Abstract
African swine fever (ASF) is originally reported in East Africa as an acute hemorrhagic fever. African swine fever virus (ASFV) is a giant and complex DNA virus with icosahedral structure and encodes a variety of virulence factors to resist host innate immune response. S273R protein (pS273R), as a SUMO-1 specific cysteine protease, can affect viral packaging by cutting polymeric proteins. In this study, we found that pS273R was an important antagonistic viral factor that suppressed cGAS-STING-mediated type I interferon (IFN-I) production. A detailed analysis showed that pS273R inhibited IFN-I production by interacting with interferon regulatory factor 3 (IRF3). Subsequently, we showed that pS273R disrupted the association between TBK1 and IRF3, leading to the repressed IRF3 phosphorylation and dimerization. Deletion and point mutation analysis verified that pS273R impaired IFN-I production independent of its cysteine protease activity. These findings will help us further understand ASFV pathogenesis.
Keywords: African swine fever virus (ASFV), cGAS-STING, S273R, IRF3, TBK1, Type I interferon (IFN-I)
Highlights
-
•
ASFV S273R is an important antagonistic viral factor that suppresses cGAS-STING-mediated type I interferon expression.
-
•
ASFV S273R modulates cGAS-STING pathway by inhibiting IRF3 phosphorylation and dimerization.
-
•
ASFV S273R interferes with the formation of the TBK1-IRF3 complex.
-
•
ASFV S273R inhibits type I interferon production, subsequently leading to the enhancement of virus replication.
1. Introduction
African swine fever (ASF) is a highly contagious disease, posing a great threat to swine industry worldwide (Dixon et al., 2020; Galindo and Alonso, 2017; Sánchez-Cordón et al., 2018; Zhou et al., 2018). ASF is caused by African swine fever virus (ASFV). ASFV is a large double-stranded DNA virus and the only member in the Asfarviridae family (Alonso et al., 2018; Rojo et al., 1999; Gacia-Beato et al., 1992). ASFV utilizes multiple mechanisms to evade host immune response to facilitate its replication (Wang et al., 2022; Dixon et al., 2019). Currently, there is no available commercial vaccine for ASF (Sanchez et al., 2019; Rock, 2017; Arias et al., 2017).
After ASFV infects pigs, the innate immune response is then activated (García-Belmonte et al., 2019). During infections with DNA viruses, cGAS senses viral DNAs to activate the cGAS-STING-TBK1-IRF3 axis to turn on the expression of type I interferons (IFN-I) and IFN-stimulated genes (ISGs) (Chen et al., 2016; Balka and De Nardo, 2021; Liang et al., 2021). The innate immune pathways are crucial for the host to resist ASFV infections (Razzuoli et al., 2020; Correia et al., 2013). However, several ASFV proteins have been identified to block or disrupt host antiviral immune responses (Ayanwale et al., 2022). For example, ASFV MGF-505-7R is shown to suppress cGAS-STING pathway by promoting autophagy-mediated STING degradation and targeting IRF3 and the IKK complex (Li et al., 2021a, 2021b). ASFV A238L, an anti-inflammatory protein with a structure similar to IκBα, inhibits NF-κB activation and therefore efficiently suppresses host inflammatory responses (Revilla et al., 1998; Granja et al., 2008). ASFV E120R directly interacts with IRF3 to block the recruitment of IRF3 to TBK1, subsequently leading to the suppression of IRF3 activation (Liu et al., 2021). ASFV DP96R is reported to negatively regulate IFN-I expression by decreasing TBK1 phosphorylation (Wang et al., 2018). These reports have suggested that inhibition of IFN-I production is an important strategy utilized by ASFV to evade host immune response and maintain its infection. Thus, deciphering mechanisms used by ASFV proteins to inhibit IFN-I production remains to be of great interest and importance for controlling ASF (Wang et al., 2021).
ASFV gene S273R encodes a 31-kDa protein that contains a “core domain” with conserved catalytic residues that have the characteristics of SUMO-1-specific cysteine proteases (Andrés et al., 2001). ASFV pS273R can cleave the viral polyproteins pp62 and pp220 in a specific way, giving rise to the same intermediates and mature products as those produced in ASFV-infected cells. Thus, pS273R is supposed to be involved in a late maturational step, which is essential for proper core assembly and infectivity of ASFV (Alejo et al., 2003). In addition, pS273R regulates host immune responses to promote viral replication (Zhao et al., 2022; Li et al., 2023). pS273R is reported to inhibit the expression of FoxJ1, a host antiviral factor (Ma et al., 2022). Recently, a study shows that pS273R negatively regulates cGAS-STING pathway by disturbing the interaction between IKKε and STING through its interaction with IKKε (Luo et al., 2022). Here, we demonstrated that pS273R inhibited IFN-I production by interfering with TBK1 and IRF3 interaction. Our findings reveal another mechanism used by pS273R to antagonize host innate immune responses.
2. Materials and methods
2.1. Cell lines and viruses
HEK293T cells (ATCC CRL-3216) were cultured in Dulbecco's minimum essential medium (DMEM) (Gibco, USA) with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin. 3D4/21 cells (ATCC CRL-2843 cells), a porcine alveolar macrophage cell line, were maintained in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin. HeLa cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were cultured and maintained in a humidified atmosphere incubator with 5% CO2 at 37 °C. The vesicular stomatitis virus (VSV) and herpes simplex virus-1 (HSV-1) were kindly provided by Dr. Xiaojia Wang (China Agricultural University, China). Pseudorabies virus (PRV) was kindly provided by Dr. Jun Tang (China Agricultural University, China). Virus titer was determined using Reed-Muench method, and expressed as the median tissue culture infective dose (TCID50). Viruses were stored at −80 °C until use.
2.2. Plasmids
Porcine IFNβ-, ISRE-, and IRF3-luciferase reporter plasmids were constructed using pGL3-Basic vector (Promaga, USA) as described elsewhere (Huang et al., 2014). pRL-TK containing the Renilla luciferase was used as a normalization control. The plasmids pRK5-Flag-cGAS, pRK5-Flag-STING, pRK5-Flag-TBK1, and pRK5-Flag-IRF3 were constructed as described previously (Wang et al., 2018). The ASFV S273R wild type (WT) and mutants, including Arm, Core, S273R-3M, and S273R-Core-3M were constructed by using pCMV-Myc-N vector. The S273R and IRF3 genes were amplified from pCMV-Myc-S273R and Flag-IRF3, and were then cloned into pGEX-6P-1 and pet30a, respectively. The IRF3 mutants, including Flag-IRF3-M1, M2, M3, M4, and M5, were cloned into pRK5-Flag. All the primers were listed in Supplementary Table S1.
2.3. ELISA
Cell supernatants were collected and assayed for porcine or human IFN-β using a porcine or human IFN-β ELISA kit (Gene-Protein Link, Beijing, China). The measured value was compared with the standard according to the manufacturer's instructions.
2.4. Antibodies and reagents
Antibodies against TBK1/NAK (38066S), phospho-TBK1/NAK (Ser172) (5483S), IRF3 (10949S), GST-Tag (2624S), His-Tag (12698S), Flag-Tag (14793S) and HA-Tag (3724S) were purchased from Cell Signaling Technology (USA). Rabbit anti-IRF3 (11312-1-AP) and Myc-Tag (16286-1-AP) polyclonal antibodies were from Proteintech (USA). Rabbit anti-phospho-IRF3 (Ser396) (SAB5700435) and α-tubulin (SAB4500087) polyclonal antibodies were purchased from Sigma (USA). Mouse monoclonal anti-β-actin (A0101) antibody was from Lablead (China). Antibody against histone 3 (sc-56616) was purchased from Santa-Aldrich (USA). Goat anti-mouse IgG (QYB001) or anti-rabbit IgG (QYB002) secondary antibodies were purchased from Qualityard Biotechnology (China). The cysteine protease inhibitor (HY-15282, E-64) was from MedChemExpress (USA). JetPRIME kit (101000046) was obtained from Polyplus Transfection (France). Double-Luciferase Reporter Assay Kit (E1960) was purchased from Promaga (USA). 2′3′-cGAMP (tlrl-nacga23-02), poly(dA:dT) (86828-69-5), and Lipofectamine 3000 (L3000015) were purchased from Invitrogen (USA).
2.5. Dual-luciferase reporter assay
HEK293T and 3D4/21 cells were cultured in 48-well plates to 80% confluence. pGL3-IFNβ-luc, pGL3-ISRE-luc, or pGL3-IRF3-luc was then co-transfected with pRL-TK, cGAS, STNG, S273R, and empty plasmids using Lipofectamine 3000 (Invitrogen, USA) into the cells. At 24 h post transfection (hpt), cell samples were harvested to analyze luciferase activity using a Double-Luciferase Reporter Assay Kit according to the manufacturer's protocol (Promega, USA).
2.6. RNA extraction and RT-PCR analysis
Total RNAs were extracted from cells with TRIzol Reagent (CWBIO, China) and were then reverse transcribed to cDNA using HiFiScript cDNA Synthesis Kit (CWBIO, China). Quantitative real-time PCR (qRT-PCR) analysis was performed using a ViiA7 real-time PCR System with SYBR Green real-time PCR Master Mix (Mei5 Biotechnology, China). The 2–ΔΔCT Ct method was used to calculate the relative expression of the target gene. Gene-specific primers for IFN-β, ISG54, ISG56 and GAPDH were designed and listed in Supplementary Table S2. The IFN-I expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward: 5′-CCTTCCGTGTCCCTACTGCCAAC-3′and reverse: 5′- GACGCCTGCTTCACCACCTTCT-3′) and presented as the change (n-fold) in induction relative to the control.
2.7. Immunofluorescence microscopy
3D4/21 cells were transfected with the HA-vector or HA-S273R for 24 h. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature. After 3 washes with ice-cold PBS, cells were permeabilized with 0.1% Triton X-100 for 3 min; 5% bovine serum albumin (BSA) in PBST was used as the blocking buffer. After blocking at room temperature for 1 h, cells were incubated with mouse anti-IRF3 (10949S, Cell Signaling Technology, USA) and rabbit anti-HA tag (12698S, Cell Signaling Technology, USA) antibody at 4 °C overnight and then with Alexa Fluor 488-conjugated affinipure goat anti-mouse IgG (H + L) (1:100, P03S19S, Gene-Protien Link, China) and Alexa Fluor 594-conjugated affinipure goat anti-rabbit IgG (H + L) (1:100, P03S19L, Gene-Protien Link, China). The cells were observed by Nikon A1 laser confocal microscopy (Japan) after DAPI (P36941, Invitrogen) nuclear staining.
2.8. IRF3 dimerization assay
IRF3 dimerization in native PAGE and Western blotting analysis were conducted as described in a previous study (Robitaille et al., 2016). Briefly, HEK293T cells in 6-well plates were transfected with HA-S273R or pRK5-HA for 12 h and then infected with VSV (MOI = 1) for 12 h. Cells were subsequently lysed with lysis buffer [50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA and 0.05% NP-40] containing 20 μmol/L NaF and 0.1 mmol/L phenylmethylsulfonyl fluoride (PMSF). A native gel (8%) was pre-run in a native running buffer (25 mmol/L Tris and 192 mmol/L glycine, pH 8.4 with or without 1% dexycholate (DOC) in cathode and anode chamber) for 30 min at 40 mA, and then the samples in the native buffer (10 μg protein, 62.5 mmol/L Tris-Cl, pH 6.8, 15% glycerol and 1% DOC) were loaded and run at 60 V. Monomeric and dimerized IRF3 molecules were detected by Western blotting analysis.
2.9. Western blotting analysis
Cells were collected and lysed with ice-cold lysis buffer, and were then incubated on ice for 15 min. Protein levels were quantified by using bicinchoninic acid (BCA) assay. The amount of protein from each extract was separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, USA). Membranes were blocked with 5% skim milk in phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBST) for 30 min, and then incubated for 2 h at room temperature with antibodies at a suitable dilution as recommended (anti-p-TBK1, -TBK1, -p-IRF3, -IRF3, -Flag, -HA, and -Myc at 1:1000; anti-β-actin at 1:2000). The membranes were then incubated with the appropriate secondary antibody for 1 h at a dilution of 1:5000. After washing, signals were visualized using enhanced chemiluminescence (CWBIO, China) according to the manufacturer's protocols. Densitometry analysis of the indicate protein levels have been assessed by Image J software, and normalized to β-actin.
2.10. Co-immunoprecipitation (Co-IP)
HEK293T cells were seeded on 6-well plates, and on the next day, cells were transfected with Flag-IRF3 and HA-S273R. At 24 hpt, cells were collected, washed with PBS, and subsequently lysed with 200 μL of the whole-cell extract buffer [50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA and 0.05% NP-40] containing 20 μmol/L NaF and 0.1 mmol/L PMSF. After a 20-min centrifugation with 12,000×g, total proteins of cell extract were incubated with 10 μL of anti-FLAG M2 magnetic beads (Sigma, Germany) overnight at 4 °C. After a 5-min centrifugation at 3000 ×g, the beads were thoroughly washed four times and the immuneprecipitates were subjected to Western blotting analysis with specific antibodies.
2.11. Glutathione S-transferase (GST) pull-down assay
pET-30a-IRF3 was transformed into Escherichia coli (E. coli) strain BL21. The transformed cells were grown at 37 °C until the concentration at A600 reached to 0.6–0.8 and then induced with IPTG (1 mmol/L) for 5 h. Cells were collected after incubation at 18 °C for 8h. His-fused IRF3 was purified using Ni-nitrilotriacetic acid (NTA) His-Bind resin (catalogue no. 70666; Novagen). The plasmid pGEX-6p-1-S273R was transformed into E. coli strain BL21. The GST and GST-S273R were induced by IPTG with a final concentration of 0.5 mmol/L. The purified GST or GST-S273R protein was incubated with glutathione Sepharose beads (Novagen, Beijing, China) and washed three times with PBS. Purified GST-S273R (10 μg) or GST protein (10 μg) was incubated with 10 μg purified His-IRF3 for 4 h at 4 °C, and the protein complexes were precipitated with glutathione sepharose beads and detected with anti-GST and anti-His antibodies.
2.12. Viral growth curve
The supernatants of virus-infected HeLa cells were collected and 10-fold serially diluted in DMEM medium containing 2% FBS with each dilution four to eight replications. HeLa cells were cultured in 96-well plates for 12 h, and then incubated with the diluted supernatants at 37 °C for 36 h. TCID50 was calculated with Reed-Muench method (Elikaei et al., 2017).
2.13. Statistical analysis
Experiments were performed independently three times. Statistical analysis was performed by using Student's t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; ns indicates no significance).
3. Results
3.1. pS273R inhibits cGAS-SITNG-mediated IFN-I production
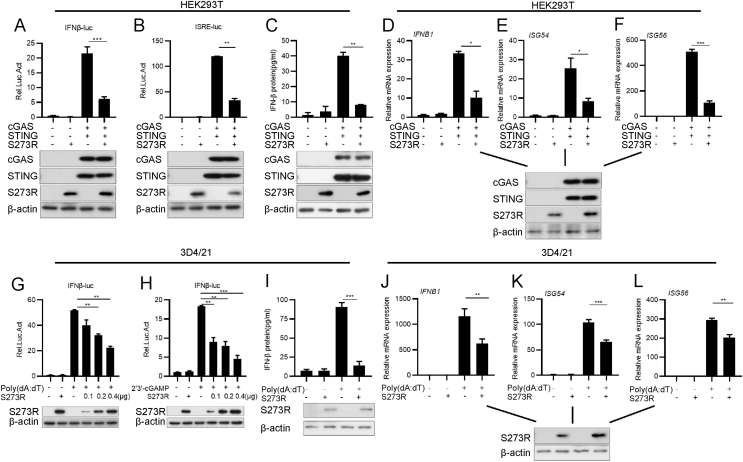
To confirm whether ASFV pS273R regulates IFN-I induction, we first evaluated the effect of pS273R on the activation of cGAS-STING-mediated IFN-β or ISRE luciferase reporter by dual-luciferase reporter assay. Flag-cGAS, Flag-STING, and IFN-β or IFN-stimulated response element (ISRE) luciferase reporter plasmids were co-transfected into HEK293T cells along with HA-S273R or pRK5-HA for 24 h, and then cells were harvested for the analysis of luciferase activities. As shown in Fig. 1A and B, pS273R significantly suppressed IFN-β and ISRE promoter activation. To verify that pS273R modulates cGAS-STING-mediated signaling pathway, we analyzed the effect of pS273R on the expression of IFN-β and ISGs. HEK293T cells were co-transfected with cGAS, STING, and S273R expression plasmids for 24 h. We harvested the supernatants for the analysis of IFN-β production using ELISA. As shown in Fig. 1C, IFN-β secretion was suppressed by pS273R. Cells were then collected to analyze IFN-β, ISG54 and ISG56 expressions by qRT-PCR. Our results showed that pS273R significantly inhibited cGAS-STING-mediated IFN-β, ISG54 and ISG56 expressions (Fig. 1D–F).
Fig. 1.
pS273R inhibits cGAS-STING-mediated IFN-β production. A, B Flag-S273R was co-transfected with IFNβ-Luc (20 ng) or ISRE-Luc (20 ng), pRL-TK (2 ng), Flag-cGAS (10 ng) and Flag-STING (40 ng) into HEK293T cells for 24 h followed by luciferase reporter assay. The expression of cGAS, STING, or pS273R was analyzed by Western blotting with β-actin as an internal control. C–F HEK293T cells were transfected with Flag-cGAS, Flag-STING, and S273R (0.2 μg) or empty vector for 24 h. The supernatants were collected to measure IFN-β using an ELISA kit (C), and the mRNA levels of IFN-β (D), ISG54 (E), and ISG56 (F) were quantified by qRT-PCR. G, H 3D/21 cells were transfected with IFNβ-Luc, pRL-TK plasmids, and increasing doses of Flag-S273R expression plasmid (0, 50, 100, and 200 ng). At 24 hpt, cells were treated with poly(dA:dT) (1 μg/mL) or 2′3′-cGAMP (1 μg/mL) for 12 h. The promoter activation of IFN-β was evaluated by the luciferase assay. The expression of pS273R was analyzed by Western blotting with β-actin as an internal control. J–L 3D4/21 cells cultured in 24-well plates were transfected with 200 ng of empty vector or Flag-S273R expression plasmid. At 24 hpt, cells were transfected with poly(dA:dT) (1 μg/mL) for 12 h, and the mRNA levels of IFN-β (J), ISG54 (K), and ISG56 (L) were analyzed by qRT-PCR. I Transfection experiments were performed as described in the legend to panel (J). The supernatants were collected, and the amount of secreted IFN-β was measured by using an ELISA kit. Data are presented as mean ± SD of three independent experiments. Statistical significance between groups was determined using a t-test with GraphPad Prism software. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
To further confirm the inhibitory effect of pS273R on IFN-β production, we used porcine alveolar macrophage cell line 3D4/21 cells. 3D4/21 cells were co-transfected with IFN-β luciferase reporter plasmid, pRL-TK, HA vector or HA-S273R expression plasmid. At 24 hpt, cells were treated with the synthetic double-stranded DNA (dsDNA)-mimetic poly(dA:dT) or 2′3′-cGAMP. At 12 h later, cells were harvested for the analysis of IFN-β promoter activation. Our results showed that pS273R significantly suppressed IFN-β promoter activation induced by poly(dA:dT) and 2′3′-cGAMP in a dose-dependent manner, respectively (Fig. 1G and H). Next, we analyzed the effect of pS273R on IFN-β expression. 3D4/21 cells were transfected with HA vector or HA-S273R expression plasmid and then stimulated with poly(dA:dT) for 12 h. IFN-β secretion induced by poly(dA:dT) was also suppressed by pS273R (Fig. 1I). The expressions of porcine IFN-β, ISG54, and ISG56 induced by poly(dA:dT) were all significantly repressed by pS273R (Fig. 1J–L). Taken together, these results indicate that ASFV pS273R inhibits IFN-β production mediated by cGAS-STING signaling pathway.
3.2. pS273R inhibits IRF3 activation
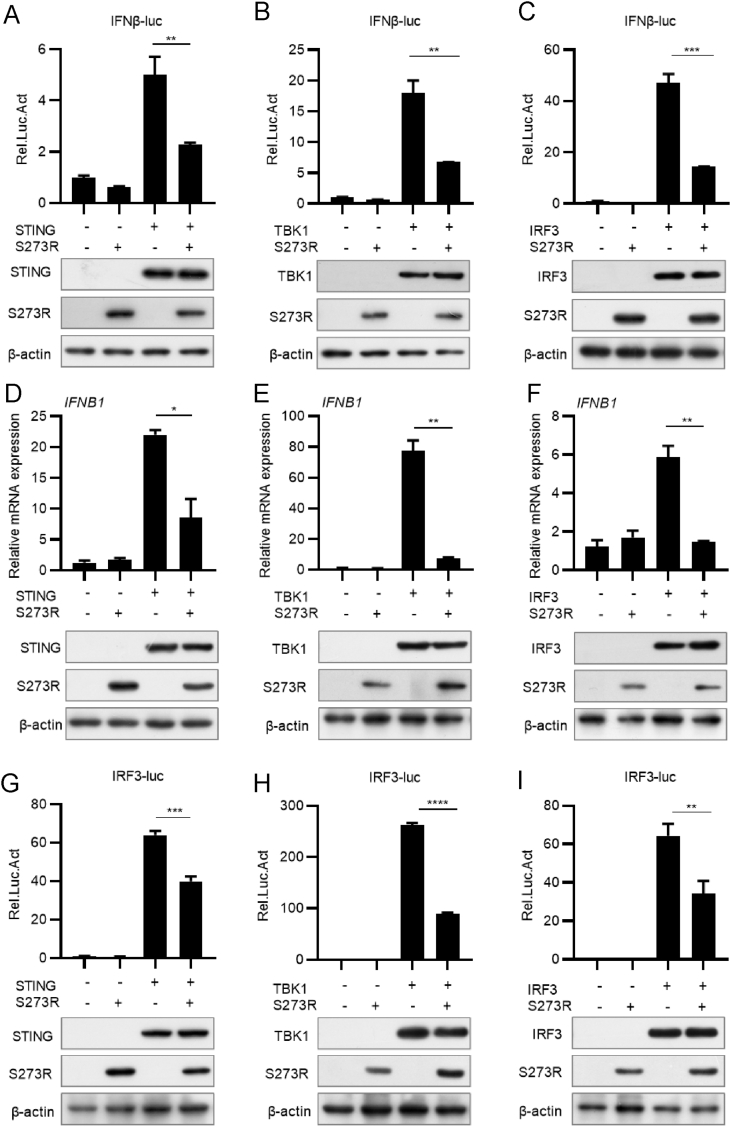
The observed inhibition of IFN-I production by ASFV pS273R raises the possibility that pS273R may target one or several components in cGAS-STING signaling pathway. To identify the potential component(s) targeted by pS273R, we transfected S273R expression plasmid into HEK293T cells with each plasmid expressing the component (including STING, TBK1, and IRF3) in the cGAS-STING signaling pathway, IFNβ-Luc and pRL-TK plasmids. At 24 hpt, cells were collected to analyze the activation of the IFN-β promoter. As shown in Fig. 2A–C, all the signaling components (STING, TBK1, and IRF3) activated IFN-β promoter. However, their activities were downregulated by pS273R. To investigate the effect pS273R on IFN-β expression, we transfected HEK293T cells with S273R, and STING, TBK1 or IRF3 expressing plasmids. Cells were harvested for the analysis of IFN-β expression by qRT-PCR. As expected, pS273R significantly downregulated IFN-β expression induced by STING (Fig. 2D), TBK1 (Fig. 2E), or IRF3 (Fig. 2F). To further verify that pS273R disrupts IRF3 activation, we co-transfected IRF3-Luc (which contains four IRF3 responsive elements of the IFN-β promoter) and S273R plasmids into HEK293T cells with STING, TBK1, or IRF3 plasmid. As shown in Fig. 2G–I, IRF3-Luc was activated by STING, TBK1, and IRF3, respectively. However, the luciferase activity of IRF3-Luc was suppressed in the presence of pS273R.
Fig. 2.
pS273R suppresses IFN-β induction mediated by STING, TBK1, or IRF3. A–C The expression plasmid of the signaling molecule STING (50 ng), TBK1 (10 ng), or IRF3 (50 ng) was transfected into HEK293T cells with IFN-β-Luc (20 ng), pRL-TK (2 ng), and S273R (100 ng) expression plasmids. At 24 hpt, cells were harvested for luciferase activity analysis. The expression of STING, TBK1, IRF3, pS273R and β-actin was analyzed by Western blotting. D–F Experiments were conducted as in (A) to (C) except that transfection was performed without IFN-β-Luc and pRL-TK. The mRNA level of IFN-β was analyzed by qRT-PCR. The expression of STING, TBK1, IRF3, pS273R and β-actin was analyzed by Western blotting. G–I The dual-luciferase reporter assay was performed as in panels (A) to (C) except that IRF3-Luc plasmid was used instead of IFN-β-Luc. The expression of all proteins was analyzed by Western blotting with β-actin as an internal control. Data are presented as mean ± SD of three independent experiments. Statistical significance between groups was determined using a t-test with GraphPad Prism software. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
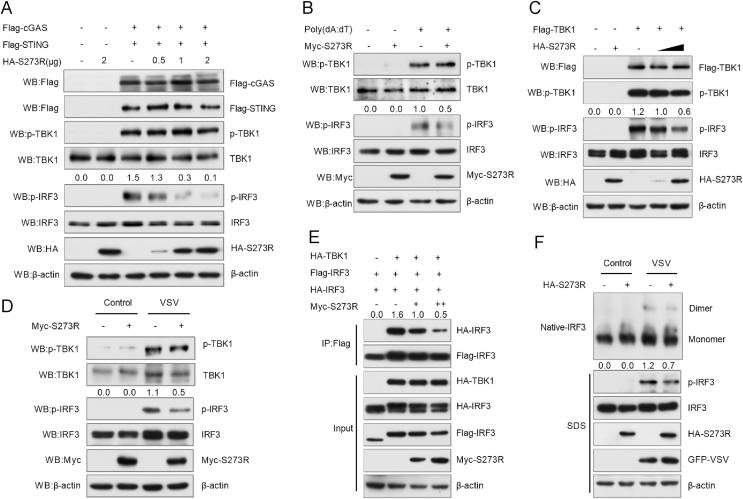
Next, we analyzed the effect of pS273R on TBK1 and IRF3 phosphorylation. pS273R expression plasmid was co-transfected with Flag-cGAS and Flag-STING into HEK293T cells. At 24 hpt, cells were collected for the analysis of TBK1 and IRF3 phosphorylation. Western blotting results showed that pS273R did not inhibit TBK1 phosphorylation, while it inhibited cGAS-STING induced IRF3 phosphorylation in a dose-dependent manner (Fig. 3A). Meanwhile, pS273R did not affect the expression of TBK1 and IRF3. To confirm this result, we transfected 3D4/21 cells with pS273R. At 24 hpt, cells were stimulated with poly(dA:dT). Similarly, pS273R downregulated poly(dA:dT)-induced IRF3 phosphorylation (Fig. 3B). To further verify the impact of pS273R on IRF3 activation, HEK293T cells were co-transfected with TBK1 and pS273R. Our data showed that pS273R also significantly inhibited TBK1-induced IRF3 phosphorylation in a dose-dependent manner (Fig. 3C). Previous studies have shown that VSV can induce IFN-β by activating IRF3 signaling pathway. Thus, we also evaluated the effect of pS273R on VSV triggered IRF3 phosphorylation. Myc-S273R or pCMV-Myc plasmid was transfected into HEK293T cells for 12 h, followed by infection with VSV (MOI = 1) for 12 h. Cells were then harvested to analyze IRF3 phosphorylation. Our results indicated that pS273R inhibited VSV-induced IRF3 phosphorylation (Fig. 3D). These results suggest that pS273R interferes with IRF3 activation.
Fig. 3.
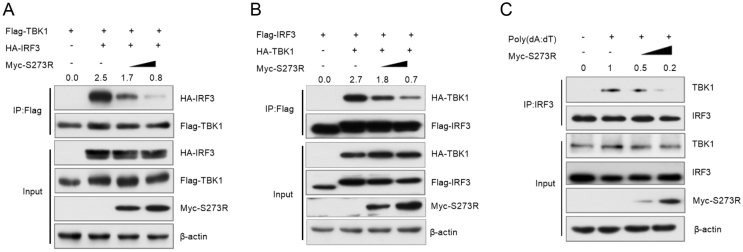
pS273R suppresses IRF3 phosphorylation and dimerization. A HEK293T cells were transfected with Flag-cGAS (100 ng) and Flag-STING (320 ng) plasmids, and different doses of HA-S273R (0, 0.5, 1, and 2 μg) or empty vector. Cells were harvested at 24 hpt. Western blotting was used to examine cGAS, STING, TBK1, p-TBK1, IRF3, p-IRF3, pS273R, and β-actin. B 3D4/21 cells were transfected with empty vector or Myc-S273R (1 μg) for 24 h and then stimulated with poly(dA:dT) (1 μg/mL) for 12 h. The cells were harvested for Western blotting analysis. C HA-S273R plasmid (500 ng or 1000 ng) was co-transfected with Flag-TBK1 (200 ng) into HEK293T cells. After 24 h, cells were harvested for Western blotting analysis. D HEK293T cells were transfected with Myc-S273R for 12 h and then infected with VSV (MOI = 1) for 12 h. Cells were lysed for Western blotting analysis to determine the levels of p-TBK1, TBK1, p-IRF3, IRF3, pS273R, and β-actin. E HEK293T cells were transfected with Flag-IRF3, HA-IRF3, HA-TBK1, and Myc-S273R (1 μg or 2 μg). At 24 hpt, cells were harvested for co-immunoprecipitation analysis. F HEK293T cells were transfected with pRK5-HA (1 μg) or HA-S273R for 12 h and then infected with VSV (MOI = 1) for 12 h. Whole cell extracts were subjected to native PAGE. Dimers or monomers of IRF3, pS273R, and β-actin were detected with the respective antibodies.
IRF3 phosphorylation is required for the formation of IRF3 dimer. To test whether pS273R affects IRF3 dimerization, we co-transfected Myc-S273R expression plasmid together with Flag-IRF3, HA-IRF3, and HA-TBK1 plasmids into HEK293T cells. At 24 hpt, cells were harvested for Co-IP analysis with anti-Flag monoclonal antibody (MAb). The results showed that pS273R significantly inhibited TBK1-induced IRF3 dimerization (the interaction of IRF3-Flag/IRF3-HA) (Fig. 3E). In addition, we also analyzed IRF3 dimerization by using native PAGE. HEK293T cells were transfected with HA-S273R expression plasmid for 12 h, and then infected with VSV (MOI = 1). At 12 h post infection (hpi), cells were harvested for native PAGE analysis. Our results showed that pS273R strikingly repressed IRF3 dimerization induced by VSV infection (Fig. 3F).
Taken together, these data suggest that ASFV pS273R disrupts cGAS-STING pathway by inhibiting IRF3 phosphorylation and dimerization.
3.3. pS273R interacts with IRF3
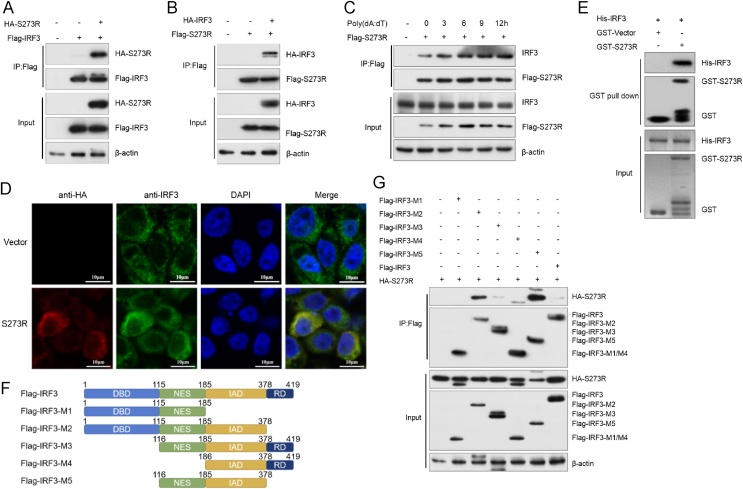
The specific inhibition of IRF3 by pS273R prompts us to investigate the possible interaction between the two proteins. HEK293T cells were transfected with Flag-IRF3 and HA-S273R expression plasmids. At 24 hpt, cells were harvested for Co-IP analysis using anti-Flag antibody. As shown in Fig. 4A, pS273R was immunoprecipitated with IRF3 by the anti-Flag antibody. And reciprocally, HA-IRF3 was also immunoprecipitated with Flag-S273R (Fig. 4B), suggesting that pS273R interacts with IRF3. To verify that pS273R interacts with IRF3, we analyzed the interaction between pS273R and endogenous IRF3. Flag-S273R was transfected into 3D4/21 cells for 24 h, and cells were then stimulated by poly(dA:dT). Next, cells were collected at 0, 3, 6, 9, and 12 h after stimulation for co-immunoprecipitation. As shown in Fig. 4C, endogenous IRF3 was co-precipitated with pS273R. To further confirm the interaction between pS273R and IRF3, we performed indirect immunofluorescence microscopy assay (IFA) to test whether pS273R and IRF3 share the similar subcellular locations. 3D4/21 cells were transfected with HA-vector or HA-S273R plasmid. At 24 hpt, cells were subjected to confocal microscopy analysis. As shown in Fig. 4D, pS273R mainly colocalized with IRF3 in the cytoplasm. To explore whether there is a direct physical interaction between S273R and IRF3, a GST pull-down assay was performed. As shown in Fig. 4E, His-IRF3 was effectively pulled down by GST-S273R, but not GST.
Fig. 4.
pS273R interacts with IRF3. A, B HEK293T cells were transfected with HA-S273R (1 μg) and Flag-IRF3 (1 μg) or HA-IRF3 and Flag-S273R expression plasmids for 24 h. The interaction of IRF3 and pS273R was analyzed by co-immunoprecipitation assay and Western blotting. C 3D4/21 cells were transfected with Flag-S273R (1 μg) for 24 h and then transfected with poly(dA:dT) (1 μg/mL). Cells were harvested at 0, 3, 6, 9, and 12 h after stimulation. Cell lysates were immunoprecipitated with anti-Flag antibody and subjected to Western blotting using the indicated antibodies. D 3D4/21 cells were transfected with empty vector or HA-S273R (500 ng) expression plasmids for 24 h. Cells were then fixed and processed for dual labeling. IRF3 (green) and pS273R (red) were visualized by immunostaining with rabbit anti-HA and mouse anti-IRF3 antibodies. Cell nuclei was counterstained with DAPI (blue). The areas of colocalization in merged images were shown in yellow. Scale bar, 10 μm. E Purified GST or GST-S273R protein was incubated with the purified His-IRF3 and analyzed by Western blotting using anti-GST or -His monoclonal antibody. F Schematic of full-length IRF3 and its truncated mutants. G HEK293T cells were transfected with full-length Flag-IRF3 (1 μg) or each of its truncated mutants, and HA-S273R (1 μg) expression plasmids. At 24 hpt, cell lysates were immunoprecipitated with anti-Flag antibody, and the whole-cell lysates were analyzed by Western blotting.
In addition, to map the domain(s) of IRF3 required for its interaction with pS273R, we generated five IRF3 truncated mutants: M1 (aa 1 to 185), M2 (aa 1 to 378), M3 (aa 116 to 419), M4 (aa 186 to 419), and M5 (aa 116 to 378) (Fig. 4F). Each of the Flag-tagged IRF3 mutants or full-length IRF3 was co-transfected with HA-S273R into HEK293T cells. At 24 hpt, cell lysates were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting. As shown in Fig. 4G, pS273R interacted with IRF3-M2, IRF3-M3, and IRF3-M5, but not with IRF3-M1 and IRF3-M4. These results reveal that the region of amino acids 116 to 378 in IRF3 is crucial for IRF3-pS273R interaction.
3.4. pS273R interferes with the interaction between IRF3 with TBK1
The interaction between TBK1 and IRF3 is critical for IRF3 phosphorylation and IFN-I production (Zhao, 2013). Our initial results showed that pS273R interacted with IRF3 and suppressed its phosphorylation, but did not affect TBK1 phosphorylation. Therefore, it is reasonable to infer that pS273R might disrupt the interaction between TBK1 and IRF3. To test this assumption, we transfected HEK293T cells with TBK1 and IRF3 expression plasmids along with S273R plasmid. At 24 hpt, cells were harvested for Co-IP analysis with the anti-Flag MAb. As shown in Fig. 5A, IRF3 was pulled down by TBK1. However, the amount of IRF3 pulled down by TBK1 gradually decreased as the amount of pS273R increased. Conversely, when pS273R was present, the amount of TBK1 pulled down by IRF3 was also significantly decreased (Fig. 5B). Furthermore, we used 3D4/21 cells to test whether pS273R has an effect on the interaction between endogenous TBK1 and IRF3. Cells were transfected with pS273R for 24 h, and then were treated with poly(dA:dT). At 12 h after stimulation, cells were harvested for immunoprecipitation. As shown in Fig. 5C, TBK1 was pulled down by anti-IRF3 antibody, and pS273R inhibited the interaction between IRF3 and TBK1. Overall, our findings suggest that pS273R disrupts the formation of the TBK1-IRF3 complex.
Fig. 5.
pS273R impairs the interaction of IRF3 and TBK1. A, B Myc-S273R (1 μg or 2 μg) was co-transfected with Flag-TBK1 (800 ng) and HA-IRF3 (1 μg) or HA-TBK1 and Flag-IRF3 as indicated. At 24 hpt, cells were harvested. The lysates were immunoprecipitated with anti-Flag antibody and then subjected to Western blotting analysis. C 3D4/21 cells were transfected with Myc vector or different doses of Myc-S273R (1 μg or 2 μg) expression plasmid. At 24 hpt, cells were transfected with poly(dA:dT) (1 μg/mL) for 12 h. Cells were lysed, and the lysates were immunoprecipitated with anti-IRF3 antibody and subjected to Western blotting using the indicated antibodies.
3.5. pS273R represses IRF3 activation independently of its cysteine protease activity
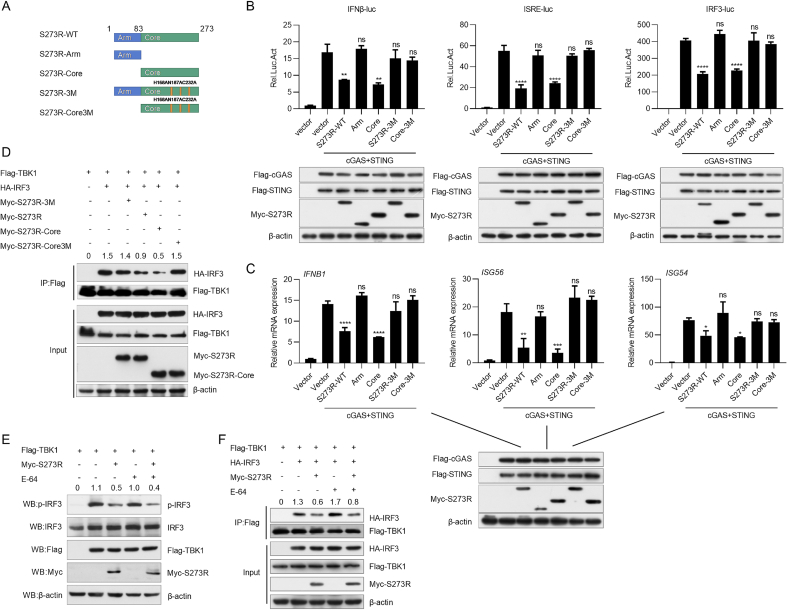
pS273R has two domains. The “arm domain” plays an important role in maintaining its enzyme activity, and the “core domain” contains the catalytic triad (C232-H168-N187) (Li et al., 2020). To explore the key domains and amino acids in S273R required for the suppression of IFN-I production, we constructed four S273R mutants: S273R-Arm (aa 1 to 83), S273R-Core (aa 84 to 273), S273R-3M (C232A, H168A, N187A), and S273R-Core-3M (C232A, H168A, N187A). Based on its structure, all these pS273R mutants have no enzyme activity (Fig. 6A). Then, we investigated the effect of these mutants on IFN-I production. All the mutants were co-transfected into HEK293T cells with cGAS and STING expression plasmids, and IFN-β-, ISRE-, or IRF3-luciferase reporter. As shown in Fig. 6B, S273R-Arm, S273R-3M, and S273R-Core3M, but not the S273R-Core mutant, almost completely lost the ability to inhibit the activation of IFN-β, ISRE, and IRF3 luciferase reporters mediated by cGAS and STING. In addition, S273R-Arm, S273R-3M, and S273R-Core3M mutants had no significant effect on IFN-β, ISG54, and ISG56 expressions, respectively (Fig. 6C). These data suggest that the core domain of pS273R is required for pS273R to inhibit IFN-I production. To examine whether these mutants affected the interaction between IRF3 and TBK1, we transfected HEK293T cells with Myc vector, Myc-S273R, Myc-S273R-Core, Myc-S273R-3M, or Myc-S273R-Core3M expression plasmid along with Flag-TBK1 and HA-IRF3 expression plasmids. At 24 hpt, cell lysates were immunoprecipitated with anti-Flag antibody and analyzed by Western blotting. As expected, S273R-3M and S273R-Core3M did not affect the interaction between IRF3 and TBK1 (Fig. 6D). These results suggest that the core domain and the catalytic triad (C232-H168-N187) are essential for pS273R to interfere with IRF3 and TBK1 interaction. It has been reported that E-64 effectively inhibits the enzyme activity of the pS273R protease (Liu et al., 2021; Lu et al., 2023). To verify whether pS273R inhibits IFN-β production dependently of its cysteine protease activity, we transfected S273R and TBK1 into HEK293T cells. At 12 h later, cells were treated with cysteine protease inhibitor E-64. Cells were then harvested to analyze IRF3 phosphorylation at 36 h after the treatment. As shown in Fig. 6E and F, pS273R still inhibited TBK1-induced IRF3 phosphorylation and the formation of TBK1-IRF3 complex in the presence of the inhibitor E-64. These results imply that pS273R inhibits IRF3 activation independent of its cysteine protease activity.
Fig. 6.
pS273R affects IRF3 activation independently of its cysteine protease activity. A Schematic of full-length pS273R and its truncated mutants. B cGAS (10 ng), STING (40 ng), pRL-TK (2 ng) plasmid, and IFNβ-Luc (20 ng), ISRE-Luc (20 ng), or IRF3-Luc (20 ng) reporter plasmid were co-transfected into HEK293T cells with empty vector, pS273R wild type (WT) (100 ng), Arm, Core, S273R-3M, or Core-3M expression plasmid. At 24 hpt, cells were harvested to analyze the luciferase activity. The expression of Flag-cGAS, Flag-STING and various S273R mutants were determined by Western blotting. C Flag-cGAS and Flag-STING were co-transfected with empty vector, pS273R WT, Arm, Core, S273R-3M, or Core-3M expression plasmid into HEK293T cells. At 24 hpt, the mRNA levels of IFN-β, ISG56, and ISG54 were analyzed by qRT-PCR. D Flag-TBK1 (800 ng) and HA-IRF3 (1 μg) were co-transfected with pS273R WT (1 μg), Core, S273R-3M, or Core-3M expression plasmid in HEK293T cells for 24 h. Cells were harvested and the lysates were immunoprecipitated with anti-Flag antibody and subjected to Western blotting analysis. E HEK293T cells were transfected with Flag-TBK1 (500 ng) and Myc-S273R (1 μg) for 12 h and then treated with E-64 (4 mmol/L) for 36 h. The cells were harvested for Western blotting analysis. F HEK293T cells were transfected with Flag-TBK1 (800 ng), HA-IRF3 (1 μg), and Myc-S273R (1 μg) for 12 h and then treated with E-64 (4 mmol/L) for 36 h. Cells were then harvested for co-immunoprecipitation analysis. Data are presented as mean ± SD of three independent experiments. Statistical significance between groups was determined using a t-test with GraphPad Prism software. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ns, not significant.
3.6. pS273R promotes HSV-1 and VSV replication by inhibiting IFN-I production
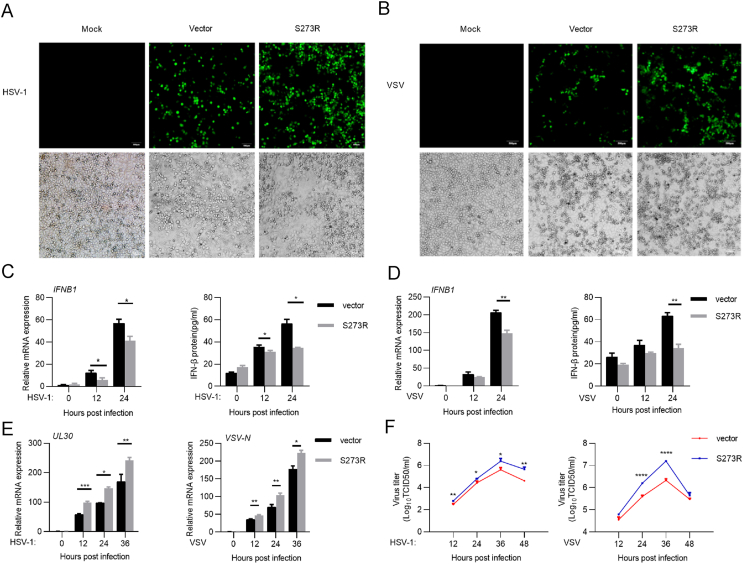
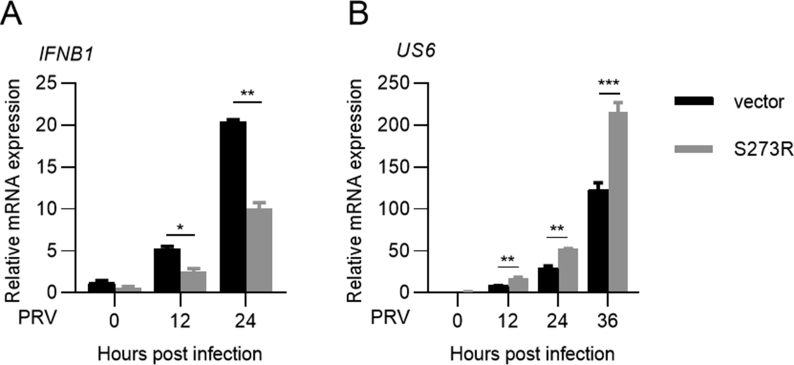
The innate immune system is capable of sensing virus infection and induces antiviral response (Ma and Damania, 2016). To determine the role of pS273R in regulating the innate immune response against pathogen invasion, we used two model viruses, HSV-1, a DNA virus, and VSV, an RNA virus. HeLa cells were transfected with S273R for 12 h, and then infected with HSV-1 or VSV (MOI = 0.001). As shown in Fig. 7A and B, the number of HSV-1 or VSV infected cells increased in the presence of pS273R. To investigate the effects of pS273R on IFN-I production during virus infection, we transfected HeLa cells with control vector or pS273R expression plasmid, and then infected cells with HSV-1 or VSV (MOI = 0.01) 12 h later. At 0 h, 12 h, and 24 hpi, cells and supernatants were collected to analyze IFN-β expression by qRT-PCR and ELISA, respectively. The results showed that pS273R suppressed IFN-β production stimulated by HSV-1 (Fig. 7C) or VSV infection (Fig. 7D). To detect whether pS273R promotes HSV-1 and VSV replication, we measured the expression of HSV-1 DNA polymerase (UL30) and VSV N protein by qRT-PCR. As shown in Fig. 7E, HSV-1 UL30 or VSV N transcription was significantly upregulated by pS273R compared to the control. In addition, we investigated pseudorabies virus (PRV) infection in pS273R transfected 3D4/21 cells. pS273R also significantly repressed the production of IFN-β, and promoted PRV replication (Supplementary Fig. S1). Besides, the virus titers in the supernatants of HeLa cells infected with HSV-1 or VSV at an MOI 0.01 was increased when pS273R was present (Fig. 7F). Taken together, these data demonstrate that ASFV pS273R inhibits IFN-I production, subsequently leading to the enhancement of virus replication.
Fig. 7.
pS273R promotes HSV-1 and VSV replication by inhibiting IFN-I production. A, B HeLa cells were transfected with empty or Myc-S273R plasmid. At 12 hpt, cells were infected with HSV-1 or VSV at an MOI of 0.001 for 24 h and then were analyzed by fluorescence microscope. Scale bar, 500 μm. C–F HeLa cells were transfected with empty or Myc-S273R plasmid for 12 h and then infected with HSV-1 (MOI = 0.01) or VSV (MOI = 0.01) for the indicated times. The infected cells and supernatants were harvested to analyze IFN-β expression (C, D). HSV-1 UL30 and VSV N gene transcriptions were measured by qRT-PCR (E). The viral titer in the supernatant from HSV-1 or VSV infected cells was analyzed by TCID50 assay (F). Data are presented as mean ± SD of three independent experiments. Statistical significance between groups was determined using a t-test with GraphPad Prism software. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001.
4. Discussion
In the present study, we demonstrated that ASFV pS273R suppressed cGAS-STING-mediated IFN-I production by interfering with IRF3 and TBK1 interaction.
IRF3 is critical for various pattern recognition receptors (PRRs) to trigger IFN production (Canivet et al., 2018). During viral infection, IRF3 undergoes phosphorylation, dimerization, and translocation into the nucleus, leading to the expression of IFN-I (Xue et al., 2018). Thus, many viruses have evolved to encode proteins to interfere with IRF3 activation (Petro, 2020). Influenza NS1 protein of the 2009H1N1 and H5N1 is reported to block IRF3 activation (Kuo et al., 2010). HSV-2 ICP27 protein and ICP0 protein are found to inhibit IRF3 activity by promoting IRF3 degradation (Melroe et al., 2004). Varicella zoster ORF61 protein is also reported to promote IRF3 degradation. West Nile Virus NS1 protein blocks IRF3 translocation, thereby abrogating its transcriptional activity (Wilson et al., 2008). Porcine reproductive and respiratory syndrome virus NSP1 protein is reported to inhibit IRF3 phosphorylation (Beura et al., 2010). Both hepatitis A virus (Qu et al., 2011) and mouse Theiler virus (Ricour et al., 2009) encode leader proteins to block IRF3 dimerization, thereby preventing its activation. Here, we showed that ASFV pS273R targeted IRF3 to inhibit its interaction with TBK1, confirming that IRF3 is a common target for many viruses.
pS273R belongs to SUMO-1-specific cysteine protease and is a multifunctional protein in ASFV replication (Alejo et al., 2003). Crystal structures reveal that pS273R contains an arm domain and a core domain. A canonical catalytic triad composed of Cys232, His168, and Asp187 is located in the core domain (Li et al., 2020). A report shows that pS273R inhibits cGAS-STING pathway by interfering with IKK sumoylation dependently on pS273R catalytic activity (Luo et al., 2022). To examine whether the cysteine protease activity is also essential for pS273R to interfere with IRF3 activation, we constructed pS273R point (Cys232, His168, and Asp187) and deletion mutants without enzymatic activity. We found that all four mutants are protease inactive, and all except core mutant lose the inhibition ability, thus it appears that the protease activity of pS273R plays a role in the inhibition of IRF3 activation. However, when cysteine protease inhibitor was present, pS273R still inhibited the interaction between TBK1 and IRF3, implying that its cysteine protease activity might not be essential for pS273R to interfere with the complex formation of IRF3 and TBK1. Interestingly, S273R-Core-3M (in which the C232, H168 and N187 were mutated to alanine) did not repress IFN-I production and the formation of TBK1-IRF3 complex, suggesting that the three amino acids, C232, H168, and N187 play an essential part in antiviral immune responses. Therefore, our results are complementary to previous reports (Luo et al., 2022; Zhao et al., 2022).
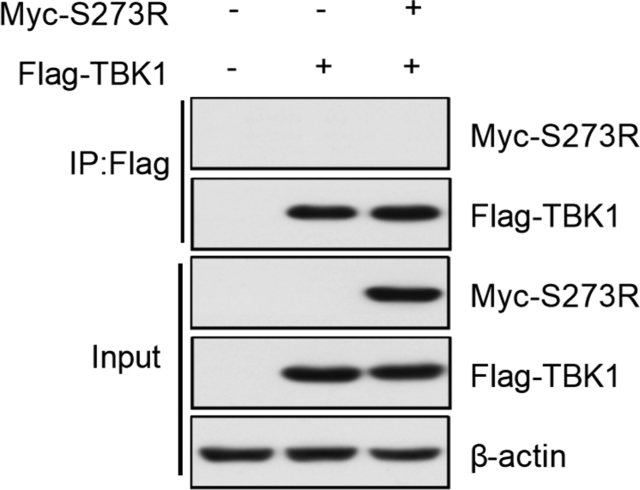
IRF3 contains a conserved DNA binding domain (DBD), an IRF association domain (IAD), a C-terminal regulatory domain (RD), and a nuclear export sequence (NES) (Zhang et al., 2019; Gottipati et al., 2016). Since pS273R does not interact with TBK1 (Supplementary Fig. S2), the interaction between pS273R and IRF3 seems to block the interaction between TBK1 and IRF3. TBK1 interacts with the IAD domain of IRF3, and then activates IRF3 phosphorylation, dimerization and translocation to the nucleus (Ikeda; et al., 2007). The 116–378 aa region of IRF3 includes the IAD domain, and we find that this region is critical for IRF3 to interact with pS273R. Thus, we assume that pS273R competes with TBK1 to bind to the IAD domain of IRF3.
In summary, our data demonstrate that pS273R interferes with the formation of the TBK1-IRF3 complex by interacting with IRF3, resulting in the suppression of IFN-I production. These findings suggest a novel strategy used by ASFV to subvert IFN-I production and evade host innate immune response.
5. Conclusions
In conclusion, we demonstrate that ASFV pS273R is an important antagonistic viral factor, which can be used by ASFV to subvert IFN-I production to evade host immune response.
Data availability
All the data generated during the current study are included in the manuscript. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any authors.
Author contributions
Hui Li: conceptualization, formal analysis, data curation, investigation, writing-original draft. Xiaojie Zheng: methodology, and investigation. You Li: methodology. Yingqi Zhu: methodology. Yangyang Xu: methodology. Zilong Yu: methodology. Wen-Hai Feng: funding acquisition, resources, conceptualization, supervision, project administration, writing-review and editing. All authors have read and agreed to the version of the manuscript.
Conflict of interest
Prof. Wen-Hai Feng is an editorial board member for Virologica Sinica and was not involved in the editorial review or the decision to publish this article. The authors declare that they have no conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 32172869), China.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2023.08.009.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
References
- Alejo A., Andres G., Salas M.L. African Swine Fever virus proteinase is essential for core maturation and infectivity. J. Virol. 2003;77:5571–5577. doi: 10.1128/JVI.77.10.5571-5577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Borca M., Dixon L., Revilla Y., Rodriguez F., Escribano J.M., Ictv, Report C. ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 2018;99:613–614. doi: 10.1099/jgv.0.001049. [DOI] [PubMed] [Google Scholar]
- Andrés G., Alejo A., Simón-Mateo C., Salas M.L. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001;276:780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- Arias M., de la Torre A., Dixon L., Gallardo C., Jori F., Laddomada A., Martins C., Parkhouse R.M., Revilla Y., Rodriguez F.A.J., Sanchez V. Approaches and perspectives for development of african swine fever virus vaccines. Vaccines. 2017;5:35. doi: 10.3390/vaccines5040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayanwale A., Trapp S., Guabiraba R., Caballero I., Roesch F. New insights in the interplay between african swine fever virus and innate immunity and its impact on viral pathogenicity. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.958307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balka K.R., De Nardo D. Molecular and spatial mechanisms governing STING signalling. FEBS J. 2021;288:5504–5529. doi: 10.1111/febs.15640. [DOI] [PubMed] [Google Scholar]
- Beura L.K., Sarkar S.N., Kwon B., Subramaniam S., Jones C., Pattnaik A.K., Osorio F.A. Porcine reproductive and respiratory syndrome virus nonstructural protein 1β modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canivet C., Rheaume C., Lebel M., Piret J., Gosselin J., Boivin G. Both IRF3 and especially IRF7 play a key role to orchestrate an effective cerebral inflammatory response in a mouse model of herpes simplex virus encephalitis. J. Neurovirol. 2018;24:761–768. doi: 10.1007/s13365-018-0666-9. [DOI] [PubMed] [Google Scholar]
- Chen Q., Sun L., Chen Z.J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Correia S., Ventura S., Parkhouse R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013;173:87–100. doi: 10.1016/j.virusres.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Dixon L.K., Islam M., Nash R., Reis A.L. African swine fever virus evasion of host defences. Virus Res. 2019;266:25–33. doi: 10.1016/j.virusres.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.K., Stahl K., Jori F., Vial L., Pfeiffer D.U. African swine fever epidemiology and control. Annu Rev Anim Biosci. 2020;8:221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- Elikaei A., Hosseini S.M., Sharifi Z. Inactivation of model viruses and bacteria in human fresh frozen plasma using riboflavin and long wave ultraviolet rays. Zh. Mikrobiol. Epidemiol. Immunobiol. 2017;9:50–54. [PMC free article] [PubMed] [Google Scholar]
- Gacia-Beato R., Salas M.L., Vinuela E., Salas J. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology. 1992;188:637–649. doi: 10.1016/0042-6822(92)90518-t. [DOI] [PubMed] [Google Scholar]
- Galindo I., Alonso C. African swine fever virus: a review. Viruses. 2017;9:103. doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Belmonte R., Pérez-Núñez D., Pittau M., Richt J.A., Revilla Y., Shisler J.L. African swine fever virus Armenia/07 virulent strain controls interferon beta production through the cGAS-STING pathway. J. Virol. 2019;93 doi: 10.1128/JVI.02298-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati K., Holthauzen L.M., Ruggli N., Choi K.H. Pestivirus npro directly interacts with interferon regulatory factor 3 monomer and dimer. J. Virol. 2016;90:7740–7747. doi: 10.1128/JVI.00318-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja A.G., Perkins N.D., Revilla Y. A238L inhibits NF-ATc2, NF-kappa B, and c-Jun activation through a novel mechanism involving protein kinase C-theta-mediated up-regulation of the amino-terminal transactivation domain of p300. J. Immunol. 2008;180:2429–2442. doi: 10.4049/jimmunol.180.4.2429. [DOI] [PubMed] [Google Scholar]
- Huang C., Zhang Q., Guo X.K., Yu Z.B., Xu A.T., Tang J., Feng W.H. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 antagonizes beta interferon expression by targeting the NF-kappaB essential modulator. J. Virol. 2014;88:10934–10945. doi: 10.1128/JVI.01396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., Hecker C.M., Rozenknop A., Dietrich R., Nordmeier Involvement of the ubiquitin-like domain of TBK1 IKK-i kinases in regulation of IFN-inducible genes. EMBO J. 2007;26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo R.-L., Zhao C., Malur M., Krug R.M. Influenza A virus strains that circulate in humans differ in the ability of their NS1 proteins to block the activation of IRF3 and interferon-β transcription. Virology. 2010;408:146–158. doi: 10.1016/j.virol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu X., Yang M., Zhang G. Crystal structure of african swine fever virus pS273R protease and implications for inhibitor design. J. Virol. 2020;94 doi: 10.1128/JVI.02125-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang J., Yang W., Li P., Ru Y., Kang W., Li L., Ran Y., Zheng H. African swine fever virus protein MGF-505-7R promotes virulence and pathogenesis by inhibiting JAK1- and JAK2-mediated signaling. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Song J., Kang L., Huang L., Zhou S., Hu L., Zheng J., Li C., Zhang X., He X., Zhao D., Bu Z., Weng C. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Li X., Wang X., Chen X., Zhao G., Liu C., Bao M., Song J., Li J., Huang L., Rong J., Tian K., Deng J., Zhu J., Cai X., Bu Z., Zheng J., Weng C. African swine fever virus pS273R antagonizes stress granule formation by cleaving the nucleating protein G3BP1 to facilitate viral replication. J. Biol. Chem. 2023 doi: 10.1016/j.jbc.2023.104844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Hong Z., Sun B., Guo Z., Wang C., Zhu J. The alternatively spliced isoforms of key molecules in the cGAS-STING signaling pathway. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.771744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Cui Y., Lu G., Wei S., Yang Z., Du F., An T., Liu J., Shen G., Chen Z. Small molecule inhibitor E-64 exhibiting the activity against African swine fever virus pS273R. Bioorg. Med. Chem. 2021;35 doi: 10.1016/j.bmc.2021.116055. [DOI] [PubMed] [Google Scholar]
- Liu H., Zhu Z., Feng T., Ma Z., Xue Q., Wu P., Li P., Li S., Yang F., Cao W., Xue Z., Chen H., Liu X., Zheng H. African swine fever virus E120R protein inhibits interferon beta production by interacting with IRF3 to block its activation .pdf. J. Virol. 2021;95 doi: 10.1128/JVI.00824-21. e00824-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Ou K., Jing Y., Zhang H., Feng S., Yang Z., Shen G., Liu J., Wu C., Wei S. The structural basis of african swine fever virus pS273R protease binding to E64 through molecular dynamics simulations. Molecules. 2023;28:1435. doi: 10.3390/molecules28031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Zhang J., Ni J., Jiang S., Xia N., Guo Y., Shao Q., Cao Q., Zheng W., Chen N., Zhang Q., Chen H., Chen Q., Zhu H., Meurens F., Zhu J. The African swine fever virus protease pS273R inhibits DNA sensing cGAS-STING pathway by targeting IKKepsilon. Virulence. 2022;13:740–756. doi: 10.1080/21505594.2022.2065962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe. 2016;19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Li S., Yang F., Cao W., Liu H., Feng T., Zhang K., Zhu Z., Liu X., Hu Y., Zheng H. FoxJ1 inhibits African swine fever virus replication and viral S273R protein decreases the expression of FoxJ1 to impair its antiviral effect. Virol. Sin. 2022;37:445–454. doi: 10.1016/j.virs.2022.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melroe G.T., DeLuca N.A., Knipe D.M. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petro T.M. IFN regulatory factor 3 in health and disease. J. Immunol. 2020;205:1981–1989. doi: 10.4049/jimmunol.2000462. [DOI] [PubMed] [Google Scholar]
- Qu L., Feng Z., Yamane D., Liang Y., Lanford R.E., Li K., Lemon S.M. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzuoli E., Franzoni G., Carta T., Zinellu S., Amadori M., Modesto P., Oggiano A. Modulation of type I interferon system by african swine fever virus. Pathogens. 2020;9:361. doi: 10.3390/pathogens9050361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla Y., Callejo M., Rodríguez J.M., Culebras E., Nogal María L., Salas María L., uela E.V., Fresno M. Inhibition of nuclear factor kappaB activation by a virus-encoded IkappaB-like protein. J. Biol. Chem. 1998;273:5405–5411. doi: 10.1074/jbc.273.9.5405. [DOI] [PubMed] [Google Scholar]
- Ricour C., Delhaye S., Hato S.V., Olenyik T.D., Michel B., van Kuppeveld F.J.M., Gustin K.E., Michiels T. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein. J. Gen. Virol. 2009;90:177–186. doi: 10.1099/vir.0.005678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille A.C., Mariani M.K., Fortin A., Grandvaux N. A high resolution method to monitor phosphorylation-dependent activation of IRF3. J. Vis. Exp. 2016;107 doi: 10.3791/53723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock D.L. Challenges for African swine fever vaccine development-"... perhaps the end of the beginning". Vet. Microbiol. 2017;206:52–58. doi: 10.1016/j.vetmic.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Rojo G., García-Beato R., Viñuela E., Salas M.L., Salas J. Replication of african swine fever virus DNA in infected cells. Virology. 1999;257:524–536. doi: 10.1006/viro.1999.9704. [DOI] [PubMed] [Google Scholar]
- Sanchez E.G., Perez-Nunez D., Revilla Y. Development of vaccines against African swine fever virus. Virus Res. 2019;265:150–155. doi: 10.1016/j.virusres.2019.03.022. [DOI] [PubMed] [Google Scholar]
- Sánchez-Cordón P.J., Montoya M., Reis A. African swine fever: a re-emerging viral disease threatening the global pig industry. Vet. J. 2018;233:41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wu J., Wu Y., Chen H., Zhang S., Li J., Xin T., Jia H., Hou S., Jiang Y., Zhu H., Guo X. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 2018;506:437–443. doi: 10.1016/j.bbrc.2018.10.103. [DOI] [PubMed] [Google Scholar]
- Wang Y., Kang W., Yang W., Zhang J., Li D., Zheng H. Structure of african swine fever virus and associated molecular mechanisms underlying infection and immunosuppression: a review. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.715582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ai Q., Huang S., Ou Y., Gao Y., Tong T., Fan H. Immune escape mechanism and vaccine research progress of african swine fever virus. Vaccines. 2022;10:344. doi: 10.3390/vaccines10030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.R., Sessions P.F.d., Leon M.A., Scholle F. West nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 2008;82:8262–8271. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Q., Liu H., Zhu Z., Yang F., Ma L., Cai X., Xue Q., Zheng H. Seneca Valley Virus 3C(pro) abrogates the IRF3- and IRF7-mediated innate immune response by degrading IRF3 and IRF7. Virology. 2018;518:1–7. doi: 10.1016/j.virol.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Zhang K., Zhang Y., Xue J., Meng Q., Liu H., Bi C., Li C., Hu L., Yu H., Xiong T., Yang Y., Cui S., Bu Z., He X., Li J., Huang L., Weng C. DDX19 inhibits type I interferon production by disrupting TBK1-IKKepsilon-IRF3 interactions and promoting TBK1 and IKKepsilon degradation. Cell Rep. 2019;26:1258–1272 e4. doi: 10.1016/j.celrep.2019.01.029. [DOI] [PubMed] [Google Scholar]
- Zhao W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 2013;587:542–548. doi: 10.1016/j.febslet.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Li T., Liu X., Zhang T., Zhang Z., Kang L., Song J., Zhou S., Chen X., Wang X., Li J., Huang L., Li C., Bu Z., Zheng J., Weng C. African swine fever virus cysteine protease pS273R inhibits pyroptosis by noncanonically cleaving gasdermin D. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2021.101480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li N., Luo Y., Liu Y., Miao F., Chen T., Zhang S., Cao P., Li X., Tian K., Qiu H.J., Hu R. Emergence of african swine fever in China, 2018. Transbound Emerg Dis. 2018;65:1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript. The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.