Significance
P. aeruginosa and S. aureus are two important human pathogens that often cause co-infections. Understanding their polymicrobial interactions is key to treating such infections. HQNO secreted by P. aeruginosa is known to antagonize the growth of S. aureus. However, we identified a role for sub-lethal levels of HQNO in inducing S. aureus STX production. This facilitates a cooperative behavior between the two pathogens, resulting in resistance to host ROS and increased bacterial burden during co-infections in vivo. Our findings contribute to the understanding of complex polymicrobial interactions and suggest a critical role for HQNO in controlling the balance between cooperative and competitive behaviors between P. aeruginosa and S. aureus.
Keywords: Staphyloxanthin, Pseudomonas aeruginosa, Staphylococcus aureus, polymicrobial, innate immunity
Abstract
Bacterial infections are often polymicrobial. Pseudomonas aeruginosa and Staphylococcus aureus cause chronic co-infections, which are more problematic than mono-species infections. Understanding the mechanisms of their interactions is crucial for treating co-infections. Staphyloxanthin (STX), a yellow pigment synthesized by the S. aureus crt operon, promotes S. aureus resistance to oxidative stress and neutrophil-mediated killing. We found that STX production by S. aureus, either as surface-grown macrocolonies or planktonic cultures, was elevated when exposed to the P. aeruginosa exoproduct, 2-heptyl-4-hydroxyquinoline N-oxide (HQNO). This was observed with both mucoid and non-mucoid P. aeruginosa strains. The induction phenotype was found in a majority of P. aeruginosa and S. aureus clinical isolates examined. When subjected to hydrogen peroxide or human neutrophils, P. aeruginosa survival was significantly higher when mixed with wild-type (WT) S. aureus, compared to P. aeruginosa alone or with an S. aureus crt mutant deficient in STX production. In a murine wound model, co-infection with WT S. aureus, but not the STX-deficient mutant, enhanced P. aeruginosa burden and disease compared to mono-infection. In conclusion, we identified a role for P. aeruginosa HQNO mediating polymicrobial interactions with S. aureus by inducing STX production, which consequently promotes resistance to the innate immune effectors H2O2 and neutrophils. These results further our understanding of how different bacterial species cooperatively cause co-infections.
Pseudomonas aeruginosa and Staphylococcus aureus are two common microorganisms colonizing cystic fibrosis (CF) airways and chronic wounds (1–4). Co-infection correlates with increased disease severity, compared to mono-infections caused by either species (4–6).
P. aeruginosa and S. aureus have an intriguingly complicated relationship and have been used as model organisms to investigate polymicrobial interactions. In vitro, P. aeruginosa outcompetes S. aureus. Many antagonistic mechanisms have been determined in P. aeruginosa (7–10). However, of relevance to this study is the P. aeruginosa quorum-sensing (QS) system PQS (Pseudomonas quinolone signal) which is crucial for antagonizing S. aureus (11). The PQS system is involved in the production of 2-heptyl-4-hydroxyquinoline n-oxide (HQNO) which inhibits respiration (12) and promotes small colony variant formation in S. aureus (13). This operon is responsible for synthesizing the HQNO precursor HHQ (14), which is converted to HQNO by the PqsL enzyme. In contrast to antagonism in vitro, both pathogens can co-exist in vivo. In fact, interaction with S. aureus can benefit P. aeruginosa by increased biofilm formation (15), host immune evasion (16), and antibiotic resistance (17, 18). One of the keys to understanding this polymicrobial relationship is the subtle balance between the competitive and cooperative behaviors of these two organisms.
Staphyloxanthin (STX) is a membrane-bound yellow pigment, synthesized by the crt operon in S. aureus (19–21) and widely produced among clinical and environmental isolates (22). Strains deficient in STX production appear as white colonies on solid media. By functioning as an antioxidant to resist oxidative stress and altering membrane fluidity to combat antimicrobial peptides (APs), STX mediates S. aureus resistance to host defense mechanisms (23, 24). Interestingly, a P. aeruginosa wound isolate was observed to induce STX production in a co-isolated white variant of S. aureus (25). This implies that STX mediates interactions between P. aeruginosa and S. aureus in vivo.
Given the importance of P. aeruginosa and S. aureus co-infections and the implication of STX in polymicrobial interactions, here we investigated the role of STX during P. aeruginosa and S. aureus co-infections. We found that STX production is induced by P. aeruginosa HQNO and affords cross-species protection against H2O2- and neutrophil-mediated killing.
Results
P. aeruginosa Exoproduct HQNO Induces STX Production in Both Surface- and Planktonic-grown S. aureus.
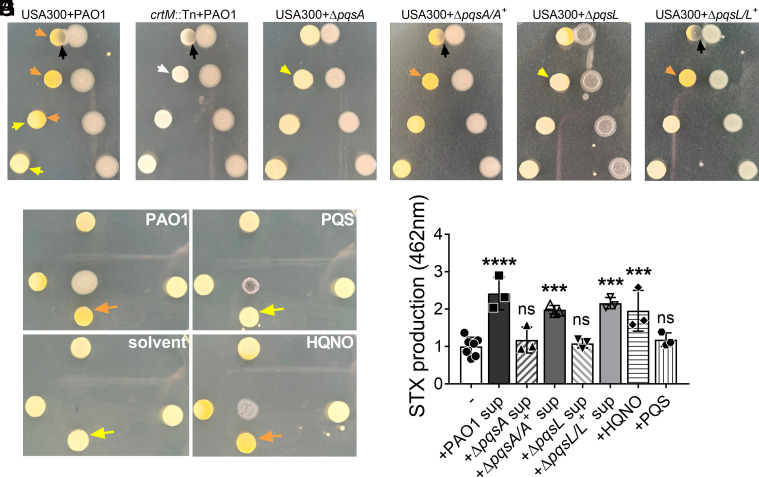
In our previous study, we examined the contribution of P. aeruginosa factors, especially released exopolysaccharide Psl (10), in antagonizing the growth of S. aureus in a macrocolony proximity assay. S. aureus USA300 was grown at increasing distances from P. aeruginosa PAO1 on solidified media (Fig. 1A). As expected, the growth of USA300 adjacent to PAO1 was inhibited. Interestingly, we also observed a difference in USA300 pigmentation. USA300 macrocolonies furthest from PAO1 were light yellow, whereas the ones closer to PAO1 were more pigmented, suggestive of elevated STX production. In contrast, macrocolonies of an S. aureus crtM transposon mutant (crtM::Tn, SAUSA300_2499) deficient in STX production, remained non-pigmented regardless of the distance from PAO1 macrocolonies (Fig. 1B). This suggests that an exoproduct released by PAO1 can diffuse through the solidified media to induce STX production in the nearby USA300.
Fig. 1.
P. aeruginosa HQNO induces S. aureus STX production. (A–G) USA300 or crtM::Tn was grown at increasing distances from designated P. aeruginosa strains, PQS, HQNO, or the solvent in which the molecules were dissolved, on solidified media in a macrocolony proximity assay. Yellow arrows point to USA300 with no pigment change, orange arrows point to USA300 with increased yellow pigmentation, and the white arrow indicates white crtM::Tn colonies. The black arrows point to S. aureus growth inhibition by P. aeruginosa. (H) STX production in LB-grown USA300 treated with (+) or without (−) 5% filter-sterilized spent media (sup) of designated P. aeruginosa strains, or 5 μM of HQNO or PQS. STX production was quantified by absorbance at OD462. The results were normalized to the untreated group. Data are presented as mean ± SD from the results of at least three biological replicates, each with two technical replicates. ***P < 0.001; ****P < 0.0001; ns, not significant, compared to the untreated group, determined by one-way ANOVA.
STX production is a stress response of S. aureus (26). Since there was increased STX production along with growth inhibition of USA300 when adjacent to PAO1, we investigated whether STX induction by P. aeruginosa was a response to general growth inhibition. USA300 macrocolonies were grown at increasing distances from filter disks soaked in ciprofloxacin or daptomycin antibiotics, or PAO1 (SI Appendix, Fig. S1). STX induction was only observed for USA300 macrocolonies grown in proximity to PAO1, but not the antibiotics, despite similar levels of growth inhibition. This implies that S. aureus STX is induced in response to released P. aeruginosa factor(s) and not a general response to growth inhibition.
P. aeruginosa can secrete many antagonistic factors, including Psl (10), rhamnolipid (9), HQNO (13), LasA (7), and pyoverdine (8), to inhibit the growth of S. aureus. We used the corresponding mutants in PAO1 to determine whether any of these mechanisms were responsible for inducing S. aureus STX production. When USA300 and these PAO1 mutants were grown together in the macrocolony proximity assay, ΔpqsA was the only mutant that failed to antagonize S. aureus and not induce STX production (Fig. 1C), while others still antagonized the growth of USA300 and induced STX production (SI Appendix, Fig. S2). Chromosomal complementation of ΔpqsA (ΔpqsA/A+) was able to revert the phenotype to the level of PAO1 (Fig. 1D). pqsA is one of the P. aeruginosa PQS biosynthetic operon enzymes responsible for the synthesis of HHQ, the precursor of HQNO (14, 27, 28). To investigate whether either or both products were sufficient for STX induction, USA300 macrocolonies were grown at increasing distances from commercially acquired HQNO and PQS, the solvent used to dissolve the chemicals, or PAO1 (Fig. 1D). Increased STX production of USA300 macrocolonies was only observed when grown adjacent to HQNO and PAO1, but not PQS or the solvent. pqsL is directly responsible for HQNO synthesis (29). Interestingly, ΔpqsL poorly induced STX production in adjacently grown USA300 (Fig. 1E). Chromosomal complementation (ΔpqsL/L+) fully reverted the phenotype comparable to PAO1 (Fig. 1F). The above data suggest that the P. aeruginosa PQS system, particularly the synthesis of HQNO, is responsible for inducing STX production in surface- grown S. aureus.
Since exoproducts are secreted into the spent media during bacterial planktonic growth, we also examined whether S. aureus STX could be induced by P. aeruginosa exoproducts when grown in planktonic culture. 50% PAO1 spent media demonstrated a strong bactericidal effect on S. aureus within 4 h of treatment (10). High concentrations of HQNO (e.g., 400 μM) also inhibit the growth of S. aureus (30). To eliminate complications due to potential growth inhibition, USA300 planktonic cultures were grown in LB supplemented with 5% PAO1 spent media, 5 μM of HQNO, or 5 μM of PQS for 16 h. STX was then extracted and quantified as described (19, 20). The presence of PAO1 spent media and HQNO, but not ΔpqsA and ΔpqsL spent media or PQS, induced S. aureus STX production (Fig. 1H).
Synthetic CF sputum media (SCFM2) mimics the CF sputum composition and has been used to culture both P. aeruginosa and S. aureus (31, 32). USA300 was also grown planktonically in SCFM2 supplemented with either PAO1 spent media or HQNO. In both conditions, STX production was significantly higher, compared to USA300 grown in SCFM2 alone, implying that HQNO can induce STX production under in vivo-like conditions (SI Appendix, Fig. S3). Overall, the above data indicate that P. aeruginosa HQNO is sufficient to induce S. aureus STX production, regardless of the mode of growth.
P. aeruginosa Induction of S. aureus STX Production Is Prevalent among Clinical Isolates.
STX production varies across S. aureus laboratory strains and clinical isolates (33, 34). We screened a collection of 61 S. aureus clinical isolates, from CF lung and bloodstream infections (SI Appendix, Table S1), and found that the majority of them (78.7%) had increased yellow pigmentation when grown in proximity to PAO1, suggesting STX induction (SI Appendix, Fig. S4A and Table S2). A similar phenotype was observed with methicillin-sensitive S. aureus (MSSA; SI Appendix, Fig. S5). P. aeruginosa clinical isolates synthesize varying levels of HQNO (35, 36). We also screened 29 P. aeruginosa clinical isolates, derived from CF lung and wound infections (SI Appendix, Table S1). 72.4% of them induced STX production in USA300, with the representative strains producing HQNO varying from 13.6 μM to 119.6 μM. The remaining isolates did not induce STX production, and the representative strains produced no detectable HQNO (SI Appendix, Fig. S4 B and C and Table S3). Overall, P. aeruginosa induction of S. aureus STX production was observed in the majority of the examined clinical isolates.
Mucoid conversion, defined by the overproduction of the exopolysaccharide alginate, occurs frequently in P. aeruginosa clinical CF strains (37). Mucoid P. aeruginosa can co-exist with S. aureus better than non-mucoid counterparts, due to reduced production of antagonistic factors, including HQNO (32). Since some of the P. aeruginosa clinical isolates tested above were mucoid and induced USA300 STX production (SI Appendix, Fig. S4B), we examined whether mucoid P. aeruginosa could induce STX production in an HQNO-dependent manner. To test this, we used a laboratory P. aeruginosa mucoid strain (PAO1 mucA22; PDO300) and created a ΔpqsA allele in this background. In the macrocolony proximity assay, PDO300 induced STX production in the adjacent USA300 macrocolonies, although without growth inhibition (SI Appendix, Fig. S4D). However, for the PDO300ΔpqsA mutant, STX induction was abolished. USA300 was also grown planktonically in media supplemented with spent media from either PDO300, PDO300ΔpqsA, or PAO1ΔpqsA, followed by STX extraction and quantification. When grown in the presence of PDO300 spent media, USA300 produced significantly more STX, compared to growth in media alone (SI Appendix, Fig. S4E). Neither PAO1ΔpqsA nor PDO300ΔpqsA spent media was able to induce STX production. This suggests that mucoid P. aeruginosa also induces S. aureus STX production in an HQNO-dependent manner.
P. aeruginosa–Induced STX Production Protects Both S. aureus and P. aeruginosa from H2O2-mediated Killing.
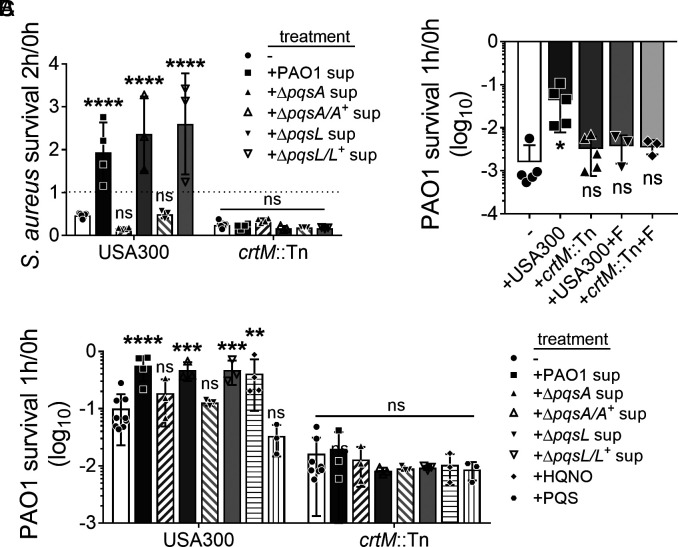
STX provides S. aureus resistance to oxidative stress, as a crtM mutant deficient in STX production was more sensitive to H2O2-mediated killing than WT S. aureus (23). However, the survival of S. aureus to oxidative stress upon STX induction has not been described. We therefore wanted to determine whether P. aeruginosa–induced STX production could be beneficial for S. aureus survival in the presence of H2O2. As previously described, USA300 and crtM::Tn were grown overnight in media supplemented with PAO1, ΔpqsA, or ΔpqsL spent media to induce STX production. The cultures were then subjected to 3% H2O2–mediated killing for up to 2 h. S. aureus survival was quantified by colony-forming units (CFUs) at designated time points, normalized to that of 0h. At 1h of H2O2 treatment, little difference in survival was observed for USA300 or crtM::Tn (SI Appendix, Fig. S6A). At 2 h, the survival of H2O2-treated USA300 and crtM::Tn, grown without P. aeruginosa spent media, reduced to 47% and 25%, respectively (Fig. 2A). Prior growth in media supplemented with PAO1, ΔpqsA/A+, or ΔpqsL/L+ spent media, but not ΔpqsA or ΔpqsL, significantly increased USA300 survival by >fourfold, compared to USA300 grown in media alone (Fig. 2A). There was no significant difference in crtM::Tn survival under these conditions. The above data indicate that P. aeruginosa–induced STX production can further protect S. aureus from H2O2-mediated killing.
Fig. 2.
STX induction protects both S. aureus and P. aeruginosa from H2O2-mediated killing. (A) S. aureus USA300 and crtM::Tn were pre-treated with or without 5% filter-sterilized spent media (sup) from designated P. aeruginosa strains overnight and then subjected to 3% H2O2-mediated killing for 2 h. The dotted line indicates 100% survival. (B) PAO1, alone (−) or mixed with an equal amount of S. aureus, was subjected to 3% H2O2-mediated killing for 1 h. USA300 and crtM::Tn were pre-treated with 50 μg/mL flavone (+F) to inhibit STX production and serve as controls. (C) PAO1 mixed with an equal amount of S. aureus with various treatments was subjected to 3% H2O2-mediated killing for 1h in LB. USA300 and crtM::Tn were pre-treated with or without (−) 5% filter-sterilized P. aeruginosa spent media (sup), or 5 μM HQNO or PQS overnight. Bacterial survival is presented as CFUs normalized to the starting CFUs at 0 h. Data are presented as mean ± SD from the results of at least three biological replicates, each with three technical replicates. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant, compared to the P. aeruginosa alone group (B) or the no treatment group (A and C). Statistical differences are determined by two-way ANOVA (A and C) or one-way ANOVA (B).
STX is a potent antioxidant and scavenges free radicals via conjugated double bonds (23, 38). We therefore hypothesized that in co-culture, STX may benefit P. aeruginosa by protecting it from H2O2-mediated killing. PAO1 alone, or mixed with an equal amount of S. aureus, was treated with 3% H2O2 for 1 h (Fig. 2B). PAO1 survival increased >10-fold when mixed with USA300, compared to PAO1 alone. However, co-culture with crtM::Tn did not significantly change PAO1 survival in the presence of H2O2. We then wanted to confirm that the protection conferred to P. aeruginosa from H2O2-mediated killing by USA300 was due to STX production, rather than other potential functions of crtM. To test this, USA300 and crtM::Tn were grown overnight in media supplemented with flavone, a plant flavonoid that inhibits S. aureus STX production without affecting growth (39). The S. aureus cultures were then mixed with PAO1 and treated as above, and bacterial survival quantified (Fig. 2B). Flavone completely abolished the protection that USA300 conferred on PAO1 against H2O2. No difference in the survival of PAO1 was observed when mixed with crtM::Tn grown with or without flavone. As expected, with 1 h treatment conditions, survival for both USA300 and crtM::Tn was not affected (SI Appendix, Fig. S6B), suggesting that the difference in PAO1 survival is attributed to STX, rather than the amount of S. aureus present. Together, the above data suggest that USA300 protects PAO1 from H2O2-mediated killing, and this protection is STX-dependent.
Since HQNO induces S. aureus STX production, we further tested if S. aureus with induced STX production affords better protection to PAO1 from H2O2. USA300 STX production was induced when grown in LB supplemented with PAO1 spent media or HQNO (Fig. 1H). When mixed with the above cultures, the survival of PAO1 challenged with H2O2 was increased >fivefold, compared to PAO1 mixed with USA300 grown in LB alone (Fig. 2C). Supplementing with ΔpqsA or ΔpqsL spent media or PQS during growth did not increase the ability of USA300 to protect PAO1. This was reversed upon complementation of the deleted genes with corresponding wild-type alleles. No significant difference was found in the survival of PAO1 when mixed with crtM::Tn grown with or without any P. aeruginosa exoproducts. The survival of USA300 and crtM::Tn remained unchanged for all conditions (SI Appendix, Fig. S6C). The above data indicate that induced STX production in S. aureus affords better protection to PAO1 from H2O2-mediated killing.
In addition to growth in LB, the presence of STX-producing USA300 also protected PAO1 from H2O2-mediated killing in SCFM2. Moreover, USA300 with increased STX production, induced by PAO1 spent media or HQNO, afforded better protection to PAO1 in SCFM2 compared to USA300 grown in only LB (SI Appendix, Figs. S6D and S7). The above data suggest that S. aureus STX also protects P. aeruginosa from H2O2 killing in environments mimicking the CF airway.
We also examined whether STX can protect mucoid PDO300 from H2O2 killing (SI Appendix, Fig. S8). Compared to PDO300 alone, the presence of USA300 increased the survival of PDO300 by 10-fold. PDO300 survival was further increased when the mixed USA300 had induced STX production due to growth in PDO300 spent media. This was not observed with PDO300ΔpqsA or PAO1ΔpqsA spent media. The above data indicate that STX protection from H2O2-mediated killing extends to P. aeruginosa mucoid strains.
STX Protects P. aeruginosa from Killing by Human Neutrophils.
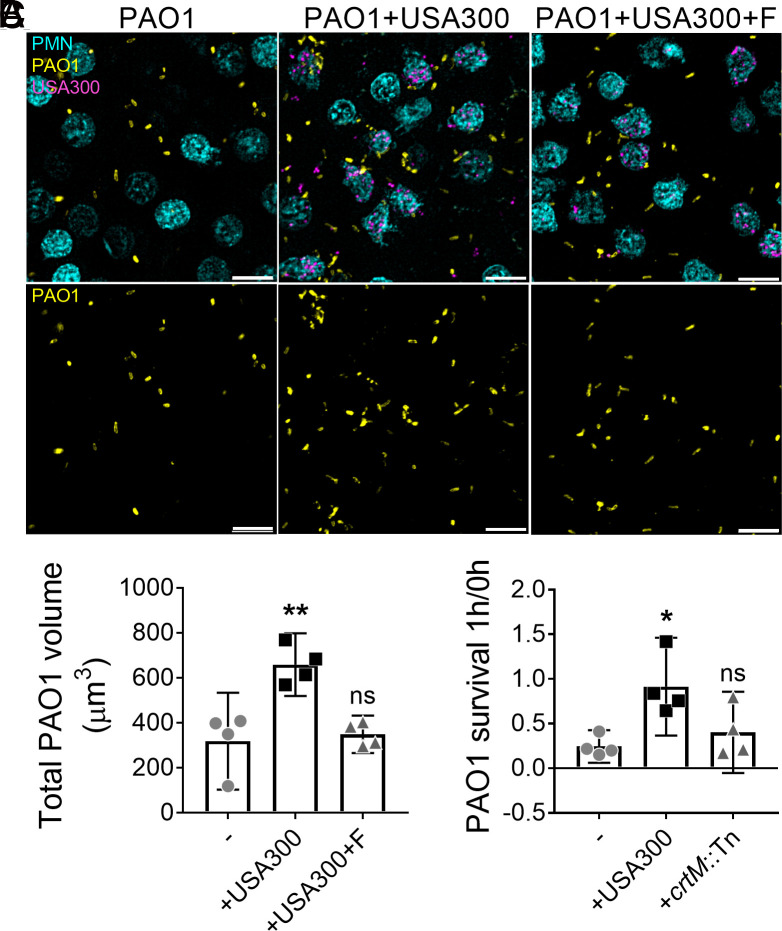
Neutrophils can combat pathogens by generating both intra- and extra-cellular reactive oxygen species (ROS) (40). STX mediates S. aureus resistance to neutrophil killing by serving as an antioxidant (23, 38). In the experiments outlined above, we demonstrated that STX protected P. aeruginosa from oxidative stress generated by H2O2 (Fig. 2 B and C). We wanted to determine if S. aureus STX could also protect P. aeruginosa from killing by human neutrophils. To test this, human peripheral blood–derived neutrophils were infected with fluorescently tagged USA300 and PAO1. To inhibit STX production, USA300 was grown in the presence of flavone as described (Fig. 2B). After 1 h incubation, the infection was visualized by wide-field fluorescent microscopy (Fig. 3A). Imaging analysis was performed to quantify the volume of PAO1 fluorescent signal, as an indication of PAO1 survival. This revealed that PAO1 survival was higher in the co-infection with USA300, compared to PAO1 mono-infection (Fig. 3B). PAO1 survival in the presence of flavone-treated USA300 was equivalent to that of PAO1 mono-infection (Fig. 3B). To support these observations, human peripheral blood–derived neutrophils were infected with PAO1 alone, or with USA300 or crtM::Tn for 1 h, and bacterial survival quantified by CFU (Fig. 3C). A significant increase was found in the survival of PAO1 when co-infected with USA300, but not with crtM::Tn, compared to PAO1 alone. S. aureus survival remained unchanged in mono-infections or co-infections with PAO1 (SI Appendix, Fig. S9). The above data suggest that S. aureus STX protects P. aeruginosa from killing by human neutrophils.
Fig. 3.
STX can protect P. aeruginosa from killing by human neutrophils. (A and B) PAO1-TdTomato, either alone or mixed with an equal amount of USA300-GFP, was subjected to adhered human neutrophil (PMN) for 1 h to assess PAO1 survival (MOI = 10 for each species). USA300-GFP was pre-treated with 50 μg/mL flavone (+F) to inhibit STX production. (A) Representative images of PAO1 and USA300 infected neutrophils. (Scale bar: 40 μm.) (B) Total PAO1 volume was quantified by measuring fluorescence intensity. Data are presented as mean ± 95%CI from the results of four biological replicates, each with six technical replicates. (C) PAO1, either alone or mixed with an equal amount of USA300 or crtM::Tn, was subjected to human neutrophil killing for 1 h (MOI = 10 for each species). PAO1 survival is presented as CFUs normalized to the starting CFUs at 0 h. Data are presented as mean ± 95%CI from the results of four biological replicates, each with three technical replicates. *P < 0.05; **P < 0.01; ns, not significant, compared to PAO1 mono-infection determined by one-way ANOVA. S. aureus survival is quantified in SI Appendix, Fig. S9.
STX Enhances P. aeruginosa Infection In Vivo.
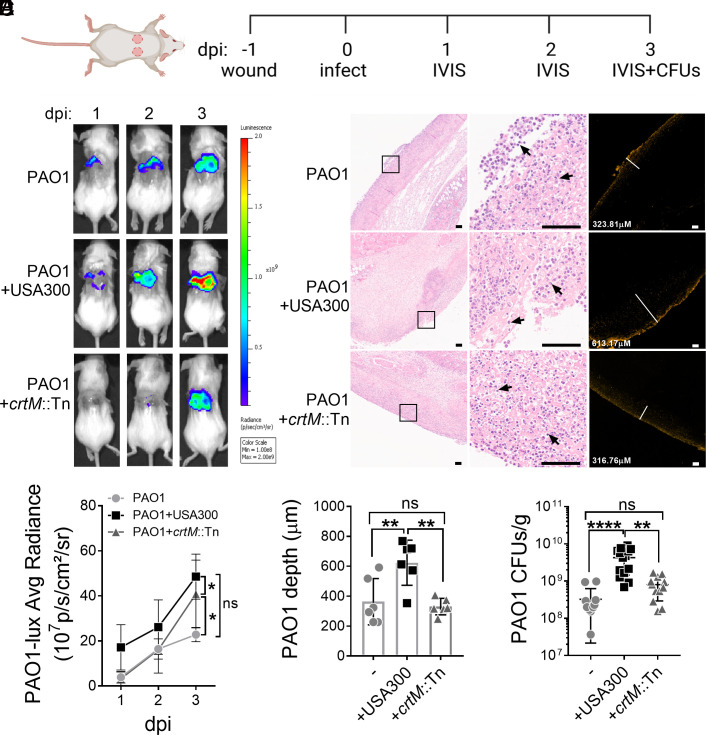
Our findings so far have demonstrated that STX protects P. aeruginosa from killing by H2O2 and neutrophils (Figs. 2 and 3). Since neutrophils are one of the first innate immune cells recruited to the infection site (41), we hypothesized that STX could also protect PAO1 during the early stages of infection. To test this, a dermal full-thickness murine wound model was used to examine P. aeruginosa and S. aureus co-infection (42). Briefly, two dorsal wounds were generated by punch biopsies and infected with either a luminescent tagged PAO1 strain, USA300, crtM::Tn, or both species. The infection was allowed to progress for 3 d to focus on early bacterial infection and innate immunity (Fig. 4A). By using an in vivo imaging system (IVIS), we monitored the burden of the bioluminescent PAO1 daily by measuring signal intensity. Throughout the 3-d infection period, PAO1 burden was significantly higher in mice co-infected with USA300, than those only infected with PAO1 or co-infected with crtM::Tn (Fig. 4 B and D and SI Appendix, Fig. S10A). No significant difference was found between PAO1 mono-infection and crtM::Tn co-infection. The above data suggest that S. aureus STX increases the PAO1 burden throughout infection.
Fig. 4.
STX promotes the establishment of P. aeruginosa infection in vivo. (A) Schematic of the murine wound model and course of infection. Two identical full-thickness dorsal wounds were generated using 6 mm punch biopsies. After 24 h, the wounds were mono-infected with luminescent PAO1, USA300, or crtM::Tn, or co-infected with PAO1 and USA300 or crtM::Tn. On 1, 2, and 3 days post-infection (dpi), IVIS was used to monitor PAO1 burden among all groups. Wounds were harvested and homogenized to plate for both PAO1 and S. aureus CFUs on 3 dpi. (B) Representative images of luminescent PAO1 detected using IVIS on murine wounds throughout the 3 d of infection. (C) Representative images of H&E (Left and Middle panels) and IF (Right panel) stained adjacent wound sections (4 μm). The Left panel shows the wound beds in low magnifications. Magnified boxed areas are shown in the Middle panel, with black arrows pointing to neutrophil infiltration. The Right panel shows the presence of immunofluorescently labeled PAO1 and the white lines measure the depth of PAO1 penetration (labeled in the bottom left corner). (Scale bar: 100 μm.) (D) Luminescent signal intensity of PAO1-lux was quantified by the average radiance. Significant differences were determined by comparing the area under the curve (SI Appendix, Fig. S10A). Data presented as mean ± 95%CI from the results of >12 biological replicates. (E) The depth of PAO1 penetration into the wound with PAO1 mono-infection or co-infection with USA300 or crtM::Tn. Data are presented as mean ± 95%CI from the results of six biological replicates. (F) PAO1 CFUs per gram of wound tissue (CFUs/g) among all groups was quantified. Data are presented as mean ± 95%CI from the results of >12 biological replicates with three technical replicates. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant, determined by one-way ANOVA (D and E) or the Kruskal–Wallis test (F). S. aureus survival is quantified in SI Appendix, Fig. S10.
On day 3 post infection, wound tissues were harvested and processed for either hematoxylin and eosin (H&E) staining or immunofluorescence (IF) labeling of PAO1, to assess inflammation and PAO1 localization respectively. We observed infiltration of mixed inflammatory cells, which predominantly consisted of neutrophils, into the wound bed for all groups (Fig. 4 C, Left and Middle panels). IF staining showed that PAO1, when co-infected with USA300, penetrated significantly deeper into the tissue (623.8 ± 150.9 μm) compared to mono-infection (363.5 ± 154.8 μm) or co-infection with crtM::Tn (330.2 ± 54.81 μm) (Fig. 4 C and E). Total PAO1 fluorescent signal from immunofluorescent images was quantified as another indication of PAO1 burden and was higher when PAO1 was co-infected with USA300, but not crtM::Tn or PAO1 alone (SI Appendix, Fig. S10B). The above results were corroborated by enumerating bacteria by CFU, after 3 d of infection (Fig. 4F). When PAO1 was co-infected with USA300, but not crtM::Tn, there was a significant 10-fold increase in PAO1 burden, compared to PAO1 mono-infection. There was no significant difference in the bacterial burden between USA300 or crtM::Tn (SI Appendix, Fig. S10D). Overall, the above data indicate that STX-producing S. aureus promotes P. aeruginosa wound colonization during the early stages of infection, demonstrating a clear role for STX in enhancing P. aeruginosa infection in vivo.
Discussion
In this study, we identified a role for P. aeruginosa HQNO in inducing S. aureus STX production (Fig. 1). The well-studied influence of HQNO on S. aureus is that, at high concentrations (400 μM), it antagonizes growth by inhibiting respiration (30, 43, 44), and promotes the formation of S. aureus small colonies, slow-growing, and non-pigmented variants (13, 30, 44, 45). However, we observed increased STX production in the presence of sub-lethal HQNO concentrations (5 μM). It is likely that varying concentrations of HQNO may elicit different responses in S. aureus. Many factors contribute to the local concentration of HQNO. The spatial distribution of bacteria can alter local QS signal concentration (46). In Fig. 1A, USA300 growth was inhibited by the adjacent PAO1 with high local HQNO concentration. The antagonism disappeared and STX induction was observed as the HQNO concentration decreased, due to the increased distance between the two bacteria macrocolonies. As the distance between the two colonies further increased, HQNO concentration was likely too low to induce STX production. In addition, different P. aeruginosa laboratory and clinical strains can produce varying amounts of HQNO, ranging from 0 to 50 μM (35, 36). Mucoid P. aeruginosa can co-exist with S. aureus better than the non-mucoid counterparts (32). Exogenous alginate can mitigate the killing of S. aureus by down-regulating HQNO production and other antagonistic factors in both mucoid and non-mucoid strains (47). Consistent with this, we observed reduced USA300 antagonism and induction of STX in some mucoid clinical isolates and PDO300 in an HQNO-dependent manner (SI Appendix, Fig. S4 B–E). The above evidence suggests that mucoid P. aeruginosa can create a low-HQNO environment that promotes S. aureus niche compatibility. This allows for the induction of STX which is beneficial for both species. In addition to mucoid strains, some non-mucoid P. aeruginosa clinical isolates can also induce STX production without inhibiting the growth of adjacent USA300 (SI Appendix, Fig. S4B), possibly due to reduced HQNO production in these isolates. Indeed, the clinical isolates that we examined produced varying levels of HQNO, which positively correlated with their ability to induce STX (SI Appendix, Fig. S4 B and C).
Apart from spatial distribution and strain variations, nutrient availability and host factors may also alter HQNO concentration (46, 48, 49). Overall, we predict that different concentrations of HQNO may be responsible for controlling the balance between cooperative and competitive behaviors among P. aeruginosa and S. aureus. Interestingly, Ibberson et al. recently discovered that HQNO can mediate the spatial structure of P. aeruginosa and S. aureus in wound infections (50). Collectively, both of our findings support the critical role of HQNO in modulating P. aeruginosa and S. aureus interactions during co-infections.
The mechanism(s) of how HQNO induces STX remains unclear. It seems to correlate with, but is not dependent on growth inhibition, as most of the STX-inducing P. aeruginosa clinical isolates can inhibit S. aureus growth (SI Appendix, Fig. S4B). This implies that STX may be induced in response to HQNO-mediated antagonism. HQNO can interfere with the electron transfer system in bacteria and mitochondria, which results in the production of ROS (12, 51–53). It is possible that S. aureus produces more STX to counteract HQNO-generated ROS. However, results from our experiments suggest otherwise. PAO1-mediated growth inhibition of USA300 and crtM::Tn were comparable in both surface-grown colonies (Fig. 1A) and planktonic co-cultures (SI Appendix, Fig. S11). Moreover, no STX induction was found in USA300 when treated with a sublethal concentration of H2O2, or ciprofloxacin which results in ROS production (54) (SI Appendix, Figs. S1A and S12). In addition, we examined the specificity of HQNO-induced STX production. Burkholderia cenocepacia produces 4-hydroxy-3-methyl-2-alkyquinolines which are structurally similar to HQNO (55). Neither of the two B. cenocepacia strains that we tested inhibited the growth of USA300, nor significantly induced STX production (SI Appendix, Fig. S13). One of them, however, demonstrated a modest induction ability, though much lower than PAO1 (SI Appendix, Fig. S13 A and B, c2), warranting future investigations into the potential cooperative behaviors between S. aureus and B, cenocepacia. Overall, we speculate that the induction of STX by HQNO may be a result of S. aureus specifically sensing P. aeruginosa–derived HQNO signals. Interspecies signaling is a key factor contributing to polymicrobial interaction and spatial distribution. There is evidence that S. aureus may have membrane receptors for another P. aeruginosa QS molecule, acyl homoserine lactone (56, 57), but little is known about HQNO. Interestingly, STX is produced by some S. aureus clinical isolates but is not induced by the adjacent PAO1 (SI Appendix, Fig. S4A). This suggests that these isolates may be blind to sensing HQNO. By examining the clinical isolates’ genomes and using STX induction as an output to screen for HQNO-unresponsive variants, future studies will uncover the mechanism(s) of how S. aureus senses P. aeruginosa HQNO and elevates STX production.
One of the key findings of this study was that STX afforded cross-species protection to P. aeruginosa from H2O2- and neutrophil-mediated killing. Both S. aureus and P. aeruginosa can colonize the same niche in vivo, within several μm of each other (2, 50). Since the membrane-bound STX has the capability to scavenge free radicals (38), we hypothesize that this creates a low-ROS sink around S. aureus cells. This unique microenvironment may, in turn, confer benefits to neighboring P. aeruginosa (SI Appendix, Fig. S14). In accord with this, STX not only protected S. aureus but also P. aeruginosa from H2O2- and neutrophil-mediated killing (Figs. 2 and 3). In addition, co-infection of murine wounds with STX-producing USA300 promoted increased PAO1 burden than the STX-deficient crtM::Tn (Fig. 4), despite little difference observed in pathology (SI Appendix, Fig. S10C). We speculate that the in vivo fitness afforded to PAO1, by USA300, is due to the antioxidant nature of STX. However, we acknowledge that other host innate immune effectors, excluding neutrophil and H2O2, may play a role in STX-mediated protection. Furthermore, we cannot exclude the possibility that additional STX-independent factor(s) produced by USA300 may enhance PAO1 infection.
Apart from PAO1, we also quantified S. aureus survival. In accord with the findings by Liu et al., crtM::Tn survival, compared to USA300, was modestly lower when subjected to H2O2 killing for 2 h (Fig. 2A) and in the murine wound infection (SI Appendix, Fig. S10D). Since HQNO can induce STX production (Figs. 1 and 2), we also examined whether the presence of PAO1 was beneficial for USA300 in vivo. The survival of USA300 and crtM::Tn was reduced during co-infection with PAO1, compared to mono-infections (SI Appendix, Fig. S10D). However, the S. aureus burden remained high (107 ~ 108 CFU/g). This is consistent with previous findings that the in vivo wound environment can promote co-existence, despite an antagonistic relationship (3, 49, 50, 58). Interestingly, the ratio of crtM::Tn to that of USA300, during co-infection with PAO1, was lower compared to S. aureus mono-infections (SI Appendix, Fig. S10E). Given that crtM::Tn was not more sensitive to antagonism by PAO1 than USA300 (SI Appendix, Fig. S11), we speculate that this difference may be attributed to the relatively higher survival of USA300 than crtM::Tn when co-infected with PAO1. The presence of PAO1 may induce STX production in USA300 which promotes resistance to host ROS and better survival, but not in crtM::Tn. Unfortunately, we were unable to extract and directly quantify STX levels from the homogenized wounds, due to contamination by host debris.
In summary, we identified a role for P. aeruginosa HQNO in inducing S. aureus STX production which is prevalent among clinical isolates. We also identified a cooperative behavior between the two pathogens during co-infection, resulting in resistance to H2O2- and neutrophil-mediated killing and increased P. aeruginosa burden in vivo (SI Appendix, Fig. S14). Overall, our findings add another layer to the already complex interaction between P. aeruginosa and S. aureus in vivo and highlight the need for future investigation in treating polymicrobial infections.
Materials and Methods
Full and detailed materials and methods can be found in SI Appendix.
Bacterial Strains and Growth Conditions.
All bacterial strains and plasmids are listed in SI Appendix, Table S1. Gene deletion constructs were incorporated into the P. aeruginosa genome using homologous recombination (59). Chromosomal complementation of gene deletions in P. aeruginosa was performed as previously described (60). The presence of transposon insertion for S. aureus crtM::Tn was verified by PCR (61). All planktonic cultures were grown at 37 °C with 200-rpm shaking for 16 h. S. aureus planktonic culture was grown in either lysogeny broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl; LB) or SCFM2 (62). P. aeruginosa planktonic culture was grown in either LB with no salt (LBNS) or SCFM2. For macrocolony proximity assay, P. aeruginosa and S. aureus were grown on lysogeny agar (LB supplemented with 1.5% agar; LA).
Macrocolony Proximity Assay.
Macrocolonies were grown by inoculating 5 µL of overnight bacterial culture onto the surface of LA in two sets of experiments. First, P. aeruginosa or B. cenocepacia and S. aureus were spotted on opposite sides of the plate with distances of 0 cm, 1 cm, 2 cm, and 3 cm in between. Second, 5 µL of either PAO1 culture, 50 µM HQNO or PQS, the solvent (methanol and ethanol), or antibiotics (10 mg/mL daptomycin or 1 mg/mL ciprofloxacin) was spotted onto the center of the plate, and the USA300 macrocolonies were spotted 0.5 cm, 1 cm, 2 cm and 3 cm away from the center. The plate was incubated at 37 °C overnight, and the S. aureus colonies were examined for their survival and pigment production.
STX Production in Planktonic Culture.
P. aeruginosa overnight cultures were normalized to OD600 2.5, and filter sterilized to collect cell-free spent media. To induce STX production, early-stationary phase S. aureus cultures (OD600 = 1) were supplemented with either 5% or 20% (v/v) P. aeruginosa spent media, 5 µM of PQS or HQNO (Cayman Chemicals) or sublethal concentrations of H2O2. For PAO1 and its variants, 5% spent media (v/v) was added to S. aureus. Since PDO300 produces less HQNO (32), 20% spent media (v/v) of PDO300 and its variants was added to S. aureus. To inhibit STX production, S. aureus was grown in LB supplemented with 50 μg/mL flavone (Sigma-Aldrich) overnight.
STX Extraction.
Extraction and quantification of STX were carried out as previously described with modifications (19). In brief, overnight cultures of S. aureus were normalized to an OD600 of 3. After centrifugation, the pellet was resuspended in 250 µL of methanol and incubated at 55 °C for 3 min. The samples were centrifuged to collect the supernatant and measured for absorbance at OD462 using a plate reader (SpectraMax® i3x; Molecular Device).
H2O2-Mediated Killing Assay.
This assay was carried out as previously described (23) with modifications. Overnight cultures of P. aeruginosa and S. aureus were diluted to OD600 0.5 in fresh LB. They were either combined at a 1:1 ratio or separately subjected to 3% H2O2 (Spectrum Chemical) and incubated at 37 °C with 200 rpm shaking for up to 2 h. Aliquots were taken at every hour, treated with 2,000 U/mL catalase (Sigma-Aldrich), serially diluted, and plated on Difco™ Pseudomonas Isolation Agar (PIA) and BBL™ Mannitol Salt Agar (MSA) to enumerate for CFUs of P. aeruginosa and S. aureus, respectively. Bacterial survival at each time point was normalized to the CFUs at 0 h.
Neutrophil Isolation.
Informed written consent was obtained from all four healthy donors before the collection of peripheral blood for isolating primary human neutrophils. All procedures were approved by the Ohio State University Institutional Review Board (IRB-2009H0314). Neutrophils were isolated as previously described (63).
Neutrophil Killing Assay.
This assay was carried out as previously described with modifications (64). P. aeruginosa and S. aureus overnight cultures were normalized to an OD600 of 0.5 and opsonized with 20% human serum (CompTech) for 30 min at 37 °C. The two bacteria were then either mixed at a 1:1 ratio or separately incubated with neutrophils statically for 1 h at 37 °C (MOI = 10 for each bacterial species). The samples were centrifuged at 18,000 × g for 10 min to lyse the neutrophils and release internalized bacteria. The pellets were resuspended in HBSS, serially diluted, and plated on PIA and MSA to enumerate CFUs. Bacterial survival was normalized to the CFUs at 0 h.
For microscopy analysis, neutrophils were seeded on poly-l-lysine coated coverslips in HBSS supplemented with 100 μM CellTracker™ Blue (Invitrogen) for 30 min at 37 °C, 5% CO2. USA300 was grown overnight in LB supplemented with or without 50 μg/mL flavone. Attached neutrophils were infected with fluorescently tagged PAO1, USA300, or both species for 1 h at 37 °C, 5% CO2 (MOI = 10 for each bacterial species). Unattached cells were washed away with HBSS. Coverslips were fixed in 4% paraformaldehyde for 30 min at room temperature, mounted to slides using Prolong™ Gold antifade reagent (Invitrogen), and visualized using a Nikon Ti2 wide field microscope fitted with a 60× oil objective. Total volume of bacteria was quantified as described in SI Appendix.
Dermal Full-Thickness Murine Wound Infection.
This assay was carried out as previously described with modifications (42). Six-week-old female BALB/c mice were used in this experiment. For each mouse, two identical full-thickness dorsal wounds were generated with a 6-mm punch biopsy tool (Integra™ Miltex®) and bandaged with a Tegaderm dressing (3M). After 24 h, each wound was infected with mid-log bacterial cultures containing 5 × 106 cells of either PAO1 containing a constitutively expressed luminescent marker (65), USA300 or crtM::Tn, or both species. A total of seven animals were used for each group. To assess PAO1 burden throughout infection, the wound luminescence was imaged daily with an IVIS Lumina II optical imaging system (PerkinElmer Inc.). The average radiance of PAO1-lux on each animal was used to access the PAO1 burden throughout infection. Three days post infection, the wounded tissues were collected, homogenized, serially diluted, plated on PIA and MSA, and incubated at 37 °C overnight. CFUs were calculated per gram of tissue.
H&E and IF Staining and Pathology Analysis on the Wound Tissues.
Three days post infection, wounds were harvested, fixed in 4% paraformaldehyde for a week, transferred into 100% ethanol, and sent to HistoWiz. The tissues were embedded in paraffin, sectioned longitudinally (4 μm), and stained with H&E. As for the IF staining, the slides were deparaffinized, blocked with 3% bovine serum albumin, incubated with primary P. aeruginosa antibody (66) (1:500 dilution) and secondary antibody (Alexa FluorTM 647 chicken anti-rabbit IgG, Invitrogen; 1:500 dilution). They were visualized by microscopy (Nikon ECLIPSE Ti2) using a 4× objective. Six wounds were imaged for each group. The depth of PAO1 penetration into the wound and total pixel count were measured by NIS-elements AR software.
Statistical Analysis.
Statistical significance was determined using either ANOVA or the Kruskal–Wallis test after the Shapiro–Wilk test for normality. Analyses were performed using GraphPad Prism v.7 (GraphPad Software). Statistical significance was determined using a P-value < 0.05.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was supported by the NIH R01AI143916, R01AI077628, and R01AI134895 to D.J.W. and M.R.P., and a fellowship program for Advancing Research in Infection and Immunity to Y.L. This work was supported in part by the Cure CF Columbus Translational Core (C3TC). C3TC is supported by the Division of Pediatric Pulmonary Medicine, the Biopathology Center Core, and the Data Collaboration Team at Nationwide Children’s Hospital. Grant support was provided by The Ohio State University Center for Clinical and Translational Science (National Center for Advancing Translational Sciences, Grant UL1TR002733) and by the Cystic Fibrosis Foundation (Research Development Program, Grant MCCOY19RO). We thank Dr. Traci Wilgus for helping us with the paraffin removal of wound sections. We also thank the Mass Spectrometry and Proteomics Facility, supported by NIH Award P30 CA016058, at the Ohio State University for its help with HQNO quantification.
Author contributions
Y.L., E.S.G., and D.J.W. designed research; Y.L., E.A.M., and P.S.J.B.R. performed research; Y.L., E.S.G., and D.J.W. analyzed data; and Y.L., P.S.J.B.R., E.S.G., M.R.P., and D.J.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Surette M. G., The cystic fibrosis lung microbiome. Ann. Am. Thorac. Soc. 11, S61–S65 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Wakeman C. A., et al. , The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun. 7, 11951 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pastar I., et al. , Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8, e56846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serra R., et al. , Chronic wound infections: The role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert. Rev. Anti. Infect. Ther. 13, 605–613 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Limoli D. H., et al. , Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 35, 947–953 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Seth A. K., et al. , Quantitative comparison and analysis of species-specific wound biofilm virulence using an in vivo, rabbit-ear model. J. Am. Coll. Surg. 215, 388–399 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Kessler E., Safrin M., Olson J. C., Ohman D. E., Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268, 7503–7508 (1993). [PubMed] [Google Scholar]
- 8.Mashburn L. M., Jett A. M., Akins D. R., Whiteley M., Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187, 554–566 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bharali P., Saikia J. P., Ray A., Konwar B. K., Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: A novel chemotaxis and antibacterial agent. Colloids Surf. B Biointerfaces 103, 502–509 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Gloag E. S., Hill P. J., Parsek M. R., Wozniak D. J., Interbacterial antagonism mediated by a released polysaccharide. J. Bacteriol. 204, e0007622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reen F. J., et al. , The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 77, 413–428 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Lightbown J. W., Jackson F. L., Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem. J. 63, 130–137 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman L. R., et al. , Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 103, 19890–19895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deziel E., et al. , Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 101, 1339–1344 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves P. M., et al. , Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 76, fty003 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Armbruster C. R., et al. , Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tognon M., et al. , Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 11, 2233–2243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaudoin T., et al. , Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 3, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall J. H., Wilmoth G. J., Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J. Bacteriol. 147, 900–913 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieland B., et al. , Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4’-diaponeurosporene of Staphylococcus aureus. J. Bacteriol. 176, 7719–7726 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G. Y., Nizet V., Color me bad: Microbial pigments as virulence factors. Trends Microbiol. 17, 406–413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., et al. , Genetic and virulent difference between pigmented and non-pigmented Staphylococcus aureus. Front. Microbiol. 9, 598 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu G. Y., et al. , Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202, 209–215 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra N. N., et al. , Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob. Agents Chemother. 55, 526–531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonic V., Stojadinovic A., Zhang B., Izadjoo M. J., Alavi M., Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect. Drug Resist. 6, 175–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranganathan N., Johnson R., Edwards A. M., The general stress response of Staphylococcus aureus promotes tolerance of antibiotics and survival in whole human blood. Microbiol. (Reading) 166, 1088–1094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman J. P., et al. , Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J. Bacteriol. 190, 1247–1255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepine F., Milot S., Deziel E., He J., Rahme L. G., Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J. Am. Soc. Mass. Spectrom. 15, 862–869 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Drees S. L., et al. , PqsL uses reduced flavin to produce 2-hydroxylaminobenzoylacetate, a preferred PqsBC substrate in alkyl quinolone biosynthesis in Pseudomonas aeruginosa. J. Biol. Chem. 293, 9345–9357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell G., et al. , Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10, 33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barraza J. P., Whiteley M., A Pseudomonas aeruginosa antimicrobial affects the biogeography but not fitness of Staphylococcus aureus during coculture. mBio 12, e00047-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limoli D. H., et al. , Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8, e00186-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieneke M. K., et al. , Association of diverse Staphylococcus aureus populations with Pseudomonas aeruginosa coinfection and inflammation in cystic fibrosis airway infection. mSphere 6, e0035821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Land A. D., Hogan P., Fritz S., Levin P. A., Phenotypic variation is almost entirely independent of the host-pathogen relationship in clinical isolates of S. aureus. PLoS One 10, e0129670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fugere A., et al. , Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One 9, e86705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collalto D., et al. , In vitro activity of antivirulence drugs targeting the las or pqs quorum sensing against cystic fibrosis Pseudomonas aeruginosa isolates. Front. Microbiol. 13, 845231 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Govan J. R., Deretic V., Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539–574 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clauditz A., Resch A., Wieland K. P., Peschel A., Gotz F., Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 74, 4950–4953 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J. H., Park J. H., Cho M. H., Lee J., Flavone reduces the production of virulence factors, staphyloxanthin and alpha-hemolysin, in Staphylococcus aureus. Curr. Microbiol. 65, 726–732 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Nguyen G. T., Green E. R., Mecsas J., Neutrophils to the ROScue: Mechanisms of NADPH oxidase activation and bacterial resistance. Front. Cell Infect. Microbiol 7, 373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra F. E., Borgogna T. R., Patel D. M., Sward E. W., Voyich J. M., Epic immune battles of history: Neutrophils vs. Staphylococcus aureus. Front. Cell Infect. Microbiol. 7, 286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestrak M. J., et al. , Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob. Agents Chemother. 63, e00234-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machan Z. A., Taylor G. W., Pitt T. L., Cole P. J., Wilson R., 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30, 615–623 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Filkins L. M., et al. , Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J. Bacteriol. 197, 2252–2264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proctor R. A., et al. , Staphylococcus aureus Small Colony Variants (SCVs): A road map for the metabolic pathways involved in persistent infections. Front. Cell Infect. Microbiol. 4, 99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer K. L., Aye L. M., Whiteley M., Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price C. E., Brown D. G., Limoli D. H., Phelan V. V., O’Toole G. A., Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J. Bacteriol. 202, e00559-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao T., Sweedler J. V., Bohn P. W., Shrout J. D., Spatiotemporal distribution of Pseudomonas aeruginosa Alkyl quinolones under metabolic and competitive stress. mSphere 5, e00426-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith A. C., et al. , Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect. Immun. 85, e00116-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ibberson C. B., Barraza J. P., Holmes A. L., Cao P., Whiteley M., Precise spatial structure impacts antimicrobial susceptibility of S. aureus in polymicrobial wound infections. Proc. Natl. Acad. Sci. U.S.A. 119, e2212340119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Ark G., Berden J. A., Binding of HQNO to beef-heart sub-mitochondrial particles. Biochim. Biophys. Acta 459, 119–127 (1977). [DOI] [PubMed] [Google Scholar]
- 52.Hacker B., Barquera B., Crofts A. R., Gennis R. B., Characterization of mutations in the cytochrome b subunit of the bc1 complex of Rhodobacter sphaeroides that affect the quinone reductase site (Qc). Biochemistry 32, 4403–4410 (1993). [DOI] [PubMed] [Google Scholar]
- 53.Hazan R., et al. , Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol. 26, 195–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Becerra M. C., Albesa I., Oxidative stress induced by ciprofloxacin in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 297, 1003–1007 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Coulon P. M. L., Groleau M. C., Deziel E., Potential of the Burkholderia cepacia complex to produce 4-Hydroxy-3-Methyl-2-Alkyquinolines. Front. Cell Infect. Microbiol. 9, 33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Highlander S. K., et al. , Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7, 99 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qazi S., et al. , N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 74, 910–919 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalton T., et al. , An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6, e27317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi K. H., Schweizer H. P., An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5, 30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P., Integration-proficient plasmids for Pseudomonas aeruginosa: Site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Fey P. D., et al. , A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner K. H., Wessel A. K., Palmer G. C., Murray J. L., Whiteley M., Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 112, 4110–4115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nauseef W. M., Isolation of human neutrophils from venous blood. Methods Mol. Biol. 412, 15–20 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Mishra M., et al. , Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 14, 95–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lochab V., et al. , Ultrastructure imaging of Pseudomonas aeruginosa lawn biofilms and eradication of the tobramycin-resistant variants under in vitro electroceutical treatment. Sci. Rep. 10, 9879 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy S., et al. , Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J. Pathol. 233, 331–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.