Abstract
The genetic background of inflammatory bowel disease, both Crohn’s disease and ulcerative colitis, has been known for more than 2 decades. In the last 20 years, genome-wide association studies have dramatically increased our knowledge on the genetics of inflammatory bowel disease with more than 200 risk genes having been identified. Paralleling this increasing knowledge, the armamentarium of inflammatory bowel disease medications has been growing constantly. With more available therapeutic options, treatment decisions become more complex, with still many patients experiencing a debilitating disease course and a loss of response to treatment over time. With a better understanding of the disease, more effective personalized treatment strategies are looming on the horizon. Genotyping has long been considered a strategy for treatment decisions, such as the detection of thiopurine S-methyltransferase and nudix hydrolase 15 polymorphisms before the initiation of azathioprine. However, although many risk genes have been identified in inflammatory bowel disease, a substantial impact of genetic risk assessment on therapeutic strategies and disease outcome is still missing. In this review, we discuss the genetic background of inflammatory bowel disease, with a particular focus on the latest advances in the field and their potential impact on management decisions.
Key Points
| Inflammatory bowel disease has a strong genetic background with more than 240 identified susceptibility loci. |
| Despite the identification of various risk genes, a substantial impact of genetic risk assessment on therapeutic strategies and disease outcome is still missing. |
| Precision medicine might become a guidance tool for inflammatory bowel disease drug dosing or drug sparing if patients with a low risk of relapse could be identified adequately. |
Introduction
Inflammatory bowel disease (IBD) refers to a chronic inflammation of the gastrointestinal tract with its main representatives Crohn’s disease (CD) and ulcerative colitis (UC). While UC is limited to the mucosal layer of the colon, CD is a transmural inflammation that may affect any part of the gastrointestinal tract between the mouth and anus [1]. Both forms present with a chronic relapsing pattern of abdominal pain and diarrhea. Diagnosis is made based on clinical, endoscopic, histological, laboratory, and eventually radiologic findings [1]. Both sexes appear to be equally affected [2]. Inflammatory bowel disease can develop at any age with a clustering between the age of 15–30 years, and a second peak in the elderly population [2]. Historically, IBD was considered a disease of the industrialized Western world, including North America, Europe, and Oceania [2]. However, while prevalence remains highest in Western civilizations with over 3 million people with a diagnosis of IBD in the USA and Europe, latest analyses show increasing incidences in South America, Asia, and Africa [3, 4]. In the twenty-first century, IBD can therefore be considered a global epidemic. Contrasting a tremendous progress in the understanding of IBD, its exact pathogenesis remains at best partially explained. Inflammatory bowel disease with its subtypes UC and CD is likely driven by a complex interaction of environmental factors with a dysregulation of the immune system in response to an altered intestinal flora in genetically predisposed individuals [5]. Since the first introduction of an anti-tumor necrosis factor (TNF) agent in 1998, the armamentarium of IBD therapeutics has dramatically expanded in the last 20 years, with the recent approval of the first IBD-specific small molecules, the sphingosine-1-P modulator ozanimod and the Janus kinase inhibitors tofacitinib, filgotinib, and upadacitinib [6–8].
With more available therapeutic options, treatment decisions become more complex, with still many patients experiencing debilitating disease course and a loss of response to treatment over time. With a better understanding of the disease, more effective personalized treatment strategies are looming on the horizon. Genotyping has long been considered a strategy for treatment decisions, such as the detection of thiopurine S-methyltransferase and nudix hydrolase 15 polymorphisms before the initiation of azathioprine [9, 10]. However, although many risk genes have been identified in IBD, a substantial impact of a genetic risk assessment on therapeutic strategies and disease outcome is still missing [11]. In this review, we discuss the genetic background of IBD, with a particular focus on the latest advances in the field and their potential impact on management decisions.
Genetic Background of IBD
Analyzing the Swedish twin registry, Tysk and colleagues set a milestone with regard to the genetic basis of IBD [12]. This landmark study was followed by various twin and family studies further supporting the genetic background of both UC and CD. However, despite numerous studies performed, an accurate genetic risk assessment is still difficult as rates vary widely in the literature with a positive family history being reported between 1.5 and 28% in CD and between 1.5 and 24% in UC [12–14]. Simplified, first-degree relatives of patients with CD have an eight-fold higher risk of developing IBD, while first-degree relatives of patients with UC have an approximately four-fold higher risk, making a positive family history of IBD the strongest risk factor for IBD [15]. Concordance rates in monozygotic and dizygotic twins reach up to 50% and 10% in CD compared with up to 15% and 4% in UC, suggesting a lower heritability in UC than CD [16, 17]. Interestingly, a nationwide and population-based study from Denmark demonstrated a relevant impact of non-genetic exposures, such as exposure to antibiotics and prior ankylosing spondylitis, on the development of new-onset IBD in families with pre-existing IBD [18]. Wide variations between genetic studies might be explained by their heterogeneity with regard to study design and population selection. Of note, results from twin studies contrast the much lower heritability estimated from genome-wide association studies (GWAS, see below). This might be partly explained by the shortcomings of heritability estimation, where genetic and environmental variance are considered as separate unities and epigenetics are neglected. Nevertheless, early family and twin studies were able to unravel the relative contribution of genetic and environmental influences in the etiology of IBD, with none of the genetic background explaining more than 50% of disease prevalence [16, 17, 19, 20]. Data from the UK Biobank even show that in patients with a high genetic risk of adult-onset IBD, the risk may be reduced by half in patients with adherence to a favorable lifestyle [21].
GWAS
Whole genome linkage analyses highlight co-segregation of certain genetic loci for common complex diseases, thereby uncovering Mendelian traits within families or ethnicities [22]. In IBD, application of whole genome linkage enabled the detection of nucleotide-binding oligomerization domain-containing protein 2 (NOD2) as a highly susceptible gene for CD [22]. In 2005, GWAS emerged representing a very efficient method to identify alleles with weak and inconsistent replication that are commonly seen in complex diseases such as IBD [22, 23]. Genome-wide association studies explore the genome focusing on single-nucleotide polymorphisms (SNPs) that are defined genome variations in DNA sequences. Several millions of SNPs are genotyped on platforms to uncover specific SNP variants that more often occur in a person with a trait than in one without [24]. As mentioned above, both GWAS and twin studies have been used to calculate heritability showing remarkably different results [25]. This gap, also known as missing or hidden heritability and not only seen in IBD, might have multiple explanations, such as the over-simplification of genetic identity, environmental assumptions, and not considering epigenetics [23].
Since its first implementation, numerous different study populations varying in ethnicity and geographic location have been investigated using GWAS, resulting in the detection of roughly 240 susceptibility loci in IBD. Of these SNPs, the majority were associated with both UC and CD, assuming similar disease liability and common signaling pathways. Identification of mechanistic pathways was succeeded by expansion of disease knowledge of intestinal homeostasis, most importantly, regulations in epithelial and barrier function, adaptive immunity, a host-pathogen interaction such as an immunity response, as well as autophagy [23, 26, 27]. Furthermore, over 60% of the susceptibility loci are associated with other immune-mediated diseases such as psoriasis, celiac disease, and ankylosing spondylitis [1, 28]. The strongest genetic effects have been seen for interleukin-23 receptor (IL-23R) in IBD, NOD2 in CD, and human leukocyte antigen (HLA) complex in UC [29].
Genome-wide association studies have several limitations. One limiting factor is linkage disequilibrium of the human genome [19]. Linkage disequilibrium refers to alleles (two or more) of SNPs that are non-randomly associated with each other and therefore tend to be passed on together [30]. Genome-wide association studies are thought to identify SNPs through linkage disequilibrium in the case where a significant gene does not lie within the examined region of the genome. If linkage disequilibrium is insufficient, SNPs may not be inherited with the coded gene and therefore key genes might not be captured. Indeed, not every polymorphism can be covered by GWAS. Roughly 30% of the genes are not detected; in particular, rare SNPs with high penetrance or common SNPs with low penetrance might be missed [19, 30]. In addition, it is known that the contribution of common variations to disease liability is small, with odds ratios mostly between 1.05 and 1.5 [22]. Furthermore, even highly associated GWAS SNPs might not be causative. As a consequence, the question of functionality and causality remains unresolved. Nevertheless, GWAS expanded our knowledge far beyond a basic genetic influence on human diseases and their potential has probably not been fully unlocked yet.
NOD2
NOD2 is located on chromosome 16p21 [31]. NOD2 wild type acts as an intracellular bacterial sensor regulating immune response such as defense or tolerance against/to pathogens. It is expressed in different cell types such as epithelial cells, endothelial cells, macrophages, T cells, and in particular, ileal Paneth cells, thereby approaching immune response in various ways. NOD2 mutations were the first to be described as associated with CD. Until now, NOD2 confers the greatest risk in the development of CD, out of all genetic variations known so far [31–33]. Moreover, studies suggest that disease characteristics, particularly earlier onset of CD and an increased severity of disease course, are influenced by NOD2 polymorphisms [34, 35]. Disease-related NOD2 is remarkably less active and can even result in complete unresponsiveness to bacterial pathogens [33]. The most frequent variants in NOD2 are R702W, G908R, and 1007fs. All of these variants appear to influence protein function and signaling inflammatory pathways through impaired activation of the NF-kB pathway or reduced/absent cytokine production in response to pathogens [36]. Carriers of one of these disease-related NOD2 alleles have an approximate two- to four-fold increased risk of developing CD, while individuals with homozygous or compound heterozygous mutations have a 15- to 40-fold increase in CD risk. However, mutated NOD2 alleles occur in 0.5–2% of healthy individuals and about 60% of patients with CD carry no NOD2 mutation, suggesting synergistic effects of more than one factor in the development of CD [37–39]. Moreover, the common genetic variants in NOD2 confer a disease risk specific to European and African-American populations, but not the Asian population, as it could not be reproduced in studies with Japanese and Chinese individuals [40–42].
IL23R
IL23R is located on chromosome 1p31 encoding a subunit of the proinflammatory cytokine IL-23R [43]. Variants in IL23R have been associated with a protection against IBD, presumably with a loss of function of the receptor itself or at least through mediated cell function impairment [44]. In addition, IL-23 as the ligand of IL23R plays a key role in the pathogenesis of intestinal inflammation: IL-23 and its closely related cytokine IL-12 with their common subunit p40 have become an attractive target of neutralizing antibodies in the treatment of IBD [45]. However, the main anti-inflammatory effect is achieved by blocking IL-23 or more specifically its p19 subunit (instead of p40), which has led to development of more selective IL-23 antibodies such as guselkumab, risankizumab, mirikizumab, and brazikumab, which are currently being evaluated in phase II or phase III trials [46, 47]. Their efficacy can be explained by the disruption of a wide variety of signaling pathways including JAK-STAT, NF-κB, interleukin-17, as well as a loss of cell function in T helper-17, CD4+, and CD8+ T cells [48].
HLA Complex
The HLA complex, also known as the major histocompatibility complex, is located on chromosome 6p21.3. HLA genes encode specific cell surface proteins of antigen-representing cells to initiate a T-cell-mediated immune response [49]. The HLA region plays a key role in most chronic inflammatory diseases. The exact pathophysiological mechanisms of HLA involvement in IBD remain elusive [50]. It is hypothesized that molecular mimicry is one of the main mechanisms by which a foreign antigen causes autoimmunity. Cross-reactive epitopes present through the major histocompatibility complex and trigger an immune response by T-cell activation [51]. This proinflammatory response, which is essential to clear pathogens, may persist if there is sequential or structural homology between foreign and self-antigens [52]. Not surprisingly, HLA is the genetic region where the most significant risk variants with the largest effect sizes are located, for the development of IBD and other immune-mediated diseases such as psoriasis, rheumatoid arthritis, or spondyloarthropathies [53]. In the case of IBD, classical HLA genes are most strongly associated with disease development. Those can be divided into two groups: (I) class I gene region with its HLA traits A, B, and C and (II) class II genes with the HLA traits DR, DQ, and DP [54]. The most consistent association of HLA class II alleles in IBD are seen with HLA-DRB1 and HLA-DQB1. DRB1 has been the most extensively studied gene and confers the greatest risk out of all HLA genes in IBD. HLA-DRB1*0103 is strongly associated with both entities, UC and CD. Nevertheless, it is considered a rare allele with a less than 2% frequency in European, White North American, and Jewish populations. The variant shows a particularly strong association in patients with severe extensive UC and those with colonic CD [49]. Further associations have been seen with HLA-DRB1*1502 in UC, especially affecting the Japanese population, with a high prevalence of about 20% [49]. In CD, most reproducible associations have been found with HLA-DRB1*07, particularly with ileal disease, and HLA-DRB1*04, predominantly in the Japanese population [49]. Overall, the influence of HLA has been found to be greater in UC than CD [29, 55]. A genome-wide association study showed that the carriage of the HLA-DQA1*05 allele (approximately 40% of European individuals) doubled the risk of immunogenicity to anti-TNF therapy [56]. Taken together, the association of HLA alleles and IBD remains complex and difficult to resolve, particularly because of the high density of genetic variants/candidate genes and a strong linkage disequilibrium.
Genetics in EIM and Perianal Disease
It is well known that IBD is a systemic disease with many patients presenting with extraintestinal manifestations (EIM) [57]. Prevalence of these EIM ranges from 6 to 47% [58–65]. The classic EIM involve the following four organ system: (1) joints; (2) skin; (3) eyes; and (4) hepatobiliary system [57, 65]. The pathogenesis of EIM in the context of IBD is only partially understood, but several mechanisms have been proposed, among which are aberrant lymphocyte homing, cross-reactivity (also called molecular mimicry), or a simple proinflammatory state with upregulation of specific cytokines and chemokines resulting in a disbalance of components of the immune system [66]. The overlapping genetic background between EIM and IBD further supports a common pathophysiology.
Association studies show concordance rates of 70–80% in parent-child pairs or sibling pairs with EIM [67], while a positive family history has been known as an independent predictor for the presence of EIM, at least in patients with CD [65]. Differences in EIM prevalence among different ethnicities further support the concept of a genetic background of EIM: (1) EIM appears more frequently in Western countries compared with Eastern countries [68, 69], which is particularly true for primary sclerosing cholangitis; (2) joint manifestations are more frequent in African-American individuals and Asian individuals; and (3) EIM are more often diagnosed in Indian individuals compared with Malay or Chinese subjects [70, 71]. Several genes have been identified in the pathogenesis of EIM. The following HLA genotypes have been implicated as genetic risk factors: HLA-B27 in joint, skin, and ophthalmologic manifestations, particularly axial spondyloarthropathy; HLA-B8/DR3 in primary sclerosing cholangitis; HLA-B35 in type 1 arthritis; HLA-B44 in type 2 arthritis; HLA-B58 in joint, skin, and ophthalmologic manifestations; HLA-DRB1*0103 in joint, skin, and ophthalmologic manifestations. HLA-A2, HLA-DR1 and HLA-DQw5 are risk factors for EIM in general [66]. Genome-wide association studies for axial spondyloarthropathy and IBD revealed the following risk genes: IL23R, IL12B, STAT3, PTERG4, CARD9, IL1R2, and ORMDL3. Genome-wide association studies for psoriasis and IBD showed the involvement of IL12B, IL23R, JAK2, and STAT3, while GWAS for erythema nodosum and IBD revealed CLCA2, LY75, and 2q24.1 as risk loci. Furthermore, NOD2 has been shown to be a risk factor for ileal CD, sacroiliitis, and uveitis [72] However, the role of NOD2 in perianal disease remains unclear. Newer data point to an important role of a SNP in Complement Factor B. The non-synonymous variant rs4151651 in Complement Factor B has been associated with perianal CD in three independent cohorts [73]. Presumably, the genetic contribution to the pathogenesis of EIM and IBD is a combination of overlapping and independent gene loci, reflecting the occurrence of EIM that parallel and do not parallel intestinal inflammation [74].
Pharmacogenomics
The field of pharmacogenomics studies how genetics influence an individual’s response to drug therapy potentially opening the door to individualized precision medicine. Its value in clinical practice has not yet been exhausted and may grow as knowledge increases [75]. While the identification of patients who will or will not respond to a specific treatment, based on genetic testing, is still an unmet need, only a few pre-therapeutic genetic tests have made it into clinical practice, largely those that identify patients at risk for drug toxicity. Such drug toxicity affected by genotypes has been most extensively studied in the context of thiopurine treatment. Genotypes of the two enzymes thiopurine methyltransferase and nudix hydrolase 15 slow down thiopurine metabolism leading to elevated drug concentrations and therefore increased toxicity (e.g., severe myelosuppression) [9, 10]. Pre-treatment genotyping is endorsed by the European Crohn’s and Colitis Organisation guidelines [76], but does not substitute monitoring of drug toxicity under treatment.
Apart from pre-treatment assessment of thiopurine toxicity, no genetic test has made it into clinical practice. Nevertheless, several genetic polymorphisms have been linked with response to treatment with anti-TNF, particularly those within the TNF and TNFR genes (rs1800629, rs1799724, rs767455, rs1061624, and rs976881 showing an inferior outcome, and rs4149570, rs361525, and rs3397 showing a superior outcome) [77], but also in innate immunity genes (TLR4, IL6, IL1, IL17, TLR2, TLR9) [77, 78], and genes involved in autophagy and apoptosis (Fas-L, CASP9, and ATG16L1) [77]. Further data are evolving with regard to responses to other treatments such as anti-IL-12/23 ustekinumab (e.g., rs7234029) [79].
Epigenetic Mechanisms
As previously mentioned, only a part of the IBD risk can be explained by genetic factors. Additional epigenetic mechanisms have long been proposed and considered among pathogenic factors contributing to IBD, potentially accounting for the “hidden heritability”. Epigenetics is considered the link between environmental factors and the genetic background. Epigenetic studies analyze the heritable changes in gene function that cannot be explained by changes in DNA (e.g., through mutation, recombination, or translocation) [80]. The three major epigenetic modifications are: (1) DNA methylation, which is the most studied; (2) post-translational modifications of histones, such as acetylation of H3K27; and (3) the expression of non-coding RNAs.
DNA methylation has been shown to play a key role in cell processes such as cell differentiation or gene expression, eventually defining cell phenotypes. Of note, DNA methylation is dependent on several cofactors, one of them being nutrition [24, 81, 82], thereby linking genetic with environmental factors. In mouse models, a low-methyl diet has been shown to be associated with increased rates of obesity and diabetes [83–85]. In IBD, DNA global hypomethylation was found in the rectal mucosa from patients with UC, but not healthy controls [86]. In addition, there is evidence that local inflammation (such as seen in uncontrolled IBD) can accelerate DNA methylation changes, thereby resulting in cancer development [87].
Post-translational modification refers to a process that stimulates alteration in nucleosome composition [88]. Nucleosomes consist of DNA segments and the histones they wrap around. They form the smallest packaging unit of chromatin. Molecular differences between different nucleosomes result from numerous chemical modifications of the histone proteins [80]. As an example, G9a is a lysine methyltransferase and catalyzes demethylation of histone H3 lysine 9 (H3K9me2). Lysine methylation alters transcriptional activity without affecting the DNA sequence itself and thereby regulates gene expression in response to environmental stimuli [89]. T-cell-intrinsic G9a has recently been associated with altered T-cell differentiation inducing colitis [90].
Non-coding RNA refers to RNA that is not translated into proteins [91]. It regulates gene expression at transcriptional and post-transcriptional levels. Non-coding RNA can be divided into two classes by the number of nucleotides, short with < 200 nucleotides or long with >200 nucleotides. Both classes have received increasing attention as most of the IBD-associated SNPs are linked to non-coding genetic sequences [92]. They account for several changes in the adaptive immune system (e.g., T-cell and T-helper regulation, T-helper differentiation) as well as the innate immune system (e.g., NOD2- and Toll-like receptors), potentially influencing disease activity and disease course [93, 94].
Clinical Implications: The Road to Precision Medicine
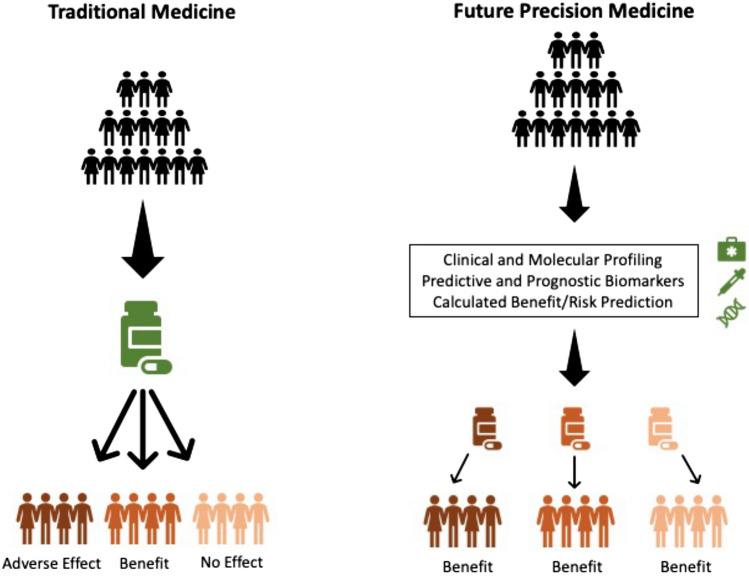
A wide armamentarium for the treatment of IBD is available that ranges from topical formulations, systemic steroids, and immunomodulators to more specific therapeutics such as anti-TNF, anti-integrins, anti-IL-12/23, anti-IL-23p19, and more recently, small molecules. Currently, clinicians, together with the patients, face the difficult challenge of where to position all the available options in the treatment algorithm. While many drugs appear to be equally effective, loss of response in the long term is a universal problem. The question remains as to which patient might respond best to the chosen treatment and which patient does not lose this response over time. A genetic risk assessment appears to be an appealing strategy. In fact, polygenic risk scores have been used not only to predict IBD in the general population, but also to stratify patients, performing better in CD than UC [95–98]. However, the universal use of genotyping has never (or not yet) made it into clinical practice given the high costs, but low benefits [99]. Among potential implications are: assessment of disease risk in relatives (diagnostic biomarker), prediction of disease complications/progression (predictive biomarker), or prediction of therapeutic response or side effects to biological treatment regimens (response biomarker, safety biomarker) [100, 101]. Precision medicine might become a guidance tool for drug dosing or drug sparing if patients with a low risk of relapse could be identified adequately. Unnecessary immunosuppression, at least for a certain time period, could be avoided in such patients (Fig. 1).
Fig. 1.
Illustration of the potential evolution from the current therapeutic strategy, in which one drug is assigned to all patients, to future medicine, where multiple details are considered to create an individualized therapeutic regimen [102]
Adapted from Vieujean and Louis
Conclusions
The progress made in the genetics of IBD over the last 20 years is impressive with over 200 different gene loci identified. However, the contribution of a single polymorphism to the development of IBD or its disease course is small, and most risk alleles are rare. The pathophysiology of IBD appears to be too complex in order to be explained by a few genes or pathways. The most appealing advantage of a genetic risk assessment will lay in the field of precision medicine where the presence or absence of one or several risk genes might help in the therapeutic decision-making process. However, many more studies are needed to fill this important gap.
Declarations
Funding
Open access funding provided by University of Zurich. This work was supported by a grant from the Swiss National Science Foundation to Thomas Greuter (Grant no. P2ZHP3_168561). This work was further supported by a young investigator award from the Swiss Society of Gastroenterology to Thomas Greuter, a research grant from the Novartis Foundation for Medical-Biological Research to Thomas Greuter, a research award from the Swiss IBDnet to TG, and a research grant from the EoE Foundation to Thomas Greuter.
Conflicts of Interest/Competing Interests
Philipp Schreiner has received fees for consulting from AbbVie, Galapagos, Lilly, Sanofi-Regeneron, Falk Pharma, Ferring, Pfizer, Janssen, and Takeda. Stephan R. Vavricka has received consulting fees and unrestricted research grants from Abbott, Falk Pharma GmbH, Ferring Pharmaceuticals, Janssen, MSD, Pfizer Inc, Sanofi-Aventis, Takeda, Tillotts, UCB, and Vifor. Thomas Greuter has consulting contracts with Sanofi-Regeneron, Bristol-Myers Squibb, Takeda, AbbVie, and Falk Pharma GmbH, received travel grants from Falk Pharma GmbH and Vifor, speaker’s fees from Norgine and Amgen, and an unrestricted research grant from Novartis. Jasmina El Hadad has no conflicts of interest that are directly relevant to the content of this article. No company representative was involved in the conception, writing, or financing of this study.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Code Availability
Not applicable.
Authors’ Contributions
JE: drafting manuscript; PS: drafting manuscript and critical review; SRV: drafting manuscript and critical review; TG: drafting manuscript and critical review, responsible investigator.
References
- 1.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan GG, Ng SC. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology. 2017;152(2):313–21.e2. doi: 10.1053/j.gastro.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 5.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandborn WJ, Feagan BG, D'Haens G, Wolf DC, Jovanovic I, Hanauer SB, et al. Ozanimod as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2021;385(14):1280–1291. doi: 10.1056/NEJMoa2033617. [DOI] [PubMed] [Google Scholar]
- 7.Sandborn WJ, Su C, Sands BE, D'Haens GR, Vermeire S, Schreiber S, et al. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2017;376(18):1723–1736. doi: 10.1056/NEJMoa1606910. [DOI] [PubMed] [Google Scholar]
- 8.Danese S, Vermeire S, Zhou W, Pangan AL, Siffledeen J, Greenbloom S, et al. Upadacitinib as induction and maintenance therapy for moderately to severely active ulcerative colitis: results from three phase 3, multicentre, double-blind, randomised trials. Lancet. 2022;399(10341):2113–2128. doi: 10.1016/S0140-6736(22)00581-5. [DOI] [PubMed] [Google Scholar]
- 9.Dickson AL, Daniel LL, Zanussi J, Dale Plummer W, Wei WQ, Liu G, et al. TPMT and NUDT15 variants predict discontinuation of azathioprine for myelotoxicity in patients with inflammatory disease: real-world clinical results. Clin Pharmacol Ther. 2022;111(1):263–271. doi: 10.1002/cpt.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relling MV, Schwab M, Whirl-Carrillo M, Suarez-Kurtz G, Pui CH, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin Pharmacol Ther. 2019;105(5):1095–1105. doi: 10.1002/cpt.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49(2):256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tysk C, Lindberg E, Järnerot G, Flodérus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins: a study of heritability and the influence of smoking. Gut. 1988;29(7):990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006;12(23):3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos MPC, Gomes C, Torres J. Familial and ethnic risk in inflammatory bowel disease. Ann Gastroenterol. 2018;31(1):14–23. doi: 10.20524/aog.2017.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moller FT, Andersen V, Wohlfahrt J, Jess T. Familial risk of inflammatory bowel disease: a population-based cohort study 1977–2011. Am J Gastroenterol. 2015;110(4):564–571. doi: 10.1038/ajg.2015.50. [DOI] [PubMed] [Google Scholar]
- 16.Orholm M, Binder V, Sørensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins: results of a nationwide study. Scand J Gastroenterol. 2000;35(10):1075–1081. doi: 10.1080/003655200451207. [DOI] [PubMed] [Google Scholar]
- 17.Thompson NP, Driscoll R, Pounder RE, Wakefield AJ. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ. 1996;312(7023):95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres J, Gomes C, Jensen CB, Agrawal M, Ribeiro-Mourão F, Jess T, et al. Risk factors for developing inflammatory bowel disease within and across families with a family history of IBD. J Crohns Colitis. 2023;17(1):30–36. doi: 10.1093/ecco-jcc/jjac111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye BD, McGovern DP. Genetic variation in IBD: progress, clues to pathogenesis and possible clinical utility. Expert Rev Clin Immunol. 2016;12(10):1091–1107. doi: 10.1080/1744666X.2016.1184972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12(4):205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 21.Sun Y, Yuan S, Chen X, Sun J, Kalla R, Yu L, et al. The contribution of genetic risk and lifestyle factors in the development of adult-onset inflammatory bowel disease: a prospective cohort study. Am J Gastroenterol. 2023;118(3):511–522. doi: 10.14309/ajg.0000000000002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brant SR. Promises, delivery, and challenges of inflammatory bowel disease risk gene discovery. Clin Gastroenterol Hepatol. 2013;11(1):22–26. doi: 10.1016/j.cgh.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Mirkov MU, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol. 2017;2(3):224–234. doi: 10.1016/S2468-1253(16)30111-X. [DOI] [PubMed] [Google Scholar]
- 24.Wawrzyniak M, Scharl M. Genetics and epigenetics of inflammatory bowel disease. Swiss Med Wkly. 2018;148:w14671. doi: 10.4414/smw.2018.14671. [DOI] [PubMed] [Google Scholar]
- 25.Chen GB, Lee SH, Brion MJ, Montgomery GW, Wray NR, Radford-Smith GL, et al. Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data. Hum Mol Genet. 2014;23(17):4710–4720. doi: 10.1093/hmg/ddu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellinghaus D, Ellinghaus E, Nair RP, Stuart PE, Esko T, Metspalu A, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90(4):636–647. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140(6):1704–1712. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon H, Trier Moller F, Andersen V, Harbord M. Heritability in inflammatory bowel disease: from the first twin study to genome-wide association studies. Inflamm Bowel Dis. 2015;21(6):1428–1434. doi: 10.1097/MIB.0000000000000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 32.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 33.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140(6):1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abreu MT, Taylor KD, Lin YC, Hang T, Gaiennie J, Landers CJ, et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123(3):679–688. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 35.Adler J, Rangwalla SC, Dwamena BA, Higgins PD. The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am J Gastroenterol. 2011;106(4):699–712. doi: 10.1038/ajg.2011.19. [DOI] [PubMed] [Google Scholar]
- 36.Sidiq T, Yoshihama S, Downs I, Kobayashi KS. Nod2: a critical regulator of ileal microbiota and Crohn's disease. Front Immunol. 2016;7:367. doi: 10.3389/fimmu.2016.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonen DK, Ogura Y, Nicolae DL, Inohara N, Saab L, Tanabe T, et al. Crohn's disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124(1):140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad T, Armuzzi A, Bunce M, Mulcahy-Hawes K, Marshall SE, Orchard TR, et al. The molecular classification of the clinical manifestations of Crohn's disease. Gastroenterology. 2002;122(4):854–866. doi: 10.1053/gast.2002.32413. [DOI] [PubMed] [Google Scholar]
- 39.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99(12):2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 40.Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123(1):86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 41.Brant SR, Okou DT, Simpson CL, Cutler DJ, Haritunians T, Bradfield JP, et al. Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology. 2017;152(1):206–17.e2. doi: 10.1053/j.gastro.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leong RW, Armuzzi A, Ahmad T, Wong ML, Tse P, Jewell DP, et al. NOD2/CARD15 gene polymorphisms and Crohn's disease in the Chinese population. Aliment Pharmacol Ther. 2003;17(12):1465–1470. doi: 10.1046/j.1365-2036.2003.01607.x. [DOI] [PubMed] [Google Scholar]
- 43.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivanesan D, Beauchamp C, Quinou C, Lee J, Lesage S, Chemtob S, et al. IL23R (interleukin 23 receptor) variants protective against inflammatory bowel diseases (IBD) display loss of function due to impaired protein stability and intracellular trafficking. J Biol Chem. 2016;291(16):8673–8685. doi: 10.1074/jbc.M116.715870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abreu MT, Rowbotham DS, Danese S, Sandborn WJ, Miao Y, Zhang H, et al. Efficacy and safety of maintenance ustekinumab for ulcerative colitis through 3 years: UNIFI long-term extension. J Crohns Colitis. 2022;16(8):1222–1234. doi: 10.1093/ecco-jcc/jjac030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13(5):715–725. doi: 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 47.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116(5):1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52(1):65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmad T, Marshall SE, Jewell D. Genetics of inflammatory bowel disease: the role of the HLA complex. World J Gastroenterol. 2006;12(23):3628–3635. doi: 10.3748/wjg.v12.i23.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satsangi J, Welsh KI, Bunce M, Julier C, Farrant JM, Bell JI, et al. Contribution of genes of the major histocompatibility complex to susceptibility and disease phenotype in inflammatory bowel disease. Lancet. 1996;347(9010):1212–1217. doi: 10.1016/S0140-6736(96)90734-5. [DOI] [PubMed] [Google Scholar]
- 51.Rojas M, Restrepo-Jiménez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramírez-Santana C, et al. Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–123. doi: 10.1016/j.jaut.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 52.Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42(1):102–111. doi: 10.1007/s12016-011-8294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5(12):889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 54.Yap LM, Ahmad T, Jewell DP. The contribution of HLA genes to IBD susceptibility and phenotype. Best Pract Res Clin Gastroenterol. 2004;18(3):577–596. doi: 10.1016/j.bpg.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4(4):e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sazonovs A, Kennedy NA, Moutsianas L, Heap GA, Rice DL, Reppell M, et al. HLA-DQA1*05 carriage associated with development of anti-drug antibodies to infliximab and adalimumab in patients with Crohn's disease. Gastroenterology. 2020;158(1):189–199. doi: 10.1053/j.gastro.2019.09.041. [DOI] [PubMed] [Google Scholar]
- 57.Vavricka SR, Schoepfer A, Scharl M, Lakatos PL, Navarini A, Rogler G. Extraintestinal manifestations of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21(8):1982–1992. doi: 10.1097/MIB.0000000000000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bernstein CN, Blanchard JF, Rawsthorne P, Yu N. The prevalence of extraintestinal diseases in inflammatory bowel disease: a population-based study. Am J Gastroenterol. 2001;96(4):1116–1122. doi: 10.1111/j.1572-0241.2001.03756.x. [DOI] [PubMed] [Google Scholar]
- 59.Bernstein CN, Wajda A, Blanchard JF. The clustering of other chronic inflammatory diseases in inflammatory bowel disease: a population-based study. Gastroenterology. 2005;129(3):827–836. doi: 10.1053/j.gastro.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 60.Mendoza JL, Lana R, Taxonera C, Alba C, Izquierdo S, Díaz-Rubio M. Extraintestinal manifestations in inflammatory bowel disease: differences between Crohn's disease and ulcerative colitis. Med Clin (Barc) 2005;125(8):297–300. doi: 10.1157/13078423. [DOI] [PubMed] [Google Scholar]
- 61.Ricart E, Panaccione R, Loftus EV, Tremaine WJ, Harmsen WS, Zinsmeister AR, et al. Autoimmune disorders and extraintestinal manifestations in first-degree familial and sporadic inflammatory bowel disease: a case-control study. Inflamm Bowel Dis. 2004;10(3):207–214. doi: 10.1097/00054725-200405000-00005. [DOI] [PubMed] [Google Scholar]
- 62.Rankin GB, Watts HD, Melnyk CS, Kelley ML. National Cooperative Crohn's Disease Study: extraintestinal manifestations and perianal complications. Gastroenterology. 1979;77(4 Pt 2):914–920. doi: 10.1016/0016-5085(79)90391-3. [DOI] [PubMed] [Google Scholar]
- 63.Su CG, Judge TA, Lichtenstein GR. Extraintestinal manifestations of inflammatory bowel disease. Gastroenterol Clin North Am. 2002;31(1):307–327. doi: 10.1016/S0889-8553(01)00019-X. [DOI] [PubMed] [Google Scholar]
- 64.Veloso FT, Carvalho J, Magro F. Immune-related systemic manifestations of inflammatory bowel disease: a prospective study of 792 patients. J Clin Gastroenterol. 1996;23(1):29–34. doi: 10.1097/00004836-199607000-00009. [DOI] [PubMed] [Google Scholar]
- 65.Vavricka SR, Brun L, Ballabeni P, Pittet V, Prinz Vavricka BM, Zeitz J, et al. Frequency and risk factors for extraintestinal manifestations in the Swiss inflammatory bowel disease cohort. Am J Gastroenterol. 2011;106(1):110–119. doi: 10.1038/ajg.2010.343. [DOI] [PubMed] [Google Scholar]
- 66.Greuter T, Vavricka SR. Extraintestinal manifestations in inflammatory bowel disease: epidemiology, genetics, and pathogenesis. Expert Rev Gastroenterol Hepatol. 2019;13(4):307–317. doi: 10.1080/17474124.2019.1574569. [DOI] [PubMed] [Google Scholar]
- 67.Satsangi J, Grootscholten C, Holt H, Jewell DP. Clinical patterns of familial inflammatory bowel disease. Gut. 1996;38(5):738–741. doi: 10.1136/gut.38.5.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thia KT, Loftus EV, Sandborn WJ, Yang SK. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008;103(12):3167–3182. doi: 10.1111/j.1572-0241.2008.02158.x. [DOI] [PubMed] [Google Scholar]
- 69.Prideaux L, Kamm MA, De Cruz PP, Chan FK, Ng SC. Inflammatory bowel disease in Asia: a systematic review. J Gastroenterol Hepatol. 2012;27(8):1266–1280. doi: 10.1111/j.1440-1746.2012.07150.x. [DOI] [PubMed] [Google Scholar]
- 70.Hou JK, El-Serag H, Thirumurthi S. Distribution and manifestations of inflammatory bowel disease in Asians, Hispanics, and African Americans: a systematic review. Am J Gastroenterol. 2009;104(8):2100–2109. doi: 10.1038/ajg.2009.190. [DOI] [PubMed] [Google Scholar]
- 71.Walker DG, Williams HR, Kane SP, Mawdsley JE, Arnold J, McNeil I, et al. Differences in inflammatory bowel disease phenotype between South Asians and Northern Europeans living in North West London. UK. Am J Gastroenterol. 2011;106(7):1281–1289. doi: 10.1038/ajg.2011.85. [DOI] [PubMed] [Google Scholar]
- 72.Martin TM, Smith JR, Rosenbaum JT. Anterior uveitis: current concepts of pathogenesis and interactions with the spondyloarthropathies. Curr Opin Rheumatol. 2002;14(4):337–341. doi: 10.1097/00002281-200207000-00001. [DOI] [PubMed] [Google Scholar]
- 73.Akhlaghpour M, Haritunians T, More SK, Thomas LS, Stamps DT, Dube S, et al. Genetic coding variant in complement factor B (CFB) is associated with increased risk for perianal Crohn's disease and leads to impaired CFB cleavage and phagocytosis. Gut. 2023;72(11):2068–2080. doi: 10.1136/gutjnl-2023-329689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hedin CRH, Vavricka SR, Stagg AJ, Schoepfer A, Raine T, Puig L, et al. The pathogenesis of extraintestinal manifestations: implications for IBD research, diagnosis, and therapy. J Crohns Colitis. 2019;13(5):541–554. doi: 10.1093/ecco-jcc/jjy191. [DOI] [PubMed] [Google Scholar]
- 75.Weinshilboum R, Wang L. Pharmacogenomics: bench to bedside. Nat Rev Drug Discov. 2004;3(9):739–748. doi: 10.1038/nrd1497. [DOI] [PubMed] [Google Scholar]
- 76.Ruemmele FM, Veres G, Kolho KL, Griffiths A, Levine A, Escher JC, et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn's disease. J Crohns Colitis. 2014;8(10):1179–1207. doi: 10.1016/j.crohns.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 77.Lauro R, Mannino F, Irrera N, Squadrito F, Altavilla D, Squadrito G, et al. Pharmacogenetics of biological agents used in inflammatory bowel disease: a systematic review. Biomedicines. 2021;9(12):1748. doi: 10.3390/biomedicines9121748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bek S, Nielsen JV, Bojesen AB, Franke A, Bank S, Vogel U, et al. Systematic review: genetic biomarkers associated with anti-TNF treatment response in inflammatory bowel diseases. Aliment Pharmacol Ther. 2016;44(6):554–567. doi: 10.1111/apt.13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamilton JP. Epigenetics: principles and practice. Dig Dis. 2011;29(2):130–135. doi: 10.1159/000323874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jenke AC, Zilbauer M. Epigenetics in inflammatory bowel disease. Curr Opin Gastroenterol. 2012;28(6):577–584. doi: 10.1097/MOG.0b013e328357336b. [DOI] [PubMed] [Google Scholar]
- 82.Xu J, Xu HM, Yang MF, Liang YJ, Peng QZ, Zhang Y, et al. New insights into the epigenetic regulation of inflammatory bowel disease. Front Pharmacol. 2022;13:813659. doi: 10.3389/fphar.2022.813659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J. 2007;21(12):3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
- 84.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23(3):314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 85.Natasha G, Zilbauer M. Epigenetics in IBD: a conceptual framework for disease pathogenesis. Frontline Gastroenterol. 2022;13(e1):e22–7. [DOI] [PMC free article] [PubMed]
- 86.Glória L, Cravo M, Pinto A, de Sousa LS, Chaves P, Leitão CN, et al. DNA hypomethylation and proliferative activity are increased in the rectal mucosa of patients with long-standing ulcerative colitis. Cancer. 1996;78(11):2300–2306. doi: 10.1002/(SICI)1097-0142(19961201)78:11<2300::AID-CNCR5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 87.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61(9):3573–3577. [PubMed] [Google Scholar]
- 88.Bowman GD, Poirier MG. Post-translational modifications of histones that influence nucleosome dynamics. Chem Rev. 2015;115(6):2274–2295. doi: 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Z, Connolly J, Biggar KK. Beyond histones: the expanding roles of protein lysine methylation. FEBS J. 2017;284(17):2732–2744. doi: 10.1111/febs.14056. [DOI] [PubMed] [Google Scholar]
- 90.Ramos GP, Bamidele AO, Klatt EE, Sagstetter MR, Kurdi AT, Hamdan FH, et al. G9a modulates lipid metabolism in CD4 T cells to regulate intestinal inflammation. Gastroenterology. 2023;164(2):256–71.e10. doi: 10.1053/j.gastro.2022.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yarani R, Mirza AH, Kaur S, Pociot F. The emerging role of lncRNAs in inflammatory bowel disease. Exp Mol Med. 2018;50(12):1–14. doi: 10.1038/s12276-018-0188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Braga-Neto MB, Gaballa JM, Bamidele AO, Sarmento OF, Svingen P, Gonzalez M, et al. Deregulation of long intergenic non-coding RNAs in CD4+ T cells of lamina propria in Crohn's disease through transcriptome profiling. J Crohns Colitis. 2020;14(1):96–109. doi: 10.1093/ecco-jcc/jjz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu XM, Zhang HJ. miRNAs as new molecular insights into inflammatory bowel disease: crucial regulators in autoimmunity and inflammation. World J Gastroenterol. 2016;22(7):2206–2218. doi: 10.3748/wjg.v22.i7.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, et al. MicroRNAs: new players in IBD. Gut. 2015;64(3):504–517. doi: 10.1136/gutjnl-2014-307891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garcia-Etxebarria K, Merino O, Gaite-Reguero A, Rodrigues PM, Herrarte A, Etxart A, et al. Local genetic variation of inflammatory bowel disease in Basque population and its effect in risk prediction. Sci Rep. 2022;12(1):3386. doi: 10.1038/s41598-022-07401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gettler K, Levantovsky R, Moscati A, Giri M, Wu Y, Hsu NY, et al. Common and rare variant prediction and penetrance of IBD in a large, multi-ethnic, health system-based Biobank cohort. Gastroenterology. 2021;160(5):1546–1557. doi: 10.1053/j.gastro.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen GB, Lee SH, Montgomery GW, Wray NR, Visscher PM, Gearry RB, et al. Performance of risk prediction for inflammatory bowel disease based on genotyping platform and genomic risk score method. BMC Med Genet. 2017;18(1):94. doi: 10.1186/s12881-017-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20(10):1122–1130. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 100.Fiocchi C, Dragoni G, Iliopoulos D, Katsanos K, Ramirez VH, Suzuki K, et al. Results of the Seventh Scientific Workshop of ECCO: Precision medicine in IBD: what, why, and how. J Crohns Colitis. 2021;15(9):1410–1430. doi: 10.1093/ecco-jcc/jjab051. [DOI] [PubMed] [Google Scholar]
- 101.Verstockt B, Noor NM, Marigorta UM, Pavlidis P, Deepak P, Ungaro RC, et al. Results of the Seventh Scientific Workshop of ECCO: precision medicine in IBD-disease outcome and response to therapy. J Crohns Colitis. 2021;15(9):1431–1442. doi: 10.1093/ecco-jcc/jjab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vieujean S, Louis E. Precision medicine and drug optimization in adult inflammatory bowel disease patients. Therap Adv Gastroenterol. 2023;16:17562848231173331. doi: 10.1177/17562848231173331. [DOI] [PMC free article] [PubMed] [Google Scholar]