1.0 μm), represented by isolates B, C, and F1; and those with the largest capsule size (from 3.9 ± 1.3 to 4.2 ± 0.8 μm), represented by isolates D1 and D2. Statistical analysis by the Bonferroni t test revealed significant differences (P < 0.05) among these groups.
The MICs of amphotericin B, fluconazole, and 5-fluorocytosine for each isolate are shown in Table 3. All isolates had the same amphotericin B MIC, whereas MICs of fluconazole and 5-flucytosine were within a narrow range of 0.5 to 2.0 μg/ml.
MAb 2H1 agglutinated all nine isolates at concentrations ranging from 0.78 to 12.5 μg/ml (Table 3). The majority of isolates (six of nine) were agglutinated at MAb 2H1 concentrations of 3.12 to 6.25 μg/ml. However, isolate D2 required a higher concentration of MAb 2H1 than the others (12.5 μg/ml), while isolates A and G agglutinated at lower antibody concentrations (1.56 and 0.78 μg/ml, respectively). There was a positive correlation between capsule size and agglutination indices for strains D2, A, and G; i.e., strain D2 had a higher agglutination titer and a bigger capsule, whereas strains A and G had lower titers and were small-capsule strains. Overall, the MAb 2H1 epitope was present in all isolates, but differences in 2H1 agglutination end points implied differences in epitope availability among some isolates. Phagocytosis studies revealed that MAb 2H1 was opsonic for all isolates, and there were no differences in the phagocytic index for the various isolates in the presence and absence of MAb 2H1. Addition of MAb 2H1 produced a significant increase in the number of attached and ingested C. neoformans cells for all the isolates (Table 3). No significant differences in the phagocytic index were detected for pairwise comparisons among the isolates.
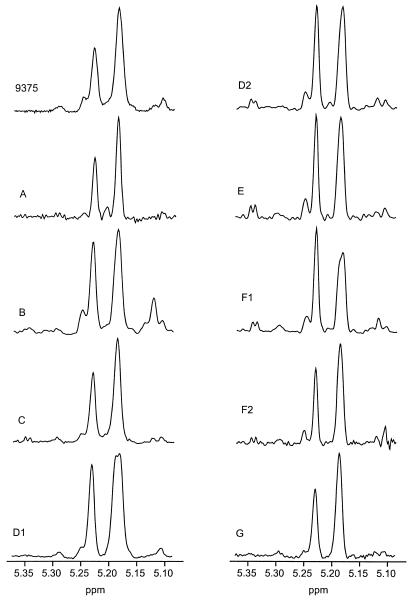
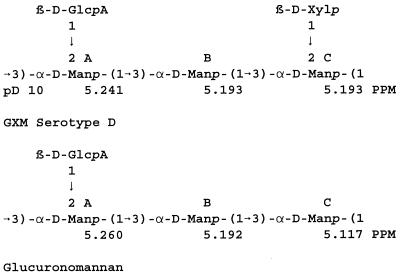
Analysis of the structure reporter group region for the nine ATCC 24067 isolates showed the presence of typical serotype D GXM structures (Fig. 5). The microheterogeneity showed by the NMR profiles illustrated in Fig. 5 is not considered to be significant. The structural elements of serotype D GXM are diagrammed in Fig.6. The low-intensity peaks at 5.260 and 5.117 ppm (Fig. 5) are characteristic of mannosyl residues A and C of glucuronomannan (Fig. 6). The resonance of mannosyl residue B of glucuronomannan at 5.192 ppm is degenerate with the resonances of mannosyl residues B and C, which occur at 5.193 ppm for serotype D GXM at pD 10.
FIG. 5.
1H-NMR spectra of resolution-enhanced anomeric regions of the de-O-acetylated GXMs for the various isolates of ATCC 24067 used in this study. The 1H-NMR data for the serotype D isolate 9375 is included as a reference (42).
FIG. 6.
Repeating units of the reference serotype D GXM and for glucuronomannan with the observed chemical shifts for the mannosyl residues of the structure reporter group region at pD 10.
Virulence studies.
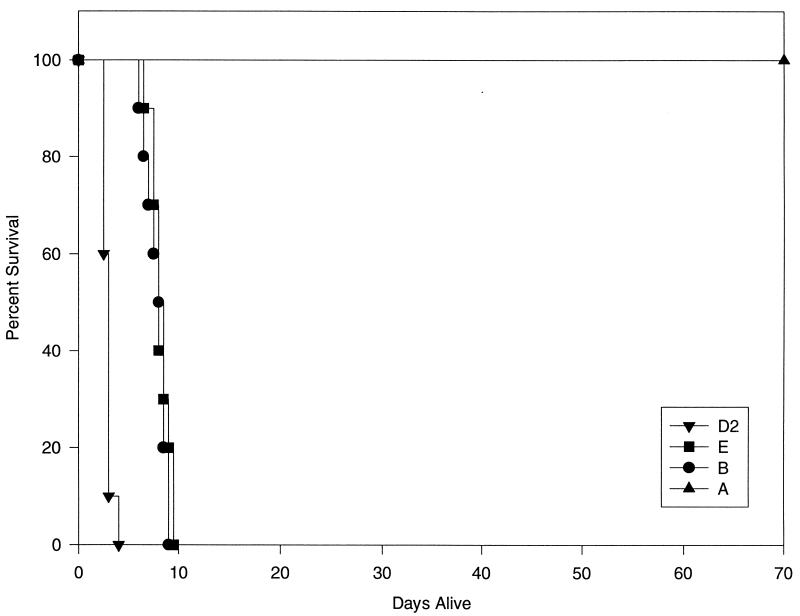
Our initial observation was that isolate 24067-A had significantly reduced virulence relative to our historical experience with strain ATCC 24067. To formally evaluate this observation, we compared the relative virulences of isolates A, B, E, and D2 in a murine model of intravenous infection (Fig. 7). The average survival of mice infected with D2, B, and E was 2.9 ± 0.9, 7.85 ± 1.0, and 8.2 ± 0.9 days, respectively. All mice infected with isolate A were still alive at 70 days postinfection, with only low numbers of organisms detected in their lungs (4,520 ± 10,994 per lung). The survival times were significantly different (P < 0.00013) between all groups of mice, except those infected with isolates B and E.
FIG. 7.
Survival curves for adult A/J mice infected intravenously with 5 × 105 C. neoformans organisms via the lateral tail vein. The average survival of mice infected with isolates D2, E, B, and A was 2.9 ± 0.9487, 8.2 ± 0.9487, 7.85 ± 1.0554, and 70 days, respectively (n = 10 per group). All mice infected with strain A were alive at the termination of the experiment at 70 days postinfection, and thus this data set contains censored values.
DISCUSSION
C. neoformans ATCC 24067 (serotype D) has been widely used for cryptococcal research (Table 1). This strain was first isolated by J. E. Bennett (49) from the cerebrospinal fluid of a patient with cryptococcal meningitis and was deposited at the American Type Culture Collection (ATCC) in 1972. After observing a dramatic attenuation of virulence in one isolate of ATCC 24067, we examined the genetic and phenotypic characteristics of nine isolates of strain ATCC 24067 from six different laboratories. We found major differences in virulence, growth rate, capsule size, extracellular proteinase production, melanization, and colony morphology for a set of isolates whose ancestry is traced to an ATCC strain. However, all the isolates investigated retained the typical serotype D GXM structure as determined by NMR spectroscopy analysis of the purified polysaccharides, and all were clearly related by RFLP and CHEF analyses. Phagocytic indices, agglutination titers, carbon assimilation patterns, and drug susceptibility patterns were also similar among the isolates included in this study.
The phenomenon of in vitro attenuation, similar to that described in the present study for isolate A, is a well known phenomenon for bacterial pathogens. For example, clinical isolates of Neisseria meningitidis are known to lose virulence under laboratory conditions (3). Moreover, changes in virulence after in vitro maintenance have also been described for the eukaryotic pathogens Leishmania donovani (34), Toxoplasma gondii (28), and Naegleria fowleri (50). For these organisms, loss of virulence is presumably a result of adaptation to the in vitro culture conditions. In the case of N. fowleri, it has been suggested that the physical characteristics and possible nutritional deficiencies associated with in vitro culture methods resulted in the selection of variants with decreased virulence (50). Attenuation in vitro has been reported for another pathogenic fungus, Paracoccidioides brasiliensis, after continuous subcultivation in the laboratory (22).
The ATCC 24067 isolate that became attenuated had been kept in liquid broth at room temperature for an undetermined time. This is not a standard method to maintain strains, since most laboratories (including our own) keep their isolates at either 4 or −70°C. However, it is noteworthy that significant differences were apparent in our study even among isolates maintained in standard conditions. Storage of C. neoformans cultures frozen or in the lyophilized state is likely to best preserve the original characteristics of the strain, but this is not necessarily a guarantee of stability of all phenotypic markers. Lyophilizing or freezing and thawing may conceivably select for variants which are more resistant to drying and/or low temperatures. Freezing and thawing can kill the majority of cells in a culture (41). A review of papers dealing with C. neoformans published in Infection and Immunity in the past 3 years revealed 40 articles from 17 cryptococcal research groups. Eighteen of these (45%) do not state the storage conditions for the C. neoformans isolates; nine (22.5%) state that the isolates are kept in artificial media, such as Sabouraud dextrose agar, yeast extract-peptone-dextrose, or brain heart glucose agar, but do not mention the temperature of storage; five (12.5%) state that the isolates are kept in Sabouraud dextrose agar slants at 4°C; four (10%) report maintenance of the isolates in distilled water or saline; and another four (10%) state that the isolates are kept by serial passages in Sabouraud dextrose agar. Hence, there is considerable interlaboratory variability in the maintenance of C. neoformans strains.
Our results demonstrate large phenotypic differences among ATCC 24067 isolates kept in different laboratory environments, despite being maintained under laboratory conditions for only a few years. Genetic differences were found for two isolates in the form of karyotype changes. Chromosome rearrangements have been documented for isolates passaged in mice (16), but this appears to be the first report of chromosome changes in vitro. These findings imply that C. neoformans has the ability to undergo rapid changes in vitro. The mechanisms responsible for these rapid changes are not understood. It is possible that conditions associated with in vitro maintenance (e.g., low temperatures, decreased nutrient availability with time, and artificial media, etc.) select for variants that are different from the original population. Plasticity in strain characteristics may reflect an inherent ability of C. neoformans to adapt to environmental changes. This process, if it occurs during infection, could promote persistence of infection by the emergence of variants that are able to escape host immune mechanisms.
The less virulent ATCC 24067-A isolate had a smaller capsule size, grew more slowly, and produced melanin at a lower rate than the highly virulent isolate D2. For C. neoformans, the growth rate, the presence of a capsule, and the ability to make melanin have all been associated with virulence (27). Hence, it appears that the emergence of a low-virulence phenotype for 24067-A was accompanied by multiple changes in phenotypes that have been linked to virulence. Isolate G was similar to isolate A with regard to melanin production and capsule size but differed in its ability to produce extracellular proteinases. Since isolate 24067-G had no proteolytic activity in azoalbumin plates and also lacked a protein of approximately 29 to 36 kDa, we tentatively assign the C. neoformans extracellular proteinase activity to this protein. The finding of multiple phenotypic variations in isolates that differ in virulence is consistent with the recent proposal by Perfect (36) for regulation of virulence traits by coordinate changes in genes.
In summary, we have described the occurrence of phenotypic and genotypic variation with time among isolates of a commonly studied C. neoformans ATCC strain and have provided a detailed description of this strain. Our results indicate that strain ATCC 24067 can change in different laboratory environments over time. This observation raises the possibility that inconsistencies in experimental results within and between laboratories involved in cryptococcal research could be the result of microevolution of C. neoformans strains. We propose that the maintenance of strains be limited to frozen or lyophilized states with the caveat that neither storage technique has been rigorously proven to ensure strain stability. Additional research into storage, maintenance, and upkeep of C. neoformans strains is needed.
ACKNOWLEDGMENTS
A.C. is supported by a Burroughs Wellcome Fund Development Therapeutics Award and NIH grants AI22774 and AI13342. R.C. is supported by NIH grant AI31769. J.S.H. is supported by grants from CNPq, FAPEMIG, and PRPq-UFMG (Brazil).
We are grateful to all the laboratories that supplied samples of ATCC 24067 for this study.
REFERENCES
- 1.Block E R, Jennings A E, Bennett J E. 5-Fluorocytosine resistance in Cryptococcus neoformans. Antimicrob Agents and Chemother. 1973;3:649–656. doi: 10.1128/aac.3.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt M E, Pfaller M A, Hajjeh R A, Graviss E A, Ress J, Spitzer E D, Pinner R W, Mayer L W the Cryptococcal Disease Active Surveillance Group. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated cryptococcosis. J Infect Dis. 1996;174:812–820. doi: 10.1093/infdis/174.4.812. [DOI] [PubMed] [Google Scholar]
- 3.Branham S E. The meningococcus (Neisseria intracellularis) Bacteriol Rev. 1940;17:175–188. doi: 10.1128/br.4.2.59-96.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall A, Freundlich L F, Marsh L, Scharff M. Extensive allelic variation in Cryptococcus neoformans. J Clin Microbiol. 1992;30:1080–1084. doi: 10.1128/jcm.30.5.1080-1084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L-C, Blank E S, Casadevall A. Extracellular proteinase activity of Cryptococcus neoformans. Clin Diagn Lab Immunol. 1996;3:570–574. doi: 10.1128/cdli.3.5.570-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L-C, Pirofiski L, Casadevall A. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect Immun. 1997;65:2599–2605. doi: 10.1128/iai.65.7.2599-2605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherniak R, Morris L C, Belay T, Spitzer E D, Casadevall A. Variation in the structure of glucuronoxylomannan in isolates from patients with recurrent cryptococcal meningitis. Infect Immun. 1995;63:1899–1905. doi: 10.1128/iai.63.5.1899-1905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherniak R, Valafar H, Morris L C, Valafar F. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Cryptococcus neoformans chemotyping by quantitative analysis of 1H-NMR spectra of glucuronoxylomannans using a computer based neural network, abstr. F39. [Google Scholar]
- 9.Currie B, Sanati H, Ibrahim A S, Edwards J E, Jr, Casadevall A, Ghannoum M. Sterol compositions and susceptibilities to amphotericin B of environmental Cryptococcus neoformans isolates are changed by murine passage. Antimicrob Agents Chemother. 1995;39:1934–1937. doi: 10.1128/aac.39.9.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie B P, Freundlich L F, Casadevall A. Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. J Clin Microbiol. 1994;32:1188–1192. doi: 10.1128/jcm.32.5.1188-1192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis J L, Huffnagle G B, Chen G H, Warnock M L, Gyetko M, McDonald R, Scott P, Towes G, B. Experimental murine pulmonary cryptococcosis. Lab Invest. 1994;71:113–126. [PubMed] [Google Scholar]
- 12.Fan M, Currie B P, Gutell R R, Ragan M A, Casadevall A. The 16S-like, 5.8S, and 23S-like rRNAs of the two varieties of Cryptococcus neoformans: sequence, secondary structure, phylogenetic analysis, and restriction fragment polymorphisms. J Med Vet Mycol. 1994;32:163–180. doi: 10.1080/02681219480000231. [DOI] [PubMed] [Google Scholar]
- 13.Fan M, Chen L C, Ragan M A, Gutell R R, Warner J R, Currie B P, Casadevall A. The 5S rRNA and the rDNA intergenic spacer of the two varieties of Cryptococcus neoformans: sequence, structure, and phylogenetic implications. J Med Vet Mycol. 1995;33:215–221. doi: 10.1080/02681219580000451. [DOI] [PubMed] [Google Scholar]
- 14.Feldmesser M, Casadevall A. Effect of serum IgG1 to Cryptococcus neoformans glucuronoxylomannan on murine pulmonary infection. J Immunol. 1997;158:790–799. [PubMed] [Google Scholar]
- 15.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fries B C, Chen F, Currie B P, Casadevall A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman D L, Franzot S, Casadevall A. Abstracts of the 97th General Meeting of the American Society for Microbiology, 1997. Washington, D.C: American Society for Microbiology; 1997. Phenotypic switching in Cryptococcus neoformans, abstr. F44. [Google Scholar]
- 18.Hoag K A, Street N E, Huffnagle G B, Lipscomb M F. Early cytokine production in pulmonary Cryptococcus neoformans infections distinguishes susceptible and resistant mice. Am J Respir Cell Mol Biol. 1995;13:487–495. doi: 10.1165/ajrcmb.13.4.7546779. [DOI] [PubMed] [Google Scholar]
- 19.Huffnagle G B, Chen G-H, Curtis J L, McDonald R A, Strieter R M, Toews G B. Down-regulation of the afferent phase of T cell-mediated pulmonary inflammation and immunity by a high melanin-producing strain of Cryptococcus neoformans. J Immunol. 1995;155:3507–3516. [PubMed] [Google Scholar]
- 20.Huffnagle G B, Yates J L, Lipscomb M F. T-cell-mediated immunity in the lung: a Cryptococcus neoformans pulmonary infection model using SCID and athymic nude mice. Infect Immun. 1991;59:1423–1433. doi: 10.1128/iai.59.4.1423-1433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito W, Iba Y, Kurosawa Y. Effects of substitutions of closely related amino acids at the contact surface in an antigen-antibody complex on the thermodynamic parameters. J Biol Chem. 1993;268:16639–16047. [PubMed] [Google Scholar]
- 22.Kashino S S, Singer-Vermes L M, Calich V L, Burger E. Alterations in the pathogenecity of one Paracoccidioides brasiliensis isolate do not correlate with its in vitro growth. Mycopathologia. 1990;111:173–180. doi: 10.1007/BF02282801. [DOI] [PubMed] [Google Scholar]
- 23.Kozel T R. Dissociation of a hydrophobic surface from phagocytosis of encapsulated and non-encapsulated Cryptococcus neoformans. Infect Immun. 1983;39:1214–1219. doi: 10.1128/iai.39.3.1214-1219.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozel T R, Gotschlich E. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J Immunol. 1982;129:1675–1680. [PubMed] [Google Scholar]
- 25.Kozel T R, Highison B, Stratton C J. Localization on encapsulated Cryptococcus neoformans of serum components opsonic for phagocytosis by macrophages and neutrophils. Infect Immun. 1984;43:574–579. doi: 10.1128/iai.43.2.574-579.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozel T R, Pfrommer G S T, Redelman D. Activated neutrophils exhibit enhanced phagocytosis of Cryptococcus neoformans opsonized with normal human serum. Clin Exp Immunol. 1987;70:238–246. [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin D-B, Su K-E, Yu J-C. Studies on cryopreservation of Toxoplasma gondii and its antigenicity in mice. Am J Trop Med Hyg. 1995;53:392–396. doi: 10.4269/ajtmh.1995.53.392. [DOI] [PubMed] [Google Scholar]
- 29.Lovchik J A, Lyons C R, Lipscomb M F. A role for gamma interferon-induced nitric oxide in pulmonary clearance of Cryptococcus neoformans. Am J Respir Cell Mol Biol. 1995;13:116–124. doi: 10.1165/ajrcmb.13.1.7598935. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee J, Scharff M, Casadevall A. Protective murine monoclonal antibodies to Cryptococcus neoformans. Infect Immun. 1992;60:4534–4541. doi: 10.1128/iai.60.11.4534-4541.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee S, Lee S C, Casadevall A. Antibodies to Cryptococcus neoformans glucuronoxylomannan enhance antifungal activity of murine macrophages. Infect Immun. 1995;63:573–579. doi: 10.1128/iai.63.2.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing for yeasts: proposed standard M27-P. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 34.Nolan T J, Herman R. Effects of long-term in vitro cultivation on Leishmania donovani promastigotes. J Protozool. 1985;32:70–75. doi: 10.1111/j.1550-7408.1985.tb03015.x. [DOI] [PubMed] [Google Scholar]
- 35.Nosanchuk J D, Casadevall A. Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccahride, melanin, and monoclonal antibody binding. Infect Immun. 1997;65:1836–1841. doi: 10.1128/iai.65.5.1836-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perfect J R. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob Agents Chemother. 1996;40:1577–1583. doi: 10.1128/aac.40.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perfect J R, Magee B B, Magee P T. Separation of chromosomes of Cryptococcus neoformans by pulsed-field gel electrophoresis. Infect Immun. 1989;57:2624–2627. doi: 10.1128/iai.57.9.2624-2627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perfect J R, Hobbs M M, Granger D L, Durack D T. Cerebrospinal fluid macrophage response to experimental cryptococcal meningitis: relationship between in vivo and in vitro measurements of cytotoxicity. Infect Immun. 1988;56:849–854. doi: 10.1128/iai.56.4.849-854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perfect J R, Ketabchi N, Cox G M, Ingram C W, Beiser C. Karyotyping of C. neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polacheck I, Lebens G A. Electrophoretic karyotype of the pathogenic yeast Cryptococcus neoformans. J Gen Microbiol. 1989;134:1037–1041. doi: 10.1099/00221287-135-1-65. [DOI] [PubMed] [Google Scholar]
- 41.Rosas A L, Casadevall A. Melanization affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol Lett. 1997;153:265–272. doi: 10.1111/j.1574-6968.1997.tb12584.x. [DOI] [PubMed] [Google Scholar]
- 42.Skelton M A, Cherniak R, Poppe L, van Halbeek H. Structure of the de-O-acetylated glucuronoxylomannan from Cryptococcus neoformans serotype D, as determined by 2D NMR spectroscopy. Magn Reson Chem. 1991;29:786–793. [Google Scholar]
- 43.Spitzer E D, Spitzer S G. Use of a dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J Clin Microbiol. 1992;30:1094–1097. doi: 10.1128/jcm.30.5.1094-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan D, Haynes K, Moran G, Shanley D, Coleman D. Persistence, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J Clin Microbiol. 1996;34:1739–1744. doi: 10.1128/jcm.34.7.1739-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varma A, Kwon-Chung K J. Restriction fragment polymorphism in mitochondrial DNA of Cryptococcus neoformans. J Gen Microbiol. 1989;135:3353–3362. doi: 10.1099/00221287-135-12-3353. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Aisen P, Casadevall A. Melanin, melanin “ghosts,” and melanin composition in Cryptococcus neoformans. Infect Immun. 1996;64:2420–2424. doi: 10.1128/iai.64.7.2420-2424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickes B L, Moore D E, Kwon-Chung K J. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology. 1994;140:543–555. doi: 10.1099/00221287-140-3-543. [DOI] [PubMed] [Google Scholar]
- 49.Wilson D E, Bennett J E, Bailey J W. Serologic grouping of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1968;127:820–823. doi: 10.3181/00379727-127-32812. [DOI] [PubMed] [Google Scholar]
- 50.Wong M M, Karr S L, Chow C K. Changes in the virulence of Naegleria fowleri maintained in vitro. J Parasitol. 1977;5:872–877. [PubMed] [Google Scholar]