Abstract
A large gene cluster associated with the biosynthesis of the serotype-specific polysaccharide antigen (SPA) of Actinobacillus actinomycetemcomitans Y4 (serotype b) was cloned and characterized. Western blot analysis showed that Escherichia coli DH5α, containing a plasmid carrying this cluster, produced a polysaccharide which reacted with a monoclonal antibody directed against the SPA of A. actinomycetemcomitans Y4. High-performance liquid chromatography analysis indicated that the polysaccharide produced by an E. coli transformant, as well as A. actinomycetemcomitans Y4 SPA, was composed of rhamnose and fucose. Furthermore, using various derivatives of the plasmid, we demonstrated that the cloned 13-kb BssHII-BspHI fragment was indispensable for SPA synthesis in E. coli DH5α. The 24,909-bp nucleotide sequence, which included this fragment and its flanking regions, was determined. In the sequenced area, 24 open reading frames (ORFs) with the same orientation were found. Most of these were located sequentially within a short distance of each other. Many of the deduced amino acid sequences were similar to the gene products of the polysaccharide synthetic genes of other bacteria. The average G+C content (37.7%) of all 24 ORFs in the sequenced area was lower than that (45.6%) of the whole chromosome of A. actinomycetemcomitans Y4. It is noteworthy the average G+C content of the nine ORFs in the 8.5-kb central region of the 13-kb BssHII-BspHI fragment indispensable for SPA synthesis in E. coli was found to be especially low (27.0%).
Actinobacillus actinomycetemcomitans is a nonmotile, gram-negative, capnophilic, fermentative coccobacillus which has previously been implicated in the etiology and pathogenesis of localized juvenile periodontitis (3, 37, 55), adult periodontitis (36), and severe nonoral human infections (14). A. actinomycetemcomitans strains isolated from the human oral cavity are divided into five serotypes, a, b, c, d, and e (10, 30, 56). Of these serotypes, serotype b is most frequently isolated from subjects with localized juvenile periodontitis (3, 56) who exhibit elevated serum antibody levels to serotype b-specific polysaccharide antigen (SPA) of A. actinomycetemcomitans (5, 35). SPA has previously been shown to be one of the immunodominant antigens in this organism (5, 24). Page et al. (24) and Perry et al. (26) claimed that SPA is a constituent of the polysaccharide region of lipopolysaccharide.
We reported previously that the SPA of A. actinomycetemcomitans Y4 is a capsular polysaccharide-like antigen consisting of two deoxyhexoses, d-fucose and l-rhamnose (1). We recently demonstrated that this antigen plays an important role in resistance to phagocytosis and killing by human polymorphonuclear leukocytes (51). Moreover, SPA has the ability to induce the release of interleukin-1 by murine macrophages (44) and to promote osteoclast-like cell formation in mouse marrow cultures (23). Little is known, however, about the structural genes responsible for SPA biosynthesis in A. actinomycetemcomitans.
In general, the clustering of exopolysaccharide synthetic genes is a common feature of almost all bacterial polysaccharide loci studied so far. Indeed, it has previously been shown that the exopolysaccharide synthetic genes of Salmonella enterica (13), Shigella flexneri (27), Erwinia amylovora (4), Escherichia coli K1, K5, K7, and K-12 (29), Haemophilus influenzae (17), Klebsiella pneumoniae (2), and Neisseria meningitidis (9) are clustered on segments of DNA from 10 to 25 kb in length. In gram-negative bacteria, there appears to be a considerable degree of sequence homology and a conserved genetic organization within these loci. Therefore, it may be that the SPA biosynthetic genes of A. actinomycetemcomitans are clustered in the same fashion as are the capsular polysaccharide biosynthetic genes of other bacteria and that they are similar to genes responsible for exopolysaccharide synthesis in other organisms. On the basis of such genetic predictions, we tried to clone and express the A. actinomycetemcomitans SPA gene cluster in E. coli DH5α. Here, we report the isolation and characterization of a DNA fragment which contains the SPA biosynthetic genes of A. actinomycetemcomitans and its flanking regions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A. actinomycetemcomitans Y4 (serotype b) was obtained from Y. Yamamoto (Sunstar Corp., Osaka, Japan). A. actinomycetemcomitans Y4 was grown in Trypticase soy broth (BBL Microbiology Systems, Cockeysville, Md.) containing 0.6% yeast extract (Difco Laboratories, Detroit, Mich.) and 0.04% sodium bicarbonate at 37°C in a 5% CO2 atmosphere (39). E. coli DH5α [supE44 ΔlacU169 (ς80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (31) was used in DNA manipulations. E. coli DH5α was grown aerobically in 2× TY broth at 37°C (31). When required, antibiotics were added at concentrations of 50 μg per ml for ampicillin and 20 μg per ml for chloramphenicol.
MAb.
Monoclonal antibodies (MAb) directed against A. actinomycetemcomitans Y4 SPA (MAb S5) and lipopolysaccharide (LPS) (MAb L2) were prepared and purified by the method of Koga et al. (15).
DNA manipulations.
DNA fragment preparation, agarose gel electrophoresis, DNA labeling, ligation, bacterial transformation, and colony immunoblotting were performed by the methods of Sambrook et al. (31).
Southern hybridization and colony hybridization.
Southern hybridization and colony hybridization were performed overnight under stringent conditions (hybridization fluid with 50% formamide at 25°C). Posthybridization washes were performed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate (SDS) at room temperature for 15 min per wash and twice with 0.1× SSC–0.1% (wt/vol) SDS at room temperature for 15 min per wash. All other procedures that involved Southern hybridization and colony hybridization were performed by the methods of Sambrook et al. (31).
Cloning of the SPA gene cluster.
To detect the gene homologous to the rfbA gene of S. flexneri (one of the S. flexneri rhamnose biosynthetic genes) (27), we constructed a digoxigenin (DIG)-labeled PCR probe with a nonradioactive DIG DNA labeling and detection kit (Boehringer GmbH, Mannheim, Germany) in accordance with the instructions of the supplier. The probe was amplified by PCR with pSBA85, which contains the rfbA gene in pUC18 (52), and with primers synthesized by using published sequences (27) (forward primer, 5′-ATTCTGGCTGGTGGTTCCGGC-3′, and reverse primer, 5′-CAGCAGATACTGACCATAAGC-3′). To construct a cosmid gene bank of A. actinomycetemcomitans Y4, chromosomal DNA from this organism was completely digested with SalI. Cosmid vector pMBLcos (22) was digested with the same enzyme. Equal molar amounts of vector and insert fragments were ligated (T4 DNA ligase), packaged into bacteriophage λ (Gigapack II XL; Stratagene, La Jolla, Calif.), and transfected into E. coli DH5α. The A. actinomycetemcomitans clone bank was screened for the gene which hybridized with the rfbA gene-specific DIG-labeled PCR probe by colony hybridization. Confirmation of the reactivity of screened clones with MAb S5 was made by colony immunoblotting (31).
DNA sequencing and data analysis.
The SPA gene cluster was recloned into seven subclones (data not shown). Unidirectional deletions were generated by using exonuclease III and mung bean nuclease (Takara Shuzo Co., Kyoto, Japan). Nucleotide sequencing was performed by the dideoxy chain termination technique of Sanger et al. (32) with a Taq dye primer cycle sequencing kit and an ABI 373A DNA sequencer (Perkin-Elmer Japan, Urayasu, Japan). The nucleotide sequence was assembled with the DNASIS sequence analysis program (Hitachi Software Engineering Co., Yokohama, Japan). Database searching was performed with the FASTA program (19) of the DDBJ e-mail server in the National Institute of Genetics, Mishima, Japan.
Immunodiffusion analysis.
Cell suspensions of A. actinomycetemcomitans Y4 and E. coli DH5α transformants in phosphate-buffered saline (0.12 M NaCl, 0.01 M Na2HPO4, 5 mM KH2PO4 [pH 7.5]) were autoclaved at 121°C for 20 min and centrifuged at 4°C. After centrifugation, supernatants were collected and used as autoclaved extracts. Immunodiffusion analysis was carried out in 1.0% agarose (Gibco-BRL, Gaithersburg, Md.) in phosphate-buffered saline.
Western blotting (immunoblotting).
Autoclaved extracts from A. actinomycetemcomitans Y4 and E. coli DH5α transformants were mixed with an equal volume of 0.2 mM Tris-HCl buffer (pH 6.8) containing 2% (wt/vol) SDS, 2% (vol/vol) 2-mercaptoethanol, and 40% (vol/vol) glycerol and heated at 100°C for 5 min. The mixtures were electrophoresed at 25 mA per gel at room temperature for 1.5 h on 12.5% (wt/vol) resolving and 3% (wt/vol) stacking polyacrylamide gels (90 by 80 by 1 mm) containing 0.1% (wt/vol) SDS and subjected to immunoblot analysis by the method of Towbin et al. (47). After blocking with Tris-buffered saline (0.01 M Tris-HCl, 0.15 M NaCl [pH 7.5]) containing 3% (wt/vol) skim milk, blots were treated with MAb S5 or L2 at a 1:400 dilution in TBST-BSA (Tris-buffered saline containing 0.05% [vol/vol] Tween 20 and 1% [wt/vol] bovine serum albumin). The antibody bound to immobilized replica antigens on blots was detected by a solid-phase immunoassay with alkaline phosphatase-conjugated goat anti-mouse immunoglobulins (Zymed Laboratories, South San Francisco, Calif.) diluted 1:1,000 in TBST-BSA.
Sugar composition analysis.
Component sugars in the partially purified polysaccharides from A. actinomycetemcomitans Y4 and E. coli DH5α transformants were analyzed by high-performance liquid chromatography (HPLC) with fluorescence labeling. Lyophilized-cell suspension (80 mg/ml) in DNase buffer (0.1 M sodium acetate, 5 mM MgSO4 [pH 5.0]) was autoclaved at 121°C for 20 min. After being autoclaved, the suspension was cooled and centrifuged. The supernatant was treated with DNase (10 μg/ml) and RNase (10 μg/ml) at 37°C for 3 h and successively extracted with phenol-chloroform and chloroform. Low-molecular-weight molecules were removed from the extract in a NAP-10 column (Pharmacia Biotech Inc., Uppsala, Sweden), and the column eluate containing partially purified high-molecular-weight molecules was evaporated. The pellet was dissolved in 10 μl of distilled water, and 40 μl of 5 M trifluoroacetic acid was added. After the tube was sealed under vacuum, the mixture was heated at 100°C for 3 h and dried at 50°C. Free amino groups were acetylated by adding 50 μl of a 3:6:2 mixture of pyridine-methanol-water and 2 μl of acetic anhydride. The solution was left standing for 30 min at room temperature and dried at 35°C. Sugar components in the hydrolyzed and acetylated solution were coupled with 2-aminopyridine, and the pyridylamino sugars were analyzed by HPLC with an anion-exchange column by the method of Suzuki et al. (43). After the hydrolyzed solution had been dried at 50°C, 10 μl of a coupling reagent (0.67 g of 2-aminopyridine per ml in acetic acid) was added. The mixture was heated at 90°C for 20 min, and excess reagents were removed by evaporation. Then 10 μl of a reducing reagent (60 mg of borane-dimethylamine complex per ml in acetic acid) was added. The mixture was reduced at 90°C for 35 min and dried under a stream of nitrogen gas at 50°C for 10 min. The dried sample was analyzed by HPLC with a PALPAK type A column (Takara Shuzo Co.) and a mixture of 0.7 M boric acid (pH 9.0) and acetonitrile (9:1) at 0.3 ml/min. An excitation wavelength of 310 nm and an emission wavelength of 380 nm were used to detect pyridylamino sugars.
Nucleotide sequence accession number.
The sequence reported here was submitted to the EMBL and GenBank databases through DDBJ and assigned accession no. AB002668.
RESULTS
Isolation of plasmids carrying an SPA gene cluster.
As SPA of A. actinomycetemcomitans consists of two deoxyhexoses, d-fucose and l-rhamnose (1), we predicted that A. actinomycetemcomitans Y4 chromosomal DNA includes the rhamnose biosynthetic genes. We used a fragment of the S. flexneri rfbA gene, encoding a glucose-1-phosphate-tymidylyltransferase (one of four dTDP-rhamnose biosynthetic enzymes) (27), as a probe for Southern hybridization analysis of chromosomal DNA of A. actinomycetemcomitans Y4. Southern blotting with the S. flexneri rfbA gene-specific probe suggested that a 38-kb chromosomal SalI fragment of A. actinomycetemcomitans Y4 contained an rfbA homolog (data not shown). Based on this result, a cosmid gene bank of A. actinomycetemcomitans Y4 was constructed with complete SalI digests of chromosomal DNA from the organism and intermediate-copy-number cosmid vector pMBLcos (22). This cosmid vector was chosen in order to avoid the instability of high-copy-number plasmids containing large inserts in E. coli DH5α. Two colonies hybridized with the S. flexneri rfbA gene-specific probe and were isolated from 800 colonies in the library. The production of SPA in these colonies was confirmed by colony immunoblotting with MAb S5. One colony carried a 42-kb plasmid, designated pARF100, whereas the other colony carried another plasmid, designated pARF200, which contained the same fragment in the opposite orientation to that of pARF100. The promoter utilized to express the genes responsible for SPA synthesis seems to be located on the cloned fragment since both E. coli DH5α containing pARF100 and E. coli DH5α containing pARF200 produced SPA.
Localization of the region indispensable for SPA synthesis in E. coli.
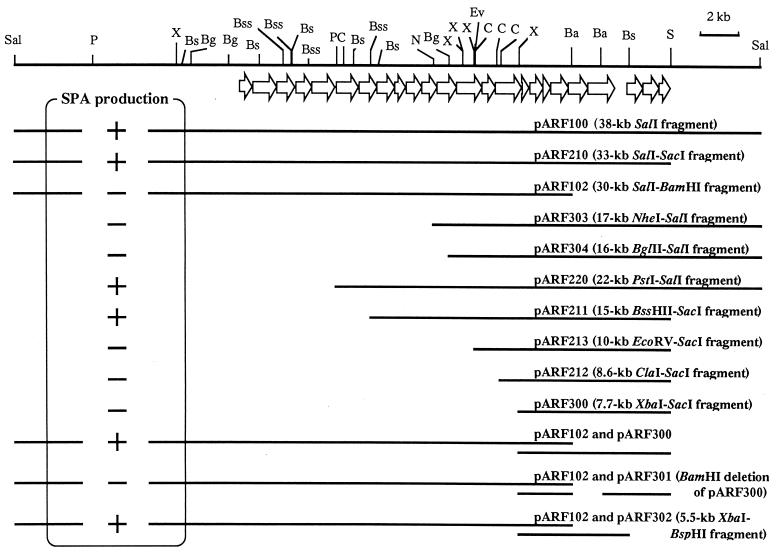
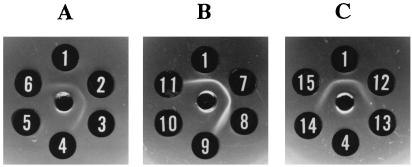
To locate the genes responsible for SPA synthesis, a restriction map of the 38-kb fragment in pARF100 was constructed with several restriction endonucleases and deletion analysis of pARF100 was carried out. E. coli DH5α was transformed with nine deletion derivatives of pARF100 (pARF210, pARF102, pARF303, pARF304, pARF220, pARF211, pARF212, pARF213, and pARF300) (Fig. 1). The genes on all these plasmids, except for pARF102, were expressed under lac promoter control, whereas the expression of genes on pARF102 seems to be controlled by the same promoter as that on pARF100 or pARF200. Only four of these nine transformants (pARF100, pARF210, pARF220, and pARF211) produced polysaccharides which reacted with MAb S5 (Fig. 1). The production of A. actinomycetemcomitans SPA in each transformant was determined by immunodiffusion analysis (Fig. 2). Moreover, E. coli DH5α containing both pARF102 and pARF300 produced SPA. To ascertain that the 3′ end of this region is indispensable for SPA synthesis in E. coli DH5α, E. coli DH5α containing pARF102 was transformed with two deletion derivatives of pARF300 (pARF301 and pARF302). pARF300, pARF301, and pARF302 had the ColE1 origin and the chloramphenicol resistance gene, whereas pARF102 had the ColE1-compatible origin p15A and the ampicillin resistance gene. E. coli DH5α containing both pARF102 and pARF302 produced SPA, but E. coli DH5α containing both pARF102 and pARF301 did not (Fig. 1). These results indicate that the region involved in SPA synthesis in E. coli DH5α lies within the 13-kb BssHII-BspHI fragment combined with pARF211.
FIG. 1.
Restriction map and deletion analysis of pARF100. A linearized restriction map of the chromosomal 38-kb SalI fragment containing the SPA gene cluster is shown. Open arrows indicate the positions of ORFs. Horizontal lines below the map show the DNA inserts carried by the indicated recombinant plasmids. A. actinomycetemcomitans SPA production in E. coli DH5α is shown to the left of each fragment as follows: +, positive production of SPA; −, undetectable production of SPA. Restriction enzyme abbreviations: Ba, BamHI; Bg, BglII; Bs, BspHI; Bss, BssHII; C, ClaI; Ev, EcoRV; N, NheI; P, PstI; S, SacI; Sal, SalI; X, XbaI.
FIG. 2.
Immunodiffusion reactions of MAb S5 with autoclaved extracts prepared from A. actinomycetemcomitans Y4 and E. coli transformants. Center wells contained MAb S5. Outer wells contained autoclaved extracts from A. actinomycetemcomitans Y4 (well 1), E. coli containing pARF100 (well 2), E. coli containing pARF210 (well 3), E. coli containing pARF102 (well 4), E. coli containing pARF304 (well 5), E. coli containing pARF303 (well 6), E. coli containing pARF220 (well 7), E. coli containing pARF211 (well 8), E. coli containing pMBLcos (well 9), E. coli containing pARF213 (well 10), E. coli containing pARF212 (well 11), E. coli containing both pARF102 and pARF300 (well 12), E. coli containing pARF300 (well 13), E. coli containing both pARF102 and pARF301 (well 14), and E. coli containing both pARF102 and pARF302 (well 15).
DNA sequencing and computational analysis of the SPA gene cluster.
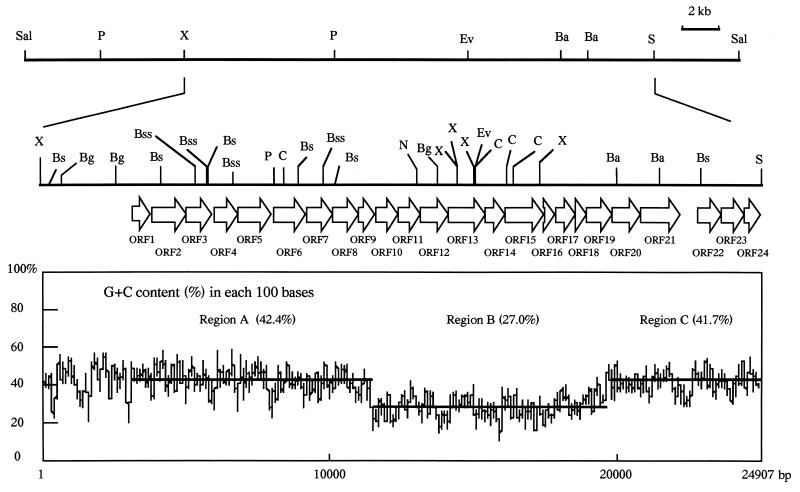
To analyze the genes required for SPA synthesis, seven genomic fragments were subcloned from the 25-kb XbaI-SacI fragment into pMCL200, pMCL210 (22), and pHSG399 (45) and subsequently sequenced. (The sequence data were deposited in international DNA databases [EMBL and GenBank] through DDBJ.) Twenty-four possible open reading frames (ORFs) were identified in the sequence area (Fig. 3). All of these ORFs had the same orientation. With the exception of ORF22, all ORFs were located one after the other, separated by short distances. No ORF in the opposite strand of this fragment was more than 300 bp in length.
FIG. 3.
ORFs in A. actinomycetemcomitans SPA region and its flanking regions and G+C contents of the fragments responsible for A. actinomycetemcomitans SPA. ORFs are represented by open arrows. The percent G+C content in the area sequenced was calculated for each block of 100 nucleotide residues and is shown below the restriction map. Three hypothetical segments are shown as bars over the plot of the SPA region. Parenthetical data are the average G+C contents of regions A, B, and C. The restriction enzyme abbreviations used are the same as those identified in the legend to Fig. 1.
Possible Shine-Dalgarno sequences (34) were identified just upstream of the potential initiation codons of all 24 ORFs. Of the putative start codons for all these ORFs, eight (ORF2, ORF7, ORF8, ORF12, ORF13, ORF18, ORF19, and ORF23) overlapped the stop codon of the previous ORF and nine (ORF3, ORF5, ORF9, ORF11, ORF14, ORF15, ORF16, ORF17, and ORF20) were located within 10 bases of the stop codon of the previous ORF. However, ORF22 was separated from ORF21 by a much greater distance (541 bp). Although GTG was used as the start codon once (ORF4), ATG was used the other 23 times (all other ORFs). The usage of terminator codons agreed with the usual E. coli preferences. TAA was used 18 times as a single stop codon, and TAG was used 5 times; however, TGA was used only once. The average G+C content of all the sequenced regions was 37.7%. An especially low G+C content (27.0%) was observed in the region containing ORF10 through ORF19 (Fig. 3).
The amino acid sequence of each putative ORF was deduced, and a homology search was made. Sixteen amino acid sequences with considerable similarities to previously reported sequences were detected, and most of them (ORF3, ORF4, ORF5, ORF6, ORF7, ORF8, ORF9, ORF10, ORF11, ORF12, ORF13, ORF19, ORF20, ORF21, and ORF24) were similar to the products of polysaccharide synthetic genes of other bacteria. On the other hand, the deduced amino acid sequences of proteins encoded by eight ORFs (ORF1, ORF2, ORF14, ORF15, ORF16, ORF17, ORF18, and ORF23) showed identities of <10% with proteins previously identified (Table 1). It is very interesting that the amino acid sequence of ORF12 showed considerable similarity to that of ORF20 (23.7%). The former showed 26.6% identity to the amino acid sequence of a rhamnosyltransferase from Salmonella enterica (13, 20), whereas the latter showed 54.4% identity (Fig. 4).
TABLE 1.
Profiles of ORFs in the region responsible for SPA synthesis and flanking regions
| Potential ORF | No. of amino acids in protein | G+C content (%) | Homologous gene | Potential function | Bacterium | Protein sequence identity (%) | Reference |
|---|---|---|---|---|---|---|---|
| ORF1 | 256 | 44.4 | —a | ||||
| ORF2 | 398 | 45.1 | — | ||||
| ORF3 | 293 | 42.8 | amsB | Glycosyltransferase | Erwinia amylovora | 22.6 | 4 |
| ORF4 | 267 | 44.3 | amsE | Unknown | Erwinia amylovora | 44.8 | 4 |
| ORF5 | 376 | 44.3 | mltB | Lytic transglycosylase | E. coli | 23.5 | 7 |
| ORF6 | 355 | 41.3 | ORF2 (rmlBb) | dTDP-d-glucose-4,6-dehydratase | N. meningitidis | 79.6 | 11 |
| ORF7 | 290 | 41.0 | ORF1 (rmlAb) | Glucose-1-phosphate-thymidylyltransferase | N. meningitidis | 79.2 | 11 |
| ORF8 | 292 | 43.3 | rfbC (rmlDb) | dTDP-4-keto-l-rhamnose reductase | E. coli | 43.8 | 40 |
| ORF9 | 179 | 38.2 | rfbD (rmlCb) | dTDP-4-keto-6-deoxy-d-glucose-3,5-epimerase | S. flexneri | 58.6 | 27 |
| ORF10 | 263 | 25.2 | tagG | ABC transport protein | B. subtilis | 24.7 | 18 |
| ORF11 | 245 | 31.6 | abcA | ABC transport protein | Aeromonas salmonicida | 26.6 | 6 |
| ORF12 | 323 | 27.3 | rfbN | Rhamnosyltransferase | Salmonella enterica | 26.6 | 13 |
| ORF13 | 418 | 29.3 | rfbH | Unknown | Y. enterocolitica | 23.5 | 57 |
| ORF14 | 230 | 25.1 | — | ||||
| ORF15 | 446 | 26.9 | — | ||||
| ORF16 | 122 | 25.1 | — | ||||
| ORF17 | 234 | 30.2 | — | ||||
| ORF18 | 126 | 26.7 | — | ||||
| ORF19 | 289 | 34.3 | rfbG | Unknown | S. flexneri | 21.0 | 21 |
| ORF20 | 318 | 42.7 | rfbN | Rhamnosyltransferase | Salmonella enterica | 54.4 | 13 |
| ORF21 | 452 | 42.5 | wbaP | Galactosyltransferase | Salmonella enterica | 43.3 | 50 |
| ORF22 | 267 | 43.9 | xth | Exodeoxynuclease | E. coli | 69.7 | 33 |
| ORF23 | 264 | 39.5 | — | ||||
| ORF24 | 180 | 46.4 | rfbB (rmlBb) | dTDP-d-glucose-4,6-dehydratase | Salmonella enterica | 59.6 | 13 |
FIG. 4.
Alignment of ORF20 and ORF12 products with RfbN from Salmonella enterica (13). Amino acids identical in all three sequences are indicated by asterisks. Dashes indicate gaps.
Western blotting analysis.
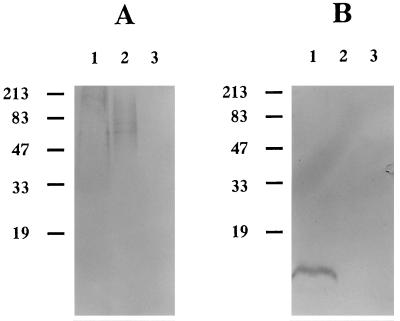
Autoclaved extracts from A. actinomycetemcomitans Y4 and E. coli DH5α containing pARF100 or pMBLcos were analyzed by Western blotting with MAb S5 directed against A. actinomycetemcomitans Y4 SPA or MAb L2 directed against A. actinomycetemcomitans Y4 LPS (Fig. 5). In autoclaved extract from A. actinomycetemcomitans Y4, high- and low-molecular-weight bands reacted with MAb S5 and L2, respectively. On the other hand, in autoclaved extract from E. coli DH5α containing pARF100, a high-molecular-weight band reacted with MAb S5; however, there was no reaction with MAb L2. The size of the MAb S5-reactive polymer in autoclaved extract from A. actinomycetemcomitans Y4 was greater than that in autoclaved extract from E. coli DH5α containing pARF100 (Fig. 5A, lanes 1 and 2, respectively). Autoclaved extract from E. coli DH5α containing pMBLcos did not react with any MAb. These results indicate that E. coli DH5α containing pARF100 has the ability to produce high-molecular-weight SPA but not low-molecular-weight LPS.
FIG. 5.
Western blotting analysis of autoclaved extracts of A. actinomycetemcomitans Y4 and E. coli transformants. (A) Immobilized antigens transferred to a nitrocellulose sheet by an electrophoretic blotting procedure were allowed to react with MAb S5. (B) Antigens were allowed to react with MAb L2. Lanes: 1, A. actinomycetemcomitans Y4 (positive control); 2, E. coli DH5α containing pARF100; 3, E. coli DH5α containing pMBLcos (negative control). The molecular size markers (in kilodaltons) shown on the left of each gel are protein standards.
Sugar compositions of partially purified polysaccharides.
The sugar compositions of partially purified polysaccharides from A. actinomycetemcomitans Y4 and E. coli containing pMBLcos, pARF102, or pARF211 were determined. Rhamnose, fucose, glucose, galactose, N-acetylglucosamine, and some unidentified sugars were detected in the polysaccharide preparations from A. actinomycetemcomitans Y4 and E. coli transformants (Table 2). The hydrolysate of partially purified polysaccharide preparation from E. coli containing either pMBLcos or pARF102 contained no detectable amounts of rhamnose or fucose, whereas that from E. coli containing pARF211 contained detectable amounts of rhamnose and fucose. However, the amounts of rhamnose and fucose from E. coli containing pARF211 were approximately one-fifth of those from A. actinomycetemcomitans Y4.
TABLE 2.
Sugar compositions of polysaccharide preparations from A. actinomycetemcomitans Y4 and E. coli transformants
| Strain | Sugar content (nmol/g [dry wt] of whole cells)a
|
||||
|---|---|---|---|---|---|
| Rhamnose | Fucose | Glucose | Galactose | N-Acetylglucosamine | |
| A. actinomycetemcomitans Y4 | 5,881 ± 263 | 5,460 ± 229 | 728 ± 27 | 644 ± 30 | 127 ± 3 |
| E. coli | |||||
| DH5α(pMBLcos) | NDb | ND | 2,630 ± 53 | 1,435 ± 29 | 467 ± 9 |
| DH5α(pARF102) | ND | ND | 2,723 ± 91 | 3,937 ± 114 | 908 ± 27 |
| DH5α(pARF211) | 916 ± 23 | 902 ± 26 | 2,003 ± 50 | 1,278 ± 33 | 291 ± 7 |
Data are means ± standard deviations of three independent experiments.
ND, not detectable by HPLC.
DISCUSSION
Although we tried to clone the genes responsible for SPA synthesis in A. actinomycetemcomitans Y4, we failed to clone an SPA gene cluster, probably because we used high-copy-number vectors such as Charomid 9-28 and 9-20 (Nippon Gene Co., Ltd., Toyama, Japan). The genes associated with SPA synthesis seemed to be unstable in E. coli DH5α when they were ligated into Charomid 9-28 or 9-20. Therefore, we constructed pMBLcos, which is an intermediate-copy-number cosmid vector based on pACYC177 (22). This cosmid vector proved very helpful in our isolation of clones containing the SPA gene cluster of A. actinomycetemcomitans.
Western blotting analysis showed that autoclaved extract from E. coli DH5α containing pARF100 reacted with MAb S5 directed against A. actinomycetemcomitans Y4 SPA (Fig. 5), indicating that pARF100 contains the locus involved in the synthesis of A. actinomycetemcomitans SPA. In this regard, the size of the MAb S5-reactive band in autoclaved extract from A. actinomycetemcomitans Y4 was different from that in autoclaved extract from E. coli DH5α containing pARF100. The length of the polymer may be affected by growth conditions and the activities of enzymes involved in SPA synthesis. HPLC analysis revealed that both rhamnose and fucose were included in the polysaccharide preparation from E. coli DH5α containing pARF211, whereas neither of these sugars was detected in the polysaccharide preparation from E. coli DH5α containing pMBLcos or pARF102. E. coli DH5α containing pARF211 produced SPA, but E. coli DH5α containing pMBLcos or pARF102 did not (Fig. 1). These results suggest that both SPA synthesized in an E. coli DH5α transformant and SPA produced by A. actinomycetemcomitans Y4 are composed of rhamnose and fucose. Furthermore, our deletion analysis showed that the genes indispensable to SPA synthesis were located within the 13-kb BssHII-BspHI fragment of pARF100. Sequence analysis showed that this BssHII-BspHI fragment contained 13 ORFs (from ORF9 to ORF21). To determine whether the cloned genes are sufficient for SPA synthesis on the surface of E. coli, we carried out immunofluorescence analysis of intact cells of recombinant E. coli with MAb S5. The binding of MAb S5 to intact cells of E. coli DH5α containing pARF211 was observed by confocal fluorescence microscopy with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G antibody (54). Moreover, MAb S5 induced strong aggregation of intact cells of E. coli DH5α containing pARF211 (54). The 13 cloned genes (from ORF9 to ORF21) may be sufficient for SPA synthesis on the surface of E. coli.
The protein encoded by ORF22 showed high homology to exonuclease III from E. coli. This enzyme has five catalytic activities. (i) It is an apurinic-apyrimidinic endonuclease, (ii) it is a 3′-to-5′ exonuclease specific for bihelical DNA, (iii) it can remove a number of 3′ termini from duplex DNA, (iv) it has an RNase H activity, and (v) it can act endonucleolytically at urea-N-glycosides in duplex DNA (33). It seems reasonable to suppose that none of these functions is required for SPA synthesis. Indeed, neither ORF22, ORF23, nor ORF24 was indispensable for SPA synthesis (Fig. 1).
The N-terminal sequence of 126 amino acid residues of the protein encoded by ORF24 was very similar to that of the same region of glucose-4,6-dehydratase (RfbB) from Salmonella enterica (13), but the 54 C-terminal amino acid residues were not similar to any sequence previously reported. The ORF24 product does not seem to function as a glucose-4,6-dehydratase, because this ORF was not included in the region responsible for SPA synthesis in E. coli DH5α (Fig. 1) and this ORF was not as long as rfbB.
The proteins encoded by ORF3 and ORF4 had high homologies to AmsB and AmsE, respectively, in Erwinia amylovora (4). Two genes that code for these enzymes are located on the ams operon, which is required for exopolysaccharide synthesis. The AmsB enzyme is a putative glycosyltransferase, but the function of the AmsE enzyme is unknown (4). The ORF5 product is similar to Slt35, which is a 35-kDa soluble lytic transglycosylase involved in peptidoglycan metabolism in E. coli (7). Although ORF3, ORF4, and ORF5 were not required for SPA synthesis in E. coli DH5α, the possibility that these genes play a role in SPA synthesis in A. actinomycetemcomitans cannot be ruled out because E. coli DH5α may have enzymes that correspond to ORF3, ORF4, and ORF5 products.
ORF6, ORF7, ORF8, and ORF9 showed strong homology to the rml genes involved in dTDP-rhamnose biosynthesis in N. meningitidis, E. coli, and S. flexneri (11, 27, 40). dTDP-l-rhamnose is known to be synthesized from dTTP and d-glucose-1-phosphate by the combined action of four rml gene products in these bacteria. It is possible that the dTDP-rhamnose biosynthetic genes are responsible for A. actinomycetemcomitans Y4 SPA, since SPA in this organism consists of two deoxyhexoses, d-fucose and l-rhamnose (1). Deletion analysis, however, showed that the region upstream of ORF8 was not essential for SPA synthesis in E. coli DH5α. Nevertheless, ORF6 and ORF7 may be essential for SPA synthesis in A. actinomycetemcomitans. It is possible that the rml genes of E. coli also participate in SPA synthesis (40).
In the exopolysaccharide transport systems of gram-negative bacteria, several specific components are necessary for polymer translocation. In general, the transport system includes at least two cytoplasmic membrane proteins that belong to the ATP-binding-cassette (ABC) superfamily of active transporters (12). One of these proteins is a translocase, and the other is an ATP-hydrolase that provides energy for the process. The ORF10 product strongly resembled TagG from Bacillus subtilis (18) and membrane-spanning domain proteins of Vibrio cholerae (41), Yersinia enterocolitica (57), E. coli K5 (38) and K1 (25), H. influenzae (16), and N. meningitidis (8). The deduced amino acid sequence for the protein encoded by ORF11 revealed the ATP-binding motif GXXGXGKS (49) and significant homology with the ABC protein of Aeromonas salmonicida (6). Hence, it is likely that the ORF10 and ORF11 products belong to the ABC superfamily of active transporters. Deletion analysis showed that ORF11 was indispensable for SPA synthesis in E. coli DH5α.
Deletion analysis showed that ORF12, ORF20, and ORF21 were indispensable for SPA synthesis in E. coli DH5α. The amino acid sequence of the protein encoded by ORF12 showed 26.6% identity with RfbN, which is a rhamnosyltransferase of Salmonella enterica (13, 20), whereas the ORF20 product showed 54.4% identity with the same protein. Alignment of the sequences of the ORF12 and ORF20 products with RfbN is shown in Fig. 4. A number of identical and conserved amino acids in these three sequences are observed along the entire length of the polypeptide chain, suggesting that both proteins encoded by ORF12 and ORF20 are sugar transferases. However, the differences in sequence between the two enzymes may create a difference in substrate specificity and different linkage of the polysaccharide synthesized. The protein encoded by ORF21 was similar to WbaP from Salmonella enterica. This protein is believed to be involved in the transfer of galactosyl-1-phosphate from GDP-galactose to undecaprenyl phosphate and in the inversion of the O-antigen subunit on undecaprenyl pyrophosphate from the cytoplasmic face to the periplasmic face of the cytoplasmic membrane (50), suggesting that the product of ORF21 is involved in the first step of SPA synthesis.
The protein encoded by ORF13 was similar to the Y. enterocolitica rfbH gene product required for O-antigen biosynthesis (57). However, the function of this protein is unclear. The protein encoded by ORF19 was similar to S. flexneri RfbG, whose function is also unknown (21). Both of these ORFs were indispensable for SPA synthesis in E. coli DH5α.
All 24 genes in the 25-kb XbaI-SacI fragment had low G+C contents (25.1 to 46.4%) compared with the G+C content (45.6%) of the entire A. actinomycetemcomitans Y4 chromosome (46) or with the average G+C content (46.6 to 53.3%) of the eight genes we had previously cloned from A. actinomycetemcomitans Y4 chromosomal DNA (53) (Table 1). Based on the average G+C content, all of the genes in this fragment can be divided into three groups. As shown in Fig. 3, the average G+C content of ORF20, ORF21, ORF22, ORF23, and ORF24 (region C) was 41.7%. The average G+C content of 10 ORFs (region B; ORF10 to ORF19) upstream from region C was considerably lower (27.0%). The average G+C content of nine ORFs (ORF1 to ORF9) in the region furthest upstream (region A) was 42.4%. The average G+C contents of the genes involved in exopolysaccharide synthesis in K. pneumoniae, S. flexneri, Salmonella enterica, and E. coli are known to be low (2, 13, 21, 40). Interestingly, the average G+C content of region B (8.5 kb) is much lower than that of genes involved in exopolysaccharide synthesis in other organisms. The proteins encoded by ORF14, ORF15, ORF16, ORF17, and ORF18 did not show significant homology to any protein previously reported. Furthermore, these ORFs (ORF14 to ORF18) are indispensable for SPA synthesis (Fig. 1). The genes in this region are novel polysaccharide synthetic genes, and the functions of these five ORFs could be peculiar to SPA synthesis in A. actinomycetemcomitans.
In general, determination of the G+C ratio is useful in predicting a degree of genetic relatedness among bacterial species. The divergence in G+C ratio between species is thought to be attributable to the variation in the mutation rates of (A/T) to (G/C) and (G/C) to (A/T) base pairs (42). Therefore, the discrepancy in the G+C content within the SPA gene cluster strongly suggests that some subsets of the genes in this cluster have different origins or histories.
In conclusion, we cloned the SPA gene cluster of serotype b A. actinomycetemcomitans. Knowledge of this SPA gene cluster will be useful in elucidating the mechanism of SPA synthesis by A. actinomycetemcomitans. In addition, we found that E. coli DH5α containing the SPA biosynthetic genes produced a polysaccharide which was composed of rhamnose and fucose and reacted with a MAb directed to A. actinomycetemcomitans SPA. Further functional analysis of SPA synthetic genes and their products is in progress.
ACKNOWLEDGMENTS
We thank K. Rajakumar for providing us with pSBA85.
This work was supported in part by grants-in-aid of scientific research no. 07557136 and 08457572 from the Ministry of Education, Science, Sports and Culture, Tokyo, Japan, and by a research grant from the Fund for Comprehensive Research on Aging and Health.
REFERENCES
- 1.Amano K, Nishihara T, Shibuya N, Noguchi T, Koga T. Immunochemical and structural characterization of a serotype-specific polysaccharide antigen from Actinobacillus actinomycetemcomitans Y4 (serotype b) Infect Immun. 1989;57:2942–2946. doi: 10.1128/iai.57.10.2942-2946.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asikainen S, Lai C-H, Alaluusua S, Slots J. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 1991;6:115–118. doi: 10.1111/j.1399-302x.1991.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 4.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 5.Califano J V, Schenkein H A, Tew J G. Immunodominant antigen of Actinobacillus actinomycetemcomitans Y4 in high-responder patients. Infect Immun. 1989;57:1582–1589. doi: 10.1128/iai.57.5.1582-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu S, Trust T J. An Aeromonas salmonicida gene which influences A-protein expression in Escherichia coli encodes a protein containing an ATP-binding cassette and maps beside the surface array protein gene. J Bacteriol. 1993;175:3105–3114. doi: 10.1128/jb.175.10.3105-3114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra A J, Hermann F, Keck W. Cloning and controlled overexpression of the gene encoding the 35 kDa soluble lytic transglycosylase from Escherichia coli. FEBS Lett. 1995;366:115–118. doi: 10.1016/0014-5793(95)00505-4. [DOI] [PubMed] [Google Scholar]
- 8.Frosch M, Edwards U, Bousset K, Krauße B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 9.Frosch M, Weisgerber C, Meyer T F. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc Natl Acad Sci USA. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gmür R, McNabb H, van Steenbergen T J M, Baehni P, Mombelli A, van Winkelhoff A J, Guggenheim B. Seroclassification of hitherto nontypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol Immunol. 1993;8:116–120. doi: 10.1111/j.1399-302x.1993.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 11.Hammerschmidt S, Birkholz C, Zähringer U, Robertson B D, van Putten J, Ebeling O, Frosch M. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 12.Higgins C F, Hiles I D, Salmond G P C, Gill D R, Downie J A, Evans I J, Holland I B, Gray L, Buckel S D, Bell A W, Hermodson M A. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986;323:448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X-M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan A H, Weber D J, Oddone E Z, Perfect J R. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev Infect Dis. 1989;11:46–63. doi: 10.1093/clinids/11.1.46. [DOI] [PubMed] [Google Scholar]
- 15.Koga T, Senpuku H, Nakashima K, Ishihara Y, Nishihara T. Monoclonal antibody-coated latex agglutination assay for identification of Actinobacillus actinomycetemcomitans. Zentralbl Bakteriol. 1990;274:91–99. doi: 10.1016/s0934-8840(11)80978-3. [DOI] [PubMed] [Google Scholar]
- 16.Kroll J S, Loynds B, Brophy L N, Moxon E R. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 17.Kroll J S, Zamze S, Loynds B, Moxon E R. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J Bacteriol. 1989;171:3343–3347. doi: 10.1128/jb.171.6.3343-3347.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 19.Lipman D J, Pearson W R. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Haase A M, Lindqvist L, Lindberg A A, Reeves P R. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: identification and characterization of transferase genes of groups B, C2, and E1. J Bacteriol. 1993;175:3408–3413. doi: 10.1128/jb.175.11.3408-3413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano Y, Yoshida Y, Yamashita Y, Koga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 23.Nishihara T, Ueda N, Amano K, Ishihara Y, Hayakawa H, Kuroyanagi T, Ohsaki Y, Nagata K, Noguchi T. Actinobacillus actinomycetemcomitans Y4 capsular-polysaccharide-like polysaccharide promotes osteoclast-like cell formation by interleukin-1α production in mouse marrow cultures. Infect Immun. 1995;63:1893–1898. doi: 10.1128/iai.63.5.1893-1898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page R C, Sims T J, Engel L D, Moncla B J, Bainbridge B, Stray J, Darveau R P. The immunodominant outer membrane antigen of Actinobacillus actinomycetemcomitans is located in the serotype-specific high-molecular-mass carbohydrate moiety of lipopolysaccharide. Infect Immun. 1991;59:3451–3462. doi: 10.1128/iai.59.10.3451-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavelka M S, Jr, Wright L F, Silver R P. Identification of two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1991;173:4603–4610. doi: 10.1128/jb.173.15.4603-4610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry M B, MacLean L L, Gmür R, Wilson M E. Characterization of the O-polysaccharide structure of lipopolysaccharide from Actinobacillus actinomycetemcomitans serotype b. Infect Immun. 1996;64:1215–1219. doi: 10.1128/iai.64.4.1215-1219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajakumar K, Jost B H, Sasakawa C, Okada N, Yoshikawa M, Adler B. Nucleotide sequence of the rhamnose biosynthetic operon of Shigella flexneri 2a and role of lipopolysaccharide in virulence. J Bacteriol. 1994;176:2362–2373. doi: 10.1128/jb.176.8.2362-2373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves P, Hobbs M, Valvano A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 29.Roberts I S, Mountford R, Hodge R, Jann K B, Boulnois G J. Common organization of gene clusters for production of different capsular polysaccharides (K antigens) in Escherichia coli. J Bacteriol. 1988;170:1305–1310. doi: 10.1128/jb.170.3.1305-1310.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saarela M, Asikainen S, Alaluusua S, Pyhälä L, Lai C-H, Jousimies-Somer H. Frequency and stability of mono- or poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol Immunol. 1992;7:277–279. doi: 10.1111/j.1399-302x.1992.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saporito S M, Smith-White B J, Cunningham R P. Nucleotide sequence of the xth gene of Escherichia coli K-12. J Bacteriol. 1988;170:4542–4547. doi: 10.1128/jb.170.10.4542-4547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sims T J, Moncla B J, Darveau R P, Page R C. Antigens of Actinobacillus actinomycetemcomitans recognized by patients with juvenile periodontitis and periodontally normal subjects. Infect Immun. 1991;59:913–924. doi: 10.1128/iai.59.3.913-924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slots J, Bragd L, Wikström M, Dahlén G. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J Clin Periodontol. 1986;13:570–577. doi: 10.1111/j.1600-051x.1986.tb00849.x. [DOI] [PubMed] [Google Scholar]
- 37.Slots J, Reynolds H S, Genco R J. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith A N, Boulnois G J, Roberts I S. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990;4:1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- 39.Sreenivasan P K, LeBlanc D J, Lee L N, Fives-Taylor P. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect Immun. 1991;59:4621–4627. doi: 10.1128/iai.59.12.4621-4627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stroeher U H, Karageorgos L E, Morona R, Manning P A. Serotype conversion in Vibrio cholerae O1. Proc Natl Acad Sci USA. 1992;89:2566–2570. doi: 10.1073/pnas.89.7.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci USA. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki J, Kondo A, Kato I, Hase S, Ikenaka T. Analysis by high-performance anion-exchange chromatography of component sugars as their fluorescent pyridylamino derivatives. Agric Biol Chem. 1991;55:283–284. [Google Scholar]
- 44.Takahashi T, Nishihara T, Ishihara Y, Amano K, Shibuya N, Moro I, Koga T. Murine macrophage interleukin-1 release by capsularlike serotype-specific polysaccharide antigens of Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59:18–23. doi: 10.1128/iai.59.1.18-23.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 46.Tanner A C R, Visconti R A, Socransky S S, Holt S C. Classification and identification of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus by cluster analysis and deoxyribonucleic acid hybridizations. J Periodontal Res. 1982;17:585–596. doi: 10.1111/j.1600-0765.1982.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 47.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol. 1997;179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J Bacteriol. 1996;178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi N, Kawasaki M, Yamashita Y, Nakashima K, Koga T. Role of the capsular polysaccharide-like serotype-specific antigen in resistance of Actinobacillus actinomycetemcomitans to phagocytosis by human polymorphonuclear leukocytes. Infect Immun. 1995;63:4589–4594. doi: 10.1128/iai.63.12.4589-4594.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida Y, Nakano Y, Yamashita Y, Koga T. The gnd gene encoding a novel 6-phosphogluconate dehydrogenase and its adjacent region of Actinobacillus actinomycetemcomitans chromosomal DNA. Biochem Biophys Res Commun. 1997;230:220–225. doi: 10.1006/bbrc.1996.5917. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida, Y., and N. Yamaguchi. Unpublished data.
- 55.Zambon J J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 56.Zambon J J, Slots J, Genco R J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Al-Hendy A, Toivanen P, Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-l-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol. 1993;9:309–321. doi: 10.1111/j.1365-2958.1993.tb01692.x. [DOI] [PubMed] [Google Scholar]