Abstract
Quiescent human hematopoietic stem cells (HSC) are ideal targets for gene therapy applications due to their preserved stemness and repopulation capacities; however, they have not been exploited extensively because of their resistance to genetic manipulation. We report here the development of a lentiviral transduction protocol that overcomes this resistance in long-term repopulating quiescent HSC, allowing their efficient genetic manipulation. Mechanistically, lentiviral vector transduction of quiescent HSC was found to be restricted at the level of vector entry and by limited pyrimidine pools. These restrictions were overcome by the combined addition of cyclosporin H (CsH) and deoxynucleosides (dNs) during lentiviral vector transduction. Clinically relevant transduction levels were paired with higher polyclonal engraftment of long-term repopulating HSC as compared with standard ex vivo cultured controls. These findings identify the cell-intrinsic barriers that restrict the transduction of quiescent HSC and provide a means to overcome them, paving the way for the genetic engineering of unstimulated HSC.
Keywords: hematopoietic stem cells, gene therapy, lentiviral vectors, innate immunity, antiviral restriction factors
Graphical abstract
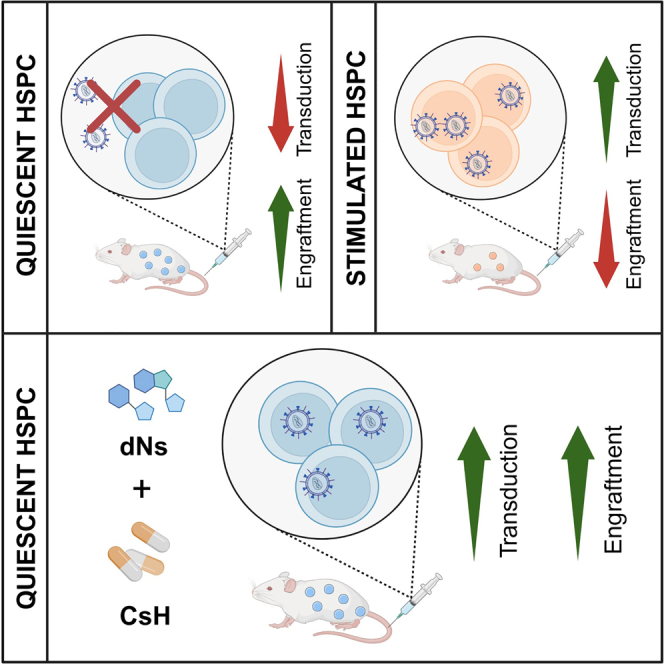
Kajaste-Rudnitski and colleagues identify multiple innate immune barriers that restrict gene transfer into quiescent HSCs and show that they can be overcome by the combination of dNs and CsH. This novel transduction protocol yields higher and polyclonal long-term engraftment of efficiently transduced HSC.
Introduction
Ex vivo gene therapies using hematopoietic stem and progenitor cells (HSPC) have become effective therapeutic options for the treatment of several monogenic diseases, including blood and metabolic disorders.1,2 However, current HSPC gene therapy protocols require the use of high vector doses and prolonged ex vivo culture to reach clinically relevant transduction levels, imposing large-scale vector production and potentially compromising HSPC preservation in culture. In agreement, extensively cultured HSPC fail to maintain multipotency, showing rapid and progressive loss of stem cell activity required for good engraftment and homing capacity, underscoring the need for novel protocols aimed at combining high levels of gene transfer with minimal HSPC manipulation.3,4,5
Unstimulated HSPC, thus, represent the ideal gene therapy target, but their quiescence and expression of innate antiviral restriction factors render them resistant to lentiviral vectors (LV).6,7,8,9 We have previously shown that counteracting the restriction factor interferon inducible transmembrane protein 3 (IFITM3) by cyclosporin H (CsH) improves transduction in unstimulated, quiescent HSPC, but the absolute levels of gene correction remain limited, potentially highlighting the presence of other antiviral blocks in these cells.8 The sterile alpha motif domain and HD domain-containing protein 1 (SAMHD1) has been hypothesized to contribute to the low LV transduction efficiency in HSPC as it impairs HIV reverse transcription in non-dividing myeloid cells by lowering the levels of intracellular deoxynucleoside triphosphates (dNTPs).10,11 However, counteracting SAMHD1 through the accessory viral protein Vpx or the addition of exogenous dNs did not significantly improve transduction of stimulated HSPC.12
As the antiviral activity of SAMHD1 is limited to non-cycling cells where dNTP availability is already restricted, we hypothesized that SAMHD1 restriction of LV transduction could be more relevant in the context of unstimulated HSPC. Exogenous administration of dNTPs in murine unstimulated HSPC improved LV transduction by 2-fold, with absolute levels of gene transfer remaining limiting from a translational perspective.13 The role of SAMHD1 in unstimulated, quiescent human HSPC has not been explored and combinatorial approaches relieving the earlier block to LV entry imposed by IFITM3 remain to be tested. Here, we show that combined counteraction of these restrictions enables the transduction of clinically relevant unstimulated HSPC, resulting in an increased transduction frequency and the polyclonal engraftment of minimally manipulated, long-term repopulating cells.
Results
Combined relief of antiviral restrictions allows efficient transduction of quiescent HSPC
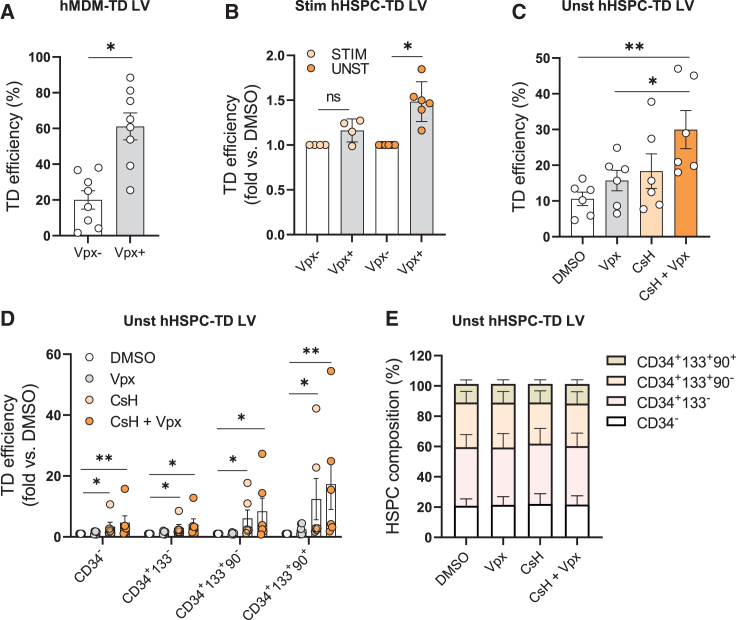
To address the role of SAMHD1 in human HSPC, we used LV that directly incorporate the SIV macaque (SIVmac) accessory protein Vpx into the LV particle.14 We first validated the system in primary human monocyte-derived macrophages (MDMs) (Figure 1A) and tested it in human HSPC, deriving either from cord blood (CB) or mobilized peripheral blood (mPB). While Vpx incorporation did not improve transduction efficiency in HSPC pre-stimulated with growth-promoting cytokines, a slight advantage was observed when transducing freshly isolated, quiescent HSPC (Figure 1B). The benefit of Vpx may be masked by IFITM3 that we have shown to restrict LV entry into HSPC upstream of the SAMHD1-imposed block to reverse transcription (RT).8 Therefore, we evaluated the effect of Vpx incorporation in combination with CsH, a compound that overcomes IFITM3 restriction.8 The combination resulted in an additive increase in gene transfer efficacy over control transduced HSPC (Figures 1C–S1A) supporting a role for both IFITM3 and SAMHD1 in the resistance of quiescent HSPC to LV transduction, despite the lower expression of these restriction factors in quiescent cells with respect to their stimulated counterparts (Figures S1B and S1C). This benefit was evident in the most primitive CD34+CD133+CD90+ subset of HSPC (Figure 1D) with Vpx delivery not altering subpopulation composition (Figure 1E). In line with our transduction data, Vpx exposure increased dNTP levels in quiescent but not stimulated HSPC, with a more prominent increase in deoxyadenosine triphosphate (dATP) and deoxyguanosine triphosphate (dGTP) levels (Figure S1D), as previously reported for human MDM and SAMHD1-deficient mice.15,16 Together, these results suggest that gene transfer into quiescent HSPC is hampered by both a block at entry and by limiting dNTP pools.
Figure 1.
Combined relief of antiviral restrictions allows efficient transduction of quiescent HSPC
(A) Human MDM (hMDM) were transduced with a Vpx incorporating LV, at MOI of 1. Percentages of transduced cells were assessed at 5 days post-transduction by fluorescence-activated cell sorting (FACS) (mean ± SEM; n = 8; Mann Whitney test). (B) Stimulated (Stim) and unstimulated (Unst) human HSPC (hHSPC) were transduced with LV ± Vpx (MOI = 1 for stimulated; MOI = 5 for unstimulated). Percentages of transduced cells were assessed at 5 days post-transduction by FACS and expressed as fold increase vs. Vpx-control (mean ± SEM; n = 4 stimulated hHSPC; n = 6 unstimulated hHSPC; Wilcoxon signed rank test vs. Vpx- = 1). (C) Transduction efficiencies (MOI = 25) in unstimulated hHSPC ± Vpx ±8 μM CsH (mean ± SEM; n = 6; Mann Whitney test). (D) Transduction efficiencies in the different subpopulations of unstimulated hHSPC expressed as fold increase vs. DMSO control (mean ± SEM; n = 6; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). (E) The composition of unstimulated hHSPC was evaluated 5 days post transduction by FACS (mean ± SEM; n = 5). Statistical significance is for ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns indicates non-significance.
Exogenous dNs synergize with CsH to improve transduction and gene editing in quiescent HSPC
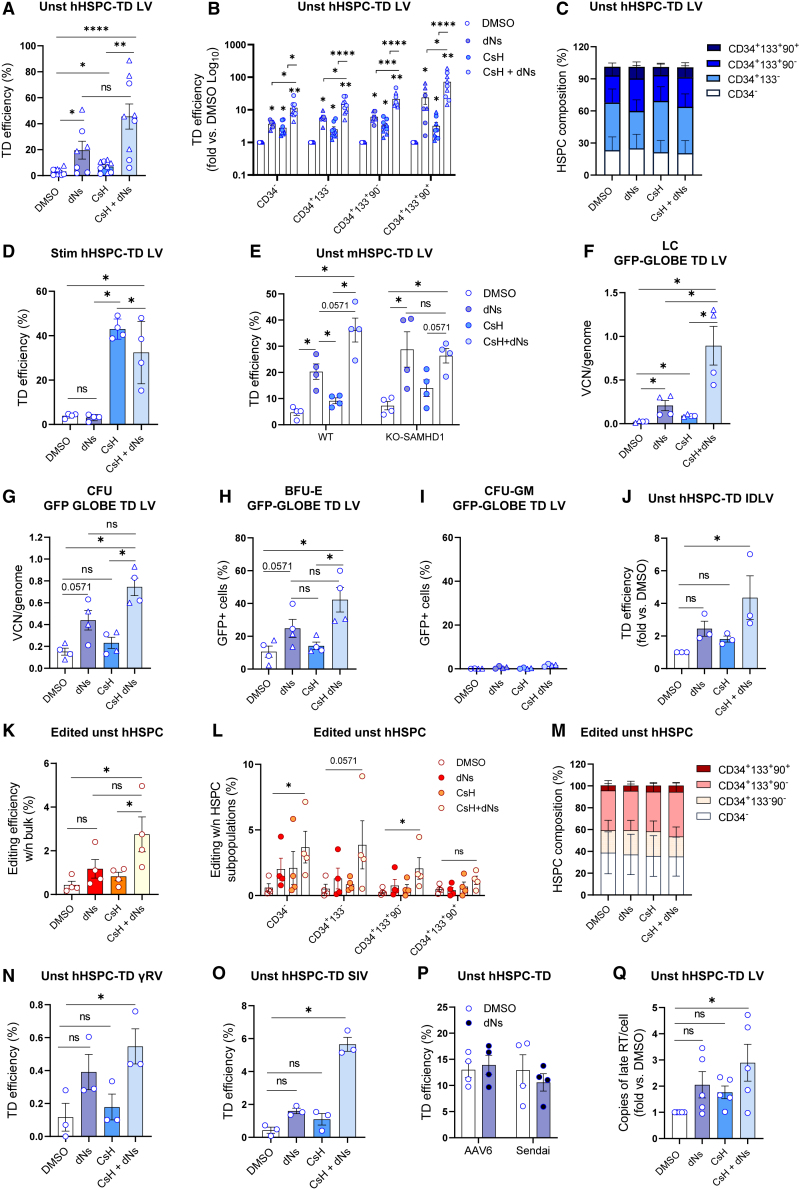
As the increase in intracellular dNTPs upon Vpx delivery might still be insufficient for an optimal RT to occur, we evaluated the impact of excess dNTPs on gene transfer efficacy as an alternative strategy to overcome SAMHD1-mediated restriction.17,18 To this end, we provided dNTPs precursors, dNs, during transduction, alone or in combination with CsH. dNs increased transduction of quiescent HSPC more than Vpx or CsH alone (Figures 1C and 2A), with a striking benefit observed when provided together with CsH, yielding up to a 15-fold increase in the percentage of GFP+ cells over the DMSO control (Figure 2A). This benefit was significant in all fractions of HSPC, with the strongest effect observed in the most primitive CD34+CD133+CD90+ compartment, resulting in an average 70-fold increase in transduction over the control (Figures 2B–S2A), without impacting subpopulation composition (Figure 2C). Of note, dNs enhanced transduction in unstimulated HSPC over a dose range of 25–1,000 μM, with 500 μM providing the highest benefit in transduction (Figure S3A) and maintained a similar effect if provided before or during transduction and CsH addition (Figure S3B). A positive impact of exogenous dNs on the intracellular dNTP levels was confirmed (Figure S3C), with CsH not altering intracellular dNTP pools in quiescent HSPC (Figure S3C). Similar to Vpx, adding dNs did not benefit transduction in pre-stimulated HSPC, alone or in combination with CsH (Figure 2D), further suggesting that the intracellular dNTP pools of stimulated HSPC are not limiting for viral RT. A dN-mediated increase in transduction was confirmed also in unstimulated murine HSPC. Although a slightly higher basal level of transduction was observed in murine HSPC from SAMHD1 KO compared with those from SAMHD1 WT mice (Figure 2E), the effect of dNs on transduction was SAMHD1 independent (Figure 2E), suggesting that the intracellular dNTP pools are rate limiting also in murine HSPC and that the lack of SAMHD1 is not sufficient to maximally remove this restriction.
Figure 2.
Exogenous dNs synergize with CsH to improve transduction and gene editing in quiescent HSPC
(A) Unstimulated hHSPC were transduced with an LV (pool of MOI = 5 and 25) ±8 μM CsH after pre-exposure or not to a mix of dNs (500 μM each). Percentages of transduced cells were assessed 5 days post-transduction by FACS (mean ± SEM; n = 7–9; Mann Whitney test; circles represent CB-derived HSPC; triangles represent mPB-derived HSPC). (B) Transduction efficiencies in the different subpopulations of unstimulated hHSPC (mean ± SEM; n = 7–9; Wilcoxon signed rank test vs. DMSO = 1 or Mann Whitney test for other comparisons; circles represent CB-derived HSPC; triangles represent mPB-derived HSPC). (C) The composition of unstimulated hHSPC was evaluated 5 days post transduction by FACS (mean ± SEM; n = 7–9). (D) Transduction efficiencies (MOI = 1) in stimulated hHSPC ± dNs ± CsH (mean ± SEM; n = 4, Mann Whitney test). (E) Unstimulated murine HSPC from WT or SAMHD1 KO mice were transduced with an LV (MOI = 10) ± 8 μM CsH ± dNs (mean ± SEM; n = 4; Mann Whitney test). (F) Unstimulated hHSPC were transduced with a GFP-GLOBE LV (pool of MOI = 25 and 50) ± CsH ± dNs. VCNs were evaluated 14 days post-transduction in the LC (mean ± SEM; n = 4; Mann Whitney test; circles represent CB-derived HSPC; triangles represent mPB-derived HSPC). (G–I) Unstimulated hHSPC were plated the day after transduction with GFP-GLOBE LV in methocult to perform CFU assay. VCN were evaluated in total colonies (G), and percentages of GFP+ cells were evaluated in burst forming units-erythroid (BFU-E) (H) and CFU- granulocyte–macrophage (CFU-GM) (I) by FACS (mean ± SEM; n = 4; Mann Whitney test; circles represent CB-derived HSPC; triangles represent mPB-derived HSPC). (J) Unstimulated hHSPC were transduced with an IDLV vector at MOI = 200 ± CsH ± dNs. Transduction efficiency was evaluated 3 days post transduction by FACS (mean ± SEM; n = 3; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). (K and L) Percentages of edited cells at AAVS1 locus measured within the bulk (K) or within the indicated HSPC subpopulations (L) (mean ± SEM; n = 4, Mann Whitney test). (M) The composition of edited unstimulated hHSPC was evaluated 3 days post editing by FACS (mean ± SEM; n = 4). (N and O) Unstimulated hHSPC were transduced with γRV (MOI = 50) or SIV (MOI = 10) ± CsH ± dNs (mean ± SEM; n = 3; Dunn’s adjusted Kruskal-Wallis test vs. DMSO). (P) Unstimulated hHSPC were transduced with AAV6 (MOI = 10000) or SENDAI vector (MOI = 10) ± dNs (mean ± SEM; n = 3 AAV6; n = 4 SENDAI). (Q) Late-RT replication intermediates were measured 6 h post-transduction in unstimulated hHSPC transduced with an LV at MOI = 100 and expressed as fold increase vs. DMSO (mean ± SEM; n = 5; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). Statistical significance is for ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns indicates non-significance.
Synergistic enhancement of transduction by the combination of CsH and dNs was confirmed also in the context of a more complex GFP-GLOBE LV, derived from the therapeutic GLOBE LV, currently used for treatment of β-thalassemia.19 The combination of CsH and dNs led to a robust increase in transduction as compared with the DMSO vehicle control in liquid culture (LC) (Figure 2F), as well as in the colony-forming unit (CFU) assay both in terms of vector copy number (VCN) (Figure 2G) and of transgene expression in burst forming units-erythroid colonies (Figure 2H). Of note, lack of transgene expression in CFU granulocyte-macrophage confirmed the erythroid specificity of this vector (Figure 2I).
dNs+CsH enhanced transduction also of an integrase-defective LV (IDLV) (Figure 2J). Given that IDLV can be used as donor DNA delivery vehicle in the context of gene editing applications, we tested the effect of dNs+CsH addition on the efficiency of IDLV-mediated HSPC gene editing.20 Despite overall low efficiency due to the quiescence of these cells, CsH and dNs enhanced CRISPR-Cas9-based genome editing in unstimulated HSPC, without altering the relative composition of the hematopoietic subpopulations (Figures 2K–2M). Moreover, to further exclude the dNTP-independent effects of exogenous dNs on LV transduction, we confirmed that dNs addition improved transduction only of vectors that undergo RT. While dNs increased γRV and SIV transduction of unstimulated human HSPC alone or in combination with CsH (Figures 2N and 2O), no effect was observed for adeno-associated vector (AAV) or SENDAI vectors (Figure 2P) that have single-strand DNA or RNA genome, respectively. The CsH+dNs combination increased late RT products (Figure 2Q), further supporting an early RT-dependent effect.
Together, these data identify the combination of dNs and CsH as able to overcome innate immune barriers to efficient gene transfer and editing in quiescent human HSPC.
Pyrimidine pools are limiting for lentiviral transduction in quiescent HSPC
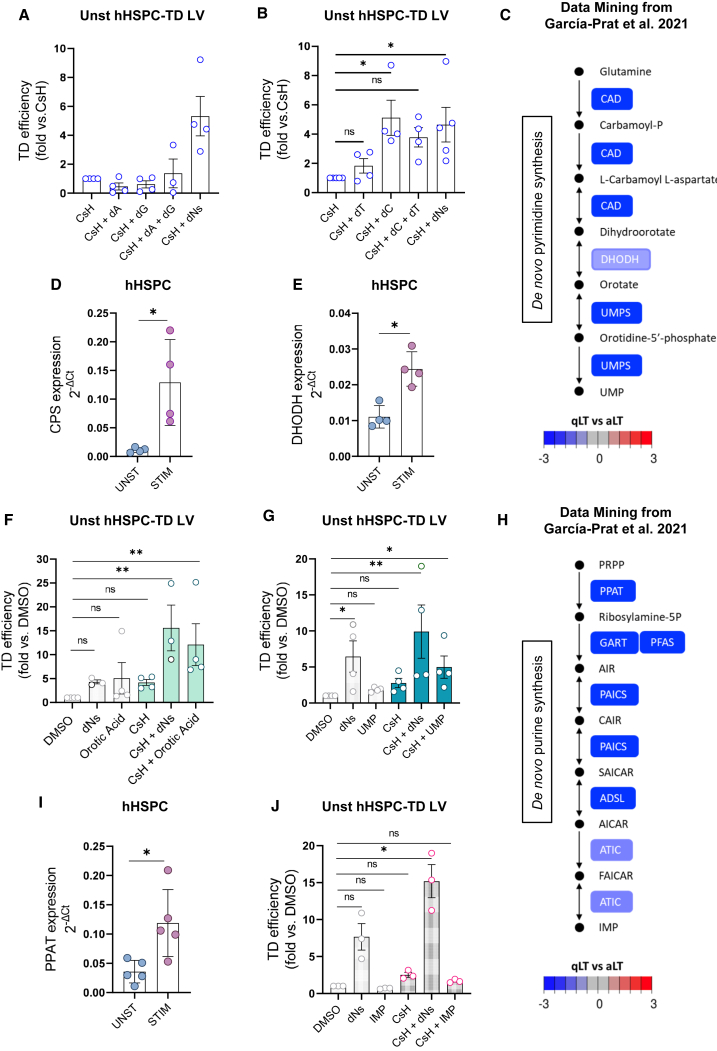
To further explore the role of individual dNs in limiting gene transfer into quiescent HSPC, cells were transduced in the presence of CsH and the single dNs or the combination of purine or pyrimidine dNTPs precursors. The addition of dA, dG, or the combination of both did not improve transduction of quiescent HSPC (Figure 3A). Instead, this effect was mainly mediated by the addition of the two pyrimidine dNTPs precursors, with dC alone, leading to gene marking levels comparable with the combination of all dNs (Figure 3B). dNs effect was not related to increased levels of the ribonucleotide reductase enzyme upon their delivery. Indeed, while the expression of the two enzyme subunits ribonucleotide reductase catalytic subunit M1 (RRM1) and subunit M2 (RRM2) was lower in unstimulated vs. stimulated cells (Figures S3D and S3E), no increase in their expression was observed upon single dNs addition or their combination (Figures S3F and S3G).
Figure 3.
Pyrimidine pools are limiting for lentiviral transduction in quiescent HSPC
(A) Unstimulated hHSPC were transduced (MOI = 5) in presence of CsH upon exposure to dA, dG or dA + dG mix. Percentages of transduced cells were assessed 5 days post transduction by FACS and expressed as fold increase vs. CsH (mean ± SEM; n = 3–4). (B) Unstimulated hHSPC were transduced (MOI = 5) in presence of CsH upon exposure to dC, dT or dC + dT mix. Percentages of transduced cells expressed as fold increase vs. CsH (mean ± SEM; n = 4–5; Dunn’s adjusted Kruskal-Wallis test vs. CsH = 1). (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway illustration of the de novo pyrimidine synthesis pathway. Genes are colored based on the log2/(fold change) of expression values retrieved from publicly available dataset between quiescent long-term (qLT)-HSC vs. activated long-term (aLT)-HSC. Arrows between enzymes indicate the direction of the reaction according to the KEGG database. (D and E) Relative gene expression levels of CPS (D) and DHODH (E) in unstimulated and stimulated hHSPC from the same donors (mean ± SEM; n = 4; Mann Whitney test). (F) Unstimulated hHSPC were transduced with an LV (MOI = 5) ± 8 μM CsH after exposure or not to dNs or 7.5 μM orotic acid. Percentages of transduced cells expressed as fold increase vs. DMSO control (mean ± SEM; n = 3–4; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). (G) Unstimulated hHSPC were transduced with an LV (MOI = 5) ± 8 μM CsH after exposure or not to dNs or 1 mM uridine 5′-monophosphate (UMP). Percentages of transduced cells expressed as fold increase vs. DMSO (mean ± SEM; n = 4; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). (H) KEGG pathway illustration of the de novo purine synthesis pathway. Genes are colored based on the log2/(fold change) of expression values retrieved from publicly available dataset between qLT-HSC vs. aLT-HSC. Arrows between enzymes indicate the direction of the reaction according to the KEGG database. (I) Relative gene expression levels of PPAT in unstimulated and stimulated hHSPC from the same donors (mean ± SEM; n = 5; Mann Whitney test). (J) Unstimulated hHSPC were transduced with an LV (MOI = 5) ± 8 μM CsH after exposure or not to dNs or 5 mM inosine monophosphate (IMP). Percentages of transduced cells expressed as fold increase vs. DMSO (mean ± SEM; n = 3; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). Statistical significance is for ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns indicates non-significance.
To dissect the mechanisms behind the differential effects of pyrimidine and purine dNTPs precursors on transduction of quiescent HSPC, we assessed the endogenous levels of the de novo pyrimidine synthesis pathway components in quiescent vs. activated HSPC from publicly available datasets.21 As expected for low metabolically active cells, enzymes involved in the de novo pyrimidine synthesis pathway were downregulated in quiescent long-term repopulating HSC as compared with their activated counterparts (Figure 3C), as confirmed by gene expression analysis of the two rate-limiting enzymes, carbamoyl-P synthetase (CPS) and dihydro-orotate dehydrogenase (DHODH), in stimulated and unstimulated HSPC (Figures 3D and 3E). In agreement, fueling the pathway with intermediate precursors of the pyrimidine synthesis such as orotic acid or uridine 5′-monophosphate increased LV transduction, and further benefitted from the addition of CsH (Figures 3F and 3G). Also, the levels of the enzymes of the de novo purine synthesis pathway resulted downregulated in quiescent vs. activated HSPC (Figure 3H) with phosphoribosyl pyrophosphate amidotransferase (PPAT), the first rate-limiting enzyme of the pathway, confirmed to be less expressed in unstimulated HSPC (Figure 3I). However, addition of the inosine monophosphate precursor did not have any effect on the transduction level of quiescent HSPC, alone or in combination with CsH (Figure 3J). These data are in line with the lack of impact of dA and/or dG addition on transduction (Figure 3A) despite increased levels of dATP upon dNs delivery (Figure S3C).
Overall, these findings suggest that dNs addition enhances LV transduction in unstimulated HSPC by overcoming limiting pyrimidine pools that are required for efficient viral RT.
Delivery of dNs does not affect unstimulated HSPC properties in vitro
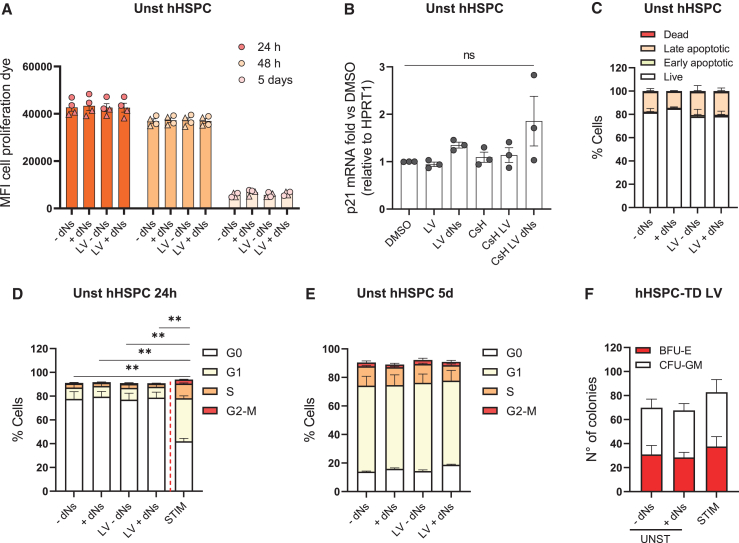
We next performed in vitro assays to evaluate potential effects of dNs on proliferation, apoptosis or cell-cycle status of HSPC that were kept unstimulated for 16–20 h before cytokines addition. Neither dNs addition nor LV transduction had an impact on the proliferation rate of unstimulated HSPC (Figures 4A–S2B) and no marked increase in the levels of p21 mRNA, a surrogate marker of DNA damage responses, was observed except for a slight effect of the CsH+dNs combination (Figure 4B). In addition, no increased signs of apoptosis were detected in unstimulated HSPC 48 h after dNs exposure, regardless of transduction (Figure 4C).
Figure 4.
Delivery of dNs does not affect unstimulated HSPC properties in vitro
(A) Impact of dNs on cell proliferation was assessed in transduced (pool of MOI = 25 and 50) or untransduced quiescent hHSPC. Mean fluorescent intensity (MFI) of the cell proliferation dye was evaluated at the indicated time points by FACS (mean ± SEM; n = 4; circles represent CB-derived HSPC; triangles represent mPB-derived HSPC). (B) p21 mRNA levels were evaluated by qPCR in unstimulated HSPC 48 h post transduction (MOI 50) ± CsH ± dNs (mean ± SEM; n = 3; Dunn’s adjusted Kruskal-Wallis test vs. DMSO = 1). (C) Impact of dNs on apoptosis was assessed in transduced (MOI = 50) or untransduced quiescent hHSPC at 48 h post transduction (mean ± SEM; n = 3–4). (D and E) Impact of dNs on the cell-cycle status of transduced (MOI = 50) or untransduced quiescent hHSPC was evaluated at 24 h (D) and 5 days (E) post transduction. Analysis of the cell-cycle status of the stimulated counterpart at 24 h was included as reference (mean ± SEM; n = 4; Mann Whitney test). (F) The number of total, myeloid, and erythroid CFU was assessed in vitro 2 weeks after cells plating for both quiescent hHSPC exposed or not to dNs and stimulated hHSPC (pool of MOI = 50 and 100) (mean ± SEM; n = 4). Statistical significance is for ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns indicates non-significance.
As maintenance of HSPC quiescence contributes to preserve their stem cell properties, we evaluated whether dNs alter the cell-cycle status or clonogenic potential of unstimulated HSPC. As expected, unstimulated cells showed a greater percentage of cells in the G0 phase of the cell cycle with respect to their stimulated counterparts at 24 h, with almost complete absence of cycling cells (Figure 4D). dNs delivery did not alter the quiescence status of these cells at 24 h (Figure 4D) or cell-cycle distribution at longer time points (Figure 4E), regardless of transduction, and no difference in CFU was observed between transduced unstimulated cells that received dNs or not (Figure 4F). Of note, unstimulated HSPC showed a slightly lower colony output than transduced stimulated HSPC, likely reflecting the lower percentage of progenitors due to the lack of ex vivo culture (Figure 4F).
Overall, our data indicate a safe in vitro profile for dNs addition in unstimulated HSPC, suggesting they could be exploited to enhance transduction while preserving the quiescence and stem cell properties of unstimulated HSPC.
CsH+dNs-transduced unstimulated HSPC show superior and polyclonal long-term engraftment
Ex vivo culture of HSPC impacts their engraftment potential, due to cell-cycle progression that drives lineage commitment and differentiation, as well as loss of adhesion molecules, which impact their homing capacity into the bone marrow (BM) niche.4,22,23 On these premises, we reasoned that an efficient gene transfer into quiescent HSPC should allow better preservation of their biological properties, including long-term repopulation capacity. In agreement, unstimulated human CB-derived HSPC transduced in the presence of CsH and dNs (Figure S4A) maintained high and stable gene marking levels from in vitro (Figures S4B–S4D) to in vivo (Figures S4E–S4I) and engrafted similarly to their stimulated counterpart transduced in presence of CsH alone (Figures S4K–S4P), despite almost one-half of the initial cellular input (Figure S4J).
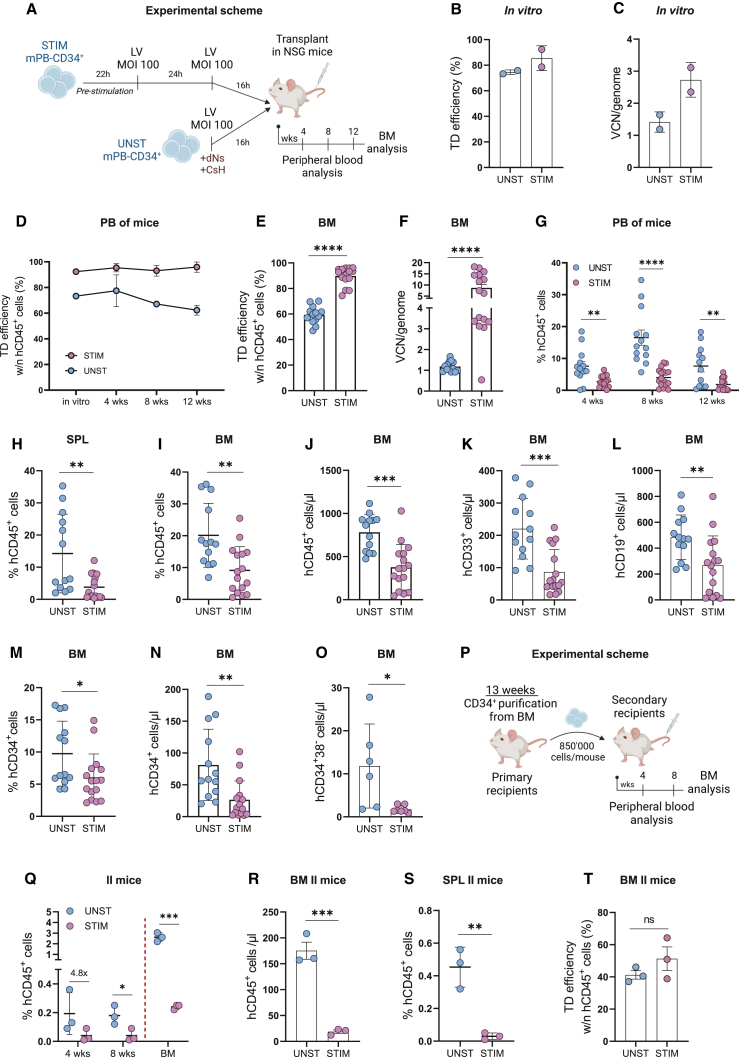
To validate the potential of this transduction protocol in a more clinically relevant setting, we compared the performance of the CsH+dNs transduction of unstimulated mPB-derived HSPC to the gold standard two-hit transduction protocol in which stimulated HSPC are transduced twice in the absence of transduction enhancers (Figure 5A). To better compare the engraftment potential, cell numbers were normalized between stimulated and unstimulated conditions at the time of transplant. The CsH+dNs combination resulted in around 70% of transgene expression at one VCN per genome in vitro (Figures 5B and 5C) within all the different HSPC subpopulations (Figure S5A) and in the long-term repopulating HSC in vivo (Figures 5D and 5E). Albeit remaining lower than copies reached in the stimulated group, a stable and clinically relevant VCN of 1 was maintained for the unstimulated HSPC transduced with the CsH+dNs combination (Figure 5F). Better preservation of the percentage of CD34+ cells and the most primitive CD34+CD133+CD90+ fraction was observed for unstimulated HSPC in vitro (Figure S5B). This was reflected as a significantly enhanced in vivo engraftment potential of the quiescent HSPC as compared with their stimulated counterparts in the periphery (Figure 5G), in the spleen (Figure 5H), and in the BM with increased percentages and absolute counts of human CD45+ cells (Figures 5I and 5J) and myeloid (CD33+) and lymphoid (CD19+) progenitors (Figures 5K and 5L), as well as of the CD34+ and most primitive HSPC fraction (Figures 5M–5O, S5C, and and S5D). Importantly, secondary transplantation assays confirmed superior engraftment of cells coming from mice transplanted with the unstimulated HSPC in the peripheral blood, BM, and spleen (Figures 5P–5S) with maintenance of similar transduction levels compared with the stimulated counterparts (Figure 5T). Of note, the superior engraftment capacity of unstimulated HSPC transduced in presence of CsH and dNs was confirmed also in comparison with stimulated cells transduced following a short clinical 36-h transduction protocol in which LentiBOOST (LB) or a combination of LB and prostaglandin E2 (PGE2) were exploited as transduction enhancers (Figures S5I–S5M). Altogether, these results confirm the dual benefit of the CsH+dNs transduction protocol in allowing efficient gene transfer into long-term repopulating HSC while significantly preserving their biological properties.
Figure 5.
CsH+dNs-transduced unstimulated HSPC show superior long-term engraftment
(A) Experimental scheme of the in vivo experiment. Human CD34+ cells from mPB were pre-stimulated 22h with a cocktail of early active cytokines and transduced twice with a GFP-LV (MOI = 100) or kept unstimulated and transduced immediately after thawing with a GFP-LV (MOI = 100) in presence of CsH and 500 μM mix of all dNs. Then, 4–5 × 105 cells were injected 16 h post transduction into sub lethally irradiated NSG mice (n = 2 independent experiments; n = 13 mice for unstimulated group, n = 16 mice for stimulated group). (B) In vitro transduction efficiency was assessed 5 days post transduction in the bulk population of HSPC (mean ± SD; n = 2). (C) In vitro VCN/genome were measured 14 days post transduction (mean ± SD; n = 2). (D) Transduction efficiencies, measured as percentages of GFP+ cells within the hCD45+ cells, in the peripheral blood of mice from the two experimental groups (mean ± SD; n = 13–16). (E) Transduction efficiencies, measured as percentages of GFP+ cells within the hCD45+ cells, in the BM at 13 weeks post transplant (mean ± SD; n = 13–16, Mann Whitney test). (F) VCN/genome were measured in the BM at 13 weeks post transplant (mean ± SD; n = 13–16, Mann Whitney test). (G) Engraftment levels in the peripheral blood of mice from the two experimental groups (mean ± SD; n = 13–16; Mann Whitney test). (H) Engraftment levels in the spleen at 13 weeks post-transplant (mean ± SD; n = 13–16, Mann Whitney test). (I) Engraftment levels in the BM at 13 weeks post transplant (mean ± SD; n = 13–16; Mann Whitney test). (J) Absolute counts of hCD45+ cells in the BM at 13 weeks post-transplant (mean ± SD; n = 13–16; Mann Whitney test). (K and L) Absolute counts of hCD33+ (K) and hCD19+ (L) cells in the BM at 13 weeks (mean ± SD; n = 13–16, Mann Whitney test). (M) Percentages of hCD34+ cells within hCD45+ in the BM at 13 weeks post-transplant (mean ± SD; n = 13–16; Mann Whitney test). (N) Absolute counts of hCD34+ cells in the BM at 13 weeks (mean ± SD; n = 13–16; Mann Whitney test). (O) Absolute counts of the hCD34 + 38- fraction in the BM at 13 weeks (mean ± SD; n = 6–7 from one experiment; Mann Whitney test). (P) Experimental scheme of the secondary transplant. CD34+ cells were purified from BM cells of primary mice from one of the two experiments described in A, and pooled for each condition. 850000 cells were then injected into sub lethally irradiated secondary NSG recipients (n = 1 experiment; n = 3 mice per group). (Q) Engraftment levels in the peripheral blood and BM of secondary recipients (mean ± SD; n = 3, unpaired t test). (R) Absolute counts of hCD45+ cells in the BM of secondary mice at 9 weeks post-transplant (mean ± SD; n = 3, unpaired t test). (S) Engraftment levels in the spleen of secondary mice at 9 weeks post-transplant (mean ± SD; n = 3, unpaired t test). (T) Transduction efficiencies within the hCD45+ cells in the BM of secondary mice at 9 weeks post-transplant (mean ± SD; n = 3, unpaired t test). Statistical significance is for ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns indicates non-significance.
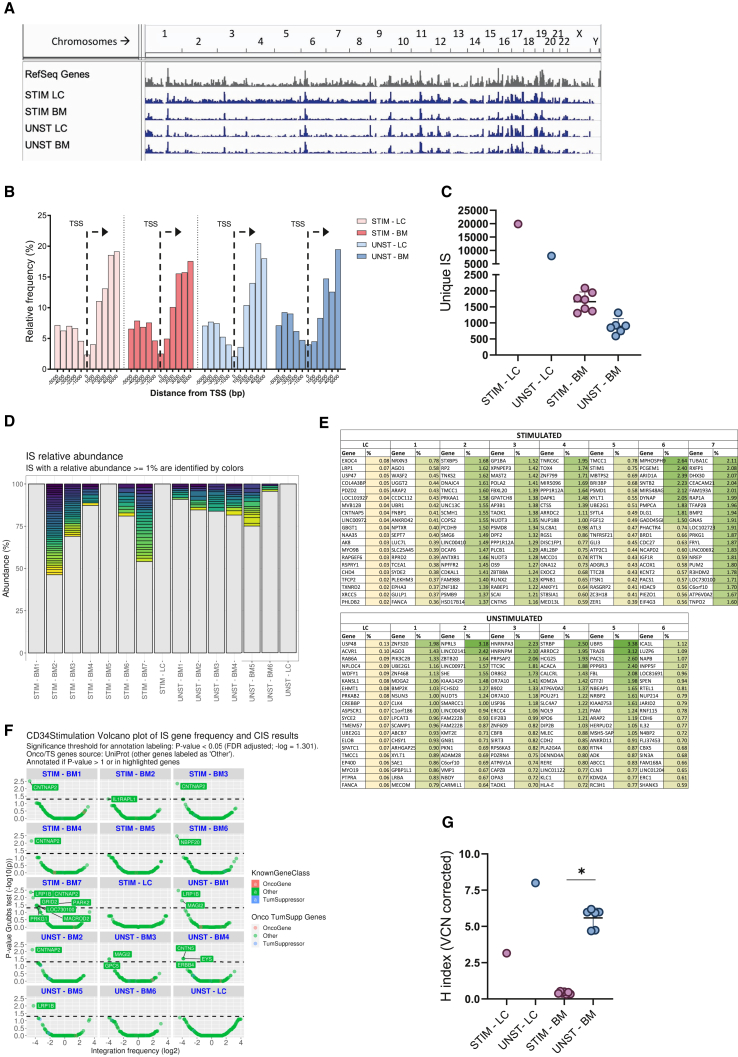
To address the safety and impact of the CsH+dNs transduction protocol on HSPC clonality, we performed integration site analysis on genomic DNA isolated from BM cells of NSG mice transplanted with the mPB-derived HSPC transduced when quiescent with the CsH+dNs protocol or upon stimulation with the two-hit standard protocol.24,25 The global integration patterns were similar between the two transduction protocols and were in line with those typically observed in LV-based gene therapy trials with no genotoxic events observed (Figures 6A and 6B).19,26,27,28,29 An abundance of unique integration sites reflected the efficiency of transduction, with more integrations retrieved from the stimulated HSPC in which the overall VCN remained higher (Figures 6C–5F). No evidence of clonal expansion could be observed (Figure 6D). Moreover, the top 20 most abundant clones showed a maximum abundance of 3.4%, with no evidence of enrichment for genes involved in clonal expansion (Figure 6E). In agreement, common insertion site analysis did not show any significantly over targeted gene (Figure 6F).30 Importantly, when assessing the clonal diversity of the engrafted cell population through the Shannon diversity index corrected by the VCN, the cells that underwent transduction using the CsH+dNs combination, while quiescent showed a higher index as compared with the stimulated counterparts (Figure 6G). These results indicate that transduction of quiescent HSPC in presence of CsH+dNs yields a polyclonal population of cells that maintain a better engraftment capacity over the stimulated counterparts.
Figure 6.
Integration site analysis in unstimulated HSPC supports a polyclonal engraftment
(A) Frequency distribution in chromosomes (indicated on top) of Ref Seq genes (in gray) and LV-GFP insertions retrieved from in vitro cultured HSPC transduced following a two-hit standard protocol or unstimulated HSPC transduced in presence of CsH and dNs (indicated as STIM-LC and UNST-LC, respectively) and SIN-LV-GFP insertions retrieved from BM cells harvested from NSG mice 13 weeks after transplant with HSPC from the above mentioned experimental groups (indicated as STIM-BM and UNST-BM, respectively). (B) Frequency distribution of LV integration sites around gene transcription start sites (x axis, in bp) from the two experimental groups for both the in vitro and the in vivo condition. (C) Number of unique integration sites retrieved for each treatment group, defined as in (A) (mean ± SD; n = 1 for LC, n = 7 for STIM-BM; n = 6 for UNST-BM). (D) Bar-plot representation of clonal abundance. Integration sites with an abundance <1% were merged and represented in gray at the bottom of each bar. Integration sites with a relative abundance ≥1% are identified by colors. (E) The genes targeted in the 20 most abundant clones are listed in the table (most abundant ranked from top to bottom). (F) Volcano plot representation of the common insertion site (CIS) analysis results. CIS analysis was performed on the integration site (IS) identified for each sample using the Grubbs test for outliers. All genes targeted by IS are tested and plotted with dots of size proportional to the gene length; on the x axis is represented as negative Z score of the Log2 of the gene integration frequency normalized by gene length. Values <0 or >0 represent integration frequencies below or above the average respectively. The y axis shows the p value determined by Grubbs test for outliers (-log10 of p-value). Dots with significant p values (alfa threshold of 0.05) are above the dashed horizontal line and labeled with the closest gene name (RefSeq). (G) Shannon diversity index, corrected by VCN, was calculated to measure the clonal diversity of HSPC for each treatment group, defined as in A (mean ± SD; n = 1 for LC, n = 7 for STIM-BM; n = 6 for UNST-BM; Mann Whitney test; statistical significance is for ∗p < 0.05).
Overall, our data support the use of the CsH+dNs combination to transduce quiescent, unstimulated HSPC with the benefit of obtaining a better engrafting and polyclonal cell product.
Discussion
The genetic engineering of quiescent HSPC remains challenging as these cells are highly refractory to viral vector-mediated gene transfer. The method we have developed here overcomes these limitations, allowing efficient transduction of quiescent human HSPC. Efforts to improve LV transduction in HSPC have led to the development of transduction enhancers that allow more efficient gene transfer into these therapeutic targets.7,8,9,31,32,33 Nevertheless, few studies have addressed the poor permissiveness of quiescent HSPC. We have previously shown that CsH can increase LV transduction in pre-stimulated and in quiescent HSPC.8 However, the overall transduction levels remained below the levels required for clinical translation in quiescent HSPC, suggesting that additional blocks beyond IFITM3-mediated restriction of LV entry persist in these cells. SAMHD1 has been suggested to play an important role in restricting HIV RT in quiescent cells, but Vpx-mediated depletion of SAMHD1 had little effect on LV in pre-stimulated HSPC.12,18 We show here that Vpx improved, albeit modestly, the LV transduction of quiescent HSPC when combined with CsH, consistent with sequential antiviral blocks mediated by IFITM3 and SAMHD1 on the lentiviral life cycle.
Consistent with the SAMHD1-mediated block to RT through the regulation of cellular dNTP levels, the addition of exogenous dNTPs has been shown to moderately improve LV transduction in quiescent murine HSPC.13,17 Here, exogenously supplied dNs significantly improved LV transduction in unstimulated human HSPC. However, as opposed to Vpx, the combination of exogenous dNs with CsH led to enhanced LV transduction, suggesting that the Vpx-mediated relief of SAMHD1 restriction is suboptimal in quiescent HSPC. Alternatively, dNTP pools may remain limiting for viral transduction in HSPC despite SAMHD1 removal. Indeed, dNs improved LV transduction independent of SAMHD1 in the context of quiescent murine HSPC.
We found that this rescue could be ascribed to pyrimidine dNTPs precursors, in particular to dC, in quiescent HSPC, despite no major increase in intracellular dCTP levels being registered. Each dNTP is differently modulated within the cells and the apparent lack of intracellular dCTP increase upon dNs addition in quiescent HSPC could be related to a different consumption or degradation kinetic that does not allow to adequately capture in a single time point analysis the dNs-mediated increase, and further experiments would be needed to better address this aspect.16
Quiescent cells have low dNTPs requirements and mainly rely on the salvage pathways to recycle nucleic acids to regenerate dNTPs. In line, we show and confirm that key enzymes of the de novo synthesis pathways are strongly downregulated in quiescent vs. activated HSPC.21 Interestingly, rescue of the de novo pyrimidine, but not of the purine synthesis, pathway through the addition of key intermediates molecules had a similar effect of providing exogenous dNs, further confirming a major role of the pyrimidine pool in restricting quiescent HSPC.
It has been shown that purine or pyrimidine imbalances may negatively impact cell proliferation and viability, and cellular pyrimidines deficiency has been linked to mitochondrial DNA-dependent innate immune activation.34,35,36 Moreover, excess intracellular dNTP levels have been linked to cell-cycle progression and genomic instability.37 Thus, despite observing that pyrimidine dNTPs precursors are sufficient to enhance LV transduction, all four dNs were included for the in vivo validation of the CsH+dNs protocol and the impact of dNs delivery on HSPC properties was carefully evaluated. Importantly, exogenous dNs addition did not affect the proliferation rate of unstimulated HSPC or alter their quiescent phenotype. Moreover, we did not observe activation of the DNA damage response with only a slight increase in p21 expression levels when combining CsH and dNs, which may be explained by activation of p53-dependent responses by higher vector copies within the cells as previously observed for stimulated HSPC.38
The unstimulated HSPC engrafted and yielded hematopoietic output at higher levels compared with their stimulated counterparts, with higher percentages and absolute counts of the most primitive, long-term engrafting HSC retrieved when quiescent HSPC were transduced. This supports better preservation of their biological properties and repopulation capacity due to the lack of ex vivo culture. Importantly, albeit remaining lower than levels reached in stimulated HSPC, the dNs+CsH transduction protocol enabled high transgene expression and clinically relevant levels of integrated copies long term in vivo without affecting HSC function. This is extremely important if we consider some of the limitations encountered in current clinical trials. Thus far, X-linked chronic granulomatous disease (X-CGD) LV gene therapy has failed to fully restore functional defects and several patients have lost engraftment of gene-marked HSC.39,40 While the etiology of graft loss remains unclear, increased oxidative DNA damage and inflammation have been suggested to compromise X-CGD HSC.41,42 Similarly, current gene therapy protocols for Fanconi anemia rely on very short, and thus less efficacious, transduction protocols aimed at limiting the ex vivo manipulation of these extremely fragile cells.43 On these premises, we predict that our CsH+dNs transduction protocol that does not require prolonged ex vivo culture of HSPC will provide a significant improvement for such settings in which the disease background impacts HSC biological properties.
Because of concerns about potential genotoxicity, the safety of LV-based ex vivo gene therapies is actively monitored across trials worldwide. However, vector integration has never been evaluated in unstimulated quiescent HSPC. Our analysis revealed that the dNs+CsH-based transduction of quiescent HSPC did not alter the overall distribution of LV integrations, but captured a higher Shannon diversity index for the quiescent HSPC, further suggesting safe and polyclonal engraftment of the transduced cells.
Our results demonstrate that LV-mediated gene transfer to quiescent HSPC is restricted by multiple innate immune barriers that can be experimentally removed. These findings pave the way for the genetic engineering of unstimulated HSPC in cell and gene therapies.
Material and methods
Vectors
LV and SIN-RV vectors were produced and titered as previously described.44,45,46 GFP-GLOBE LV is a derivative of the GLOBE LV, developed for gene therapy clinical trial of β-thalassemia.47 Briefly, the expression of EGFP is under the control of the β-globin promoter and locus control region elements. The β-globin mini-gene cassette containing the EGFP gene in place of β-globin exon-1 was released by digestion with EcoRI from MA951 (kindly provided by Michael Antoniou, Department of Medical and Molecular Genetics, King’s College London, Guy’s Hospital, London, UK). The fragment was cloned into EcoRI sites of pCCL-GLOBE-Kana LV.47 IDLV was produced as previously described substituting the packaging plasmid pMDLg/pRRE with pMD.Lg/pRRE.D64VInt.48 Vpx-containing LV were produced as previously described with Vpx sequence of SIVmac239 strain.14 SIVmac-based vector was produced as previously described.14,49 AAV6 donor template was generated from a construct containing AAV2 inverted terminal repeats, produced at the TIGEM vector core by a triple-transfection method and purified by ultracentrifugation on a cesium chloride gradient, as previously described.50 An AAV6 donor template with homologies for AAVS1 locus and encompassing a PGK.GFP reporter cassette was utilized.51 The Sendai viral vector was kindly provided by ID Pharma Co., Ltd.
VCN analysis
DNA was extracted using the QIAamp DNA micro kit (Qiagen). Vector copies per diploid genome (VCN) of the integrated LV and Late-RT copies were quantified by quantitative droplet digital-PCR using the following primers: ΔU3 sense 5′-TCACTCCCAACGAAGACAAGATC-3′, PBS antisense 5′-GAGTCCTGCGTCGAGAGAG-3′ as described.38 The amount of human DNA loaded in the reaction was quantified using primers designed to amplify the hTERT gene as described.48
Cells
Human CD34+ cells were either freshly purified from umbilical CB collected upon informed consent from healthy volunteers according to the Institutional Ethical Committee approved protocol (TIGET01/09), or purchased frozen from Lonza as for G-CSF mPB CD34+ cells. Human CB-derived HSPC were cultured in serum-free StemSpan medium (STEMCELL Technologies) supplemented with penicillin (100 IU/mL), streptomycin (100 mg/mL), 100 ng/mL recombinant human stem cell factor (rhSCF), 20 ng/mL recombinant human thrombopoietin (rhTPO), 100 ng/mL recombinant human Flt3 ligand (rhFlt3), and 20 ng/mL recombinant human IL6 (all from Peprotech) for 16–24 h prior to transduction. mPB-derived HSPC were cultured in CellGro medium (CellGenix) supplemented with penicillin (100 IU/mL), streptomycin (100 mg/mL), 300 ng/mL rhSCF, 100 ng/mL rhTPO, 300 ng/mL rhFlt3, and 50 ng/mL recombinant human IL3 for 16–24 h prior to transduction.8,38 Unstimulated HSPC were kept in the same media in the absence of cytokines during transduction for 16–20 h and then supplemented with the above described human cytokines and 10 μM of the reverse transcriptase inhibitor 3TC (Sigma) to avoid subsequent transduction due to cytokines stimulation. When not clearly indicated, HSPC derived from CB or mPB were used matching the transduction levels between the two sources by adjusting the multiplicity of infection (MOI). Unstimulated murine Lin- HSPC were purified from littermate Samhd1+/+ (WT) and Samhd1−/− (KO)52 bone marrow (BM) cells using the Lineage Cell Depletion Kit (Miltenyi Biotec) and kept in serum-free StemSpan medium containing penicillin (100 IU/mL), streptomycin (100 mg/mL), and 2% glutamine without cytokines during transduction for 16–20 h and then supplemented with a combination of mouse cytokines (20 ng/mL IL-3, 100 ng/mL SCF, 100 ng/mL Flt-3L, and 50 ng/mL TPO, all from Peprotech) and 10 μM of 3TC.8 MDM were differentiated from isolated CD14+ monocytes in DMEM supplemented with 10% fetal bovine serum, penicillin (100 IU/mL), streptomycin (100 mg/mL), 2% glutamine, and 5% human serum AB (Lonza) for 7 days.
All cells were maintained in a 5% CO2 humidified atmosphere at 37°C.
Transduction and compounds
All cells were transduced at the indicated MOI. Lower MOIs were used in most of the in vitro experiments, and higher MOIs were used in experiments in which clinically relevant transduction levels were sought. CsH (Sigma) was added during transduction at 8 μM concentration. dA (D8668), dC (D0776), dG (D0901), and dT (T1895) (Sigma) were added during transduction at 500–1000 μM each. Uridine 5′-monophosphate 1 mM (Selleckchem), orotic acid 7.5 μM (Sigma), and inosine 5′-monophosphate 5 mM (Sigma) were added 2–4 h before transduction. Prostaglandin E2 (Cayman Chemicals) was added 2 h before transduction at 10 μM concentration. LB (SIRION biotech) was added during transduction at 1:100 dilution from the stock. All compounds were washed out with the vector 16–20 h later.
Gene editing
For gene editing, HPSCs were transduced while quiescent with IDLV at an MOI of 200, in the presence or not of CsH and dNs. An IDLV donor template with homologies for AAVS1 locus (encoding for a PGK.GFP reporter cassette) was utilized.53 At 24 h after transduction, cells were washed with PBS and electroporated using P3 Primary Cell 4D-Nucleofector X Kit and Nucleofector 4D device (program EO-100) (Lonza) according to the manufacturer’s instructions with ribonucleoprotein particles (RNPs) at a final concentration of 1.25–2.5 μM together with 0.1 nmol Alt-R Cas9 Electroporation Enhancer (Integrated DNA Technologies).54 RNP complexes were assembled by incubating at 1:1.5 M ratio Streptococcus pyogenes Cas9 protein (Aldevron) with pre-annealed synthetic Alt-R crRNA:tracrRNA (Integrated DNA Technologies) for 10 min at 25°C. The genomic target sequence of guide RNA was previously reported.51 After electroporation, cells were maintained in medium with human cytokines as previously reported for stimulated HSPC, supplemented with 10 μM 3TC. Editing efficiency was evaluated after 3 days by fluorescence-activated cell sorting (FACS), looking at the percentage of cells expressing the GFP marker.
Immunofluorescence
We seeded 5 × 104 mPB-CD34+ cells in Multitest slide glass 10-well 8 mm (MP Biomedicals) precoated with poly-L-lysine solution (Sigma) for 20 min. Cells were fixed with 4% paraformaldehyde (in 1× PBS) for 20 min at room temperature, washed with 1× PBS, and permeabilized with 0.1% Triton X-100 for 20 min at room temperature. Cells were then incubated 30 min in PBG (5% BSA, 2% gelatin from cold water fish skin; Sigma) and then stained over night at 4°C with rabbit anti-IFITM3 polyclonal antibody (1:200 dilution, Proteintech 11714-1-AP) and mouse anti-SAMHD1 polyclonal antibody (1:200 dilution, Abcam ab67820). After three washes with 1× PBS, cells were incubated with donkey anti-Rabbit IgG, Alexa Fluor 555 (1:500 dilution, Thermo Fisher Scientific A-31572), and anti-mouse IgG, Alexa Fluor 488 (1:500 dilution, Thermo Fisher Scientific A-21202) for 1 h at room temperature. Nuclei were stained with 4′,6-diamidino-2-phenylindole (10236276001 Roche) for 5 min at room temperature. Images were recorded using the TCS SP5 Leica confocal microscope, 40× with oil.
dNTP measurements
dNTPs were extracted from a known number of cells and measured by RT-based dNTP assay as previously described.55 dNTP amounts were normalized by the numbers of the cells used, and the fold differences in the normalized dNTP amounts were determined.
RNA extraction and gene expression analysis
RNA extraction from cells was performed using the RNeasy Plus micro Kit (Qiagen) according to manufacturer’s instructions. The extracted RNA was reverse transcribed using SuperScript IV VILO Master Mix (Invitrogen). RT-qPCR analyses were performed using TaqMan probes from Applied Biosystems to detect endogenous mRNA levels. The following probes were used: P21 (Hs00355782_m1), DHODH (Hs00361406_m1), CPS II (Hs00983188_m1), PPAT (Hs00601264_m1) RRM1 (Hs01040698_m1), and RRM2 (Hs00357247_g1). The expression was normalized using the housekeeping genes HPRT1 (Hs01003267_m1) or B2M (Hs99999907_m1).
Cell cycle, apoptosis, and cell proliferation
Cell-cycle analysis was performed at the indicated time points with Ki67 (BD Pharmingen) and Hoechst (Invitrogen) as previously described.38 Apoptosis assay was performed with the Annexin V Apoptosis Detection Kit I (BD Pharmingen) according to the manufacturer’s instructions at 48 h after dNs addition and/or transduction. For proliferation assay cells were stained with Cell Proliferation Dye eFluor 670 (Invitrogen) immediately after thawing and the mean fluorescent intensity of the dye was analyzed at different time points after dNs addition/transduction by FACS.
Colony-forming cell assay
Colony-forming cell assays were performed by plating 800 human HSPC in in MethoCult H4434 Classic medium (STEMCELL Technologies) supplemented with streptomycin (0.1 mg/mL). Fifteen days later, colonies were scored by light microscopy for colony counts and morphology as erythroid or myeloid, collected as a pool and lysed for molecular analysis to evaluate transduction efficiencies.
Markers expression analysis
Expression analysis of quiescent and activated HSC markers genes for de novo pyrimidines and purines synthesis pathway was performed by exploiting the publicly available RNA-Seq dataset (GSE153911) from the Gene Expression Ominbus database. The raw counts matrices from qLT-HSCs and aLT-HSCs samples were retrieved, processed, and compared using the R/Bioconductor package DESeq2, following the standard analysis workflow.56 Mapping of gene expression levels to corresponding Kyoto Encyclopedia of Genes and Genomes pathways was performed using the R/Bioconductor package Pathview.57
Flow cytometry
All cytometric analyses were performed using the FACS Canto II and analyzed with the FACS Express De Novo Software. Samples were collected, processed, and analyzed as previously described.8 HSPC subpopulation composition was determined using the following antibodies: CD34 (RRID: AB_2868855), CD133 (RRID: AB_2726012), and CD90 (RRID: AB_398677). Human engraftment in the peripheral blood and in the spleen of mice was evaluated with the following antibodies: CD45 (RRID: AB_1944368), CD19 (RRID: AB_2868815), CD13 (RRID: AB_2737672), and CD3 (RRID: AB_398591). The BM was analyzed using the following antibodies: CD45 (RRID: AB_1944368), CD34 (RRID: AB_2868855), CD38 (RRID: AB_256218), CD19 (RRID: AB_2868815), CD90 (RRID: AB_398677), and CD33 (RRID: AB_2737405). CFU were analyzed using the following antibodies: CD235a (RRID: AB_398499) and CD33 (RRID: AB_2737405). We added 7-aminoactinomycin D to exclude dead cells from the analysis.
Transplantation of human HSPC
Human HSPC were kept unstimulated or pre-stimulated with human cytokines for 24 h and transduced with a GFP-expressing LV at the indicated MOI and timing. The day after transduction cells were infused into the tail vein of sublethally irradiated (200 cGy) 8-to 10-week-old female NSG mice (NOD scid gamma, The Jackson Laboratory). Peripheral blood was analyzed at the indicated time points and at indicated times mice were sacrificed by CO2 for organ analysis. For secondary transplant experiments, BM of primary recipients were pooled per condition and CD34+ cells were isolated through positive magnetic beads selection (Miltenyi). We injected 850,000 cells into the tail vein of sublethally irradiated 8- to 10-week-old NSG mice. Peripheral blood and organs were analyzed at indicated times. All animal procedures were performed according to protocols approved by the Animal Care and Use Committee of the Ospedale San Raffaele (IACUC 782 and 1220) and communicated to the Ministry of Health and local authorities according to Italian law.
Integration site retrieval and bioinformatics analyses
The genomic DNA extracted form LC and BM samples was subjected to Sonication and Linker Mediated (SLiM)-PCR as described.58 Briefly, the genomic DNA was physically sheared by sonication using a Covaris E220 ultrasonicator, end-repaired and 3′-adenylated using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs) and then ligated to a synthetic linker cassette previously described. The PCR procedure applied for integration site retrieval and mapping is equal to what previously described.58 Sequencing reads were processed using a custom bioinformatics pipeline VISPA2 that isolates genomic sequences flanking the vector LTR and maps them to the human genome (hg19).25 Downstream analyses of vector integration sites were executed with ISAnalytics to evaluate the clonal diversity by Shannon diversity index corrected by the VCN24,58; clonal abundance, estimated as relative percentage of genome counts determined by the R-package “sonicLength,” and common insertion sites, calculated by the Grubbs test for outliers.30,59
Statistical analysis
Statistical analyses were conducted with GraphPad Prism 9.0 version. Values are expressed as mean ± SEM or SD and all n numbers represent biological repeats. The number of samples analyzed and the statistical test used are indicated in the figure legends. Differences were considered statistically significant at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns indicates non-significance.
Data and code availability
Integration site analysis Matrix data (Data S1) will be disclosed in a data supplement available with the online version of this article. Relative fastQ data will be deposited and available through the DDBJ Sequence Read Archive (DRA) public repository (https://www.ddbj.nig.ac.jp/dra/index-e.html). For all other original data, please contact anna.kajaste@unipv.it.
Acknowledgments
This work was supported by grants from the European Research Council (ERC-CoG 819815-ImmunoStem), The Italian Ministry of Health (GR-2018-12366006), and the Telethon Foundation (TELE20-C3) to A.K.R. and (TELE20-A4) to G.F.. This work was also supported by NIH AI136581 and AI162633 to B.K. by the German Research Foundation — Project-ID 369799452-TRR237 B19 to R.B. and by grants from the National Institutes of Health to N.R.L. (DA046100, AI122390 and AI120898). E.V. and G.U. conducted this study as partial fulfilment of their PhD in Molecular Medicine, Program in Cellular and Molecular Physiopathology, International PhD School, Vita-Salute San Raffaele University, Milan, Italy. Experimental schemes in the figures were created with BioRender.com. We wish to thank Roberta Caccia and all members of the A.K.R. lab for help with the in vivo sample collections, Simone Cardaci for helpful discussions on the role of pyrimidines in HSPC, and Cesare Covino from Alembic for help with confocal imaging acquisition and analysis.
Author contributions
E.V., G.U., F.P., and I.Ca. conducted experiments and analyzed data; I.Cu. provided technical assistance; M.A.A. and I.M. performed bioinformatic meta-analysis; A.O. and B.K. performed dNTP quantifications; G.P., F.B., S.A., G.S., and E.M. performed and analyzed vector integration sites; M.L., G.F., R.B., and N.R.L. provided reagents and intellectual input; E.V., G.U., F.P., and A.K.R. designed the research study, analyzed data, and wrote the manuscript.
Declaration of interests
E.V., G.U., F.P., and A.K.-R. are inventors on pending and issued patents on lentiviral gene transfer filed by the Telethon Foundation and the San Raffaele Scientific Institute.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.11.020.
Supplemental information
References
- 1.Naldini L. Genetic engineering of hematopoiesis: current stage of clinical translation and future perspectives. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201809958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari G., Thrasher A.J., Aiuti A. Gene therapy using haematopoietic stem and progenitor cells. Nat. Rev. Genet. 2021;22:216–234. doi: 10.1038/s41576-020-00298-5. [DOI] [PubMed] [Google Scholar]
- 3.Millington M., Arndt A., Boyd M., Applegate T., Shen S. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PloS One. 2009;4:e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glimm H., Oh I.H., Eaves C.J. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0) Blood. 2000;96:4185–4193. [PubMed] [Google Scholar]
- 5.Barquinero J., Segovia J.C., Ramírez M., Limón A., Güenechea G., Puig T., Briones J., García J., Bueren J.A. Efficient transduction of human hematopoietic repopulating cells generating stable engraftment of transgene-expressing cells in NOD/SCID mice. Blood. 2000;95:3085–3093. [PubMed] [Google Scholar]
- 6.Santoni de Sio F.R., Gritti A., Cascio P., Neri M., Sampaolesi M., Galli C., Luban J., Naldini L. Lentiviral vector gene transfer is limited by the proteasome at postentry steps in various types of stem cells. Stem Cells. 2008;26:2142–2152. doi: 10.1634/stemcells.2007-0705. [DOI] [PubMed] [Google Scholar]
- 7.Petrillo C., Cesana D., Piras F., Bartolaccini S., Naldini L., Montini E., Kajaste-Rudnitski A. Cyclosporin a and rapamycin relieve distinct lentiviral restriction blocks in hematopoietic stem and progenitor cells. Mol. Ther: J. Am. Soc. Gene Ther. 2015;23:352–362. doi: 10.1038/mt.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrillo C., Thorne L.G., Unali G., Schiroli G., Giordano A.M.S., Piras F., Cuccovillo I., Petit S.J., Ahsan F., Noursadeghi M., et al. Cyclosporine H Overcomes Innate Immune Restrictions to Improve Lentiviral Transduction and Gene Editing In Human Hematopoietic Stem Cells. Cell Stem Cell. 2018;23:820–832.e9. doi: 10.1016/j.stem.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C.X., Sather B.D., Wang X., Adair J., Khan I., Singh S., Lang S., Adams A., Curinga G., Kiem H.P., et al. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124:913–923. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrecka K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M.P., Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li D., Schlaepfer E., Audigé A., Rochat M.A., Ivic S., Knowlton C.N., Kim B., Keppler O.T., Speck R.F. Vpx mediated degradation of SAMHD1 has only a very limited effect on lentiviral transduction rate in ex vivo cultured HSPCs. Stem Cell Res. 2015;15:271–280. doi: 10.1016/j.scr.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkola H., Woods N.B., Sjögren M., Helgadottir H., Hamaguchi I., Jacobsen S.E., Trono D., Karlsson S. Lentivirus gene transfer in murine hematopoietic progenitor cells is compromised by a delay in proviral integration and results in transduction mosaicism and heterogeneous gene expression in progeny cells. J. Virol. 2000;74:11911–11918. doi: 10.1128/jvi.74.24.11911-11918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bobadilla S., Sunseri N., Landau N.R. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 2013;20:514–520. doi: 10.1038/gt.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behrendt R., Schumann T., Gerbaulet A., Nguyen L.A., Schubert N., Alexopoulou D., Berka U., Lienenklaus S., Peschke K., Gibbert K., et al. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 2013;4:689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollenbaugh J.A., Tao S., Lenzi G.M., Ryu S., Kim D.H., Diaz-Griffero F., Schinazi R.F., Kim B. dNTP pool modulation dynamics by SAMHD1 protein in monocyte-derived macrophages. Retrovirology. 2014;11:63. doi: 10.1186/s12977-014-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E.C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldauf H.M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marktel S., Scaramuzza S., Cicalese M.P., Giglio F., Galimberti S., Lidonnici M.R., Calbi V., Assanelli A., Bernardo M.E., Rossi C., et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ss-thalassemia. Nat. Med. 2019;25:234–241. doi: 10.1038/s41591-018-0301-6. [DOI] [PubMed] [Google Scholar]
- 20.Ferrari S., Jacob A., Cesana D., Laugel M., Beretta S., Varesi A., Unali G., Conti A., Canarutto D., Albano L., et al. Choice of template delivery mitigates the genotoxic risk and adverse impact of editing in human hematopoietic stem cells. Cell Stem Cell. 2022;29:1428–1444.e9. doi: 10.1016/j.stem.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Prat L., Kaufmann K.B., Schneiter F., Voisin V., Murison A., Chen J., Chan-Seng-Yue M., Gan O.I., McLeod J.L., Smith S.A., et al. TFEB-mediated endolysosomal activity controls human hematopoietic stem cell fate. Cell Stem Cell. 2021;28:1838–1850.e10. doi: 10.1016/j.stem.2021.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Larochelle A., Gillette J.M., Desmond R., Ichwan B., Cantilena A., Cerf A., Barrett A.J., Wayne A.S., Lippincott-Schwartz J., Dunbar C.E. Bone marrow homing and engraftment of human hematopoietic stem and progenitor cells is mediated by a polarized membrane domain. Blood. 2012;119:1848–1855. doi: 10.1182/blood-2011-08-371583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallinikou K., Anjos-Afonso F., Blundell M.P., Ings S.J., Watts M.J., Thrasher A.J., Linch D.C., Bonnet D., Yong K.L. Engraftment defect of cytokine-cultured adult human mobilized CD34(+) cells is related to reduced adhesion to bone marrow niche elements. Br. J. Haematol. 2012;158:778–787. doi: 10.1111/j.1365-2141.2012.09219.x. [DOI] [PubMed] [Google Scholar]
- 24.Pais G., Spinozzi G., Cesana D., Benedicenti F., Albertini A., Bernardo M.E., Gentner B., Montini E., Calabria A. ISAnalytics enables longitudinal and high-throughput clonal tracking studies in hematopoietic stem cell gene therapy applications. Brief. Bioinform. 2023;24 doi: 10.1093/bib/bbac551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spinozzi G., Calabria A., Brasca S., Beretta S., Merelli I., Milanesi L., Montini E. VISPA2: a scalable pipeline for high-throughput identification and annotation of vector integration sites. BMC Bioinformatics. 2017;18:520. doi: 10.1186/s12859-017-1937-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentner B., Tucci F., Galimberti S., Fumagalli F., De Pellegrin M., Silvani P., Camesasca C., Pontesilli S., Darin S., Ciotti F., et al. Hematopoietic Stem- and Progenitor-Cell Gene Therapy for Hurler Syndrome. N. Engl. J. Med. 2021;385:1929–1940. doi: 10.1056/NEJMoa2106596. [DOI] [PubMed] [Google Scholar]
- 27.Sessa M., Lorioli L., Fumagalli F., Acquati S., Redaelli D., Baldoli C., Canale S., Lopez I.D., Morena F., Calabria A., et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase ½ trial. Lancet. 2016;388:476–487. doi: 10.1016/S0140-6736(16)30374-9. [DOI] [PubMed] [Google Scholar]
- 28.Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S., et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 29.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C., et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biffi A., Bartolomae C.C., Cesana D., Cartier N., Aubourg P., Ranzani M., Cesani M., Benedicenti F., Plati T., Rubagotti E., et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 31.Zonari E., Desantis G., Petrillo C., Boccalatte F.E., Lidonnici M.R., Kajaste-Rudnitski A., Aiuti A., Ferrari G., Naldini L., Gentner B. Efficient Ex Vivo Engineering and Expansion of Highly Purified Human Hematopoietic Stem and Progenitor Cell Populations for Gene Therapy. Stem Cell Rep. 2017;8:977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heffner G.C., Bonner M., Christiansen L., Pierciey F.J., Campbell D., Smurnyy Y., Zhang W., Hamel A., Shaw S., Lewis G., et al. Prostaglandin E2 Increases Lentiviral Vector Transduction Efficiency of Adult Human Hematopoietic Stem and Progenitor Cells. Mol. Ther: J. Am. Soc. Gene Ther. 2018;26:320–328. doi: 10.1016/j.ymthe.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozog S., Timberlake N.D., Hermann K., Garijo O., Haworth K.G., Shi G., Glinkerman C.M., Schefter L.E., D'Souza S., Simpson E., et al. Resveratrol trimer enhances gene delivery to hematopoietic stem cells by reducing antiviral restriction at endosomes. Blood. 2019;134:1298–1311. doi: 10.1182/blood.2019000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprenger H.G., MacVicar T., Bahat A., Fiedler K.U., Hermans S., Ehrentraut D., Ried K., Milenkovic D., Bonekamp N., Larsson N.G., et al. Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity. Nat. Metab. 2021;3:636–650. doi: 10.1038/s42255-021-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diehl F.F., Miettinen T.P., Elbashir R., Nabel C.S., Darnell A.M., Do B.T., Manalis S.R., Lewis C.A., Vander Heiden M.G. Nucleotide imbalance decouples cell growth from cell proliferation. Nat. Cell Biol. 2022;24:1252–1264. doi: 10.1038/s41556-022-00965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davenne T., Klintman J., Sharma S., Rigby R.E., Blest H.T.W., Cursi C., Bridgeman A., Dadonaite B., De Keersmaecker K., Hillmen P., et al. SAMHD1 Limits the Efficacy of Forodesine in Leukemia by Protecting Cells against the Cytotoxicity of dGTP. Cell Rep. 2020;31:107640. doi: 10.1016/j.celrep.2020.107640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohnken R., Kodigepalli K.M., Wu L. Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Mol. Cancer. 2015;14:176. doi: 10.1186/s12943-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piras F., Riba M., Petrillo C., Lazarevic D., Cuccovillo I., Bartolaccini S., Stupka E., Gentner B., Cittaro D., Naldini L., et al. Lentiviral vectors escape innate sensing but trigger p53 in human hematopoietic stem and progenitor cells. EMBO Mol. Med. 2017;9:1198–1211. doi: 10.15252/emmm.201707922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohn D.B., Booth C., Kang E.M., Pai S.Y., Shaw K.L., Santilli G., Armant M., Buckland K.F., Choi U., De Ravin S.S., et al. Lentiviral gene therapy for X-linked chronic granulomatous disease. Nat. Med. 2020;26:200–206. doi: 10.1038/s41591-019-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnani A., Brosselin P., Beauté J., de Vergnes N., Mouy R., Debré M., Suarez F., Hermine O., Lortholary O., Blanche S., et al. Inflammatory manifestations in a single-center cohort of patients with chronic granulomatous disease. J. Allergy Clin. Immunol. 2014;134:655–662.e8. doi: 10.1016/j.jaci.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Yahata T., Takanashi T., Muguruma Y., Ibrahim A.A., Matsuzawa H., Uno T., Sheng Y., Onizuka M., Ito M., Kato S., et al. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood. 2011;118:2941–2950. doi: 10.1182/blood-2011-01-330050. [DOI] [PubMed] [Google Scholar]
- 42.Weisser M., Demel U.M., Stein S., Chen-Wichmann L., Touzot F., Santilli G., Sujer S., Brendel C., Siler U., Cavazzana M., et al. Hyperinflammation in patients with chronic granulomatous disease leads to impairment of hematopoietic stem cell functions. J. Allergy Clin. Immunol. 2016;138:219–228.e9. doi: 10.1016/j.jaci.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Rio P., Navarro S., Wang W., Sanchez-Dominguez R., Pujol R.M., Segovia J.C., Bogliolo M., Merino E., Wu N., Salgado R., et al. Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat. Med. 2019;25:1396–1401. doi: 10.1038/s41591-019-0550-z. [DOI] [PubMed] [Google Scholar]
- 44.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Follenzi A., Ailles L.E., Bakovic S., Geuna M., Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 46.Montini E., Cesana D., Schmidt M., Sanvito F., Ponzoni M., Bartholomae C., Sergi Sergi L., Benedicenti F., Ambrosi A., Di Serio C., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 47.Lidonnici M.R., Paleari Y., Tiboni F., Mandelli G., Rossi C., Vezzoli M., Aprile A., Lederer C.W., Ambrosi A., Chanut F., et al. Multiple Integrated Non-clinical Studies Predict the Safety of Lentivirus-Mediated Gene Therapy for beta-Thalassemia. Mol. Ther. Methods Clin. Dev. 2018;11:9–28. doi: 10.1016/j.omtm.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D., et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 49.Mangeot P.E., Duperrier K., Negre D., Boson B., Rigal D., Cosset F.L., Darlix J.L. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol. Ther: J. Am. Soc. Gene Ther. 2002;5:283–290. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- 50.Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W.L., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schiroli G., Conti A., Ferrari S., Della Volpe L., Jacob A., Albano L., Beretta S., Calabria A., Vavassori V., Gasparini P., et al. Precise Gene Editing Preserves Hematopoietic Stem Cell Function following Transient p53-Mediated DNA Damage Response. Cell Stem Cell. 2019;24:551–565.e8. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behrendt R., Schumann T., Gerbaulet A., Nguyen L.A., Schubert N., Alexopoulou D., Berka U., Lienenklaus S., Peschke K., Gibbert K., et al. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 2013;4:689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C., et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrari S., Jacob A., Beretta S., Unali G., Albano L., Vavassori V., Cittaro D., Lazarevic D., Brombin C., Cugnata F., et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol. 2020;38:1298–1308. doi: 10.1038/s41587-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diamond T.L., Roshal M., Jamburuthugoda V.K., Reynolds H.M., Merriam A.R., Lee K.Y., Balakrishnan M., Bambara R.A., Planelles V., Dewhurst S., et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cesana D., Calabria A., Rudilosso L., Gallina P., Benedicenti F., Spinozzi G., Schiroli G., Magnani A., Acquati S., Fumagalli F., et al. Retrieval of vector integration sites from cell-free DNA. Nat. Med. 2021;27:1458–1470. doi: 10.1038/s41591-021-01389-4. [DOI] [PubMed] [Google Scholar]
- 59.Berry C.C., Gillet N.A., Melamed A., Gormley N., Bangham C.R.M., Bushman F.D. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28:755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Integration site analysis Matrix data (Data S1) will be disclosed in a data supplement available with the online version of this article. Relative fastQ data will be deposited and available through the DDBJ Sequence Read Archive (DRA) public repository (https://www.ddbj.nig.ac.jp/dra/index-e.html). For all other original data, please contact anna.kajaste@unipv.it.