Abstract
Background
Heart failure (HF) is a progressive condition with high global incidence. HF has two main subtypes: HF with preserved ejection fraction (HFpEF) and HF with reduced ejection fraction (HFrEF). There is an inherent need for simple yet effective electrocardiogram (ECG)-based artificial intelligence (AI; ECG-AI) models that can predict HF risk early to allow for risk modification.
Objective
The main objectives were to validate HF risk prediction models using Multi-Ethnic Study of Atherosclerosis (MESA) data and assess performance on HFpEF and HFrEF classification.
Methods
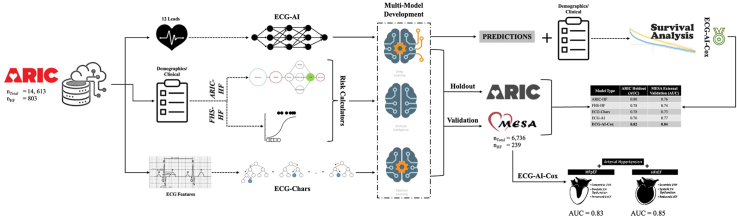
There were six models in comparision derived using ARIC data. 1) The ECG-AI model predicting HF risk was developed using raw 12-lead ECGs with a convolutional neural network. The clinical models from 2) ARIC (ARIC-HF) and 3) Framingham Heart Study (FHS-HF) used 9 and 8 variables, respectively. 4) Cox proportional hazards (CPH) model developed using the clinical risk factors in ARIC-HF or FHS-HF. 5) CPH model using the outcome of ECG-AI and the clinical risk factors used in CPH model (ECG-AI-Cox) and 6) A Light Gradient Boosting Machine model using 288 ECG Characteristics (ECG-Chars). All the models were validated on MESA. The performances of these models were evaluated using the area under the receiver operating characteristic curve (AUC) and compared using the DeLong test.
Results
ECG-AI, ECG-Chars, and ECG-AI-Cox resulted in validation AUCs of 0.77, 0.73, and 0.84, respectively. ARIC-HF and FHS-HF yielded AUCs of 0.76 and 0.74, respectively, and CPH resulted in AUC = 0.78. ECG-AI-Cox outperformed all other models. ECG-AI-Cox provided an AUC of 0.85 for HFrEF and 0.83 for HFpEF.
Conclusion
ECG-AI using ECGs provides better-validated predictions when compared to HF risk calculators, and the ECG feature model and also works well with HFpEF and HFrEF classification.
Keywords: Heart failure, Artificial intelligence, ECG-AI, ECG-AI-Cox, HFpEF, HFrEF
Graphical abstract
Introduction
Heart failure (HF) is a progressive and complex condition with high morbidity, high mortality, and increasing global incidence.1 HF results from structural and/or functional cardiac changes2 and manifests as at least 2 major phenotypes of left ventricular ejection fraction, namely HF with reduced ejection fraction (HFrEF) and HF with preserved ejection fraction (HFpEF). Early diagnosis of HF is an important component for precision medicine and early treatment of HF.
Artificial intelligence (AI) models have been developed to predict risk of abnormal heart conditions3,4 using several risk factors. Research also tried to improve the models by incorporating electrocardiogram (ECG) data.5, 6, 7 Most AI research in the prediction of HF does not specify the time window of prediction, or it is within a short time period.2,8,9 Risk calculators based on demographic and comorbidity data have been developed to predict risk of developing HF.2,10 Studies have also used traditional ECG features/characteristics, translated from an ECG waveform.5 ECG features provide information on aspects of ECGs, eg, amplitude, duration, angles, and axes of waves.5,11 Kwon and colleagues6 showed that using a large number of ECG characteristics can be important in detecting HF using AI. However, this study was limited to the detection of prevalent HF rather than time-dependent risk prediction, which is necessary for early assessment.12 Therefore, using AI on raw waveform ECG as a time-voltage can reliably predict risk of HF.13,14
In our previous research,7 we used the Atherosclerosis Risk in Communities (ARIC) study data within an AI framework to show using raw 12-lead ECGs, which are readily collected in most healthcare settings, can predict 10-year HF risk. A Cox proportional hazards (CPHs) model, called ECG-AI-Cox, was also developed and showed improvement in risk prediction when compared to other available calculators based on demographic and clinical variables (eg, ARIC and Framingham Heart Study [FHS]).10
In addition, we aimed to develop (on ARIC data), validate (on Multi-Ethnic Study of Atherosclerosis [MESA] data), and compare a model called ECG-Chars, built using ECG features (rather than waveform ECG data). Research by Kwon and colleagues6 and Khurshid and colleagues15 provided insight into the use of a large number of ECG characteristics, in combination with clinical variables, and their importance in detecting heart failure using both deep and machine learning algorithms. Since such features are captured within the signal of the ECG, it is plausible that using AI on raw waveform ECG as a time-voltage can be an asset to the reliable prediction of HF, reducing the overall burden of extracting and selecting features from ECGs.
We hypothesized that ECG-based AI models are generalizable and can predict HF risk within 10 years with similar or better accuracy than current HF risk calculators or those that use traditional ECG features. This research has multiple aims. Primarily, we aimed to validate the developed models (ECG-AI, ECG-AI-Cox, ECG-Chars, ARIC-HF, and FHS-HF) using data obtained from the MESA cohort study.16
Furthermore, this study aimed to assess the prediction performance for HF subtypes, such as HFpEF and HFrEF.
Methods
ARIC and MESA cohorts
The ARIC study is an ongoing prospective epidemiologic study initiated in 1987 and conducted in 4 communities in the United States (Forsyth County, NC; Jackson, MS; Washington County, MD; and the northwest suburbs of Minneapolis, MN). The initial cohort size was 15,792. From the 15,792 participants in the ARIC cohort study, 1179 (7.5%) participants were excluded from the study. From these, 739 had HF at baseline and 440 (2.8% of the total number of participants) had missing ECGs. After exclusion, the dataset contained 14,613 participants.
MESA is an ongoing research study that includes 6814 participants16 over, to date, 6 examinations. Exam 1, data from which was used in this study, occurred between July 2000 and August 2002. MESA participants were free of clinically recognized cardiovascular disease at exam 1. In the MESA dataset, 78 participants (1.1% of the total number of participants) had missing baseline ECGs and were therefore excluded from the study. After exclusion, the final cohort included 6736 participants. For both the ARIC and MESA cohorts, incident HF was confirmed by an adjudication committee. Within the MESA study, ejection fraction (EF) values to define HFrEF (EF <50%; n = 119) and HFpEF (EF ≥50%; n = 103) were recorded at the time of HF diagnosis from a clinical echocardiogram or medical record review.17
Outcome
The main aims of this study were to compare the ECG-AI deep learning models to risk calculators (ARIC-HF, FHS-HF, and CPH) and a model using ECG features (ECG-Chars) in predicting 10-year risk for HF, externally validate all the models, and assess the best-performing model’s prediction performance for HFpEF and HFrEF classification. Within the ARIC study, HF was defined when diagnostic symptoms, such as onset/worsening of shortness of breath, edema, hypoxia, etc, were apparent at first hospitalization or from information extracted from a death certificate specifying HF (ICD-9/10 codes).10,18 In the MESA study, HF was sub-categorized as probable or definite HF.3 Probable HF required diagnosis by physicians of symptoms, including shortness of breath or edema, while definite HF required diagnosis of 1 or more objective criteria, including pulmonary edema, dilated ventricle, or poor left ventricular (LV) function or evidence of LV diastolic dysfunction.3 Following HF adjudication, HFpEF and HFrEF are typically adjudicated based on the EF obtained during hospitalization with incident HF.
Summary of previously developed ECG-AI, ECG-AI-Cox, and clinical models ARIC-HF and FHS-HF
ECG-AI7 used raw digital supine 12-lead ECG data at 500 Hz of 10 seconds (time-voltage) obtained from the ARIC study was used to develop a convolutional neural network by adapting the ResNet architecture, which was originally developed for 2-dimensional image data, for 1-dimensional ECG signals.19 From the 10-second ECG, the first second was removed to reduce introduction of noise (eg, when setting up the electrodes or movement by the participant). Briefly, the ECG-AI model is built on 64 layers and uses the Leaky Rectified Linear Unit activation function20 to optimize the model by activating neurons with negative inputs in the data. Dropout layers with a rate of 0.1 were introduced in blocks to develop an architecture with more generalized performance and reduce risk of overfitting. The Adam21 optimizer was used with its default hyper-parameters (beta_1 = 0.9, beta_2 = 0.999, and learning rate = 0.001) and a batch size of 128.
Training of the ECG-AI model was originally performed on the ARIC dataset, with 80% of the data used, with 5-fold cross-validation, while the remaining 20% was used as a holdout dataset. Results from this 20% holdout are used to compare performance on the MESA dataset as part of this study.
The ARIC-HF and FHS-HF risk calculators used demographic and clinical variables. ARIC-HF used a Cox regression model12 and FHS-HF used a standard pooled logistic regression model.2 For full comparison, we also developed another Cox regression model (called CPH) that included all 12 variables used in ARIC-HF and FHS-HF. The variables inputted into each developed model are provided in Table 1. The ECG-AI-Cox model was developed by incorporating the ECG-AI risk prediction output plus 12 clinical variables, listed in Table 1, within a CPH regression to assess HF hazard and survivability.
Table 1.
Model details: summary of the models developed, method used, and input types
| Model type | Method | Input data |
|---|---|---|
| ECG-Chars | Light Gradient Boosting Machine | 288 ECG characteristics as inputs |
| ARIC-HF risk calculator | Cox regression | Age, sex, race, BMI, heart rate, systolic blood pressure, diabetes mellitus, smoking status, hypertension |
| FHS-HF risk calculator | Standard pooled logistic regression | Age, BMI, heart rate, systolic blood pressure, diabetes mellitus, coronary heart disease, left ventricular hypertrophy, valvular disease |
| CPH | Cox regression | Age, sex, race, BMI, heart rate, systolic blood pressure, diabetes mellitus, coronary artery disease, smoking status, hypertension, left ventricular hypertrophy, valvular disease |
| ECG-AI | Convolutional neural network | 12-Lead raw ECG data |
| ECG-AI-Cox | Cox regression | Risk factors from CPH plus ECG-AI output |
BMI = body mass index; ECG = electrocardiogram.
ECG characteristics data
The ECG-Chars Light Gradient Boosting Machine model included 288 waveform ECG features related to duration and amplitude of the P, Q, R, S, and T waves for each of 12 leads, from the ARIC data. Furthermore, the duration of the QRS complex; the minimum and maximum amplitude of the ST segment; the ST elevation; ST level at the J point; middle of the ST segment range; the P-, R-, and T-wave axis; the PR, QT interval (and corrected QT), and the QT index (derived as the % of QT prolongation) were among the features included. These features are represented within the waveform and, therefore, including such features within a machine learning model makes for a good comparative model.
Both waveform data and the used ECG features were direct exports from GE MUSE v7. In addition, these 288 features are the more commonly exported features and were available from both the ARIC and MESA studies.
Study design
Validation of the ECG-AI, ARIC-HF, FHS-HF, and ECG-AI-Cox models was performed on the MESA dataset. The ECG-AI model was first deployed on the MESA data and the outputted predicted HF risk was included with the 12 clinical variables extracted from the MESA dataset. The ECG-AI-Cox model was then validated. Separately, these 12 variables were included in the Cox regression model (CPH) to compare accuracy of models with and without the ECG component. The ECG-Chars model was built on the ARIC data following the same methodology, ie, 80% data used for training, with 5-fold cross-validation, and 20% retained as holdout data,7 and then validated on the MESA data. A variable importance analysis was performed on ECG-Chars using SHAP (Shapley Additive Explanations).22 DeLong tests were performed to statistically compare all the models. Subgroup analyses were also performed (described below).
The Python programming language was used for all analyses. The Python code associated with this research is available on GitHub (https://github.com/ikarabayir/ECG_AI_HF).
Validation of the developed models using the MESA dataset
Validation evaluation was based on area under the receiver operating characteristics curve. The validation process required that all the MESA participant data, ie, demographics, comorbidities/clinical variables, and ECGs, were available from exam 1 and in the same format as the data used to build the original models. Only good-quality ECGs from the MESA dataset were used. The ECG quality evaluation and ranking at the ECG Reading Center (EPICARE) is conducted using an automated system that also includes visual confirmation. Four quality grades are used: 0 (excellent quality); 1 and 2 (ECGs with some quality issues but not significant to affect reading), which are automatically assigned by the GE-MUSE system; and 5, which indicates significant quality issues that can interfere with accurate automatic reading and is manually decided by EPICARE staff. No quality grades 3 or 4 are used. In cohort studies, such as ARIC and MESA, annotation of poor-quality ECG is triggered when over 5% of the ECGs have a quality grade of 5.
Subgroup analyses
A subgroup analysis of sex, race, HF subtypes (HFpEF and HFrEF), and, for those with HF, risk factors vs risk-free groups was performed on the MESA dataset using the best-performing prediction model. For the latter, the HF risk-free category was defined as participants who did not have any of the following risk factors or clinical conditions: smoking, coronary heart disease, diabetes, hypertension, LV hypertrophy, and valvular disease.
Since this research aimed to develop a generalizable AI-based model, we wanted to ensure that it performs well on all sub-demographic groups of sex and race, especially since, for the latter, the ARIC participants were of White and African American race while the MESA dataset also included participants of Chinese-American and of Hispanic ethnicity. In addition, because the models were trained on HF subgroup analysis on a composite HF outcome, subtypes would also indicate whether the model is better at classifying one subtype over the other, or whether it can accurately classify both HFrEF and HFpEF, increasing the clinical applicability of this model. From a total of 239 MESA participants who developed HF, ∼48% of participants had HFrEF and ∼43% had HFpEF. From a total of 239 MESA participants who developed HF, ∼48% of participants had HFrEF and ∼43% had HFpEF. Although there is a slight imbalance between participants with HFrEF and HFpEF, HFpEF is typically diagnosed at approximately 50% of all HF cases while HFrEF is approximately 40% of HF cases.23,24 Therefore, this imbalance was deemed acceptable and no further strategies were taken to counteract this.
Exploratory analysis on only lead I ECG
There is a fast-growing industry around ECG-enabled wearables and literature suggests the utility of remotely collected single-lead ECG. As an exploratory study, we also built a lead I–only version of the original 12-lead-based ECG-AI model on ARIC data and validated on MESA data. In this exploratory work, lead I was selected as it is the ECG lead that is typically mimicked by wearable devices or single-lead ECG devices, such as smartwatches (eg, Apple Watch and Samsung Watch).
The Institutional Review Board approval was obtained at all participating institutions. All ARIC and MESA participants had given written informed consent.
Results
Clinical characteristics
In the ARIC cohort, 45% were male, 36% were African American, and 64% were White with a mean age ± standard deviation of 54.1 ± 5.8 years. In this cohort, 5.5% of the participants had developed HF within the first 10 years of assessment. The MESA cohort included 47.2% male, 27.8% African American, 38.5% White, 11.8% Chinese American, and 22.0% Hispanic ethnicity. The mean age ± standard deviation of MESA participants was 62.2 ± 10.2 years. In this cohort, 239 (3.6%) participants had developed HF within the first 10 years from exam 1. The demographics and clinical characteristics of both ARIC and MESA datasets are provided and statistically compared in Table 2. Overall, the MESA cohort was more racially diverse, with a higher mean age across the population.
Table 2.
ARIC and MESA cohort demographics
| ARIC |
MESA |
P† | |||
|---|---|---|---|---|---|
| No HF in 10 years (n = 13,810) | HF in 10 years(n = 803) | No HF in 10 years(n = 6497) | HF in 10 years(n = 239) | ||
| Sex (male) | 6179 (44.7) | 456 (57.2) | 3037 (46.7) | 144 (60.3) | <.05 |
| Race (African American) | 3559 (25.8) | 289 (36.0) | 1790 (27.6) | 80 (33.5) | <.05 |
| Age at visit 1 (years) | 53.9 (5.7) | 57.2 (5.2) | 61.91 (10.2) | 68.68 (8.8) | <.05 |
| BMI (kg/m2) | 27.4 (5.2) | 29.5 (6.3) | 28.3 (5.5) | 30.0 (6.0) | <.05 |
| Smoking status | <.05 | ||||
| Former | 4407 (31.9) | 284 (35.4) | 2361 (36.3) | 100 (41.8) | |
| Current | 3485 (25.2) | 304 (37.9) | 844 (13.0) | 36 (15.1) | |
| Prevalent coronary heart disease | 458 (3.3) | 138 (17.2) | NA‡ | NA‡ | - |
| Diabetes mellitus | 1326 (9.6) | 286 (35.6) | 591 (9.1) | 64 (26.8) | <.05 |
| Systolic blood pressure (mm Hg) | 120.5 (18.4) | 131.2 (22.9) | 127.6 (21.5) | 139.6 (24.6) | <.05 |
| Hypertension medication | 3566 (25.8) | 420 (52.3) | 2361 (36.3) | 144 (60.3) | <.05 |
| Left ventricular hypertrophy | 253 (1.9) | 50 (6.4) | 55 (0.8) | 12 (5.0) | <.05 |
| Valvular disease | 33 (0.2) | 9 (1.1) | NA‡ | NA‡ | - |
| Heart rate (ventricular, beats/min) | 66.4 (10.0) | 70.5 (12.3) | 63.0 (9.6) | 65.3 (10.6) | <.05 |
Data are n (%) or mean (SD).
BMI = body mass index.
P values are results from the comparison of the entire ARIC study data to the entire MESA study data.
Variable imputed with 0, representing that this factor was not present at baseline.
HF prediction using ECG characteristics (ECG-Chars) and comparison with the ECG-AI model
The area under the receiver operating characteristic curve (AUC) of the ECG-Chars model on the ARIC holdout data was 0.78 (0.74–0.83), with a sensitivity of 0.66 and specificity 0.70. This model performed slightly better on the ARIC holdout data compared to the ECG-AI model, which had an AUC of 0.76 (0.72–0.80; Table 3). The DeLong test comparing the AUCs of the ECG-AI and the ECG-Chars model resulted in a significant difference, with a P value = .523 between the 2 models.
Table 3.
Area under receiver operating characteristic curve results for each model tested on ARIC data and validated on MESA data
| Model type | ARIC holdout data | MESA external validation |
|---|---|---|
| ECG-Chars† | 0.78 (0.74–0.83) | 0.73 (0.70–0.77) |
| ARIC-HF risk calculator‡ | 0.80 (0.75–0.85) | 0.76 (0.72–0.80) |
| FHS-HF risk calculator§ | 0.78 (0.74–0.83) | 0.74 (0.70–0.78) |
| CPH‖ | 0.81 (0.78–0.84) | 0.78 (0.75–0.80) |
| ECG-AI¶ | 0.76 (0.72–0.80) | 0.77 (0.74–0.79) |
| ECG-AI-Cox# | 0.82 (0.78–0.86) | 0.84 (0.81–0.87) |
Comparison of HF prediction models AUC (95% CI) developed on ARIC holdout data7 and the corresponding AUC (95% CI) results from validation on the MESA dataset.
Light Gradient Boosting Machine model using 288 ECG characteristics as inputs (called ECG-Chars).
ARIC-HF risk calculator using age, body mass index, heart rate, systolic blood pressure, diabetes mellitus, sex, race, smoking status, hypertension as inputs.
FHS-HF risk calculator using age, body mass index, heart rate, systolic blood pressure, diabetes mellitus, coronary heart disease, left ventricular hypertrophy, valvular disease as inputs.
Cox regression model using 12 risk factors from ARIC-HF and FHS-HF.
Convolutional neural network model using raw electrocardiogram data as inputs.
Cox regression model using the output of ECG-AI and 12 risk factors from ARIC-HF and FHS-HF risk calculators as inputs.
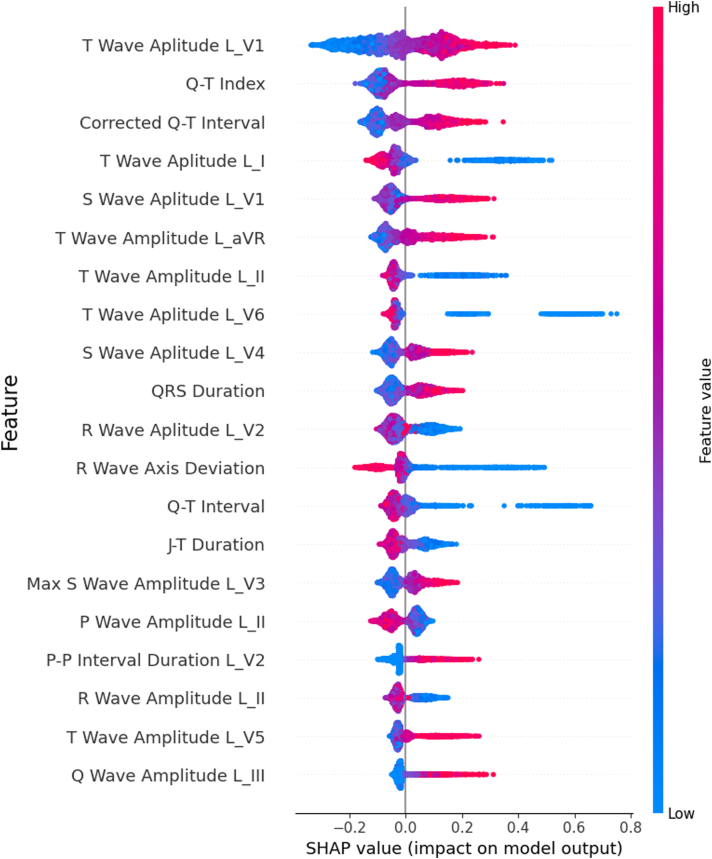
SHAP feature importance analysis of ECG-Chars on the ARIC holdout dataset
Figure 1 shows that the amplitude of the lead V1 T wave, the QT index, corrected QT interval, the amplitude of the lead I T-wave I, and the lead V1 S-wave amplitude are among the most important predictors. A higher T-wave amplitude in lead V1, QT interval, and lead V1 S-wave amplitude show association with higher influence on prediction of 10-year HF. On the other hand, lower values of T-wave amplitude from lead I, lead II, and lead V6 are associated with higher predictive importance.
Figure 1.
SHAP (Shapley Additive Explanations) variable importance analysis for the ECG-Chars model: variable importance analysis for the top 20 variables that attribute to the prediction of heart failure using 288 electrocardiogram features. L denotes “Lead,” followed by the lead type. “QRS” refers to the QRS complex.
Validation of the developed models using the MESA dataset
Table 3 provides AUCs of each model on the ARIC holdout data and the MESA data validation results. The ARIC-HF and FHS-HF risk calculators resulted in AUCs of 0.80 (0.75–0.85) and 0.78 (0.74–0.83), respectively, on the ARIC holdout data. On the MESA data, ARIC-HF and FHS-HF resulted in AUC of 0.76 (0.72–0.80) and 0.74 (0.70–0.78), respectively. The CPH model showed slightly higher AUC on both the ARIC holdout data and the MESA validation data (AUC = 0.81 and 0.78, respectively). The accuracy of the ECG-AI model was higher on the MESA validation data with AUC = 0.77 (0.74–0.79), compared to ARIC-HF, FHS-HF, and CPH. Furthermore, MESA validation of the ECG-Chars model showed lowest accuracy (AUC = 0.73). ECG-AI and ECG-Chars on the MESA dataset showed significant difference (DeLong test P < .013). ECG-AI-Cox (t = 10) resulted in the highest accuracy on both the holdout data, with AUC of 0.82 (0.78–0.86), and MESA external validation data, with an AUC of 0.84 (0.81–0.87). DeLong tests resulted in P < .001 between the ECG-AI-Cox model and all the other models. A confusion matrix for the ECG-AI-Cox model is provided in Table 4.
Table 4.
ECG-AI-Cox confusion matrix
| Predicted |
||||
|---|---|---|---|---|
| No heart failure | Heart failure | |||
| Actual | No heart failure | 4,676 | 1,821 | Specificity = 0.72 |
| Heart failure | 60 | 179 | Sensitivity = 0.75 | |
| Negative predictive value = 0.99 | Positive predictive value = 0.09 | |||
Confusion matrix for the ECG-AI-Cox model was implemented on the MESA cohort data. Threshold was selected based on the balanced output between specificity and sensitivity.
Subgroup analysis
A subgroup analyses for sex, race, and heart failure subtypes (Table 5) using the ECG-AI-Cox model was carried out on the MESA data. The AUCs for HFrEF and HFpEF were 0.85 (0.82–0.87) and 0.83 (0.80–0.85), respectively. In the sensitivity analysis, the false-negative rate was lower in HFrEF (22.7%) compared to HFpEF (28.1%), yet not statistically significant (χ2 test, P = .430).
Table 5.
Subgroup analysis on MESA cohort data using the ECG-AI-Cox model
| Subgroup | AUC (95% CI) |
|---|---|
| Sex | |
| Male | 0.83 (0.80–0.85) |
| Female | 0.84 (0.81–0.87) |
| Race/ethnicity | |
| White | 0.84 (0.82–0.87) |
| African American | 0.81 (0.79–0.85) |
| Chinese American | 0.85 (0.75–0.93) |
| Hispanic ethnicity | |
| No | 0.83 (0.81–0.85) |
| Yes | 0.87 (0.83–0.89) |
| Has any HF risk factor† | |
| Yes | 0.81 (0.79–0.83) |
| No | 0.77 (0.66–0.91) |
| HF type | |
| HFrEF | 0.85 (0.82–0.87) |
| HFpEF | 0.83 (0.80–0.85) |
| HF with missing EF | 0.87 (0.82–0.92) |
AUC = area under the receiver operating characteristic curve; EF = ejection fraction; HF = heart failure; HFpEF = heart failure with preserved ejection fraction; HFrEF = heart failure with reduced ejection fraction.
No HF risk factor category was defined as participants who did not have any of the following risk factors or clinical conditions: smoking, coronary heart disease, diabetes, hypertension, left ventricular hypertrophy, and valvular disease.
Exploratory analysis on only lead I ECG
With minor alterations to the ECG-AI architecture, we developed a lead I–only version using ARIC data. This version of ECG-AI yielded an AUC of 0.73 (0.69–0.76) on ARIC holdout data and 0.78 (0.74–0.82) on the MESA data. The performance of the lead I–only ECG-AI was not statistically different when compared to the 12-lead ECG-AI model (DeLong test P > .100).
Discussion
External validation on the MESA cohort data showed that the ECG-AI model for 10-year HF risk prediction (AUC = 0.77) works at par or better compared to the ARIC-HF (AUC = 0.76), the FHS-HF (AUC = 0.74), and the CPH model (AUC = 0.78). The ECG-AI-Cox model using the ECG-AI output plus 12 risk factors resulted in the best validation accuracy (AUC = 0.84), with statistically significant difference. This shows that ECGs have an added value to clinical risk factors in HF risk prediction. The accuracy of the original 12-lead ECG–based ECG-AI model is no different than the performance of its single-lead (lead I) version.
Similar to the ARIC-HF, FHS-HF, and CPH models, ECG-Chars reduced in accuracy when validated on the MESA cohort. The moderate-high validation accuracy of ECG-AI and ECG-AI-Cox, which includes participants of multiple races/ethnicities, suggests that deep learning–based analyses of ECGs provide robust HF risk prediction for multiple participant subgroups. Despite that ECG-AI provided similar results to clinical risk factor–based risk calculators, it has more applicability owing to its dependency on a minimal amount of input data. ECGs are routinely collected at the clinical stage and are increasing in availability in smart wearable technologies. The use of ECGs within an AI framework can be of benefit in HF risk prediction to facilitate preventive strategies. Such risk prediction tools relying on low-cost data modalities may guide preventive strategies such as lifestyle changes to reduce overall healthcare utilization owing to heart failure treatment.
Literature shows that ECG features provide information on the development of HF.25, 26, 27 Results from the variable importance analysis (Figure 1) are consistent with this, showing that changes in the T-wave morphology can detect risk of cardiac arrhythmias28,29 and abnormal QT dispersion can identify people at risk of HF.30 Although the utilization of these ECG characteristics can be beneficial, they are complex to assess, require full extraction of rhythmic waves, and can run the risk of data gaps when these data are unavailable from different institutes. More importantly, the calculation of ECG features requires very precise detection of onset and offset of each wave/complex, eg, T wave, P wave, QRS complex, etc. Yet, such calculations may change from vendor to vendor as well as be affected largely with noise. On the other hand, when analyzing an entire waveform within deep learning frameworks, such problems do not exist, as the only input is time to voltage data (or raw ECG data). In addition, besides the convenience of raw ECG data, our research shows that waveform data can be more effective in predicting HF risk when compared to the use of numerous ECG features.
A subgroup analysis on the MESA data using ECG-AI-Cox (best model) was performed to assess whether this model is biased toward sex, races/ethnicities, and HF subtypes (HFrEF and HFpEF). Results show high AUCs in predicting HF for the different subgroups of race and sex (AUCs >0.80) with no significant differences despite that the ARIC derivation data included mostly African American and White participants. This suggests that the ECG-AI model and its derivation is not biased toward different demographics. Furthermore, ECG-AI-Cox performed well in detecting HFpEF and HFrEF, with high accuracy and nonsignificance between the 2 subtypes. Interestingly, this model maintained moderate-high accuracy for HF risk–free participants compared to those with at least 1 HF risk factor (AUC = 0.77 vs 0.81; Table 5), suggesting that AI can detect silent markers of HF within an ECG.
Because ECG-AI resulted in higher validation accuracy compared to the ARIC-HF and FHS-HF risk calculators, this model has high potential and, following real-world testing, can be easily and reliably applied and implemented within clinical workflows. The novel ECG-AI and ECG-AI-Cox models can be assets to potentially predict the risk of HF within 10 years, especially for people who are at high risk of developing this disease. The implementation of both models may also help identify people who may benefit from more advanced cardiac healthcare in a timely manner, with potential follow-ups. This can be of major benefit to clinicians, especially for follow-up consultation and precision medicine treatments.
With the increase in smart devices, eg, smartphones and smartwatches, there is a possibility of developing a smart application that directly communicates with ECG-reading devices or can extract ECG data from electronic health servers. Since our results show no statistically significant differences in using 1 lead or 12 leads, there is the potential of using a single lead from an ECG recorded from a smartwatch, and the models incorporated into a smartphone via mobile applications.31 This can be used by clinicians as a prescreening tool and/or by concerned people when the model accuracies are increased enough to prevent alarm fatigue.32
However, there are some additional steps needed to bring such tools into clinical practice. In terms of technology and infrastructure, raw ECG data is typically encrypted and stored in cardiology information systems and not within the electronic health record (EHR). Automation of ECG data export from cardiology information systems, decryption, implementation of AI to decrypted ECG, and returning the results to the EHR and/or the research data warehouse is needed. Despite the fact that building infrastructure requires some investment and expertise to build, clinical incorporation is seamless, since it can be incorporated and not interfere with existing clinical workflow, but, rather, standard-of-care data will be processed in the background and the models will return additional information to assist the clinicians. However, responding to such AI-generated evidence by providers to change or modify their current care approach may need prospective assessment as well as initiation of AI-assisted clinical trials to generate more evidence on the clinical utility of ECG-AI.
Study Limitations
This research has some limitations. Both ARIC derivation and MESA external validation data are from NIH-funded studies with very high-quality ECG data and HF adjudication. Furthermore, such ECGs, as is the nature of cohort studies, are not necessarily recorded like those in standard clinical care. In a real clinical setup, linked to an EHR, ECGs may not have high quality, and ICD-based HF annotations may be not as accurate. Therefore, there is a need for future work to assess the performance of the proposed AI models on real-world data as well as continue testing the 1-lead ECG-AI model collected from smartwatches with ECG functionality.
Conclusion
We conclude that our proposed ECG-AI model using solely ECG data can predict risk for HF within 10 years with the same accuracy as the established clinical factors–based risk calculators. Combining ECG data with other clinical risk factors further significantly increases prediction accuracy. Furthermore, considering the increasing availability of wearables with ECG functionality, such AI models may lead to cost-effective and remote monitoring of certain at-risk populations to facilitate timely interventions and support clinical decision making.
Acknowledgments
We would like to thank the Atherosclerosis Risk in Communities Study and Multi-Ethnic Study of Atherosclerosis Study consortiums for providing data for this study (ARIC Proposal ID: 3678, MESA Proposal ID: 558). We also thank the staff and participants of the ARIC and MESA studies for their important contributions.
Funding Sources
MESA was supported by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, and N01-HC-95169 from the National Heart, Lung, and Blood Institute, and by grants UL1-TR-000040, UL1-TR-001079, and UL1-TR-001420 from the National Center for Advancing Translational Sciences (NCATS). The Atherosclerosis Risk in Communities study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under contract nos. HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, and HHSN268201700004I. Additional funding was supported by grant K24HL148521.
Disclosures
Dr Akbilgic is a co-founder of 9+1AI, LLC. The rest of the authors have no conflict of interest or relationship with industry to declare.
Authorship
All authors attest they meet the current ICMJE criteria for authorship.
Institutional Review Board Statement
The research reported in this paper adhered to Human research: Helsinki Declaration as revised in 2013 guidelines.
References
- 1.Akintoye E., Briasoulis A., Egbe A., et al. Effect of hospital ownership on outcomes of heart failure hospitalization. Am J Cardiol. 2017;120:831–837. doi: 10.1016/j.amjcard.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Kannel W.B., D’Agostino R.B., Silbershatz H., Belanger A.J., Wilson P.W., Levy D. Profile for estimating risk of heart failure. Arch Intern Med. 1999;159:1197–1204. doi: 10.1001/archinte.159.11.1197. [DOI] [PubMed] [Google Scholar]
- 3.Chahal H., Bluemke D.A., Wu C.O., et al. Heart failure risk prediction in the Multi-Ethnic Study of Atherosclerosis. Heart. 2015;101:58–64. doi: 10.1136/heartjnl-2014-305697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pocock S.J., Ariti C.A., McMurray J.J., et al. Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur Heart J. 2013;34:1404–1413. doi: 10.1093/eurheartj/ehs337. [DOI] [PubMed] [Google Scholar]
- 5.Hammad M., Maher A., Wang K., Jiang F., Amrani M. Detection of abnormal heart conditions based on characteristics of ECG signals. Measurement. 2018;125:634–644. [Google Scholar]
- 6.Kwon J-m, Kim K.-H., Jeon K.-H., et al. Development and validation of deep-learning algorithm for electrocardiography-based heart failure identification. Korean Circ J. 2019;49:629–639. doi: 10.4070/kcj.2018.0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbilgic O., Butler L., Karabayir I., et al. ECG-AI: electrocardiographic artificial intelligence model for prediction of heart failure. Eur Heart J Digit Health. 2021;2:626–634. doi: 10.1093/ehjdh/ztab080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tohyama T., Funakoshi K., Kaku H., et al. Artificial intelligence-based analysis of payment system data can predict one-year mortality of hospitalized patients with heart failure. Eur Heart J. 2020;41 [Google Scholar]
- 9.Nakajima K., Nakata T., Matsuo S., Doi T., Jacobson A. 239 Machine learning model for predicting sudden cardiac death and heart failure death using 123I-metaiodobenzylguanidine. Eur Heart J Cardiovasc Imaging. 2019;20 [Google Scholar]
- 10.Agarwal S.K., Chambless L.E., Ballantyne C.M., et al. Prediction of incident heart failure in general practice: the Atherosclerosis Risk in Communities (ARIC) Study. Circ Heart Fail. 2012;5:422–429. doi: 10.1161/CIRCHEARTFAILURE.111.964841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borleffs C.J.W., Scherptong R.W., Man S.-C., et al. Predicting ventricular arrhythmias in patients with ischemic heart disease: clinical application of the ECG-derived QRS-T angle. Circ Arrhythm Electrophysiol. 2009;2:548–554. doi: 10.1161/CIRCEP.109.859108. [DOI] [PubMed] [Google Scholar]
- 12.Mastoris I., Sauer A.J. Opening the “black box” of artificial intelligence for detecting heart failure. ASAIO J. 2021;67:322–323. doi: 10.1097/MAT.0000000000001401. [DOI] [PubMed] [Google Scholar]
- 13.Rahimi K., Bennett D., Conrad N., et al. Risk prediction in patients with heart failure: a systematic review and analysis. JACC Heart Fail. 2014;2:440–446. doi: 10.1016/j.jchf.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee A., Chen S., Fatemifar G., et al. Machine learning for subtype definition and risk prediction in heart failure, acute coronary syndromes and atrial fibrillation: systematic review of validity and clinical utility. BMC Med. 2021;19:1–14. doi: 10.1186/s12916-021-01940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khurshid S., Friedman S., Reeder C., et al. ECG-based deep learning and clinical risk factors to predict atrial fibrillation. Circulation. 2022;145:122–133. doi: 10.1161/CIRCULATIONAHA.121.057480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bild D.E., Bluemke D.A., Burke G.L., et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 17.Almahmoud M.F., Soliman E.Z., Bertoni A.G., et al. Fibroblast growth factor-23 and heart failure with reduced versus preserved ejection fraction: MESA. J Am Heart Assoc. 2018;7 doi: 10.1161/JAHA.117.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosamond W.D., Chang P.P., Baggett C., et al. Classification of heart failure in the Atherosclerosis Risk in Communities (ARIC) study: a comparison of diagnostic criteria. Circ Heart Fail. 2012;5:152–159. doi: 10.1161/CIRCHEARTFAILURE.111.963199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016:770–778. [Google Scholar]
- 20.Maas A.L., Hannun A.Y., Ng A.Y. Rectifier nonlinearities improve neural network acoustic models. Proceedings of the 30th International Conference on Machine Learning. 2013;28:3. [Google Scholar]
- 21.Kingma D, Ba J. Adam: a method for stochastic optmization. arXiv 2014;arXiv:1412.6980.
- 22.Lundberg S.M., Lee S.-I. Curran Associates Inc; Long Beach, CA: 2017. A unified approach to interpreting model predictions In: Proceedings of the 31st International Conference on Neural Information Processing Systems; pp. 4768–4777. [Google Scholar]
- 23.Simmonds S.J., Cuijpers I., Heymans S., Jones E.A. Cellular and molecular differences between HFpEF and HFrEF: a step ahead in an improved pathological understanding. Cells. 2020;9:242. doi: 10.3390/cells9010242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark K.A., Velazquez E.J. Heart failure with preserved ejection fraction: time for a reset. JAMA. 2020;324:1506–1508. doi: 10.1001/jama.2020.15566. [DOI] [PubMed] [Google Scholar]
- 25.Hendry P.B., Krisdinarti L., Erika M. Scoring system based on electrocardiogram features to predict the type of heart failure in patients with chronic heart failure. Cardiol Res. 2016;7:110. doi: 10.14740/cr473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porumb M., Iadanza E., Massaro S., Pecchia L. A convolutional neural network approach to detect congestive heart failure. Biomedical Signal Processing and Control. 2020;55 [Google Scholar]
- 27.Tymińska A., Ozierański K., Balsam P., et al. The prevalence and association of major ECG abnormalities with clinical characteristics and the outcomes of real-life heart failure patients—Heart Failure Registries of the ESC. Kardiol Pol. 2021;79:980–987. doi: 10.33963/KP.a2021.0053. [DOI] [PubMed] [Google Scholar]
- 28.Klingenheben T., Zabel M., D’Agostino R., Cohen R., Hohnloser S. Predictive value of T-wave alternans for arrhythmic events in patients with congestive heart failure. Lancet. 2000;356:651–652. doi: 10.1016/s0140-6736(00)02609-x. [DOI] [PubMed] [Google Scholar]
- 29.Speerschneider T., Thomsen M. Physiology and analysis of the electrocardiographic T wave in mice. Acta Physiol (Oxf) 2013;209:262–271. doi: 10.1111/apha.12172. [DOI] [PubMed] [Google Scholar]
- 30.Galinier M., Vialette J.-C., Fourcade J., et al. QT interval dispersion as a predictor of arrhythmic events in congestive heart failure: importance of aetiology. Eur Heart J. 1998;19:1054–1062. doi: 10.1053/euhj.1997.0865. [DOI] [PubMed] [Google Scholar]
- 31.McCraw C.A., Karabayir I., Akbilgic O., ECG A.I.R. An AI platform for remote smartwatch ECG-based cardiovascular disease detection and prediction. Cardiovasc Digit Health J. 2022;3:S7. [Google Scholar]
- 32.Goto S., Mahara K., Beussink-Nelson L., et al. Artificial intelligence-enabled fully automated detection of cardiac amyloidosis using electrocardiograms and echocardiograms. Nat Commun. 2021;12:1–12. doi: 10.1038/s41467-021-22877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]