Abstract
The human leukocyte antigen (HLA) locus is associated with more complex diseases than any other locus in the human genome. In many diseases, HLA explains more heritability than all other known loci combined. In silico HLA imputation methods enable rapid and accurate estimation of HLA alleles in the millions of individuals that are already genotyped on microarrays. HLA imputation has been used to define causal variation in autoimmune diseases, such as type I diabetes, and in human immunodeficiency virus infection control. However, there are few guidelines on performing HLA imputation, association testing, and fine mapping. Here, we present a comprehensive tutorial to impute HLA alleles from genotype data. We provide detailed guidance on performing standard quality control measures for input genotyping data and describe options to impute HLA alleles and amino acids either locally or using the web-based Michigan Imputation Server, which hosts a multi-ancestry HLA imputation reference panel. We also offer best practice recommendations to conduct association tests to define the alleles, amino acids, and haplotypes that affect human traits. Along with the pipeline, we provide a step-by-step online guide with scripts and available software (https://github.com/immunogenomics/HLA_analyses_tutorial). This tutorial will be broadly applicable to large-scale genotyping data and will contribute to defining the role of HLA in human diseases across global populations.
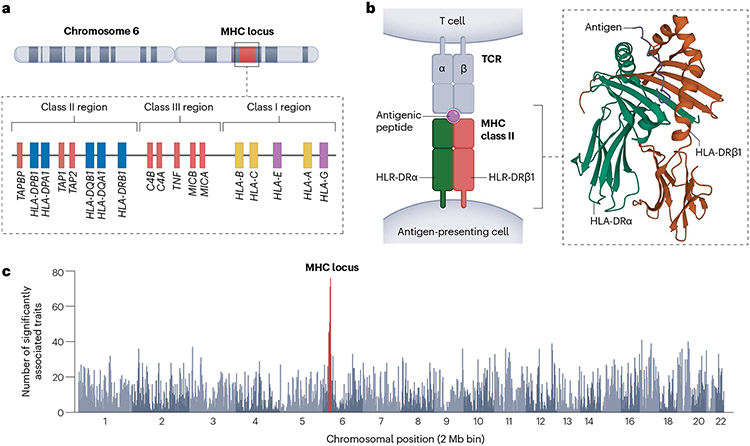
More than 50 years ago, some of the earliest complex human disease genetic associations were reported within the major histocompatibility complex (MHC) locus1,2. This locus has since been mapped to the short arm of chromosome 6. Sequencing of the human genome has revealed that the MHC locus consists of a cluster of more than 200 genes, including many with immune functions3. The MHC locus is broadly divided into three subclasses: the class I region (e.g., HLA-A, HLA-B and HLA-C genes), the class II region (e.g., HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DRA, HLA-DRB1, HLA-DRB2, HLA-DRB3, HLA-DRB4 and HLA-DRB5 genes) and the class III region, which contains additional genes implicated in immune and inflammatory responses (e.g., complement genes)4 (Fig. 1a). MHC class I and II genes encode proteins that form complexes that present antigenic peptides to T cells, thereby influencing thymic selection and T-cell activation4 (Fig. 1b). MHC class I molecules are expressed in nearly all nucleated somatic cells and present self- or cytosolic pathogens to CD8 T cells. The MHC class I molecule consists of an α-chain (encoded in the MHC class I region) and a β2-microglobulin chain (encoded on chromosome 15). MHC class I’s antigen-binding groove is closed at both ends, restricting the size of the presented peptides. By contrast, MHC class II molecules are expressed primarily on antigen presenting cells and present processed extracellular pathogens to CD4 T cells. The MHC class II molecule consists of α- and β-chains, both encoded within the MHC (e.g., HLA-DRA and HLA-DRB1). MHC class II’s antigen-binding groove is open ended and accommodates peptides of variable length5. The functional importance of the human leukocyte antigen (HLA) genes and the highly polymorphic nature of this locus have contributed to the MHC region having the largest number of disease associations of any locus, genome wide (Fig. 1c). Disease risk associated with the MHC is modulated by several underlying mechanisms. For example, in rheumatoid arthritis, polymorphisms in the amino acid sequence of HLA-DRB1 change the capability to present autoantigens6 or increase the number of autoreactive T cells during thymic selection7. In another example, the HLA-C*06:02 allele is associated with psoriasis, probably owing to an increase in CD8+ T-cell-mediated inflammatory reactions8. Schizophrenia’s association within the MHC locus was explained in part by structural variation in C4 (complement component 4; a critical component of the classical complement cascade, an immune pathway that recognizes and eliminates pathogens and cellular debris), which might modulate synaptic elimination during development9.
Fig.1 ∣. Location and structure of HLA genes on human chromosome 6 and their associations with human traits.
a, A schematic representation of the human MHC locus highlighting the three main classes of the region, and the genes within them. The classical class I HLA genes are shown in yellow, the classical class II HLA genes in blue, the nonclassical HLA genes in purple and the genes other than the HLA genes within the MHC region in red. b, Presentation of an antigenic peptide by an antigen-presenting cell to a T cell through interaction between an MHC class II molecule and a TCR. The inset shows the protein structure of the MHC class II complex composed of HLA-DRA and DRB1 bound to an antigenic peptide (PDB ID: 3L6F). c, The number of traits associated with any variants within a 2 Mb genomic window with P < 5 × 10−8 among the 198 diseases and biomarkers in the UK Biobank and FinnGen96. The MHC region is highlighted in red. The GWAS data for 198 diseases and biomarkers were obtained and analyzed as previously described96.
The HLA genes within the MHC have been difficult to study because of their highly polymorphic nature, their long history of pathogen-driven natural selection and the MHC’s unique long-range linkage disequilibrium (LD) structure. The long-range LD spans across the whole MHC region, and in particular, variants between the α- and β-chain genes of the HLA molecule are in tight LD (e.g., HLA-DRA and HLA-DRB1; Extended Data Fig. 1). The highly polymorphic nature of HLA genes renders traditional probe-based genotyping challenging. In addition, the genetic diversity at HLA genes is highly population-specific, necessitating efforts to accurately genotype HLA alleles and investigate phenotypic associations in global populations. Accurate HLA typing, especially for HLA-DR genes10 is also essential to ensure the safety and prognosis of organ transplantation.
These challenges have driven high interest in the genetics community to develop and deploy statistical techniques for HLA alleles. While the direct typing of HLA alleles (e.g., Sanger sequence-based typing (SBT) and sequence-specific oligonucleotide probe hybridization (SSOP)11-15) continues to be costly, labor-intensive and unscalable16,17, in silico HLA imputation has recently enabled rapid and accurate estimation of HLA alleles in individuals already genotyped on microarrays18-20. However, there are few guidelines for HLA imputation and fine-mapping; these methods are necessary to define HLA effects on human diseases, especially in biobank-scale data from multiple populations.
Here, we provide detailed guidelines for imputing HLA alleles and testing for their association with human diseases and traits in large-scale cohorts and global biobanks. We also provide a step-by-step online guide with scripts and available software (https://github.com/immunogenomics/HLA_analyses_tutorial). Definitions of key terms used throughout this article can be found in Table 1.
Table 1 ∣.
Key terms used in the tutorial
| Term | Definition |
|---|---|
| MHC region | The genomic region that harbors the MHC. In GRCh37, it corresponds to chr6:28,477,797-33,448,354 (6p22.1-21.3).In GRCh38, it corresponds to chr6:28,510,020-33,480,577 |
| Linkage disequilibrium | A nonrandom association or dependence of alleles at different loci in a given population, making the frequencies of the alleles deviate from the expected frequency if the alleles were independent |
| Imputation | A statistical method of estimating the missing genotypes at loci that are not assayed in the target dataset |
| Reference panel | A panel of densely genotyped haplotypes to be referred to when predicting the missing genotypes in the target cohort through imputation |
| Haplotype | A stretch of DNA sequences (including multiple polymorphic loci) along one chromosome that tend to be inherited together due to LD |
| Allele | One of two versions of a DNA sequences. An individual inherits two alleles (maternal and paternal) for any genomic location |
| HLA allele | One of the possible sequence variations at a given HLA gene |
| Genotype | An individual’s pattern of DNA sequence at a given location. Two alleles, one from the mother and one from the father, comprise a genotype |
| Haplotype phasing | Estimation of haplotypes from genotype data that usually do not provide phase information. Computational haplotype phasing can be done by statistical methods such as expectation maximization algorithm and hidden Markov models (HMM) |
| Fine-mapping | A procedure to narrow down and define potentially causal genetic variation(s) affecting the trait of interest, from all the associated genetic variations at a given locus in genome-wide association studies (GWAS) by using statistical methods |
| Homozygous | A state where the two alleles at the genetic variation of interest (e.g., an HLA gene) are the same |
| Heterozygous | A state where the two alleles at the genetic variation of interest (e.g., an HLA gene) are different |
| Association test | A statistical test to determine whether a given genotype frequency differ between two groups of individuals (e.g., cases and controls) or a genotype is correlated with a given quantitative phenotype |
| Allele divergence | A proxy for the functional difference in antigen binding between two HLA alleles based on the divergence of the amino acid sequences they encode |
Overview of the tutorial
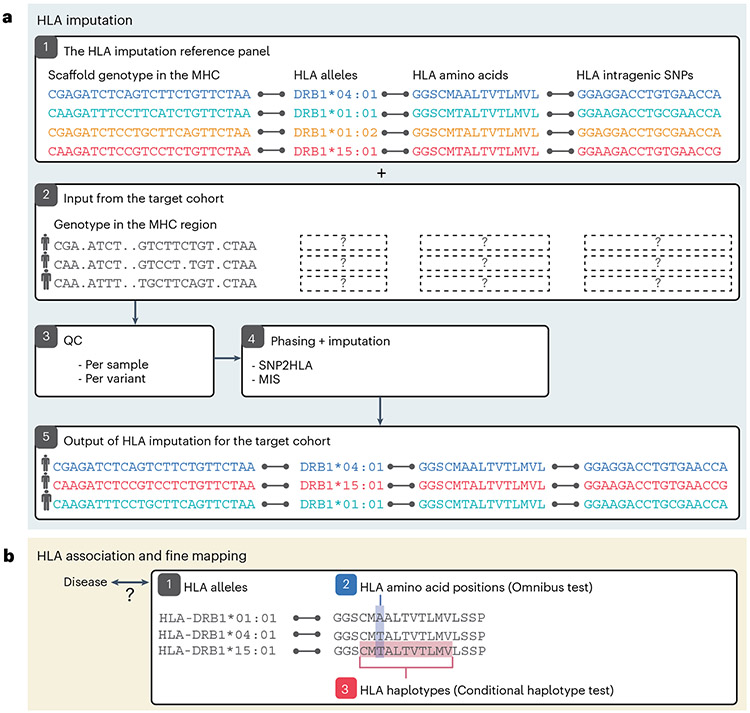
The workflow described in this tutorial is summarized in Fig. 2. It is composed of two sections: HLA imputation (Fig. 2a) and HLA association testing (Fig. 2b). HLA imputation is a method to infer HLA alleles, amino acids and single-nucleotide polymorphisms (SNPs) from a microarray-based genotype within the MHC region. We first introduce the concept of the HLA reference panel (1), which is used as a dictionary to search for similar haplotypes (keyword) to infer unknown HLA types (definition). We highlight specifically our multi-ancestry HLA reference panel, which we recently constructed to enable accurate HLA inference in diverse global populations21. We next provide specific instructions to perform quality control (QC) of the input genotype data (2), per individual and per variant (3). The quality of genotype data is critical for achieving accurate imputation, and special caution should be taken given the extremely complex and polymorphic nature of genetic variants within MHC. We then introduce options to phase and impute HLA (4), either (i) on a user’s local server or (ii) by using the Michigan Imputation Server (MIS)22, which is a publicly available, web-based imputation platform we jointly support with the University of Michigan. We finally describe the quality metrics and post-QC of the imputed variants (5).
Fig.2 ∣. Overview of HLA imputation, association and fine mapping.
a, A toy example illustrating the workflow for HLA imputation. The process begins with (1) either using an existing HLA imputation reference panel or creating a custom one, (2) collecting the input genotype in the MHC region from the target cohort without HLA types, (3) performing QC of the target genotype, (4) genotype phasing and imputation to predict the untyped HLA alleles in the target cohort (locally using the SNP2HLA software or online using the MIS), and results in (5) predicted HLA alleles in the target cohort. b, Statistical methods to investigate and fine-map association of (1) individual HLA alleles, (2) amino acid positions comprising multiple residues (highlighted in blue) and (3) their haplotypes (highlighted in red) with a trait of interest. See Fig. 3 for an overview of HLA allele nomenclature and structure.
We next describe statistical methods to perform comprehensive association tests between HLA genotype and human traits (Fig. 2b). Since HLA associations are often explained by amino acid sequences in the peptide binding groove of HLA molecules23, we describe strategies to fine-map associations with the aim of pinpointing causal variation. We start from a simple single-marker test (1), which is similar to that commonly used in genome-wide association studies (GWAS), and then elaborate on the HLA-specific fine-mapping methods (e.g., an omnibus test (2) and a conditional haplotype test (3)). We also introduce statistical tests to define nonadditive, interactive and multitrait contributions of HLA alleles.
HLA nomenclature
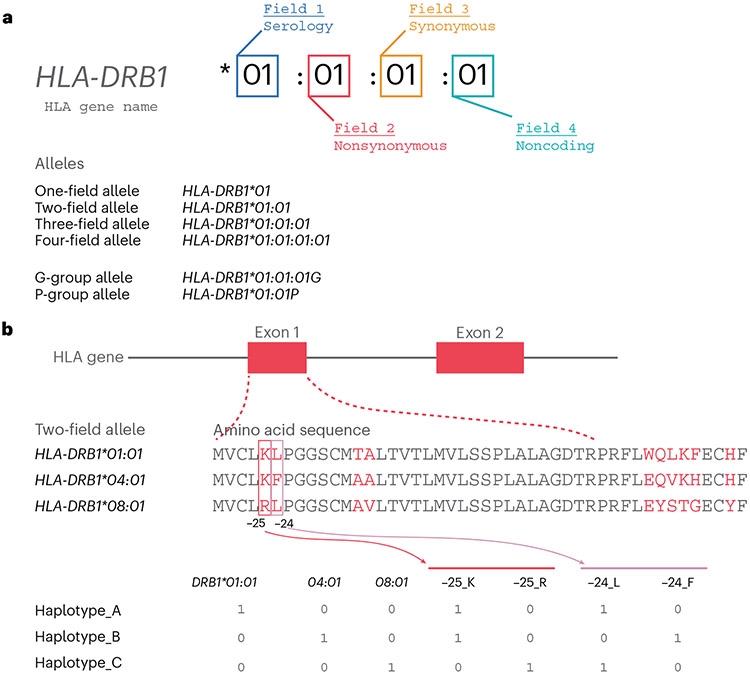
Sequence variation within the HLA genes is organized by the International Immunogenetics database (IMGT)24, which has documented and named 33,490 unique HLA alleles (URL: https://www.ebi.ac.uk/ipd/imgt/hla/about/statistics/). Within each of the HLA alleles, there are nucleotide variants that cause amino acid changes (nonsynonymous nucleotide substitutions) and those that do not (synonymous, intronic and intergenic nucleotide substitutions). A detailed nomenclature system at IMGT has been developed to organize the polymorphisms in HLA genes into four fields (Fig. 3a) (ref. 25). Field 1 (the first two digits, e.g., HLA-DRB1*01) describes the serological type, which was historically defined on the basis of similar seroreactivity to immunological reagents. Field 2 (the next set of digits, e.g., HLA-DRB1*01:01) corresponds to the unique amino acid sequence of the gene; all the nonsynonymous changes are reflected in this set. Field 3 (e.g., HLA-DRB1*01:01:01) reflects synonymous nucleotide substitutions within the coding sequences, and field 4 (e.g., HLA-DRB1*01:01:01:01) reflects polymorphisms within the intronic or noncoding regions. Thus, whereas nucleotide variants define HLA alleles at up to a four-field resolution, most disease associations are captured by a two-field HLA resolution, as amino acid sequence captures most of the structural differences between the alleles.
Fig.3 ∣. Nomenclature, structure and encoding of HLA alleles.
a, The nomenclature of HLA alleles consists of four fields, with each field corresponding to the types and consequences of nucleotide variations. b, (Top) The amino acid sequences defining each of three example HLA-DRB1 alleles. The amino acids colored in red indicate the positions where they have variations among the alleles. The numbers (−25 and −24) at the bottom indicate the relative position of those amino acids within a coding region of HLA-DRB1. The negative positions indicate amino acids within a signal peptide, which is not part of the HLA protein presented on a cell surface. (Bottom) A procedure to code each of the HLA alleles and amino acid polymorphisms as binary markers: 1 if that marker is present within a haplotype and 0 otherwise. Each of the residues is coded separately for a given amino acid position in the corresponding HLA protein.
The four-field naming system is the current and most widely used standard, but alternative nomenclatures are sometimes seen in practice. Before the current four-field naming system was introduced, the IMGT used the same nomenclature where each field must have two digits, but without a field separator (:). Therefore, one-field alleles were called two-digit alleles, and two-field alleles were called four-digit alleles. However, as the number of two-field alleles belonging to a given one-field allele began to exceed 100 (e.g., HLA-A*02101 and HLA-B*15101), the ‘four-digit’ designation became inappropriate. Thus, the IMGT updated the previous nomenclature system by introducing the field separator (e.g., HLA-A*02:101 and HLA-B*15:101) and four-field naming system26.
In this same update, the IMGT introduced two additional nomenclature schemes to facilitate practical reporting of HLA typing: G group and P group. Current classical HLA typing technologies such as SBT sometimes cannot resolve an HLA allele at a four-field resolution13 and instead define a group of similar alleles on the basis of the variations within peptide binding domains (exon 2 and 3 for class I HLA genes and exon 2 for class II HLA genes)26. The G group nomenclature represents HLA alleles that share the same nucleotide sequence in the peptide-binding domain. For instance, HLA-A*01:02:01G includes HLA-A*01:02:01:01, HLA-A*01:02:01:02, HLA-A*01:02:01:03 and HLA-A*01:412, but not HLA-A*01:02:02. The P group nomenclature represents HLA alleles that share the same protein sequence in the peptide binding domain. For example, HLA-A*01:02P includes HLA-A*01:02:01:01, HLA-A*01:02:01:02, HLA-A*01:02:01:03, HLA-A*01:02:02 and HLA-A*01:412.
HLA imputation
Genotype imputation is the term used to describe estimation of missing genotypes that are not assayed in the target dataset. Most imputation methods use data from densely genotyped samples as a reference dataset in which haplotypes have been inferred27. These methods typically use statistical approaches such as hidden Markov models (HMM) to fill in missing genotypes in a dataset of interest with incomplete genotype data. The genotype data reflects the observed states, while the template haplotypes are represented as the unknown hidden states. Most imputation algorithms produce a probabilistic prediction of each imputed genotype. These probabilities can be used to calculate either (1) a probabilistic dosage, which is a simple sum of the expected probabilistic allele counts, or (2) a best-guess genotype, which is a combination of the alleles that have the largest probability. These values can then be used in the downstream analyses. Dosages inferred from imputed results are continuous values between 0 and 2, whereas best-guess genotypes are discrete values of 0, 1 or 2 alleles. Genotype imputation can boost the power of subsequent association studies, help fine map the signal and enable meta-analysis of multiple cohorts27.
It is essential to understand the accuracy of imputation before using the imputed genotypes for downstream analyses. The quality of predictions can be technically measured by masking the genotypes, imputing them and deriving the correlation between the true (masked) genotype and the predicted genotype. We favor using this correlation as a metric, as opposed to accuracy (percent of concordance between true genotype and imputed genotype calls), since accuracy can be upwardly biased for rare alleles. In practice, true genotype data is often missing. In these instances, we can also estimate the quality of imputation, Rsq, by the ratio of the empirically observed variance of the allele dosage to the expected binomial variance at Hardy–Weinberg equilibrium (HWE).
HLA imputation is a natural extension of the genotype imputation. The HLA imputation infers HLA alleles, amino acid polymorphisms and intragenic SNPs within HLA (hidden state). Due to the excessive variation in HLA genes, these variants generally cannot be accurately assayed with popular probe-based genotyping arrays. HLA alleles are inferred indirectly by using surrounding genotyped SNPs in the MHC region (‘scaffold’ variants; Fig. 2a). Reference haplotypes are constructed from samples with both genotyped SNPs and HLA alleles genotyped by either classical SBT11 or inference from untargeted sequencing data, such as whole-genome sequencing (WGS) data28,29. The HLA amino acid sequences and intragenic SNPs within HLA genes can also be included in the reference haplotypes to enable imputation. There are many widely used statistical software tools to perform the HLA imputation, such as SNP2HLA18, HIBAG20, HLA*IMP19, HLA-IMPUTER30 and GRIMM31. SNP2HLA and HLA*IMP use the same HMM algorithm used in genome-wide imputation, whereas HIBAG uses a machine-learning technique called a bagging method32. Imputation performance is often related to the size, quality and suitability of the reference panel rather than the statistical software used. The output of the HLA imputation is a posterior probability as well as an effective dosage (ranging from 0 to 2) for each HLA allele in a given sample. Subsequent association tests usually account for the uncertainty of the imputation by using the estimated dosage as an explanatory variable of interest, which is used to test the association between the genotype and the trait.
HLA imputation reference panel.
There have been many efforts to construct haplotype reference panels in the MHC region to enable HLA imputation. Since the haplotype structure within the MHC region differs significantly among populations21, it is important that the target dataset is well represented by the reference haplotype panel. The current availability of published HLA reference panels is summarized in Table 2.
Table 2 ∣.
Available HLA imputation reference panels
| Imputation software | Name | Ancestry/population | Number of samples | Availability |
|---|---|---|---|---|
| SNP2HLA18 | T1DGC | European | 5,225 | Upon registration |
| SNP2HLA | Pan Asian91 | Han Chinese, Southeast Asian Malay, Tamil Indian ancestries and Japanese | 530 | Publicly available |
| SNP2HLA | Okada et al.92 | Japanese | 908 | Upon registration |
| SNP2HLA | Hirata et al.34 | Japanese | 1,120 | Upon request |
| SNP2HLA | Zhou et al.93 | Han Chinese | 20,635 | Publicly available |
| SNP2HLA | Kim et al.94 | Korean | 413 | Publicly available |
| SNP2HLA | 1KG | Global populations in 1000 Genomes Project (Africans, East Asian, European, South Asian and Americas) | 2,504 | Publicly available |
| HLA-TAPAS21 | 1KG | Global populations in 1000 Genomes Project (Africans, East Asian, European, South Asian and Americas) | 2,504 | Publicly available |
| MIS (Minimac) | Multi-ancestry | Multi-ancestry (admixed African, East Asian, European and Latino) | 20,349 | Limited public accessibility on web |
| HIBAG20 | HLARES | Multi-ancestry (European, Asian, Hispanic and African) | 4,000 | Publicly available |
| HIBAG | IKMB | Multi-ancestry (African American, European, East Asian, Indian and Iranian) | 1,360 | Publicly available |
| HIBAG | Degenhardt et al.95 | Multi-ancestry (African American, European, East Asian, Indian and Iranian) | ~1,300 | Upon request |
| HLA*IMP19 | 1958 Birth Cohort + HapMapCEU | European | ~2,500 | Limited public accessibility on web |
Currently available HLA imputation reference panels, the sample ancestry, the number of samples and whether they are publicly available. ‘Limited public accessibility’ means that while the raw reference panel (individual-level genetic data) is not accessible, users can use it for imputation via a web-based imputation service.
It is also possible to construct a custom HLA reference panel using tools such as SNP2HLA18, HLA-TAPAS21 and HIBAG. Starting with a SNP genotyped cohort (‘scaffold variants’), we can either (1) obtain the gold standard SBT of HLA alleles (such as SSOP11) if DNA is available or (2) infer HLA alleles from WGS (e.g., HLA*PRG and HLA*LA)28,29,33. Reference panels can include alleles of classical HLA genes21 that are most polymorphic and disease associated, or both classical and nonclassical HLA genes34. In the SNP2HLA algorithm, HLA alleles are converted to biallelic markers (e.g., one indicates the presence of the allele and 0 indicates the absence of the allele). Classical SBT, such as SSOP, is the most accurate approach to HLA genotyping13-15. Incorporation of sequence-based HLA genotypes into reference panels results in highly accurate imputation; however, since SBT is costly and labor intensive, it cannot be easily used to build large reference panels. Graph-based inference of HLA alleles from WGS is a potential alternative method that can be easily applied to large sequencing datasets that are increasingly available28,29,33, including low-coverage datasets. However, an important caveat is that the accuracy of HLA typing by those graph-based methods can be variable. Imputation performance is affected by (i) quality of the sequencing data, (ii) read coverage and length, (iii) representation of the population in reference databases such as IMGT and (iv) the degree of sequence variation within the targeted HLA gene. For example, the HLA*LA algorithm, one of the currently available graph-based HLA inference software tools, showed relatively accurate HLA typing at 15× coverage but no benchmarking data has been shown below 15× (ref. 29). For studying underrepresented populations or highly polymorphic genes, gold-standard SSOP might still be necessary to construct a suitably accurate reference panel. Use of long-read sequencing or relatively longer read sequencing beyond 150 bp could also enable more unambiguous HLA typing and sequencing-based haplotype determination35,36.
To enable imputation of amino acid polymorphisms and intragenic HLA SNPs, we can encode all these variants as binary markers on the basis of the reference amino acid and nucleotide sequences of each observed HLA allele from the IMGT HLA Database (https://www.ebi.ac.uk/ipd/imgt/hla/) (Fig. 3b). The scaffold genetic variants within the MHC region are usually obtained by either genotyping with a SNP microarray or WGS. Stringent SNP QC is essential for accurate haplotype phasing and, ultimately, accurate imputation. In constructing and updating a multi-ancestry HLA reference panel, we optimized this QC process to maximize imputation accuracy. Specifically, we started with QCing each of the global cohorts separately, with genotype call rate (for a given variant, the percentage of individuals for which the corresponding variant information is not missing and confidently called; >95%) and sample call rate (for a given sample, the percentage of genetic variants that were not missing and confidently called; >90%). We then retained the variants that were present in the 1000 Genomes Project37 and excluded any variants that were not included in commonly used genotyping arrays (Illumina Multi-Ethnic Genotyping Array, Global Screening Array, OmniExpressExome and Human Core Exome). Since these variants are not included in the target genotype data, they are more likely to result in phasing switch errors (i.e., the number of required recombination events in inferred phased haplotypes to obtain the true haplotype phase divided by the number of possible opportunities for switch error, as a metric for phasing inaccuracy38) without improving imputation accuracy. When combining all the cohorts to construct the multi-ancestry panel, we cross-imputed all the variants together to avoid excluding population-specific variants that are polymorphic in a specific cohort but monomorphic and thus not called in the other cohorts (Extended Data Fig. 2). The final reference panel includes the HLA alleles, amino acids, intragenic HLA SNPs and the ‘scaffold’ variants (i.e., SNP variants outside an HLA gene but within the extended MHC region), which are then haplotype-phased statistically or by using trios (genotype data from a mother, a father and a child).
Imputed HLA alleles and variants are often used for subsequent association testing and meta-analyses to fine-map disease risk. Such studies potentially include data from multiple cohorts, datasets or populations. To avoid spurious associations due to batch effects and population stratification, it is essential to perform HLA imputation on all datasets using the same reference panel, ideally with all case and control samples genotyped together. Given that such case-control cohorts may originate from multiple populations to increase the fine-mapping resolution, we constructed an HLA reference panel covering multiple global populations21.
With the publication of this tutorial, we present an updated version of this multi-ancestry panel (version 2). In brief, we added samples from European (n = 2,233) and Japanese (n = 723) ancestry for a total of 20,349 individuals. This panel represents admixed African, East Asian, European and Latino populations. We also updated HLA allele calls and a set of scaffold variants. We plan to maintain and update the panel further to increase representation of globally diverse populations, improve the HLA allele calls and refine the selection of scaffold variants to achieve the most accurate imputation.
Recommendations for collecting genotype and phenotype information
When designing a study to investigate the effect of HLA variation on human traits, it is important to be strategic when collecting genotype and phenotype data. For genotype data collection, one should ensure that the genotyping array used for the target cohort has a high coverage in the MHC region to adequately include variants that are in LD with HLA alleles, which contributes to accurate imputation. While most currently used genotyping arrays include a sufficient number of SNPs to tag HLA alleles for accurate imputation, some arrays have limited SNP coverage of the MHC region (Supplementary Table 1) (ref. 39). We and others have shown that lower MHC coverage results in inaccurate imputation18,40. Furthermore, all study participants should ideally be genotyped together with the same genotyping array, to avoid introducing any structure that could cause a bias in imputation and the subsequent association testing and fine-mapping.
Careful phenotype curation is very important when fine-mapping disease-associated variants. On one hand, discovery of HLA association signals can be enhanced by the addition of samples with less rigorously curated phenotypes (e.g., billing codes in large-scale biobanks). However, fine-mapping accuracy can be negatively affected by including misclassified samples. For example, studies of autoimmune disease including different categories of patients with heterogeneous clinical phenotypes or pathological pathways can obscure efforts to localize disease alleles. This has, for instance, been observed in rheumatoid arthritis, where patients with positive antibody status are phenotypically and genetically different from those with negative antibody status41,42. Recently, many efforts have been made to curate the phenotypes in large-scale biobanks43 using self-reported disease status or billing codes (e.g., ICD-10) (ref. 44). While the large number of samples in these biobanks may enable discovery of disease-relate alleles, imprecise phenotype labeling inherent to these forms of phenotyping may confound HLA fine mapping. In contrast, physician-curated cohorts, which require more efforts on accurate phenotyping and thus are inevitably smaller than biobanks, may be important for accurate fine mapping of the alleles.
In addition to disease phenotypes, one must exercise caution when measuring HLA-related molecular phenotypes, such as HLA gene and protein expression. It is well established that HLA gene and protein expression is affected by cis-regulatory genetic variants (i.e., expression quantitative trait loci (eQTL; genetic loci that affect the expression levels of genes) and protein eQTL (pQTL; genetic loci that affect the protein levels))45-47. When conducting eQTL studies, measuring HLA expression in RNA-sequencing data is particularly challenging owing to the high degree of genetic polymorphism among individuals. Standard expression quantification pipelines rely on a single human reference genome to align sequencing reads. The number of reads mapping to each HLA gene might be biased for two reasons: (1) the reads may fail to map to the reference due to the high degree of sequence variation (i.e., a large number of mismatches) and (2) the reads may not uniquely map to a single gene in the reference due to the similarity among nearby HLA genes (i.e., multimapping)47. To address this, more accurate gene expression estimates can be obtained by using an HLA-personalized reference47; instead of using a standard single human reference genome, we can supply customized HLA sequences for each target individual for each HLA gene (on the basis of either classical HLA typing or HLA imputation) to minimize the degree of variation between the RNA-sequencing reads and the reference, and hence reduce the possibility of mapping failures and multimapping. Similarly, caution should be taken for HLA pQTL studies. HLA protein expression is often measured by antibody-based methods (e.g., antibody-derived tags; antibodies against a protein conjugated with oligonucleotides that can be captured by PCR amplification and detected by sequencing) at single-cell resolution48. However, these antibodies may have differing binding affinities to the protein products of different HLA alleles. We should take caution when conducting pQTL studies, since this differing affinity might cause a bias toward specific HLA alleles when measuring the abundance of HLA proteins across individuals.
QC of the target genotype data
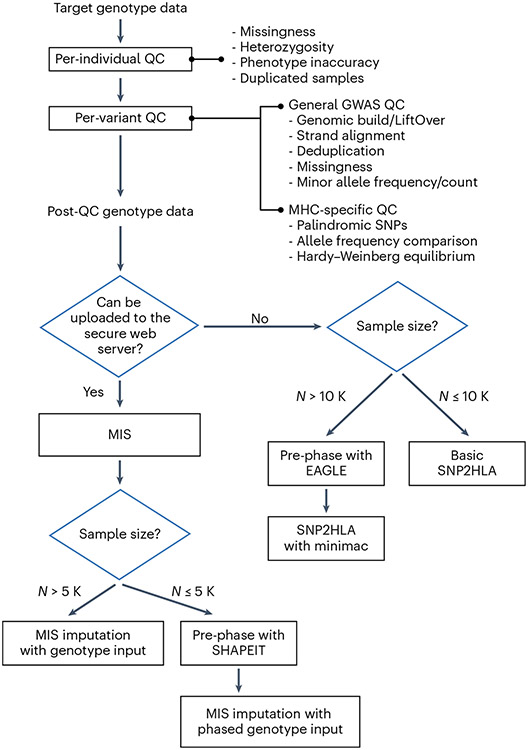
QC of genotype data before HLA imputation is extremely important. We next outline the basic QC measures commonly used in GWAS49, as well as specific instructions to handle genetic variants within the MHC region. These QC measures are typically performed once for each genotyping batch, followed by data integration and final QC for the combined dataset (Fig. 4). We assume that the target genotype data is conventionally constructed from cost-effective microarray-based genotyping, in which a limited number of genetic variants (approximately hundreds of thousands to a million) are typed that tag untyped variants by LD to cover genome-wide variants through imputation50. However, low-pass WGS after rigorous QC can be used to obtain SNP genotypes (e.g., HELIC study51,52). This could be used as an alternative cost-effective strategy especially in biobanks from populations underrepresented in GWAS studies (e.g., African populations), since commonly used microarrays may not sufficiently cover specific variants for these populations53.
Fig.4 ∣. A flow chart of suggested analytical steps for genotype QC and HLA imputation.
A best-practice guideline to impute HLA alleles by using SNP2HLA algorithm, depending on the characteristics of the target genotype data.
Per-individual QC.
We follow established guidelines43,49,54 to perform standard per-individual QC in GWAS. Typically, we remove (i) individuals with high missingness (e.g., >0.02), (ii) individuals with outlier high heterozygosity on suspicion of sample contamination, (iii) individuals with discordant sex information between the metadata and genotype and (iv) individuals suspected to be duplicate samples on the basis of genotype relatedness. We note that the threshold for each QC measure could be data dependent, and thus we recommend reviewing the distributions of those metrics for each of the datasets.
Per-variant QC.
It is important to select high-quality variants to achieve accurate imputation. We will describe the variant QC that is generally recommended for GWAS as well as specific considerations for the MHC region. As part of standard GWAS QC, we recommend ensuring that the target genotype data has genomic positions based on the same genome build as the reference panel. LiftOver software55 can re-map the genomic position from the build used in the genotype data (e.g., GRCh38) over to the desired build used in the reference panel (e.g., GRCh37). Next, genomic variants are typically aligned to the forward strand to be consistent with the reference panel. We also identify duplicated variants within the dataset on the basis of genomic position and alleles, and de-duplicate them by removing ones with higher missingness. We then remove (i) variants with high missingness (e.g., >0.01), (ii) variants demonstrating a significant deviation from the HWE and (iii) variants with very low minor allele frequency (MAF). Specifically, we remove variants an MAF lower than 0.01 or 0.005, or small minor allele count (e.g., <5), assuming low accuracy in genotype calling from clustering. The sample size should be accounted for when we use minor allele count. The estimated ancestry should also be accounted for in order to retain informative population-specific variants. For example, if the data consists of a mixture of different ancestries and one ancestry is underrepresented, we might calculate MAF separately for each ancestry and retain the union of common variants in each ancestry so that we do not lose variants specific to the underrepresented population. We usually only keep biallelic variants and remove multi-allelic variants for simplicity in the imputation.
Specific caution should be taken when performing per-variant QC in the MHC region, owing to (i) highly variable allele frequency (AF) of variants within MHC across populations and (ii) expected HWE deviation in the MHC variants due to natural selection. For example, the AF variability could be an issue when performing strand alignment. We usually align target genotype alleles to the forward strand as used in the reference panel. To do so, we have to consider whether the SNP is ‘palindromic’. A palindromic SNP (or an ‘ambiguous’ SNP) is a SNP in which the two alleles for that variant are complementary each other (i.e., SNPs with A/T or G/C alleles) and the reverse-stranded alleles yield the same genotype as the forward-stranded alleles. When we align nonpalindromic SNPs, we can simply look up the alleles with the same position in the reference human genome sequence on the forward strand. If the alleles between the target and the reference genome are different (e.g., A/C in the reference but T/G in the target), we flip the alleles in the target dataset (swap alleles from T to A and from G to C in the target). On the other hand, in handling palindromic SNPs, we usually compare population-derived AF and the AF in the target dataset to eliminate allele ambiguity. If the AFs between these datasets are largely different (e.g., A: 20% and T: 80% in the population reference but A: 78% and T: 22% in the target), we can flip the alleles in the target to be consistent with the population-derived AF (swap alleles from A to T and from T to A). However, this strategy might be ineffective within the MHC since the reference AF for those SNPs might be different from the target samples when the study population is different, when there are large AF differences between cases and controls in case-control studies, or when the study sample size is too small to estimate AF accurately. Therefore, when the strand information of those palindromic SNPs is ambiguous in the target genotyping array or the genotyped data, it may be preferable to exclude all the palindromic SNPs. Second, we may compare the AF of the variants after QC in the target data with the AF in the population-frequency database (e.g., 1000 Genomes Project37 and gnomAD56) or the AF in the reference panel as a sanity check. When the AFs are very different between the two, those variants could be subject to genotyping error and should probably be removed. However, when the population does not exactly match between the target and the database or the reference, this strategy might be ineffective within the MHC. Thus, we could consider using a liberal threshold when removing variants on the basis of the AF differences. Third, extreme deviation from HWE is usually indicative of a genotyping or genotype-calling error that results in poor clustering57-59 and is used as a metric to exclude poor-quality variants. However, the deviation from HWE is, to some extent, expected in the MHC region owing to natural selection60 or to the difference in AF between cases and controls. The expected deviation will be greater when we study a cohort from multiple populations or of admixed ancestry, or when the effect of HLA on the disease is large or has a nonadditive nature. Therefore, for the purpose of per-variant QC, we could consider (1) calculating HWE P values only within control individuals (as is generally recommended in GWAS), (2) calculating HWE P values within individuals from the most common ancestry or (3) using a liberal threshold such as HWE P < 1 × 10−10 when removing variants suspected of poor clustering while retaining the important markers for HLA imputation that could inherently deviate from HWE due to natural selection60. When we are unsure about the threshold, an appropriate value can be identified by visually inspecting the genotype cluster plots (e.g., in GenomeStudio by Illumina).
Tools for genotype phasing and HLA imputation
Once we prepare the optimal HLA reference panel and QC the target genotype data, we start HLA imputation for the target data. Table 3 summarizes the main software programs for HLA imputation. Of note, some imputation programs take as input the genotype files directly after the QC as described above, while others require users to pre-phase the genotypes to obtain haplotypes19,22 before imputation (Fig. 4).
Table 3 ∣.
Representative software programs for HLA imputation and their requirements
| Imputation software | Pre-phasing | Input file format | Local | Output | Amino acid imputation |
|---|---|---|---|---|---|
| SNP2HLA18 | Unnecessary | plink | Yes | VCF | Yes |
| SNP2HLA + Minimac | Necessary | phased VCF | Yes | VCF | Yes |
| MIS (Minimac)22 | Recommended when the sample size of the genotype data (N) is small (e.g., N < 5,000) | VCF | No | VCF | Yes |
| HIBAG20 | Unnecessary | plink | Yes | R object | Yes |
| HLA*IMP19 | Necessary | phased Oxford haps/sample | No | CSV | No |
HLA imputation software programs and their specifications and details about the input and output.
SNP2HLA, developed by our group, and its extensions21,61, are among the most widely used algorithms for HLA phasing and imputation; therefore, here, we focus on HLA imputation using the SNP2HLA algorithm along with its cloud-based implementation at the MIS (https://imputationserver.sph.umich.edu/index.html).
SNP2HLA.
The SNP2HLA18 program can phase and impute HLA alleles, amino acids and intragenic SNPs with HMM implemented in BEAGLE62 by taking the target genotype file after QC in the PLINK format as an input. The input file is internally processed to extract variants within the MHC (29–34 Mb), and then to correct or remove strand errors when possible on the basis of the genotype and AF of palindromic SNPs. In addition to the original bash scripts (http://software.broadinstitute.org/mpg/snp2hla/), there are several extensions to the SNP2HLA algorithm such as HLA-TAPAS21 (with association test function) and CookHLA61 (with improved imputation algorithm). We also provide a step-by-step guide to SNP2HLA implementation, along with a script that allows users to specify all the QC thresholds as optional parameters to handle various target cohorts (e.g., the target populations, the number of samples) on our tutorial website (https://github.com/immunogenomics/HLA_analyses_tutorial).
We note that the original implementation using BEAGLE scales to fewer than 10,000 samples in the target dataset. To address this, we also provide a pipeline using another imputation software, Minimac22, which can scale to hundreds of thousands to millions of individuals. To use Minimac for imputation, we first pre-phase the genotype by using methods such as SHAPEIT63 or EAGLE64. EAGLE has an advantage of accurate and fast phasing when the number of samples is large (e.g., N >10,000). The pre-phased output file must be converted into the Variant Call Format (VCF), and then used as an input to the Minimac software.
MIS.
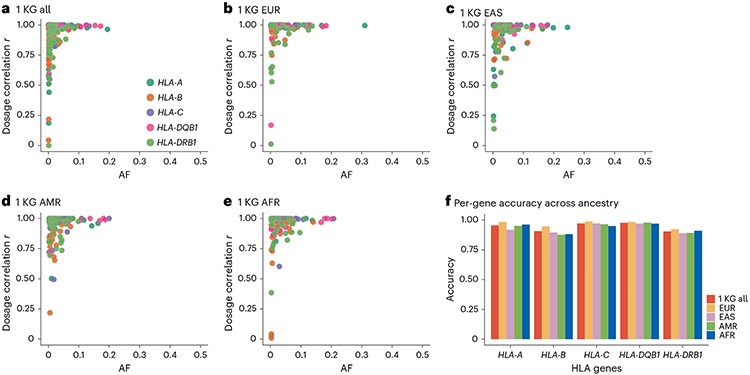
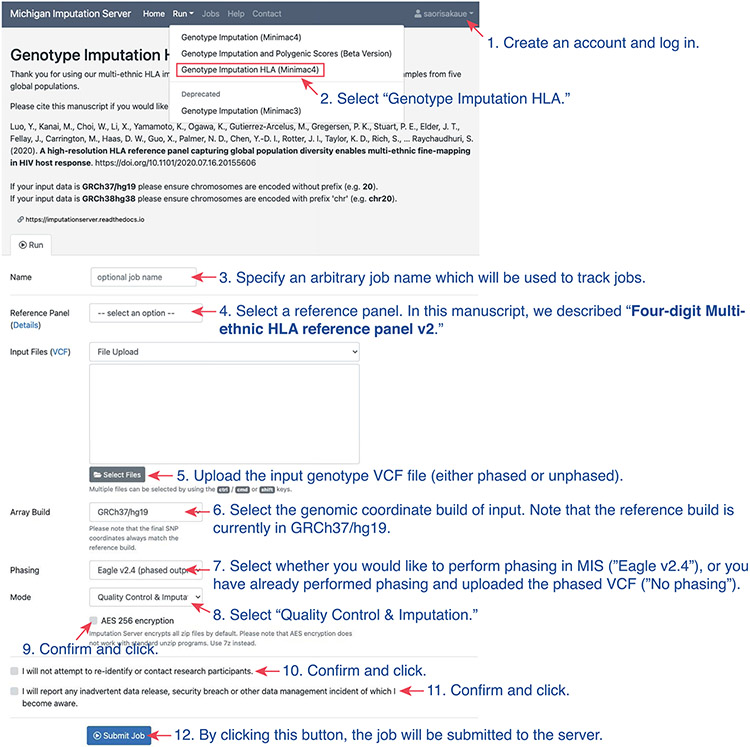
While HLA imputation using the SNP2HLA algorithm can be conducted locally using publicly available HLA reference panels, not all the HLA reference panels are available due to data sharing and privacy restrictions. Our latest multi-ancestry HLA reference panel is one such restricted-access panel21. To enable widespread access, we implemented HLA imputation on the MIS (https://imputationserver.sph.umich.edu/index.html), which is a cloud-based imputation server with a user-friendly interface (Extended Data Fig. 3). We host the multi-ancestry HLA reference panel at the MIS and implement the HLA imputation using Minimac as described above. In brief, the user first creates an account online, and securely uploads either a phased or unphased VCF genotype file if the file can be uploaded to the secure web server. If the uploaded genotypes are unphased, the uploaded genotype file will be phased within the MIS using the EAGLE algorithm. As noted above, we recommend to pre-phase the genotype (with the reference haplotype when possible) using SHAPEIT or other software when the sample size is small (e.g., N < 5,000) to achieve accurate phasing before imputation. The MIS automatically performs basic QC of the input VCF file for the strand orientation and alleles in accordance with the reference. If the input passes the QC steps, the MIS seamlessly performs the HLA imputation. The user will be notified with a download link for the imputed VCF file encrypted with a one-time password via email once the imputation is completed. The MIS has been used to impute more than 6 million genomes since we started the web-based HLA imputation service in 2021. We benchmarked the performance of HLA imputation on the MIS using individuals with both SNPs and (masked) gold-standard HLA alleles identified by Sanger SBT65,66 in the 1000 Genomes Project. We confirmed that the imputation accuracy measured by dosage correlation with true HLA alleles was very high across populations (mean dosage correlation r = 0.981 for two-field alleles with MAF >0.05; Fig. 5).
Fig.5 ∣. HLA Imputation quality in MIS.
a–e. Dosage correlation r (y axis) between the MIS imputed dosage and true genotypes of all two-field alleles in 1 KG samples as a function of AF (x axis), colored by HLA gene, for all 1 KG individuals (a) or per ancestry (b–e). f, The accuracy (concordance) of the imputed dosage of all two-field alleles in 1 KG samples in the MIS and the true genotype of those per HLA gene and per ancestry. The accuracy metric was calculated as previously described18. EUR, Europeans; EAS, East Asians; AMR, admixed Americans; AFR, Africans.
Postimputation QC
The output from the HLA imputation software is accompanied by a quality metric conveying the confidence or estimated accuracy of imputation per allele. A thorough review of these imputation metrics and their correspondence to imputation accuracy is described in Marchini and Howie27. We typically QC the imputed HLA alleles, amino acids and intragenic SNPs on the basis of imputation metrics before association testing. SNP2HLA, Minimac and MIS all include Rsq as a quality metric. The appropriate Rsq threshold for QC may depend on the study design; for example, we commonly use Rsq >0.7 in single cohort studies and Rsq >0.5 in multicohort meta-analyses. By removing imputed alleles that are below this Rsq threshold, some individuals might end up having an HLA gene for which the total number of two-field alleles does not sum up to exactly 2. Those individuals might bias the fine-mapping of disease-causing alleles, which we will explain in the subsequent sections. Thus, we recommend removing any individuals that do not have two two-field alleles for a given gene when conducting conditional haplotype tests using two-field alleles.
We recommend calculating true imputation accuracy from classical HLA typing if it is available for a subset of study individuals. While the estimated imputation accuracy generally corelates well with the true accuracy, having the ability to internally benchmark with classical allele typing for a subset of the cohort is useful for evaluating the true imputation performance, especially if the reference panel imperfectly represents the genetic ancestry of the imputed cohort.
HLA association and fine-mapping
In the following sections, we introduce basic and advanced methods for testing the significance of the imputed HLA variants associated with a trait or a risk of a disease. In addition to the statistical models presented in the mathematics formulas below, we also provide example command-line scripts to perform all these statistical tests on our website (https://github.com/immunogenomics/HLA_analyses_tutorial).
Single-marker tests.
Single-marker genetic association tests are used to investigate whether a specific HLA allele, amino acid or SNP is statistically associated with a risk of a disease or a trait of interest. Similar to the approach used in GWAS, we perform a logistic regression (for traits in case-control studies) or a linear regression (for quantitative traits) for the imputed binary makers that indicate the presence (coded as T in the imputed VCF file) or absence (coded as A in the imputed VCF file) of the selected HLA allele, amino acid or intragenic SNP. For the markers, we typically use the imputed probabilistic dosage genotypes to account for any imputation uncertainty. We include study-specific covariates that could independently explain the trait of interest, such as sex, age and genotype batches, as well as genotype principal components (PCs) to account for population stratification, and an indicator variable of cohorts when combining multiple cohorts.
The logistic regression can be formulated as:
where log () is the natural log of the odds ratio for case-control status in individual , indicates the specific allele being tested and is the imputed dosage of allele in individual . The allele could be either a single HLA allele, a single amino acid polymorphism or a single SNP. The parameter represents the additive effect per allele. For all covariates, , and are the covariate ’s value in individual and the effect size for the covariate , respectively. Similarly, and are the first th genotype PC value in individual and the effect size for the first th genotype PC, respectively, to control for genetic ancestry. The is the logistic regression intercept.
Quantitative traits that follow continuous distributions (e.g., antibody levels, blood cell counts, etc.) can be analyzed by using linear regression similarly:
where is a quantitative trait of interest and is normalized by a score or an inverse-normal transformation when the trait does not follow the Gaussian distribution implicitly assumed in the linear regression model.
These association tests can be conducted using conventional GWAS software, such as PLINK67, SAIGE68 or BOLT69, by directly using the output VCF files from imputation generated by either SNP2HLA or MIS. We use the dosage values designated as ‘DS’ in the imputed VCF files to conduct dosage-based association tests. The imputed genotype VCF file encodes the HLA alleles or amino acid residues as binary markers as we explained in the ‘HLA imputation reference panel’ section (Fig. 3b). ‘P’ or ‘T’ denotes the presence (or number of copies) of the allele in the variant name, whereas ‘A’ denotes the absence of the allele. When interpreting results from an association analysis, the effect estimate indicates the effect of having one copy of the effect allele on the log odds ratio of the disease. Therefore, the effect estimate should correspond to the presence of the effect allele (denoted with ‘P’ or ‘T’) rather than the absence of the allele (denoted with ‘A’).
Also, we note that the association of rare alleles might be spurious due to both the limited accuracy in imputation and the noise in the estimate in the regression. Thus, we might QC the association statistics by MAF to exclude rare alleles (e.g., MAF <1% or 0.5%).
The OR calculated from the beta () is the estimated risk explained by having one copy of the HLA allele of interest, and the P value indicates its significance of the association. Given the strength of LD in the MHC region, trait associations to multiple HLA alleles, amino acid polymorphisms or intragenic SNPs may yield significant results. Further analysis is then required to identify which allele(s) most significantly explain(s) the disease risk within the HLA region.
Omnibus tests for fine-mapping amino acid position.
To narrow down the causal position within amino acid sequences encoded by a specific HLA gene, we perform an omnibus test. This analysis is particularly useful when we seek to define mechanisms for the HLA association with a particular disease, for example, by changing the amino-acid compositions at the peptide binding groove of the HLA molecule. In the omnibus test, we estimate the total effect on our trait of interest of all amino acid content variation at a given amino acid position, rather than the separate effects of individual amino acids that appear at that position, as we did in the single-marker test. For an amino acid position that has possible amino acid residues, we assess the significance of the improvement in fit for the full model that includes amino acid dosages as explanatory variables when compared with a reduced model without those amino acid dosages. We usually select one amino acid residue that is most common in the studied cohort as the reference allele, and use all the other amino acid residues () as the explanatory variables. We assess the improvement in model fit by the delta deviance (sum of squares) using an F-test with degrees of freedom and derive the statistical significance of the improvement.
where is one amino acid residue at this position, is the total number of observed amino acid residues at this position, and and are the amino acid dosage of the residue in individual and the effect size for the residue , respectively.
We may use the permutation procedure to determine whether the observed association at a single-marker test is primarily driven by HLA alleles (e.g, HLA-DRB1*04:01) or amino acid polymorphisms (e.g., (HLA-DRβ1 positions 11, 71 and 74) (ref. 23). To do so, we shuffle the correspondence between amino acid sequences and each of the two-field HLA alleles that were originally defined in the IMGT database as described above. Then, in each permutation, we select each amino acid polymorphism and assess the improvement in deviance after including this amino acid polymorphism in the model. We typically perform >10,000 permutations. If the observed improvement using the actual data is significantly larger than the improvements using these permutations, we can infer that an amino acid polymorphism is driving the signal, instead of observing the ‘synthetic’ association in which the effect of the causal amino acid on a trait propagates to the marginal association statistics of the noncausal HLA allele merely in LD with the causal amino acid.
Conditional haplotype tests to define a risk sequence of amino acids.
Defining the exact stretches of HLA amino acid sequences driving the association with disease allows us to understand the mechanism by which amino acid change affects disease risk23. Importantly, to model combinations of positions, we must use phased genotyping information, rather than encoding each position separately. We perform a conditional haplotype test, where we combine the imputation results of both two-field alleles and amino acid polymorphisms to obtain phased information. We start from the most significant position in the amino acid sequence on the basis of the omnibus test we described in the previous section. If there are possible amino acid residues at this position, we can group all possible two-field alleles for this HLA gene into groups on the basis of the amino acid residue property at our selected position (Fig. 6a). Recall that each two-field allele at a given HLA gene corresponds to a unique sequence of amino acids in this gene. In the same way as we did in the omnibus test on the basis of the amino acid residues, we can estimate the effect of each of the groups using a logistic regression model (including covariates, as described above). We assess the improvement in model fit over a reduced model without including those groups by the delta deviance (sum of squares) using an F-test with degrees of freedom and derive the statistical significance of the improvement.
where is the sum of the dosage of two-field alleles from a group , explained by the th amino acid residue. We note that we recommend removing any individuals that do not have two two-field alleles for a given gene, as we explained in ‘Postimputation QC’.
Fig.6 ∣. Grouping of two-field alleles using the conditional haplotype test.
a,b, An example illustration of the conditional haplotype test for the HLA-DRB1 gene. In the first round of conditional haplotype test, (a), we group all two-field alleles (32 alleles in total) into six groups on the basis of the amino acid residues at position +11 and ask whether those groups significantly explain the disease risk by using the omnibus test. In the second round of conditional haplotype test (b; position +71 as an example), we group the two-field alleles into ten groups on the basis of the amino acid residues at positions +11 and +71. Then, we ask whether the full model with those ten groups explains the disease risk better than the reduced model with the six groups that we defined in the first round by the delta deviance using an F-test.
Once we define the most significant individual amino acid position at a given HLA gene on the basis of the statistical significance of improvement, we next seek to identify which amino acid position other than this significant position best improves the model over the model only including this significant position (Fig. 6b). Let be the most significant position in the primary analysis, which has possible amino acid residues. We sequentially test each amino acid position () other than , to ask whether haplotypes defined by the amino acid combination of positions and () explain the disease risk more than those defined only by the position . To do so, we recategorize all two-field alleles at this HLA gene into groups, where is the total number of observed haplotypes defined by the amino acid positions and . The value of must be at least if no new haplotypes are defined. We again assess the significance of the improvement in model fit of the full model (covariation at positions and ) over the reduced model (variation at position alone) by the delta deviance (sum of squares) using an F-test with () degrees of freedom.
where is the sum of the dosages of two-field alleles in a group by a given combination of the amino acid residues at positions and .
Thus, we define the next most significant amino acid position which additionally and independently explains the disease risk from the position . If the model improvement in this second round is statistically significant, we iterate the same analyses to identify amino acid position(s) other than the previously identified positions that best improve the model over the model including those previous positions, until we obtain no further significant improvement from any of the remaining positions.
Tests for nonadditivity.
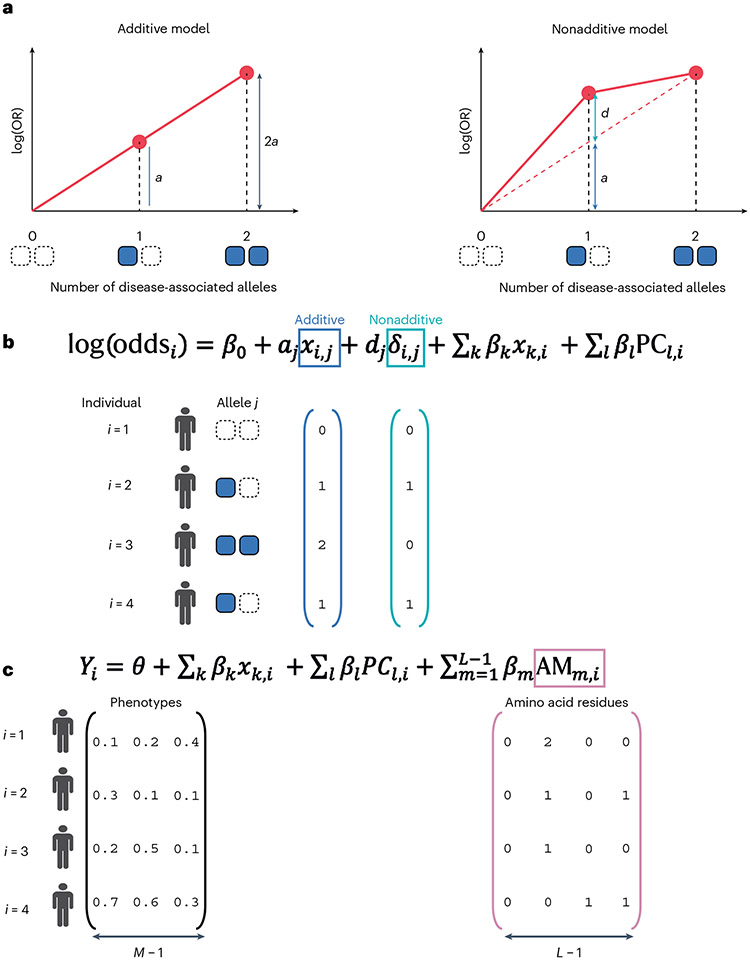
The dosage effect of HLA (having one copy or two copies of a given HLA allele) on disease risk is not purely additive in infectious diseases and autoimmune diseases70-78. All the analyses we have described above assume the additive risk model, in which the risk (i.e., log(OR)) for acquiring a disease due to carrying one copy of the allele (heterozygous state) is half the risk (log(OR)) conferred by carrying two copies (homozygous state). A nonadditive effect represents a deviation from this linear relationship between the dosage and the risk (Fig. 7a). For instance, a dominant effect might be indicated when the effect of carrying one copy is more than half the effect of carrying two copies. A biological explanation for such a dominant effect might be that (1) having one copy is enough to express the MHC variant with the disease-relevant antigen-binding properties on the cell surface, or that (2) there are synergistic interactions with another HLA allele at the same locus. Lenz et al.77,79 showed that such nonadditive effects are pervasive in a spectrum of autoimmune diseases.
Fig.7 ∣. Nonadditive test and multitrait analysis.
a, Schematic illustrations of the additive and nonadditive models using the log odds ratio (log(OR)) according to the dosage of the genotype of interest. a denotes the purely additive effect by having one copy of the disease or trait-associated allele, and d denotes any departure from additivity for a heterozygous genotype. b, A logistic regression model to assess both the additive and nonadditive effect of the allele j (see main text for details). c, Multitrait analysis using a multiple linear regression model to test the association between the multidimensional phenotype Y and the amino acid polymorphism.
To test for the nonadditive effect, we construct a logistic regression model that captures both additive and nonadditive contributions of the allele to the disease risk (Fig. 7b) (refs. 77,80). We first define the additive term as either the best-guess genotype or the dosage genotype of allele in an individual that we are interested in.
We next define the nonadditive term as the heterozygous status of the allele in an individual , which should capture any deviation of the effect from the additivity.
Using those two terms and , we construct a full model by including both the additive and nonadditive term with covariates, and a reduced model including only the additive term with covariates.
where denotes an additive effect and denotes a nonadditive (dominance if positive) effect.
We finally assess the significance of the improvement in model fit of the full model over the reduced model by the delta deviance (sum of squares) using an F-test.
Tests for interactions among HLA alleles.
Once we identify an allele harboring a possible nonadditive effect, we may also be interested in understanding whether this is due to an interaction effect between the identified allele and the other alleles at the same HLA locus. In other situations, we may want to assess an interaction effect between a pair of alleles of functional interest. If the disease risk from a combination of those two alleles deviates from the expected disease risk by multiplying the disease risk (i.e., adding the log(OR)) of each of the two alleles), that combination can be regarded as having an interaction effect. To test this hypothesis, we construct a reduced model that only includes an additive term for each of the two alleles, and a full model that includes an interaction term between the two alleles in addition to the additive term for each of the two alleles. Let be the dosage genotype of the allele in a given individual nominated by a significant nonadditive test, and let be the dosage genotype of the other allele in an individual to be tested for an interaction effect with the allele .
where is the effect size of the interaction between the alleles and . We again assess the significance of the improvement in model fit in the full model over reduced model by the delta deviance (sum of squares) using an F-test. We note that the observed interaction effects can be spurious when the frequencies of the tested alleles are relatively low, which results in noisy effect estimate. We recommend performing conservative QC of the tested alleles on the basis of MAF (e.g., only considering alleles with an MAF >0.05 or 0.10), or performing permutation analyses to test whether the observed statistics could occur by chance, in cases when the MAF is lower than these suggested thresholds.
We also note that HLA molecules form a complex three-dimensional structure to present specific antigens. The interaction analyses presented in this section in a regression framework may not be sufficient to capture higher-order interactions among the amino acid sequences encoded by the HLA81. Indeed, recent studies use a deep-learning framework for accurate prediction of antigen presentation by specific HLA alleles82. Such effort might be necessary for phenotypic association with higher-order HLA structure in future, while substantially larger number of samples might also be necessary to achieve this goal.
HLA evolutionary allele divergence.
A potential source for nonadditive interaction effects among HLA alleles is the extent to which their encoded HLA molecule variants differ functionally (i.e., in their bound antigen repertoires). Since HLA genes are generally co-dominantly expressed, both HLA variants of a heterozygous individual are presenting antigens at the cell surface. If two HLA alleles are very similar in their sequence, their encoded HLA molecules on average will bind similar sets of antigens and thus exhibit a substantial overlap in their presented antigen repertoires, while the opposite will be true for two alleles with very divergent sequences83. The concept that carrying two divergent HLA alleles will allow HLA presentation of a wider range of antigens, and by extension increase the likelihood of pathogen detection by the adaptive immune system, has been termed divergent allele advantage, as an extension of the classical heterozygote advantage84,85. Divergent allele advantage has already been shown to drive HLA allele frequencies and contribute to human immunodeficiency virus control78,83, but might have broader implications in HLA-mediated complex diseases. For instance, it was shown that cancer patients whose HLA class I alleles exhibit a higher HLA evolutionary divergence respond better to cancer immunotherapy, possibly because a greater number of mutated neoantigens are presented by their HLA86. The HLA evolutionary divergence score between two HLA alleles at a given HLA locus is based on the Grantham distance between their amino acid sequences, and is applicable to both HLA class I and class II alleles. It can be calculated using publicly available scripts83, and its effect on a given phenotype can then be estimated by adding it as a quantitative parameter in a regression model and testing for improvement in model fit with an F-test.
Multitrait analysis.
Our group recently showed that the amino acid frequencies at complementarity-determining region 3 (CDR3) of the T-cell receptor (TCR) are highly influenced by HLA alleles and amino acids, possibly through thymic selection7. This type of analysis is an extension of the analyses we described in the previous sections. One notable difference is that the response variable represents not a single trait (e.g., a disease) but multiple traits: in this case the frequencies of each amino acid residue at the position of interest within CDR3, which we call cdr3–QTL analysis. We test which amino acid position in HLA has a significant association with those frequencies in CDR3 overall, using an extended framework of the omnibus test that we described above (Fig. 7c).
In this case, the response variable is not a vector of one phenotype, but a matrix (multidimensional vector) of frequency phenotypes where each row represents an individual and each column represents a frequency of a given amino acid residue at a given position of CDR3. Let be this frequency matrix with rows and columns, and be the frequency phenotypes in an individual . denotes the number of individuals, and denotes the number of observed amino acid residues at this position in TCR. We use a multivariate multiple linear regression model to test the association between and HLA alleles or amino acid positions of interest
where is an M-dimensional parameter that represents the intercept, is the total number of observed amino acid polymorphisms at this position in HLA, and are the amino acid dosage of the residue in an individual and the M-dimensional effect sizes for the residues on , respectively.
We assess the significance of the improvement in model fit between the full model and reduced model with the multivariate analysis of variance test for quantitative traits. As spurious associations again arise when the frequencies of the tested alleles are relatively low7, we recommend performing permutation analyses to confirm the calibration of the test statistics.
By using this multitrait framework, we can assess any combination of multiple phenotypes. One potential application is to investigate multiple disease phenotypes by using rich phenotype data in biobanks. This framework could disentangle pleiotropic HLA alleles that simultaneously affect a spectrum of diseases of interest. Another interesting application might be using multiple molecular phenotypes such as expression of multiple genes or proteins, and a combination of multiple modalities (e.g., gene expression and chromatin accessibility) to determine how variation in the HLA alleles affects transcription and translation, or gene expression and epigenetic changes. We can also assess these phenotypes across multiple cell types (e.g., expression of a gene in T cells, B cells, monocytes, etc.) to investigate the effect of the HLA alleles on gene expression across multiple cell types.
Concluding remarks
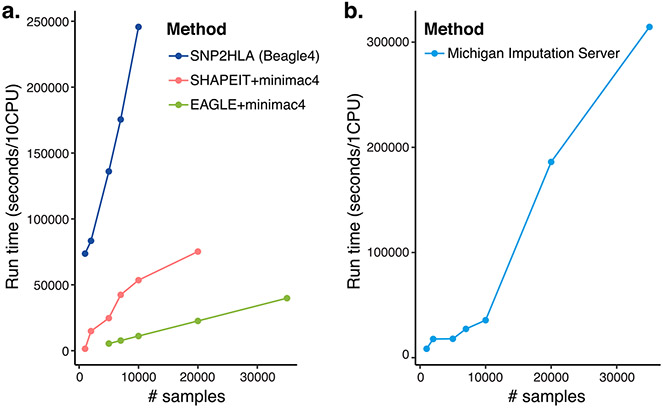
Given the increasing number of associations between the HLA region and human complex traits that have been identified through large-scale GWAS, accurate imputation and fine mapping of the causal HLA alleles and amino acids will continue to be important as the data size continues to grow. We present a strategy that can lead investigators to fine-mapped alleles. By leveraging HLA fine-mapped alleles with the variants outside of MHC region, it may be possible to construct an efficient genetic risk score to stratify people on the basis of their genetic risk for those diseases. We have publicized this imputation pipeline through the user-friendly MIS, which hosts the HLA reference panel representing multiple populations and enables web-based automatic HLA imputation for global cohorts. Another advantage of the implementation using Minimac4 (ref. 22) is the computational efficiency: HLA imputation of a cohort of millions of individuals is computationally scalable by EAGLE and minimac4 (for example, for a cohort of size 20,000, HLA imputation runs in 6 h with 10 central processing units; for benchmarking with different algorithms and platforms, see Extended Data Fig. 4). We hope this tutorial will empower the field of statistical genetics to more comprehensively define the effect of HLA variation in a spectrum of human diseases.
Despite the well-established performance of our approach, we can still improve our HLA imputation reference panel further. First, we note that the currently available HLA reference panels are still underrepresented for African populations and South Asian populations. We need to expand the reference panel to better represent global populations. Emerging biobanks for these populations87,88 could be a potential resource to this end. Similarly, the scope of genes included in the panel can be expanded to include, for example, nonclassical HLA genes (e.g., HLA-DO and HLA-DM) and C4 copy number. Second, the imputation accuracy is currently satisfactory in association testing but not yet as high as the gold-standard HLA typing. We aim to further improve the accuracy by updating the HLA calls and scaffold variants used in the reference panel as well as improving the imputation algorithms.
While fine mapping of HLA alleles has provided deeper insights into disease pathogenesis, we need more mechanistic and structural understanding of how these alleles contribute to disease biology. Why do certain HLA alleles cause a diverse spectrum of diseases? How do those alleles characterize our inherited composition of T-cell repertoires? What are the auto-antigens that are being presented by those alleles? Recent advances in experimental and computational modeling of protein structures and complexes89,90 offer promise. We need both experimental and computational approaches to answer all these important questions.
Extended Data
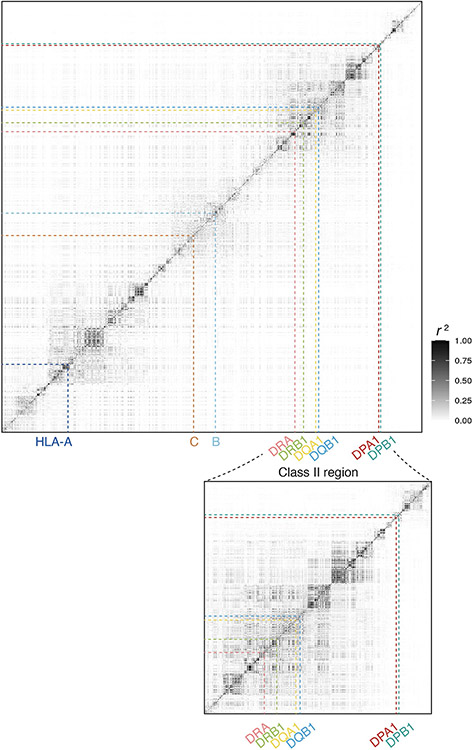
Extended Data Fig.1 ∣. The linkage disequilibrium (LD) patterns across the extended MHC region.
A heatmap of LD r2 for pairwise variants across the extended MHC region. We used biallelic markers in our HLA reference panel within European populations and calculated LD r2 values for exhaustive pairs of these variants. The variants are ordered (both on x-axis and y-axis) and annotated by HLA gene names (on x-axis) based on their genomic coordinates on chromosome 6. The bottom plot shows the detailed LD pattern in the class II region.
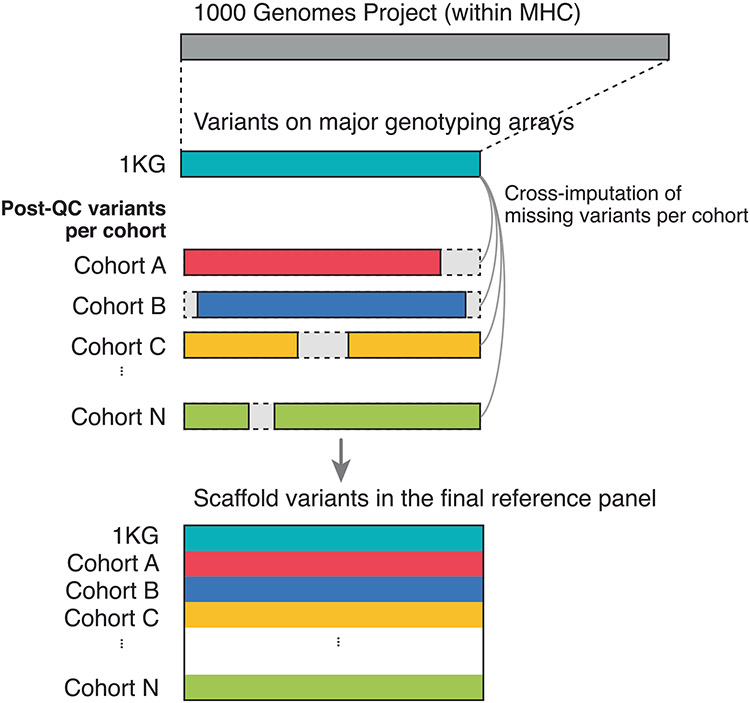
Extended Data Fig.2 ∣. Schematic illustration of method used to construct scaffold variants within multi-ancestry HLA reference panel.
We extracted SNP variants within MHC region in 1000 Genomes Project (1KG) samples. We only retained variants that were included in major genotyping arrays (Illumina Multi-Ethnic Genotyping Array, Global Screening Array, OmniExpressExome, and Human Core Exome), colored in teal. We then quality controlled each of the participating cohorts’ MHC SNPs separately, retained overlapping variants with selected SNPs in 1KG, and cross-imputed each cohorťs missing variants by using 1KG genotypes. We finally concatenate all cohorts together to construct scaffold variants for multi-ancestry reference panel.
Extended Data Fig.3 ∣. Michigan Imputation Server.
Example usage of Michigan Imputation Server for HLA imputation at https://imputationserver.sph.umich.edu/index.html.
Extended Data Fig.4 ∣. The runtime benchmark for HLA imputation using different platforms.
a. For SNP2HLA, we used BEAGLE4 for phasing and imputation algorithm (Luo et al. Nat Genet. 2021) with using 10 CPUs. For Minmac4, we used SHAPEIT2 as phasing algorithm with samples <10,000 and EAGLE2 as phasing algorithm with samples > 5,000 as we described in the manuscript both with using 10 CPUs. b. For Michigan Imputation Server, we uploaded the unphased genotype data and standard imputation pipeline was performed with default setting (with 1CPU).
Supplementary Material
Acknowledgements
This work is supported in part by funding from the National Institutes of Health (R01AR063759, U01HG012009, UC2AR081023). S.Sakaue was in part supported by the Manabe Scholarship Grant for Allergic and Rheumatic Diseases, the Uehara Memorial Foundation, and the Osamu Hayaishi Memorial Scholarship. J.B.K. was supported by NIH/NIGMS T32GM007753 and NIH/NIAID F30AI172238. A.J.D. was funded by NIH/NIDDK T32DK007028. T.L.L. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 437857095. Y.O. is supported by AMED (JP22km0405211, JP22km0405217).
Footnotes
Competing interests
B.H. is a CTO of Genealogy Inc. T.L.L. is a co-inventor on a patent application for using HLA evolutionary divergence in predicting cancer immunotherapy success. S.R. is a founder for Mestag, Inc, a scientific advisor for Sonoma, Jannsen and Pfizer, and serves as a consultant for Sanofi and Abbvie.
Extended data is available for this paper at https://doi.org/10.1038/s41596-023-00853-4.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41596-023-00853-4.
Data availability
We have summarized the availability of HLA imputation reference panels in Table 2. Our HLA imputation pipeline using a multi-ancestry HLA reference panel is publicly available at the MIS (https://imputation-server.sph.umich.edu/index.html).
Code availability
The computational scripts and instructions for their usage related to this tutorial are available at https://github.com/immunogenomics/HLA_analyses_tutorial (https://doi.org/10.5281/zenodo.7373439).
References
- 1.Trowsdale J & Knight JC Major histocompatibility complex genomics and human disease. Ann. Rev. Genomics Hum. Genet 14, 301–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amiel J. in Histocompatibility Testing (ed. Teraski PI) 79–81 (Munksgaard, 1967). [Google Scholar]
- 3.Murphy K & Weaver C Janeway’s immunology. America 1–277 (2017). [Google Scholar]
- 4.Dendrou CA, Petersen J, Rossjohn J & Fugger L HLA variation and disease. Nat. Rev. Immunol 18, 325–339 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Murphy K Kenneth M & Weaver C Janeway’s Immunobiology (Garland Science, 2016). [Google Scholar]
- 6.Scally SW et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med 210, 2569–2582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishigaki K. et al. HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat. Genet 54, 393–402 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGonagle D, Aydin SZ, Gül A, Mahr A & Direskeneli H ‘MHC-I-opathy’-unified concept for spondyloarthritis and Behçet disease. Nat. Rev. Rheumatol 11, 731–740 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Sekar A. et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery RA, Tatapudi VS, Leffell MS & Zachary AA HLA in transplantation. Nat. Rev. Nephrol 14, 558–570 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Fleischhauer K, Zino E, Bordignon C & Benazzi E Complete generic and extensive fine-specificity typing of the HLA-B locus by the PCR-SSOP method. Tissue Antigens 46, 281–292 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Cereb N, Maye P, Lee S, Kong Y & Yang SY Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens 45, 1–11 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens 80, 1–11 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Cereb N, Kim HR, Ryu J & Yang SY Advances in DNA sequencing technologies for high resolution HLA typing. Hum. Immunol 76, 923–927 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Smith AG et al. Comparison of sequence-specific oligonucleotide probe vs next generation sequencing for HLA-A, B, C, DRB1, DRB3/B4/B5, DQA1, DQB1, DPA1, and DPB1 typing: toward single-pass high-resolution HLA typing in support of solid organ and hematopoietic cell transplant programs. HLA 94, 296–306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schöfl G. et al. 2.7 million samples genotyped for HLA by next generation sequencing: lessons learned. BMC Genomics 18, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao Y. et al. High-sensitivity HLA typing by saturated tiling capture sequencing (STC-Seq). BMC Genomics 19, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8, e64683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dilthey AT, Moutsianas L, Leslie S & McVean G HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27, 968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X. et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 14, 192–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y. et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet 53, 1504–1516 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das S. et al. Next-generation genotype imputation service and methods. Nat. Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raychaudhuri S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet 44, 291–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson J. et al. IPD-IMGT/HLA database. Nucleic Acids Res. 48, D948–D955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh SGE et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75, 291 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsh SGE et al. An update to HLA nomenclature, 2010. Bone Marrow Transplant. 45, 846–848 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Marchini J & Howie B Genotype imputation for genome-wide association studies. Nat. Rev. Genet 11, 499–511 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Dilthey AT et al. High-accuracy HLA type inference from whole-genome sequencing data using population reference graphs. PLoS Comput. Biol 12, e1005151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dilthey AT et al. HLA*LA—HLA typing from linearly projected graph alignments. Bioinformatics 35, 4394–4396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen JJ et al. HLA-IMPUTER: an easy to use web application for HLA imputation and association analysis using population-specific reference panels. Bioinformatics 35, 1244–1246 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Maiers M. et al. GRIMM: GRaph IMputation and matching for HLA genotypes. Bioinformatics 35, 3520–3523 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Breiman L. Bagging predictors. Mach. Learn 24, 123–140 (1996). [Google Scholar]
- 33.Dilthey A, Cox C, Iqbal Z, Nelson MR & McVean G Improved genome inference in the MHC using a population reference graph. Nat. Genet 47, 682–688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirata J. et al. Genetic and phenotypic landscape of the major histocompatibilty complex region in the Japanese population. Nat. Genet 51, 470–480 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Hu T, Chitnis N, Monos D & Dinh A Next-generation sequencing technologies: an overview. Hum. Immunol 82, 801–811 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Hosomichi K, Jinam TA, Mitsunaga S, Nakaoka H & Inoue I Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics 14, 1–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbs RA et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Browning SR & Browning BL Haplotype phasing: existing methods and new developments. Nat. Rev. Genet 12, 703–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verlouw JAM et al. A comparison of genotyping arrays. Eur. J. Hum. Genet 29, 1611 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vince N. et al. SNP-HLA Reference Consortium (SHLARC): HLA and SNP data sharing for promoting MHC-centric analyses in genomics. Genet. Epidemiol 44, 733–740 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klareskog L, Catrina AI & Paget S Rheumatoid arthritis. Lancet 373, 659–672 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Padyukov L. et al. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis 70, 259–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bycroft C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu P. et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med. Inform https://medinform.jmir.org/2019/4/e14325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez-Arcelus M. et al. Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. Nat. Genet 52, 247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Antonio M. et al. Systematic genetic analysis of the MHC region reveals mechanistic underpinnings of HLA type associations with disease. eLife 8, e48476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aguiar VRC, César J, Delaneau O, Dermitzakis ET & Meyer D Expression estimation and eQTL mapping for HLA genes with a personalized pipeline. PLoS Genet. 15, e1008091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoeckius M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson CA et al. Data quality control in genetic case-control association studies. Nat. Protoc 5, 1564–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uffelmann E. et al. Genome-wide association studies. Nat. Rev. Methods Prim 1, 1–21 (2021). [Google Scholar]
- 51.Gilly A. et al. Very low-depth whole-genome sequencing in complex trait association studies. Bioinformatics 35, 2555–2561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilly A. et al. Very low-depth sequencing in a founder population identifies a cardioprotective APOC3 signal missed by genome-wide imputation. Hum. Mol. Genet 25, 2360–2365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin AR et al. Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations. Am. J. Hum. Genet 108, 656–668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marees AT et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int. J. Methods Psychiatr. Res 27, e1608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hinrichs AS et al. The UCSC genome browser database: update 2006. Nucleic Acids Res. 34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gomes I. et al. Hardy–Weinberg quality control. Ann. Hum. Genet 63, 535–538 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Hosking L. et al. Detection of genotyping errors by Hardy–Weinberg equilibrium testing. Eur. J. Hum. Genet 12, 395–399 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Wittke-Thompson JK, Pluzhnikov A & Cox NJ Rational inferences about departures from Hardy–Weinberg equilibrium. Am. J. Hum. Genet 76, 967 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galinsky KJ et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet 98, 456–472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cook S. et al. Accurate imputation of human leukocyte antigens with CookHLA. Nat. Commun 12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Browning SR & Browning BL Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delaneau O, Zagury JF & Marchini J Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Loh P-R et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gourraud PA et al. HLA diversity in the 1000 Genomes Dataset. PLoS One 9, e97282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abi-Rached L. et al. Immune diversity sheds light on missing variation in worldwide genetic diversity panels. PLoS One 13, e0206512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet 50, 1335–1341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loh PR et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wordsworth P. et al. HLA heterozygosity contributes to susceptibility to rheumatoid arthritis. Am. J. Hum. Genet 51, 585 (1992). [PMC free article] [PubMed] [Google Scholar]
- 71.Koeleman BPC et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun. 5, 381–388 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Thomson G. et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens 70, 110–127 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Woelfing B, Traulsen A, Milinski M & Boehm T Does intra-individual major histocompatibility complex diversity keep a golden mean? Philos. Trans. R. Soc. Lond. B Biol. Sci 364, 117–128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lipsitch M, Bergstrom CT & Antia R Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med. Genet 4, 2 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai S & Santamaria P MHC class II polymorphisms, autoreactive T-cells, and autoimmunity. Front. Immunol 4, 321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goyette P. et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet 47, 172–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lenz TL et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet 47, 1085–1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arora J. et al. HLA heterozygote advantage against HIV-1 is driven by quantitative and qualitative differences in HLA allele-specific peptide presentation. Mol. Biol. Evol 37, 639–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet 47, 898–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reynolds EGM et al. Non-additive association analysis using proxy phenotypes identifies novel cattle syndromes. Nat. Genet 53, 949–954 (2021). [DOI] [PubMed] [Google Scholar]
- 81.Segal MR, Cummings MP & Hubbard AE Relating amino acid sequence to phenotype: analysis of peptide-binding data. Biometrics 57, 632–643 (2001). [DOI] [PubMed] [Google Scholar]
- 82.Chen B. et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol 37, 1332–1343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pierini F & Lenz TL Divergent allele advantage at human MHC genes: signatures of past and ongoing selection. Mol. Biol. Evol 35, 2145–2158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wakeland EK et al. Ancestral polymorphisms of MHC class II genes: divergent allele advantage. Immunol. Res 9, 115–122 (1990). [DOI] [PubMed] [Google Scholar]
- 85.Radwan J, Babik W, Kaufman J, Lenz TL & Winternitz J Advances in the evolutionary understanding of MHC polymorphism. Trends Genet. 36, 298–311 (2020). [DOI] [PubMed] [Google Scholar]
- 86.Chowell D. et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med 25, 1715–1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choudhury A. et al. High-depth African genomes inform human migration and health. Nature 586, 741–748 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wall JD et al. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576, 106–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakane T. et al. Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jumper J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pillai NE et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum. Mol. Genet 23, 4443–4451 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Okada Y. et al. Construction of a population-specific HLA imputation reference panel and its application to Graves’ disease risk in Japanese. Nat. Genet 47, 798–802 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Zhou F. et al. Deep sequencing of the MHC region in the Chinese population contributes to studies of complex disease. Nat. Genet 48, 740–746 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Kim K, Bang SY, Lee HS & Bae SC Construction and application of a Korean reference panel for imputing classical alleles and amino acids of human leukocyte antigen genes. PLoS One 9, e112546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Degenhardt F. et al. Construction and benchmarking of a multi-ethnic reference panel for the imputation of HLA class I and II alleles. Hum. Mol. Genet 28, 2078–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sakaue S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet 53, 1415–1424 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have summarized the availability of HLA imputation reference panels in Table 2. Our HLA imputation pipeline using a multi-ancestry HLA reference panel is publicly available at the MIS (https://imputation-server.sph.umich.edu/index.html).
The computational scripts and instructions for their usage related to this tutorial are available at https://github.com/immunogenomics/HLA_analyses_tutorial (https://doi.org/10.5281/zenodo.7373439).