Abstract
Objective
Pre-treatment enhanced CT image data were used to train and build models to predict the efficacy of non-small cell lung cancer after conventional radiotherapy and chemotherapy using two classification algorithms, Logistic Regression (LR) and Gaussian Naive Baye (GNB).
Methods
In this study, we used pre-treatment enhanced CT image data for region of interest (ROI) sketching and feature extraction. We utilized the least absolute shrinkage and selection operator (LASSO) mutual confidence method for feature screening. We pre-screened logistic regression (LR) and Gaussian naive Bayes (GNB) classification algorithms and trained and modeled the screened features. We plotted 5-fold and 10-fold cross-validated receiver operating characteristic (ROC) curves to calculate the area under the curve (AUC). We performed DeLong's test for validation and plotted calibration curves and decision curves to assess model performance.
Results
A total of 102 patients were included in this study, and after a comparative analysis of the two models, LR had only slightly lower specificity than GNB, and higher sensitivity, accuracy, AUC value, precision, and F1 value than GNB (training set accuracy: 0.787, AUC value: 0.851; test set accuracy: 0.772, AUC value: 0.849), and the LR model has better performance in both the decision curve and the calibration curve.
Conclusion
CT can be used for efficacy prediction after radiotherapy and chemotherapy in NSCLC patients. LR is more suitable for predicting whether NSCLC prognosis is in remission without considering the computing speed.
1. Introduction
According to data published in the Global Cancer Statistics report, lung cancer is currently the malignancy with the highest risk of morbidity and mortality [1]; 85 % of them are non-small cell lung cancer (NSCLC), mainly adenocarcinoma and squamous carcinoma [2]. The National Comprehensive Cancer Network (NCCN) practice guideline 2022.5 for NSCLC recommends that the primary treatment options for patients with NSCLC include surgery, radiotherapy therapy, chemotherapy, molecular targeted therapy, and immunotherapy [3], and the current treatment modality for all patients with NSCLC for whom radical surgery is not preferred is often a combination of radiation therapy and chemotherapy. However, in clinical practice, due to the insidious nature of NSCLC, most patients have missed the opportunity of radical surgery at the time of diagnosis, or are unable to undergo surgery due to the location of the tumor close to the vital organs or main bronchus, so a large proportion of patients take combined radiotherapy and chemotherapy.
In clinical practice, even at the same disease stage, heterogeneity exists in different individuals, and there is also heterogeneity within the tumor, which does not manifest as homogeneous. Current diagnostic modalities, such as pathological diagnosis and imaging diagnosis, have difficulty in reflecting the heterogeneity within the tumor that is not visible to the naked eye, so the treatment outcome after radiotherapy and chemotherapy may also vary considerably [4]. Suppose reliable efficacy predictors are available to classify patients before treatment or early in the disease. In that case, they can help physicians develop individualized treatment plans, provide a basis for decision-making on drug therapy, adjust the dose of radiotherapy and chemotherapy regimens, or extend the duration of treatment appropriately, which can be used to improve patient prognosis [5].
Radiomics mines the vast amount of information inherent in CT images and performs quantitative analysis to explore potential diagnostic, therapeutic, and prognostic applications, which have the advantages of non-invasiveness, reproducibility, convenience, and ease of operation occupy an important position in precision medicine. Currently, with the continuous development and improvement of chest Computed Tomography (CT), CT is the most commonly used noninvasive examination modality in lung cancer diagnosis and efficacy assessment, and CT has the advantages of a lower price, fewer scanning parameters, more stable image quality, and more standard image data [6]. Therefore, this study was conducted to predict and validate the prognosis of NSCLC patients after conventional radiotherapy and chemotherapy by radiomics based on the enhanced CT images before treatment.
2. Materials and methods
This study was approved by the Ethics Review Committee of the Hospital of Chuanbei Medical College, File No. 2022ER581-1, and the patient gave consent for his images and data to be used for the experiment and study. The study was conducted in accordance with established ethical guidelines and complied with all regulations.
3. Data and methods
3.1. Study population
Inclusion criteria: ① pathological histological puncture biopsy confirmed the diagnosis of NSCLC (including squamous cell carcinoma, adenocarcinoma, and other types); ② stage II and III patients who were inoperable; ③ stage IV patients without multiple metastases and distant organ metastases; ④ receiving radical radiotherapy and 4–6 cycles of platinum-containing chemotherapy; ⑤ patients in good general condition with a Karnofsky (KPS) score ≥70 scores (Table 1).
Table 1.
Karnofsky (kps, percentage method) functional status rating scale.
| Physical status | Score* |
|---|---|
| Normal, no signs and symptoms | 100 |
| Able to perform normal activities with minor signs and symptoms | 90 |
| Barely able to perform normal activities with some signs and symptoms | 80 |
| Self-care, but unable to maintain normal life and work | 70 |
| Mostly self-care, but occasionally needs help | 60 |
| Needs frequent care | 50 |
| Cannot take care of himself/herself, needs special care and assistance. | 40 |
| Severely unable to care for self | 30 |
| Severely ill, requiring hospitalization and active, supportive care | 20 |
| Critically ill, near death | 10 |
| Death | 0 |
*:The higher the score, the better the health status, the more tolerable the side effects of the treatment, and therefore the more likely it is that the treatment will be complete. The lower the score, the worse the health status; if the score is below 60, many effective anti-tumor treatments cannot be implemented.
Exclusion criteria: (i) patients with incomplete clinical or image information of low quality; (ii) patients without efficacy assessment information after treatment; (iii) those with other primary tumors; (iv) those who did not follow up regularly according to the follow-up requirements.
The data of patients diagnosed with NSCLC in our hospital between December 2018 and July 2022 and meeting the above criteria were retrospectively analyzed, and 102 cases were finally included.1.2 Treatment options.
3.1.1. Radiotherapy protocols
The total dose was 40–60 Gy, with single irradiation of 2.0 Gy, once a day, five days a week treatment.
3.1.2. Chemotherapy regimen
Four to six treatment cycles of platinum-based chemotherapy drugs were used. The drug doses were calculated based on the patient's body surface area, dosing regimen, and liver and kidney function.
3.2. Efficacy assessment and grouping
3.2.1. Efficacy assessment
All cases were rated according to the WHO Response Evaluation Criteria In Solid Tumors [7] (RECIST1.1) as complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD).
3.2.2. Case grouping
Patients were divided into the remission group (including CR and PR) and the non-remission group (including PD and SD).
3.3. Image Acquisition and processing
3.3.1. Acquisition of images
In order to ensure the timeliness of obtaining image information that can reflect the closest treatment phase, all images were obtained from the radiotherapy center's images of the radiation target area plan developed within 1–3 days prior to radiotherapy. For patients who could cooperate with breathing instructions, we instructed patients to hold their breath during the scanning period to minimize the impact of respiratory motion. Specifically, patients held their breath at maximal inspiration upon hearing the breath-hold command from the CT technologist prior to initiating non-table motion scanning.All patients were trained to familiarize themselves with the breath-holding process before the scan. For patients with potential inadequate breath-hold times, scanning was performed in segments. Depending on patient positioning, the scanning direction was adjusted to minimize respiratory motion of the tumor. Scanning equipment: CT (Siemens Emotion 16-slice configuration) Scanning parameters: Tube Voltage: 120 kV; Tube Current: 200 mA; Layer Thickness: 3 mm; Pitch: 0.625 mm; FOV = 700 mm × 700 mm. In this study, we used an iodixanol contrast agent manufactured by Yangzijiang Pharmaceutical Group Co., Ltd. with a specification of 74.1 g of iodine in 100 ml, i.e., 350 mg of iodine per 100 ml.
3.3.2. Image preprocessing
in order to provide a high level of detail, we resample the image using a 3d silcer, the resampled voxel size is set to 1 × 1 × 1 mm. reconstruction matrix size: 64 x 256, reconstruction kernel size: 2 × 3 × 4.
3.3.3. Image segmentation
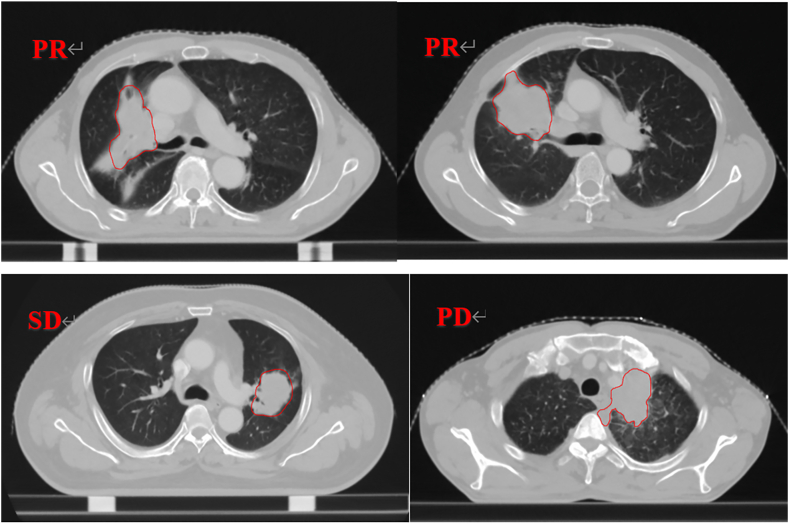
Export the images in DICOM format and upload them to the Monaco system for image segmentation. When uploading the images, the images were renamed to achieve the effect of removing the blended names. In this study, the Gross Tumor Volume (GTV) was manually segmented layer by layer using 3D Slicer software by two radiation therapists with more than five years of experience in thoracic tumor therapy. After the initial segmentation, the results were reviewed and modified by a radiation therapist with more than ten years of experience in thoracic tumor radiotherapy. The whole segmentation process strictly followed the recommendations of the National Comprehensive Cancer Network (NCCN) [8]Guidelines for the Treatment of Non-Small Cell Lung Cancer (NCCN Guidelines Version 3.2022) (Fig. 1). [9].
Fig. 1.
Illustration of ROI Contour
PR, SD and PD respectively represent three types of therapeutic effects after evaluation based on RECIST 1.1 criteria; the contour in the figure represents GTV, where A corresponds to PR (Partial Response), B corresponds to SD (Stable Disease), and C corresponds to PD (Progressive Disease) in terms of total tumor volume. ROI refers to Region of Interest.
3.4. Feature extraction
The image and the outlined ROI structure are imported into 3D Slicer (4.11) for feature extraction. The features extracted by 3D Slicer are mainly performed through the open-source plug-in (Slicer Radiomics). The extracted features include the mean, minimum, maximum, standard deviation, skewness, kurtosis, etc., of the first-order statistical features of the original image; the surface area, volume, surface area to volume ratio, sphericity, compactness, and 3D diameter of the morphological features and five types of texture features [10]: Gray Level Cooccurence Matrix (GLCM), Gray Level Run Length Matrix (GLRLM), Gray Level Size Zone Matrix (GLSZM), Neighboring Gray Tone Difference Matrix (NGTDM), Gray Level Dependence Matrix (GLDM); and the features after filtering and transforming the images using Laplacian of Gaussian (LoG) with LoG-sigma sizes of 4.0 mm, 5.0 mm, and 6.0 mm, respectively, and the eight filter functions of LLL, LLH, LHL, LHH, HLL, HHL, HLH, HHH, and HHH in the wavelet transform for a total of 11 filtering methods, each first-order, second-order and texture features mentioned above after the transformation [11]. In this case, the wavelet transform image was created using the default parameters of 3D Slicer. The wavelet sequence used was Daubechies (Db4). A 3-layer decomposition was performed. The fill mode was symmetric filling. Quantization of 64 bins was applied. The sampling mode was custom sampling, with specific sampling densities of x-, y- and z-axis sampling intervals of 4, 5, and 6 voxels, respectively.
3.5. Model training and validation
3.5.1. Screening of features
Radiomic features were extracted using the Python module in 3D Slicer 4.10.2 with Anaconda Python 3.7. Downstream data processing and analysis were performed in R 4.2.2. Patients were categorized into mitigation and non-mitigation groups, labeled as 0 and 1 respectively. The data were then normalized and standardized. Features with confidence coefficients greater than 0.1 were filtered out using the confidence method. The mutual information coefficient threshold was set to 0.1 for relatively loose initial filtering, which retained more potential predictive features. This prepared for the next stage of more stringent LASSO regression selection. Finally, useful features were filtered again using LASSO. Next, the data were randomized and randomly split into training (70 %) and test sets (30 %).
3.5.2. Model selection and training
The training set data were input into two classification algorithms - Logistic Regression (LR) , Gaussian Naive Bayes (GNB), Support Vector Machine (SVM)and K-Nearest Neighbors (KNN), The above models are commonly used in the binary classification problem of lung cancer and have been supported by studies to have better prediction effects [[12], [13], [14]]. The training set was further randomly divided into a training subset (70 %) and validation subset (30 %). The training subset data were fed into models for training. During the model training process, the training set data are randomly divided into a training set and validation set [15], brought into the model for training and adjusting the model, and then substituted into the test set data and plotted the training curve together. Pre-screening based on AUC results to exclude SVM and KNN models. In this study, we set the parameters of LR and GNB models as follows. For the LR model, we use LogisticRegression in sklearn, and the main parameters are. penalty = 'l1'; C = 0.1; solver = 'liblinear'; max_iter = 1000. For the GNB model, we use GaussianNB in sklearn, and the main parameters are: var_smoothing = 1e-9,; priors are empty; fit_prior = False.The model training dynamics are observed for overfitting or underfitting. The training curve is used to select the best training depth of the model, i.e., the best model for that data for that algorithm. After the model is trained and brought into the test set data, the corresponding evaluation index values are generated by (i) bringing in the data and then calculating the average value after getting some evaluation index values, such as the accuracy of the model by ten times five-fold cross-validation, (i) bringing in the data and then calculating the average value after getting some evaluation index values, such as the accuracy of the model by ten times five-fold cross-validation. In this study, 5-fold cross-validation was performed 10 times to evaluate model performance. In each cross-validation iteration, the training data was randomly split into 5 equal folds. 4 folds were used for model training and the remaining fold was used for validation. This process was repeated 10 times so that each fold served as the validation set once, The cross-validation accuracy was recorded for each iteration. The final reported cross-validation accuracy is the average of the 5 × 10 = 50 accuracy values obtained during cross-validation.; (ii) building the confusion matrix and the classification report of the model with the model prediction results to evaluate the model; (iii) then plotting the ROC curve with each model training and prediction results and calculating the AUC values [16]. We adhered to the Image Biomarker Standardization Initiative(IBSI) [17]guidelines during image processing procedures to ensure standardization.
3.5.3. Validation of the model
For the LR and GNB models constructed in this paper, the calibration degree of model classification was assessed by calibration curves on the training set and test set, respectively. The calibration curves indicate the extent to which the predicted probabilities match the actual observed probabilities. Decision curves were plotted to assess the performance of model classification on the training set and test set, respectively. The threshold was set to 0.5, reflecting the model's false positive rate versus the true positive rate, and thus indicating the net benefit rate.
3.6. Statistical analysis
The t-test was used for the measurement data; the χ2 test or Fisher's exact test was used for the one-way analysis of the count data. The test levels α = 0.05 and P < 0.05 were considered statistically significant differences.
4. Results
4.1. General information of patients
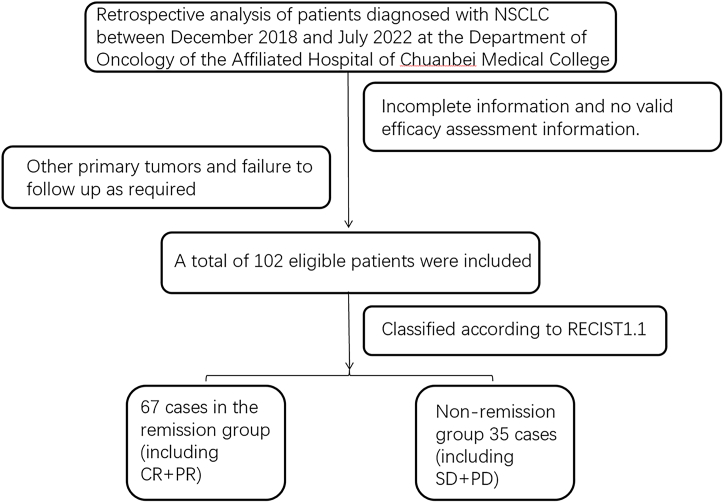
Following the flow of Fig. 2, a total of 102 patients were included in this study, according to the clinical efficacy evaluation index of solid tumors, including 67 cases of PR, 33 cases of SD, 2 cases of PD, 67 cases in the remission group, and 35 cases in the non-remission group. The overall mean age was (62.18 ± 8.50) years, the mean age of the influential group was (62.46 ± 8.47) years, and the mean age of the non-remission group was (61.63 ± 8.57) years.、
Fig. 2.
Inclusion of exclusion flow chart CR,complete response; PR,partial response; SD,stable disease; PD,progressive disease; NSCLC, non-small cell lung cancer.
4.2. Statistical analysis of patients' clinical data and efficacy
The efficacy of 102 patients was evaluated according to the RECIST 1.1 evaluation index. The results showed that the current remission rate of patients who received 4–6 cycles of chemotherapy and radical radiotherapy was 65.69 % (67/102). The general clinical data of the patients are described in Table 2; the measurement data are expressed as "x ± s," and the counting data are expressed as [n(%)]. Except for the age of the patients, the pairwise results between the groups showed (p > 0.05), demonstrating no significant difference between the two groups of patients, which was beneficial for the following analysis.
Table 2.
General clinical data non-remission group remission group and non-remission group description.
| Group | remission group | non-remission group | P |
|---|---|---|---|
| Quantity | 67(65.69 %) | 35(34.31 %) | |
| Age (mean ± standard deviation) | 62.46 ± 8.47 | 61.63 ± 8.57 | 0.2961 |
| Gender | 60(89.55 %) | 0.0062 | |
| Man | 7(10.45 %) | 23(65.71 %) | |
| Woman | 12(34.29 %) | ||
| Smoke | 48(71.64 %) | 0.0515 | |
| Yes | 19(28.36 %) | 18(51.43 %) | |
| No | 17(48.57 %) | ||
| History of chronic lung disease | 8(11.94 %) | >0.9999 | |
| Yes | 59(88.06 %) | 4(11.43 %) | |
| No | 31(88.57 %) | ||
| Pathological pattern | 39(58.21 %) | 0.0971 | |
| squamous carcinoma | 28(41.79 %) | 14(40.00 %) | |
| Adenocarcinoma and others | 21(60.00 %) | ||
| Staging | 2(2.99 %) | 0.6590 | |
| II | 44(65.67 %) | 2(5.71 %) | |
| III | 21(31.34 %) | 20(57.14 %) | |
| IV | 13(37.14 %) |
4.3. Results of radiomics feature screening
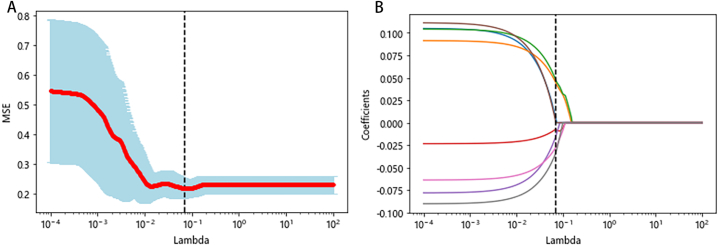
The general clinical data were converted into digital form and normalized, and after standardization, the features were screened together with radiomics features. The features with confidence coefficients greater than 0.1 were screened by the confidence method, and then eight useful features were finally screened by LASSO, at which time the best λ = 0.0693 (as in Fig. 3). It can be seen that all are filtered features: two features after filtering with an LoG-sigma kernel size of 4.0 mm, one feature after filtering with 6.0 mm, and the filtered features of LLH, HLH, and HHH filter functions in wavelet transform. Among the five selected features, only the features of the LLH filter function in wavelet transform are first-order features. The rest are texture features, of which three texture features of the wavelet-HHH filter function in wavelet transform are selected, and the specific features are named in Table 3.
Fig. 3.
LASSO Feature Selection Results (λ = 0.0693)
LASSO regression was employed for feature selection. The parameter (λ) was adjusted using ten rounds of five-fold cross-validation, and bias curves were plotted. The dashed lines indicate the minimum criterion (B) and the 1-SE of the minimum criterion (A). Applying the 1-SE criterion resulted in the selection of eight features, with the optimal value of λ being 0.0693.
Table 3.
Radiomics feature screening.
| Feature name | Source | Type |
|---|---|---|
| log-sigma-4-0-mm-3D:glszm:SmallAreaLowGrayLevelEmphasis | Gauss-Laplace transform | Texture features |
| log-sigma-6-0-mm-3D:firstorder:RootMeanSquared | Wavelet Transform | Texture features |
| log-sigma-6-0-mm-3D:glcm:Imc1 | Wavelet Transform | Texture features |
| wavelet-LLH:firstorder:Median | Wavelet Transform | Texture features |
| wavelet-HLH:glszm:SmallAreaEmphasis | Wavelet Transform | First-order features |
| wavelet-HHH:firstorder:Kurtosis | Wavelet Transform | Texture features |
| wavelet-HHH:glcm:ClusterShade | Wavelet Transform | Texture features |
| wavelet-HHH:glszm:LowGrayLevelZoneEmphasis | Wavelet Transform | Texture features |
4.4. Model tuning
4.4.1. Model screening
Four classifiers - logistic regression (LR), support vector machine (SVM), K-nearest neighbors (KNN), and Gaussian naive Bayes (GNB) - were trained on the dataset using eight selected feature values. The AUC value for each model was calculated. The AUC values for SVM and KNN models were relatively low at 0.62 and 0.59, respectively. Comparatively, the LR and GNB models achieved higher AUC values of 0.85 and 0.79. Based on these results, the LR and GNB classifiers were selected for further analysis in this study due to their better performance.
4.4.2. Training of the model
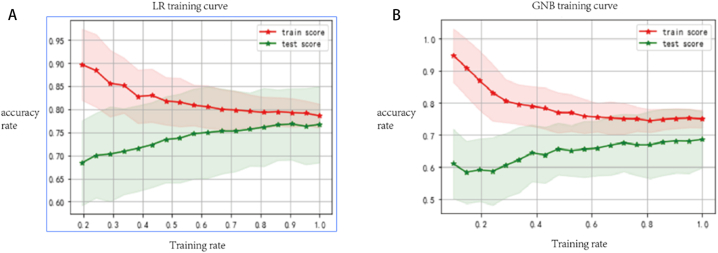
The selected eight features and the corresponding labeled training set data are brought into the LR, GNB two classifiers to train the tuning model, and the training curves of the corresponding models are obtained (as in Fig. 4).
Fig. 4.
Training Curves of LR and GNB Algorithms for Mitigation and Non-Mitigation Groups (horizontal axis represents training rates, vertical axis represents the model's accuracy on the training and testing sets at that moment, solid line denotes the average accuracy, and shaded region indicates the standard deviation generated by validation). A corresponds to the training curve of the LR model, and B corresponds to the training curve of the GNB model.
From the above training curves, we can see that when the training rate increases, i.e., the input training data keeps increasing, the training set scores of both models show a decreasing trend. The test set scores show an increasing trend with some fluctuations, and GNB and LR have a similar trend. When the data are fully input, i.e., up to 1.0 LR training set and test set scores are closest, the test set score is slightly higher than the training set.
4.5. Model test results
Table4. The remission and non-remission groups were tested using both models, and all results were arithmetically averaged as the corresponding final values after ten times five-fold cross-validation. The corresponding data were entered into the model to generate a confusion matrix to calculate further the sensitivity, specificity, precision, and F1 values.
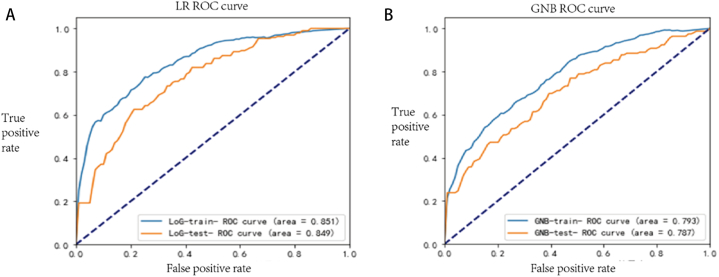
The model was brought into the test set data to generate the ROC curve (Fig. 5), and its AUC value was measured after ten times five-fold cross-validation. The arithmetic mean of the AUC value was taken as the AUC value of the ROC curve of this model (Table 4). The two algorithms were subjected to Delong'test corresponding to the model. The AUC difference between the two algorithms was statistically significant when comparing P = 0.045 < 0.05 between the training set ROC and P = 0.012 < 0.05 between the test set ROC.
From the accuracy and AUC values, we can see that for the LR model, the values of the training set and the test set are very close to each other, regardless of the accuracy or AUC values, and the accuracy, AUC values, sensitivity, precision, and F1 values are higher than those of the GNB model; for the specificity, the GBN model is higher than the LR model.
Fig. 5.
AUC Values of ROC Curves for Two Algorithms in Mitigation and Non-Mitigation Groups, where Panel A represents the LR model and Panel B represents the GNB model.
Table 4.
Two algorithm model results.
| model | accuracy rate |
AUC |
sensitivity | specificity | precision | F1 | P* | |||
|---|---|---|---|---|---|---|---|---|---|---|
| training group | test group | training group | test group | |||||||
| LR | 0.787 | 0.772 | 0.851 | 0.849 | 0.909-0.667 | 0.542 | 0.861 | 0.686 | 0.570 | 0.012 |
| GNB | 0.745 | 0.712 | 0.793 | 0.787 | 0.449 | 0.902 | 0.708 | 0.549 | ||
P*: P-value resulting from the Delong'test performed on the test set ROC of the corresponding model for both algorithms. AUC, Area Under Curve; LR, Logistic Regression; GNB, Gaussian Naive Bayes.
4.6. Model validation results
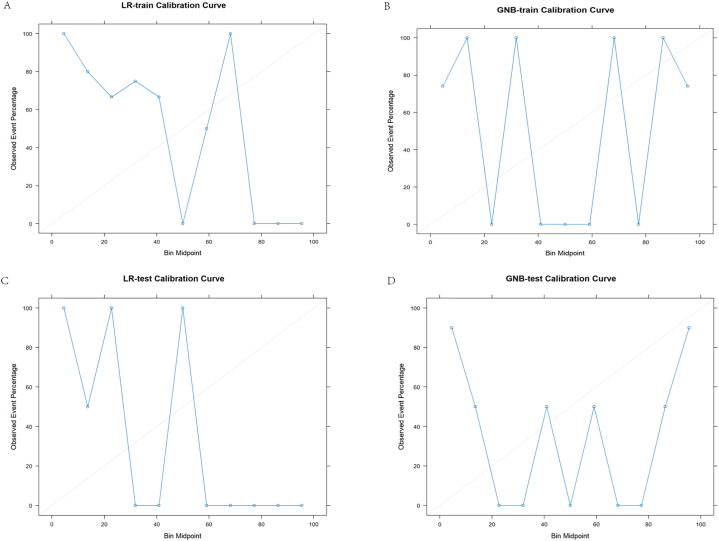
The calibration curves for the training set and test set of the LR and GNB models are shown in Fig. 6. The curves indicate that both models have large fluctuations in calibration stability. However, the LR model demonstrates better calibration than the GNB model on both the training set and test set. Additionally, the difference between the LR calibration curves on the training and test sets is smaller compared to the GNB model. This suggests the LR model has less variance in performance across datasets.
Fig. 6.
These are the calibration curves for the logistic regression (LR) and Gaussian naive Bayes (GNB) models, where the closer the curve is to the diagonal line, the more accurate the model is.A is the calibration curve for the training set of the LR model; B is the calibration curve for the test set of the LR model; C is the calibration curve for the training set of the GNB model; And D is the calibration curve for the test set of the GNB model.
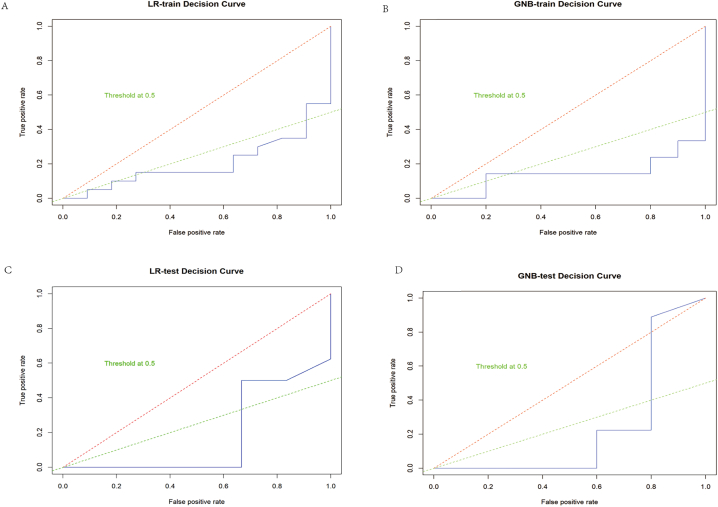
The decision curves for the training and test sets of the LR and GNB models are presented in Fig. 7. The curves show superior performance of the LR model over GNB on both datasets, with higher net benefit across threshold probabilities.
Fig. 7.
These are the decision curves for the logistic regression (LR) and Gaussian naive Bayes (GNB) models. The area below the decision curves represents all the net benefits brought by the model when combining different thresholds. The larger the area is, the stronger the model's ability to improve the overall decision-making effect. The green dashed line is the 0.5 threshold line.A is the decision curve for the training set of the LR model; B is the decision curve for the test set of the LR model; C is the decision curve for the training set of the GNB model; D is the decision curve for the test set of the GNB model.
In summary, the LR model achieves better calibration and clinical utility compared to the GNB model based on the calibration and decision curve analysis.
5. Discussion
The prerequisite for achieving precision medicine is the ability to further classify similar patients based on the current classification. Patients with driver-negative inoperable NSCLC are currently treated with radical radiotherapy combined with platinum-containing chemotherapy as the treatment of choice, but this modality lacks individuality. Conventional pathology and classification factors such as clinical staging do not allow further exploration of patient tumor heterogeneity. They can lead to different prognostic outcomes in patients with seemingly identical pathology and consistent staging, especially in more severe cases. Therefore, non-invasive and easily accessible predictors are needed to delineate patient populations further and are used to estimate the remission rate of NSCLC [18]. With the increasing development of big data and artificial intelligence, various histology concepts have been proposed one after another, and radiomics technology has emerged, accompanied by the mission to address precision medicine (also known as personalized medicine) [19]. Radiomics has been a hotspot and focus of medical research recently and was first conceptualized by Dutch scholar Lambin et al., in 2012. Defined as the high-throughput extraction of a large amount of image information through medical images, which enables image segmentation, feature extraction, and model building and can quantify image heterogeneity caused by changes that are not observed by the human eye, and then through deep data mining, prediction, and analysis thus used to analyze specific information [[20], [21], [22]]. Radiomics aims to break through the limits of the application value of traditional imaging examinations in early diagnosis, efficacy assessment, and prognosis prediction of tumors [23], to provide individualized treatment plans, and to achieve precision medicine. Radiomics has been applied to many types of tumors, but lung cancer is one of the most widely studied and applied malignancies through radiomics [24,25].
In the image segmentation, the ROI area were sketched using 3d slicers software, and two radiation therapists semi-automatically outlined the GTV. After initial segmentation, a chief physician experienced in radiation therapy for thoracic tumors reviewed and modified the results. Multiple physicians checked each other, so the accuracy of the outline could be guaranteed, and the error caused by manual outline could be effectively controlled [26]. we resampled all images to 1 × 1 × 1 mm cubic voxels using trilinear interpolation in 3D Slicer software prior to feature extraction. This voxel size was chosen to achieve isotropic voxels in order to enable standard 3D wavelet-based feature computation across patients. The trilinear interpolation method utilizes neighboring voxels in three dimensions to perform interpolation, which helps minimize resampling artifacts and maintain image continuity compared to lower order interpolation methods. Resampling to isotropic cubic voxels using trilinear interpolation allowed consistent wavelet feature calculation in 3D while preserving image information. For radiomics feature extraction, this study extracts first-order, second-order, and high-order texture features of the original image but also transforms first-order, second-order, and high-order texture features by including wavelet and LoG filter functions. The number of features extracted is as high as 1130, which increases selection possibilities and improves prediction accuracy in feature selection and phase relationship studies. In most articles, the Gaussian filtering parameters of CT images of lung cancer were chosen in the range of 2–6 mm [[27], [28]], and 4–6 mm standard deviation was matched with it to extract details and global features. Fixed three standard deviation parameters can avoid the risk of data overfitting, which is conducive to model generalization. The interval between the three parameters is 1 mm, which can balance the feature richness and the computational amount.
For data processing, this study standardized and normalized the data. The study showed that normalized and normalized processing could eliminate the differences in magnitudes between different data and make the prediction results show a better performance [29]. In the feature screening, this study applied the multiple screening of mutual trust and LASSO regression [30,31] in turn to screen the features required for the model, and according to the "one-tenth" principle proposed by Babyak [32], each feature requires at least 10–15 patients, so finally eight features with high correlation coefficients were selected for model training to reduce the risk of overfitting. In this study, we selected 8 radiomic features through multivariate statistical analysis. These features reflect information regarding gray level distribution, texture, and spatial distribution of the lesions, providing an effective feature basis for further pathological grading modeling. In summary, the selected radiomic features comprehensively reflect the imaging characteristics of the lesions, and provide a key basis of feature selection for pathological grading modeling based on CT images. In model construction, both LR and GNB algorithms were used in this study, and the ROC curves of both models were subjected to Delong'test [33] with P < 0.05. The AUC differences between them were significantly different. All evaluation indexes were averaged after ten times of five-fold cross-validation as the final This avoids over- or under-scores of the model due to a random assignment of the training set test set [34]. The 5 × 10 cross-validation provides a reliable estimate of model performance by reducing variability while avoiding excessive computation compared to other methods like leave-one-out cross-validation. The multiple repetitions help average out randomness in the fold splits. 5 folds was chosen as it offers a good balance between bias and variance. The results showed that LR had only slightly lower specificity than GNB. The sensitivity, accuracy, AUC value, precision, and F1 value were all higher than GNB. After combining the above evaluation indexes, it can be seen that LR is more suitable for predicting whether NSCLC prognosis is in remission without considering the speed of computing (training set accuracy: 0.787, AUC value: 0.851; test set accuracy: 0.772, (AUC value: 0.849). We also plotted calibration curves and decision curves for both the logistic regression (LR) and Gaussian naive Bayes (GNB) models on both the test and validation sets to assess the calibration and net benefit of the comparative models. The results showed that LR performed better in terms of model consistency, calibration, and net benefit. Yuto Sugai [35] et al. used COX to construct a prediction model based on CT images by LASSO screening features and c-dex and Kaplan-Meier curves to model and evaluate the prognosis of NSCLC patients, which is similar to the present study. However, the study used one physician to perform manual segmentation, making reducing the impact generated by subjective errors challenging. The study used COX to construct a prediction model, which often produces overfitting due to the Cox model that directly predicts the time to event with a simple regression equation. This study established two arithmetic prediction models, LR and GNB, using multi-person outlining and checking. ROC curves were drawn based on the enhanced CT of patients before treatment, and the sensitivity, specificity, accuracy, and F1 values of the two models were calculated, which can help reduce the errors generated by manual outlining and adopt a more stable prediction model with a more complex evaluation mechanism and a better evaluation effect, which can be used before patient treatment. The prediction model is used to screen out patients with less than ideal treatment effects and guide the clinic to add subsequent treatment to this part of patients based on the current radiotherapy and chemotherapy mode of treatment.
However, there are still some limitations in this study. First, this study uses a dichotomous classification method, which is more limited [36,37], with a small sample size and a single-center research scheme. It is intended to add multicenter data in later studies to increase the sample size and improve the model's accuracy.
Based on this study, the next intention is to add several algorithms currently commonly used at home and abroad for multi-classification comparison, and also to introduce deep learning methods for comparison, and then expand to multimodality (e.g., adding MRI or PET/CT) [[38], [39], [40], [41]] for a more comprehensive and in-depth study, in anticipation of developing clinical translation tools with better results.
6. Conclusion
In conclusion, predicting the efficacy of radiotherapy and chemotherapy in NSCLC patients based on pre-treatment-enhanced CT radiomics has some predictive value. This study successfully constructed a radiomics-based efficacy prediction model through which the efficacy of NSCLC patients can be predicted before radiotherapy and chemotherapy. If patients were predicted to be in non-remission by the prediction model, the current radiotherapy and chemotherapy modalities were considered to have a limited therapeutic effect, and possible further treatment modalities could be explored to improve the outcome.
Data availability
Given that the data in this study originate from a hospital, and in accordance with relevant laws and hospital policies, we are obliged to protect the privacy and confidentiality of patient data. Consequently, these data cannot be deposited into a publicly accessible repository. However, we have made every effort to ensure that the study results and analyses described in the article are understandable and verifiable by readers, and that all results are supported by the references listed.
CRediT authorship contribution statement
Hanjing Zhang: Writing - review & editing, Writing - original draft, Validation, Software, Resources, Investigation, Data curation, Conceptualization. Yu Deng: Writing - original draft, Investigation, Data curation, Conceptualization. M.A. Xiaojie: Supervision. Qian Zou: Software, Data curation. Huanhui Liu: Data curation. Ni Tang: Data curation. Yuanyuan Luo: Data curation. Xuejing Xiang: Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer Statistics 2020: GLOBOCAN estimates of Incidence and mortality Worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Fave X., Zhang L., Yang J., Mackin D., Balter P., Gomez D., Followill D., Jones A.K., Stingo F., Liao Z., Mohan R., Court L. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci. Rep. 2017 Apr 3;7(1):588. doi: 10.1038/s41598-017-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mithoowani H., Febbraro M. Non-small-cell lung cancer in 2022: a review for general Practitioners in oncology. Curr. Oncol. 2022 Mar 9;29(3):1828–1839. doi: 10.3390/curroncol29030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Hua-Chun, Zhi-Chao Fu, Cheng Hui-Hua, et al. Prostate cancer treated with reduced-volume intensity-modulated radiation therapy: report on the 5-year outcome of a prospective series. Medicine. 2017;96(52) doi: 10.1097/MD.0000000000009450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L., Zhan C., Gu J., et al. Role of Skip Mediastinal lymph node metastasis for patients with Resectable non-small-cell lung cancer: a Propensity score matching analysis. Clin. Lung Cancer. 2019;20(3):346–355. doi: 10.1016/j.cllc.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi R., Akiyoshi T. ASO Author. Reflections: CT-based radiomics model to predict lateral lymph node metastasis after Neoadjuvant (Chemo)Radiotherapy in advanced low rectal cancer. Ann. Surg Oncol. 2020;27:4284–4285. doi: 10.1245/s10434-020-08977-7. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Zhang Z., Tao P., Reyila M., Qi X., Yang J. The Abnormal expression of miR-205-5p, miR-195-5p, and VEGF-A in human cervical cancer is related to the treatment of Venous Thromboembolism. BioMed Res. Int. 2020 Aug 8 doi: 10.1155/2020/3929435. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ettinger D.S., Wood D.E., Aisner D.L., Akerley W., Bauman J.R., Bharat A., Bruno D.S., Chang J.Y., Chirieac L.R., D'Amico T.A., DeCamp M., Dilling T.J., Dowell J., Gettinger S., Grotz T.E., Gubens M.A., Hegde A., Lackner R.P., Lanuti M., Lin J., Loo B.W., Lovly C.M., Maldonado F., Massarelli E., Morgensztern D., Ng T., Otterson G.A., Pacheco J.M., Patel S.P., Riely G.J., Riess J., Schild S.E., Shapiro T.A., Singh A.P., Stevenson J., Tam A., Tanvetyanon T., Yanagawa J., Yang S.C., Yau E., Gregory K., Hughes M. Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022 May;20(5):497–530. doi: 10.6004/jnccn.2022.0025. [DOI] [PubMed] [Google Scholar]
- 9.Avanzo M., Stancanello J., El Naqa I. Beyond imaging: the promise of radiomics. Phys. Med. 2017 Jun;38:122–139. doi: 10.1016/j.ejmp.2017.05.071. [DOI] [PubMed] [Google Scholar]
- 10.Yang H., Yan S., et al. Prediction of acute versus chronic osteoporotic vertebral fracture using radiomics-clinical model on CT. Eur. J. Radiol. 2022 Apr;149 doi: 10.1016/j.ejrad.2022.110197. [DOI] [PubMed] [Google Scholar]
- 11.Churchill I.F., Sullivan K.A., Simone A.C., et al. Thoracic imaging radiomics for staging lung cancer: a systematic review and radiomic quality assessment. Clin Transl Imaging. 2022;10:191–216. doi: 10.1007/s40336-021-00474-5. [DOI] [Google Scholar]
- 12.Liu A., Wang Z., Yang Y., Wang J., Dai X., Wang L., Lu Y., Xue F. Preoperative diagnosis of malignant pulmonary nodules in lung cancer screening with a radiomics nomogram. Cancer Commun. 2020 Jan;40(1):16–24. doi: 10.1002/cac2.12002. Epub 2020 Mar 3. PMID: 32125097; PMCID: PMC7163925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marentakis P., Karaiskos P., Kouloulias V., Kelekis N., Argentos S., Oikonomopoulos N., Loukas C. Lung cancer histology classification from CT images based on radiomics and deep learning models. Med. Biol. Eng. Comput. 2021 Jan;59(1):215–226. doi: 10.1007/s11517-020-02302-w. PMID: 33411267. [DOI] [PubMed] [Google Scholar]
- 14.Chetan M.R., Gleeson F.V. Radiomics in predicting treatment response in non-small-cell lung cancer: current status, challenges and future perspectives. Eur. Radiol. 2021 Feb;31(2):1049–1058. doi: 10.1007/s00330-020-07141-9. Epub 2020 Aug 18. PMID: 32809167; PMCID: PMC7813733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., et al. In: Computational Mathematics Modeling in Cancer Analysis. CMMCA 2022. Qin W., Zaki N., Zhang F., Wu J., Yang F., editors. vol. 13574. Springer; Cham: 2022. Repeatability of radiomic features against simulated scanning position stochasticity across imaging modalities and cancer subtypes: a retrospective multi-institutional study on head-and-neck cases. (Lecture Notes in Computer Science). [DOI] [Google Scholar]
- 16.Park D., Oh D., Lee M., et al. Importance of CT image normalization in radiomics analysis: prediction of 3-year recurrence-free survival in non-small cell lung cancer. Eur. Radiol. 2022;32:8716–8725. doi: 10.1007/s00330-022-08869-2. [DOI] [PubMed] [Google Scholar]
- 17.Imaging Biomarker Standardisation Initiative. Retrieved from 10.48550/arXiv.1612.07003. [DOI]
- 18.Abbaspour S., Abdollahi H., Arabalibeik H., et al. Endorectal ultrasound radiomics in locally advanced rectal cancer patients: despeckling and radiotherapy response prediction using machine learning. Abdom Radiol. 2022;47:3645–3659. doi: 10.1007/s00261-022-03625-y. [DOI] [PubMed] [Google Scholar]
- 19.Zhao W., Yang J., Sun Y., Li C., Wu W., Jin L., Yang Z., Ni B., Gao P., Wang P., Hua Y., Li M. 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018 Dec 15;78(24):6881–6889. doi: 10.1158/0008-5472.CAN-18-0696. [DOI] [PubMed] [Google Scholar]
- 20.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R., Granton P., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambin P., van Stiphout R., Starmans M., et al. Predicting outcomes in radiation oncology, multifactorial decision support systems. Nat. Rev. Clin. Oncol. 2013;10:27–40. doi: 10.1038/nrclinonc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aerts H., Velazquez E., Leijenaar R., Parmar C., Grossmann P., Carvalho S., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambin P., Rios-Velazquez E., Leijenaar R., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B., Yang L., Zhang R., Luo W., Li W. Radiomics: an overview in lung cancer management-a narrative review. Ann. Transl. Med. 2020 Sep;8(18):1191. doi: 10.21037/atm-20-4589. PMID: 33241040; PMCID: PMC7576016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzo Fantini, Belli Maria Luisa, Irene Azzali, et al. Exploratory analysis of 18F-3’-deoxy-3’-fluorothymidine (18F-flt) PET/CT-Based radiomics for the early evaluation of response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. Front. Oncol. 2021:11. doi: 10.3389/fonc.2021.601053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Xijia, Liu Meiling, Sun Jianqing, et al. Feasibility of MRI-based radiomics features for predicting lymph node metastases and VEGF expression in cervical cancer. Eur. J. Radiol. 2021:134. doi: 10.1016/j.ejrad.2020.109429. [DOI] [PubMed] [Google Scholar]
- 27.Wu J., Aguilera T., Shultz D., Gudur M., Rubin D.L., Loo B.W., Jr., Diehn M., Li R. Early-stage non-small cell lung cancer: quantitative imaging characteristics of (18)F fluorodeoxyglucose PET/CT allow prediction of distant metastasis. Radiology. 2016 Oct;281(1):270–278. doi: 10.1148/radiol.2016151829. Epub 2016 Apr 5. PMID: 27046074; PMCID: PMC5047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganeshan B., Panayiotou E., Burnand K., et al. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur. Radiol. 2012;22:796–802. doi: 10.1007/s00330-011-2319-8. [DOI] [PubMed] [Google Scholar]
- 29.Song Ll, Chen Sj, Chen W., et al. Radiomic model for differentiating parotid pleomorphic adenoma from parotid adenolymphoma based on MRI images. BMC Med Imaging. 2021;21:54. doi: 10.1186/s12880-021-00581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ardila D., Kiraly A.P., Bharadwaj S., Choi B., Reicher J.J., Peng L., Tse D., Etemadi M., Ye W., Corrado G., Naidich D.P., Shetty S. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019 Jun;25(6):954–961. doi: 10.1038/s41591-019-0447-x. Epub 2019 May 20. Erratum in: Nat Med. 2019 Aug;25(8):1319. PMID: 31110349. [DOI] [PubMed] [Google Scholar]
- 31.Hawkins S.H., Korecki J.N., Balagurunathan Y., Gu Y., Kumar V., Basu S., et al. Predicting outcomes of nonsmall cell lung cancer using CT image features. IEEE Access. 2014;2:1418–1426. [Google Scholar]
- 32.Shi L., He Y., Yuan Z., et al. Radiomics for response and outcome assessment for non-small cell lung cancer. Technol. Cancer Res. Treat. 2018;17 doi: 10.1177/1533033818782788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Chen, Zhang Fang, Xiu-Gui Sheng, et al. Peripheral platelet/lymphocyte ratio predicts lymph node metastasis and acts as a superior prognostic factor for cervical cancer when combined with neutrophil:Lymphocyte. Medicine. 2016;95(32) doi: 10.1097/MD.0000000000004381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 May;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 35.Gazdar A.F., Bunn P.A., Minna J.D. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer. 2017 Dec;17(12):725–737. doi: 10.1038/nrc.2017.87. Erratum in: Nat Rev Cancer. 2017 Nov 10;: PMID: 29077690. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira C., Amstutz F., Vuong D., et al. Preselection of robust radiomic features does not improve outcome modelling in non-small cell lung cancer based on clinical routine FDG-PET imaging. EJNMMI Res. 2021;11:79. doi: 10.1186/s13550-021-00809-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimada Y., Kudo Y., Maehara S., et al. Radiomics with artificial intelligence for the prediction of early recurrence in patients with clinical stage IA lung cancer. Ann. Surg Oncol. 2022;29:8185–8193. doi: 10.1245/s10434-022-12516-x. [DOI] [PubMed] [Google Scholar]
- 38.Kirienko M., Cozzi L., Antunovic L., et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45:207–217. doi: 10.1007/s00259-017-3837-7. [DOI] [PubMed] [Google Scholar]
- 39.Tong L., Mitchel J., Chatlin K., et al. Deep learning based feature-level integration of multi-omics data for breast cancer patients survival analysis. BMC Med Inform Decis Mak. 2020;20:225. doi: 10.1186/s12911-020-01225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park D., Oh D., Lee M., et al. Importance of CT image normalization in radiomics analysis: prediction of 3-year recurrence-free survival in non-small cell lung cancer. Eur. Radiol. 2022;32:8716–8725. doi: 10.1007/s00330-022-08869-2. [DOI] [PubMed] [Google Scholar]
- 41.Babyak M.A. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom. Med. 2004;66(3):411–421. doi: 10.1097/01.psy.0000127692.23278.a9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Given that the data in this study originate from a hospital, and in accordance with relevant laws and hospital policies, we are obliged to protect the privacy and confidentiality of patient data. Consequently, these data cannot be deposited into a publicly accessible repository. However, we have made every effort to ensure that the study results and analyses described in the article are understandable and verifiable by readers, and that all results are supported by the references listed.