Abstract
In 2012, it was discovered that precise gene editing could be induced in target DNA using the reprogrammable characteristics of the CRISPR system. Since then, several studies have investigated the potential of the CRISPR system to edit various biological organisms. For the typical CRISPR system obtained from bacteria and archaea, many application studies have been conducted and have spread to various fields. To date, orthologs with various characteristics other than CRISPR-Cas9 have been discovered and are being intensively studied in the field of gene editing. CRISPR-Cas12 and its varied orthologs are representative examples of genome editing tools and have superior properties in terms of in vivo target gene editing compared with Cas9. Recently, TnpB and Fanzor of the OMEGA (obligate mobile element guided activity) system were identified to be the ancestor of CRISPR-Cas12 on the basis of phylogenetic analysis. Notably, the compact sizes of Cas12 and OMEGA endonucleases allow adeno-associated virus (AAV) delivery; hence, they are set to challenge Cas9 for in vivo gene therapy. This review is focused on these RNA-guided reprogrammable endonucleases: their structure, biochemistry, off-target effects, and applications in therapeutic gene editing.
Keywords: RNA-guided DNA endonucleases, CRISPR-Cas12 system, OMEGA system, genome editing
Graphical abstract

The recent identification of TnpB and Fanzor of the OMEGA system and their evolutionary relationship to the Cas12 endonuclease family pave the way for the development of novel and emerging gene editing tools with excellent properties that can be used for in vivo gene therapy.
Introduction
The CRISPR-Cas system is known as a system that recognizes and controls target nucleic acids on the basis of RNA guides as a defense mechanism against external viruses in bacteria or archaea.1,2,3,4 There are two different classes of CRISPR systems: class I includes types I, III, and IV, which require many Cas enzymes, whereas class II, such as types II, V, and VI, uses a single Cas enzyme that requires mature CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), discovered in some orthologs, for target recognition and cleavage.5 Among them, the type V CRISPR-Cas12 system is being developed for gene editing because of its ability to recognize T-rich or various protospacer adjacent motifs (PAMs) that are different from existing CRISPR-Cas9 effectors and its ability to induce target DNA cleavage very specifically.6,7,8,9 Recently, the CRISPR-Cas12 system has been further classified according to the form that exists in nature and the characteristics of processing associated nucleic acid components (Figure 1; Table 1).10 Interestingly, evolutionary analysis reveals that Cas12 and Fanzor protein found in eukaryotes may share the same transposon-encoded TnpB as their ancestor. Fanzor and TnpB belong to another RNA-guided endonuclease system called OMEGA (obligate mobile element guided activity). In this review, we characterize and compare the lineages of CRISPR-Cas12 and OMEGA endonuclease systems, discuss their structures and target cleavage mechanism, and present their promising capability for in vivo gene therapy.
Figure 1.
Phylogenetic classification of CRISPR and OMEGA endonucleases
The phylogeny shows the putative evolution of ISDra2 TnpB to CRISPR-Cas12 and Fanzor, respectively. Each CRISPR (Cas12) and OMEGA (TnpB, Fanzor) endonuclease was grouped according to the previously reported phylogenetic classification of TnpB, Cas12, and Fanzor, and Cas12 is further classified into its subtypes A–M. The shared domains of the Cas12 family are further distinguished by color. However, some domains, such as the ZF motif, NTSB, lid, and STP domains, are present only in specific subtypes. The numbers above the domains indicate the length (amino acids) of each domain of the endonuclease. Notably, TnpB, the supposed ancestor of Cas12 and Fanzor, displays the shortest length.
Table 1.
Classification of CRISPR-Cas12 subtype endonucleases
| Cas12 subtype | Origin | Length (aa) | PAM | Cleavage activity | Target substrates |
|---|---|---|---|---|---|
| Cas12a | Francisella tularensis subsp. novicida U112 | 1,300 | TTTV | cleavage | dsDNA and ssDNA |
| Cas12b | Alicyclobacillus acidoterrestris C2c1 | 1,129 | TTN | cleavage | dsDNA and ssDNA |
| Cas12c | uncultured archaeon | 1,218 | TG | no cleavage | dsDNA and ssDNA |
| Cas12e | Deltaproteobacteria | 986 | TTCN | cleavage | dsDNA |
| Cas12f | uncultured archaeon (Un1) | 529 | TTTR | cleavage | dsDNA and ssDNA |
| Cas12g | Metagenomic database | 767 | NONE | cleavage | ssRNA |
| Cas12i | Lachnospiraceae bacterium ND2006 | 1,093 | TTN | cleavage | dsDNA and ssDNA |
| Cas12k | Scytonema hofmanni | 639 | GGTT | no cleavage | dsDNA |
| Cas12j | Biggiephage | 766 | TTN | cleavage | dsDNA and ssDNA |
| Cas12m | Mycolicibacterium mucogenicum CCH10-A2 | 596 | TTN | no cleavage | dsDNA |
The Cas12 subtypes are classified according to their length (amino acids), PAM, cleavage activity, and target substrates.
Characteristics of the CRISPR-Cas12 and OMEGA effector system
Orthologs of the CRISPR-Cas12 effectors
The CRISPR-Cas12 system contains a variety of molecules, ranging from the relatively large Cas12a (1,200–1,500 amino acids [aa]) known as the CRISPR-Cas12 prototype to the recently discovered very small Cas12m (∼600 aa), Cas12n (∼500 aa), and Cas12f (400–700 aa), each of which is known to recognize a unique PAM sequence and bind to target DNA (Table 1).11,12,13,14,15 Most of these work as single effectors, but Cas12f binds to the target DNA as a homodimer and induces cleavage.15 In addition, the CRISPR-Cas12 system is divided into a group (Cas12a, Cas12b, and Cas12f) that induces double-strand breaks (DSBs) on target DNA and the Cas12c, Cas12m, and Cas12k families, which control transcription or induce transposition of target genes without double-strand DNA cleavage, depending on the presence or absence of nucleic acid cleavage ability (Table 1).13,16,17,18,19 CRISPR-Cas12 orthologs with new forms and functions have been discovered in various species of bacteria and archaea.12,20,21 On the basis of these characteristics, the advent of the CRISPR genome engineering technology has opened up endless potential applications for genome editing in living organisms.
OMEGA system: TnpB, Fanzor, and IscB effectors
Recently, a new family of RNA-guided endonucleases with a core domain similar to that of the CRISPR-Cas12 family was discovered (Figure 1; Tables 2 and 3).22,23,24 DNA endonucleases that use RNA as a guide are evolutionarily conserved, each performing various functions in vivo, from the TnpB effector found in prokaryotes to the Fanzor effector in eukaryotic organisms. TnpB and Fanzor effectors, known as the OMEGA system, are components of the transposable elements and contain a CRISPR-Cas12-like domain (RuvC) that acts as an RNA-guided endonuclease (Figure 1; Tables 2 and 3).22,23,24,25,26 In particular, TnpB enables the transposition of a specific locus by assisting the TnpA module, using the ωRNA complementary to target DNA.22 According to this RNA guidance, the DNA targeting can be reprogrammed and used extensively in genome editing (Table 2). Considering that these TnpBs have only a minimum core domain that provides the function of the CRISPR-Cas12 family, it is thought that the CRISPR-Cas12 system found in prokaryotes evolved from TnpB by inserting additional domains (Figure 1).25 The characteristics of target DNA recognition of TnpB, from the first classified ISDra2 TnpB, K, and racemifer TnpB types to the recently database-screened ISDge10, ISAam1, and ISYmu1, have been described (Table 2).27 TnpB recognizes a specific transposon-associated motif (TAM) sequence and binds to the target DNA by forming an RNA-DNA heteroduplex using ωRNA complementary to the target DNA (Table 2).
Table 2.
Classification of TnpB systems
| Selected TnpB system | Origin | Length (aa) | TAM | Cleavage activity | Target substrates |
|---|---|---|---|---|---|
| ISDra2 | Deinococcus radiodurans | 408 | TTGAT | cleavage | dsDNA and ssDNA |
| ISAba30 | Acinetobacter baumannii | 406 | TGAC | cleavage | dsDNA and ssDNA |
| ISTfu1 | Thermobifida fusca | 397 | TGAT | cleavage | dsDNA and ssDNA |
| ISDge10 | Deinococcus geothermalis | 391 | TTAT | cleavage | dsDNA and ssDNA |
| ISYmu1 | Youngiibacter multivorans | 382 | TTGAT | cleavage | dsDNA and ssDNA |
| ISAam1 | Anoxybacillus amylolyticus | 369 | TTTAA | cleavage | dsDNA and ssDNA |
Representative members of the vast TnpB family as characterized by their length (amino acids), TAM, cleavage activity, and target substrates.
Table 3.
Characteristics of Fanzor endonucleases
| Fanzor | Origin | Length (aa) | TAM | Cleavage activity | Target substrates |
|---|---|---|---|---|---|
| SpuFz1 | Spizellomyces punctatus | 638 | CATA | cleavage | dsDNA |
| GtFz1 | Guillardia theta | 690 | TTAAN | cleavage | dsDNA |
| NlovFz2 | Naegleria lovaniensis | 477 | CCG | cleavage | dsDNA |
| MmeFz2 | Mercenaria mercenaria | 478 | TAG | cleavage | dsDNA |
The length (amino acid), TAM, cleavage activity, and target substrates of the eukaryotic type Fanzor endonucleases show their homology to other OMEGA systems.
Fanzor effectors are found mainly in fungi, protists, arthropods, plants, and eukaryotic viruses and show a considerable degree of similarity with the TnpB system at the molecular level.23,26 Although the functions of the Fanzor effector in vivo have not been fully studied, it is thought that the TnpB system in prokaryotes evolved to perform similar functions in eukaryotes by transferring it to eukaryotes through gene transfer process using symbionts. Fanzor is primarily classified into Fanzor 1 and 2 types, and both types have been reported to form RNA-DNA heteroduplexes on target DNA using TAM sequence recognition and ωRNA complementary to target DNA, similar to TnpB (Table 3).23,26 The discovery of the Fanzor system indicates that RNA-guided endonucleases from prokaryotes to eukaryotes have acquired various functions at the molecular level. In addition, the discovery of many unknown functional orthologs of Fanzor-like endonucleases will enable future applications in gene editing. IscB (Insertion sequences Cas9-like OrfB) is another branch of the IS200/IS650 superfamily and also assembles with an approximately 200-nt-long ωRNA.28 Similar to TnpB and Fanzor endonucleases, IscB also has a compact size of 496 aa (OgeuIscB) and recognizes TAM. However, it shares similar domain organization (RuvC, BH, and HNH domains) and functionality and nucleic acid binding mechanism to Cas9, suggesting that it is the ancestor of Cas9.24 IscB also shows high target specificity and potential for gene editing applications.29
Comparison of domain structure and RNA-guided target DNA recognition of the CRISPR-Cas12 and OMEGA effectors
CRISPR-Cas12 effectors
The structure of the CRISPR-Cas12 family protein, Cas12a (Cpf1), was first elucidated by Yamano et al.30 in 2015. Since then, many studies have identified new CRISPR-Cas12 effectors through database searches and biochemical and structural studies.6 At a glance, like typical CRISPR-Cas9 systems, the CRISPR-Cas12 complex shows bilobed feature that is composed of a recognition (REC) lobe and nuclease (NUC) lobe (Figures 2A, 2D, and 2G).15,31,32 The REC lobe contains the WED domain, which shows defined PAM-interacting domains for some effectors, and the REC domain, which is further defined as REC1 and REC2 domains, respectively. In contrast, the NUC lobe comprises a RuvC domain containing conserved amino acid residues (D/E/D) and a nuclease domain containing a ZnF domain that directly acts on cleavage.
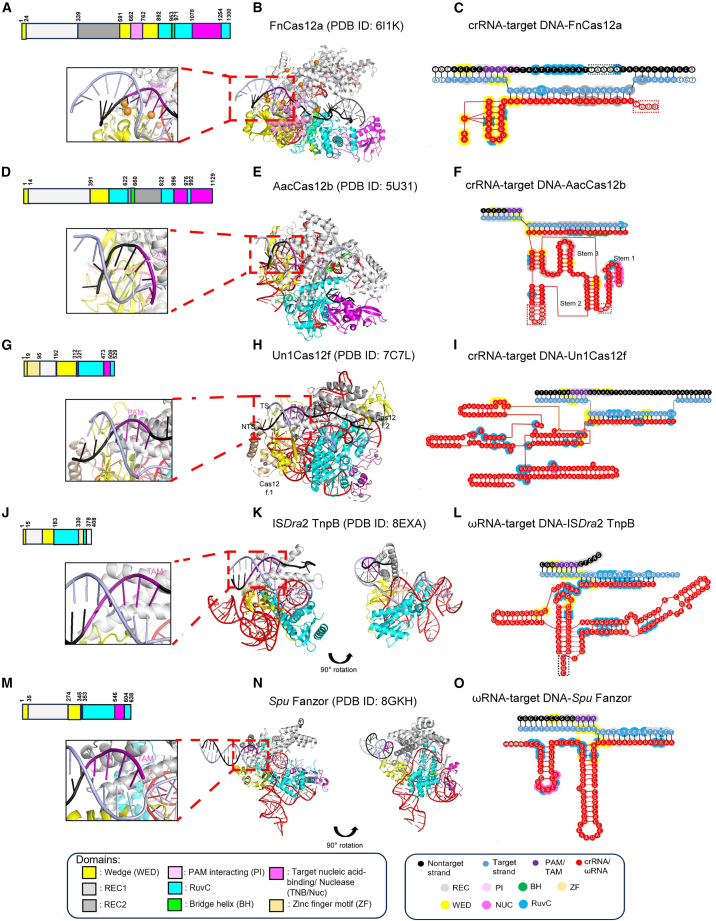
Figure 2.
Structure and interaction of RNA-target DNA complexes of CRISPR and OMEGA endonucleases
The schematics of each domain (A, D, G, J, and M), structure (B, E, H, K, and N), and interaction between DNA-RNA heteroduplex and domains (C, F, I, L, and O) of Cas12a (PDB: 6I1K), Cas12b (PDB: 5U31), Cas12f (PDB: 7C7L), ISDra2 TnpB (PDB: 8EXA), and SpuFz (PDB: 8GKH) are shown. The close-up snapshots highlight the PAM (B, E, and H, left insets) or TAM (K and N, left insets) interaction region. The structures (B, E, H, K, and N) are obtained from the PDB and illustrated using PyMOL software (PyMOL Molecular Graphics System version 2.5.4; Schrödinger, LLC.), with the colors of the domains unified across all RNA-guide endonucleases. The interactions of the DNA-RNA heteroduplex with the specific domain of the endonuclease are shown in the form of the larger circle, with colors indicating the domain (C, F, I, L, and O). The white circles indicate disordered regions in RNA or protein structure. In (K) and (N), the structures of TnpB and SpuFanzor are rotated 90° along the vertical axis to show the DNA-RNA heteroduplex interaction.
Most of the discovered Cas12 family members use the WED/REC/RuvC domain to extensively contact crRNA-tracrRNA and bind to the target DNA in a pre-ordered form.33 PAM recognition is required primarily for target DNA binding, and these Cas12 effectors are known to structurally recognize PAM sequences in target DNA using WED/REC domains.11,34,35,36,37 Cas12a is known to recognize each base of the PAM sequence using (WED/REC/PI) domains (Figures 2A–2C) through hydrogen bonding and van der Waals interactions with TTTV PAM in the target DNA (Figure 2B, inset).30 The recently identified ternary structure of Cas12b combined with the target DNA also show that the REC/WED domain region is structurally similar to that of Cas12a (Figures 2D–2F), and the positively charged groove between the REC/WED domain recognizes the TTN PAM on the target DNA (Figure 2E).32 After this binding, a structural transformation from the unlocked to the locked state occurs, and the branched form of the crRNA-DNA heteroduplex is maintained. In the recently reported case of Cas12f, PAM was recognized by the effector binding asymmetrically to the target DNA as a homodimer (Figures 2G–2I).15 At this time, only one REC.1-WED.1 domain among the homodimers was found to participate in PAM sequence recognition, and the remaining REC.2-WED.2 domains helped in the overall binding through dimer interactions (Figure 2H).
In each of the effector-crRNA-target DNA ternary complexes of the Cas12 series reported thus far, the crRNA-target DNA heteroduplex formation after PAM recognition of target DNA by each molecule showed a very similar pattern of target strand (TS) DNA branching.13,15,30,38 The DNA bending structure from the base immediately following the PAM sequence was explained by the interaction between a specific amino acid residue in the REC/WED domain and the DNA backbone (Figures 2B, 2C, 2E, 2F, 2H, and 2I). Subsequently, heteroduplex formation by complementary annealing of crRNA and the TS DNA is advantageous for R-loop propagation. The heteroduplex formed by complementary binding of crRNA and the TS DNA forms an extended form from the PAM proximal region to the distal region through interaction with the positively charged central channel domain (WED/REC/RuvC) of the Cas effector.13,38 At this time, a seed region was formed in the PAM proximal region of the heteroduplex by interaction with the amino acids constituting the channel of the Cas effector (Figures 2C, 2F, and 2I). Compared with the CRISPR-Cas9 system, the Cas12 family has been reported to show a significant decrease in cleavage when crRNA-target DNA mismatches are formed because of the introduction of mutations into these seed regions.39,40,41 This indicates that the Cas12 system has evolved to recognize the target DNA with great precision. However, as confirmed in structural studies of Cas12a and Cas12m, the Cas12 effector’s recognition of the PAM-distal region of the target DNA heteroduplex was stabilized by DNA backbone contact using the REC2 domain inserted into the WED domain (Figures 2B and 2C).42 In addition, it has been structurally shown that this type of recognition of PAM-distal region is possible even in Cas12f, which operates as a homodimer, by structural stabilization through the binding of the secondary molecule (Figures 2H and 2I).15 Overall, within the Cas12 family, there is functional conservation in a form stabilized by the interaction of the domains with the DNA backbone from the PAM proximal region to the distal region. As shown in these ternary structures (Figures 2B, 2E, and 2H), the R-loop generated by the interaction between the Cas12 effector domain and the crRNA-DNA heteroduplex appears to be very unstable.38,43 The recently identified Cas12m-crRNA-target DNA ternary complex structure explains how the non-target strand (NTS), which was repelled by the crRNA-target DNA hybridization, annealed to the TS DNA again at the PAM-distal region and closed in the form of an R-loop.42
TnpB effectors
Considering the representative ISDra2 TnpB structure among OMEGA systems, whose functions and characteristics have been identified recently, it is composed of a bilobed REC lobe and a NUC lobe, and has been reported to show the highest similarity to Cas12f of the Cas12 family (Figures 2J–2L).25,44 Through structural comparison, the endonuclease effectors of the TnpB family were found to be composed of only the core functional units of Cas12, showing a highly compact state (300–400 aa) (Table 2), and TnpB took precedence over Cas12 effectors evolutionarily (Figure 1). According to the recently identified structure of ISDra2 TnpB, the (WED/REC) domain is used to recognize the TAM sequence (5′-TTGAT-3′) of the target DNA by directly interacting with each base in the TAM sequence (Figure 2K; Table 2), and this mechanism is similar to that of the Cas12 family.25,35,36,37 In the currently identified TnpB-ωRNA-target DNA ternary structure, it can be seen that TnpB also induces R-loop formation by ωRNA-target DNA hybridization (Figure 2L).25 DNA branching is induced by the interaction of specific amino acids (K84 and Y52) in the REC domain with the base immediately after the TAM sequence, and R-loop formation is promoted by the complementary hybridization of ωRNA and target DNA. Additionally, the interaction between the WED domain and backbone phosphate stabilized the branch structure (Figure 2K, inset). The ωRNA-target DNA heteroduplex formed in this way requires a 12- to 16-bp-long ωRNA, and WED/REC/RuvC domains form 12 seed regions proximal to the TAM through hydrogen bonding and van der Waals interactions. Structural analysis revealed that the TAM distal region in this ωRNA-target DNA heteroduplex had little contact with a specific domain in the TnpB effector; therefore, it appeared highly disordered and flexible (Figure 2K). This means that compared phylogenetically, it was designed to recognize the PAM-distal region of the heteroduplex according to the acquisition of additional domains in addition to the core functional domain as it evolved from TnpB to the CRISPR-Cas12 system.42
Fanzor effectors
Structural analysis of the recently identified S. punctatus Fanzor1 (SpuFz1) endonuclease also showed that the target recognition form of RNA-guide-based endonucleases is remarkably conserved between prokaryotic and eukaryotic systems (Figures 2M–2O).23 In the structure of the SpuFz1-ωRNA-target DNA ternary complex (Figures 2N and 2O), the SpuFz1 effector is composed of a bilobed REC/NUC domain that is functionally similar to the CRISPR-Cas12 family and TnpB effectors, and uses ωRNA with a 15 nt guide and a 75 nt scaffold region to form an RNA-DNA heteroduplex within the target DNA.7,25,30,38,44 The method for recognizing the TAM nucleotide sequence (5′-CATA-3′) (Table 3) in the target DNA showed a typical TnpB-TAM sequence recognition form and induced DNA branching using the WED/REC domain. Similar to TnpB, it recognizes the TAM sequence using single-base unit interactions with specific amino acids in the WED/REC domain (Figure 2N) and stabilizes the branch structure starting from the base immediately following the base-paired TAM sequence (Figure 2O). On the basis of the structural features of these CRISPR-Cas12, TnpB, and Fanzor endonucleases, it can be seen that they recognize target DNA with a core domain conserved from prokaryotic to eukaryotic systems and have an RNA-guide-based action mechanism. They are thought to have evolved into diverse molecular forms with molecular mechanisms similar to those of various living organisms. In particular, Fanzor contains only core functional domains like TnpB in the overall domain comparison but lacks specific protein domains compared with the Cas12 effector (Figures 2A, 2D, 2G, 2J, and 2M). Evolutionarily, the functionally missing parts of these domains are supplemented with functional participation by ωRNA.
Cleavage mechanism of the CRISPR-Cas12 and OMEGA effectors
The mechanism of operation of various CRISPR systems is known to form a complex of CRISPR-Cas effectors and each crRNA-tracrRNA to have a structure favorable for target DNA recognition and to bind to a sequence complementary to an RNA guide to induce effective DNA cleavage.3,6,7,11,30,31,32,38,45,46,47,48 A series of processes in which RNA-guided endonucleases, such as the CRISPR-Cas12a system, identify target sequences in DNA and induce cleavage, have been conducted at the single-molecule level (Figure 3).49,50,51,52,53 First, the CRISPR-Cas12a effector recognizes the target sequence in DNA through an effective one-dimensional search process. Upon binding to the PAM sequence on the target DNA, crRNA triggers conformational rearrangement of the CRISPR-Cas12a effector, which consequently initiates DNA recognition and cleavage processes. In the case of CRISPR-Cas12a, the model presented for the DNA cleavage method based on the results of previous studies is unlike the CRISPR-Cas9 series, which has two HNH and RuvC cleavage domains, and uses one RuvC domain to induce double-helix cleavage by the sequential cleavage of NTS and TS (Figure 3A).41 The role of the NUC domain linked to the RuvC domain is to assist in loading the RuvC domain of the TS after NTS cleavage by the RuvC domain, so that DNA cleavage occurs sequentially.15,32,44,54 This DNA cleavage operation model is also supported by the loss of cleavage function for both the NTS and TS in studies that introduced mutations in the RuvC domain of the Cas12b effector.33 In contrast, effectors (Cas12c, Cas12m, Cas12k) that have a non-canonical form (usually bound to a single 2+ metal ion) cleavage pocket induce transcriptional silencing by binding to the target DNA instead of inducing cleavage.13,16,17,18,19,42,55,56 A recently published Cas12m showed a typical transcriptional silencing effect by inducing strong binding to the NTS using arginine-rich REC and RuvC domains.13
Figure 3.
The DNA cleavage models of the Cas12 family with targeting of dsDNA and ssDNA
(A) A sequential cis-cleavage mechanism is observed in dsDNA substrates. Target sequence recognition follows after the identification of PAM and induces conformational changes in the endonuclease. Then, the non-target strand (NTS) is cleaved by the RuvC domain (RuvC). After the release of the cleaved NTS, the NUC domain then “grabs” the target strand (TS) to bring it to the vacated RuvC domain for cleavage. (B) The indiscriminate degradation of ssDNA by the activated RuvC domain. After the cleavage of the NTS and TS, the RuvC remains active for cleaving ssDNA. NUC, nuclease domain. The figures were created using BioRender software (BioRender.com).
The recently reported RNA-guided endonucleases (ISDra2, TnpB, and SpuFanzor) of the OMEGA series also have a single RuvC domain, including typical D/E/D residues, and exhibit DNA cleavage effect.22,23,25,44 In the case of the TnpB effector, structural changes are induced by target DNA binding, resulting in the release of the RuvC domain.25 The mechanism by which the effector in the activated form cleaves the target DNA has been reported. The ternary complex of TnpB-ωRNA-target DNA shows a mechanism that enables effective DNA double-helix cleavage using the RuvC domain with a minimal functional structure compared with Cas12 family effectors.25 Specifically, the Fanzor effector is stabilized by the interaction of OMEGA RNA and the RuvC domain compared with the Cas12 family of effectors and shows an optimized form for DNA cleavage.23 This indicates that ωRNAs contribute significantly to cleavage in addition to the role of the RuvC domain evolutionarily. Interestingly, in the case of the CRISPR-Cas12 series and the ISDra2 TnpB effector, a non-specific trans-cleavage effect (non-specific single-stranded DNA [ssDNA] cleavage activity) occurred after target recognition and cleavage (Figure 3B).57,58,59 It has been reported that after guide RNA-based target recognition, structural changes are induced in each endonuclease effector, and the RuvC domain used for target DNA cleavage is still exposed to the solution used to cut ssDNA. This property has been used in many DNA-based detection technologies.59,60,61,62,63,64
Application of CRISPR-Cas12 and OMEGA effectors for genome regulation
A recent understanding of the mechanism of target DNA targeting by CRISPR-Cas12 and the OMEGA system has made it possible to edit target genomes of various living organisms. Compared with the recently discovered OMEGA system, the CRISPR-Cas12 system has relatively more research data on the targeting mechanism. Many different types of genome regulation technologies based on the CRISPR-Cas12 module have been built and applied to many living organisms. In this section, we compare the developmental aspects of gene-editing technology on the basis of CRISPR-Cas12 and OMEGA endonucleases, and the characteristics of gene editing in living organisms.
DNA-targeted editing by CRISPR-Cas12 and OMEGA effectors
CRISPR-Cas12 effectors
The CRISPR-Cas12a system, which was first studied in gene editing in the CRISPR-Cas12 family, is composed of a short crRNA and a Cas12a endonuclease protein that operates on the target DNA.11 Cas12a is relatively smaller in size than the more widely used Cas9 system, allows multiplex genome editing, and functions to expand the range of target genes by recognizing T-rich PAM instead of G-rich PAM.7,34,35,36,37,65 In fact it has been used for gene editing in various organisms successfully.21,66,67,68,69,70,71,72,73,74 Although the gene-editing efficiency of Cas12a orthologs in nature is relatively low compared with that of the Cas9 effector, it can be optimized for in vivo gene editing by specific amino acid engineering to enhance target DNA binding and activity.69,75 In addition to the effective application of the Cas12a system, various endonuclease systems of the Cas12 family have been discovered and optimized for gene editing in vivo.6
In particular, Cas12f and Cas12n effectors are very small in size and composed of bilobed forms with minimal functional domains that induce DSBs on target DNA.14,76,77,78,79,80,81 Cas12f has a robust cleavage activity although relatively lower than that of Cas9 and Cas12a.77 However, the compact size of the Cas12f effector is highly advantageous for a single AAV vector loading and results in highly efficient gene editing by effective delivery to in vivo targets.81 Delivery by adeno-associated virus (AAV) is highly effective compared with other in vivo delivery systems because it does not trigger immune response in the body. However, a single AAV can carry only up to 4.7 kb of genetic load; hence, compact Cas12 effectors are preferred than the larger SpCas9 protein.82 Recently, AAV-based gene therapy of tyrosinemia showed comparable efficacy between AAV-Cas12f and AAV-CjCas9 (previously considered the most efficient small-size effector).83 When these Cas12-type endonucleases are delivered into the cells, they specifically recognize each PAM nucleotide sequence in the target gene and induce effective indel formation (Figure 4A). In particular, if the target DNA is edited with the Cas12 endonuclease, the point at which DNA cleavage occurs appears distant from the PAM as an overhang pattern attributable to sequential cleavage induced by a single RuvC domain.81,84,85 In addition, when target DNA edited using Cas12-family endonucleases is analyzed by NGS method, a deletion pattern of random size is dominant, unlike in the Cas9 system, in which +1/−1 indels are dominant around the expected cleavage point.86 In addition to inducing indel formation in target genes, homology-directed repair can be induced through the additional transfer of a donor template using the Cas12 family of endonucleases to correct the target or insert a foreign gene.87
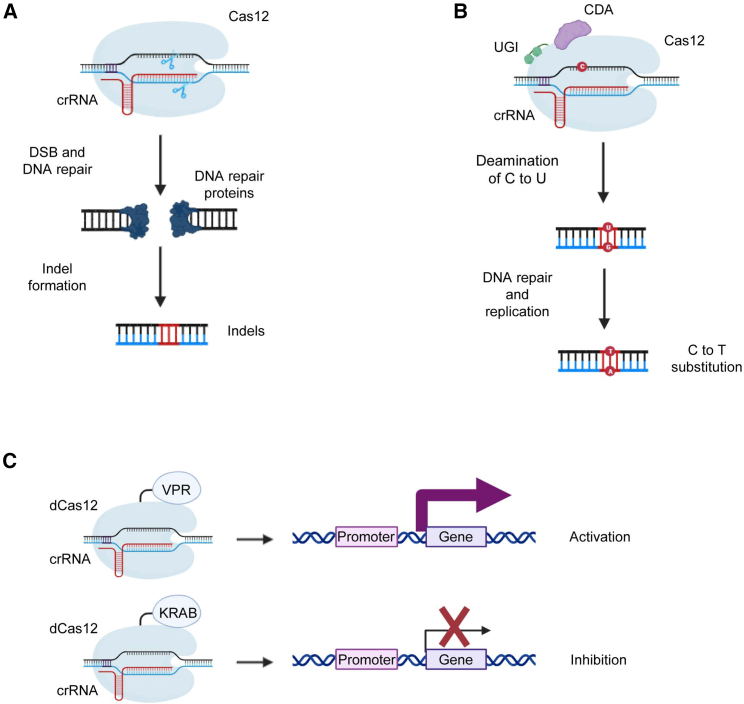
Figure 4.
Mechanisms of indel formation, base editing, and gene expression regulation that form the basis of the applications of CRISPR-Cas12
(A) Canonical Cas12a cleaves the target DNA and activates DNA double-strand break repair in the cellular system. An error-prone DNA repair system operates on the target sequence, and DNA editing in the form of insertion and deletion is induced. (B) The cytosine base editor is composed of the cytosine deaminase (CDA), uracil glycosylase inhibitor (UGI), and an effector. On the basis of the dCas12 form, which cannot induce DSB on target DNA, the base deamination domain can be conjugated and induce target base substitution within the ssDNA-exposed region of target DNA. The formation of the R-loop structure leaves the NTS susceptible to effectors such as cytosine deaminase to deaminate C to U. Because of the sequential DNA repair and replication process, the opposite nucleotide of U is converted from G to A, and finally, deaminated U is read as T. (C) Conjugating trans-activator domains such as viral protein R (VPR) or Krüppel-associated box (KRAB) to the dead form of Cas12 can enable the regulation of gene expression. VPR is a protein that, together with other transcription activators, facilitates the assembly of transcription factors and RNA polymerase. On the other hand, KRAB is an effective and widely used gene expression inhibitor. The figures were created using BioRender software (BioRender.com).
OMEGA effectors
The OMEGA system was recently discovered, and its detailed biochemical properties have been studied and applied to in vivo gene editing.24 The results of treating ISDra2 TnpB and SpuFanzor effectors in human-derived cell lines showed a typical indel pattern very similar to that of Cas12 endonucleases.22,23,25,44 Gene editing by the TnpB effector results in the formation of a typical deletion pattern induced by sticky-end DNA cleavage.22 Various deletion sizes are formed by error-prone repair centered on the cleavage point of the distal region in the TAM sequence. Among the OMEGA systems, in the case of TnpB, various ortholog effectors with gene-editing ability in the human system were recently discovered through de novo screening and characterization system.27 These TnpB effectors have a rather complex TAM sequence, but can induce chromosome-targeted DNA cleavage in human-derived cell lines, enabling effective gene editing. As most of the TnpB effector series have a small gene size, it is advantageous for in vivo delivery based on viral loading and is expected to become an excellent gene editing tool in terms of efficiency in the future through additional engineering to enhance the interaction with target DNA. Recently, Fanzor endonucleases, which are evolutionarily close and functionally similar to TnpB, were confirmed to have gene-editing abilities in human-derived cell lines.23 In particular, excellent gene-editing efficiency has been shown for various genes using Fanzor effector variants, which exhibit strong DNA interactions by introducing mutations. Because the TnpB and Fanzor endonuclease families have TAM sequences that are more complex than the PAM sequence of the CRISPR-Cas12 system, engineering the REC-WED domain required for TAM recognition is also necessary for the expansion of the targeting range.35,36,37
Base editing and transcriptional regulation without inducing DSB by using CRISPR-Cas12 effectors
With the recent development of gene-editing technology, a base editor type that substitutes only a single base without inducing double helix breaks in the target DNA or a CRISPR activation (CRISPRa)/CRISPR interference (CRISPRi) system that controls the expression level instead of gene editing has been developed (Figure 4).88,89 The CRISPR-Cas12 series has also been applied to these technologies and it is believed that the OMEGA system will be developed in this form and applied to living organisms in the near future.
Base-editing technology uses a system in which a deaminase is linked to the CRISPR module to induce single-base DNA substitutions according to target binding (Figure 4B). Starting with the cytosine (C) base editor that induces single base substitution from C to T, an adenine (A) base editor that substitutes from A to G and a trans-conversion editor have been developed and are continuously improved in terms of efficiency and applied to various living organisms.75,90,91,92,93,94,95 First, considering the case implemented in the form of base editing using the CRISPR-Cas12 system, the substitution of the target sequence was induced by linking the functional domain deaminase and uracil glycosylase inhibitor (UGI) on the basis of the Cas12a effector, such as Cas9.96 Unlike Cas9 nickase, which can induce nicking of TS and improve base substitution efficiency by base excision repair, the Cas12 base editor has no known nickase form; therefore, the base substitution efficiency was optimized in the form of a dead form that completely inhibited the activity of the RuvC domain. Starting with Cas12a-based CBE in 2018, the ABE system has also been implemented recently, enabling various base edits.97 In particular, base editors in the form of hypercompact CRISPR-Cas12 have been developed because it is difficult to develop a virus-based delivery system because of the large size of the CRISPR-Cas12a module, in which deaminase and UGI are connected.81,98 This highly compact base editor can be combined with modules with various functions, so it can be loaded into excellent delivery vehicles, such as AAV, and it is becoming a cornerstone for the development of future human-targeted treatments. In addition to the base-editing system, there is a CRISPR activator or inhibitor as an example implemented in the form of regulating the expression level of a target gene using the CRISPR-Cas12 system (Figure 4C).99,100,101,102,103 Unlike base editors that edit target gene information, trans-activator domains such as viral protein R (VPR) can be linked to the dead form CRISPR-Cas12 module to transiently regulate target gene expression.
Off-target characteristics of Cas12 and OMEGA effectors
Accuracy is an important factor for inducing gene editing in living organism.86,104,105 CRISPR-Cas12 is attracting more attention than the Cas9 system because the DNA-RNA heteroduplex region that forms the R-loop by guide RNA is very sensitive to mismatch formation, and the probability of off-target editing is low.39,40 Mismatch formation in the DNA-RNA heteroduplex region is known to form an unfavorable structure for target DNA cleavage, and most Cas12 systems discovered so far is particularly sensitive to mismatch formation in regions close to the PAM sequence, which is considered as a seed region.40,41,106 Previously, unbiased genome-wide off-target detection methods such as CIRCLE-seq, SITE-seq, CROss-seq, and Digenome-seq were used to determine the target recognition accuracy of these RNA-guide-based endonucleases.107 Using these methodologies, it is possible to accurately predict potential off-target candidates and minimize unwanted off-target editing, even for Cas12 and OMEGA systems. In particular, when using Cas12f, one of the recent studies analyzing gene editing results shows that this kind of off-target detection method effectively distinguishes small indels, deletions, and translocations generated by DSBs.77 Recently, TnpB effectors, for which biochemical studies on target DNA recognition and cleavage on the basis of endonuclease-ωRNA complex have been conducted, were surprisingly similar to CRISPR-Cas12 family members in their sensitivity to mismatch formation in the DNA-RNA heteroduplex region.25,27 In particular, TnpB effectors have seeds in the proximal region of the TAM sequence and generally have high target specificity, resulting in fewer unintended mutations at off-target sites.
Concluding remarks
A decade has passed since the discovery of mechanisms enabling the control of target genes on the basis of the reprogramming of the CRISPR-Cas9 system. The infinite potential of the CRISPR-Cas9 system for controlling biological genetic information has led to the development of next-generation gene editing tools, such as CRISPR-Cas12 and the OMEGA system, through database-based screening of bacteria and archaea. The CRISPR-Cas12 and OMEGA system discussed in this review has a high degree of target gene accuracy and unlimited potential for the future generation of transgenic animals and plants and the development of gene therapy for humans.
Currently, gene-editing technology still has room for improvement in terms of correcting genes in the context of future therapies targeting the human body, and further engineering is needed for bench-to-bedside applications.108 Especially, improvements are possible in terms of efficiency, accuracy, toxicity, and applicability in the CRISPR-Cas12 and OMEGA systems. First, when using CRISPR-Cas12 and OMEGA effectors, the efficiency of gene editing in living organisms is lower compared with the traditional Cas9 effector.23,27,44 Therefore, effective gene editing can be achieved through enhancing the target DNA binding of the effector protein itself.69,75 Furthermore, an optimized expression system can be used for effector protein and guide RNA components, tailored for efficient delivery tools such as AAV.81 Second, not only efficiency can be improved but accuracy as well. The increase in target specificity of CRISPR-Cas12 and OMEGA effectors can be possibly achieved through directed evolution, similar to the engineering of traditional CRISPR-Cas proteins.109,110 Third, addressing the issue of immunogenic control during the introduction of various organisms, including bacterial components, into the human body is essential.111,112 Introduction of engineering to effector protein and guide RNAs to control the immunity signals triggered by RNA-guided endonuclease components is possible.113 Finally, it should be possible to induce various gene edits on the basis of CRISPR-Cas12 and OMEGA effectors. For example, if an active nickase form of CRISPR-Cas12 and OMEGA effectors can be created, it would enable effective genome modifications such as base editing and prime editing, thereby making it possible to correct a variety of mutations of human diseases.88,114 In the future, thanks to the development of delivery systems such as AAV, it will be possible to create technologies that can be effectively applied to the treatment of human diseases by manipulating miniaturized and precise RNA-guided endonucleases such as CRISPR-Cas12 and OMEGA effectors. Technical advances in gene editing of this kind will provide a substantial foundation of knowledge for the development of gene therapies targeting human beings.
Acknowledgments
This research was supported by the Korean Fund for Regenerative Medicine (KFRM) grant funded by the Korea government (Ministry of Science and ICT, the Ministry of Health & Welfare) (grant 22A0203L1). This research was also supported by the Chung-Ang University Research Grants in 2022.
Author contributions
Conceptualization, I.W.B., Y.O., H.-J.K., and S.H.L.; methodology, I.W.B., Y.O., and S.H.L.; software, I.W.B., Y.O., and S.H.L.; data curation, I.W.B., Y.O., and S.H.L.; writing – original draft, I.W.B., H.-J.K., and S.H.L.; writing – review & editing, I.W.B., H.-J.K., and S.H.L.; visualization, I.W.B., Y.O., and S.H.L.; supervision, H.-J.K. and S.H.L.; project administration, H.-J.K. and S.H.L.; funding acquisition, H.-J.K. and S.H.L.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ho-Joong Kim, Email: hjkim@chosun.ac.kr.
Seung Hwan Lee, Email: lsh080390@cau.ac.kr.
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan W.X., Hunnewell P., Alfonse L.E., Carte J.M., Keston-Smith E., Sothiselvam S., Garrity A.J., Chong S., Makarova K.S., Koonin E.V., et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363:88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Safari F., Zare K., Negahdaripour M., Barekati-Mowahed M., Ghasemi Y. CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci. 2019;9:36. doi: 10.1186/s13578-019-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I., et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Shayeb B., Skopintsev P., Soczek K.M., Stahl E.C., Li Z., Groover E., Smock D., Eggers A.R., Pausch P., Cress B.F., et al. Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell. 2022;185:4574–4586.e4516. doi: 10.1016/j.cell.2022.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Tong B., Dong H., Cui Y., Jiang P., Jin Z., Zhang D. The Versatile Type V CRISPR Effectors and Their Application Prospects. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.622103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P., Zhou J., Wan Y., Liu H., Li Y., Liu Z., Wang H., Lei J., Zhao K., Zhang Y., et al. A Cas12a ortholog with stringent PAM recognition followed by low off-target editing rates for genome editing. Genome Biol. 2020;21:78. doi: 10.1186/s13059-020-01989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W.Y., Mohanraju P., Liao C., Adiego-Pérez B., Creutzburg S.C.A., Makarova K.S., Keessen K., Lindeboom T.A., Khan T.S., Prinsen S., et al. The miniature CRISPR-Cas12m effector binds DNA to block transcription. Mol. Cell. 2022;82:4487–4502.e7. doi: 10.1016/j.molcel.2022.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Chen W., Ma J., Wu Z., Wang Z., Zhang H., Fu W., Pan D., Shi J., Ji Q. Cas12n nucleases, early evolutionary intermediates of type V CRISPR, comprise a distinct family of miniature genome editors. Mol. Cell. 2023;83:2768–2780.e6. doi: 10.1016/j.molcel.2023.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Takeda S.N., Nakagawa R., Okazaki S., Hirano H., Kobayashi K., Kusakizako T., Nishizawa T., Yamashita K., Nishimasu H., Nureki O. Structure of the miniature type V-F CRISPR-Cas effector enzyme. Mol. Cell. 2021;81:558–570.e3. doi: 10.1016/j.molcel.2020.11.035. [DOI] [PubMed] [Google Scholar]
- 16.Awan M.J.A., Amin I., Mansoor S. CRISPR-Cas12c: a noncleaving DNA binder with minimal PAM requirement. Trends Biotechnol. 2022;40:1141–1143. doi: 10.1016/j.tibtech.2022.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Huang C.J., Adler B.A., Doudna J.A. A naturally DNase-free CRISPR-Cas12c enzyme silences gene expression. Mol. Cell. 2022;82:2148–2160.e4. doi: 10.1016/j.molcel.2022.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Chen W., Ren Z.H., Tang N., Chai G., Zhang H., Zhang Y., Ma J., Wu Z., Shen X., Huang X., et al. Targeted genetic screening in bacteria with a Cas12k-guided transposase. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109635. [DOI] [PubMed] [Google Scholar]
- 19.Xiao R., Wang S., Han R., Li Z., Gabel C., Mukherjee I.A., Chang L. Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition. Mol. Cell. 2021;81:4457–4466.e5. doi: 10.1016/j.molcel.2021.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zetsche B., Abudayyeh O.O., Gootenberg J.S., Scott D.A., Zhang F. A Survey of Genome Editing Activity for 16 Cas12a Orthologs. Keio J. Med. 2020;69:59–65. doi: 10.2302/kjm.2019-0009-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng F., Li J., Cui T., Xu K., Guo L., Gao Q., Feng G., Chen C., Han D., Zhou Q., Li W. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biol. 2019;20:15. doi: 10.1186/s13059-019-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karvelis T., Druteika G., Bigelyte G., Budre K., Zedaveinyte R., Silanskas A., Kazlauskas D., Venclovas Č., Siksnys V. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature. 2021;599:692–696. doi: 10.1038/s41586-021-04058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito M., Xu P., Faure G., Maguire S., Kannan S., Altae-Tran H., Vo S., Desimone A., Macrae R.K., Zhang F. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature. 2023;620:660–668. doi: 10.1038/s41586-023-06356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altae-Tran H., Kannan S., Demircioglu F.E., Oshiro R., Nety S.P., McKay L.J., Dlakić M., Inskeep W.P., Makarova K.S., Macrae R.K., et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science. 2021;374:57–65. doi: 10.1126/science.abj6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasnauskas G., Tamulaitiene G., Druteika G., Carabias A., Silanskas A., Kazlauskas D., Venclovas Č., Montoya G., Karvelis T., Siksnys V. TnpB structure reveals minimal functional core of Cas12 nuclease family. Nature. 2023;616:384–389. doi: 10.1038/s41586-023-05826-x. [DOI] [PubMed] [Google Scholar]
- 26.Bao W., Jurka J. Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob. DNA. 2013;4:12. doi: 10.1186/1759-8753-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang G., Li Y., Sun J., Huo Y., Cao S., Cao Y., Guo Y., Yang L., Cai Y., Zhang Y.E., Wang H. Evolutionary mining and functional characterization of TnpB nucleases identify efficient miniature genome editors. Nat. Biotechnol. 2023 doi: 10.1038/s41587-023-01857-x. [DOI] [PubMed] [Google Scholar]
- 28.Schuler G., Hu C., Ke A. Structural basis for RNA-guided DNA cleavage by IscB-omegaRNA and mechanistic comparison with Cas9. Science. 2022;376:1476–1481. doi: 10.1126/science.abq7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han D., Xiao Q., Wang Y., Zhang H., Dong X., Li G., Kong X., Wang S., Song J., Zhang W., et al. Development of miniature base editors using engineered IscB nickase. Nat. Methods. 2023;20:1029–1036. doi: 10.1038/s41592-023-01898-9. [DOI] [PubMed] [Google Scholar]
- 30.Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V., et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165:949–962. doi: 10.1016/j.cell.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H., Gao P., Rajashankar K.R., Patel D.J. PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell. 2016;167:1814–1828.e12. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., Chen P., Wang M., Li X., Wang J., Yin M., Wang Y. C2c1-sgRNA Complex Structure Reveals RNA-Guided DNA Cleavage Mechanism. Mol. Cell. 2017;65:310–322. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 34.Tóth E., Czene B.C., Kulcsár P.I., Krausz S.L., Tálas A., Nyeste A., Varga É., Huszár K., Weinhardt N., Ligeti Z., et al. Mb- and FnCpf1 nucleases are active in mammalian cells: activities and PAM preferences of four wild-type Cpf1 nucleases and of their altered PAM specificity variants. Nucleic Acids Res. 2018;46:10272–10285. doi: 10.1093/nar/gky815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O. Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1. Mol. Cell. 2017;67:633–645.e3. doi: 10.1016/j.molcel.2017.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimasu H., Yamano T., Gao L., Zhang F., Ishitani R., Nureki O. Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1. Mol. Cell. 2017;67:139–147.e2. doi: 10.1016/j.molcel.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao L., Cox D.B.T., Yan W.X., Manteiga J.C., Schneider M.W., Yamano T., Nishimasu H., Nureki O., Crosetto N., Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stella S., Alcón P., Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017;546:559–563. doi: 10.1038/nature22398. [DOI] [PubMed] [Google Scholar]
- 39.Kim D., Kim J., Hur J.K., Been K.W., Yoon S.H., Kim J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 40.Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swarts D.C., van der Oost J., Jinek M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell. 2017;66:221–233.e4. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omura S.N., Nakagawa R., Südfeld C., Villegas Warren R., Wu W.Y., Hirano H., Laffeber C., Kusakizako T., Kise Y., Lebbink J.H.G., et al. Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme. Nat. Struct. Mol. Biol. 2023;30:1172–1182. doi: 10.1038/s41594-023-01042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cofsky J.C., Karandur D., Huang C.J., Witte I.P., Kuriyan J., Doudna J.A. CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. Elife. 2020;9 doi: 10.7554/eLife.55143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa R., Hirano H., Omura S.N., Nety S., Kannan S., Altae-Tran H., Yao X., Sakaguchi Y., Ohira T., Wu W.Y., et al. Cryo-EM structure of the transposon-associated TnpB enzyme. Nature. 2023;616:390–397. doi: 10.1038/s41586-023-05933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirano H., Gootenberg J.S., Horii T., Abudayyeh O.O., Kimura M., Hsu P.D., Nakane T., Ishitani R., Hatada I., Zhang F., et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimasu H., Cong L., Yan W.X., Ran F.A., Zetsche B., Li Y., Kurabayashi A., Ishitani R., Zhang F., Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Globyte V., Lee S.H., Bae T., Kim J.S., Joo C. CRISPR/Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J. 2019;38 doi: 10.15252/embj.201899466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naqvi M.M., Lee L., Montaguth O.E.T., Diffin F.M., Szczelkun M.D. CRISPR-Cas12a-mediated DNA clamping triggers target-strand cleavage. Nat. Chem. Biol. 2022;18:1014–1022. doi: 10.1038/s41589-022-01082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon Y., Choi Y.H., Jang Y., Yu J., Goo J., Lee G., Jeong Y.K., Lee S.H., Kim I.S., Kim J.S., et al. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 2018;9:2777. doi: 10.1038/s41467-018-05245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui Y., Tang Y., Liang M., Ji Q., Zeng Y., Chen H., Lan J., Jin P., Wang L., Song G., Lou J. Direct observation of the formation of a CRISPR-Cas12a R-loop complex at the single-molecule level. Chem. Commun. 2020;56:2123–2126. doi: 10.1039/c9cc08325a. [DOI] [PubMed] [Google Scholar]
- 52.Son H., Park J., Hwang I., Jung Y., Bae S., Lee S. Mg(2+)-dependent conformational rearrangements of CRISPR-Cas12a R-loop complex are mandatory for complete double-stranded DNA cleavage. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2113747118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O., Bailey S., Ha T. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a) Proc. Natl. Acad. Sci. USA. 2018;115:5444–5449. doi: 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J.J., Orlova N., Oakes B.L., Ma E., Spinner H.B., Baney K.L.M., Chuck J., Tan D., Knott G.J., Harrington L.B., et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566:218–223. doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui Y., Dong H., Tong B., Wang H., Chen X., Liu G., Zhang D. A versatile Cas12k-based genetic engineering toolkit (C12KGET) for metabolic engineering in genetic manipulation-deprived strains. Nucleic Acids Res. 2022;50:8961–8973. doi: 10.1093/nar/gkac655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z., Zhong C. Cas12c-DETECTOR: A specific and sensitive Cas12c-based DNA detection platform. Int. J. Biol. Macromol. 2021;193:441–449. doi: 10.1016/j.ijbiomac.2021.10.167. [DOI] [PubMed] [Google Scholar]
- 57.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swarts D.C., Jinek M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell. 2019;73:589–600.e4. doi: 10.1016/j.molcel.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossetti M., Merlo R., Bagheri N., Moscone D., Valenti A., Saha A., Arantes P.R., Ippodrino R., Ricci F., Treglia I., et al. Enhancement of CRISPR/Cas12a trans-cleavage activity using hairpin DNA reporters. Nucleic Acids Res. 2022;50:8377–8391. doi: 10.1093/nar/gkac578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen L.T., Smith B.M., Jain P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020;11:4906. doi: 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leung R.K.K., Cheng Q.X., Wu Z.L., Khan G., Liu Y., Xia H.Y., Wang J. CRISPR-Cas12-based nucleic acids detection systems. Methods. 2022;203:276–281. doi: 10.1016/j.ymeth.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen L.T., Rananaware S.R., Pizzano B.L.M., Stone B.T., Jain P.K. Clinical validation of engineered CRISPR/Cas12a for rapid SARS-CoV-2 detection. Commun. Med. 2022;2:7. doi: 10.1038/s43856-021-00066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freije C.A., Sabeti P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe. 2021;29:689–703. doi: 10.1016/j.chom.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campa C.C., Weisbach N.R., Santinha A.J., Incarnato D., Platt R.J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods. 2019;16:887–893. doi: 10.1038/s41592-019-0508-6. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y., Long C., Li H., McAnally J.R., Baskin K.K., Shelton J.M., Bassel-Duby R., Olson E.N. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hur J.K., Kim K., Been K.W., Baek G., Ye S., Hur J.W., Ryu S.M., Lee Y.S., Kim J.S. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 2016;34:807–808. doi: 10.1038/nbt.3596. [DOI] [PubMed] [Google Scholar]
- 68.Kim Y., Cheong S.A., Lee J.G., Lee S.W., Lee M.S., Baek I.J., Sung Y.H. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 2016;34:808–810. doi: 10.1038/nbt.3614. [DOI] [PubMed] [Google Scholar]
- 69.Zhang L., Zuris J.A., Viswanathan R., Edelstein J.N., Turk R., Thommandru B., Rube H.T., Glenn S.E., Collingwood M.A., Bode N.M., et al. AsCas12a ultra nuclease facilitates the rapid generation of therapeutic cell medicines. Nat. Commun. 2021;12:3908. doi: 10.1038/s41467-021-24017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abdulrachman D., Champreda V., Eurwilaichitr L., Chantasingh D., Pootanakit K. Efficient multiplex CRISPR/Cpf1 (Cas12a) genome editing system in Aspergillus aculeatus TBRC 277. J. Biotechnol. 2022;355:53–64. doi: 10.1016/j.jbiotec.2022.06.011. [DOI] [PubMed] [Google Scholar]
- 71.Hong W., Zhang J., Cui G., Zhou Q., Wang P., Wang Y. Highly Efficient Genome Editing in Clostridium difficile Using the CRISPR-Cpf1 System. Methods Mol. Biol. 2022;2479:175–187. doi: 10.1007/978-1-0716-2233-9_12. [DOI] [PubMed] [Google Scholar]
- 72.Alok A., Sandhya D., Jogam P., Rodrigues V., Bhati K.K., Sharma H., Kumar J. The Rise of the CRISPR/Cpf1 System for Efficient Genome Editing in Plants. Front. Plant Sci. 2020;11:264. doi: 10.3389/fpls.2020.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang X., Lowder L.G., Zhang T., Malzahn A.A., Zheng X., Voytas D.F., Zhong Z., Chen Y., Ren Q., Li Q., et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants. 2017;3 doi: 10.1038/nplants.2017.103. [DOI] [PubMed] [Google Scholar]
- 74.Kim H., Kim S.T., Ryu J., Kang B.C., Kim J.S., Kim S.G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017;8 doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I., et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu T., Liu C., Zou S., Lyu R., Yang B., Yan H., Zhao M., Tang W. An engineered hypercompact CRISPR-Cas12f system with boosted gene-editing activity. Nat. Chem. Biol. 2023;19:1384–1393. doi: 10.1038/s41589-023-01380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xin C., Yin J., Yuan S., Ou L., Liu M., Zhang W., Hu J. Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption. Nat. Commun. 2022;13:5623. doi: 10.1038/s41467-022-33346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Z., Zhang Y., Yu H., Pan D., Wang Y., Wang Y., Li F., Liu C., Nan H., Chen W., Ji Q. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 2021;17:1132–1138. doi: 10.1038/s41589-021-00868-6. [DOI] [PubMed] [Google Scholar]
- 79.Xiao R., Li Z., Wang S., Han R., Chang L. Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease. Nucleic Acids Res. 2021;49:4120–4128. doi: 10.1093/nar/gkab179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karvelis T., Bigelyte G., Young J.K., Hou Z., Zedaveinyte R., Budre K., Paulraj S., Djukanovic V., Gasior S., Silanskas A., et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020;48:5016–5023. doi: 10.1093/nar/gkaa208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim D.Y., Lee J.M., Moon S.B., Chin H.J., Park S., Lim Y., Kim D., Koo T., Ko J.H., Kim Y.S. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 2022;40:94–102. doi: 10.1038/s41587-021-01009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo R., Li Z., Li G., Zhang H., Zhang C., Huo X., Zhang X., Yang X., Yang R., Liu Y., et al. In vivo treatment of tyrosinaemia with hypercompact Cas12f1. Cell Discov. 2023;9:73. doi: 10.1038/s41421-023-00554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim H., Lee W.J., Oh Y., Kang S.H., Hur J.K., Lee H., Song W., Lim K.S., Park Y.H., Song B.S., et al. Enhancement of target specificity of CRISPR-Cas12a by using a chimeric DNA-RNA guide. Nucleic Acids Res. 2020;48:8601–8616. doi: 10.1093/nar/gkaa605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim H., Lee W.J., Kim C.H., Oh Y., Gwon L.W., Lee H., Song W., Hur J.K., Lim K.S., Jeong K.J., et al. Highly specific chimeric DNA-RNA-guided genome editing with enhanced CRISPR-Cas12a system. Mol. Ther. Nucleic Acids. 2022;28:353–362. doi: 10.1016/j.omtn.2022.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang S.H., Lee W.J., An J.H., Lee J.H., Kim Y.H., Kim H., Oh Y., Park Y.H., Jin Y.B., Jun B.H., et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat. Commun. 2020;11:3596. doi: 10.1038/s41467-020-17418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vu T.V., Sivankalyani V., Kim E.J., Doan D.T.H., Tran M.T., Kim J., Sung Y.W., Park M., Kang Y.J., Kim J.Y. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol. J. 2020;18:2133–2143. doi: 10.1111/pbi.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Porto E.M., Komor A.C., Slaymaker I.M., Yeo G.W. Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 2020;19:839–859. doi: 10.1038/s41573-020-0084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kampmann M. CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 2018;13:406–416. doi: 10.1021/acschembio.7b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A∗T to G∗C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tong H., Wang X., Liu Y., Liu N., Li Y., Luo J., Ma Q., Wu D., Li J., Xu C., Yang H. Programmable A-to-Y base editing by fusing an adenine base editor with an N-methylpurine DNA glycosylase. Nat. Biotechnol. 2023;41:1080–1084. doi: 10.1038/s41587-022-01595-6. [DOI] [PubMed] [Google Scholar]
- 93.Musunuru K., Chadwick A.C., Mizoguchi T., Garcia S.P., DeNizio J.E., Reiss C.W., Wang K., Iyer S., Dutta C., Clendaniel V., et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 94.Chiesa R., Georgiadis C., Syed F., Zhan H., Etuk A., Gkazi S.A., Preece R., Ottaviano G., Braybrook T., Chu J., et al. Base-Edited CAR7 T Cells for Relapsed T-Cell Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2023;389:899–910. doi: 10.1056/NEJMoa2300709. [DOI] [PubMed] [Google Scholar]
- 95.Koblan L.W., Erdos M.R., Wilson C., Cabral W.A., Levy J.M., Xiong Z.M., Tavarez U.L., Davison L.M., Gete Y.G., Mao X., et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li X., Wang Y., Liu Y., Yang B., Wang X., Wei J., Lu Z., Zhang Y., Wu J., Huang X., et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 2018;36:324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 97.Chen F., Lian M., Ma B., Gou S., Luo X., Yang K., Shi H., Xie J., Ge W., Ouyang Z., et al. Multiplexed base editing through Cas12a variant-mediated cytosine and adenine base editors. Commun. Biol. 2022;5:1163. doi: 10.1038/s42003-022-04152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y., Qi T., Liu J., Yang Y., Wang Z., Wang Y., Wang T., Li M., Li M., Lu D., et al. A highly specific CRISPR-Cas12j nuclease enables allele-specific genome editing. Sci. Adv. 2023;9 doi: 10.1126/sciadv.abo6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang X., Wang J., Cheng Q., Zheng X., Zhao G., Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3 doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramesh A., Ong T., Garcia J.A., Adams J., Wheeldon I. Guide RNA Engineering Enables Dual Purpose CRISPR-Cpf1 for Simultaneous Gene Editing and Gene Regulation in Yarrowia lipolytica. ACS Synth. Biol. 2020;9:967–971. doi: 10.1021/acssynbio.9b00498. [DOI] [PubMed] [Google Scholar]
- 101.Choi J., Bae T., Byambasuren N., Park S.H., Jo C.H., Kim D., Hur J.K., Hwang N.S. CRISPR-Cpf1 Activation of Endogenous BMP4 Gene for Osteogenic Differentiation of Umbilical-Cord-Derived Mesenchymal Stem Cells. Mol. Ther. Methods Clin. Dev. 2020;17:309–316. doi: 10.1016/j.omtm.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Magnusson J.P., Rios A.R., Wu L., Qi L.S. Enhanced Cas12a multi-gene regulation using a CRISPR array separator. Elife. 2021;10 doi: 10.7554/eLife.66406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu Y., Wan X., Wang B. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat. Commun. 2019;10:3693. doi: 10.1038/s41467-019-11479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shi L., Tang X., Tang G. GUIDE-Seq to Detect Genome-wide Double-Stranded Breaks in Plants. Trends Plant Sci. 2016;21:815–818. doi: 10.1016/j.tplants.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 105.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pacesa M., Lin C.H., Clery A., Saha A., Arantes P.R., Bargsten K., Irby M.J., Allain F.H., Palermo G., Cameron P., et al. Structural basis for Cas9 off-target activity. Cell. 2022;185:4067–4081.e4021. doi: 10.1016/j.cell.2022.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tao J., Bauer D.E., Chiarle R. Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing. Nat. Commun. 2023;14:212. doi: 10.1038/s41467-023-35886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zaib S., Saleem M.A., Khan I. CRISPR-Cas9 Genome Engineering: Trends in Medicine and Health. Mini Rev. Med. Chem. 2022;22:410–421. doi: 10.2174/1389557521666210913112030. [DOI] [PubMed] [Google Scholar]
- 109.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mehta A., Merkel O.M. Immunogenicity of Cas9 Protein. J. Pharm. Sci. 2020;109:62–67. doi: 10.1016/j.xphs.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tang X.Z.E., Tan S.X., Hoon S., Yeo G.W. Pre-existing adaptive immunity to the RNA-editing enzyme Cas13d in humans. Nat. Med. 2022;28:1372–1376. doi: 10.1038/s41591-022-01848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim S., Koo T., Jee H.G., Cho H.Y., Lee G., Lim D.G., Shin H.S., Kim J.S. CRISPR RNAs trigger innate immune responses in human cells. Genome Res. 2018;28:367–373. doi: 10.1101/gr.231936.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen P.J., Liu D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2023;24:161–177. doi: 10.1038/s41576-022-00541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]