Abstract
The epitranscriptomic mark N6-methyladenosine (m6A), which is the predominant internal modification in RNA, is important for plant responses to diverse stresses. Multiple environmental stresses caused by the tea-withering process can greatly influence the accumulation of specialized metabolites and the formation of tea flavor. However, the effects of the m6A-mediated regulatory mechanism on flavor-related metabolic pathways in tea leaves remain relatively uncharacterized. We performed an integrated RNA methylome and transcriptome analysis to explore the m6A-mediated regulatory mechanism and its effects on flavonoid and terpenoid metabolism in tea (Camellia sinensis) leaves under solar-withering conditions. Dynamic changes in global m6A level in tea leaves were mainly controlled by two m6A erasers (CsALKBH4A and CsALKBH4B) during solar-withering treatments. Differentially methylated peak-associated genes following solar-withering treatments with different shading rates were assigned to terpenoid biosynthesis and spliceosome pathways. Further analyses indicated that CsALKBH4-driven RNA demethylation can directly affect the accumulation of volatile terpenoids by mediating the stability and abundance of terpenoid biosynthesis-related transcripts and also indirectly influence the flavonoid, catechin, and theaflavin contents by triggering alternative splicing-mediated regulation. Our findings revealed a novel layer of epitranscriptomic gene regulation in tea flavor-related metabolic pathways and established a link between the m6A-mediated regulatory mechanism and the formation of tea flavor under solar-withering conditions.
Keywords: Camellia sinensis, RNA methylation, Epitranscriptome, Secondary metabolite, Withering
Introduction
As the predominant internal modification in eukaryotic mRNAs, N6-methyladenosine (m6A) is a critical transcriptional regulator in a novel molecular mechanism that profoundly affects various biological processes by modulating multiple aspects of mRNA processing and metabolism [1], including mRNA abundance [2], [3], stabilization [4], and splicing [5]. Along with the well-studied epigenetic regulation via modifications to DNA and histones, the dynamic and reversible m6A modification is mediated by m6A methyltransferases (writers) and demethylases (erasers). In addition to these two crucial components, m6A readers are responsible for the localization and recognition of m6A-containing RNA sequences, which are necessary for the implementation of the biological effects of m6A modifications. Consequently, m6A writers, erasers, and readers collaboratively orchestrate a complex regulatory network that governs m6A modifications. Although there has been considerable progress in the research conducted to clarify the potential functions of m6A modifications in animals, the effects of the m6A regulatory mechanism in the plant kingdom are just beginning to be determined. The m6A-mediated regulatory mechanism reportedly influences the normal growth of Arabidopsis thaliana roots and shoots [6], [7]. Furthermore, there is growing evidence that m6A modifications are also involved in regulating responses to diverse environmental stresses, including drought [8], cold [9], and ultraviolet (UV) radiation [10]. More recently, a few studies have started to precisely functionally characterize the m6A regulatory mechanism or m6A regulatory genes in the plant kingdom [11], [12], [13], [14]. Compared with model plants, there has been relatively little research on the regulatory mechanisms as well as the functions of m6A modifications in horticultural crops.
Tea (Camellia sinensis) is an important and traditional economically valuable crop cultivated on a large scale in many developing and developed countries. Its tender buds and leaves are mostly used to produce highly consumed and popular beverages. Oolong tea, which is one of the six tea types in China, is famous for its elegant floral and fruity aroma, as well as its unique brisk-smooth and mellow taste. The development of the flavor and sensory characteristics of oolong tea largely depends on the postharvest manufacturing process [15]. During the manufacturing of oolong tea, withering is the first essential stage affecting the palatability and commercial value of the final product. The harvested leaves are still in a live state and continue to be exposed to various environmental stresses during the withering stage, in which there are obvious changes to multiple tastes and aroma compounds, as well as endogenous phytohormones, which directly or indirectly endow oolong tea with its characteristic flavor and health benefits [16], [17], [18]. Among these taste compounds in tea plants, flavonoids are a large group of secondary metabolites that are closely associated with tea palatability. More specifically, catechins are the dominant flavonoid components and comprise 12%–24% of the dry weight in tea leaves [19]. Flavonoids and catechins are the major contributors to the astringency and bitterness of tea [20]. Moreover, catechins can be further oxidized to high-molecular-weight polymeric compounds (i.e., theaflavins) that provide tea with beneficial health-promoting properties and a characteristic mellow taste [21]. As the most representative aroma compounds in oolong tea, volatile terpenoids are critical components of high-quality oolong tea because of their contributions to the pleasant floral and fruity fragrance. Decades of studies have demonstrated that multiple environmental stresses caused by withering treatments induce several multidimensional responses (e.g., at the genetic and metabolic levels) that further modulate the formation of the oolong tea flavor [22], [23]. However, information regarding the upstream mechanism regulating the flavor-related genes and relevant metabolites remains limited.
Previous research has indicated that solar-withering, which is a conventional method for producing tea, may be used to improve the flavor of high-quality oolong tea [24], [25]. Light serves as an energy source and signaling molecule required for the metabolic changes that occur during solar-withering. Typically, light quality and intensity are crucial parameters that determine the effects of solar-withering on tea quality. Earlier studies largely focused on the mechanism underlying the effects of light quality on the tea flavor [26], [27]. Few researchers have investigated the effects of different shading rates on the metabolism of tea flavor compounds and the formation of tea flavors during the solar-withering stage. The establishment of traditional solar-withering methods relies heavily on the subjective experiences of tea producers and random trials, leading to fluctuations in tea quality. To overcome this limitation, characterizing the metabolic pathways that affect flavor formation as well as the regulatory mechanisms under different shading rates during the solar-withering stage is vital for optimizing the shading rate to produce high-quality oolong tea efficiently. There has recently been accumulating evidence that epigenetic changes, including DNA methylation and histone modifications, are crucial for the biosynthesis of secondary metabolites and tea aroma formation [28], [29]. Unfortunately, the effects of RNA methylation, which is another epigenetic modification, and the precise regulatory mechanisms underlying the m6A-mediated flavor formation in the tea-withering stage remain largely unknown. The availability of a chromosome-level tea genome has laid a solid foundation for investigations on m6A modifications in tea plants [30].
In the present study, an integrated RNA methylome and transcriptome analysis was conducted to elucidate the effects of m6A modifications on the formation of flavor-related compounds in tea leaves in response to different shading rates during the solar-withering stage. We systematically identified the differentially methylated peak (DMP)-associated genes and revealed that the changes to m6A in many DMP-associated genes are inextricably associated with the solar-withering conditions, especially the shading rate. We further demonstrated that CsALKBH4-mediated RNA demethylation alters the extent of the m6A modifications within the 3′ untranslated region (3′ UTR) and near the stop codon and regulates the expression levels of m6A-modified RNAs, thereby affecting the accumulation of flavor metabolites and tea palatability. The effects of the m6A-mediated alternative splicing (AS) regulatory mechanism during the solar-withering stage were also explored. These findings described herein provide important insights into the regulatory effects of m6A modifications on tea plants and the contribution of the m6A-mediated regulatory mechanism to the development of high-quality oolong tea via a solar-withering method.
Results
Significant changes in global abundance and distribution of m6A modifications in tea leaves in response to solar-withering treatments
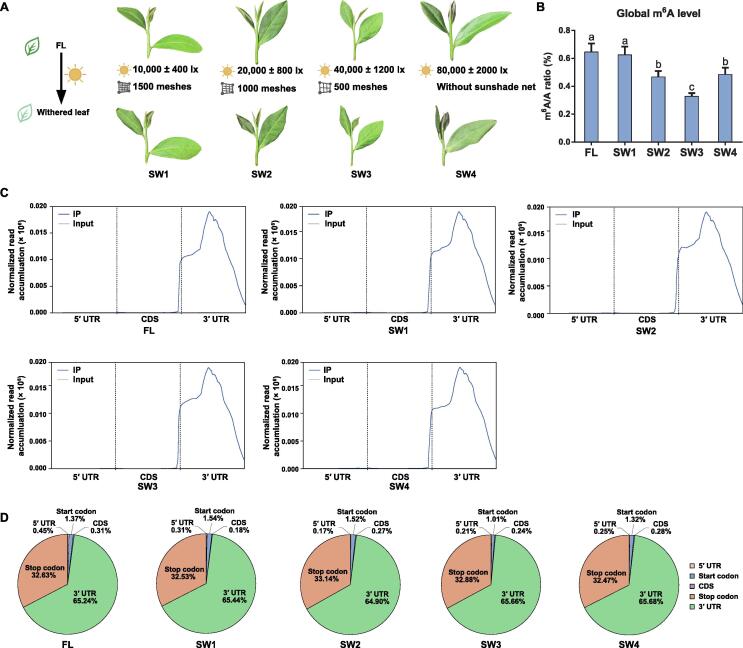
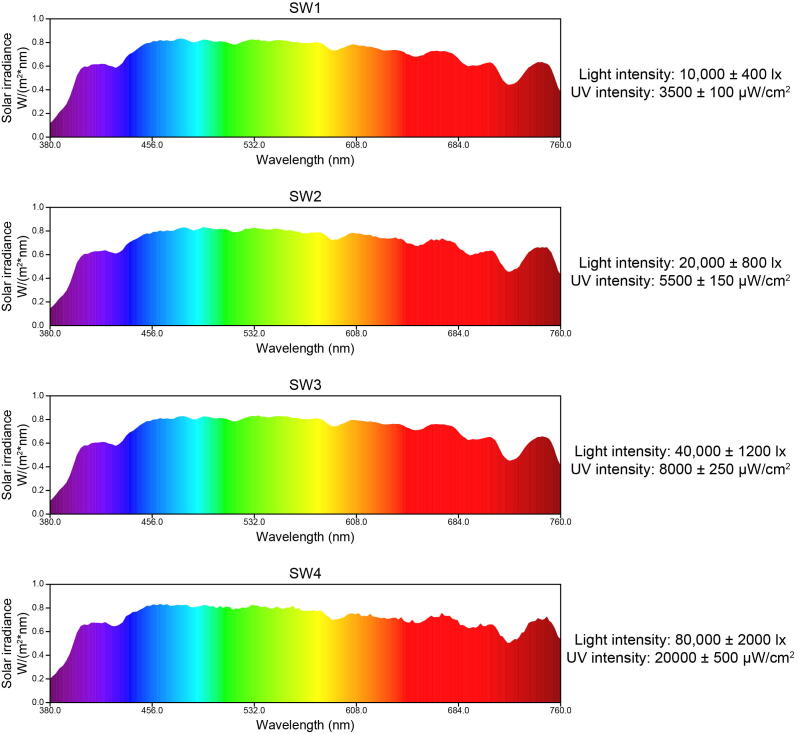
The light intensity, spectrum, and UV intensity of solar-withering treatments with different shading rates were monitored (Figure S1). The withering treatments were performed as follows: solar-withering with a high shading rate (SW1), solar-withering with a moderate shading rate (SW2), solar-withering with a low shading rate (SW3), and solar-withering with natural sunlight (SW4). Notably, the light intensity and UV intensity under the sunshade net decreased as the shading rate increased. However, the use of the sunshade net had little effect on the spectral composition, light quality, and wavelength. There were considerable differences in the UV intensity between the solar-withering with and without the shading net. The shading treatment substantially decreased the UV intensity. From SW1 to SW3, the different shading rates had relatively little effect on the UV intensity. The external characteristics of fresh leaves (FLs) and the solar-withered leaves produced using different shading rates were recorded (Figure 1A). FLs were straight, glossy, and jade green. Decreases in the shading rate during the solar-withering treatments resulted in increasingly shrunken, deformed, and darker-colored withered leaves. We then calculated the global m6A/A ratio of these tea leaves (Figure 1B). The solar-withering treatments with different shading rates decreased the global m6A level. The comparison of the various leaves detected a gradually decreasing trend in the overall m6A level from FLs to the SW3 leaves, but the m6A level rebounded following the SW4 treatment.
Figure 1.
Global abundance anddistributionof m6A modificationsin tea leaves under solar-withering with different shading rates
A. The external phenotypes of withered leaves before and after solar-withering with different shading rates. B. Dynamics of global m6A level in tea leaves under solar-withering with different shading rates. Data are presented as mean ± SD. Different lowercase letters over the error bars indicate significantly different groups via one-way ANOVA and Tukey’s post hoc test (P < 0.05). C. Metagenomic profiles of m6A distribution along transcripts. D. Pie charts showing the fractions of m6A peaks falling into five transcript segments (5′ UTR, start codon, CDS, stop codon, and 3′ UTR). m6A, N6-methyladenosine; IP, immunoprecipitation; FL, fresh leaf; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight; 5′ UTR, 5′ untranslated region; CDS, coding sequence; 3′ UTR, 3′ untranslated region; SD, standard deviation; ANOVA, analysis of variance.
Overview of the m6A MeRIP-seq
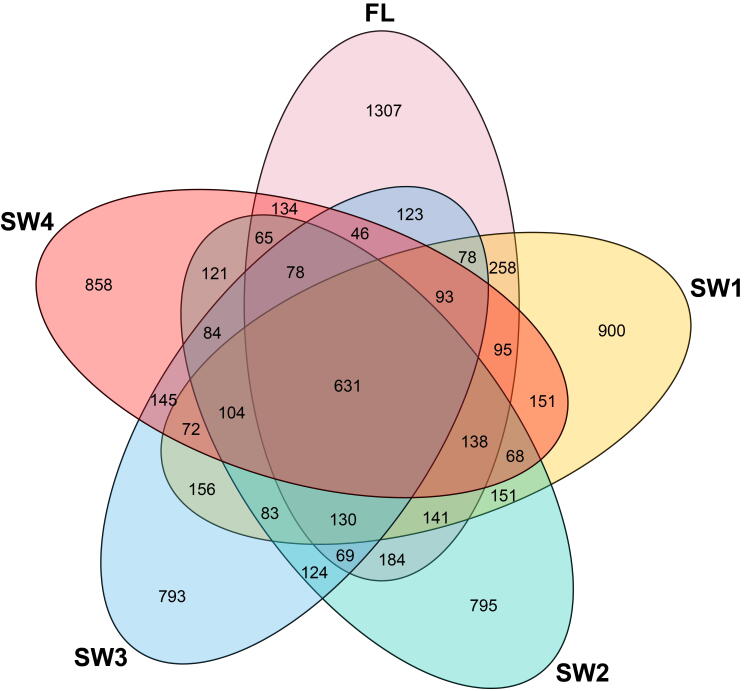
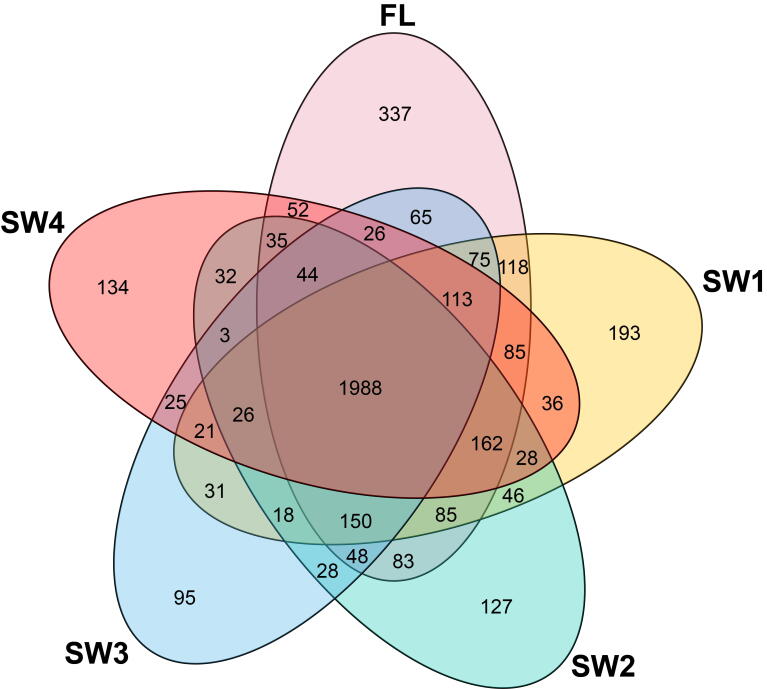
To ascertain the effects of m6A-mediated regulatory mechanisms under solar-withering conditions, we performed an m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq) experiment to analyze the m6A modifications at the mRNA level in tea leaves that underwent solar-withering with different shading rates (Figure S2). A total of 34–50 and 63–76 million high-quality clean reads were obtained for the immunoprecipitated (IP) libraries and input libraries, respectively, among which 28–41 and 55–67 million reads were mapped to the tea genome (Table S1). The base quality score of 20 (Q20) and base quality score of 30 (Q30) indices for each library exceeded 93% and 88%, respectively. Only the m6A peaks in all biological replicates with high-confidence coefficients were selected for further analyses. A total of 3570, 3249, 2966, 2809, and 2883 high-confidence m6A peaks were identified for the FL, SW1, SW2, SW3, and SW4 samples, respectively (Figure S3). Among these m6A peaks, 4605 were newly generated after the solar-withering treatment. Additionally, 31,720, 31,958, 32,146, 32,215, and 31,988 transcripts were obtained from the FL, SW1, SW2, SW3, and SW4 samples, respectively. Next, we identified 5277 high-confidence m6A peaks in 4289 transcripts. A total of 1988 m6A-marked genes were present in all samples (Figure S4). The solar-withering treatment induced the production of 843 new m6A-marked genes. The changes in the number of m6A-marked genes in response to solar-withering with different shading rates were consistent with the total m6A abundance dynamics. These observations suggest that the m6A abundance is closely related to these changes in the number of m6A-marked genes, which is in accordance with the findings of a previous study on tomato [11]. The number of m6A-marked genes gradually decreased from FLs to the SW3 leaves, whereas there were more m6A-marked genes in the SW4 leaves than in the SW3 leaves. In addition, most of the m6A-marked genes (2229 genes) were common to the FL vs. SW2 and FL vs. SW4 comparisons, whereas 219 and 216 m6A-marked genes were unique to the FL vs. SW2 and FL vs. SW4 comparisons, respectively. These results imply that the m6A-marked genes did not differ significantly between these two comparisons, with most m6A-marked genes common to both comparisons. Most of the 4289 m6A-marked genes in the five samples (average of 96.90%) contained a single m6A peak, with only a few containing more than three m6A peaks (Table S2). The distribution of m6A peaks in the tea leaves during the solar-withering treatment was then investigated. Major m6A modifications within transcripts were primarily near the stop codon and 3′ UTR (Figure 1C). Only 1.46%–2.13% of the m6A modifications were located in the coding sequence (CDS) and 5′ UTR or near the start codon (Figure 1D). From FLs to the SW3 leaves, the proportion of m6A modifications increased in the 3′ UTR and near the stop codon, but decreased in the 5′ UTR and CDS and near the start codon. Surprisingly, the proportion of m6A modifications in the 3′ UTR and around the stop codon was lower in the SW4 leaves than that in the SW3 leaves, whereas the opposite trend was observed for the m6A peaks in the 5′ UTR and CDS and near the start codon. Next, the m6A peaks were normalized by the enrichment algorithm [31]. The results revealed that m6A modifications accumulated preferentially in the 3′ UTR and around the stop codon (Figure S5). Overall, the distribution of m6A peaks in tea leaves during the solar-withering treatment changed slightly.
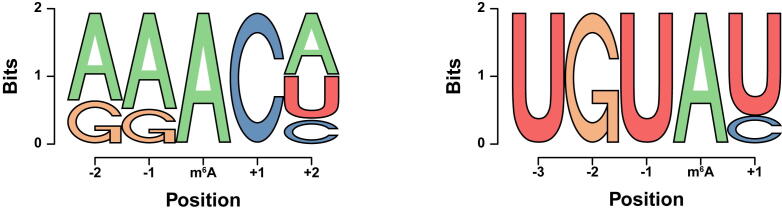
To identify the enriched motifs within the m6A peaks in tea plants, all of the m6A peaks were scanned using the MEME suite [32] and HOMER tool [33]. As expected, the canonical motif RRACH (R = G/A and H = A/U/C) was enriched in most of the m6A peaks (Figure S6). Moreover, another enriched motif, UGUAY (Y = C/U), was similar to the plant-specific motif URUAY, which can be recognized by m6A readers [34]. Although the UGUAY motif is relatively common in the m6A peaks of tomato [11], it has not been detected in other plant species. Hence, the enriched motifs at m6A-modified sites were relatively conserved in two horticultural plant species (i.e., tea and tomato).
Analysis of DMP-associated genes
To clarify the potential effects of m6A modifications during the solar-withering stage, we first searched for DMPs in the m6A methylome. The DMPs were identified according to the following criteria: |fold change (FC)| ≥ 2 and P < 0.05. A total of 265 DMPs were detected in the FL vs. SW1 comparison, of which 203 and 62 were hypermethylated and hypomethylated peaks, respectively (Table S3). The 215 DMPs identified in the FL vs. SW4 comparison were similar to the number of DMPs detected in the FL vs. SW2 comparison (i.e., 217), but more than the 183 DMPs identified in the FL vs. SW3 comparison. These results reflected the apparent changes in the global m6A status in the withered leaves following the solar-withering treatments with different shading rates. The number of detected DMPs decreased from the FL vs. SW1 comparison to the FL vs. SW3 comparison, whereas it increased substantially in the FL vs. SW4 comparison. Intriguingly, the change in the number of DMPs under solar-withering conditions was in accordance with the overall change in the total m6A level. Hence, the m6A modifications in tea leaves were tightly associated with the solar-withering treatment conditions, especially the shading rate.
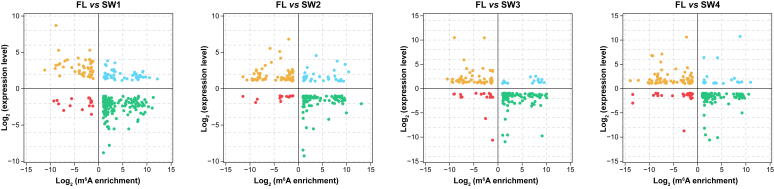
To evaluate the overall correlation between m6A modifications and gene expression levels in response to solar-withering treatments, 1137 DMP-associated genes with varying expression levels between samples were selected (Figure S7). The results showed that 74.9% of the DMP-associated genes in the FL vs. SW1 comparison, 79.6% of the DMP-associated genes in the FL vs. SW2 comparison, 83.2% of the DMP-associated genes in the FL vs. SW3 comparison, and 79.6% of the DMP-associated genes in the FL vs. SW4 comparison with increased or decreased m6A levels displayed negatively regulated gene expression. In contrast, only 25.1% of the DMP-associated genes in the FL vs. SW1 comparison, 20.4% of the DMP-associated genes in the FL vs. SW2 comparison, 16.8% of the DMP-associated genes in the FL vs. SW3 comparison, and 20.4% of the DMP-associated genes in the FL vs. SW4 comparison with increased or decreased m6A levels displayed positively regulated gene expression. Thus, the m6A modifications in most DMP-associated genes were negatively correlated with gene expression levels during the solar-withering treatment. Interestingly, combined with the increasing proportion of m6A modifications in the 3′ UTR and near the stop codon from FLs to the SW3 leaves, m6A modifications within these regions tended to decrease the mRNA abundance. Furthermore, the proportion of m6A modifications within the 3′ UTR and around the stop codon was lower in the SW4 leaves than that in the SW3 leaves, which was consistent with the obvious decrease in the proportion of DMP-associated genes with expression levels that were negatively correlated with m6A abundance. The m6A peaks distributed within the 3′ UTR and around the stop codon were negatively correlated with the mRNA abundance, whereas the m6A peaks located in the 5′ UTR and CDS and near the start codon tended to be positively correlated with gene expression. Therefore, the multifaceted effects of m6A modifications on mRNA expression may depend on the position of the m6A peaks. The specific distribution of m6A peaks is closely related to gene expression in tea leaves as well as in several other important crops, including rice [35], strawberry [13], and apple [36].
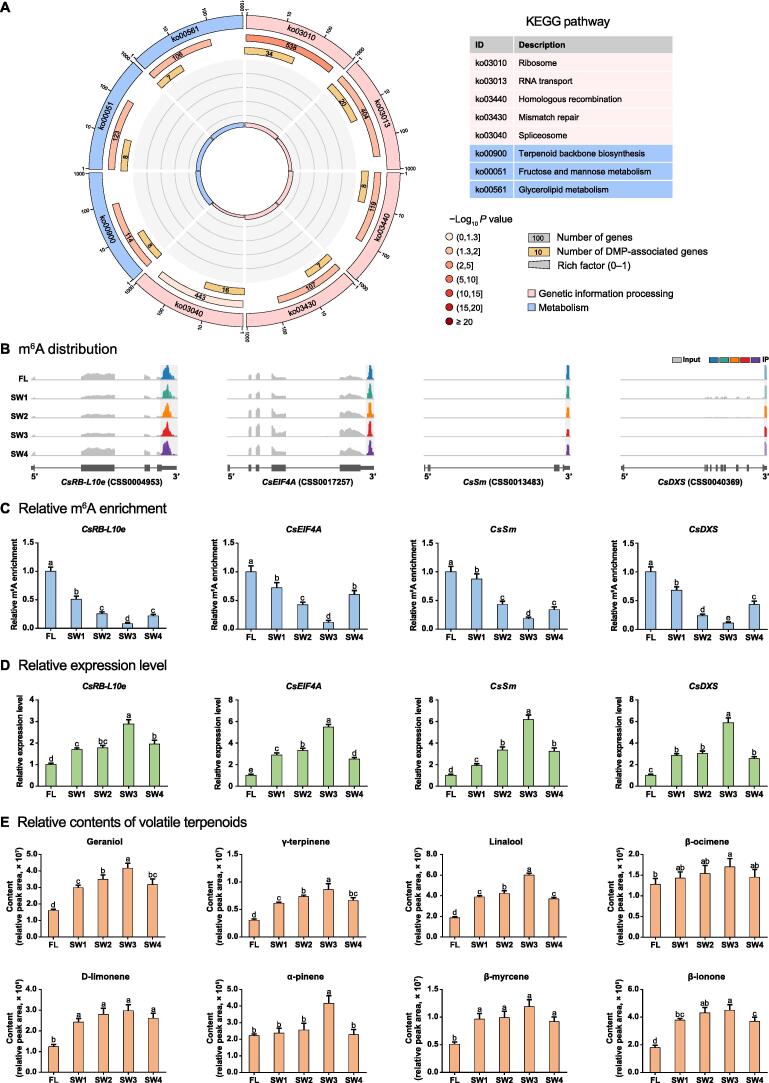
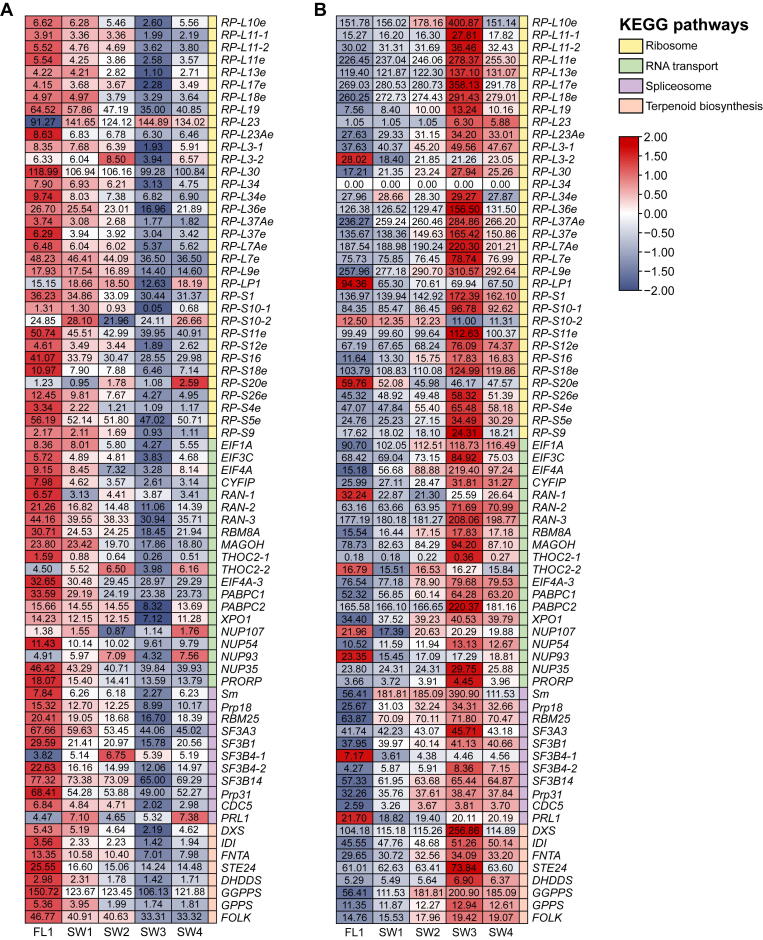
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed to decipher the biological functions of the identified DMP-associated genes. Consistent with the fact that withering affects the formation of flavor compounds in tea, the enriched KEGG pathways among the m6A-labeled genes were associated with genetic information processing (Figure 2A). More than half of the DMP-associated genes were assigned to the ribosome, RNA transport, and spliceosome pathways. Moreover, several DMP-associated genes were associated with three metabolism-related pathways. Among these pathways, the rich factor was highest for the terpenoid backbone biosynthesis pathway, which is closely related to the formation of the oolong tea aroma. We focused on the ribosome, RNA transport, spliceosome, and terpenoid backbone biosynthesis pathways in the subsequent analyses.
Figure 2.
m6A abundances of representative DMP-associated genes werenegatively correlated with their expression levelsas well asthe accumulation of volatile terpenoids under solar-withering with different shading rates
A. KEGG enrichment analysis of DMP-associated genes. From the outside to the inside, the first circle indicates enriched KEGG pathways and the number of genes corresponds to the outer circle. The second circle indicates the number of genes in the genome background and the P value for the enrichment of the genes in the specified KEGG pathway. The third circle indicates the number of DMP-associated genes. The fourth circle indicates the enrichment factor of each KEGG pathway. B. The distribution of m6A reads in CsRB-L10e, CsEIF4A, CsSm, and CsDXS. Exons and introns in the gene structures are represented by thick boxes and lines, respectively. C. Relative m6A enrichment of CsRB-L10e, CsEIF4A, CsSm, and CsDXS under solar-withering with different shading rates determined by m6A-IP-qPCR. D. Relative expression levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS under solar-withering with different shading rates determined by qRT-PCR. E. Relative contents of volatile terpenoids under solar-withering with different shading rates determined by GC–MS. Data are presented as mean ± SD. Different lowercase letters over the error bars indicate significantly different groups via one-way ANOVA and Tukey’s post hoc test (P < 0.05). DMP, differentially methylated peak; KEGG, Kyoto Encyclopedia of Genes and Genomes; m6A-IP-qPCR, m6A-immunoprecipitation-quantitative polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction; GC–MS, gas chromatography–mass spectrometry.
Negative correlation between the m6A abundances and the expression levels of most DMP-associated genes under solar-withering conditions
The m6A-seq analysis showed that the m6A abundances and the expression levels of several core genes involved in the four aforementioned KEGG pathways were significantly altered during the solar-withering process (Figure S8A and B). In the ribosome pathway, the m6A abundances of 34 ribosomal protein-encoding (RP) genes were affected by the solar-withering treatment (Table S4). These RP genes are indispensable for ribosome formation and protein biosynthesis and have important roles in the transcription–translation process. We also determined that 20 and 16 DMP-associated genes are involved in RNA transport and spliceosome pathways, respectively (Tables S5 and S6). RNA transport is functionally linked to several steps in the RNA processing stage, including RNA splicing, 3′-end formation, and transcription–translation processes, which are critical for various biological functions [37]. AS is a pivotal post-transcriptional event that diversifies the generated transcripts and the corresponding proteins, making it a critical molecular mechanism that regulates transcriptional abundance in the plant kingdom. Interestingly, significant changes in the m6A levels were detected for eight terpenoid biosynthesis-related genes in response to the solar-withering treatment (Table S7).
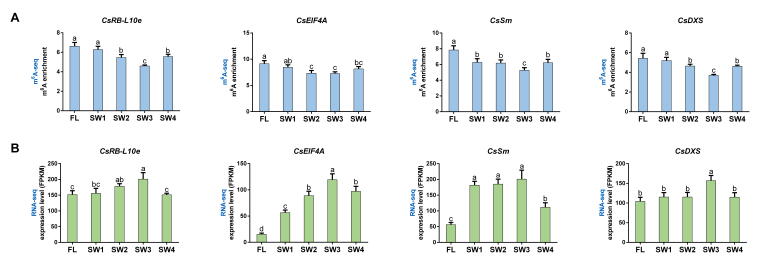
For each of the four aforementioned pathways, we selected one DMP-associated gene in which there was a dynamic change in the m6A level following the solar-withering treatments with different shading rates. The four genes encode the large subunit ribosomal protein L10e (RB-L10e), eukaryotic translation initiation factor 4A (EIF4A), core spliceosomal Sm protein (Sm), and 1-deoxy-D-xylulose-5-phosphate synthase (DXS), respectively. The m6A peaks in their mRNA sequences were mainly distributed in the 3′ UTR and around the stop codon (Figure 2B). Our m6A-seq datasets revealed significant changes in the m6A levels of these four genes (Figure S9A). We then performed an m6A-immunoprecipitation-quantitative polymerase chain reaction (m6A-IP-qPCR) analysis to further validate the m6A levels of the four aforementioned genes. As expected, the m6A levels in CsRB-L10e, CsEIF4A, CsSm, and CsDXS decreased sharply from FLs to the SW3 leaves, but increased from the SW3 leaves to the SW4 leaves (Figure 2C). The expression levels of these four genes increased from FLs to the SW3 leaves, but then decreased in the SW4 leaves, as revealed by both transcriptome datasets (Figure S9B) and the quantitative real-time polymerase chain reaction (qRT-PCR) data (Figure 2D). These results indicate that the m6A modifications in these mRNAs were negatively correlated with their corresponding expression levels, which is in agreement with the findings of a previous study that demonstrated that mRNAs with low m6A levels tend to be highly expressed in tomato [11]. We detected obvious changes in the contents of seven monoterpenoids and one apocarotenoid using gas chromatography–mass spectrometry (GC–MS) (Figure 2E). Specifically, the abundances of all eight volatiles increased markedly from FLs to the SW3 leaves before decreasing significantly in the SW4 leaves. We also determined that the accumulation of eight volatile terpenoids was positively correlated with the CsDXS expression level, but negatively correlated with the m6A level of CsDXS and the overall m6A level under solar-withering conditions.
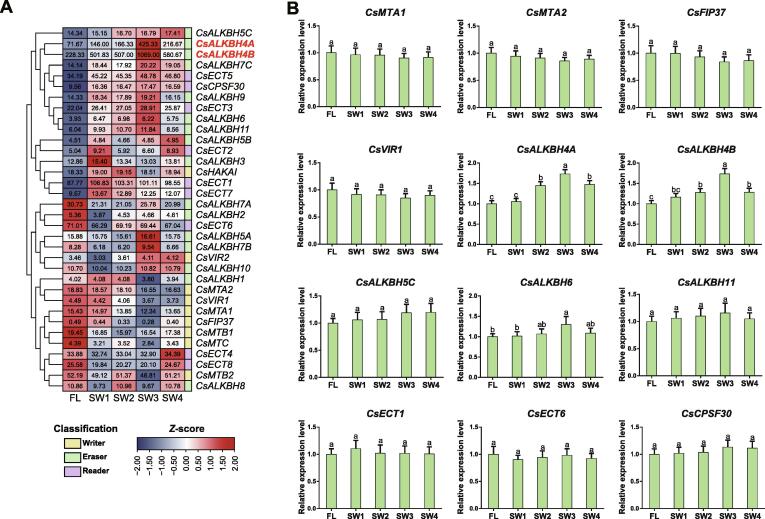
Expression profiles of m6A regulatory genes in response to solar-withering
The effects of RNA methylation are tightly associated with the transcript levels of RNA methyltransferase, demethylase, and reader genes [38]. Thus, we speculated that the m6A levels of these mRNAs are coordinately governed by m6A regulatory genes. Based on our recent report [14], 34 m6A regulatory genes were identified in the tea reference genome. We initially analyzed the expression profiles of m6A regulatory genes under solar-withering conditions (Figure 3A). The m6A regulatory genes with |FC| ≥ 2 and P < 0.05 were considered to be differentially expressed between samples. The transcript levels of all m6A writer and reader genes were not obviously altered by the solar-withering conditions. Among the 16 m6A eraser genes, only CsALKBH4A and CsALKBH4B had significant changes in their transcript levels after the solar-withering treatments. We also noticed that CsALKBH6 transcription was significantly induced only following the SW3 treatment. These results indicate that dynamic changes in m6A modifications may be mainly controlled by m6A eraser genes, with CsALKBH6 playing a particularly important role in the regulation of m6A levels in response to SW3. Next, a qRT-PCR analysis confirmed the expression patterns of m6A regulatory genes revealed by the transcriptome data, and demonstrated that the CsALKBH4A and CsALKBH4B expression levels increased continuously from FLs to the SW3 leaves, but then clearly decreased in the SW4 leaves (Figure 3B). This trend was in accordance with the expression patterns of CsRB-L10e, CsEIF4A, CsSm, and CsDXS, but was inversely correlated with the m6A abundances of these four genes and the global m6A level under solar-withering conditions. Moreover, the CsALKBH6 expression level increased significantly only following the SW3 treatment, whereas the expression of the m6A writer and reader genes was unaffected by the solar-withering treatments. These observations imply that the m6A eraser-mediated removal of m6A marks on mRNAs is closely linked with variations in m6A levels and the expression of DMP-associated genes.
Figure 3.
Expression profiles of m6A regulatory genes under solar-withering with different shading rates
A. The heatmap of m6A regulatory genes under solar-withering with different shading rates based on the transcriptome datasets. The expression values of m6A regulatory genes were normalized using z-score formula. The number in the box indicates the original FPKM value. B. Expression patterns of four m6A writer genes, five m6A eraser genes, and three m6A reader genes under solar-withering with different shading rates determined by qRT-PCR. Data are presented as mean ± SD. Different lowercase letters over the error bars indicate significantly different groups via one-way ANOVA and Tukey’s post hoc test (P < 0.05). FPKM, fragments per kilobase per million mapped reads.
CsALKBH4-mediated RNA demethylation may activate terpenoid biosynthesis
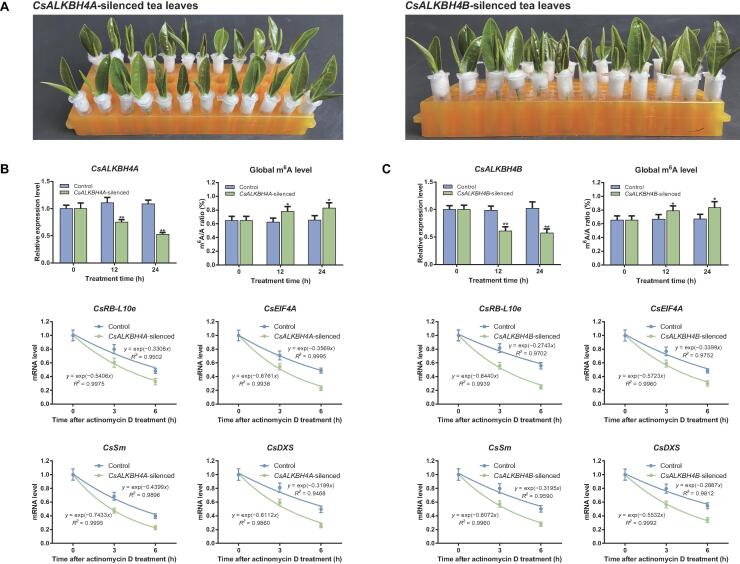
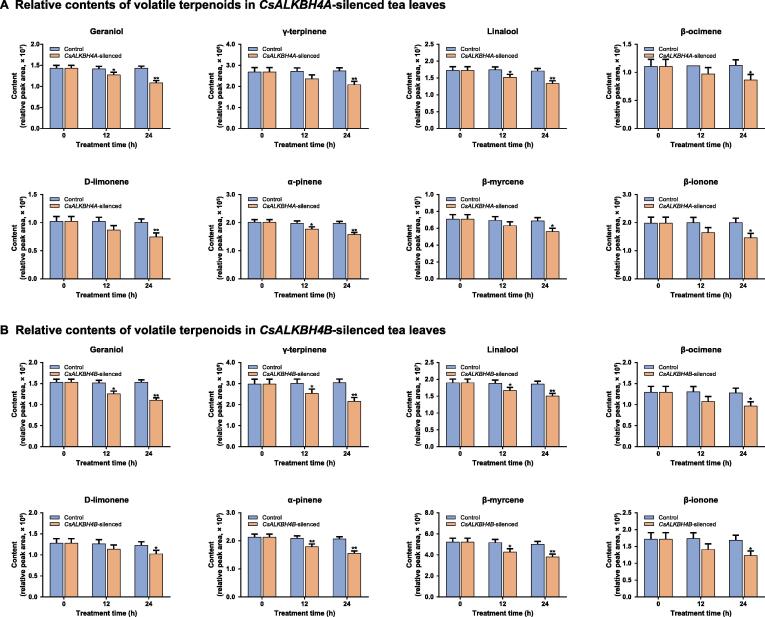
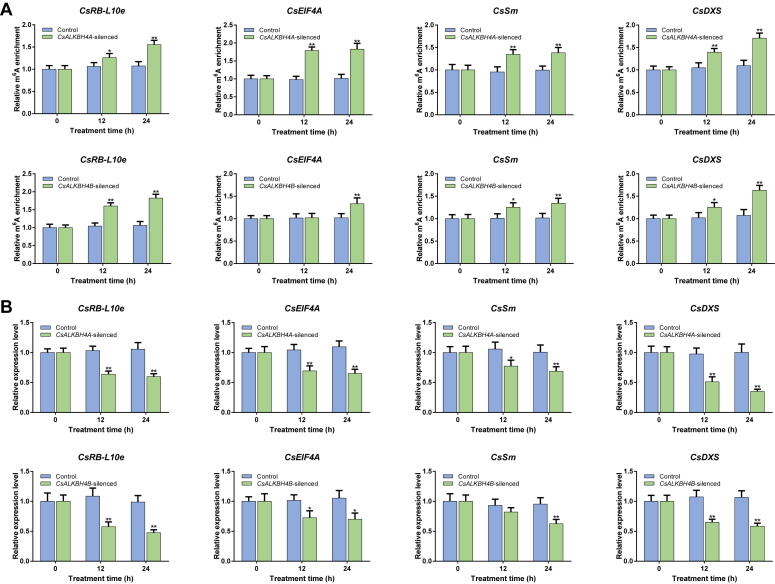
To further functionally characterize CsALKBH4A and CsALKBH4B during the solar-withering process, we transiently suppressed the expression of these two genes via an siRNA-mediated gene silencing strategy (Figure 4A). As anticipated, the expression of both CsALKBH4A and CsALKBH4B was significantly down-regulated in the gene-silenced leaves, whereas the transcription of these two genes was not substantially altered by treatments with the corresponding negative control (NC) (Figure 4B and C). The inhibited expression of CsALKBH4A and CsALKBH4B led to a marked increase in the overall m6A level in the gene-silenced tea leaves (Figure 4B and C), which occurred concomitantly with an obvious increase in the m6A levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS (Figure S10A) (compared with the respective NC). Conversely, the CsRB-L10e, CsEIF4A, CsSm, and CsDXS mRNA levels were markedly lower in the gene-silenced leaves than in the NC-treated leaves (Figure S10B). Accordingly, the diminished expression of these four genes may be mainly attributed to the suppression of CsALKBH4-mediated RNA demethylation. To further investigate how m6A demethylation influences the transcription of CsRB-L10e, CsEIF4A, CsSm, and CsDXS, we explored whether CsALKBH4-mediated RNA demethylation modulates the stability of the transcripts of these four genes by monitoring mRNA decay rates after an actinomycin D treatment (Figure 4B and C). We observed that the CsRB-L10e, CsEIF4A, CsSm, and CsDXS mRNAs degraded more rapidly in the CsALKBH4A- and CsALKBH4B-silenced leaves than in the NC-treated leaves. Notably, for CsRB-L10e, CsEIF4A, CsSm, and CsDXS, their mRNA decay rates were negatively correlated with their expression levels and positively correlated with their m6A levels. These results suggest that excessive m6A modifications in the CsRB-L10e, CsEIF4A, CsSm, and CsDXS mRNAs have a destabilizing effect, which results in a clear decrease in transcript levels. To assess the effects of RNA demethylation on the accumulation of volatile terpenoids, the terpenoid contents in CsALKBH4A- and CsALKBH4B-silenced leaves were measured. The transcriptional repression of these two genes resulted in a substantial decrease in the contents of the eight volatile terpenoids (Figure 5A and B), suggesting that CsALKBH4-mediated RNA demethylation may directly activate terpenoid biosynthesis by removing m6A marks and enhancing the stability of the corresponding mRNAs.
Figure 4.
CsALKBH4-mediated RNA demethylation contributes to the accumulation of volatile terpenoids by removing the m6A marks and enhancing the stability of corresponding mRNAs
A. Diagram of CsALKBH4A- and CsALKBH4B-silenced assay in tea leaves via an siRNA-mediated gene silencing strategy. The freshly detached tea bud and first leaf from naturally grown tea plants were incubated in 1.5-ml microcentrifuge tubes that contained 1 ml of 20 μM siRNA or siRNA-NC solution. After incubation for 12 h and 24 h, the tea bud and first leaf were harvested and then used for qRT-PCR and metabolite detection. B. Gene expression, global m6A level, and mRNA stability in CsALKBH4A-silenced tea leaves. C. Gene expression, global m6A level, and mRNA stability in CsALKBH4B-silenced tea leaves. To evaluate the mRNA stability, the leaf discs were collected from gene-silenced tea leaves and incubated in sterile water that contained 10 μg/ml actinomycin D solution. Tea leaves incubated in sterile water were used as controls. Total RNA was isolated from the leaves sampled at 6 h and 12 h, respectively. The mRNA levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS were examined by qRT-PCR. Data are presented as mean ± SD. The differences among various groups were assessed by conducting a one-way ANOVA and Tukey’s post hoc test. *, P < 0.05; **, P < 0.01. siRNA-NC, negative control siRNA.
Figure 5.
Relative contents of volatile terpenoids inCsALKBH4A/CsALKBH4B-silenced tea leaves determined by GC–MS
A. Relative contents of volatile terpenoids in CsALKBH4A-silenced tea leaves. B. Relative contents of volatile terpenoids in CsALKBH4B-silenced tea leaves. Data are presented as mean ± SD. The differences among various groups were assessed by conducting a one-way ANOVA and Tukey’s post hoc test. *, P < 0.05; **, P < 0.01.
RNA demethylation modulates AS events by influencing m6A modifications and the expression of spliceosome-related genes
The spliceosome pathway was one of the main enriched KEGG pathways among the DMP-associated genes. The m6A modifications of the core genes Sm, Prp18 (encoding pre-mRNA-splicing factor 18), and Prp31 (encoding pre-mRNA-splicing factor 31) in the spliceosome pathway were significantly altered by the solar-withering treatment, which was consistent with the obvious expression changes detected for these genes. Thus, we hypothesized that RNA demethylation might contribute to the regulation of AS events by controlling the m6A levels and the expression levels of spliceosome-related genes. To test this hypothesis, we first investigated the AS events during the solar-withering treatments with different shading rates. A total of 18,188 AS events were detected in 13,606 genes in five tea samples. There were substantially more AS events in the solar-withered leaves than in FLs, suggesting that solar-withering conditions may influence the occurrence of AS events (Table S8). Within a certain shading rate range (i.e., SW1–SW3), the frequency of AS events was closely correlated with the decreases in the shading rate. In contrast, solar-withering without shading substantially decreased the number of AS events. In the examined samples, the predominant AS events resulted in retained introns. This observation is in accordance with the results of earlier investigations on maize [39], cotton [40], and tea [41]. We next comprehensively identified the differentially expressed alternative splicing genes (DAGs) among the solar-withering treatments with different shading rates. There were more DAGs in the FL vs. SW3 comparison than in the FL vs. SW1 and FL vs. SW2 comparisons, implying that increasing the light intensity of the solar-withering treatment may promote the differential expression of AS genes (Table S9). However, there were fewer DAGs in the FL vs. SW4 comparison than in the FL vs. SW3 comparison. These findings indicate that the shading rate during the solar-withering treatment affects the number of AS events, possibly by modulating the expression of AS genes.
Association between flavonoid, catechin, and theaflavin contents and the AS gene transcript levels under solar-withering conditions
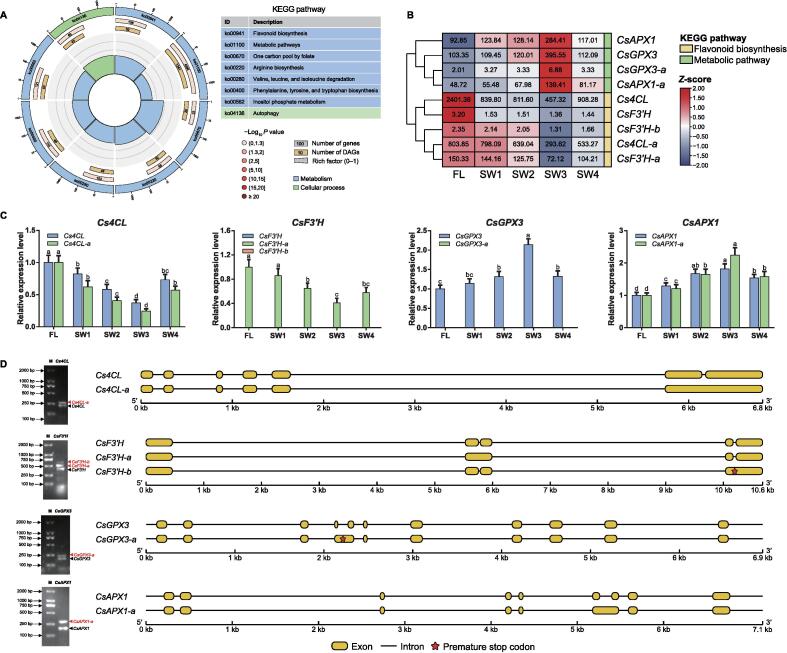
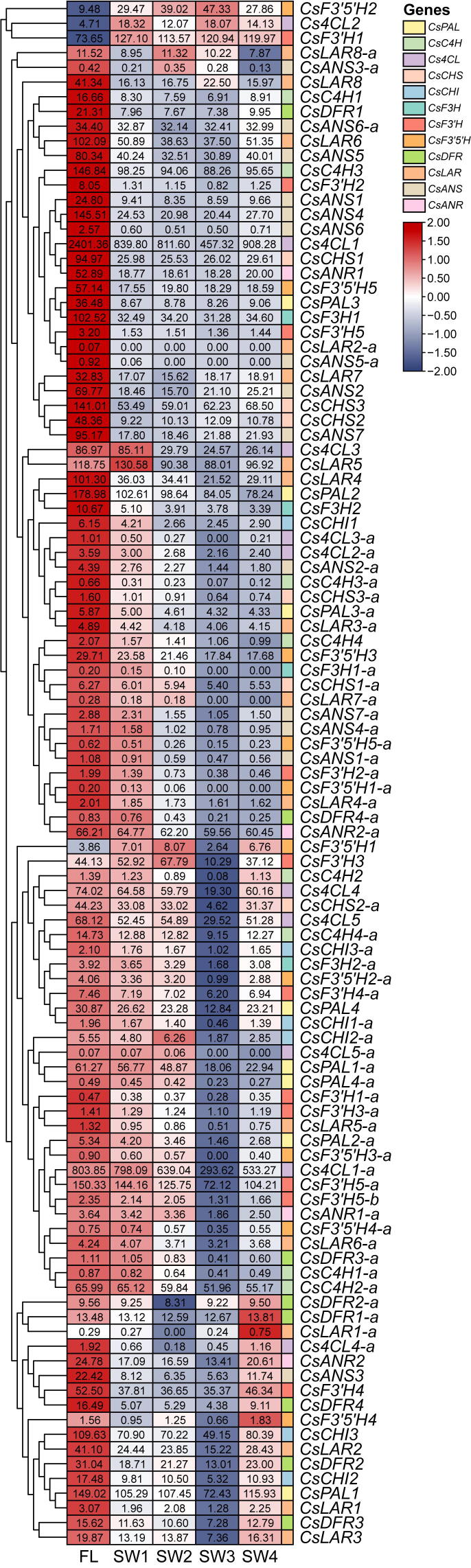
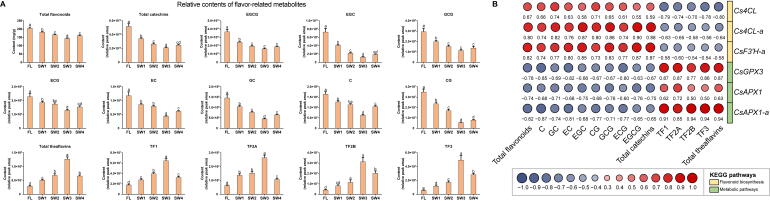
To further explore the putative effects of DAGs under solar-withering conditions, we performed a KEGG analysis of all identified DAGs. Metabolic pathways were the most enriched pathways among the DAGs, followed by the flavonoid biosynthesis pathway (Figure 6A). Therefore, structural genes involved in flavonoid biosynthesis may be affected by the m6A-mediated AS regulatory mechanism (Figure S11; Table S10). We selected 4CL (encoding 4-coumarate CoA ligase) and F3′H (encoding flavonoid 3′-hydroxylase) as two flavonoid biosynthesis-related DAGs for further analyses. AS events generated two and three splicing variants for 4CL and F3′H, respectively. On the basis of the fragments per kilobase per million mapped reads (FPKM) values obtained from the transcriptome datasets, the expression profiles of these AS transcripts under solar-withering conditions were analyzed (Figure 6B). The expression of the full-length Cs4CL transcript decreased substantially from FLs to the SW3 leaves, but then obviously increased in the SW4 leaves. The expression profile of the AS transcript Cs4CL-a was similar to that of the full-length transcript after the solar-withering treatment, whereas the Cs4CL-a transcript abundance was lower than that of Cs4CL following the same solar-withering treatment. Solar-withering also strongly inhibited the transcription of CsF3′H-a. Because of the high cycle threshold value > 35 for CsF3′H and CsF3′H-b in the qRT-PCR assay (Table S11), the expression levels for these two transcripts were extremely low in FLs and the solar-withered leaves. Hence, CsF3′H-a may be the predominant transcript involved in flavonoid biosynthesis. The qRT-PCR analysis confirmed the transcriptome datasets were reliable (Figure 6C). To identify the AS transcripts related to the biosynthesis of flavonoids and catechins during solar-withering with different shading rates, the flavonoid and catechin contents in all five samples were compared (Figure S12A). There was a sharp decrease in the total flavonoid content from FLs to the SW3 leaves, but there was a distinct rebound from SW3 to SW4.
Figure 6.
AS-mediated regulatory mechanism influences the accumulation of flavor-related metabolites
A. KEGG enrichment analysis of DAGs. From the outside to the inside, the first circle indicates enriched KEGG pathways and the number of genes corresponds to the outer circle. The second circle indicates the number of genes in the genome background and the P value for the enrichment of the genes in the specified KEGG pathway. The third circle indicates the number of DAGs. The fourth circle indicates the enrichment factor of each KEGG pathway. B. The heatmap of DAGs and their AS transcripts under solar-withering with different shading rates based on the transcriptome datasets. The expression values of DAGs and their AS transcripts were normalized using the z-score formula. The number in the box indicates the original FPKM value. C. Expression patterns of Cs4CL, CsF3′H, CsGPX3, and CsAPX1 as well as their AS transcripts under solar-withering with different shading rates determined by qRT-PCR. Data are presented as mean ± SD. Different lowercase letters over the error bars indicate significantly different groups via one-way ANOVA and Tukey’s post hoc test (P < 0.05). D. RT-PCR validation and gene structure of the full-length transcripts and AS transcripts. M indicates the lane with DNA size markers. The full-length transcripts and AS transcripts on the gel images are denoted with black and red triangles, respectively. AS, alternative splicing; DAG, differentially expressed alternative splicing gene; RT-PCR, reverse transcription-polymerase chain reaction.
Excessive and insufficient amounts of catechins have detrimental effects on tea flavor and the health benefits of tea. In this study, we examined whether the solar-withering process increases the palatability of oolong tea, while also decreasing the health benefits of tea. Intriguingly, AS events were detected for APX1 (encoding ascorbate peroxidase 1) and GPX3 (encoding glutathione peroxidase 3), which are involved in metabolic pathways. These two genes are reportedly responsible for the conversion of catechins into theaflavins [42], [43]. We hypothesized that the solar-withering treatments strongly inhibited catechin biosynthesis, resulting in an obvious increase in the theaflavin content. To assess this hypothesis, the total theaflavin content and the contents of four theaflavin components were measured using liquid chromatography–mass spectrometry (LC–MS). As expected, the solar-withering treatments caused the individual theaflavin contents and the total theaflavin content to increase substantially (Figure S12A). Additionally, the theaflavin contents were inversely correlated with the catechin contents under solar-withering conditions. Next, we observed that the CsGPX3, CsAPX1, and CsAPX1-a expression levels increased dramatically from FLs to the SW3 leaves, but then clearly decreased in the SW4 leaves. One AS transcript (CsGPX3-a) was unaffected by the solar-withering treatments. We then detected a premature stop codon (PTC) in the CsGPX3-a mRNA (Figure 6D).
To further elucidate the possible effects of AS transcripts on the accumulation of flavonoids, catechins, and theaflavins, the relationships between the abundances of flavonoid-related AS transcripts and metabolites were evaluated via Pearson’s correlation analysis (Figure S12B). Three transcripts (CsF3′H, CsF3′H-b, and CsGPX3-a) were excluded from this analysis because they were not expressed in FLs and the solar-withered leaves. The expression levels of the full-length Cs4CL transcript and the Cs4CL-a AS transcript were positively correlated with the total flavonoid, total catechin, and eight individual catechin contents. Positive correlations were also detected between the expression level of the CsF3′H-a AS transcript and the abundances of the aforementioned metabolites. Moreover, we observed that the accumulation of four individual theaflavin components and the total theaflavin content were positively correlated with the expression of CsGPX3, CsAPX1, and CsAPX1-a. We also examined the effects of AS transcripts (Cs4CL-a, CsF3′H-a, and CsAPX1-a) on the accumulation of the associated metabolites (Figure S13A). As expected, the Cs4CL-a, CsF3′H-a, and CsAPX1-a transcript levels were considerably down-regulated in the gene-silenced leaves, whereas they were not obviously altered by the corresponding NC treatments. Next, the flavonoid, catechin, and theaflavin contents in the gene-silenced leaves were analyzed (Figure S13B). The total flavonoid and total catechin contents as well as the accumulation of eight individual catechins decreased sharply following the silencing of Cs4CL-a or CsF3′H-a, which was in contrast to the lack of any significant changes to the total theaflavin content and the abundances of four individual theaflavins. The transcriptional repression of CsAPX1-a did not affect the flavonoid and catechin contents in the gene-silenced leaves, but it inhibited the accumulation of theaflavins. These observations imply that some AS transcripts are likely critical for the accumulation of flavonoids, catechins, and theaflavins.
Discussion
Dynamic changes in global m6A level in tea leaves are mainly controlled by m6A erasers under solar-withering conditions
In the past few decades, several studies on the environmental stresses during tea manufacturing (e.g., withering stage) have clarified the molecular basis of specific metabolic activities related to tea flavor formation [15], [17]. Recent research confirmed epigenetic modifications, such as DNA methylation and histone modifications, influence the production of tea flavor-related substances [28], [29]. However, it remains unclear whether RNA methylation (epitranscriptome-level changes) also regulates flavor-related metabolic pathways and tea flavor formation.
In the present study, we observed that m6A modifications are widely distributed among mRNAs in tea plants, with dynamic changes in the m6A levels induced by solar-withering treatments. More specifically, in response to solar-withering conditions, the overall m6A level decreased dramatically. From FLs to the SW3 leaves, the overall m6A level gradually decreased, but the m6A level increased in the SW4 leaves. Thus, solar-withering treatments significantly decreased the m6A levels. Moreover, there was a positive correlation between the shading rate and the global m6A abundance within a certain range. According to earlier investigations [44], [45], the overall DNA methylation level is mediated by DNA methyltransferases and demethylases. In addition, the corresponding RNA methyltransferases and demethylases have been identified in tea plants [14]. Therefore, we speculated that the overall m6A level may be controlled by RNA methyltransferases and demethylases under solar-withering conditions. To evaluate this hypothesis, we comprehensively examined the expression profiles of m6A regulatory genes following solar-withering treatments. Notably, obvious transcript-level changes were detected in two m6A eraser genes (CsALKBH4A and CsALKBH4B) during the solar-withering treatments with different shading rates, whereas the expression levels of other m6A eraser and reader genes were only slightly affected by solar-withering. These findings are consistent with the reported phenomena in A. thaliana [46] and tomato [11]. Similarly, during the solar-withering stage, tea leaves are subjected to multiple environmental stresses, including UV radiation [15]. Thus, our results imply that the removal of m6A marks, rather than the addition and decoding of these marks, may be vital for tea plant responses to multiple stresses during the tea-withering stage. The inclusion of sunshade nets in solar-withering treatments prevents the excessive UV irradiation of tea leaves, thereby minimizing the damages to the living tea leaves. The CsALKBH4A and CsALKBH4B transcription levels increased continuously as the shading rate increased (from FLs to the SW3 leaves). Furthermore, the global m6A level was negatively correlated with the expression of these two m6A eraser genes. Additionally, the overall m6A level increased in the CsALKBH4-silenced leaves. These observations suggest that m6A demethylation is primarily responsible for the decrease in the global m6A level from FLs to the SW3 leaves. However, tea leaves were exposed to high UV doses during the solar-withering without the sunshade net (SW4). Exposures to high UV doses may lead to irreversible damages to the osmotic regulatory activities of plasma membranes [47], [48], [49], ultimately leading to ruptured cells and further disruptions to the normal expression of the nuclear-localized genes CsALKBH4A and CsALKBH4B [14]. In the current study, low UV doses induced CsALKBH4 expression within a certain range during the solar-withering treatments with the sunshade net, whereas high UV doses strongly inhibited CsALKBH4 expression when the solar-withering treatment was completed without a sunshade net. The down-regulated expression of CsALKBH4 in response to the SW4 treatment may have impaired the ability of m6A erasers to remove m6A modifications, which may help to explain the observed increase in the overall m6A level from SW3 to SW4. Collectively, these results indicate that the dynamic changes in global m6A level in tea leaves undergoing solar-withering treatments with different shading rates are mainly controlled by m6A erasers. In addition, CsALKBH4-mediated RNA demethylation likely affects critical processes during the tea-withering stage.
RNA demethylation directly contributes to the accumulation of volatile terpenoids and the formation of tea aromas
Environmental stresses can considerably affect the accumulation of specialized metabolites in tea leaves, thereby affecting tea quality [50]. During postharvest processing, tea leaves are exposed to various environmental stresses, which induce obvious alterations to many flavor-related compounds, leading to the production of teas with unique flavors [17]. Although the effects of specific metabolites on tea flavor formation have been investigated at the transcriptional, translational, and metabolic levels, the functional roles of epigenetic modifications, especially RNA methylation, and the regulatory mechanisms underlying the m6A-mediated flavor formation in the tea-withering stage remain unclear.
In the current study, an integrated RNA methylome and transcriptome analysis revealed that the variations in the number of DMPs in response to solar-withering with different shading rates were in accordance with the changes in the total m6A level. The number of DMPs decreased between the FL vs. SW1 and FL vs. SW3 comparisons, but it increased in the FL vs. SW4 comparison. This increase may be associated with the inhibition of CsALKBH4 expression due to high UV doses during solar-withering without a sunshade net. The down-regulated expression of CsALKBH4 in the SW4 leaves hindered RNA demethylation, thereby increasing the differences in the m6A levels between the SW4 leaves and the leaves that underwent the other treatments and finally leading to an increase in the number of DMPs. A total of 1137 genes with DMPs under solar-withering conditions were functionally characterized. The KEGG pathway analysis showed that the identified DMP-associated genes were mainly associated with the terpenoid biosynthesis pathway. Volatile terpenoids are major quality-related compounds in oolong tea. Specifically, they significantly influence the formation of floral and honey-like aromas in high-quality oolong tea because of their low odor thresholds [15]. Moreover, several lines of evidence suggest that environmental stresses can alter the m6A levels of transcripts [12], [51]. In our study, the expression levels of seven terpenoid biosynthesis-related genes increased significantly under solar-withering conditions, which may be related to the observed considerable accumulation of volatile terpenoids. The positive correlations between the expression levels of these structural genes related to terpenoid biosynthesis and terpenoid contents imply that multiple stresses induced by solar-withering promote terpenoid accumulation by up-regulating the expression of terpenoid biosynthesis-related genes. This is in accordance with the results of another study [18]. In addition, the m6A levels in these terpenoid biosynthesis-related genes were negatively correlated with the corresponding expression levels. Similarly, an earlier investigation has demonstrated that the expression of genes with low m6A levels tends to be up-regulated [52]. These results suggest that the expression levels of m6A-containing genes may be governed by RNA methylation. According to recent reports, knocking out m6A writer genes can dramatically decrease the m6A levels of m6A-modified genes, which results in marked increases in gene expression levels [6], [53]. In an earlier study, the overaccumulation of m6A marks on mRNAs adversely affected gene expression in the AtALKBH10B mutant line, suggesting that m6A modifications may negatively affect mRNA expression [54]. In our study, the suppression of CsALKBH4A and CsALKBH4B expression resulted in a significant decrease in the stability of CsDXS mRNA and the transcripts of three DMP-associated genes. Furthermore, the abundances of eight volatile terpenoids decreased in the CsALKBH4A- or CsALKBH4B-silenced leaves. These findings along with the CsALKBH4-mediated RNA demethylation mentioned above suggest that RNA demethylation may regulate the overall m6A level by modulating the expression of m6A eraser genes, while also stabilizing the mRNAs of DMP-associated genes, thereby increasing their expression. However, some reports indicate that mRNA abundance is positively correlated with m6A modifications [13], [55], which is inconsistent with our data. Therefore, the mechanism underlying the regulatory effects of m6A modifications on mRNAs in the solar-withering stage will need to be more thoroughly characterized. A recent study concluded that abiotic stress can alter the location of m6A peaks in transcripts [9]. The data generated in the present study showed that m6A peaks in CsDXS and three other DMP-associated genes were distributed within the 3′ UTR and around the stop codon under solar-withering conditions, indicating that solar-withering can affect the m6A levels in these DMP-associated genes, but it cannot induce the redistribution of m6A marks. Upon reviewing the reports that m6A modifications positively affect mRNA abundance, we noted that these m6A marks are mainly concentrated in the CDS region. This observation along with our other findings indicates that m6A modifications may have distinct regulatory effects on mRNA expression depending on their distribution in the transcript structure. Considered together, these findings suggest that CsALKBH4-mediated RNA demethylation promotes the expression of DMP-associated genes involved in terpenoid biosynthesis and the accumulation of aroma-related terpenoids by enhancing mRNA stability. Additionally, the shading rate was negatively correlated with the expression of terpenoid biosynthesis-related genes and terpenoid contents from FLs to the SW3 leaves, implying that within a certain range, moderate shading promotes terpenoid biosynthesis and the formation of a high-quality aroma in oolong tea. Inadequate shading during the SW4 treatment inhibited CsALKBH4-mediated RNA demethylation, with the detrimental effects on the expression of terpenoid biosynthesis-related genes ultimately leading to a significant decrease in the terpenoid content. This is consistent with the beneficial effects of moderate shading on flavor formation during the production of teas. Hence, solar-withering treatments of tea leaves may be optimized by controlling the shading rate, which is critical for enhancing tea aromas.
RNA modifications indirectly affect the contents of flavonoid, catechin, and theaflavin by triggering the AS regulatory mechanism
The AS regulatory mechanism is a crucial component of plant responses to diverse environmental stimuli [56]. The generation of many AS transcripts leads to the expression of various proteins that alleviate the adverse effects of multiple stresses. Although many AS events related to secondary metabolism have been detected, the specific mechanisms regulating AS events associated with flavor-related genes and tea flavor formation during the tea manufacturing process are still unclear. Recent research found that m6A modifications have important regulatory effects on RNA splicing [38]. Likewise, we observed that CsALKBH4-mediated RNA demethylation influences CsSm mRNA abundance in the spliceosome pathway by enhancing its stability. According to previous research, Sm encodes a crucial spliceosome component [57] that interacts with several small nuclear RNAs (snRNAs) and then binds to a series of additional proteins to form small nuclear ribonucleoprotein particles. Within the spliceosome, snRNAs contribute to the catalysis and recognition of splice sites during pre-mRNA splicing. Intriguingly, both the frequency of AS events and the number of DAGs were closely associated with CsSm expression. Therefore, RNA demethylation might regulate AS events by modulating the m6A abundance and the expression of spliceosome-related genes.
To further investigate AS-mediated regulatory effects under solar-withering conditions, we performed a KEGG pathway enrichment analysis of all identified DAGs. Most of the DAGs were assigned to metabolic pathways and the flavonoid biosynthesis pathway. The subsequent analysis of the expression of four DAGs involved in these pathways revealed that the AS transcripts with the PTC (CsF3′H-b and CsGPX3-a) were expressed at almost undetectable levels under solar-withering conditions. This phenomenon may reflect the introduction of PTC into the gene structure, leading to the production of loss-of-function truncated proteins, which may be degraded via the nonsense-mediated decay pathway. This possibility is supported by the results of the analysis of the correlation between metabolite accumulation and the expression of AS transcripts, which revealed a lack of correlation between CsF3′H-b expression and the accumulation of flavonoids and catechins. Additionally, CsGPX3-a expression was only marginally correlated with the theaflavin level. Notably, the expression patterns of two non-PTC-type AS transcripts (Cs4CL-a and CsF3′H-a) were consistent with those of the corresponding full-length transcripts under solar-withering conditions. Meanwhile, Cs4CL-a and CsF3′H-a were more highly expressed than their full-length transcripts regardless of the shading rate during the solar-withering treatment. These findings imply that these two AS transcripts are the predominant transcripts in the flavonoid biosynthesis pathway. Similarly, CsbHLH-2, which is an AS transcript, is commonly formed in response to exposure to cold stress, during which it enhances stress tolerance by positively regulating certain signaling pathways [58]. Another AS transcript (CsAPX1-a) and its full-length transcript may participate in the coordinated regulation of theaflavin biosynthesis. The AS-mediated regulation of flavor metabolites was supported by our analysis of the correlation between AS transcripts and metabolites under solar-withering conditions, which revealed that Cs4CL-a and CsF3′H-a are positively correlated with the accumulation of flavonoids and catechins. A positive correlation was also detected between the AS transcript CsAPX1-a and the theaflavin content. We propose that the accumulation of flavonoids, catechins, and theaflavins is mediated by the canonical full-length transcripts as well as by particular AS transcripts, which may be important for the post-transcriptional regulation induced by solar-withering. Therefore, the down-regulated expression of Cs4CL, Cs4CL-a, and CsF3′H-a as well as the up-regulated expression of CsGPX3, CsAPX1, and CsAPX1-a in solar-withered leaves synergistically modulates flavonoid biosynthesis and flavonoid catabolism to promote the conversion of compounds associated with bitterness and astringency (e.g., flavonoids and catechins) into theaflavins, which provide tea with a mellow taste. Compared with the other treatments, SW3 resulted in the lowest catechin content and the highest theaflavin content, suggesting that a moderate shading rate is ideal for decreasing bitterness and astringency and enhancing the mellow taste of tea infusions. Hence, RNA demethylation indirectly affects the accumulation of flavonoids, catechins, and theaflavins by triggering the AS-mediated regulatory mechanism, thereby improving the palatability of oolong tea.
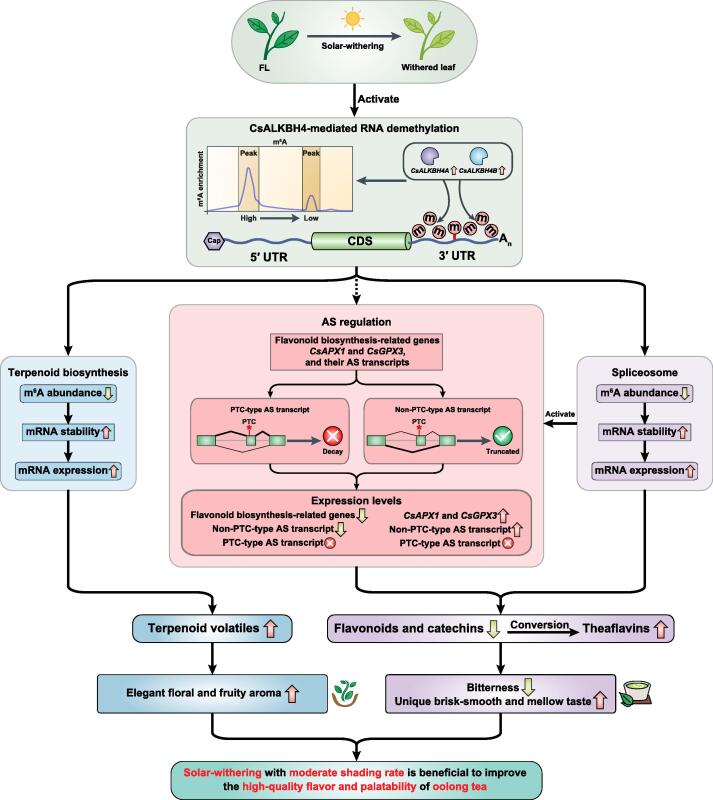
In conclusion, our integrated RNA methylome and transcriptome analysis reveal that the m6A-mediated regulatory mechanism coordinates the accumulation of specialized metabolites in tea leaves during the solar-withering stage. Moreover, the dynamic changes in global m6A level that occur in tea leaves in response to different shading rates during the solar-withering step are mainly controlled by m6A erasers. Furthermore, CsALKBH4-driven RNA demethylation directly affects the accumulation of volatile terpenoids and tea aroma formation by mediating the stability and abundance of terpenoid biosynthesis-related transcripts, while also indirectly regulating the contents of flavonoid, catechin, and theaflavin as well as the formation of tea taste-related substances by activating the AS-mediated regulatory mechanism. These findings have elucidated the effects of epigenetic modifications on the transcription of genes in the tea flavor-related metabolic pathways. They also indicate that the m6A-mediated regulatory mechanism may be targeted to enhance the high-quality flavor and palatability of oolong tea (Figure 7). Our work provides a solid foundation for future attempts at deciphering the functional effects of m6A modifications in tea plants, and it has also broadened our understanding of the regulatory mechanisms underlying m6A-mediated flavor formation during the solar-withering stage of the tea manufacturing process.
Figure 7.
Schematic model for the effects of m6A-mediated regulatory mechanism on the accumulation of flavor metabolites in tea (Camellia sinensis) leaves under solar-withering
CsALKBH4-driven RNA demethylation can not only directly affect the accumulation of volatile terpenoids and the tea aroma formation by mediating the stability and abundance of terpenoid biosynthesis-related genes, but also indirectly regulate the contents of flavonoids, catechins, and theaflavins, as well as the tea taste formation via triggering the AS-mediated regulatory mechanism. These findings uncover a novel layer of epitranscriptomic gene regulation in tea flavor-related metabolic pathways and establish a strong link between m6A-mediated regulatory mechanism and the improvement of high-quality flavor and palatability in oolong tea. Solid arrows indicate direct regulation, and dashed arrows indicate indirect regulation. PTC, premature stop codon.
Materials and methods
Plant materials and solar-withering treatments
Fresh tea buds with the first three leaves were collected from C. sinensis cv. Tieguanyin plants cultivated in a tea plantation at Fujian Agriculture and Forestry University, Fuzhou, China (E 119°14′, N 26°05′). The harvested tea leaves were evenly divided into five groups. The first group (i.e., FL) was untreated and analyzed immediately. The other four groups were laid out on bamboo sieves for the solar-withering treatments with different shading rates. Briefly, black nylon sunshade nets were positioned 0.2 m above the bamboo sieves. The solar-withering treatments were as follows: SW1 (high shading rate provided by a sunshade net with 1500 meshes; 10,000 ± 400 lx); SW2 (moderate shading rate provided by a sunshade net with 1000 meshes; 20,000 ± 800 lx); SW3 (low shading rate provided by a sunshade net with 500 meshes; 40,000 ± 1200 lx); and SW4 (no sunshade net; 80,000 ± 2000 lx). The duration of the solar-withering treatment was 45 min. The other environmental parameters were consistent with those used in our previous study [24]. Three independent biological replicates were included in each solar-withering treatment. The light intensity and spectrum were measured using a spectral irradiance colorimeter (Catalog No. SPIC-300, Everfine Corporation, Hangzhou, China), whereas the UV intensity was recorded using a UV radiometer (Catalog No. UV340B, Sanpometer Corporation, Shenzhen, China). After the solar-withering treatments, the samples were collected, frozen immediately in liquid nitrogen, and stored at −80 °C.
Quantitative analysis of the m6A/A ratio
The global m6A/A ratio for the tea leaves was determined as previously described [59]. Briefly, RNA was extracted from each sample using the TransZol UP reagent (Catalog No. ET111, TransGen, Beijing, China). Then, the quantitative analysis of the global m6A/A ratio was performed using the EpiQuik m6A RNA methylation quantification kit (Catalog No. P-9005, Epigentek, Farmingdale, NY).
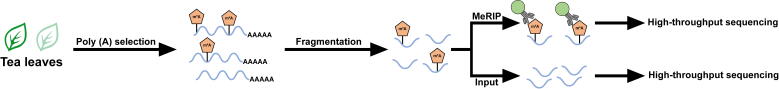
MeRIP-seq and data analysis
For MeRIP-seq, total RNA was extracted from each sample using the TransZol UP reagent (Catalog No. ET111, TransGen). The integrity and quantity of the obtained RNA were assessed by RNase-free agarose gel electrophoresis and the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). All RNA samples passed the quality checks, and there were no signs of RNA degradation (Figure S14). Next, poly(A) mRNA was isolated from the total RNA samples using the Dynabeads mRNA purification kit (Catalog No. 61,006, ThermoFisher Scientific, Waltham, CA). The enrichment of m6A was examined using the Magna MeRIP m6A kit (Catalog No. 17-10,499, Millipore, Billerica, MA). Briefly, the mRNA was fragmented into approximately 100-nt fragments using the fragmentation buffer. The fragmented RNA was divided into two groups, one of which was mixed with m6A-specific antibodies in the IP buffer. The other group of RNA was used as the input control. The m6A-containing fragments and non-IP fragments were used for constructing strand-specific libraries using the NEBNext Ultra II RNA library prep kit (Catalog No. E7770, New England Biolabs, Ipswich, MA). The average fragment size in each paired-end library was 100 ± 50 bp. Finally, m6A MeRIP-seq was performed using the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA). Three independent biological replicates were sequenced for each sample.
The raw reads containing adapter sequences and undetermined bases were removed using the Trimmomatic tool [60], after which the high-quality reads were mapped to the chromosome-level tea genome using the HISAT software [61]. The R-package exomePeak [62] was used to identify the m6A peaks in each m6A-IP sample, with the corresponding non-IP sample serving as the background. In all three independent biological replicates, peaks with an overlap of at least 50% of their length and P < 0.05 were designated as high-confidence m6A peaks. The m6A peaks were visualized using the integrative genomics viewer software [63]. According to the genomic location information, the distribution of peaks in the non-overlapping mRNA regions, including the 5′ UTR, CDS, and 3′ UTR, was determined using BEDTools [64]. The MEME suite [32] and HOMER tool [33] were used to search for motifs within the m6A peaks. The DMPs between groups were identified using the DiffBind software [65], with |FC| ≥ 2 and P < 0.05 set as the thresholds. The FPKM values were calculated to represent the mRNA expression levels in the input libraries using the StringTie software [66]. Differentially expressed genes were identified (|FC| ≥ 2 and P < 0.05) using the R package DESeq2 [67]. The expression profiles of candidate genes (standardized FPKM values) were visualized using the TBtools software [68]. The rMATS program [69] was used to detect AS events, which were considered significant if the false discovery rate was less than 0.05. The AS transcripts were validated by reverse transcription-polymerase chain reaction (RT-PCR) assays involving specific primers (Table S12) as previously described [70]. The PCR products were monitored by agarose gel electrophoresis. Differentially expressed genes associated with AS events were defined as DAGs according to previously described criteria [58]. The KEGG enrichment analysis of DMP-associated genes and DAGs was performed using a published method [25].
m6A-IP-qPCR and qRT-PCR analyses
The m6A-IP-qPCR analysis was performed according to an established procedure [11], with minor modifications. Briefly, the RNA samples used for the m6A MeRIP-seq analysis were fragmented into approximately 300-nt segments using the aforementioned fragmentation buffer. Some of the fragmented RNA samples were used for m6A-IP, which was completed using m6A-specific antibodies. The non-IP RNA was used as the input control. The m6A-containing RNA and non-IP RNA samples were reverse transcribed into cDNA using the TransScript first-strand cDNA synthesis SuperMix kit (Catalog No. AT301, TransGen). The m6A enrichment of specific mRNA regions was detected using the LightCycler 480 platform (Roche, Basel, Switzerland). The m6A abundance was quantified according to the 2−ΔΔCT method [71]. The relative abundance of specific mRNA regions in the m6A-IP sample was first normalized against that of CsActin (GenBank: HQ420251), which has no obvious m6A-modified peak and served as an internal control, and then normalized against that for the non-IP sample.
The qRT-PCR analysis was conducted using the LightCycler 480 platform as previously described [24]. The non-fragmented RNA samples were reverse transcribed into cDNA as described above. CsActin was used to normalize the mRNA expression levels. Relative mRNA levels were calculated using the 2−ΔΔCT method. The analysis was completed using three independent biological replicates. Details regarding the primers used for the m6A-IP-qPCR and qRT-PCR assays are provided in Table S12.
Analysis of volatiles by GC–MS
The volatile compounds in tea samples were extracted and analyzed as previously described [72]. The Agilent model 7890B gas chromatograph and the 7000D mass spectrometer (Agilent Technologies) were used to detect the volatile compounds. Each assay was conducted using three replicates. The detected volatiles were identified according to the retention time and mass spectra data in the National Institute of Standards and Technology Mass Spectral Library (https://www.nist.gov/srd/nist-standard-reference-database-1a/). The relative abundance of individual volatile compounds was determined on the basis of the chromatogram peak area.
Analysis of the metabolome by LC–MS
Metabolites were extracted and analyzed essentially as previously described [73]. Briefly, freeze-dried tea samples were ground into a powder using a tissue grinder (Catalog No. JXFSTPRP-24, Jingxin Company, Shanghai, China), after which 800 μl 70% methanol and 250 µl 2′,7′-dichlorofluorescein were added to the powdered material. After centrifuging the extracts at 14,000 g for 15 min, the supernatants were passed through a 0.22-µm polyvinylidene fluoride filter and analyzed using the ACQUITY two-dimensional ultra-performance liquid chromatography platform (Waters, Milford, MA) connected to a Q-Exactive quadrupole-orbitrap mass spectrometer (ThermoFisher Scientific). The compounds were separated in the Hypersil GOLD aQ column (100 mm × 2.1 mm, 1.9 μm, ThermoFisher Scientific), with 0.1% formic acid in pure water (v/v; solvent A) and 0.1% formic acid in acetonitrile (v/v; solvent B) used at a flow rate of 0.3 ml/min. The gradient elution was completed as follows: 5% solvent B for 2 min, linear increase to 95% solvent B over 22 min, 95% solvent B for 5 min, and then return to the initial condition (5% solvent B) within 3 min. The column temperature was set at 40 °C. Peaks were detected and the retention time was corrected using the compound discoverer software (version 3.1; ThermoFisher Scientific). The detected metabolites were identified by comparing their molecular mass, retention time, and mass spectrometry fragmentation patterns with those of the authentic standards, mzCloud (https://www.mzcloud.org/), and mzVault (https://mytracefinder.com/tag/mzvault/) databases. The relative abundance of each metabolite was calculated using the metaX tool [74]. The total flavonoid content of tea leaves was measured according to the aluminum chloride colorimetric method [24]. Three independent biological replicates were included in each experiment.
Gene suppression and mRNA stability assays
The gene suppression assay was performed as previously described [14]. The freshly detached tea bud and the first leaf from tea plants grown under natural conditions were added to 1.5-ml microcentrifuge tubes containing 1 ml 20 μM siRNA solution or negative control siRNA (siRNA-NC) solution. After incubating for 12 h and 24 h, the tea bud and first leaf were collected for a qRT-PCR analysis. Specific siRNAs were obtained from GenePharma (Shanghai, China). Details regarding the siRNA and siRNA-NC solutions are listed in Table S12. The overall m6A abundance and metabolite content in the gene-silenced leaves and the NC-leaves were examined as described above.
To assess mRNA stability, leaf discs were obtained from the gene-silenced tea leaves and then immersed in sterile water that was supplemented with 10 μg/ml actinomycin D (Catalog No. A1410, Sigma, St Louis, MO). Tea leaves immersed in sterile water were used as the controls. Total RNA was isolated from the leaves sampled at 6 h and 12 h, respectively. The mRNA stability was determined using a previously described qRT-PCR method [59] and specific primers (Table S12).
Statistical analysis
The correlations among the full-length transcripts, AS transcripts, and flavor metabolites were determined using the Pearson’s correlation coefficient. The differences among various groups were assessed by conducting a one-way analysis of variance (ANOVA) and Tukey’s post hoc test, with P < 0.05 set as the threshold for significance. Data are herein presented as the mean ± standard deviation (SD). The raw data for the qRT-PCR analysis are provided in Table S11.
Data availability
The raw sequencing data have been deposited in the Genome Sequence Archive [75] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: CRA006400), and are publicly accessible at https://ngdc.cncb.ac.cn/.
Conmpeting interests
The authors have declared no competing interests.
CRediT authorship contribution statement
Chen Zhu: Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Shuting Zhang: Software, Formal analysis, Data curation, Writing – original draft. Chengzhe Zhou: Validation, Investigation. Caiyun Tian: Validation, Investigation. Biying Shi: Software, Data curation. Kai Xu: Validation, Investigation. Linjie Huang: Validation, Investigation. Yun Sun: Software, Data curation. Yuling Lin: Conceptualization, Formal analysis. Zhongxiong Lai: Conceptualization, Formal analysis, Writing – review & editing, Funding acquisition, Project administration. Yuqiong Guo: Conceptualization, Methodology, Formal analysis, Investigation, Writing – review & editing, Project administration. All authors have read and approved the final manuscript.
Acknowledgments
This work was supported by the Earmarked Fund for China Agriculture Research System of Ministry of Finance and Ministry of Agriculture and Rural Affairs (Grant No. CARS-19), the Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University (Grant No. 324-1122yb070), the Scientific Research Foundation of Horticulture College of Fujian Agriculture and Forestry University (Grant No. 2019B01), the Rural Revitalization Tea Industry Technical Service Project of Fujian Agriculture and Forestry University (Grant No. 11899170145), the “Double first-class” scientific and technological innovation capacity and enhancement cultivation plan of Fujian Agriculture and Forestry University (Grant No. KSYLP004), the 6.18 Tea Industry Technology Branch of Collaborative Innovation Institute (Grant No. K1520001A), the Fujian Agriculture and Forestry University Construction Project for Technological Innovation and Service System of Tea Industry Chain (Grant No. K1520005A01), the Construction of Plateau Discipline of Fujian Province (Grant No. 102/71201801101), the Tea Industry Branch of Collaborative Innovation Institute of Fujian Agriculture and Forestry University (Grant No. K1521015A), and the Special Fund for Science and Technology Innovation of Fujian Zhang Tianfu Tea Development Foundation (Grant No. FJZTF01), China. We are grateful to Liwen Bianji (Edanz) (www.liwenbianji.cn) for its linguistic assistance.
Handled by Jianhua Yang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2023.02.003.
Contributor Information
Zhongxiong Lai, Email: 000q020029@fafu.edu.cn.
Yuqiong Guo, Email: 000q020049@fafu.edu.cn.
Supplementary material
The following are the Supplementary data to this article:
Supplementary Figure S1.
The light intensity, spectrum, and UV intensity of solar-withering with different shading rates SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight; UV, ultraviolet.
Supplementary Figure S2.
Brief workflow for MeRIP-seq The fragmented RNA was divided into two portions, one of which was enriched with an m6A-specific antibody in IP buffer. The other RNA without IP treatment was used as the input control. Finally, the two portions of RNA were sequenced using an Illumina NovaSeq 6000 platform. IP, immunoprecipitation; m6A, N6-methyladenosine; MeRIP, m6A methylated RNA immunoprecipitation.
Supplementary Figure S3.
Venn diagrams showing the overlap of m6A peaks identified from fresh leaves and four solar-withered leaves with different shading rates FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S4.
Venn diagrams showing the overlap of m6A-marked genes identified from fresh leaves and four solar-withered leaves with different shading rates FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S5.
Enrichment score of m6A peaks in five transcript segments 5′ UTR, 5′ untranslated region; CDS, coding sequence; 3′ UTR, 3′ untranslated region; FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S6.
Top sequence motifs identified within m6A peaks in tea leaves under solar-withering with different shading rates m6A, N6-methyladenosine.
Supplementary Figure S7.
Correlation between m6A modification and expression levels of the DMP-associated genes under solar-withering with different shading rates DMP-associated genes, differentially methylated peak-associated genes; m6A, N6-methyladenosine; FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S8.
m6A abundance and expression levels of DMP-associated genes under solar-withering with different shading rates based on RNA methylome and transcriptome datasets A. m6A abundance of DMP-associated genes under solar-withering with different shading rates. B. Expression levels of DMP-associated genes under solar-withering with different shading rates. The m6A abundance and expression levels of DMP-associated genes were normalized using the z-score formula, respectively. FPKM, fragments per kilobase per million mapped reads; KEGG, kyoto encyclopedia of genes and genomes; m6A, N6-methyladenosine; FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S9.
m6A abundance and expression levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS under solar-withering with different shading rates based on RNA methylome and transcriptome datasets A. m6A abundance of CsRB-L10e, CsEIF4A, CsSm, and CsDXS under solar-withering with different shading rates. B. Expression levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS under solar-withering with different shading rates. Data are presented as mean ± standard deviation. Different lowercase letters over the error bars indicate significantly different groups via one-way analysis of variance and Tukey’s post hoc test (P < 0.05). m6A, N6-methyladenosine; FPKM, fragments per kilobase per million mapped reads; FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S10.
Relative m6A enrichment and relative expression levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS in CsALKBH4A- or CsALKBH4B-silenced leaves A. Relative m6A enrichment of CsRB-L10e, CsEIF4A, CsSm, and CsDXS in CsALKBH4A- or CsALKBH4B-silenced leaves determined by m6A-IP-qPCR. B. Relative expression levels of CsRB-L10e, CsEIF4A, CsSm, and CsDXS in CsALKBH4A- or CsALKBH4B-silenced leaves determined by qRT-PCR. Data are presented as mean ± standard deviation. The differences among various groups were assessed by conducting a one-way analysis of variance and Tukey’s post hoc test. *, P < 0.05; **, P < 0.01; m6A, N6-methyladenosine; m6A-IP-qPCR, m6A-immunoprecipitation-quantitative polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction.
Supplementary Figure S11.
Expression levels of DAGs and their AS transcripts involved in flavonoid biosynthesis pathway under solar-withering with different shading rates based on transcriptome datasets Expression values of DAGs and their AS transcripts were normalized using the z-score formula. DAGs, differentially expressed alternative splicing genes; AS, alternative splicing; FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
Supplementary Figure S12.
Relative contents of flavor-related metabolites and correlation analyses of full-length transcripts, AS transcripts, and flavor metabolites A. Relative contents of flavonoids, catechins, and theaflavins under solar-withering with different shading rates determined by LC-MS. Data are presented as mean ± standard deviation. Different lowercase letters over the error bars indicate significantly different groups via one-way analysis of variance and Tukey’s post hoc test (P < 0.05). B. Correlation analyses of full-length transcripts, AS transcripts, and flavor metabolites. The correlation analyses were performed based on Pearson’s correlation coefficient. The number below the circle indicates the corresponding Pearson’s correlation coefficient. A correlation coefficient greater than 0 indicates a positive correlation, while a correlation coefficient less than 0 indicates a negative correlation. LC-MS, liquid chromatography-mass spectrometry; FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight; C, catechin; CG, catechin gallate; EC, epicatechin; ECG, epicatechin gallate; EGC, epigallocatechin; EGCG, epigallocatechin gallate; GC, gallocatechin; GCG, gallocatechin gallate; TF1, theaflavin; TF2A, theaflavin-3-gallate; TF2B, theaflavin-3′-gallate; TF3, theaflavin-3,3′-digallate; KEGG, kyoto encyclopedia of genes and genomes.
Supplementary Figure S13.
Relative expression levels of Cs4CL-a, CsF3'H-a, and CsAPX1-a, as well as the contents of flavor metabolites in AS transcript-silenced leaves A. Relative expression levels of Cs4CL-a, CsF3'H-a, and CsAPX1-a in AS transcript-silenced leaves determined by qRT-PCR. B. The contents of flavor metabolites in AS transcript-silenced leaves were determined by LC-MS. Data are presented as mean ± standard deviation. The differences among various groups were assessed by conducting a one-way analysis of variance and Tukey’s post hoc test. *, P < 0.05; **, P < 0.01; C, catechin; CG, catechin gallate; EC, epicatechin; ECG, epicatechin gallate; EGC, epigallocatechin; EGCG, epigallocatechin gallate; GC, gallocatechin; GCG, gallocatechin gallate; TF1, theaflavin; TF2A, theaflavin-3-gallate; TF2B, theaflavin-3′-gallate; TF3, theaflavin-3,3′-digallate.
Supplementary Figure S14.
RNA quality of fresh leaves and four solar-withered leaves with different shading rates Three independent biological replicates were performed for each sample. FL, fresh leaves; SW1, solar-withering with a high shading rate; SW2, solar-withering with a middle shading rate; SW3, solar-withering with a low shading rate; SW4, solar-withering with natural sunlight.
A summary of m6A-seq reads in tea leaves under solar-withering with different shading rates
The number of m6A peaks detected in m6A-marked genes
Statistics of DMPs under solar-withering with different shading rates
DMP-associated genes involved in ribosome pathway
DMP-associated genes involved in RNA transport pathway
DMP-associated genes involved in spliceosome pathway
DMP-associated genes involved in the terpenoid biosynthesis pathway
Statistics of AS events under solar-withering with different shading rates
Statistics of DAGs under solar-withering with different shading rates
DAGs involved in the flavonoid biosynthesis pathway
Raw data for qRT-PCR
Primers used in this study
References
- 1.Scarrow M., Chen N., Sun G.L. Insights into the N6-methyladenosine mechanism and its functionality: progress and questions. Crit Rev Biotechnol. 2020;40:639–652. doi: 10.1080/07388551.2020.1751059. [DOI] [PubMed] [Google Scholar]
- 2.Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 4.Wang X., Lu Z.K., Gomez A., Hon G.C., Yue Y.N., Han D.L., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pendleton K.E., Chen B.B., Liu K.Q., Hunter O.V., Xie Y., Tu B.P., et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169 doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen L.S., Liang Z., Gu X.F., Chen Y., Teo Z.W., Hou X.L., et al. N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell. 2016;38:186–200. doi: 10.1016/j.devcel.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J.Z., Manduzio S., Kang H.S. Epitranscriptomic RNA methylation in plant development and abiotic stress responses. Front Plant Sci. 2019;10:500. doi: 10.3389/fpls.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scutenaire J., Deragon J.M., Jean V., Benhamed M., Raynaud C., Favory J.J., et al. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30:986–1005. doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang D.D., Xu H.C., Liu Y., Li M.Z., Ali M., Xu X.Y., et al. RNA N6-methyladenosine responds to low-temperature stress in tomato anthers. Front Plant Sci. 2021;12:687826. doi: 10.3389/fpls.2021.687826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L.L., Tian S.P., Qin G.Z. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SIDML2 in tomato fruit ripening. Genome Biol. 2019;20:156. doi: 10.1186/s13059-019-1771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G.F., Wang J., Hou X.L. Transcriptome-wide N6-methyladenosine (m6A) methylome profiling of heat stress in pak-choi (Brassica rapa ssp. Chinensis) Plants. 2020;9:1080. doi: 10.3390/plants9091080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L.E., Tang R.K., Li X.J., Tian S.P., Li B.B., Qin G.Z. N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 2021;22:168. doi: 10.1186/s13059-021-02385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu C., Zhang S.T., Zhou C.Z., Xie S.Y., Chen G.W., Tian C.Y., et al. Genome-wide investigation of N6-methyladenosine regulatory genes and their roles in tea (Camellia sinensis) leaves during withering process. Front Plant Sci. 2021;12:702303. doi: 10.3389/fpls.2021.702303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng L.T., Zhou X.C., Su X.G., Yang Z.Y. Chinese oolong tea: an aromatic beverage produced under multiple stresses. Trends Food Sci Tech. 2020;106:242–253. [Google Scholar]
- 16.Hu C.J., Li D., Ma Y.X., Zhang W., Lin C., Zheng X.Q., et al. Formation mechanism of the oolong tea characteristic aroma during bruising and withering treatment. Food Chem. 2018;269:202–211. doi: 10.1016/j.foodchem.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Zeng L.T., Watanabe N., Yang Z.Y. Understanding the biosyntheses and stress response mechanisms of aroma compounds in tea (Camellia sinensis) to safely and effectively improve tea aroma. Crit Rev Food Sci Nutr. 2019;59:2321–2334. doi: 10.1080/10408398.2018.1506907. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y., Zeng L.T., Liu X.Y., Gui J.D., Mei X., Fu X.M., et al. Formation of (E)-nerolidol in tea (Camellia sinensis) leaves exposed to multiple stresses during tea manufacturing. Food Chem. 2017;231:78–86. doi: 10.1016/j.foodchem.2017.03.122. [DOI] [PubMed] [Google Scholar]
- 19.Lin N., Liu X.Y., Zhu W.F., Cheng X., Wang X.H., Wan X.C., et al. Ambient ultraviolet B signal modulates tea flavor characteristics via shifting a metabolic flux in flavonoid biosynthesis. J Agric Food Chem. 2021;69:3401–3414. doi: 10.1021/acs.jafc.0c07009. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L., Cao Q.Q., Granato D., Xu Y.Q., Ho C.T. Association between chemistry and taste of tea: a review. Trends Food Sci Tech. 2020;101:139–149. [Google Scholar]
- 21.Li Y., Shibahara A., Matsuo Y., Tanaka T., Kouno I. Reaction of the black tea pigment theaflavin during enzymatic oxidation of tea catechins. J Nat Prod. 2010;73:33–39. doi: 10.1021/np900618v. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Zheng P.C., Liu P.P., Song X.W., Guo F., Li Y.Y., et al. Novel insight into the role of withering process in characteristic flavor formation of teas using transcriptome analysis and metabolite profiling. Food Chem. 2019;272:313–322. doi: 10.1016/j.foodchem.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Ni T.C., Xu S.S., Wei Y.M., Li T.H., Jin G., Deng W.W., et al. Understanding the promotion of withering treatment on quality of postharvest tea leaves using UHPLC-orbitrap-MS metabolomics integrated with TMT-based proteomics. LWT. 2021;147:111614. [Google Scholar]
- 24.Zhu C., Zhang S.T., Fu H.F., Zhou C.Z., Chen L., Li X.Z., et al. Transcriptome and phytochemical analyses provide new insights into long non-coding RNAs modulating characteristic secondary metabolites of oolong tea (Camellia sinensis) in solar-withering. Front Plant Sci. 2019;10:1638. doi: 10.3389/fpls.2019.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu C., Zhang S.T., Zhou C.Z., Chen L., Zaripov T., Zhan D.M., et al. Integrated transcriptome, microRNA, and phytochemical analyses reveal roles of phytohormone signal transduction and ABC transporters in flavor formation of oolong tea (Camellia sinensis) during solar withering. J Agric Food Chem. 2020;68:12749–12767. doi: 10.1021/acs.jafc.0c05750. [DOI] [PubMed] [Google Scholar]
- 26.Ai Z.Y., Zhang B.B., Chen Y.Q., Yu Z., Chen H.C., Ni D.J. Impact of light irradiation on black tea quality during withering. J Food Sci Technol. 2017;54:1212–1227. doi: 10.1007/s13197-017-2558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y.C., He C., Yu X.L., Zhou J.T., Ran W., Chen Y.Q., et al. Effects of red-light withering on the taste of black tea as revealed by non-targeted metabolomics and transcriptomics analysis. LWT. 2021;147:111620. [Google Scholar]
- 28.Yang J., Zhou X.C., Wu S.H., Gu D.C., Zeng L.T., Yang Z.Y. Involvement of DNA methylation in regulating the accumulation of the aroma compound indole in tea (Camellia sinensis) leaves during postharvest processing. Food Res Int. 2021;142:110183. doi: 10.1016/j.foodres.2021.110183. [DOI] [PubMed] [Google Scholar]
- 29.Gu D.C., Yang J., Wu S.H., Liao Y.Y., Zeng L.T., Yang Z.Y. Epigenetic regulation of the phytohormone abscisic acid accumulation under dehydration stress during postharvest processing of tea (Camellia sinensis) J Agric Food Chem. 2021;69:1039–1048. doi: 10.1021/acs.jafc.0c07220. [DOI] [PubMed] [Google Scholar]
- 30.Xia E.H., Tong W., Hou Y., An Y.L., Chen L.B., Wu Q., et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol Plant. 2020;13:1013–1026. doi: 10.1016/j.molp.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Luo G.Z., Macqueen A., Zheng G.Q., Duan H.C., Dore L.C., Lu Z.K., et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun. 2014;5:5630. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., et al. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei L.H., Song P.Z., Wang Y., Lu Z.K., Tang Q., Yu Q., et al. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in arabidopsis. Plant Cell. 2018;30:968–985. doi: 10.1105/tpc.17.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K., Zhuang X.J., Dong Z.Z., Xu K., Chen X.J., Liu F., et al. The dynamics of N6-methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol. 2021;22:189. doi: 10.1186/s13059-021-02410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo T.L., Liu C.H., Meng F.X., Hu L., Fu X.M., Yang Z.H., et al. The m6A reader MhYTP2 regulates MdMLO19 mRNA stability and antioxidant genes translation efficiency conferring powdery mildew resistance in apple. Plant Biotechnol J. 2022;20:511–525. doi: 10.1111/pbi.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrnsberger H.F., Grasser M., Grasser K.D. Nucleocytosolic mRNA transport in plants: export factors and their influence on growth and development. J Exp Bot. 2019;70:3757–3763. doi: 10.1093/jxb/erz173. [DOI] [PubMed] [Google Scholar]
- 38.Zheng H.X., Li S.M., Zhang X.S., Sui N. Functional implications of active N6-methyladenosine in plants. Front Cell Dev Biol. 2020;8:291. doi: 10.3389/fcell.2020.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B., Tseng E., Regulski M., Clark T.A., Hon T., Jiao Y.P., et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 2016;7:11708. doi: 10.1038/ncomms11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M.J., Wang P.C., Liang F., Ye Z.X., Li J.Y., Shen C., et al. A global survey of alternative splicing in allopolyploid cotton: landscape, complexity and regulation. New Phytol. 2018;217:163–178. doi: 10.1111/nph.14762. [DOI] [PubMed] [Google Scholar]
- 41.Mi X.Z., Yue Y., Tang M.S., An Y.L., Xie H., Qiao D.H., et al. TeaAS: a comprehensive database for alternative splicing in tea plants (Camellia sinensis) BMC Plant Biol. 2021;21:280. doi: 10.1186/s12870-021-03065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G.Y., Yang J.H., Cui D.D., Zhao D.D., Benedito V.A., Zhao J. Genome-wide analysis and metabolic profiling unveil the role of peroxidase CsGPX3 in theaflavin production in black tea processing. Food Res Int. 2020;137:109677. doi: 10.1016/j.foodres.2020.109677. [DOI] [PubMed] [Google Scholar]
- 43.Zhang G.Y., Yang J.H., Cui D.D., Zhao D.D., Li Y.Y., Wan X.C., et al. Transcriptome and metabolic profiling unveiled roles of peroxidases in theaflavin production in black tea processing and determination of tea processing suitability. J Agric Food Chem. 2020;68:3528–3538. doi: 10.1021/acs.jafc.9b07737. [DOI] [PubMed] [Google Scholar]
- 44.Tong W., Li R.P., Huang J., Zhao H.J., Ge R.H., Wu Q., et al. Divergent DNA methylation contributes to duplicated gene evolution and chilling response in tea plants. Plant J. 2021;106:1312–1327. doi: 10.1111/tpj.15237. [DOI] [PubMed] [Google Scholar]
- 45.Wang L., Shi Y., Chang X.J., Jing S.L., Zhang Q.J., You C.J., et al. DNA methylome analysis provides evidence that the expansion of the tea genome is linked to TE bursts. Plant Biotechnol J. 2018;17:826–835. doi: 10.1111/pbi.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson D.C., Flematti G.R., Riseborough J.A., Ghisalberti E.L., Dixon K.W., Smith S.M. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:7095–7100. doi: 10.1073/pnas.0911635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collings E.R., Alamar M.C., Márquez M.B., Kourmpetli S., Kevei Z., Thompson A.J., et al. Improving the tea withering process using ethylene or UV-C. J Agric Food Chem. 2021;69:13596–13607. doi: 10.1021/acs.jafc.1c02876. [DOI] [PubMed] [Google Scholar]
- 48.Reape T.J., Molony E.M., Mccabe P.F. Programmed cell death in plants: distinguishing between different modes. J Exp Bot. 2008;59:435–444. doi: 10.1093/jxb/erm258. [DOI] [PubMed] [Google Scholar]
- 49.Shamala L.F., Zhou H.C., Han Z.X., Wei S. UV-B induces distinct transcriptional re-programing in UVR8-signal transduction, flavonoid, and terpenoids pathways in Camellia sinensis. Front Plant Sci. 2020;11:234. doi: 10.3389/fpls.2020.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao C.Y., Zhang C.Y., Lv Z.D., Shen C.W. Pre- and post-harvest exposure to stress influence quality-related metabolites in fresh tea leaves (Camellia sinensis) Sci Hortic. 2021;281:109984. [Google Scholar]
- 51.Huong T.T., Ngoc L.N., Kang H.S. Functional characterization of a putative RNA demethylase ALKBH6 in Arabidopsis growth and abiotic stress responses. Int J Mol Sci. 2020;21:6707. doi: 10.3390/ijms21186707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Z.H., Shi X.B., Bao M.M., Song X.Q., Zhang Y.X., Wang H.Y., et al. Transcriptome-wide analysis of RNA m6A methylation and gene expression changes among two Arabidopsis ecotypes and their reciprocal hybrids. Front Plant Sci. 2021;12:685189. doi: 10.3389/fpls.2021.685189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F., Zhang Y.C., Liao J.Y., Yu Y., Zhou Y.F., Feng Y.Z., et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019;15:e1008120. doi: 10.1371/journal.pgen.1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duan H.C., Wei L.H., Zhang C., Wang Y., Chen L., Lu Z., et al. ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell. 2017;29:2995–3011. doi: 10.1105/tpc.16.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng H.X., Sun X., Li J.L., Song Y.S., Song J., Wang F., et al. Analysis of N6-methyladenosine reveals a new important mechanism regulating the salt tolerance of sweet sorghum. Plant Sci. 2021;304:110801. doi: 10.1016/j.plantsci.2020.110801. [DOI] [PubMed] [Google Scholar]
- 56.Chen M.X., Zhang K.L., Zhang M., Das D., Fang Y.M., Dai L., et al. Alternative splicing and its regulatory role in woody plants. Tree Physiol. 2020;40:1475–1486. doi: 10.1093/treephys/tpaa076. [DOI] [PubMed] [Google Scholar]
- 57.Dvinge H. Regulation of alternative mRNA splicing: old players and new perspectives. FEBS Lett. 2018;592:2987–3006. doi: 10.1002/1873-3468.13119. [DOI] [PubMed] [Google Scholar]
- 58.Li Y.Y., Mi X.Z., Zhao S.Q., Zhu J.Y., Guo R., Xia X.B., et al. Comprehensive profiling of alternative splicing landscape during cold acclimation in tea plant. BMC Genomics. 2020;21:65. doi: 10.1186/s12864-020-6491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu J.Z., Cai J., Park S.J., Lee K., Li Y.X., Chen Y., et al. N6-methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 2021;106:1759–1775. doi: 10.1111/tpj.15270. [DOI] [PubMed] [Google Scholar]
- 60.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meng J., Lu Z.L., Liu H., Zhang L., Zhang S.W., Chen Y.D., et al. A protocol for RNA methylation differential analysis with MeRIP-seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stark R., Brown G. DiffBind: differential binding analysis of ChIP-seq peak data. Bioconductor. 2011 doi: 10.18129/B9.bioc.DiffBind. [DOI] [Google Scholar]
- 66.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C.J., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y.H., et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 69.Shen S.H., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Q.S., Zhu J.Y., Zhao S.Q., Hou Y., Li F.D., Tai Y.L., et al. Transcriptome profiling using single-molecule direct RNA sequencing approach for in-depth understanding of genes in secondary metabolism pathways of Camellia sinensis. Front Plant Sci. 2017;8:1205. doi: 10.3389/fpls.2017.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Zhang W.J., Cao J.X., Li Z.G., Li Q.H., Lai X.F., Sun L.L., et al. HS-SPME and GC/MS volatile component analysis of Yinghong No. 9 dark tea during the pile fermentation process. Food Chem. 2021;357:129654. doi: 10.1016/j.foodchem.2021.129654. [DOI] [PubMed] [Google Scholar]
- 73.Yu X.M., Xiao J.J., Chen S., Yu Y., Ma J.Q., Lin Y.Z., et al. Metabolite signatures of diverse Camellia sinensis tea populations. Nat Commun. 2020;11:5586. doi: 10.1038/s41467-020-19441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen B., Mei Z.L., Zeng C.W., Liu S.Q. MetaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics. 2017;18:183. doi: 10.1186/s12859-017-1579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen T.T., Chen X., Zhang S.S., Zhu J.W., Tang B.X., Wang A.K., et al. The Genome Sequence Archive Family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A summary of m6A-seq reads in tea leaves under solar-withering with different shading rates
The number of m6A peaks detected in m6A-marked genes
Statistics of DMPs under solar-withering with different shading rates
DMP-associated genes involved in ribosome pathway
DMP-associated genes involved in RNA transport pathway
DMP-associated genes involved in spliceosome pathway
DMP-associated genes involved in the terpenoid biosynthesis pathway
Statistics of AS events under solar-withering with different shading rates
Statistics of DAGs under solar-withering with different shading rates
DAGs involved in the flavonoid biosynthesis pathway
Raw data for qRT-PCR
Primers used in this study
Data Availability Statement
The raw sequencing data have been deposited in the Genome Sequence Archive [75] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: CRA006400), and are publicly accessible at https://ngdc.cncb.ac.cn/.