Abstract
By cloning into Escherichia coli and construction of isogenic mutants of Haemophilus ducreyi, we showed that the hemoglobin receptor (HgbA) is TonB dependent. An E. coli hemA tonB mutant expressing H. ducreyi hgbA grew on low levels of hemoglobin as a source of heme only when an intact H. ducreyi Ton system plasmid was present. In contrast, growth on heme by the E. coli hemA tonB mutant expressing hgbA was observed only at high concentrations of heme, was TonB independent, and demonstrated that H. ducreyi HgbA was not sufficient to function as a typical TonB-dependent heme receptor in E. coli. Allelic replacement of the wild-type H. ducreyi exbB, exbD, and tonB loci with the exbB, exbD, and tonB deletion resulted in an H. ducreyi isogenic mutant unable to utilize hemoglobin but able to utilize hemin at the same levels as the parent strain to fulfill its heme requirement. This finding confirms the TonB dependence of HgbA-mediated hemoglobin utilization and suggests that uptake of hemin in H. ducreyi is TonB independent. Additionally, the H. ducreyi Ton system mutant synthesized increased amounts of HgbA and other heme-regulated outer membrane proteins, consistent with derepression of these proteins due to lower intracellular heme and/or iron concentrations in the mutant. Sequencing of the Ton system genes revealed that the arrangement of the genes was exbB exbD tonB. The proximity and structure of these genes suggested that they are transcribed as an operon. This arrangement, as well as the DNA and deduced amino acid sequences of these H. ducreyi genes, was most similar to those from other pasteurellae.
Haemophilus ducreyi is the etiologic agent of chancroid, a genital ulcer disease transmitted by sexual contact (reviewed in references 2 and 66). Prevalent in Africa, Asia, and certain developing nations, chancroid has recently gained importance as an independent risk factor for the heterosexual transmission of human immunodeficiency (67). There is accumulating evidence suggesting that the rapid spread of human immunodeficiency virus type 1 in eastern and southern Africa has been due, at least in part, to the presence of chancroid and other genital ulcer diseases (13, 51). At present no vaccine or practical field diagnostic test exists for H. ducreyi.
H. ducreyi is a fastidious slow-growing, gram-negative rod with an optimum growth temperature of 33°C. An important feature of H. ducreyi is its requirement for heme, which it is unable to synthesize. This obligate requirement for heme by H. ducreyi is in contrast with H. influenzae, which is able to use its enzyme ferrochelatase to ferrate protoporphyrin IX, bypassing the need for heme (24, 53). In H. ducreyi, heme is acquired from its only known host, humans, in several possible forms, including hemoglobin, hemoglobin complexed to haptoglobin, catalase, free hemin, or heme bound to carrier proteins such as albumin (3, 39, 64). Some or all of these heme sources could be released from host cells by the action of the H. ducreyi hemolysin (45, 46, 63) or cytotoxin (16, 37, 49).
Host heme compounds represent important sources of iron (44). Host iron is sequestered by several mechanisms, and invading bacteria must gain access to these iron sources to survive and initiate disease. Some bacteria utilize iron-scavenging siderophore systems; others directly bind host iron or heme/iron-containing compounds such as transferrin, lactoferrin, heme, or hemoglobin (19, 44). No siderophores have been found in H. ducreyi (39). Since H. ducreyi requires both heme and iron, differentiation between the requirement for both of these can be problematic.
Siderophore receptors and most receptors for host iron-binding compounds belong to a family of outer membrane receptors designated TonB dependent because they require the cytoplasmic membrane protein TonB for activity (7, 11, 57). TonB-dependent receptors are related structurally and functionally; where studied, they have been shown to form specific pores for the entry of low-molecular-weight molecules (12, 40). TonB-dependent receptors for host-iron complexes such as transferrin or lactoferrin have been shown to internalize the iron only (41), whereas receptors for siderophores internalize the iron-siderophore complex (43). TonB-dependent receptors are also related at the amino acid level by virtue of several regions of distinct homology, including an N-terminal sequence termed the TonB Box (18, 28, 35, 54). In Escherichia coli, the TonB protein has been shown to directly interact with TonB-dependent receptors FepA (5, 55). TonB and its accessory proteins ExbB and ExbD comprise a system for the transfer of energy from the cytoplasmic membrane to outer membrane receptors (10). Null mutations in either tonB, exbB, or exbD can variably affect the ability of TonB-dependent receptors to internalize or utilize their cognate ligand (48). The effect of these null mutations can vary from partial to total inhibition of uptake of ligands.
Previously, we identified and purified an outer membrane hemoglobin receptor from H. ducreyi termed HgbA (22) and showed that the HgbA protein is functionally and immunologically conserved between geographically diverse isolates. Further analysis was performed by using molecular techniques (23, 56). The hgbA gene was cloned and sequenced; its deduced amino acid sequence is similar to the TonB-dependent family of outer membrane receptors (23, 56). An hgbA isogenic mutant of H. ducreyi cannot bind or utilize hemoglobin as a source of heme but can utilize free hemin, implying that the utilization of hemin does not require HgbA. It has been shown by Stevens et al. (56) that an isogenic mutant of hgbA (hup) expressed reduced virulence in an animal model of H. ducreyi infection.
In the course of these functional studies, we found that an E. coli hemA mutant expressing hgbA binds hemoglobin yet does not grow on hemoglobin as a porphyrin source (23). This result suggested that additional components are necessary for utilization of hemoglobin after the binding step. The objective of the present study was to identify the additional components necessary for removal of heme from hemoglobin and for the transport of heme across the outer membrane.
MATERIALS AND METHODS
Strains and media.
Bacterial strains used in this study are described in Table 1. For routine growth, H. ducreyi was maintained on chocolate agar plates prepared by following gonococcal medium base (GCB) instructions (Difco). No fetal bovine serum was utilized in chocolate agar plates used for routine passage. Plates containing the various heme sources were prepared by using GCB and IsoVitaeleX supplements (Difco) (termed GCB-I). Human hemoglobin (H7379; Sigma Chemical, St. Louis, Mo.) was dissolved in phosphate-buffered saline at a concentration of 10 mg/ml and rocked at room temperature for 2 h or overnight at 4°C prior to filter sterilization. Heme (Sigma) was made 10 mg/ml in 0.1 N NaOH and was used without sterilization. To bind heme to either serum or serum proteins, various amounts of heme were added to the protein source and allowed to bind for at least 10 min at room temperature prior to incorporation into GCB-I plates. Human serum albumin (HSA; Sigma) was saturated to 50% by the addition of 1/2 mol of hemin per mol of albumin. We assumed that the albumin contained no hemin to start with and that it contained a single hemin binding site. Catalase was obtained from Sigma (C-100, lot no. 106H7055; C-6655, lot no. 23H70355). Five micrograms of hemin per milliliter corresponds to 7.6 μM hemin.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype and/or phenotype | Reference(s) or Source |
|---|---|---|
| Strains | ||
| E. coli K-12 | ||
| EB53 | hemA aroB rpoB | 58 |
| IR754 | EB53, but tonB::Kanr | 59 |
| DH5αMCR | recA gyrB | Bethesda Research Laboratories |
| H. ducreyi | ||
| 35000 | Wild type | Stanley Spinola, Indiana University |
| FX504 | hgbA | 23 |
| FX514 | ΔexbB exbD tonB | This work |
| FX514 repair | Wild type | This work |
| FX514(pUNCH 1210) | Wild type | This work |
| Plasmids | ||
| pUNCH 579 | ColE1 replicon, hgbA Apr | 23 |
| pUNCH 556 | p15 replicon, hgbA Ter | This work |
| pUNCH 562 | ColE1 replicon, exbB exbD tonB Apr | This work |
| pUNCH 563 | ColE1 replicon, exbB+ exbD+ tonB+ Apr, subclone of pUNCH 562 | This work |
| pUNCH 568 | ColE1 replicon, ΔexbB exbD tonB Apr Cmr, deletion mutant of pUNCH 563 | This work |
| pBluescript | ColE1 replicon, Apr | Stratagene |
| pACYC184 | p15 replicon, Cmr Ter | 1 |
| pGJ300 | pLS88 replicon, exbB exbD tonB Kanr Sulfr Strr | 33, 34 |
| pLS88 | H. ducreyi shuttle plasmid, Kanr Sulfr Strr | 21 |
| pUNCH 1210 | pLS88 containing pUNCH 563 insert in EcoRI site, Kanr | This work |
E. coli hemA mutants EB53 and IR754 were maintained on LB agar plates containing 25 μM δ-aminolevulonic (δALA) acid with antibiotic selection where appropriate. Antibiotics were used at the following concentrations for E. coli: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 30 μg/ml; and tetracycline, 12 μg/ml. Antibiotic concentrations for H. ducreyi were 1 μg/ml (chloramphenicol) and 20 μg/ml (kanamycin).
Growth conditions for testing phenotypes.
For inoculation of plates for testing phenotypes, H. ducreyi strains were heme starved by growth anaerobically on GCB-I plates without a heme source (56). This method of inoculation reduced the possibility of heme carryover or differences in intracellular heme stores among the various strains. After H. ducreyi was grown anaerobically on GCB-I plates, inocula were standardized spectrophotometrically and 10-fold dilutions were made for each strain in a microtiter tray. A Steers replicator was used to inoculate agar medium containing dilutions of each heme source. Thus, the inoculum and heme sources were present in a checkerboard titration. Plates were incubated at 35°C in 5% CO2.
Outer membrane isolation and analysis.
Large-scale cultures of H. ducreyi were performed in Fernbach flasks with 1 liter of GCB-I broth containing 5% fetal calf serum (FCS) and heme at 2 μg/ml (heme limiting) or 50 μg/ml (heme replete) (22). It has been shown by several investigators that H. ducreyi and H. influenzae strains require less hemin when grown in liquid media compared to solid medium (3, 22, 60, 64). Therefore, these concentrations should not be compared with those used in solid medium used for testing phenotypes. Flasks were inoculated to a starting density of 2.4 × 106 CFU/ml with growth harvested from chocolate agar plates and were incubated 24 h with shaking in a 5% CO2 atmosphere at 35°C. The purity of each culture was routinely verified by streaking onto agar plates which did (chocolate agar) and did not support the growth of H. ducreyi but which supported the growth of most other bacteria (GCB-I, without hemin). This ensured that minor contamination could be detected and that the novel (previously undescribed) proteins detected (see Fig. 5A) were not the result of contamination. Outer membranes were harvested as previously described (22) except that lysozyme was added to the harvested cells prior to the French press step (200 μg/ml, 10 min, 4°C) and two solubilizations were performed with Sarkosyl rather than one. Protein concentrations were determined by using a bicinchoninic acid kit from Pierce (Rockford, Ill.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as previously described (22) except for the antiserum to HgbA. Anti-HgbA was produced by immunizing mice with 25 μg of purified HgbA (22) three times at 2-week intervals. Complete Freund’s adjuvant was used for the first immunization, and incomplete Freund’s adjuvant was used for the last two immunizations.
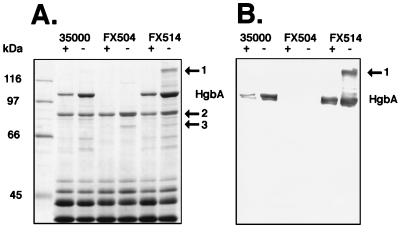
FIG. 5.
SDS-PAGE and Western blotting of outer membranes from H. ducreyi. The indicated strains were grown under heme-replete (+) or heme-limiting (−) conditions, and their outer membranes were isolated and subjected to SDS-PAGE (7.5% gel) and Coomassie staining (A) or Western blotting using anti-HgbA sera (B). Strains were H. ducreyi parent strain 35000 (wild type), FX504 (hgbA), and FX514 (ΔexbB exbD tonB). Positions of molecular mass markers are shown at the left. HgbA, hemoglobin receptor protein; protein bands 1, 2, and 3, previously undescribed heme-regulated proteins expressed by Ton mutant FX514 (protein 2 refers to the lower band of the doublet). The Western blot (B) was purposely overloaded in order to detect potential proteolytic breakdown products of HgbA as a source of proteins 2 and 3 and other, smaller proteins.
Chemicals and reagents.
All chemicals and reagents, unless otherwise noted, were from Sigma.
DNA manipulations.
Standard recombinant DNA methods were used as described in reference 52 or following manufacturers’ instructions.
The 8.5-kb EcoRI fragment of pUNCH 579 containing the entire hgbA gene and promoter was ligated into the EcoRI site of pACYC184 (15) to construct pUNCH 556. Expression of hgbA in E. coli IR754(pUNCH 556) was confirmed by Western blotting as previously described (22).
For construction of a library from strain 35000, chromosomal DNA was partially restricted with Sau3A1 under limiting enzyme conditions, and 5- to 12-kb fragments were isolated from a preparative gel. These fragments were ligated into the BamHI site of purified pBluescript SK(−) vector. To avoid restriction in subsequent hosts, E. coli DH5αMCR was transformed with the ligation mixture, the resulting ampicillin-resistant transformants were pooled, and plasmid DNA was isolated by using the Magic Miniprep procedure (Promega). To clone the Ton system, this plasmid pool was electroporated into E. coli IR754(pUNCH 556), and selection was performed on LB agar containing hemoglobin (100 μg/ml), ampicillin (100 μg/ml), and tetracycline (10 μg/ml).
DNA sequencing and analysis.
DNA sequence analysis was performed at the University of North Carolina at Chapel Hill Automated Sequencing Facility, using Taq terminator chemistry. Both strands were completely sequenced and assembled by using AssemblyLIGN software (IBI). Amino acid alignments were done with the programs PILEUP and PRETTYBOX (25, 50). The default threshold parameters for PRETTYBOX were used: 1.5, identical (black boxes); 1.0, similar (dark grey boxes); 0.5, somewhat similar (light grey); 0.0, dissimilar (white boxes) (see Fig. 2). Table 2 was constructed by using data generated from the program OLDDISTANCES (25, 50).
FIG. 2.
Comparison of H. ducreyi ExbB (A), ExbD (B), and TonB (C) proteins with selected Ton system homologs from other gram-negative bacteria, generated with the programs PILEUP and PRETTYBOX. Black boxes, identical residues; grey boxes, related conserved substitutions; white boxes, nonconservative substitutions. In panel A, the N-terminal 74 aa are not shown for Pseudomonas putida ExbB. Hd, H. ducreyi; Ph, P. haemolytica; Hi, H. influenzae; Nm, Neisseria meningitidis; Pp, Pseudomonas putida; Ec, E. coli.
TABLE 2.
Percent identities among ExbB, ExbD, and TonB homologsa
| Homolog | % Identityb
|
||||||
|---|---|---|---|---|---|---|---|
| Strain | Hd | Ph | Hi | Nm | Pp | Ec | |
| ExbB | Hd | 100 | 68 | 61 | 28 | 24 | 23 |
| Ph | 100 | 61 | 27 | 24 | 24 | ||
| Hi | 100 | 27 | 28 | 26 | |||
| Nm | 100 | 32 | 31 | ||||
| Pp | 100 | 60 | |||||
| Ec | 100 | ||||||
| ExbD | Hd | 100 | 84 | 70 | 22 | 26 | 25 |
| Ph | 100 | 59 | 19 | 21 | 21 | ||
| Hi | 100 | 19 | 23 | 21 | |||
| Nm | 100 | 33 | 35 | ||||
| Pp | 100 | 66 | |||||
| Ec | 100 | ||||||
| TonB | Hd | 100 | 41 | 24 | 19 | 17 | 23 |
| Ph | 100 | 13 | 14 | 9 | 13 | ||
| Hi | 100 | 18 | 21 | 26 | |||
| Nm | 100 | 13 | 17 | ||||
| Pp | 100 | 25 | |||||
| Ec | 100 | ||||||
Hd, H. ducreyi; Ph, P. haemolytica; Hi, H. influenzae; Nm, Neisseria meningitidis; Pp, Pseudomonas putida; Ec, E. coli.
Subcloning and construction of an H. ducreyi mutant.
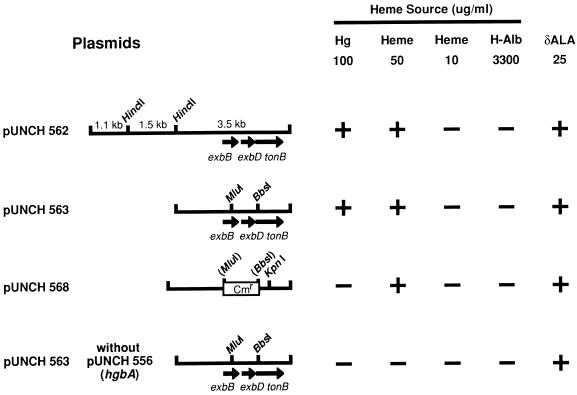
pUNCH 562 was subcloned by removal of two small HincII fragments by restriction and religation of the gel-purified 6.5-kb fragment to form pUNCH 563 (Fig. 1). pUNCH 563 was mutagenized by restriction with BbsI and MluI followed by Klenow enzyme treatment to create blunt ends (Fig. 1). A chloramphenicol-resistant (Cmr) cassette was isolated from plasmid pNC40 (61) by digestion with BglII and end repair using Klenow enzyme. This cassette was ligated to digested pUNCH 563 to form pUNCH 568 (Fig. 1). The deleted DNA between the BbsI and MluI sites of pUNCH 563 contains sequences encoding the C terminus of ExbB, all of ExbD, and the N terminus of TonB. E. coli IR754(pUNCH 556) was electroporated with pUNCH 568, and transformants were selected on LB agar containing chloramphenicol (30 μg/ml), tetracycline (10 μg/ml), and δALA (25 μg/ml). To compare the phenotypes of E. coli IR754(pUNCH 556) containing pUNCH 563 and its mutagenized counterpart pUNCH 568, single colonies of each were streaked onto LB medium containing ampicillin, tetracycline, and either hemoglobin or δALA and on free hemin or 50% heme-saturated human albumin (Fig. 1 and 3).
FIG. 1.
Restriction maps and phenotypes of relevant H. ducreyi exbB, exbD, and tonB clones. Not shown in pUNCH 568 are unique XbaI and XhoI vector sites used to release the insert prior to allelic exchange. E. coli IR754 (hemA tonB aroB) containing pUNCH 556 and the indicated plasmid was inoculated onto LB medium containing various sources of heme or δALA: hemoglobin (Hg), 100 μg/ml (6 μM hemin); heme, 50 μg/ml (77 μM) or 10 μg/ml (15 μM); heme-HSA (H-Alb), 3,300 μg/ml (50 μM), saturated at a 50% molar concentration with hemin, 16 μg/ml (25 μM); δALA, 25 μg/ml (4 μM). +, macroscopic growth at 48 h; −, no macroscopic growth at 48 h.
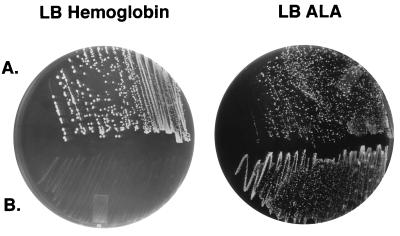
FIG. 3.
Growth of recombinant Ton system clones on LB hemoglobin agar or δALA (LB-ALA) E. coli IR754 (hemA tonB aroB)(pUNCH 556 [hgbA]) containing either pUNCH 563 (ExbB+ ExbD+ TonB+) (A) or pUNCH 568 (ExbB− ExbD− TonB−) (B) was streaked onto LB agar containing hemoglobin (100 μg/ml [5 μM hemin]) or δALA (25 μg/ml [4 μM]) with antibiotic selection and incubated at 37°C for 48 h.
An isogenic mutant (FX514 [Table 1]) was constructed by allelic replacement of the wild-type locus of strain 35000 with the mutation in pUNCH 568 (Fig. 1). Electroporation of strain 35000 was performed by using 1 μg of XbaI- and XhoI-restricted pUNCH 568 and selection on 1 μg of chloramphenicol per ml in chocolate agar plates as previously described (23, 27). Cmr isolates were tested for inability to grow on hemoglobin agar plates. Strain 35000 and FX504 hgbA were used as growth controls for the hemoglobin plates.
Southern blot analysis was used to confirm that the genetic event that occurred in construction of H. ducreyi mutant FX514 was the result of a double crossover. Chromosomal DNA was isolated, restricted with KpnI and HincII, and subjected to electrophoresis and bidirectional transfer. One blot was probed with the insert from pUNCH 563 (Ton system probe), and the other was probed with pNC40 (chloramphenicol acetyltransferase [CAT] probe). Probes were labeled with digoxigenin by using a Genius random primed kit (Boehringer Mannheim). Bound probe was detected with alkaline phosphatase-labeled antidigoxigenin antibody (Boehringer Mannheim) followed by detection with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate.
Repair of the chromosomal defect in Ton mutant FX514 was accomplished by electroporation of XhoI-XbaI-restricted pUNCH 563 into FX514 and plating on hemoglobin agar without an expression period. H. ducreyi isolates that survived multiple passages on hemoglobin agar and that were sensitive to chloramphenicol were subjected to Southern analysis. All 12 isolates regained the wild-type Southern blotting pattern of parent 35000, using pUNCH 563 and CAT probes as shown in Fig. 1 (Southern data for repaired FX514 not shown).
To complement FX514 in trans, the XhoI-XbaI insert from pUNCH 563 was Klenow enzyme treated and ligated into EcoRI-restricted, Klenow enzyme-treated vector pLS88 (21, 68). E. coli IR754(pUNCH 579) was electroporated and plated onto LB hemoglobin agar containing appropriate antimicrobials. Several complementing plasmids were then moved to E. coli DH5αMCR and confirmed by restriction. One plasmid, designated pUNCH 1210, was electroporated into FX514, and selection was done on GCB-I hemoglobin plates containing kanamycin (20 μg/ml). pLS88 was used as the negative control vector for these experiments.
Nucleotide sequence accession number.
Relevant DNA sequence of the Ton system gene locus has been submitted to GenBank (accession no. AF001034).
RESULTS
Cloning of the H. ducreyi exbB, exbD, and tonB loci.
We found that cloned hgbA was able to confer upon E. coli EB53 (hemA aroB) (Table 1) the ability to bind human hemoglobin (23), but this strain was unable to utilize hemoglobin as a source of heme (data not shown). We reasoned that HgbA expressed in E. coli EB53 might not be energized by the E. coli Ton system, since E. coli and H. ducreyi are only distantly related (2). An experiment was designed to clone the Ton system of H. ducreyi which was similar to that used to clone the Ton system of meningococci (57). A partial Sau3AI library was transformed into E. coli IR754 (tonB hemA aroB) expressing hgbA from the compatible plasmid pUNCH 556 (Table 1) and plated on hemoglobin as the sole source of heme. If the hemoglobin receptor plus gene products from certain plasmids allowed the uptake of heme from hemoglobin, then the E. coli heme synthesis mutation would be bypassed and growth on hemoglobin plates would occur. Of approximately 5 × 106 total transformants, 30 complementing clones were obtained on LB hemoglobin plates. To confirm that these clones required HgbA for growth on hemoglobin, plasmid DNA isolated from each was transformed into E. coli IR754 lacking pUNCH 556 (hgbA) and again plated onto hemoglobin agar plates. Thirteen of the 30 clones failed to grow without pUNCH 556, suggesting that the gene products expressed from those clones were insufficient to allow IR754 to utilize the heme from hemoglobin. This screen eliminated clones which allowed the heme from hemoglobin to leak in nonspecifically or directly complemented the hemA defect in IR754. Eleven of the 13 plasmids hybridized to the insert from pGJ300 containing the H. influenzae tonB operon (33, 34) (data not shown). The two plasmids which did not hybridize apparently suffered deletions during propagation since they were reduced to the size of the vector. Nine of 11 hybridizing plasmids contained a 6.5-kb hybridizing band; each conferred upon IR754 carrying pUNCH 556 (hgbA) the ability to grow on LB hemoglobin agar (data not shown). One plasmid, pUNCH 562, was chosen for further study (Fig. 1).
DNA and deduced amino acid sequence of the H. ducreyi Ton system gene cluster.
The location of the TonB gene cluster was revealed by subcloning and deletion analysis as shown in Fig. 1. The relevant regions were sequenced, and the order of the genes was exbB exbD tonB. Sequences similar to −35 and −10 consensus sequences were found 114 and 82 nucleotides (nt) upstream of the exbB start codon and were separated by 16 nt. Putative ribosome-binding sites were found at 9, 11, and 11 nt upstream of the respective exbB, exbD, and tonB ATG start codons. Just downstream of the tonB open reading frame there was an AT-rich region with dyad symmetry consistent with a transcription terminator. Notably absent were sequences corresponding to promoters upstream of the exbD and tonB structural genes.
Comparisons of the H. ducreyi deduced amino acid sequences with the ExbB, ExbD, and TonB protein sequences are shown in Fig. 2. Since the enterobacterial sequences were closely related, E. coli was chosen as the type strain to represent them for the sake of brevity.
The percent amino acid identities and similarities are shown in Table 2. The amino acid sequences for ExbB and ExbD were most similar to the proteins from other members of the pasteurellae, and this family comprised a group separate from the others. The ExbB and ExbD proteins were the most similar among all the genera, whereas the sequences of the TonB proteins were more divergent. The TonB protein of H. ducreyi was most closely related to the TonB protein from Pasteurella haemolytica (26).
Phenotypic characterization of the H. ducreyi Ton system.
Subcloning and deletion analyses were used to define regions of the original plasmid pUNCH 562 required to express a hemoglobin-utilizing phenotype. E. coli IR754(pUNCH 556) containing subclone pUNCH 563 also demonstrated a hemoglobin-utilizing phenotype (Fig. 1 and 3A), but not in the absence of pUNCH 556 (hgbA) (Fig. 1). pUNCH 563 was mutagenized by deleting the DNA between the unique BbsI and MluI sites and replacing it with a Cmr (23) cassette to form pUNCH 568 (Fig. 1). This deletion included all of exbD and portions of exbB and tonB. IR754(pUNCH 556) containing the mutagenized Ton system plasmid pUNCH 568 was unable to grow on hemoglobin (Fig. 1 and 3B).
The reconstructed H. ducreyi hemoglobin-utilizing system in E. coli IR754 was tested for its ability to utilize other heme sources. E. coli IR754(pUNCH 556) grew on hemoglobin with pUNCH 563 but not with pUNCH 568; however, both strains grew on heme but only at relatively high concentrations (50 μg/ml). Since the presence of an intact Ton system in IR754(pUNCH 556) was not required for growth on heme, its growth was TonB independent (Fig. 1). Previously, we reported that E. coli clones expressing hgbA (pUNCH 556) are somewhat leaky in that the outer membrane is perturbed (the periplasmic enzyme RNase leaks to the extracellular environment) (23, 69). We assume that this leakiness accounts for the strain’s ability to grow on high levels of hemin. Neither IR754(pUNCH 556) containing pUNCH 563 nor IR754(pUNCH 556) containing pUNCH 568 grew on 50 μM HSA (3.3 mg/ml) saturated at 50% hemin (Fig. 1). Fifty micromolar HSA is about 10% of the normal serum concentration. All strains grew on medium containing δALA, confirming viability. Thus, these data indicated that under these conditions, energized HgbA functioned as a receptor for hemoglobin but not for free heme or for heme complexed to albumin.
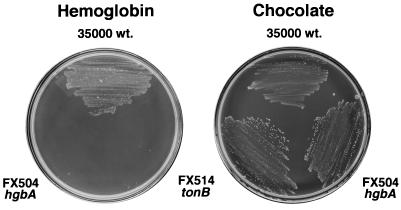
An H. ducreyi Ton system mutant was constructed to examine which sources of heme are utilized by TonB-dependent receptors. Parent H. ducreyi strain 35000 was electroporated with deletion/mutant plasmid pUNCH 568 and plated on chloramphenicol-containing chocolate agar. To screen for putative mutants, 10 Cmr transformants were tested for the ability to grow on hemoglobin agar, and none was able to. Shown in Fig. 4 is the phenotype of one transformant, FX514 (ΔexbB exbD tonB); it was used for further experiments. Positive control parent strain 35000 grew on hemoglobin, and negative control strain FX504 (hgbA) did not.
FIG. 4.
Growth of H. ducreyi Ton system isogenic mutant on hemoglobin agar. H. ducreyi parent strain 35000 (wild type [wt]), FX504 (hgbA), and FX514 (ΔexbB exbD tonB) were streaked onto GCB-I medium containing human hemoglobin (200 μg/ml [12 μM hemin]) or chocolate agar and incubated at 35°C for 48 h with 5% CO2.
Evidence that the inability of FX514 to utilize hemoglobin was due to a specific mutation in the Ton system was obtained as follows. The mutation in the FX514 chromosome was repaired by electroporating Ton system plasmid pUNCH 563 followed by selection on hemoglobin agar (Tables 1 and 3). Since plasmid pUNCH 563 cannot replicate in H. ducreyi, growth on hemoglobin selected for repair of the mutant Ton system chromosomal locus. In addition, the defect in FX514 was complemented in trans by using pUNCH 1210 (Table 3). pUNCH 1210 contains the entire Ton system insert from pUNCH 563 in shuttle vector pLS88, which is able to replicate in H. ducreyi (Table 1). Since both chromosomal repair and complementation in trans restored the ability of the H. ducreyi FX514 to grow on hemoglobin, but vector control plasmid pLS88 did not, we concluded that the inability of FX514 to grow on hemoglobin was due to a specific mutation in the Ton locus (data not shown). Furthermore, the phenotype of FX514 was not due to a polar effect on downstream genes since plasmid pUNCH 1210 fully restored hemoglobin utilization.
TABLE 3.
Growth of H. ducreyi on agar plates containing various heme sources
| Strain | Growtha with indicated hemin source
|
|||||
|---|---|---|---|---|---|---|
| Hemoglobin, 100b | Hemin
|
Hemealbumin, 2,000 | Catalase, 250 | Chocolate | ||
| 10 | 50 | |||||
| 35000 (wild type) | + | − | + | − | + | + |
| FX504 (hgbA) | − | − | + | − | + | + |
| FX514 (exbB exbD tonB) | − | − | + | − | + | + |
| FX514(pUNCH 1210) | + | − | + | − | + | + |
| FX514 repairc | + | − | + | − | + | + |
+, macroscopic growth present by 48 h; −, no macroscopic growth at 48 h.
Concentration (micrograms per milliliter).
The Ton system mutation in FX514 was repaired to wild type as described in text.
Southern blotting of chromosomal DNA from FX514 confirmed that FX514 contained the appropriately larger HincII-to-KpnI fragment (3.25 kb) compared to parent 35000 (3.0 kb) (Fig. 1 and Southern blot data not shown). The HincII-to-KpnI fragment in FX514, but not 35000, was recognized by the CAT probe. The appropriately sized HincII-to-KpnI bands were also observed in pUNCH 563 and pUNCH 568, respectively, using these probes (data not shown). These results indicated FX514 contained an allelic replacement of the Ton system. Chromosomal repair of FX514 resulted in a Southern blot pattern identical to that of 35000.
Phenotype of the H. ducreyi Ton mutant on sources of heme other than hemoglobin.
In other bacteria, Ton system mutations are impaired in the ability to use a variety of heme/iron compounds transported by TonB-dependent receptors; therefore, the growth phenotype of FX514 was examined on other heme-containing media that support the growth of H. ducreyi. H. ducreyi was first heme starved (56) and then serially diluted for use as the inoculum. There was no difference for either mutant on hemin or catalase relative to the parent (Table 3). All three strains grew with hemin at 50 μg/ml or catalase at 250 μg/ml but not lower amounts. None of the three strains grew on GCB-I agar containing 33 μM HSA (2 mg/ml) saturated with 50% hemin. Taken together, these data indicated that utilization of hemoglobin was TonB dependent whereas utilization of free hemin and catalase appeared to be TonB independent.
Novel proteins expressed by H. ducreyi Ton system mutant FX514.
Previous work in E. coli demonstrated that Ton system mutants are relatively iron starved based on the observation that genes normally regulated by Fur are derepressed (48). SDS-PAGE analysis of outer membrane proteins from H. ducreyi Ton mutant FX514 demonstrated several heme-regulated gene products present in increased amounts (Fig. 5A). The major protein present in increased amounts in FX514 was HgbA, confirming that the inability to utilize hemoglobin in this mutant was not due to the lack of synthesis of HgbA. Three other outer membrane proteins, designated 1, 2 (protein 2 refers to the lower band of the doublet), and 3 in Fig. 5A, were also present in increased amounts in certain of the mutants under heme-regulated conditions. Protein 1 was present only in FX514 grown under heme-limiting conditions. Proteins 2 and 3, although present in strain 35000, were more highly expressed in the two mutants than in strain 35000 under heme limitation. In experiments not presented here, Western blotting of H. ducreyi using specific antibodies to proteins 2 and 3 demonstrate they were clearly heme regulated, albeit at lower amounts than HgbA (data not shown). Outer membranes prepared from FX514 which had been repaired by chromosomal transformation regained the protein expression pattern of parent strain 35000 (data not shown).
In some gels, including Fig. 5A, two minor proteins of approximately 60 and 65 kDa were present in slightly increased amounts in 35000 and FX514 grown under heme limitation. It was possible that these lower-molecular-weight protein bands present only in 35000 and FX514 were breakdown products of HgbA, since they were absent in FX514. Immunologically reactive material in Western blots of H. ducreyi in this size range have been occasionally observed in assays using antisera to HgbA (data not shown). To address this issue, a Western blot of these preparations was probed with polyclonal antiserum to purified HgbA (Fig. 5B). The gel used for this blot was intentionally overloaded to visualize minor breakdown products of HgbA. Besides the expected reactivity to HgbA, reactivity to protein 1, but not protein 2, protein 3, or smaller bands present in strain 35000 or FX514, by this antiserum was observed. These data are consistent with derepression of the several genes encoding heme/iron-regulated outer membrane proteins.
DISCUSSION
Utilization of hemoglobin is TonB dependent.
Several TonB-dependent receptors for heme or hemoglobin have been cloned from gram-negative pathogens, utilizing a strategy based on the complementation of a heme synthesis defect (hemA) or iron acquisition (aroB) defect in E. coli (20, 29–31, 42, 57–59, 62). This method relies on the principle that E. coli K-12 strains are unable to transport hemin or hemoglobin across the outer membrane. However, once hemin traverses the outer membrane, E. coli K-12 strains are able transport it across the cytoplasmic membrane, where it can be used to fulfill its heme or iron requirements. Using this method, we have also attempted to clone a heme receptor from H. ducreyi in E. coli EB53 (hemA aroB), but initial experiments have been unsuccessful. This failure, coupled with our inability to demonstrate a TonB-dependent hemoglobin utilizing phenotype in E. coli EB53 with cloned hgbA, prompted us to search for an explanation. It was found that the cloned H. ducreyi hgbA receptor required its homologous Ton system in E. coli for growth on hemoglobin. Furthermore, the Ton system mutant FX514 was unable to utilize hemoglobin at wild-type levels, unambiguously demonstrating that HgbA was TonB dependent.
Since the deletion plasmid used to construct the H. ducreyi Ton system mutant contained deletions in exbB, exbD, and tonB genes, it is not possible to infer if the Ton system could function in the absence of only a single protein. However, it should be noted that in E. coli, most null mutations in any three of these genes result in the total or partial loss of function for the Ton system.
Role of HgbA in heme utilization.
In an experiment to examine the role of HgbA in heme acquisition, we tested whether cloned HgbA with its homologous Ton system could confer upon E. coli hemA tonB aroB strains the ability to grow on free hemin or hemin complexed to HSA. No growth was observed on HSA-heme agar. Growth on hemin agar was observed only at high concentrations (50 μg/ml) and growth was TonB independent, suggesting that HgbA alone does not function as a typical TonB-dependent receptor for free hemin or hemin complexed to HSA. This finding does not rule out the possibility that HgbA is involved in internalization of free hemin in H. ducreyi but may indicate that additional components are required. Alternatively, HgbA and other (heme/iron) receptors may form a complex, and the absence of HgbA may disturb the function of this complex. Similar nonspecific defects in the uptake of iron compounds have been found for TonB-dependent receptor mutants in other bacteria (1, 6).
Utilization of heme or catalase in H. ducreyi is TonB independent.
Although neither H. ducreyi FX504 nor FX514 grew on hemoglobin as a sole source of heme, both mutants exhibited no such growth defect on hemin relative to parent 35000 when prestarved for heme by growth anaerobically in the absence of heme. Further, all three strains required relatively high concentrations of free hemin (50 μg/ml) for growth on plates. To date, bacteria containing documented heme receptors require much less heme (less than 10 μg/ml) than H. ducreyi to fulfill their iron requirement. The requirement of the wild-type H. ducreyi strain for high concentrations of heme is reminiscent of certain mutants of H. influenzae. H. influenzae tonB (34) or hxuC (17) mutants require high levels of heme (50 μg/ml), whereas wild-type strains require only 10 μg/ml or less (0.1 μg/ml) (60). Most previous studies have either shown the TonB dependence or implied the TonB dependence of heme uptake (20, 29–31, 42, 57–59, 62). However, in contrast to these previous studies, in Neisseria spp., heme utilization has very recently been shown to be TonB independent (7, 57). No heme receptor has been found in Neisseria spp.
The growth of H. ducreyi on catalase was also TonB independent. We confirmed the ability of H. ducreyi to grow on catalase by using highly purified preparations (Sigma C-100), which demonstrated a single band by SDS-PAGE. It remains unclear whether a specific receptor for catalase exists or whether catalase releases hemin upon prolonged incubation; however, a comparison of the appearance (color) of catalase plates with that of hemin plates indicates that there is insufficient free heme present in the former to support the growth of H. ducreyi.
Previous studies have reported that the addition of FCS improves the growth of H. ducreyi (3). The addition of FCS to plates containing dilutions of heme or catalase reduced the requirements of strain 35000 to 1 μg/ml for both heme and catalase (data not shown). However, FCS did not lower the requirements for heme or catalase for mutants FX504 and FX514. These results are difficult to interpret and suggest several possibilities. The simplest explanation is that FCS contains contaminating hemoglobin. Another possibility is that FCS contains a heme/iron source whose utilization requires the expression of both HgbA and the Ton system. It is also possible that FCS enhances the uptake of heme or catalase or expression of heme or catalase receptors. Experiments are currently under way to address these issues.
Our inability to obtain growth of H. ducreyi on heme-albumin is in contrast to previous studies (3, 39). These differences could at least be partially explained by differences in materials and methods (percent heme saturation and inoculation differences) used in the various studies.
The H. ducreyi Ton system mutant expresses novel heme-regulated outer membrane proteins.
Regulation of most iron-repressed genes in E. coli is under the control of the global negative repressor Fur (4). In E. coli, derepression of the Fur regulon in tonB mutants is believed to be due the inability to transport iron by means of the various TonB-dependent receptors, resulting in low intracellular iron levels (47). H. ducreyi contains a functional Fur protein (14); however, the H. ducreyi genes regulated by Fur have not yet been identified. In H. ducreyi, expression of hgbA and perhaps hemolytic activity (9, 23) are regulated by the levels of heme in the medium. Since heme and hemoglobin contain iron, it is possible that these heme sources also serve as a source of intracellular iron, thereby indirectly affecting expression of the H. ducreyi Fur regulon. H. ducreyi Ton system mutant FX514 demonstrated increased expression of HgbA as well as several previously undescribed outer membrane proteins, which appeared to be regulated by the levels of heme in the medium. The possibility that these novel proteins are involved in heme/iron acquisition is consistent with observations made in other pathogens. Further work is needed in this area to understand the repertoire of receptors for heme/iron compounds in H. ducreyi and their regulation.
Structure and arrangement of the H. ducreyi Ton system.
The arrangement of genes in the H. ducreyi tonB cluster was similar to that in certain other gram-negative bacteria described elsewhere (8, 33, 34), with the order exbB exbD tonB. The arrangement, the DNA sequence, and the proximity of the structural genes suggested that only one promoter was responsible for expression of all three genes. We speculate that these genes may be transcribed as a multicistronic message; however, this remains to be proven experimentally.
Significant diversity exists between the H. ducreyi and E. coli TonB proteins. These differences could account for the inability of the H. ducreyi HgbA receptor to function together with the E. coli EB53 Ton system. However, three of four general domains previously described for E. coli TonB were present in H. ducreyi TonB (38, 65). The sequence of the N-terminal hydrophobic region (amino acids [aa] 12 to 32) (36) spans the cytoplasmic membrane in E. coli and may interact with ExbB. H. ducreyi TonB contains three of four invariant residues in this N-terminal region compared to E. coli. The second domain traverses the periplasmic space in E. coli and contains the characteristic X-Pro repeat region (aa 63 to 102) and is present in the H. ducreyi sequence. A third short sequence has been implicated in the interaction of TonB with outer membrane receptors and consists of YPARA (aa 160 to 167) in E. coli and YPARE in H. ducreyi. The fourth domain of TonB (aa 199 to 216) in E. coli, predicted to form an alpha helix, is present in all enteric species yet is so diverged in both Haemophilus species that no primary sequence homology exists. Four monoclonal antibodies against E. coli TonB (38) failed to react with H. ducreyi TonB (data not shown), providing further evidence of the dissimilarity between the TonB proteins of E. coli and H. ducreyi. Despite these dissimilarities, the E. coli proton motive force can be transduced through the H. ducreyi TonB to HgbA, suggesting that certain critical interacting domains are present. The reconstituted system in E. coli containing HgbA and the H. ducreyi Ton system also contains ExbB and ExbD proteins from E. coli. It is not clear whether it is the E. coli or H. ducreyi versions of ExbB and ExbD proteins that interact with H. ducreyi TonB.
It has recently been shown that an hgbA (hupA) mutant of strain 35000 (56) is less virulent in a temperature-dependent rabbit model of infection. In this model, lesion scores were smaller and isolation of the mutant from lesions was unsuccessful. We have confirmed this finding by using FX504 (23a) and have similar data from a swine ear model of infection (23b, 32). These results suggest that the acquisition of hemoglobin is vital for this pathogen to survive and produce disease in vivo. We predict that a Ton mutant would have similar or possibly even more profound virulence defects based on its inability to utilize hemoglobin and possibly other heme compounds transported by TonB-dependent receptors.
Our results implicate several novel outer membrane proteins which were more highly expressed in the Ton system mutant and might be additional TonB-dependent receptors to study. Mutagenesis of the genes encoding these putative novel receptors might give insights into their function for the acquisition of heme- or iron-containing compounds utilized by H. ducreyi. Lastly, assuming that additional H. ducreyi TonB-dependent receptors exist for sources of heme other than hemoglobin, we hope to clone appropriate homologous receptors by functional complementation of E. coli IR754(pUNCH 563) and selecting on appropriate heme sources.
ACKNOWLEDGMENTS
We thank P. Frederick Sparling and Cynthia Cornelissen for continued helpful comments, members of the Sparling laboratory for critiquing the manuscript, and Annice Roundtree for expert technical assistance. We thank Igor Stojiljkovic for helpful discussions, including unpublished results, and for strains used in these experiments. We thank Kathleen Postle for the generous gift of monoclonal antibodies and helpful comments. We thank Marcia Hobbs and Tom Kawula for critiquing the manuscript and their work with the swine model of infection.
This work was supported by developmental grant UO1-AI31496 from the North Carolina Sexually Transmitted Disease Infections Research Center, University of North Carolina at Chapel Hill, WHO grant SDI/94/006, and grant R29-AI40263 to C.E. and Public Health Service grant A126837 to P. Frederick Sparling.
REFERENCES
- 1.Aebi C, Stone B, Beucher M, Cope L, Maciver I, Thomas S, McCracken G J, Sparling P, Hansen E. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect Immun. 1996;64:2024–2030. doi: 10.1128/iai.64.6.2024-2030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton W L. Biology of Haemophilus ducreyi. Microbiol Rev. 1989;53:377–389. doi: 10.1128/mr.53.4.377-389.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albritton W L, Macklean I W, Bertram P D, Ronald A R. Haemophilus, Pasteurella, and Actinobacillus. New York, N.Y: Academic Press, Inc.; 1981. [Google Scholar]
- 4.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell P E, Nau C D, Brown J T, Konisky J, Kadner R J. Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J Bacteriol. 1990;172:3826–3829. doi: 10.1128/jb.172.7.3826-3829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beucher M, Sparling P F. Cloning, sequencing and characterization of the gene encoding FrpB, a major iron-regulated, outer membrane protein of Neisseria gonorrhoeae. J Bacteriol. 1995;177:2041–2049. doi: 10.1128/jb.177.8.2041-2049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas G D, Anderson J E, Sparling P F. Cloning, sequencing and genetic characterization of tonB-exbB-exbD genes of Neisseria gonorrhoeae. Mol Microbiol. 1997;24:169–179. doi: 10.1046/j.1365-2958.1997.3421692.x. [DOI] [PubMed] [Google Scholar]
- 8.Bitter W, Tommassen J, Weisbeck P J. Identification and characterization of the exbB, exbD, and tonB genes of Pseudomonas putida WCS358: their involvement in ferric-pseudobactin transport. Mol Microbiol. 1993;7:117–130. doi: 10.1111/j.1365-2958.1993.tb01103.x. [DOI] [PubMed] [Google Scholar]
- 9.Bozue J A, Hansen E J, Munson R S. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Regulation of hemolysin expression in Haemophilus ducreyi by heme and hemoglobin, B-315; p. 209. [Google Scholar]
- 10.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 12.Braun V, Killmann H, Benz R. Energy-coupled transport through the outer membrane of Escherichia coli small deletions in the gating loop convert the FhuA transport protein into a diffusion channel. FEBS Lett. 1994;346:59–64. doi: 10.1016/0014-5793(94)00431-5. [DOI] [PubMed] [Google Scholar]
- 13.Caldwell J C, Caldwell P. The African AIDS epidemic. Sci Am. 1996;3:64–68. doi: 10.1038/scientificamerican0396-62. [DOI] [PubMed] [Google Scholar]
- 14.Carson S D B, Thomas C E, Elkins C. Cloning and sequencing of a Haemophilus ducreyi fur homolog. Gene. 1996;176:125–129. doi: 10.1016/0378-1119(96)00236-3. [DOI] [PubMed] [Google Scholar]
- 15.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope L, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelissen C N, Biswas G D, Tsai J, Paruchuri D K, Thompson S A, Sparling P F. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J Bacteriol. 1992;174:5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornelissen C N, Sparling P F. iron piracy: iron acquisition from transferrin by bacterial pathogens. Mol Microbiol. 1994;14:843–850. doi: 10.1111/j.1365-2958.1994.tb01320.x. [DOI] [PubMed] [Google Scholar]
- 20.Daskaleros P A, Stoebner J A, Payne S M. Iron uptake in Plesiomonas shigelloides: cloning of the genes for heme-iron uptake system. Infect Immun. 1991;59:2706–2711. doi: 10.1128/iai.59.8.2706-2711.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon L G, Albritton W L, Willson P J. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid. 1994;32:228–232. doi: 10.1006/plas.1994.1060. [DOI] [PubMed] [Google Scholar]
- 22.Elkins C. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect Immun. 1995;63:1241–1245. doi: 10.1128/iai.63.4.1241-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkins C, Chen C J, Thomas C E. Characterization of the hgbA locus of Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Elkins, C., and P. Totten. Unpublished data.
- 23b.Elkins, C., M. Hobbs, and T. Kawula. Unpublished data.
- 24.Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J-F, Dougherty B, Merrick J, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J, Weidman J, Phillips C, Spriggs T, Hedblom E, Cotton M, Utterback T, Hanna M, Nguyen D, Saudek D, Brandon R, Fine L, Fritchman J, Fuhrmann J, Geoghagen N, Gnehm C, McDonald L, Small K, Fraser C, Smith H, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 25.Genetics Computer Group. Program manual for the Wisconsin package. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 26.Graham M, Lo R Y C. Cloning and characterization of the exbB-exbD-tonB locus of Pasteurella haemolytica A1. Gene. 1997;186:201–205. doi: 10.1016/s0378-1119(96)00703-2. [DOI] [PubMed] [Google Scholar]
- 27.Hansen E J, Latimer J L, Thomas S E, Helminen M, Albritton W L, Radolf J D. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J Bacteriol. 1992;174:5442–5449. doi: 10.1128/jb.174.16.5442-5449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heller J K, Kadner R J, Gunter K. Suppression of btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene. 1988;64:147–153. doi: 10.1016/0378-1119(88)90488-x. [DOI] [PubMed] [Google Scholar]
- 29.Henderson D P, Payne S M. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol Microbiol. 1993;7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 30.Henderson D P, Payne S M. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson D P, Payne S M. Vibrio cholerae iron transport systems: roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect Immun. 1994;62:5120–5125. doi: 10.1128/iai.62.11.5120-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobbs M M, San Mateo L, Orndorff P, Almond G, Kawula T. Swine model of Haemophilus ducreyi infection. Infect Immun. 1995;63:3094–3100. doi: 10.1128/iai.63.8.3094-3100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarosik G P, Hansen E J. Cloning and sequencing of the Haemophilus influenzae exbB and exbD genes. Gene. 1995;152:89–92. doi: 10.1016/0378-1119(94)00675-i. [DOI] [PubMed] [Google Scholar]
- 34.Jarosik G P, Sanders J D, Cope L D, Muller-Eberhard U, Hansen E J. A functional tonB gene is required for both utilization of heme and virulence expression in Haemophilus influenzae type b. Infect Immun. 1994;62:2470–2477. doi: 10.1128/iai.62.6.2470-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadner R J. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol Microbiol. 1990;4:2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 36.Koebnik R. The molecular interaction between components of the TonB-ExbBD-dependent and of the TolQRA-dependent bacterial uptake systems. Mol Microbiol. 1993;9:219. doi: 10.1111/j.1365-2958.1993.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 37.Lagergard T, Purven M. Neutralizing antibodies to Haemophilus ducreyi cytotoxin. Infect Immun. 1993;61:1589–1592. doi: 10.1128/iai.61.4.1589-1592.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen R A, Myers P S, Skare J T, Seachord C L, Darveau R P, Postle K. Identification of TonB homologs in the family Enterobacteriaceae and evidence of conservation of TonB-dependent energy transduction complexes. J Bacteriol. 1996;178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee B C. Iron sources for Haemophilus ducreyi. J Med Microbiol. 1991;34:317–322. doi: 10.1099/00222615-34-6-317. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Rutz J M, Feix J B, Klebba P E. Permeability properties of a large gated channel within the ferric enterobactin receptor, FepA. Proc Natl Acad Sci USA. 1993;90:10653–10657. doi: 10.1073/pnas.90.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna W R, Mickelsen P A, Sparling P F, Dyer D W. Iron uptake from lactoferrin and transferrin by Neisseria gonorrhoeae. Infect Immun. 1988;56:785–791. doi: 10.1128/iai.56.4.785-791.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills M, Payne S M. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J Bacteriol. 1995;177:3004–3009. doi: 10.1128/jb.177.11.3004-3009.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 44.Otto B R, Vught A M J J V-V, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 45.Palmer K L, Grass S, Munson R S. Identification of a hemolytic activity elaborated by Haemophilus ducreyi. Infect Immun. 1994;62:3041–3043. doi: 10.1128/iai.62.7.3041-3043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer K L, Munson R S. Cloning and characterization of the genes encoding haemolysin of Haemophilus ducreyi. Mol Microbiol. 1995;18:821–830. doi: 10.1111/j.1365-2958.1995.18050821.x. [DOI] [PubMed] [Google Scholar]
- 47.Postle K. TonB and the gram-negative dilemma. Mol Microbiol. 1990;4:2019–2025. doi: 10.1111/j.1365-2958.1990.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 48.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 49.Purven M, Lagergard T. Haemophilus ducreyi, a cytotoxin-producing bacterium. Infect Immun. 1992;60:1156–1162. doi: 10.1128/iai.60.3.1156-1162.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice P. Program manual for the EBCG package. 1995. Cambridge, England. [Google Scholar]
- 51.Ronald A R, Albritton W. Chancroid and Haemophilus ducreyi. In: Holmes K K, Mardh P-A, Sparling P F, Wiesner P J, editors. Sexually transmitted diseases. 2nd ed. New York, N.Y: McGraw-Hill; 1990. pp. 263–272. [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Sanders J, Cope L, Hansen E. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schramm E, Mende J, Braun V, Kamp R M. Nucleotide sequence of the colicin B activity gene cba: consensus pentapeptide among TonB-dependent colicins and receptors. J Bacteriol. 1987;169:3350–3357. doi: 10.1128/jb.169.7.3350-3357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skare J T, Ahmer B M M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 56.Stevens M K, Porcella S, Klesney-Tait J, Lumbley S, Thomas S E, Norgard M V, Radolf J D, Hansen E J. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect Immun. 1996;64:1724–1735. doi: 10.1128/iai.64.5.1724-1735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stojiljkovic I, Srinvisan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stojiljkovic I, Hantke K. Haemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in Gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stojiljkovic I, Hwa V, Martin L D S, O’Gaora P, Nassif X, Heffron F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 60.Stull T L. Protein sources of heme for Haemophilus influenzae. Infect Immun. 1987;55:148–153. doi: 10.1128/iai.55.1.148-153.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas C E, Carbonetti N H, Sparling P F. Pseudo-transposition of a Tn5 derivative in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145:371–376. doi: 10.1111/j.1574-6968.1996.tb08603.x. [DOI] [PubMed] [Google Scholar]
- 62.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1996;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 63.Totten P A, Norn D V, Stamm W E. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect Immun. 1995;63:4409–4416. doi: 10.1128/iai.63.11.4409-4416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Totten P A, Stamm W E. Clear broth and plate media for the culture of Haemophilus ducreyi. J Clin Microbiol. 1994;32:2019–2023. doi: 10.1128/jcm.32.8.2019-2023.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Traub I, Gaisser S, Braun V. Activity domains of the TonB protein. Mol Microbiol. 1993;8:409–423. doi: 10.1111/j.1365-2958.1993.tb01584.x. [DOI] [PubMed] [Google Scholar]
- 66.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wasserheit J N. Interrelationships between human immunodeficiency virus infection and other sexually transmitted diseases. Sex Transm Dis. 1991;19:61–77. [PubMed] [Google Scholar]
- 68.Willson P J, Albritton W L, Slaney L, Setlow J K. Characterization of a multiple antibiotic resistance plasmid from Haemophilus ducreyi. Antimicrob Agents Chemother. 1989;33:1627–1630. doi: 10.1128/aac.33.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young K, Silver L L. Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J Bacteriol. 1991;173:3609–3614. doi: 10.1128/jb.173.12.3609-3614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]