Abstract
Background:
The missing requirement for resection for the majority of hepatic hemangiomas (HH) and tissue scarcity for rare diseases such as hepatic epithelioid hemangioendotheliomas (HEHE) complicate the characterization of the spatial immunovascular niche of these benign and malignant vascular neoplastic diseases.
Methods:
Two tissue cohorts containing 98 HHs and 13 HEHEs were used to study entity-specific and disease stage–specific endothelial cell (EC) phenotype and immune cell abundance. Using semiquantitative assessment, annotation-based cell classifiers, digital cell detection on whole slides, and tissue microarrays, we quantified 23 immunologic and vascular niche-associated markers and correlated this with clinicopathologic data.
Results:
Both HH and HEHE ECs were characterized by a CD31high, CD34high, FVIII-related antigenhigh expression phenotype with entity-specific expression differences of sinusoidal EC markers Stabilin1, Stabilin2, CD32, and Lymphatic Vessel Endothelial Hyaluronan Receptor 1 (LYVE-1). Cell detection identified an HH margin-prevailing immunologic response dominated by Myeloperoxidase+ (MPO+) macrophages, CD3+ and CD8+ T cell subsets, and B cells (CD20+, CD79A+). In HEHE, increased CD68+ and CD20+ cell demarcation of lesion margins was observed, while CD3+ and CD8+ T cells were equally detectable both marginally and intralesionally. Stage-specific pairwise correlation analysis of HH and HEHE revealed disease entity–specific immunologic infiltration patterns as seen by high CD117+ cell numbers in HH, while HEHE samples showed increased CD3+ T cell infiltration.
Conclusions:
ECs in HH and HEHE share a continuous EC expression phenotype, while the expression of sinusoidal EC markers is more highly retained in HEHE. These phenotypic differences are associated with a unique and disease-specific immunovascular landscape.
INTRODUCTION
Pathogenesis and disease evolution in human hepatic hemangioma (HH) are still poorly understood even though HH represents the most common liver tumor, which accounts for ~70% of all benign liver tumors and has a prevalence of ~2.5%.1–3 Incidental diagnosis with absent or unspecific clinical symptoms, lack of surgical indication and a slow growth rate lead to a collection bias toward highly progressed, larger and clinically apparent lesions that may inaccurately reflect the full spectrum of HH disease development and stage.4–7 Moreover, hepatic vascular heterogeneity comprising sinusoidal and capillary vascular beds and neoplasia-driven transdifferentiation of liver sinusoidal endothelial cells (LSECs) and expansion of continuous endothelial cells (CEC) make the identification of disease-initiating ECs challenging.8–10 Lesion heterogeneity in HH explants comprising regions of restricted proliferation, senescent and regressed regions may reflect later stages of HH evolution, which comprise slow growth, involution or regression, as are analogously described in infantile hemangioma.6,11,12 As such, HH displays a CEC and non-sinusoidal vascular tumor entity, where EC intrinsic biological feedback circuits may still control and repress proliferation and induce cellular senescence—characteristics that are typically lost in malignant vascular tumor entities such as hepatic epithelioid hemangioendothelioma (HEHE).13–15 Still, it remains poorly understood to which extent vascular liver tumors share or exclusively inherit cellular characteristics that lead to immune cell evasion, even though EC immune anergy has been described in the context of angiogenesis of non-EC malignancies.16 Thus, an entity-specific analysis of immune cell infiltration patterns of HH and HEHE may shed light on EC-specific mechanisms of immune cell recruitment in the process of EC-associated phenotypic changes toward a CEC phenotype and HEHE-exclusive malignant transformation.
The hepatic CEC niche, its gene expression, signal cues and immunologic composition have been characterized in molecular detail in a steady state, yet the immunologic crosstalk of CECs with disease-modifying immune cells in CEC-expanding diseases remains poorly understood.10,17,18 For example, whether the expanded CECs within HH/HEHE would still sustain the liver CEC-specific immunologic niche that comprises tissue-resident myeloid and T cell subsets remains unknown. The hepatic endothelial expansion includes a close interplay of ECs with myeloid cells that may additionally affect the cellular composition of the CEC-associated immunologic niche. Moreover, the immunologic surveillance of HH/HEHE remains poorly studied and it is unknown whether a particular immunophenotype may be associated with unfavorable outcome and disease progression.
With reference to HH margins and their proximity to adjacent liver tissue, the disease evolution of HH tissue margins may be even more challenging to interpret. For example, liver vessels in close adjacency to HH, which are termed “hemangioma-like vessels,” may have a similar tortuous morphology as HH vessels, indicating that HH marginal zones within adjacent liver tissue may be exposed to different biomechanical and microenvironmental cues.19 Yet, the marginal zones of HH remain poorly studied, even though incomplete resection may be the basis for recurrent disease, a phenomenon that is observed in ~1–2% of patients after surgical resection.20 For HEHE, a spatial site–specific analysis may equally shed light on the pathogenesis of HEHE disease progression, since many cases are diagnosed in a multifocal setting, which associates with poor patient survival.21,22 A cross-entity comparison of benign HH with a low-grade malignant neoplasm such as HEHE could shed light on phenotypic and transformation-associated changes in ECs in disease settings, which are not as highly dysregulated as in malignant high-grade disease entities such as angiosarcoma, thereby enabling the study of the immunologic landscape in a background of limited malignancy-associated cellular dysregulation.
In this study, we aim to characterize the immunophenotype within HH/HEHE disease stages to elucidate whether intralesional or marginal immune cell responses correlate with clinicopathological characteristics. Furthermore, conserved immunological infiltration patterns across these hepatic vascular tumor entities are analyzed.
METHODS
Patient cohort and tissue microarray design
Patients with HH included in this study were resected at the Department of Surgery of the University Hospital Heidelberg from 1995 to 2017. In total, 98 HH tissues were provided by the Tissue Bank of the National Center for Tumor Diseases, Heidelberg, Germany, in accordance with the regulations of the tissue bank and the approval of the ethics committee of Heidelberg University (application number S-230/20). The HH tissue cohort contained tissues with a diagnosis of cavernous hemangioma, HH with/without signs of tissue regression, and 2 cases of capillary hemangioma. The HH cohort contained 64 female HH specimens, which were resected at an age of 54.8 ± 14.1 (SD) years with an average lesion size of 5.6 ± 5.5 cm, and 34 male HH specimens, which were resected at an age of 59.1 ± 11.5 (SD) years with an average lesion size of 5.4 ± 5.5 cm.6 The HEHE tissue microarray (TMA) cohort comprised 9 samples, which were resected at an age of 46.5 ± 14.9 (SD) years. The majority of HEHEs were resected at a progressed disease stage with an average diameter of 6.8 ± 5.4 cm. Sinusoidal marker expression analysis of HEHE whole slides contained sample materials of 13 patients. The HEHE tissues were analyzed in accordance with the regulations of the tissue bank and the approval of the ethics committee of Heidelberg University and the Technical University of Munich (application numbers S-206, 207/05 and 160/19S).
A TMA with 98 HHs (HH-TMA) with a core diameter of 1.5 mm consisting of 339 cores on 5 sections was designed. To capture lesion heterogeneity, 2 cores of HH center (n = 181), 1 core of HH/liver parenchyma interface (n = 80), and 1 core of far distant liver tissue (n = 78) were sampled, if sufficient material could be obtained. Depending on the performed staining, data points may be lower than the total calculated core number, since individual cores were consumed in the process of a total >30 stainings being performed for this and our previous study,6 thereby affecting the statistical power of our analysis. To visualize this, we plotted our data with data points using violin plots generated by ggplot using geom_jitter(). For the analysis of HEHE, a tissue cohort of 9 HEHEs in a second TMA containing 42 cores (18 HEHE centers, 9 HEHE margins, and 15 distant livers/liver controls) was generated for the cross-entity comparison with HH.
Immunohistochemistry
For immunohistochemical staining, 3 µm thick TMA sections were deparaffinized using xylene and rehydrated using ethanol in descending concentration (100%, 96%, and 70%). Antigen retrieval was performed by either using Ventana CC1 solution (Ventana Medical Systems, Oro Valley, USA; 24–48 min incubation time) or a pH6 citrate-based retrieval solution from Dako (Dako/Agilent Technologies, Santa Clara, USA). Primary antibodies were incubated for 24–40 minutes and bound antibodies were detected using the OptiView DAB IHC detection kit or the Dako Dual Vision System-HRP (Dako). After detection, tissue sections were counterstained using hematoxylin and mounted. In total, 24 immunohistochemical markers were stained to assess NK/T cell (CD3, CD4, CD8, CD56), B cell (CD20, CD79a), and myeloid cell (CD68, CD117, CD163, MPO) content. GFAP and VIM were stained to assess HSC or mesenchymal content since HSC and fibroblasts have been described to be key modulators in tissue fibrosis and angiogenesis.23–25 CD31, CD34, FVIII-related antigen (FVIII), and p16 were used to characterize ECs as described.6 Arginase 1 (Arg1) was used to quantify hepatocyte content within cores that captured the adjacent liver/HH or HEHE interface. Specific information about antibody clones, dilutions, and staining protocols can be found in Table 1. In the case different clones/dilutions were used for the staining of HH and HEHE, HH used clones and dilutions were listed first.
TABLE 1.
Antibody list
| Antibody | Clone | Company/order number | Dilution |
|---|---|---|---|
| ARG1 | SP156 | Ventana Medical Systems #760-4801 | 1:100 |
| CD3 | 2GV6 | Ventana Medical Systems #790-4341 | RTU |
| CD4 | SP35 | Ventana Medical Systems #790-4423 | RTU |
| CD8 | SP57 | Ventana Medical Systems #790-4460 | RTU |
| CD20 | L26 | Ventana Medical Systems #760-2531 | 1:2000 |
| CD31 | JC70 | Ventana Medical Systems #760-4378 | RTU |
| CD32 | 190723 | R&D #AF1330 | 1:1000 |
| CD34 | QBEnd/10 | Ventana Medical Systems #760-2927 | RTU |
| CD56 | MRQ-42 | Ventana Medical Systems #760-4596 | RTU |
| CD79A | SP18 | Ventana Medical Systems #790-4432 | 1:400 |
| CD117 | polyclonal | Dako #A450229-2 | 1:100 1:200 |
| CD163 | MRQ-26 | Ventana Medical Systems #760-4437 | RTU |
| FVIII | polyclonal | Ventana Medical Systems #760-2642 | RTU |
| GFAP | EP672Y | Ventana Medical Systems #760-4345 | RTU |
| LYVE-1 | polyclonal | Acris #DP-3500PS | 1:200 |
| MPO | polyclonal | Ventana Medical Systems #760-2659 ThermoFisher #RB-373-A |
RTU 1:100 |
| PDPN | D2-40 | Cell Marque #760-4395 Dako #M3619 |
RTU 1:50 |
| p16 | MX007 | Zeta Corporation #Z2016 | 1:100 |
| VIM | V9 | Ventana Medical Systems #790-2917 | 1:300 |
| CD68 | PG-M1 | DAKO #M0876 | 1:100 |
| ERG | EPR3864 | Ventana Medical Systems #790-4576 | RTU |
| THBD | 1009 | DAKO #M0617 | 1:40 |
Abbreviations: ARG1, Arginase 1; FVIII, FVIII-related antigen; GFAP, Glial Fibrillary Acidic Protein; LYVE-1, Lymphatic Vessel Endothelial Hyaluronan Receptor 1; MPO, Myeloperoxidase; PDPN, Podoplanin; RTU, ready to use; VIM, Vimentin.
Immunohistochemical stainings on HH/HEHE whole slides were manually performed according to standard protocols. Antibodies used for whole slides: Stabilin1 (STAB1 a-GST-hSTAB1-CT rabbit Nr.1676/01, 1:2000, provided by K. Schledzewski and C. Géraud, Department of Dermatology, Venereology, and Allergology, University Medical Center and Medical Faculty Mannheim, Germany), Stabilin2 (STAB2 IgG1 Monoclonal 3.1; 1:100, provided by C. Géraud), CD32 (Anti-Hs FCGR2 CD32; goat IgG R&D AF 1330, 1:1000), and Lymphatic Vessel Endothelial Hyaluronan Receptor 1 (DP 3500PS rabbit; 0403R02-1; Acris, 1:200).
Slide scanning and digital quantification
TMAs were scanned by a Pannoramic Scan II (3DHISTECH, Budapest, Hungary) or Aperio AT2 scanning device (Leica, Wetzlar, Germany) using 40-fold magnification and a 0.25 µm resolution. QuPath’s TMA dearrayer and positive cell detection were used to detect and quantify positively stained cells using optical density sum with a pixel size of 0.5 µm and single thresholding.26 Cores with significant artifacts such as large cracks comprising more than one-third of the core area or large tissue folds were excluded from the evaluation. Core-specific quantitative data were exported and further analyzed in R studio (see below). For the visualization of local cell density of the cellular neighborhood or of positive cells, measurement maps/density maps were calculated based on a weighted average of individual cells and neighboring cells for the specific measurement type. Smoothed data (100 µm radius) were calculated and displayed as color code in a low magnification overview.
Statistics
Data points of this study showed a non-normal distribution, as tested by the Shapiro-Wilk test, and the Wilcoxon rank-sum test was used for the calculation of p values. To correct for multiple testing, a Benjamini and Hochberg correction was applied to calculated p values (p-adjust function, R studio). p values are indicated as following:*p < 0.05, **p < 0.01, ***p < 0.001. Plots were generated in R studio (ggplot2, ggpubr, corrplot).
RESULTS
Immunologic demarcation of hepatic hemangiomas and epithelioid hemangioendotheliomas
First, we analyzed immune cell composition and its spatial context in a cohort of 61 hematoxylin eosin (HE)-stained HH whole slides to screen for HH immune cell density and immunologic demarcation at the HH/adjacent liver interface. While we found that 55% of HH centers are characterized by immunologic anergy (lack of increased numbers of immune cells and lymph follicles in HE), we found a significant fraction of HH (81%) to show a characteristic immunologic demarcation of the HH margin and 30% of the HH showing the presence of lymph follicle aggregates in the marginal liver tissue (Figure 1A, Supplemental Figure 1A, http://links.lww.com/HC9/A720). This prompted us to analyze the immune cell infiltration pattern in HEHE. In HEHE, an immunologic demarcation of the lesion margin could be observed in all samples, and 4 of 9 samples showed the presence of lymph follicle aggregates (44%), while only 2/9 samples showed an immune cell enrichment within the lesion centers (Figure 1B, Supplemental Figure 1B, http://links.lww.com/HC9/A720). To verify these findings in an automated and high-throughput manner, we next analyzed 2 tissue cohorts comprising 98 HH and 9 HEHE tissues including cores sampling the lesion/liver interface6 (Figure 1C). For this, we trained a cell classification and annotation tool to identify immune cells (Figure 1D). The cell classifier accurately predicted immune cells according to the HE morphology and confirmed increased immune cell numbers at HH margins, while cell numbers at HEHE margins were not significantly altered to HEHE centers. (Figure 1D, E). This analysis confirmed immune cell enrichment at HH margins, which was further confirmed by a measurement map-based visualization of local cell density (Figure 1D). Increased local cell density was detected in liver margins, where EC marker expression changed from LSEC-predominant to a marker expression that is indicative of CECs. This prompted us to perform immunohistochemical detection of typical LSEC markers in HH/HEHE.
FIGURE 1.
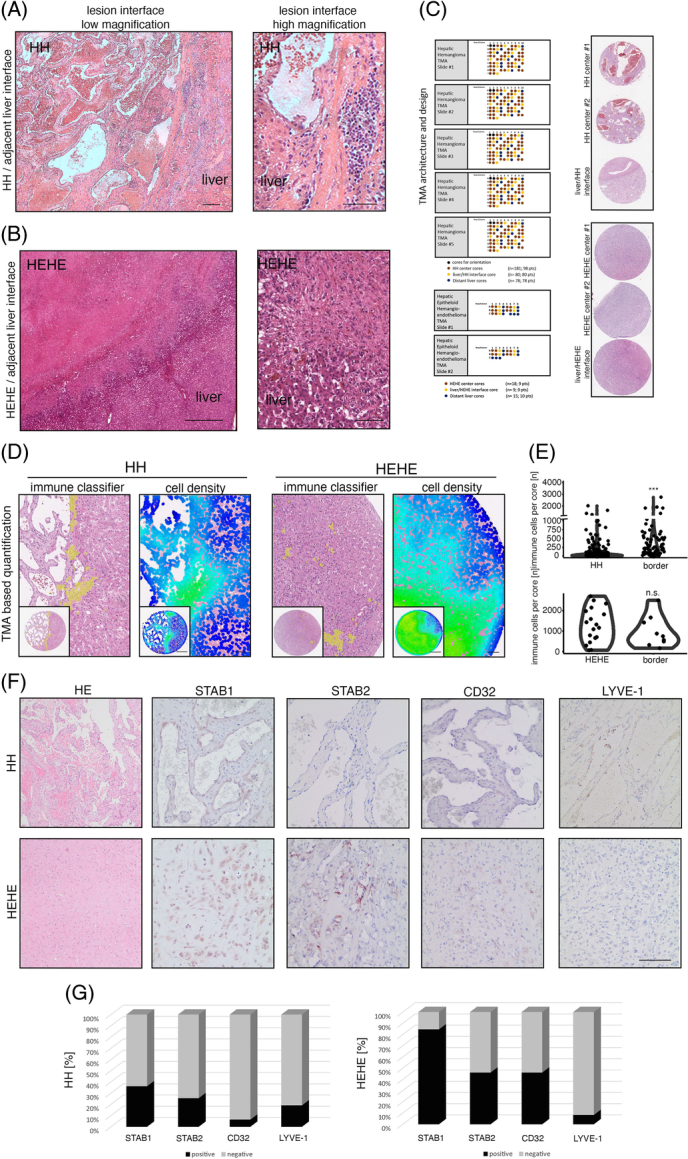
Immunological demarcation and intralesional immune cell infiltration of human hepatic hemangiomas and hepatic epithelioid hemangioendotheliomas. (A) HE stain of HH/adjacent liver interface and magnified region of interest with characteristic immunological demarcation. Scale bar: 200 µm (low) and 100 µm (high). (B) HE stain of HEHE/adjacent liver interface and magnified region of interest with indistinct lesion border. Scale bar: 800 µm (low) and 200 µm (high). (C) HH-TMA cohort as previously published.6 TMA contained patient material from 98 HH and 80 HH margins. HEHE cohort contained 9 HEHEs and 9 margins. (D) Immune cell detection within HH/HEHE cores by a trained cell classifier. Magnified regions of interest and full cores of HE, overlaid cell detections, and cell densities are displayed. Scale bar: 400 µm (low) and 100 µm (high). (E) Violin plots of quantified immune cell detections reveal significantly enriched immune cell density in HH margins but not HH centers, while HEHE margins remain statistically not significant. Wilcoxon rank-sum test (***p <0.001, n.s.). (F) Immunohistochemistry of HH/HEHE whole slides against sinusoidal EC markers STAB1, STAB2, CD32, and LYVE-1. The majority of HH ECs are negative for sinusoidal EC markers, while a higher proportion of HEHE ECs retain partial sinusoidal marker expression. Scale bar: 100 µm. (G) Barplots showing the percentual distribution of marker-positive ECs in HH/HEHE whole slides. The majority of HH ECs are negative for STAB1, STAB2, CD32, and LYVE-1, while a majority of HEHE ECs retain STAB1 (HH n = 47, HEHE n = 13). Abbreviations: EC, endothelial cell; HEHE, hepatic epithelioid hemangioendothelioma; HH, hepatic hemangioma; LYVE-1, Lymphatic Vessel Endothelial Hyaluronan Receptor 1; STAB1, Stabilin1; STAB2, Stabilin2; TMA, tissue microarray.
As expected, STAB1, STAB2, LYVE-1, and CD32 stainings revealed negative expression within ECs in the majority of HH, and the fraction of marker-positive HH ranged from 6% (CD32) to 36% (STAB1, Figure 1F, G). This loss of the LSEC marker expression was entity-specific, as could be seen by a higher fraction of marker-positive lesions in HEHE, where 84% were positive for STAB1 and 46% for STAB2 and CD32, respectively (Figure 1F, G).
These first results indicate an incomplete and heterogeneous EC marker expression toward a partial or mixed CEC phenotype within HH/HEHE ECs. Furthermore, the results propose a focal immune cell enrichment at the HH/liver parenchyma interface that could serve as an indicator regarding HH immunogenicity. For HEHE, the sampling of the TMA tissue cohort was insufficient to confirm an increased immune cell infiltrate at lesion margins, even though it was indicated by the whole slide HE analysis.
HH/adjacent liver interface classification and immune cell composition
After the first classification results based on a trained and annotation-based cell classifiers, we now aimed to further dissect the HH immune compartment using a TMA and conventional immunohistochemistry.
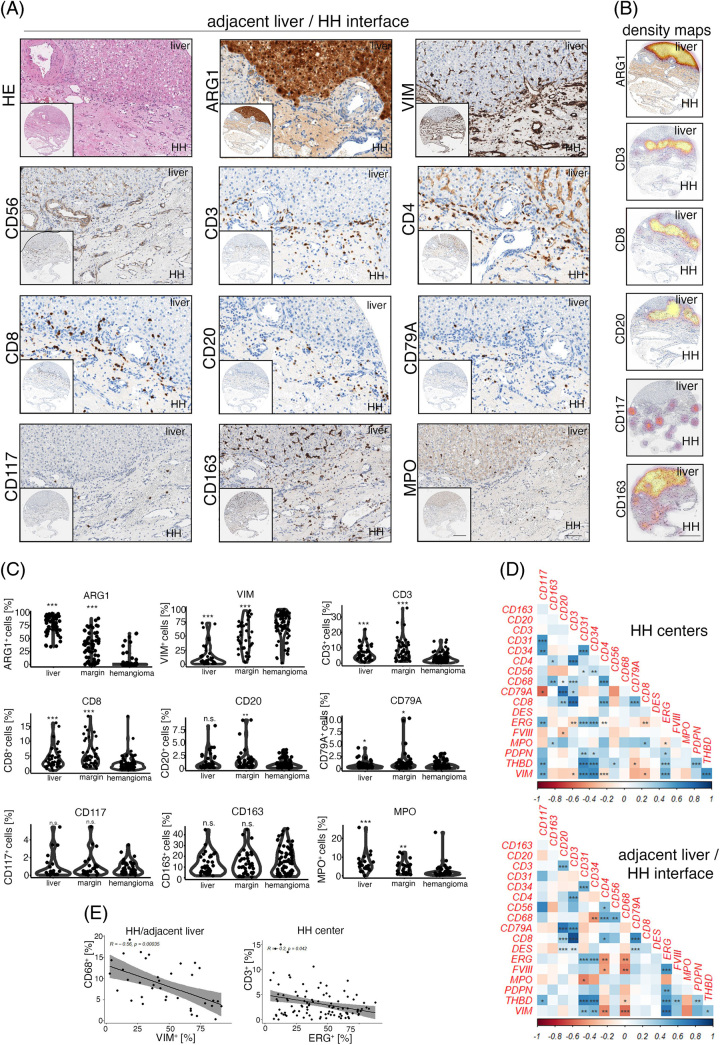
In HH, CD3+ T cells were abundant and were generally localized at the adjacent liver parenchyma/HH interface (Figure 2A). To further characterize whether these cells were of T-helper cell or cytotoxic phenotype, we stained for CD4 and CD8; however, CD4 also strongly stained liver sinusoids, but not HH ECs as has been described27 (Figure 2A). CD8+ cell distribution correlated with CD3+, as could be seen by cell density–based visualization using density maps, where Arginase 1 served as a hepatocyte-specific marker (Figure 2B). B cell marker (CD20/CD79A) distribution showed low numbers of positive cells and again an enrichment of cells at the HH lesion/liver parenchyma interface (Figure 2A, B). Exact quantification of marker-positive cells confirmed an increased cell density of CD3, CD8, CD20, CD79A, and MPO at the lesion margin, but not the HH center, while GFAP staining was negative (Figure 2C, Supplemental Figure 1C, http://links.lww.com/HC9/A720). CD117+ and CD163+ cell numbers in distant liver, margin, and HH were not significantly altered (Figure 2A-C).
FIGURE 2.
mmunologic characterization of the human hepatic hemangioma/adjacent liver parenchyma interface. (A) Low and high magnification of HE, Arginase 1 (ARG1), Vimentin (VIM), CD56, CD3, CD4, CD8, CD20, CD79A, CD117, CD163 and Myeloperoxidase (MPO) in HH margins. Immune cell accumulation at the lesion/liver interface can be detected. Scale bar: 400 µm (low) and 100 µm (high). (B) Depiction of ARG1+, CD3+, CD8+, CD20+, CD117+ and CD163+ cell density overlay at the HH/adjacent liver parenchyma interface. ARG1 highlights hepatocytes, and CD3, CD8 and CD20 show the highest cell densities at the liver/lesion interface. Spatial cell distribution of CD117 and CD163 reveals high-density foci of marker-positive cells also within HH. Scale bar: 400 µm. (C) Percentual distribution of ARG1, VIM, CD3, CD8, CD20, CD79A, CD117, CD163 and MPO in distant liver tissues, HH/adjacent liver margins (margin), and HH centers (hemangioma). Wilcoxon rank-sum test of HH centers versus tissue area of interest (*p < 0.05, **p < 0.01, ***p < 0.001, n.s. = not significant). (D) Heatmaps displaying pairwise correlation metrics of marker-positive cells (%) in HH center and HH/adjacent liver parenchyma tissues. Spearman correlation coefficient displayed as a color code from 1 (blue) to −1 (red; *p < 0.05, **p < 0.01, ***p < 0.001). (E) Correlation analysis between CD68+/VIM+ and CD3+/ETS Transcription Factor ERG+ (ERG+) percentual numbers in adjacent liver/HH interface and HH center cores (Spearman correlation analysis). Abbreviations: HE, hematoxylin eosin; HH, hepatic hemangioma.
Next, we aimed to identify HH niche-specific similarities in immune cell abundance using a pairwise correlation analysis approach (Figure 2D). This analysis revealed disease compartment–specific relationships, including CD68+ cell numbers positively correlating with CD3+/CD20+ cells in HH centers, while within HH margins, CD68+ macrophages correlated with CD56+ cell abundance. Moreover, the HH/liver parenchyma interface was characterized by an inverse correlation of the mesenchymal marker VIM with CD68, which was not detectable in HH centers. HH central tissue cores were characterized by a negative correlation of CD3/CD8 with ETS Transcription Factor ERG, indicating an inverse relationship between T cell and EC density within HH (Figure 2D, E). These data indicate a complex distribution of diverse immune cell subtypes in the HH lesion margin and center.
To test for dynamic and disease stage–specific immune cell infiltration patterns in HH, we assessed myeloid (CD68/CD163/MPO), T cell (CD3, CD4, CD8), and B cell (CD20, CD79A) abundances in dependency of HH size or patient age at diagnosis (cutoffs: 5 cm/60 y lesion diameter). However, immune cell infiltration was not significantly altered at the HH/adjacent liver parenchyma interface and HH centers with regard to patient age or HH size.
In conclusion, immunohistochemical staining and quantification of myeloid, T, and B cell marker proteins on HH margins and centers revealed a T cell-predominant immune cell demarcation, independent of patient age and lesion size.
Immunologic composition within spatial and disease-stage classified HH centers
Since our first analysis revealed a predominant accumulation of immune cells at the margin of HH, we were interested in whether within central parts of the lesion different immunophenotypes could be observed that may be linked to HH disease stages.
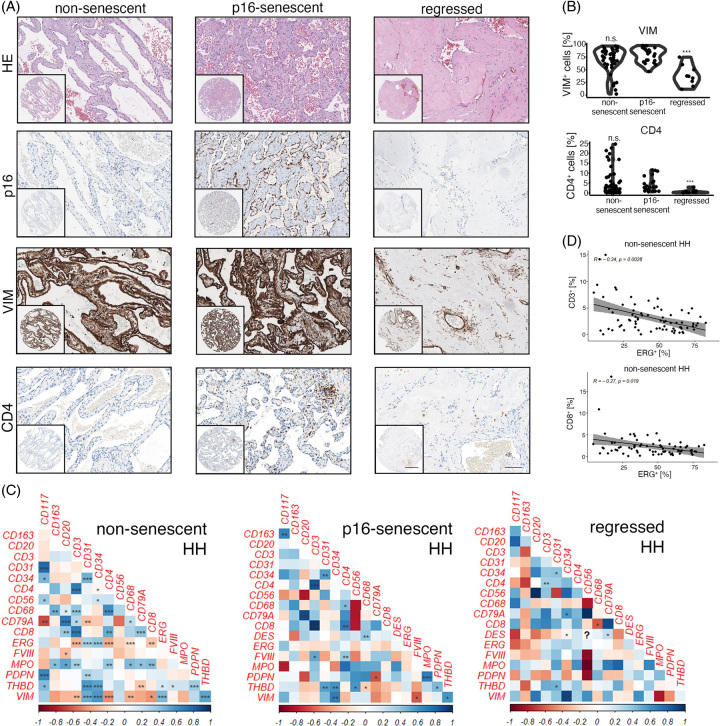
In previous work,6 we showed that HH disease stages may be subtyped based on the expression of cellular senescence markers (p16/p53) and on morphologic criteria (regression). We hypothesized that viable and non-senescent, senescent or regressed HH may represent different disease stages that associate with a stage-specific immunologic landscape. Albeit, the immune cell infiltration patterns between p16-senescent/non-senescent and p16-senescent/regressed HH disease stages were largely comparable. In fact, the concentration of CD56+, CD3+, CD8+, CD20+, CD79A+, CD117+, CD163+, and MPO+ cells was not significantly altered between p16-senescent/non-senescent and p16-senescent/regressed HH. (Supplemental Figure 2A, B, http://links.lww.com/HC9/A720). However, HH CD4+ T cell density was significantly lower in regressed hemangiomas compared to p16-senescent HH, which were also characterized by low VIM+ cell numbers, while no difference between p16-senescent and non-senescent HH could be detected for CD4 (Figure 3A, B).
FIGURE 3.
HH disease stage–specific immunologic infiltration patterns. (A) Low and high magnification of HE, p16, Vimentin (VIM) and CD4 in non-senescent, p16-senescent and regressed HH. Scale bar: 400 µm (low) and 100 µm (high). (B) Percentual distribution of VIM and CD4 in non-senescent, p16-senescent and regressed HH. Wilcoxon rank-sum test of p16-senescent HH versus HH of interest (***p < 0.001, n.s. = not significant). (C) Heatmaps displaying pairwise correlation metrics of marker-positive cells (%) in non-senescent, p16-senescent and regressed HH center tissues. Spearman correlation coefficient displayed as a color code from 1 (blue) to −1 (red; *p < 0.05, **p < 0.01, ***p < 0.001). (D) Correlation analysis between CD3+/ETS Transcription Factor ERG+ (ERG+) and CD8+/ERG+ percentual numbers in non-senescent HH center cores (Spearman correlation analysis). Abbreviations: HE, hematoxylin eosin; HH, hepatic hemangioma.
Pairwise correlation analyses of non-senescent/p16-senescent and regressed HH revealed different correlations between immune and EC abundance in these HH disease stages (Figure 3C, D). Non-senescent HH showed a negative correlation between ERG+ and CD3+ cells, but an inverse trend could be observed for regressed HH; however, it did not reach the level of significance (Figure 3C, D). p16-senescent HH showed a significant positive correlation between CD117/CD163 and MPO/Podoplanin (Figure 3C). Regressed HH was typically characterized by a low CD68+ density6 and CD68 negatively correlated with CD8+ cell counts, whereas p16-senescent/non-senescent HH showed a positive correlation trend; however, it did not reach the statistical level of significance (Figure 3C).
Together, HH immune cell infiltration patterns are largely independent between p16-senescent/non-senescent and p16-senescent/regressed HH disease stages, and correlation analysis reveals a complex interplay of EC cell state and HH immune cell density.
Comparative immunologic phenotyping of HH and HEHE
Based on the analysis of the immunologic landscape of HH and adjacent liver tissues, we were interested in whether the immunologic phenotypes observed may be transferable to other vascular tumors of the liver such as HEHE, which contains malignant ECs with a mixed CEC phenotype. A comparison of HH to a low-grade malignant neoplasm such as HEHE could thereby enable the study of the immune cell composition in dependency on phenotypic and transformation-associated changes in ECs in malignancy, while tumor endothelial cells may not be completely dysregulated as in high-grade EC malignancies.
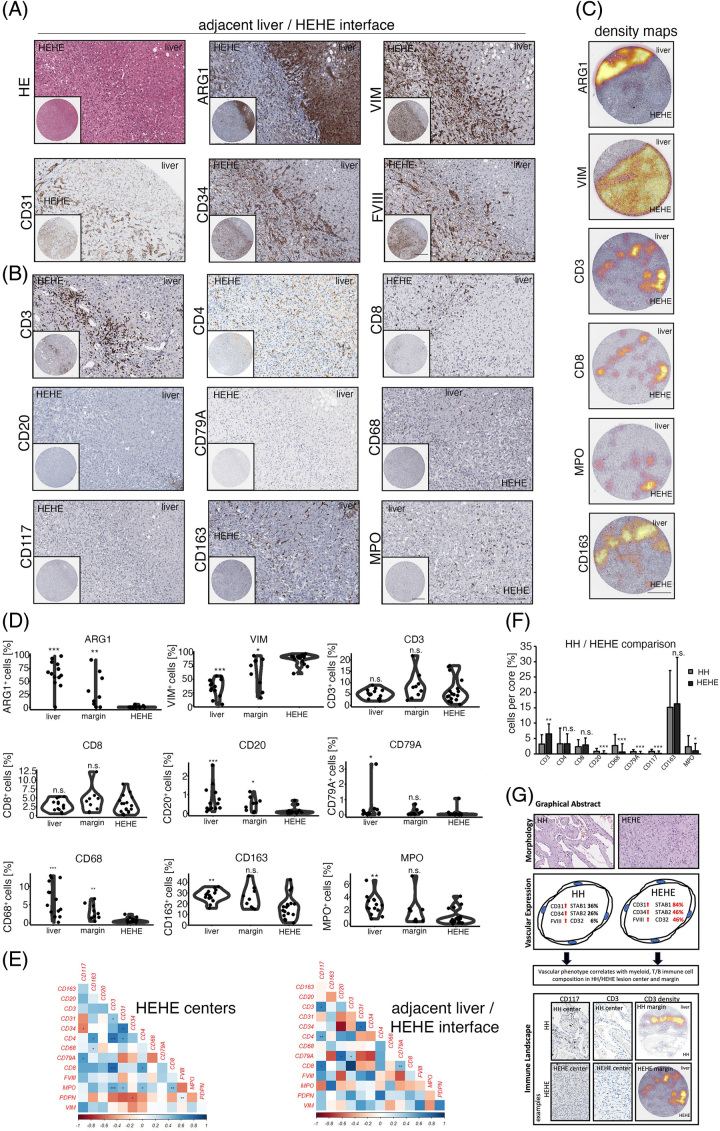
While the expression of STAB1, STAB2 and CD32 was retained in a fraction of HEHE ECs, the expression of LYVE-1 was found to be not expressed in the majority of analyzed samples (Figure 1G). Additionally, the expansion of ECs in HEHE included a CEC-associated, strong expression of CD31, CD34 and FVIII-related antigen with a simultaneous absence of CD4 expression (Figure 4A, B).
FIGURE 4.
Immunologic characterization of the human HEHE/adjacent liver parenchyma interface. (A) Low and high magnification of HE, ARG1, VIM, CD31, CD34 and FVIII at the HEHE/adjacent liver interface. Scale bar: 400 µm (low) and 100 µm (high). (B) Low and high magnification of CD3, CD4, CD8, CD20, CD79A, CD68, CD117, CD163 and MPO at the HEHE/adjacent liver parenchyma interface. Scale bar: 400 µm (low) and 100 µm (high). (C) Depiction of ARG1+, VIM+, CD3+, CD8+, MPO+ and CD163+ cell density overlay at the HEHE/adjacent liver interface. ARG1 highlights hepatocytes, and CD3, CD8 and CD163 show clear immunologic demarcation at the liver lesion interface. Spatial cell distribution of CD3, CD8 and MPO reveals high-density foci of marker-positive cells also within HEHE. Scale bar: 400 µm. (D) Percentual distribution of ARG1, VIM, CD3, CD8, CD20, CD79A, CD68, CD163 and MPO in the distant liver, HEHE/adjacent liver parenchyma interface (margin), and HEHE centers. Wilcoxon rank-sum test of HEHE centers versus tissue area of interest (*p < 0.05, **p < 0.01, ***p < 0.001, n.s. = not significant). (E) Heat maps displaying pairwise correlation metrics of marker-positive cells (%) in HEHE centers and at the HEHE/adjacent liver interface. Spearman correlation coefficient displayed as a color code from 1 (blue) to −1 (red; *p < 0.05, **p < 0.01, ***p < 0.001). (F) Barplot depicting percentual infiltration of marker-positive immune cells in HH/HEHE in an entity-specific comparison. Comparison of HH (overall) versus HEHE (overall) by Wilcoxon rank-sum test (*p < 0.05, **p < 0.01, ***p < 0.001, n.s. = not significant). (G) Graphical abstract summarizing the main results of this study. Abbreviations: ARG1, Arginase 1; FVIII, FVIII-related antigen; HE, hematoxylin eosin; HEHE, hepatic epithelioid hemangioendothelioma; HH, hepatic hemangioma. MPO, Myeloperoxidase; VIM, Vimentin.
The immunologic characterization of HH prompted us to analyze myeloid and T cell abundances in HEHE (Figure 4B). Along with the different biological behavior of HEHE showing an invasive growth pattern, we found comparable CD3+ and CD8+ cell numbers in HEHE centers, margins and distant livers, indicating site-independent homogeneous T cell counts in this lesion, while HH was characterized by a more pronounced immune infiltrate at the HH/liver parenchyma interface (Figure 4C, D; Figure 2B, C). In contrast to this, myeloid cells showed a reduced infiltration of CD68+, CD163+ and MPO+ cells in HEHE central areas compared to distant non-tumorous liver tissue, and the HEHE invasion front showed increased CD68+ counts compared to HEHE central areas (Figure 4B - D). Correlation analyses of immune cell counts in HEHE central areas revealed a positive correlation between CD31+/MPO+ and MPO+/CD3+ cells, while tissues of the HEHE invasive front showed an inverse correlation of CD20+/CD34+ cells (Figure 4E, Supplemental Figure 3A, http://links.lww.com/HC9/A720).
Finally, in a cross-entity analysis, we compared immune infiltration patterns in HH versus HEHE (overall comparison) to see whether entity-specific immunophenotypes exist. Indeed, HH was characterized by higher percentual and absolute cell counts of CD20+, CD79A+, and CD117+ cells, while HEHE showed more prominent infiltration with CD3+ cells (Figure 4F, Supplemental Figure 3B, http://links.lww.com/HC9/A720).
In conclusion, characterized HH immunophenotypes could be partially recapitulated in HEHE with significant differences in the T, B cell and CD117+ cell infiltration patterns (Figure 4G).
DISCUSSION
Neoplasia-driven angiogenesis of ECs with a partial or complete CEC phenotype has been associated with immune cell recruitment in primary and secondary liver malignancies; however, little is known about the immune cell composition of vascular tumors, such as in HH or HEHE. While the clinical implications of the immunovascular landscape in HH/HEHE remain elusive, we hypothesize that immune cell demarcation/infiltration in HH and HEHE may potentially serve as prognostic indicators of the disease course.
In this study, we identified the immune cell composition in HH as dominated by CD163+ cells, while B and T cell densities were in general low. Immune cell–mediated tissue regression and senescence surveillance have been described in various hepatic disease contexts,28,29 yet T cell infiltration was higher within HEHE, even though a fraction of HH in our cohort has been identified as p16+ senescent.6 It is tempting to speculate whether immune cell and in particular M1/T/NK cell infiltration density may be a prognostic feature of p16+ HH, which marks the susceptibility toward senolytic therapy as an alternative to conventional surgical resection.30,31 Pharmacologic substances have been identified that allow the direct targeting and the clearance of senescent ECs, which may be potentially beneficial in a subgroup of HH that is characterized by high immune cell content and p16+ EC abundance.32
A prognostic feature of HEHE may include CD3high HEHE, which may represent a subtype that may have a favorable disease course. Whether the different genomic translocations responsible for HEHE tumor development, including CAMTA1+/TFE3+ HEHE subtypes, induce different immunologic infiltration patterns requires further investigation.15,33 Additionally, HEHE spatial heterogeneity and immune cell diversity in HEHE should be further assessed in dependency on the tumor cell content and the presence of necrosis.
Our HH-TMA data are consistent with a scRNA-seq analysis of cavernous hemangioma of the skin, which implicated a high immune cell infiltration with M2 polarized macrophages,34 mirroring our CD163 immunohistochemistry in HH. Even though organ site-specific differences of hemangioma immune cell composition cannot be excluded, these results are likely to be partially applicable to the liver, and this study reported that CD4+ and CD8+ cells were almost equally abundant, while the role of CD8+ T cells in involution and regression of these lesions have been hypothesized previously.35
Our study has several limitations, including that the presented data relied on an automated analysis of consecutive sections of TMA cores, which may only partly represent the overall morphology in the liver and also may change when multiple sections have been analyzed. Additionally, this approach requires a high tissue consumption of individual cores. In that regard, more sophisticated and tissue-efficient, multiplexed imaging techniques such as CODEX, molecular cartography and other emerging technologies that enable multiplexed readout of RNA/protein will give better insight toward a more precise characterization of the hepatic vascular immune landscape both on protein and RNA levels.36–38
Altogether, a TMA-based analysis of HH/HEHE tissue cohorts showed a myeloid-predominant infiltration of HH margins and central regions, while central HH was characterized by a higher abundance of CD117+ and CD20+/CD79A+ cells compared to HEHE centers, which were characterized by a higher infiltration of CD3+ cells. These results, which require independent verification, may imply vascular neoplasm–conserved immune cell infiltration patterns in HH/HEHE, with unique entity-specific immunophenotypic differences that may be associated with divergent EC-dedifferentiation and transformation-associated events.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Carolin Kerber, Tim Rau, Nina Wilhelm, Veronika Eckel, and Dr. Alexander Brobeil (NCT Tissue Bank and Institute of Pathology, University Hospital Heidelberg). They also thank Annett Hering (Comparative Experimental Pathology, Technical University Munich) and Christina Schott (Tissue Bank of the Institute of Pathology of the Technical University of Munich) for their support. They furthermore thank Prof. Cyrill Géraud and Dr. Kai Schledzewski (Department of Dermatology, University Hospital Mannheim) for providing the antibodies against STAB1 and STAB2.
Footnotes
Abbreviations: CEC, continuous endothelial cell; EC, endothelial cell; HE, hematoxylin eosin; HEHE, hepatic epithelioid hemangioendothelioma; HH, hepatic hemangioma; L SEC, liver sinusoidal endothelial cells; LYVE-1, Lymphatic Vessel Endothelial Hyaluronan Receptor 1; MPO, Myeloperoxidase; STAB1, Stabilin1; STAB2, Stabilin2; TMA, tissue microarray.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.hepcommjournal.com.
Contributor Information
Stefan Thomann, Email: stefan.thomann@uni-wuerzburg.de.
Thomas Metzler, Email: thomas.metzler@tum.de.
Marcell Tóth, Email: marcell.toth@med.uni-heidelberg.de.
Peter Schirmacher, Email: peter.schirmacher@med.uni-heidelberg.de.
Carolin Mogler, Email: carolin.mogler@tum.de.
ACCESS TO TISSUE MICROARRAY MATERIAL
The tissue microarrays are stored at the Tissue Bank of the National Center for Tumor Diseases (NCT), Heidelberg (HH), and the Tissue Bank of the Institute of Pathology of the Technical University of Munich (HEHE). For material requests, contact the corresponding author.
ETHICS APPROVAL STATEMENT
All analyses have been approved by the ethics committee of the University of Heidelberg (application number S-230/20, S-206, 207/05) and the ethics committee of the Technical University of Munich (160/19S).
AUTHOR CONTRIBUTIONS
Stefan Thomann: Conception and design of the study; Stefan Thomann, Thomas Metzler, Marcell Tóth, and Carolin Mogler: acquisition and analysis of data; Stefan Thomann and Carolin Mogler: drafting the manuscript and the figures; and Peter Schirmacher: supervision. All authors have read and approved the final manuscript.
FUNDING INFORMATION
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG): CM project Z02 within CRC1366 “Vascular Control of Organ Function” [project number 39404578].
CONFLICTS OF INTEREST
The authors have no conflicts to report.
REFERENCES
- 1.Mocchegiani F, Vincenzi P, Coletta M, Agostini A, Marzioni M, Baroni GS, et al. Prevalence and clinical outcome of hepatic haemangioma with specific reference to the risk of rupture: A large retrospective cross-sectional study. Dig Liver Dis. 2016;48:309–314. [DOI] [PubMed] [Google Scholar]
- 2.Trotter JF, Everson GT. Benign focal lesions of the liver. Clin Liver Dis. 2001;5:17–42; v. [DOI] [PubMed] [Google Scholar]
- 3.Belli L, De Carlis L, Beati C, Rondinara G, Sansalone V, Brambilla G. Surgical treatment of symptomatic giant hemangiomas of the liver. Surg Gynecol Obstet. 1992;174:474–478. [PubMed] [Google Scholar]
- 4.Toro A, Mahfouz AE, Ardiri A, Malaguarnera M, Malaguarnera G, Loria F, et al. What is changing in indications and treatment of hepatic hemangiomas. A review. Ann Hepatol. 2014;13:327–339. [PubMed] [Google Scholar]
- 5.Hasan HY, Hinshaw JL, Borman EJ, Gegios A, Leverson G, Winslow ER. Assessing normal growth of hepatic hemangiomas during long-term follow-up. JAMA Surg. 2014;149:1266–1271. [DOI] [PubMed] [Google Scholar]
- 6.Thomann S, Tóth M, Sprengel SD, Liermann J, Schirmacher P. Digital staging of hepatic hemangiomas reveals spatial heterogeneity in endothelial cell composition and vascular senescence. J Histochem Cytochem. 2022;70:531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jing L, Liang H, Caifeng L, Jianjun Y, Feng X, Mengchao W, et al. New recognition of the natural history and growth pattern of hepatic hemangioma in adults. Hepatol Res. 2016;46:727–733. [DOI] [PubMed] [Google Scholar]
- 8.Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res. 2007;100:174–190. [DOI] [PubMed] [Google Scholar]
- 9.Géraud C, Mogler C, Runge A, Evdokimov K, Lu S, Schledzewski K, et al. Endothelial transdifferentiation in hepatocellular carcinoma: Loss of Stabilin-2 expression in peri-tumourous liver correlates with increased survival. Liver Int. 2013;33:1428–1440. [DOI] [PubMed] [Google Scholar]
- 10.Thomann S, Weiler SME, Marquard S, Rose F, Ball CR, Tóth M, et al. YAP Orchestrates heterotypic endothelial cell communication via HGF/c-MET signaling in liver tumorigenesis. Cancer Res. 2020;80:5502–5514. [DOI] [PubMed] [Google Scholar]
- 11.Léauté-Labrèze C, Harper JI, Hoeger PH. Infantile haemangioma. Lancet. 2017;390:85–94. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Mulliken JB, Kozakewich HP, Rogers RA, Folkman J, Ezekowitz RA. Cellular markers that distinguish the phases of hemangioma during infancy and childhood. J Clin Invest. 1994;93:2357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mogler C, Koschny R, Heilig CE, Frohling S, Schirmacher P, Weichert W, et al. Molecular characterization of hepatic epithelioid hemangioendothelioma reveals alterations in various genes involved in DNA repair, epigenetic regulation, signaling pathways, and cell cycle control. Genes Chromosomes Cancer. 2020;59:106–110. [DOI] [PubMed] [Google Scholar]
- 14.Wagner MJ, Ravi V, Menter DG, Sood AK. Endothelial cell malignancies: New insights from the laboratory and clinic. NPJ Precis Oncol. 2017;1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seavey CN, Hallett A, Li S, Che K, Pobbati AV, Ma S, et al. Loss of CDKN2A cooperates with WWTR1(TAZ)-CAMTA1 gene fusion to promote tumor progression in epithelioid hemangioendothelioma. Clin Cancer Res. 2023;29:2480–2493. [DOI] [PubMed] [Google Scholar]
- 16.Huijbers EJM, Khan KA, Kerbel RS, Griffioen AW. Tumors resurrect an embryonic vascular program to escape immunity. Sci Immunol. 2022;7:eabm6388. [DOI] [PubMed] [Google Scholar]
- 17.Ficht X, Iannacone M. Immune surveillance of the liver by T cells. Sci Immunol. 2020;5:eaba2351. [DOI] [PubMed] [Google Scholar]
- 18.Thomann S, Weiler SME, Wei T, Sticht C, De La Torre C, Tóth M, et al. YAP-induced Ccl2 expression is associated with a switch in hepatic macrophage identity and vascular remodelling in liver cancer. Liver Int. 2021;41:3011–3023. [DOI] [PubMed] [Google Scholar]
- 19.Kim GE, Thung SN, Tsui WMS, Ferrell LD. Hepatic cavernous hemangioma: Underrecognized associated histologic features. Liver Int. 2006;26:334–338. [DOI] [PubMed] [Google Scholar]
- 20.Tang T, Wang X, Mao Y, Li J, Wen T, Jia W, et al. Real-world data on the clinicopathological traits and outcomes of hospitalized liver hemangioma patients: A multicenter study. Ann Transl Med. 2021;9:1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makhlouf HR, Ishak KG, Goodman ZD. Epithelioid hemangioendothelioma of the liver: A clinicopathologic study of 137 cases. Cancer. 1999;85:562–582. [DOI] [PubMed] [Google Scholar]
- 22.Cardinal J. Treatment of hepatic epithelioid hemangioendothelioma: A single-institution experience with 25 cases. Arch Surg. 2009;144:1035–1039. [DOI] [PubMed] [Google Scholar]
- 23.Carotti S, Morini S, Corradini SG, Burza MA, Molinaro A, Carpino G, et al. Glial fibrillary acidic protein as an early marker of hepatic stellate cell activation in chronic and posttransplant recurrent hepatitis C. Liver Transpl. 2008;14:806–814. [DOI] [PubMed] [Google Scholar]
- 24.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bocca C, Novo E, Miglietta A, Parola M. Angiogenesis and fibrogenesis in chronic liver diseases. Cell Mol Gastroenterol Hepatol. 2015;1:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scoazec JY, Feldmann G. Both macrophages and endothelial cells of the human hepatic sinusoid express the CD4 molecule, a receptor for the human immunodeficiency virus. Hepatology. 1990;12(3 Pt 1):505–510. [DOI] [PubMed] [Google Scholar]
- 28.Eggert T, Wolter K, Ji J, Ma C, Yevsa T, Klotz S, et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30:533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HA, Ho YJ, Mezzadra R, Adrover JM, Smolkin R, Zhu C, et al. Senescence rewires microenvironment sensing to facilitate antitumor immunity. Cancer Discov. 2023;13:432–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloom SI, Islam MT, Lesniewski LA, Donato AJ. Mechanisms and consequences of endothelial cell senescence. Nat Rev Cardiol. 2023;20:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu KQ, Muratore CS, So EY, Sun C, Dubielecka PM, Reginato AM, et al. M1 macrophage-induced endothelial-to-mesenchymal transition promotes infantile hemangioma regression. Am J Pathol. 2017;187:2102–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Tchkonia T, Pirtskhalava T, Gower AC, Ding H, Giorgadze N, et al. The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell. 2015;14:644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji F, Liu Y, Shi J, Liu C, Fu S, Wang H, et al. Single-cell transcriptome analysis reveals mesenchymal stem cells in cavernous hemangioma. Front Cell Dev Biol. 2022;10:916045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun ZJ, Zhao YF, Zhang WF. Immune response: A possible role in the pathophysiology of hemangioma. Med Hypotheses. 2007;68:353–355. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Burrack RK, Li Q. Spatially resolved and highly multiplexed protein and RNA in situ detection by combining CODEX With RNAscope in situ hybridization. J Histochem Cytochem. 2022;70:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moses L, Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022;19:534–546. [DOI] [PubMed] [Google Scholar]
- 38.Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, et al. Multiplex staining by sequential immunostaining and antibody removal on routine tissue sections. J Histochem Cytochem. 2017;65:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]