Abstract
In this study, we examined the binding of Candida albicans synchronized yeast-phase cells to plastic, immobilized amino acids and bovine serum albumin (BSA) and quantified the binding by using an XTT tetrazolium salt assay and absorbance determination. Our results show that C. albicans binds efficiently and specifically to several nonpolar aliphatic amino acids and positively charged amino acids and to BSA immobilized on tissue culture plastic but not to polar uncharged, negatively charged, or aromatic amino acids. Adhesion of yeasts to immobilized amino acids was not affected by preincubation of cells with BSA, whereas binding to immobilized BSA was affected by preincubation of yeasts with alanine, proline, and leucine but not by arginine or lysine. The ability to distinguish the chirality of these amino acids was also examined by using both the d and l amino acid configurations, and the results show that C. albicans yeasts recognize only the l configuration of these amino acids. The observations that C. albicans specifically binds to certain amino acids indicate that these amino acids may prove useful tools for studying the binding interactions of C. albicans yeasts with host proteins such as components of the extracellular matrix.
Candida albicans is a prevalent and troublesome opportunistic yeast which can cause a variety of superficial and disseminated deep-seated mycoses (3, 20). The most serious complications of the disease are the development of the disseminated infections, resulting in endocarditis, nephritis, and endophthalmitis. The ability of Candida to attach to different types of host surfaces is currently undergoing extensive investigation as a potential new area for therapy (16) and is considered to be one of the initial steps in the pathogenesis of the organism (9, 18). For example, Candida has been shown to adhere to a variety of different surfaces, including plastic and epithelial and endothelial cells, and to the extracellular matrix proteins fibronectin, type IV collagen, laminin, and entactin (6, 9, 14, 15, 18, 21), of which the adhesion process is highly dependent on the nature of the Candida cell surface and that of the substratum (2, 9, 18). Furthermore, several postadhesion events, such as the synthesis of new Candida surface proteins and tyrosine phosphorylation of a number of proteins (1) and phagocytosis and damage of endothelial cells by C. albicans (5), can occur.
Several adhesion studies employing antibody, radiolabelling, or microbiological assays have demonstrated the binding of C. albicans to plastic, to human serum proteins such as albumin and transferrin, and to several extracellular matrix proteins (2, 9, 10–12, 19). The influence of Arg-Gly-Asp (RGD)-containing peptides on the binding of C. albicans to extracellular components has also been examined (9). In this study, we used XTT tetrazolium salt, which is reduced to the blue-black formazan product by metabolically active cells, and a 96-well microtiter format to study the adhesion of C. albicans to plastic or substrate-coated surfaces. We show that C. albicans yeasts can bind specifically to immobilized bovine serum albumin (BSA) and to certain immobilized amino acids and that the binding of yeasts to amino acids is stereospecific.
MATERIALS AND METHODS
Chemicals, organisms, and reagents.
Type B gelatin from bovine skin, type IV BSA, phenazine methosulfate (PMS), and 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide inner salt (XTT) tetrazolium salt were purchased from Sigma Chemical Co. (St. Louis, Mo.). Amino acids of d and l configurations were purchased from Calbiochem (La Jolla, Calif.). C. albicans CA6406, CA444, and CA74 were kindly provided by D. Kerridge (Cambridge, England), J.-P. Latgé (Pasteur Institut, Paris, France), and M. Monod (Lausanne, Switzerland), respectively.
Culture conditions.
C. albicans ATCC 10231, CA6406, CA444, CA74, and SKF 2270 were grown as synchronized yeasts and prepared for use in binding experiments as described previously (7). Briefly, to obtain synchronized stationary yeast-phase cultures, freeze-dried stock cultures were resuspended in 0.1 M phosphate-buffered saline (PBS; pH 7.2), inoculated into Minimal-40 medium (Difco Laboratories, Detroit, Mich.) at 105 cells/ml, and incubated at 25°C for 48 h without agitation. This synchronous cell yield was 1 × 108 to 2 × 108. Cells were then harvested, washed three times in 0.1 M PBS, and used immediately.
Binding assay.
One hundred microliters of either BSA (1 mg/ml) or appropriate amino acid (1 mg/ml) in 0.15 M PBS (pH 7.38), or PBS only, was added to wells of 96-well Greiner microtiter plates, which were incubated for 1 to 2 h at room temperature. At the end of the incubation period, the solution was removed from the wells and the remaining sites were blocked with a 1% (wt/vol) solution of gelatin for 1 h at room temperature. Thereafter, the wells were challenged with a 100-μl solution of PBS containing 105 C. albicans yeasts/ml. The plates were incubated on an orbital shaker at 100 rpm for 1 h at room temperature, after which time the wells were washed vigorously three times with PBS to remove nonadherent yeasts. Thereafter, adhesion of yeasts to wells was determined by using a modification of the XTT assay described by Tellier et al. (22), used to assess the activities of antifungal agents against planktonic C. albicans yeasts, as follows. Prior to each adhesion assay, XTT (1 g/liter of Ringer’s lactate) was thawed, and 40 μl of phenazine methosulfate stock solution (1.53 mg/ml of PBS) was added to 8 ml of XTT. Thereafter, 100 μl of the XTT-phenazine methosulfate solution was added to wells, the wells were incubated for up to 3.5 h at 35°C, and the absorbance of wells was measured at 492 nm, using a Titertek microtiter plate reader.
For competition experiments, 105 yeasts were preincubated, with agitation, for 1 h in 1 ml of a 1-mg/ml solution of the appropriate amino acid or BSA. Where indicated for concentration-dependent experiments, yeasts were preincubated with various concentrations of BSA ranging between 0.001 and 10 mg/ml. Free amino acid or BSA was subsequently removed by centrifugation and washing three times with PBS prior to challenging immobilized amino acid or BSA. Thereafter, adhesion of yeasts was quantified by the XTT tetrazolium salt assay described above.
Microscopy.
The numbers of yeasts bound to different substrates were also quantified in an Olympus (1×70) inverted microscope. Ten randomized fields of 360,000 μm2 per well were counted and normalized to the total surface area of the microtiter well. At least three to four wells were counted for each substrate binding experiment.
Statistical analysis.
Comparisons of data from binding experiments were carried out by Student’s t test, using the StatWorks program. Significant differences between data were expressed at P < 0.01, P < 0.05, or P < 0.001.
RESULTS
Binding of yeasts to plastic or protein-coated surfaces.
The ability of yeasts from five different C. albicans strains to bind to uncoated (plastic) or protein-coated surfaces was examined. After removal of nonadherent yeasts by washing with PBS, the yeasts which remained bound to the uncoated (plastic) or surfaces coated with BSA or gelatin were examined by light microscopy. No morphological differences were detected between the adherent and the nonadherent populations; all cells showed the typical blastospore morphology. The number of yeasts which bound to the plastic surface represented up to 50% of the initial added inoculum. Five C. albicans strains, including the four clinical isolates CA74, CA4444, CA6406, and SKF 2270, were used in these preliminary studies. All strains exhibited very similar behaviors, and consequently strain ATCC 10231 was chosen for all further studies.
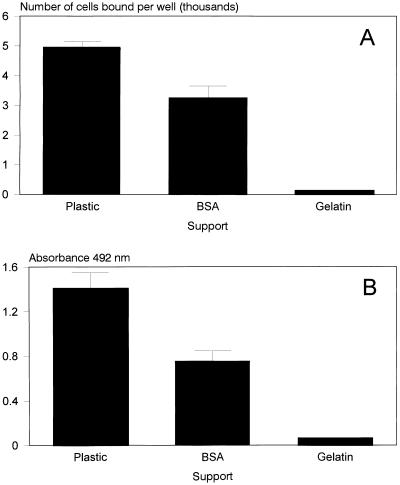
The binding of yeasts to plastic, BSA, and gelatin was also quantified. The adherent cells were counted in at least three to four wells for each condition, using light microscopy and viewing a surface area of 360,000 μm2 (see Materials and Methods). As shown in Fig. 1A, there was good binding to both plastic and BSA but minimal binding to gelatin. Similar results were obtained when the XTT tetrazolium salt procedure was used in that a large increase in absorbance was observed in the uncoated (plastic) or BSA-coated wells but absorbance in the gelatin-coated wells was similar to that in wells which had not been challenged with yeasts (Fig. 1). For example, cell counting suggests that levels of binding to BSA and gelatin were about 54 and 4%, respectively, compared with the binding of yeasts to plastic. The XTT procedure gave similar results whereby the levels of binding of yeasts to BSA and gelatin were about 65 and 2%, respectively, compared with the binding of yeasts to plastic. Untreated yeasts or yeasts preincubated with a 1% (wt/vol) solution of gelatin prior to challenging the uncoated (plastic) wells yielded similar results in that both untreated and treated yeasts bound efficiently to plastic and high absorbance at 492 nm was observed. These data suggest that under these conditions, C. albicans yeasts do not bind gelatin. These results suggest that the XTT tetrazolium salt procedure provides a useful means for the quantification of yeast cells. This procedure was therefore used in all further experiments to quantify binding of yeasts to different substrates by determination of absorbance at 492 nm.
FIG. 1.
(A) Binding of yeasts to plastic, BSA, or gelatin quantified by light microscopy (see Materials and Methods). The bars represent the means (n = 4) ± standard deviations from a typical experiment. (B) Quantification of yeasts bound to plastic, BSA, or gelatin by the XTT tetrazolium salt procedure. XTT formazan was measured after 3.5 h of incubation, and the bars represent the means ± standard deviations (n = 21).
Binding of yeasts to immobilized BSA and influence of pretreatment with BSA.
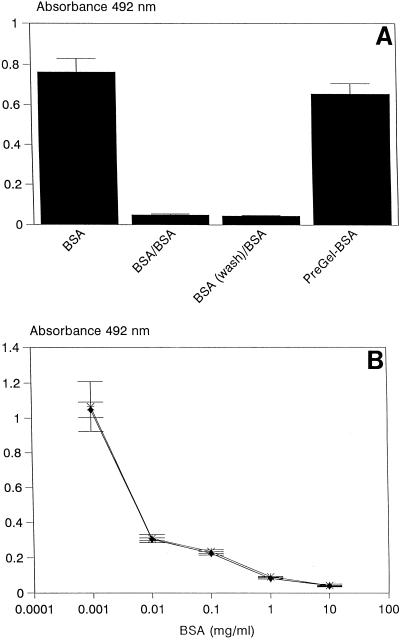
The inability of yeasts to bind to immobilized gelatin suggested that this protein can be used as a blocking agent and thus provides a useful means to study the specific interaction of yeast cells with different substrates. The interaction of yeast cells with BSA was therefore examined by immobilizing BSA in wells and by blocking remaining sites with gelatin prior to challenge with yeast cells. The presence of gelatin did not affect the ability of yeast cells to bind to immobilized BSA (Fig. 2A). Similar treatment with BSA significantly (P < 0.001) abolished the ability of the yeast cells to bind to immobilized BSA (Fig. 2A) and was concentration dependent; i.e., binding of yeast cells to immobilized BSA was significantly reduced (P < 0.001) when yeasts were preincubated with either 1 or 10 mg of BSA per ml (Fig. 2B). Preincubation of yeasts followed by removal of BSA in solution further showed that the binding to immobilized BSA was attenuated (P < 0.001), indicating the specific interaction of the yeast cells with the immobilized substrate (Fig. 2B).
FIG. 2.
Binding of yeast cells to immobilized BSA. (A) Binding of yeast cells to BSA (BSA), effects of preincubation with BSA on binding of yeast cells to immobilized BSA (BSA/BSA) and following washout of free BSA [BSA(wash)/BSA], and effects of preincubation with gelatin on the binding of yeast cells to immobilized BSA (PreGel-BSA). (B) Effects of preincubation of yeast cells with 0.001 to 10 mg of BSA per ml on binding of yeast cells to immobilized BSA (BSA pretreatment followed by removal of free BSA in solution or BSA treatment only). Data points represent the means ± standard deviation (n = 20) of yeast cell binding from a typical experiment.
Binding of yeasts to immobilized amino acids.
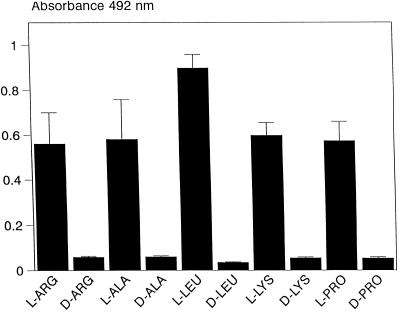
The interactions of yeast cells with immobilized amino acids was also examined. The amino acids were coated onto microtiter plates (Materials and Methods), and the nonspecific protein binding sites were blocked with a 1% (wt/vol) solution of gelatin. After blocking with gelatin, the immobilized amino acids were challenged with yeast cells, and after removal of nonadherent yeasts by washing, the specific binding was determined by using the XTT assay and measurements of absorbance at 492 nm. Of the natural amino acids tested, yeasts were able to bind efficiently only to immobilized arginine, alanine, leucine, lysine, and proline (Table 1). Typically, binding of yeasts to these immobilized amino acids was similar to the binding of yeast cells to immobilized BSA (Fig. 1 and 2). The binding of yeast cells to the d configuration of these amino acids was also determined, and as shown in Fig. 3, yeast cells failed to show any significant binding (P < 0.001) to the d configuration of these amino acids.
TABLE 1.
Binding of yeasts to immobilized amino acids
| Amino acid group | Amino acid | A492a |
|---|---|---|
| Nonpolar aliphatic | Glycine | 0.108 ± 0.013 |
| Alanine | 0.886 ± 0.082 | |
| Valine | 0.047 ± 0.002 | |
| Leucine | 1.210 ± 0.11 | |
| Isoleucine | 0.046 ± 0.001 | |
| Proline | 1.008 ± 0.067 | |
| Polar uncharged | Serine | 0.048 ± 0.005 |
| Threonine | 0.059 ± 0.008 | |
| Cysteine | 0.044 ± 0.004 | |
| Methionine | 0.040 ± 0.003 | |
| Asparagine | 0.082 ± 0.018 | |
| Glutamine | 0.099 ± 0.022 | |
| Positively charged | Lysine | 0.917 ± 0.026 |
| Arginine | 0.927 ± 0.04 | |
| Histidine | 0.040 ± 0.002 | |
| Negatively charged | Aspartate | 0.102 ± 0.02 |
| Glutamate | 0.070 ± 0.013 | |
| Aromatic | Phenylalanine | 0.044 ± 0.003 |
| Tyrosine | 0.053 ± 0.003 | |
| Tryptophan | 0.040 ± 0.002 |
Mean absorbance ± standard deviation (n = 21) after subtraction of the background in the absence of cells from a typical experiment.
FIG. 3.
Measurements of the binding of yeast cells to l and d configurations of immobilized alanine (ALA), arginine (ARG), leucine (LEU), lysine (LYS), and proline (PRO). The bars represent the means ± standard deviations (n = 20) of yeast cell binding from a typical experiment.
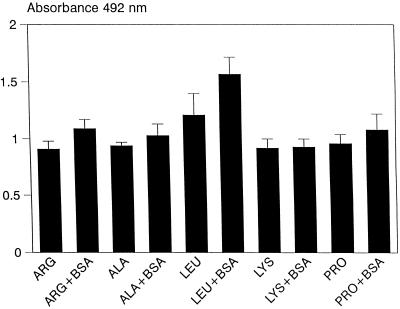
In view of the observation that preincubation with BSA inhibits the ability of yeast cells to bind to immobilized BSA (Fig. 2), we also examined the effect of preincubation of yeasts with BSA on the ability of yeast cells to bind to immobilized l-amino acids. As shown in Fig. 4, yeast cells bound equally well to immobilized amino acids in the presence or absence of BSA treatment, except that pretreatment with BSA resulted in a slight increase (P < 0.05) in binding to l-leucine (Fig. 4).
FIG. 4.
Effects of preincubation of yeast cells with BSA on the binding of yeast cells to immobilized alanine (ALA), arginine (ARG), leucine (LEU), lysine (LYS), or proline (PRO). Data points represent the means ± standard deviations (n = 20) of yeast cell binding from a typical experiment.
Specificity of binding of yeasts to immobilized amino acid or BSA.
Considering the ability of yeast cells to bind to BSA or amino acids, we examined the effects of preincubation with each amino acid on the ability of yeast cells to adhere to immobilized BSA. In these experiments, BSA was immobilized in wells, yeasts were preincubated with 1 mg of appropriate amino acid per ml, and the immobilized BSA was challenged with the amino acid-pretreated yeast cells. Preincubation of yeast cells with 1 mg of arginine or lysine per ml did not attenuate the binding of yeast cells to immobilized BSA, whereas preincubation with 1 mg of alanine, leucine, or proline per ml attenuated the binding of yeast cells to immobilized BSA (Table 2).
TABLE 2.
Competitive effects of the binding of yeasts treated with amino acids to immobilized substrates
| Substrate | % bindinga
|
||||
|---|---|---|---|---|---|
| Alanine | Proline | Leucine | Lysine | Arginine | |
| Alanine | 0.52* | 101.2 | 99.0 | 0.4* | 98.8 |
| Proline | 105.6 | 0* | 0.2* | 0.6* | 105.6 |
| Leucine | 2.70* | 95.8 | 3.9* | 92.1 | 67.5** |
| Lysine | 97.8 | 106.8 | 110.1 | 0.07* | 0.2* |
| Arginine | 76.2** | 86.2 | 88.8 | 2.5* | 1.5* |
| BSA | 45.6*** | 76.2** | 67.4** | 94.5 | 91.7 |
Yeast cells were pretreated with 1 mg of appropriate amino acid per ml. Thereafter, each immobilized substrate was challenged with pretreated yeasts. The values represent the mean percent binding (n = 12) of yeasts to each immobilized substrate from a typical experiment. Standard deviations were always <10%. *, value significantly different at P < 0.001 compared with the binding of untreated yeasts to the corresponding substrate; **, value significantly different at P < 0.05 compared with the binding of untreated yeasts to the corresponding substrate; ***, value significantly different at P < 0.01 compared with the binding of untreated yeasts to the corresponding substrate.
To assess the specificity of the binding interaction between yeast cells and each amino acid, we preincubated yeasts with 1 mg of each amino acid per ml and then challenged wells containing immobilized amino acid with yeasts. Yeast cells preincubated with the same amino acid as that immobilized in wells showed dramatically attenuated binding to the immobilized amino acid and acted as positive controls in the experiment (for statistical analysis and significance values, see Table 2). The binding of yeast cells to arginine was inhibited by lysine but not by leucine, proline, or alanine. Similarly, binding to lysine was inhibited by arginine but not by the other amino acids (Table 2). The binding of yeasts to immobilized alanine was inhibited by lysine but not by the other amino acids, whereas binding of yeasts to proline was inhibited by leucine and lysine but not by alanine and arginine (Table 2). By contrast, binding to leucine was affected by alanine but not by the other amino acids (Table 2).
DISCUSSION
Candida cells have been suggested to bind to a variety of human proteins and gelatin (2, 7, 18). Our results show that C. albicans yeasts bind to plastic and BSA-coated surfaces but fail to adhere to gelatin-coated surfaces. Quantification of adherent cells by light microscopy and use of XTT tetrazolium salt yield similar results. The XTT tetrazolium salt procedure has several important advantages over use of cell counting techniques in that (i) the method is easy to use compared to the tedious counting by light microscopy, (ii) it allows a rapid and quantitative measurement, and (iii) only metabolically active cells are determined by this method. Here we have shown that by using XTT tetrazolium salt, C. albicans yeasts bind efficiently to plastic, binding of yeasts to plastic is abolished by blocking the surface with gelatin, and yeasts bind to immobilized BSA and interestingly to several different amino acids. Gelatin therefore provides a means to diminish nonspecific binding to plastic surfaces such that the specific binding to immobilized substrates such as amino acids, peptides, and proteins can be determined.
Our data concerning gelatin do not appear to be in agreement with the observations of Klotz and Smith (12), who suggested that yeasts may bind to porcine gelatin. However, we have also used porcine gelatin and have found that, as was the case for bovine gelatin, yeasts do not bind to this substrate (8). The reasons for the difference between our data and those of Klotz and Smith (12) are unclear but may reflect differences in experimental procedures. For example, in this study we used synchronized yeast-phase cells which were prepared by growth in Minimal-40 medium and quantified binding of the yeasts to different substrates by using both light microscopy and the XTT tetrazolium salt procedure. In addition, the assay conditions involve binding of yeasts (initial inoculum of 105 cells/ml) to different substrates at 25°C for 1 h with agitation, and the assay is performed in wells of 96-well microtiter plates. Under these conditions using five C. albicans strains including four clinical isolates, we observed that all of the strains showed similar behaviors in terms of the ability to bind to plastic and BSA and the inability to bind to gelatin. The conditions used in this study appear to be quite different from those described elsewhere. Generally, most studies use lower inocula and consequently examine the adherence of significantly fewer cells. For example, Klotz and Smith (12) used about 250 cells per well in 24-well tissue culture plates, whereas we used 10,000 cells per well in 96-well tissue culture plates. The number of adherent cells in the case of the plastic surface is normally about 5,000 in our study, compared with 200 cells in the case of the study by Klotz and Smith (12). The assay conditions used by Klotz and Smith (12) involved yeasts which were previously cultured in Sabouraud dextrose broth and binding of yeasts (initial inoculum of 103 cells/ml) at 37°C for 30 min without agitation (12). Indeed, Klotz et al. (13) have previously noted that there is no standardized method for measuring adherence, making it difficult to compare results from different studies. Under the conditions used, Klotz et al. (13) have shown the presence of a fibronectin/gelatin binding protein which appears to be present on the C. albicans cells. There are different reports in the literature which demonstrate the presence of specific epitopes under specific environmental conditions (4). It is quite probable that culture conditions, inoculum size, time of contact, temperature, and the surface area to which the yeast cells are applied influence the ability of yeast cells to bind to immobilized substrates, and one or more of these variations may account for the different observations between this study and that described elsewhere (12).
The binding of yeast cells to immobilized BSA was reduced by more than 90% when yeast cells were preincubated with BSA, binding was dependent on the concentration of BSA used in preincubations, and removal of free BSA in solution also showed attenuation of the binding of yeast cells to immobilized BSA. These data, therefore, indicate that the yeast cell-BSA interaction may be a specific process and that the affinity of this interaction is high. Other studies have shown that several host proteins, notably lactoferrin and ovotransferrin (23), and complement fragments, such as C3d and iC3b (10), are known to bind to C. albicans, and C. albicans germ tubes bind human albumin with high avidity (17).
Binding studies with individual natural amino acids with C. albicans yeast cells demonstrated that yeast cells may bind to specific nonpolar aliphatic amino acids, namely, alanine, leucine, and proline, and the positively charged R-group amino acids arginine and lysine. By contrast, yeast cells were unable to bind to amino acids from polar uncharged, aromatic, or negatively charged amino acids. Interestingly, yeast cells were not able to bind to other nonpolar aliphatic amino acids such as glycine, valine, and isoleucine or to the positively charged histidine, which suggests a degree of specificity in the binding of yeast cells to amino acids from nonpolar aliphatic and positively charged groups. Moreover, unlike the binding of yeast cells to the l configuration of alanine, arginine, leucine, lysine, or proline, yeast cells were unable to bind the d configuration of these amino acids, further indicating that the yeast cell-amino acid binding interactions are specific and demonstrate stereospecificity. Furthermore, competitive binding studies showed that preincubation with each amino acid inhibited the interaction with itself when immobilized on wells.
To the best of our knowledge, this is the first report of the binding of C. albicans to immobilized amino acids. By contrast, other studies have concentrated on interactions of Candida yeasts or germ tubes with components of the extracellular matrix such as fibronectin, laminin, and collagen and the potential of different peptides, including those with an RGD sequence, on this interaction (9). Furthermore, the binding of Candida yeasts to large proteins such as BSA is likely to involve particular amino acids. Therefore, it would have been expected that pretreatment of yeast cells with BSA would inhibit the binding interaction of yeast cells to immobilized amino acids. However, binding of BSA-treated yeast cells to amino acids was similar to the binding of untreated, control yeast cells, indicating that pretreatment with BSA does not affect the binding interaction of yeast cells with immobilized amino acids. On the other hand, preincubation of yeast cells with the nonpolar aliphatic amino acids attenuated the binding of yeast cells to immobilized BSA by 25 to 55%, while preincubation with arginine or lysine did not affect the binding interaction between yeast cells and immobilized BSA. In large proteins such as BSA, one may expect a number of potential sites for binding whereby the binding interaction with the sites may be of a cooperative nature. Therefore, the effect of each amino acid on the binding of yeast cells to immobilized BSA may not necessarily cause a complete attenuation of binding. Indeed, the slight reduction in the ability of yeast cells treated with alanine, leucine, or proline to bind to immobilized BSA may indicate that such amino acids are involved in the interaction of BSA with C. albicans yeast cells.
As regards the binding of yeast cells to immobilized amino acids, lysine effectively blocked the binding of yeasts to immobilized arginine and vice versa, as would be expected for similarly charged amino acids, suggesting the presence of similar binding sites for arginine and lysine. On the other hand, the interaction of yeast cells with immobilized proline was also competed for by lysine and leucine but not by alanine or arginine, and proline did not markedly attenuate the binding interaction with immobilized alanine, arginine, leucine, or lysine. These data suggest that the proline binds to sites which partially overlap with sites for leucine and lysine but with a lower affinity, whereas binding sites for leucine and alanine may partially overlap with the binding sites for lysine and arginine or may be sterically hindered by the binding of lysine or arginine. Moreover, all of the nonpolar aliphatic amino acids, including proline, in fact inhibit the binding of yeast cells to immobilized BSA, an effect which is not seen with arginine or lysine.
Given that C. albicans yeast cells bind avidly to certain amino acids, it may prove fruitful to use these findings as tools for future research into the characteristics of the binding of yeast cells to different substrates. Indeed, we are currently using this information to study the specific interactions of C. albicans yeast cells with, for example, components of the extracellular matrix.
REFERENCES
- 1.Bailey A, Wadsworth E, Calderone R. Adherence of Candida albicans to human buccal epithelial cells: host-induced protein synthesis and signaling events. Infect Immun. 1995;63:569–572. doi: 10.1128/iai.63.2.569-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderone R A, Braun P C. Adherence and receptor relationships of Candida albicans. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emori T G, Gaynes R P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas L J. Adhesin-receptor interactions in the attachment of Candida albicans to host epithelial cells. Can J Bot. 1995;73:S1147–S1153. [Google Scholar]
- 5.Filler S G, Swerdloff J N, Hobbs C, Luckett P M. Penetration and damage of endothelial cells by Candida albicans. Infect Immun. 1995;63:976–983. doi: 10.1128/iai.63.3.976-983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawser S P, Douglas L J. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62:915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawser S P, Islam K. Spectrophotometric determination of the morphogenetic transformation by synchronous Candida albicans: effects of antifungal agents. J Antimicrob Chemother. 1996;38:67–73. doi: 10.1093/jac/38.1.67. [DOI] [PubMed] [Google Scholar]
- 8.Hawser, S. P., and K. Islam. Unpublished results.
- 9.Hostetter M K. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial surfaces. Clin Microbiol Rev. 1994;7:29–42. doi: 10.1128/cmr.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hostetter M K. An integrin-like protein in Candida albicans: implications for pathogenesis. Trends Microbiol. 1996;4:242–246. doi: 10.1016/0966-842X(96)10036-6. [DOI] [PubMed] [Google Scholar]
- 11.Klotz S A. Adherence of Candida albicans to components of the subendothelial extracellular matrix. FEMS Microbiol Lett. 1990;68:249–254. doi: 10.1016/s0378-1097(05)80049-7. [DOI] [PubMed] [Google Scholar]
- 12.Klotz S A, Smith R L. Gelatin fragments block adherence of Candida albicans to extracellular matrix proteins. Microbiology. 1995;141:2681–2684. doi: 10.1099/13500872-141-10-2681. [DOI] [PubMed] [Google Scholar]
- 13.Klotz S A, Rutten M J, Smith R L, Babcock S R, Cunningham M D. Adherence of Candida albicans to immobilized extracellular matrix proteins is mediated by calcium-dependent surface glycoproteins. Microb Pathog. 1993;14:133–147. doi: 10.1006/mpat.1993.1014. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Ribot J L, Chaffin W L. Binding of the extracellular matrix component entactin to Candida albicans. Infect Immun. 1994;62:4564–4571. doi: 10.1128/iai.62.10.4564-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negre E, Vogel T, Levanon A, Guy R, Walsh T J, Roberts D D. The collagen binding domain of fibronectin contains a high affinity binding site for Candida albicans. J Biol Chem. 1994;269:22039–22045. [PubMed] [Google Scholar]
- 16.Ofek I, Kahane I, Sharon N. Toward anti-adhesion therapy for microbial diseases. Trends Microbiol. 1996;4:297–299. doi: 10.1016/0966-842x(96)30023-1. [DOI] [PubMed] [Google Scholar]
- 17.Page S, Odds F C. Binding of plasma proteins to Candida species in vitro. J Gen Microbiol. 1988;134:2693–2702. doi: 10.1099/00221287-134-10-2693. [DOI] [PubMed] [Google Scholar]
- 18.Pendrak M L, Klotz S A. Adherence of Candida albicans to host cells. FEMS Microbiol Lett. 1995;129:103–114. doi: 10.1111/j.1574-6968.1995.tb07566.x. [DOI] [PubMed] [Google Scholar]
- 19.Penn C, Klotz S A. Binding of plasma fibronectin to Candida albicans occurs through the cell binding domain. Microb Pathog. 1994;17:387–393. doi: 10.1006/mpat.1994.1084. [DOI] [PubMed] [Google Scholar]
- 20.Pittet D, Wenzel R P. Nosocomial bloodstream infections. Secular trends in rates, mortality, and contribution to total hospital deaths. Arch Intern Med. 1995;155:1177–1184. doi: 10.1001/archinte.155.11.1177. [DOI] [PubMed] [Google Scholar]
- 21.San Millan R, Ezkurra P A, Quindos G, Robert R, Senet J M, Ponton J. Effect of monoclonal antibodies directed against Candida albicans cell wall antigens of the fungus to polystyrene. Microbiology. 1996;142:2271–2277. doi: 10.1099/13500872-142-8-2271. [DOI] [PubMed] [Google Scholar]
- 22.Tellier R, Krajden M, Grigoriew G A, Campbell I. Innovative endpoint determination system for antifungal susceptibility testing of yeasts. Antimicrob Agents Chemother. 1992;36:1619–1625. doi: 10.1128/aac.36.8.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valenti P, Visca P, Antonini G, Orsi N. Interaction between lactoferrin and ovotransferrin and Candida cells. FEMS Microbiol Lett. 1986;33:271–275. [Google Scholar]