Abstract
The cell envelope is a multilayered structure that insulates the interior of bacterial cells from an often chaotic outside world. Common features define the envelope across the bacterial kingdom, but the molecular mechanisms by which cells build and regulate this critical barrier are diverse and reflect the evolutionary histories of bacterial lineages. Intracellular pathogens of the genus Brucella exhibit marked differences in cell envelope structure, regulation, and biogenesis when compared to more commonly studied gram-negative bacteria and therefore provide an excellent comparative model for study of the gram-negative envelope. We review distinct features of the Brucella envelope, highlighting a conserved regulatory system that links cell cycle progression to envelope biogenesis and cell division. We further discuss recently discovered structural features of the Brucella envelope that ensure envelope integrity and that facilitate cell survival in the face of host immune stressors.
Keywords: Alphaproteobacteria, zoonosis, intracellular pathogen, gram negative
OVERVIEW
The Gram-Negative Envelope
The cell envelope separates the cytoplasm of a cell from the exterior environment. It determines cell shape; harbors macromolecular complexes that generate usable forms of energy to power motility and growth; and directly interacts with features of the surrounding environment, including plant and animal hosts (70, 111). The envelope of gram-negative bacteria is particularly interesting because it comprises two structurally distinct lipid bilayers with a thin periplasm and peptidoglycan cell wall in the space between them (111). The ability of micron-sized bacterial cells to maintain this structurally complex, didermic envelope while growing and dividing is a remarkable feat.
Our understanding of the gram-negative envelope is primarily based on studies of the class Gammaproteobacteria, specifically Enterobacteriaceae such as Escherichia coli. However, gram-negative bacteria are a varied group of organisms that exhibit remarkable diversity in the chemical and structural makeup of their envelopes. In recent years, cellular and molecular studies of the Alphaproteobacteria have revealed conserved molecular features that distinguish the envelope of this class. Species in the Alphaproteobacteria thus provide useful comparative models when considering which molecular processes of gram-negative envelope biogenesis and homeostasis are general, and which are specific to a particular phylogenetic group (38).
Brucella: A Brief Introduction
The Alphaproteobacteria include species that inhabit diverse niches, including the plant rhizosphere and phyllosphere; freshwater, marine, and soil ecosystems; and mammalian and insect hosts (11). Brucella is perhaps the most notorious genus in this class. Brucella abortus and Brucella melitensis were first identified in the late nineteenth century as the etiologic agents of contagious abortion in cows and Malta fever in humans, respectively (9, 18). These intracellular pathogens cause a disease now known as brucellosis, which remains among the most widespread zoonoses globally (32, 96). The Brucella genus is genetically monomorphic, and it has been proposed to be monospecific (128). However, Brucella species cluster into classifiable phylogenetic groups that align with animal host range and select molecular and physiologic characteristics, including differences in the structure and chemical makeup of the cell envelope (90). In this review we provide an overview of the Brucella cell envelope and highlight recent advances in the study of Brucella spp. envelope structure, function, and regulation.
DEVELOPMENTAL REGULATION OF BRUCELLA CELL ENVELOPE BIOGENESIS
The CtrA Regulatory Network
When viewed from afar, Brucella looks like any other gram-negative bacteria: it possesses a phospholipid bilayer inner membrane (IM), an outer membrane with a phospholipid inner leaflet and a lipopolysaccharide (LPS) outer leaflet, and a peptidoglycan cell wall in the periplasmic space between. Close inspection of single cells has provided strong evidence for an asymmetric growth mechanism. Specifically, peptidoglycan labeling experiments in B. abortus demonstrated that new cell envelope material is synthesized strictly from the new cell pole produced immediately after cell division (17, 64). At later stages of the cell cycle, a small growth zone appears at the nascent division site in B. abortus (101, 103, 127) (Figure 1a). Asymmetry in the biogenesis of new cell material during growth is reflected in asymmetric subcellular organization of key protein regulators of Brucella cell cycle and development. For example, the essential polar development sensor histidine kinase, PdhS, colocalizes with the response regulator DivK to the old cell pole during the B. abortus cell cycle; DivK localization requires the conserved aspartyl phosphorylation site in its receiver domain. Upon division, polarly localized PdhS is asymmetrically inherited by the mother cell; the newborn cell later acquires PdhS at its old pole, approximately 30 min after division (56) (Figure 1a). The developmental processes regulated by PdhS and DivK are integrated into a large regulatory network that is conserved across Alphaproteobacteria and that ensures the molecular machinery required to build new cell material is (a) localized to the appropriate polar location and (b) activated at the appropriate time during the cell cycle (77, 95, 125, 136). This elaborate molecular control system comprises multiple sensor histidine kinases, response regulators, diguanylate cyclases, and phosphodiesterases, all of which influence the expression, phosphorylation, and stability of the essential DNA-binding response regulator, CtrA. It can be reasonably argued that CtrA is the central molecular player of cell cycle and cell development regulation in many Alphaproteobacteria, including Brucella (98).
Figure 1.
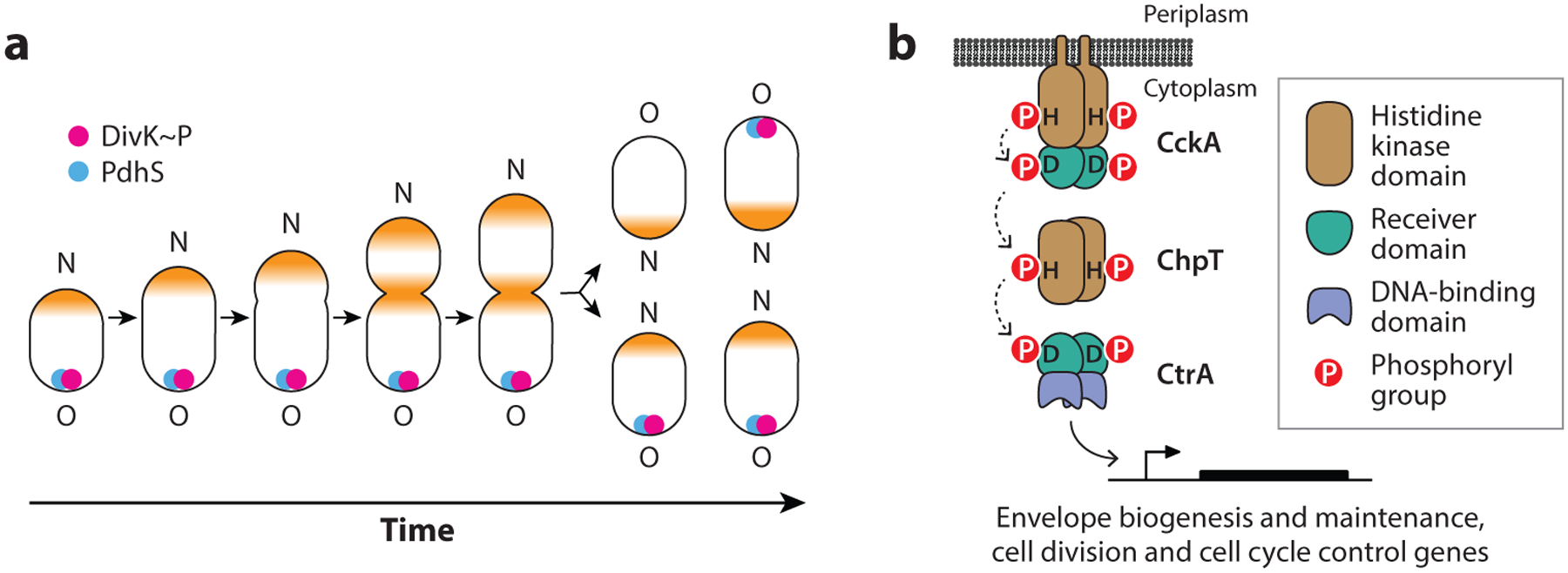
(a) Brucella spp., like other Rhizobiales, exhibit unipolar growth from the new cell pole. As the cell cycle progresses, cell growth shifts from being exclusively polar to being at both the pole and the nascent division site (growth sites colored orange). The Brucella developmental regulators, PdhS and DivK, exhibit dynamic polar localization to the inner membrane as a function of cell cycle. The new (N) pole and old (O) cell pole are marked. (b) Model of the CckA-ChpT-CtrA phosphorelay. The histidine kinase CckA autophosphorylates on a conserved histidine residue and transfers a phosphoryl group to a conserved aspartic acid residue on its C-terminal receiver domain. CckA~P transfers a phosphoryl group to the ChpT phosphotransferase, which can subsequently transfer this phosphoryl group to the receiver domain of CtrA. CtrA~P is a DNA-binding response regulator that modulates transcription of genes controlling cell polarity, division, and intracellular survival.
Like other response regulators, the activity of Brucella CtrA as a transcription factor is controlled by its phosphorylation at a single aspartic acid residue. The phosphorylation state of the CtrA protein is directly regulated by a multiprotein phosphorelay comprising the transmembrane sensor histidine kinase CckA and the histidine phosphotransferase ChpT (134) (Figure 1b). Chromatin immunoprecipitation sequencing (ChIP-seq) studies of B. abortus CtrA have demonstrated that CtrA directly binds the promoter regions of dozens of genes involved in cell division, peptidoglycan cell wall metabolism, outer membrane biogenesis, and biosynthesis of LPS (43). The importance of CtrA as a direct regulator of a diverse set of cell envelope proteins is clearly evident in studies of depletion and temperature-sensitive mutants of B. abortus (43, 134). Shifting conditional ctrA mutant strains to restrictive conditions resulted in reduced levels of select outer membrane proteins (OMPs) and the formation of filamentous branched cells, consistent with a defect in polar cell development and division. Branched cells were also observable in the mammalian intracellular niche upon depletion of ctrA expression; this phenotype was correlated with reduced intracellular viability (43). Attenuation of a conditional ctrA mutant in an in vitro infection model is expected, given that B. abortus cell cycle progression is intimately tied to intracellular trafficking in the endosomal compartments (33).
A Role for Cyclic Diguanylate?
The concentration and asymmetric localization of the second messenger cyclic-di-GMP (c-di-GMP) directly control the phosphorylation state and stability of CtrA in Caulobacter crescentus (77, 79, 112). A direct role for this signaling molecule in regulation of the Brucella CtrA pathway has not been shown, but there is evidence that c-di-GMP metabolizing enzymes play a role in Brucella envelope biology. A screen of annotated B. melitensis diguanylate cyclases and phosphodiesterases in a heterologous Vibrio system identified at least three active enzymes, including the phosphodiesterase PdeA (also known as BpdA) (97). B. melitensis pdeA mutants were attenuated in vitro and in vivo (67, 97), and deletion of B. abortus and B. melitensis pdeA resulted in cell rounding, with cells becoming shorter and wider (67, 101). This shape phenotype was independent of any phosphodiesterase activity the enzyme may have in cells, as a catalytically deficient pdeAE742A mutant retained wild-type morphology (101). Peptidoglycan analysis using fluorescent d-amino acid (HADA) labeling provided evidence that PdeA is required for localizing the site of peptidoglycan insertion to the pole (101). This result is consistent with studies of a PdeA homolog in Sinorhizobium meliloti known as RgsP, which functions at the membrane to control peptidoglycan composition (108).
Though PdeA is an important regulator of envelope biology in Brucella, a role for the c-di-GMP molecule per se has not been defined. Studies of diverse bacteria have shown that c-di-GMP levels in the cytoplasm control the transition from motile to sessile behavior via a variety of molecular mechanisms (31, 105). Brucella spp. contain assorted flagellar genes and pseudogenes (26, 36) but have long been classified as nonmotile bacteria (109). A B. melitensis strain was reported to elaborate a sheathed flagellum under select growth conditions (44), but efforts to identify actively motile cells among the classical Brucella species (B. abortus, B. melitensis, B. ovis, B. suis, B. canis, and B. neotomae) have been unsuccessful to our knowledge. However, several new Brucella species have been isolated from a range of animals (133), including amphibian isolates that are clearly motile and elaborate flagella and pilus-like structures from their envelopes (3). Whether c-di-GMP influences motility in these newly identified flagellated and motile Brucella species is not known, but it seems likely considering reported connections between pdeA and transcription from flagellar promoters in B. melitensis (97). Future efforts to define a specific role(s) for c-di-GMP as an effector of envelope processes in Brucella may be informed by studies in related free-living rhizobial species (e.g., S. meliloti) where c-di-GMP levels are linked to motility, polysaccharide production, and cell wall biosynthesis (71).
THE BRUCELLA OUTER MEMBRANE
The outer membrane is the interface between Brucella cells and their environment. This asymmetrical lipid bilayer contains primarily LPS in the outer leaflet and phospholipids in the inner leaflet. As an intracellular pathogen, Brucella must withstand a variety of host assaults, including compounds that disrupt membranes, such as antimicrobial peptides. Relative to a panel of Enterobacteriaceae, including E. coli, Brucella spp. outer membranes are more resistant to envelope stressors including polymyxin B, melittin, EDTA, and lysozyme (84). This enhanced resistance has been attributed to particular OMPs, distinct chemical features of core lipid A, and the density of O-polysaccharides of the LPS leaflet of the outer membrane, which will be discussed below.
Outer Membrane Proteins
The outer membrane is likely the best-studied of the Brucella envelope layers because it contains many antigens, including a diverse array of OMPs, that invoke host immune responses (47). Focused effort to characterize Brucella OMPs over the past several decades (129) have been motivated in part by the fact that OMPs can—in some cases—confer protective immunity, facilitate serologic classification of strains, and be developed as vaccines. We now know that Brucella spp. encode multiple families of OMPs, including many heat-stable porins and lipoproteins. A large body of literature on Brucella OMP content, structure, and immunology has been covered in previous reviews (24, 52, 106), and we encourage readers to consult these for additional information and additional perspectives on this topic.
Targeting and Assembly: Lol, Bam, and Tam
The mechanism by which lipid-modified OMPs (i.e., lipoproteins) become localized to the Brucella outer membrane remains an open question. In gram-negative bacteria, the lipoprotein localization (Lol) pathway comprises a set of proteins that traffic outer membrane lipoproteins across the periplasm to their final address (54). However, the gene encoding the LolB protein, which is necessary for lipoprotein trafficking in many gram-negative bacteria, is absent in the Alphaproteobacteria, including Brucella (52, 92). It has been proposed that LolA may serve the functions of both LolA and LolB in clades where LolB is absent (118), though this hypothesis remains untested in Brucella. Recent efforts to define the functions of uncharacterized B. abortus proteins involved in envelope stress responses identified a periplasmic domain of unknown function protein (DUF1849) conserved in a subset of Alphaproteobacteria—primarily Rhizobiales—that is now named EipB (61). The eipB chromosomal locus exhibits synteny homology across the Rhizobiales with genes that function in membrane and cell wall synthesis, LPS synthesis, and outer membrane protein assembly. The overall organization of this genomic region is highly conserved in Proteobacteria (Figure 2a), and the association of eipB with this locus supports a functional role for eipB in cell envelope biology. EipB comprises 14 antiparallel β strands, organized in a cylindrical, spiral-like shape, with three α-helical connector segments. Though EipB has no clear structural homologs in the Protein Data Bank, its β-barrel architecture resembles those of E. coli LolA and LolB, and the OMP assembly proteins TamA and BamA (Figure 2b). Deletion of eipB resulted in sensitivity to compounds that disrupt the integrity of the cell envelope and compromised B. abortus infection in a murine model of disease (61). These recent results support a functional role for EipB in determining cell envelope integrity, but it is not known whether EipB assumes a LolB-like function in lipoprotein trafficking, whether it functions as an OMP chaperone or assembly factor, or whether it has another role in the Brucella cell envelope.
Figure 2.
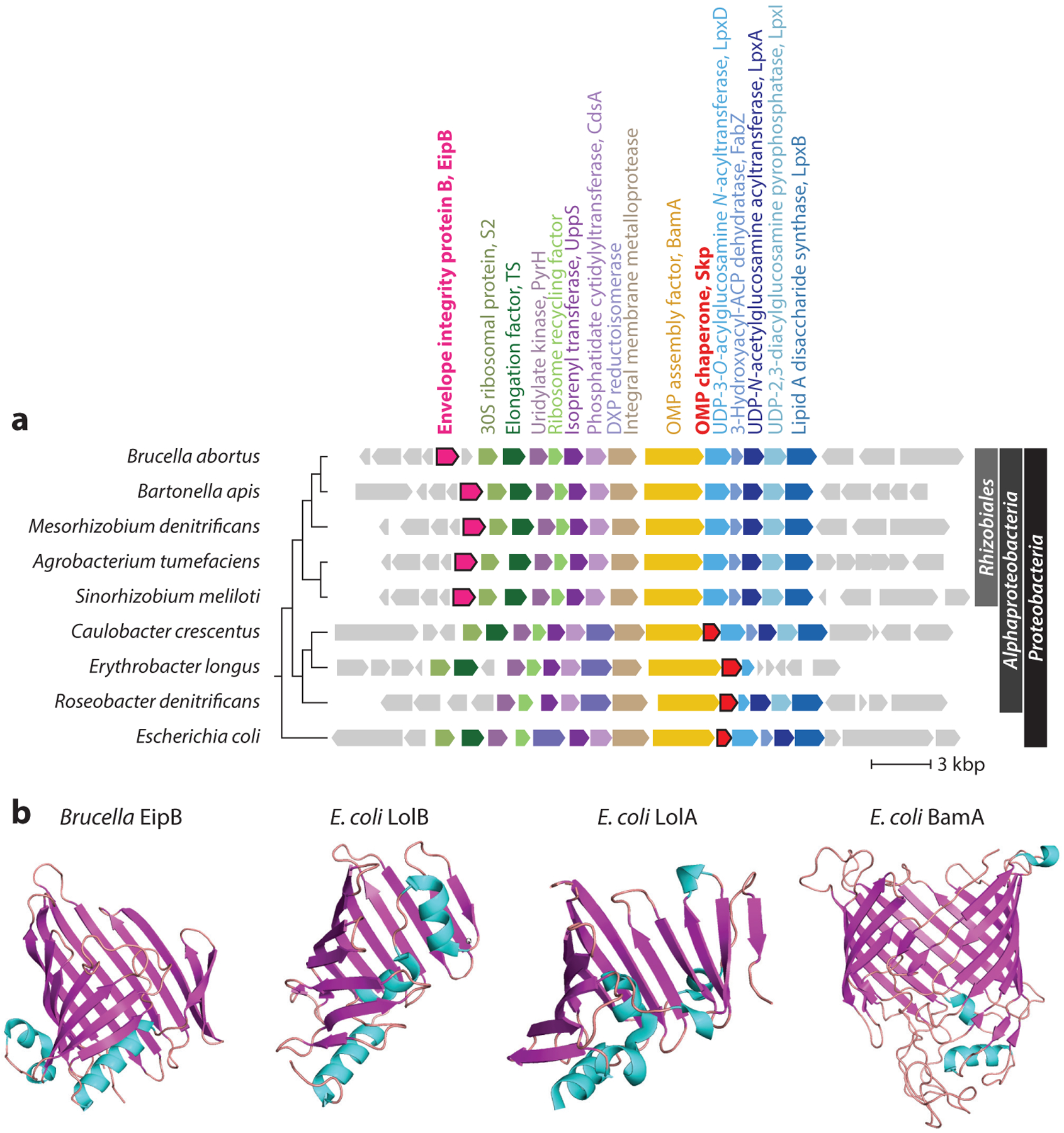
(a) Phylogenetic distribution and synteny of the envelope integrity protein B (eipB) genomic region in Proteobacteria; gene neighborhood is anchored on bamA. Brucella eipB is part of a highly conserved cell envelope gene cluster in the Proteobacteria that includes genes involved in outer membrane protein assembly (bamA), undecaprenyl phosphate biosynthesis, phospholipid synthesis (cdsA), lipopolysaccharide synthesis (lpxDAIB), and translation (ef-ts). eipB (pink) is conserved in the Rhizobiales group of the Alphaproteobacteria (phylogenetic classifications on the right). Skp is present outside the Rhizobiales. Phylogenetic tree is based on BamA protein sequence (left). (b) Crystal structures of Brucella EipB (PDB: 6NTR), Escherichia coli LolB (PDB: 1IWM), E. coli LolA (PDB: 1IWL), and E. coli BamA (PDB: 5OR1). Abbreviation: PDB, Protein Data Bank.
In addition to the Lol system, there are other molecular systems that ensure Brucella appropriately assembles OMPs into the outer membrane. Among these is the β-barrel assembly machine (BAM) complex, which facilitates protein folding into the outer membrane (81). The genus Brucella encodes homologs of the BamA, BamD, and BamE proteins and is missing the BamB and BamC proteins present in E. coli and other Gammaproteobacteria. Our application of Bayesian or HMM gene essentiality algorithms (34, 35) to published transposon sequencing (Tn-Seq) data from B. abortus 2308 (61, 62) and B. ovis 25840 (126) provides evidence that bamE, bamD, and bamA are essential in both species. Anwari and colleagues (4) identified a BAM complex component, BamF (DUF3035), that is restricted to the Alphaproteobacteria, though sequence models in the InterPro-Pfam database (15) indicate that BamF/DUF3035 is absent in the genus Brucella. The composition of the BAM complex varies across gram-negative bacteria, and it is not known what proteins—if any—fulfill the functional roles of BamB, BamC, and BamF in Brucella. The fact that the periplasmic β-barrel protein, EipB, is encoded from a locus proximal to bamA suggests that it could function with the BAM complex in OMP assembly (Figure 2a). Notably, the OMP chaperone Skp is encoded adjacent to BamA in many Proteobacteria; this gene is absent from Rhizobiales cataloged in InterPro. The presence of eipB in this conserved cell envelope cluster (instead of skp) in Rhizobiales raises the possibility that EipB and Skp have analogous chaperoning roles.
The outer membrane translocation and assembly module (TAM) is a system that is evolutionarily related to the BAM complex and plays a key role in the assembly of select OMPs (58). The system comprises two protein components: the integral OMP TamA, which is related to BamA, and the inner membrane protein TamB. Studies in B. suis have shown that the TamB homolog, known as MapB, controls translocation of a subset of OMPs to the outer membrane (13). A B. suis mapB deletion mutant had disrupted localization of select OMPs, a cell division/morphology defect, and was sensitive to multiple envelope-disrupting compounds, including polymyxin B, Triton X-100, and lysozyme (13). mapB/tamB deletion in B. suis (13) and B. melitensis (135) resulted in strain attenuation in infection models, and B. abortus strains lacking either tamA or tamB were attenuated in a mouse macrophage infection model (116). A specific role for the TAM system during infection is supported by proteomic analyses showing that the steady-state level of TamB protein in B. abortus (BAB1_0046/Omp160) was enhanced ~20–100-fold just 3 h after macrophage infection and remained high over a two-day infection time course (75).
The Outer Membrane–Peptidoglycan Connection
In E. coli and related bacteria, the hyperabundant outer membrane lipoprotein Lpp is covalently linked to the peptidoglycan (8, 16, 57), which provides important structural support for the envelope. Brucella and other related Rhizobiales lack lpp, but biochemical evidence for covalent interactions between Brucella spp. OMPs and the peptidoglycan cell wall (25, 51) has been long discussed. Recent work by Godessart and colleagues (49) has demonstrated that multiple OMPs in B. abortus and the related rhizobial species Agrobacterium tumefaciens are covalently linked to the peptidoglycan via a conserved alanyl-aspartyl motif at the protein N terminus (Figure 3a). Like the covalent linkage of E. coli Lpp to peptidoglycan, this linkage in Brucella is catalyzed by l,d-transpeptidases. This result explains long-noted observations of peptidoglycan linkages to OMPs in Brucella and provides new biochemical and structural understanding of Brucella envelope integrity. Notably, several other bacteria lacking Lpp have now been shown to anchor the outer membrane to peptidoglycan in a similar fashion to Brucella (107).
Figure 3.
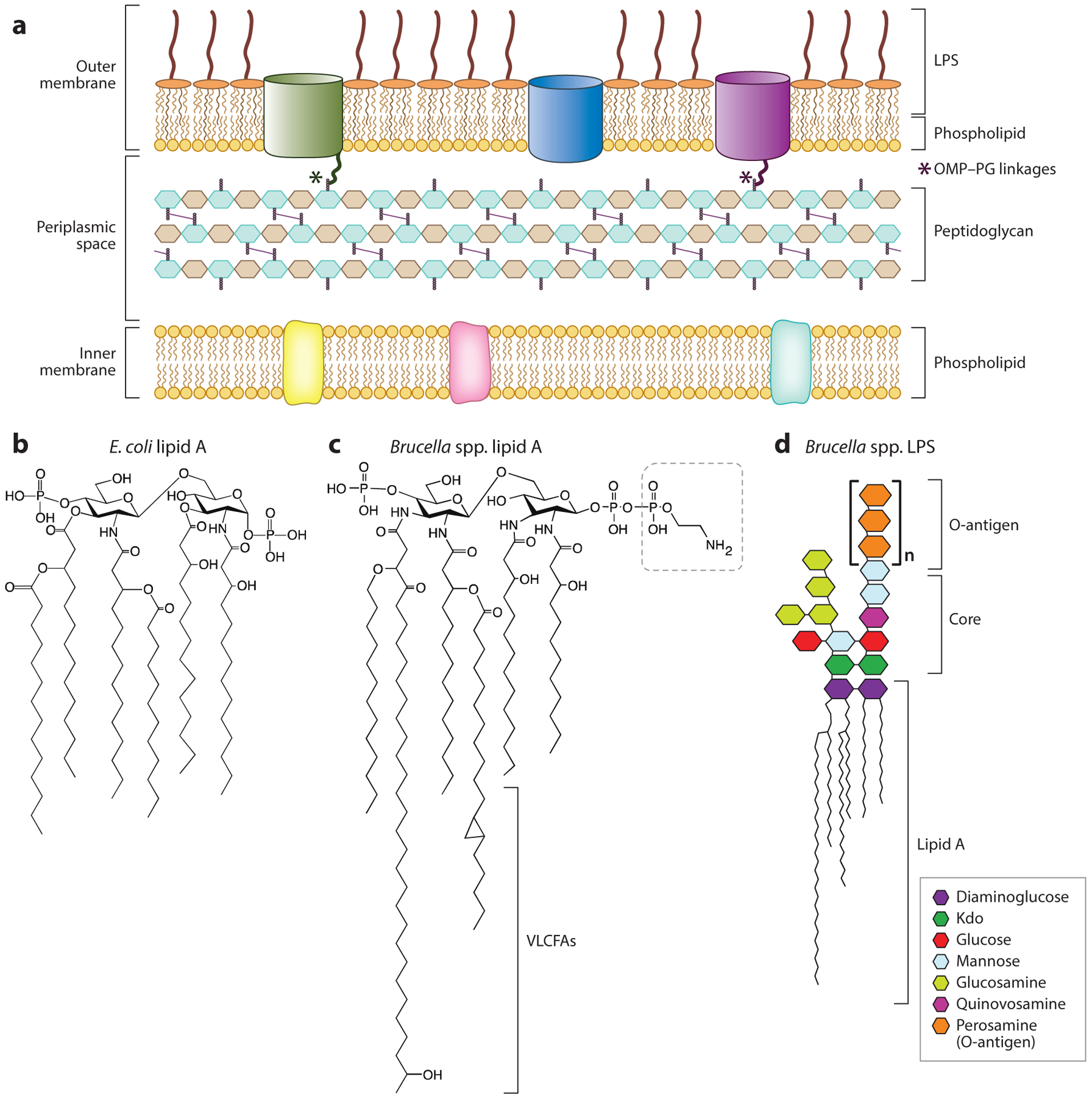
(a) Overview of the envelope layers of Brucella. Notably, a subset of Brucella outer membrane proteins are covalently linked to the peptidoglycan cell wall via a conserved sequence at the protein N terminus. (b) Chemical structure of Escherichia coli lipid A (100). (c) Chemical structure of Brucella spp. lipid A showing VLCFA tails and pyrophosphorylethanolamine modification of diaminoglucose backbone (outlined with dashed-line box). Structure adapted from model and data presented in References 20 and 29. (d) Brucella smooth LPS structure showing lipid A, core oligosaccharide, and O-polysaccharide (O-antigen), based on models and data presented in References 41, 72, and 117. Abbreviations: Kdo, 3-deoxy-d-manno-2-octulosonic acid; LPS, lipopolysaccharide; OMP, outer membrane protein; PG, peptidoglycan; VLCFA, very-long-chain fatty acid.
LIPOPOLYSACCHARIDE
LPS, the primary lipid of the outer leaflet of the outer membrane, is a major pathogen-associated molecular pattern (PAMP) recognized by the innate immune system (120). Accordingly, Brucella LPS is a highly studied molecule that has been well-reviewed (106, 117, 137), and we encourage readers to consult these reviews for more information on this topic. Here we provide an abridged overview of distinctive features of Brucella LPS structure and biosynthesis.
LPS Structure
The structure of Brucella spp. LPS has chemical features that distinguish this envelope component from the highly inflammatory LPS of enteric bacteria (Figure 3b) and that enable these pathogens to evade host recognition by the innate immune system (10, 28). Canonically, LPS consists of a hexa-acylated disaccharide called lipid A, which is linked to the core oligosaccharides. In species with smooth LPS, repeating O-polysaccharide units decorate the core polysaccharide (Figure 3). The lipid A portion of Brucella LPS consists of diaminoglucose sugars linked to C16–C18 fatty acids and very-long-chain fatty acids (VLCFAs), including a significant proportion of 27-OH-28:0 (91) (Figure 3c). This unusual feature of Brucella lipid A is restricted to the alpha-2 subgroup of Proteobacteria, and it may enable lipid A to extend through the outer membrane outer leaflet into the inner leaflet (Figure 3a), potentially increasing the structural integrity of the outer membrane (12). The level of VLCFAs on lipid A is controlled in part by the inner membrane–bound BacA protein in both B. abortus and the related rhizobial species S. meliloti (39), suggesting that this unique LPS feature can be regulated. Relative to other gram-negative bacteria, Brucella lipid A is a poor stimulant of the mammalian inflammatory response (37). The core oligosaccharide of Brucella spp. lipid A contains two 3-deoxy-d-manno-2-octulosonic acid (Kdo) sugars (89) linked to diaminoglucose, one of which has a branched linkage to a chain of glucose, mannose, and glucosamine (48) (Figure 3d). There is evidence that this branched core structure shields lipid A from recognition by the host innate immune sensor TLR4 (28) and thereby contributes to the reduced inflammatory capacity of Brucella LPS.
Among the classical Brucella species (132), the best-described difference in cell envelope composition is the presence or absence of O-polysaccharide, also known as O-antigen (117). Brucella spp. O-antigen consists primarily of N-formylated perosamine (50) (Figure 3d) of two major linkage forms: A (from B. abortus) and M (from B. melitensis) (72, 87). While Brucella spp. typically elaborate an O-antigen and are considered to have smooth LPS (S-LPS), B. ovis and B. canis harbor genetic mutations that result in an inability to synthesize O-antigen and have what is therefore considered naturally rough LPS (R-LPS). O-antigen is an important virulence determinant in Brucella (76), yet B. ovis and B. canis cause disease in their primary hosts, sheep and dogs, respectively. The pathogenicity of these rough strains in their primary hosts is notable because rough mutants of naturally smooth strains have large defects in intracellular growth and replication and can be cytotoxic to host macrophages.
The structure of O-antigen from the numerous new Brucella species identified in recent years (133) remains largely undefined, but several strains in the recently identified BO2 clade of Brucella are missing genes in the wbk region that are required for S-LPS synthesis. Experimental analyses of a BO2 strain showed that it produced an S-LPS (138) not composed of N-formyl-perosamine (130). In the place of the wbk genes, this strain contained genes for biosynthesis of a rhamnose-based O-antigen; BO2 S-LPS had biochemical properties that were distinct from those of the classical Brucella species (130). Future analyses of LPS structure across the expanded Brucella clade will likely illuminate new and diverse chemical features of Brucella LPS.
Spatial Control of LPS Biosynthesis in Brucella
An elaborate transport system ensures proper addressing of LPS to the outer membrane in gram-negative bacteria (115). In B. abortus, the biosynthesis and elaboration of LPS on the surface of the cell is spatially controlled. B. abortus has both R-LPS and S-LPS patches on its envelope that can be visualized by immunofluorescence (127). Insertion of new LPS material occurs unipolarly at the new pole of the cell (generated immediately after division), where new cell wall material is also added (127). The inner membrane LPS biosynthesis proteins LptB, LptC, LptF, and LptG are similarly localized to the new pole. There are conflicting reports of the spatial distribution of the outer membrane LPS translocase, LptD. Fluorescent protein fusions to LptD showed a polar localization pattern in wild-type B. abortus (123) consistent with the localization of LptBCFG, while staining by immunogold provides evidence that LptD is localized across the cell surface (110).
Periplasmic Factors Linked to LPS
Recent studies have identified previously uncharacterized domain of unknown function (DUF) proteins that are secreted to the periplasm and have roles in cell envelope processes. The periplasmic β-barrel protein EipB (61), which may function in OMP targeting, is described above. Studies of Brucella RomA and EipA provide evidence that these periplasmic proteins have a functional connection to LPS.
The RomA protein is a member of the DUF3126 superfamily, which is restricted to the Alphaproteobacteria; in B. abortus this protein is secreted to the periplasm (123). A B. abortus romA deletion strain (ΔromA) had multiple envelope defects including altered LPS composition: ΔromA had a higher proportion of S-LPS, with longer O-polysaccharide chains than the wild type (123). This LPS phenotype was correlated with altered localization of a fluorescent protein fusion to the LPS translocase, LptD. Specifically, the ΔromA strain showed diffuse localization of LptD across the cell, while LptD was polarly localized in the wild type. Localization of the O-polysaccharide flippase, RfbD (Bab1_0543), was not altered in ΔromA. The precise function of RomA remains undefined, but it seems likely that this protein functions in the periplasm to coordinate LPS synthesis.
The Brucella EipA protein is a member of the DUF1134 protein superfamily, which is largely restricted to the Alphaproteobacteria (Figure 4a). In the Rhizobiales, eipA is encoded in the neighborhood of ctrA and chpT (Figure 4a), and its expression is directly controlled by CtrA (62). EipA is secreted to the periplasm, and deletion of B. abortus eipA resulted in sensitivity to envelope stressors including EDTA, SDS, and ampicillin (62). As would be expected for a strain with a general envelope defect, ΔeipA was attenuated in macrophage and mouse infection models. The molecular function of EipA in the periplasm has not been determined, but a screen for genes that are synthetically lethal with eipA deletion in B. abortus showed that disruption of O-polysaccharide synthesis genes is not tolerated in the absence of eipA (62). Congruent with this genetic analysis in B. abortus, eipA is essential in the naturally rough species B. ovis, which lacks the O-polysaccharide. A high-resolution crystal structure of EipA revealed a novel protein fold (62) composed of 10 antiparallel β strands, with β1–β5 and β8–β10 forming a small β barrel (Figure 4b). Though EipA interaction partners have not been defined, a recent solution NMR structure of the lipid/phosphoinositide-binding SYLF domain protein BPSL1445 revealed significant structural similarity to Brucella EipA, suggesting that EipA is a lipid-interacting protein (99).
Figure 4.
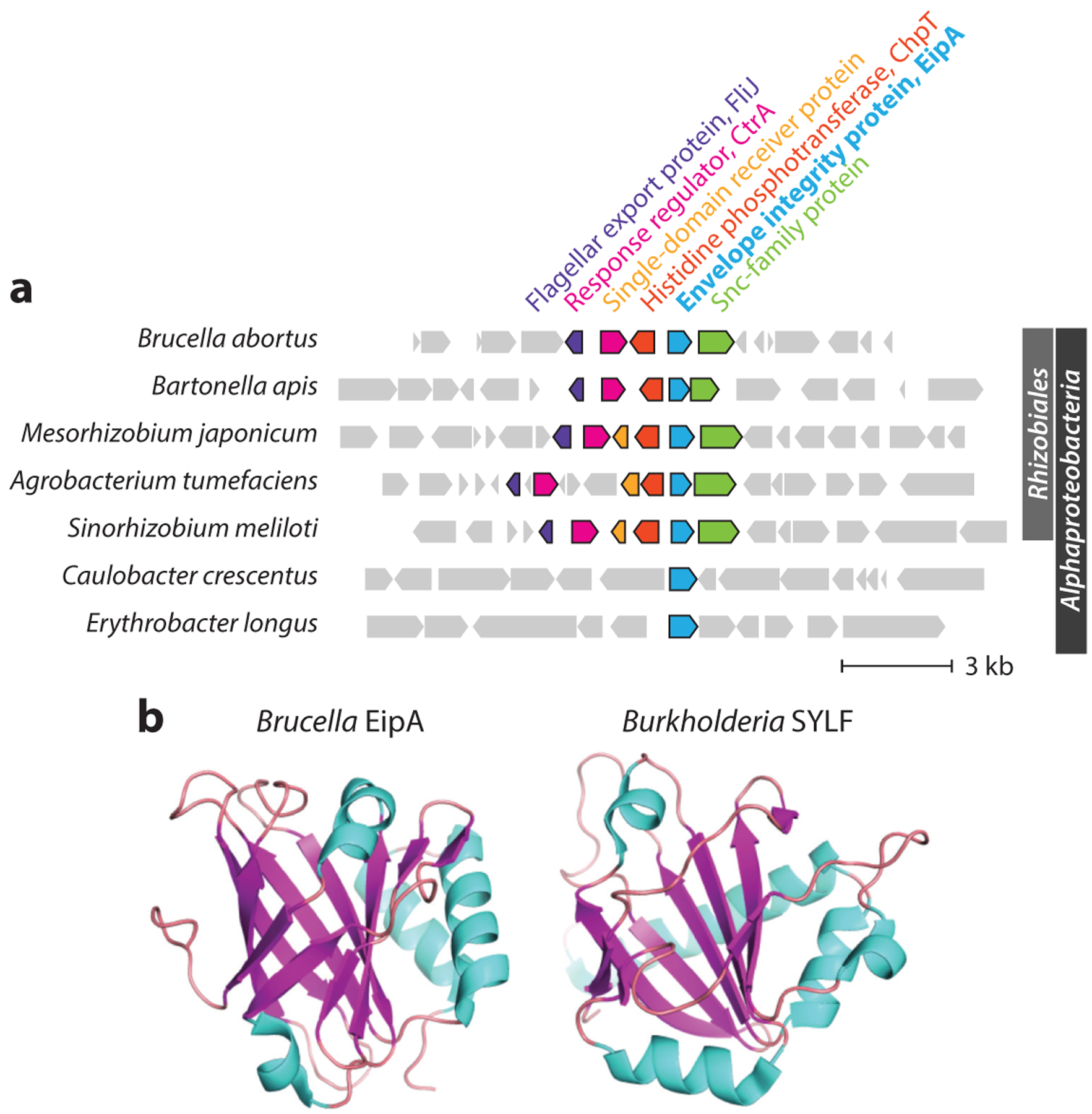
(a) Phylogenetic distribution and synteny of the envelope integrity protein A (eipA) genomic region in Proteobacteria; gene neighborhood is anchored on eipA. In Brucella and other Rhizobiales, eipA is part of a genetic locus that encodes the essential cell cycle regulatory proteins CtrA and ChpT. (b) Crystal structures of Brucella EipA (PDB 5UC0) and a Burkholderia SYLF domain protein (PDB 7OFN). Abbreviation: PDB, Protein Data Bank.
BRUCELLA PHOSPHOLIPIDS
Phospholipids are a key molecular component of the cell envelope, and Brucella has interesting lipid membrane features that merit discussion. Phosphatidylethanolamine (PE) is the dominant phospholipid in most gram-negative bacteria, but Brucella membranes contain comparatively low PE and a high proportion of phosphatidylcholine (PC), a phospholipid usually associated with eukaryotic membranes (45, 113, 122). PE synthesis in B. abortus is apparently dispensable; deletion of phosphatidylserine synthase (PssA), which catalyzes the first step in PE synthesis, is compensated by increased production of PC and ornithine lipids (OLs) (19). It has been proposed that an elevated PC level may provide a mechanism to mask pathogens, including Brucella, from their mammalian hosts (2, 27, 30). This idea is supported by studies of the phospholipase A1, BveA, of B. melitensis. Analysis of lipid extracts of a bveA mutant revealed elevated levels of PE. Enhanced PE levels were associated with higher susceptibility to polymyxin B and reduced survival in in vitro and in vivo infection models (66).
The notable skew in the PC:PE ratio in Brucella spp. relative to other gram-negative bacteria has motivated studies of the mechanism of PC biosynthesis. PC can be synthesized through one of two pathways: the methylation pathway and the choline pathway. In the methylation pathway, PmtA mediates successive methylation of phosphatidylethanolamine, using S-adenosylmethionine (SAM) as a methyl donor. In the choline pathway, the enzyme Pcs synthesizes PC from choline and cytidine diphosphate-diacylglycerol precursors (113). The B. abortus genome contains both pcs and pmtA homologs, but genetic analysis of Δpcs and ΔpmtA mutants showed that only pcs was required for normal growth and synthesis of PC in complex medium (27, 30). However, both Δpcs and ΔpmtA mutants were attenuated for replication in the mouse spleen at different stages of infection, and a Δpcs ΔpmtA double mutant (a) had a larger growth defect in defined medium than single mutants and (b) was more attenuated in mouse spleen than either single mutant (30). These results provide evidence that both pathways to PC are operational in B. abortus and that the nutritional environment influences the requirement for these genes. A related series of experiments on PC synthesis in B. melitensis 16M extracts led to the conclusion that Pcs was the sole route to PC production (85), while the conclusion based on experiments in B. abortus was that PC was synthesized from host-derived choline exclusively via the choline (Pcs) pathway (27). Acquisition of choline from the host is consistent with a report that B. abortus requires the high-affinity choline transporter ChoXWV for PC synthesis when choline concentrations are low (59). Polymorphisms in the SAM-binding site of PmtA across the Brucella genus likely influence the enzymatic routes to PC production at the species level and may explain why select B. abortus, B. melitensis, and B. suis strains have differing requirements for pcs and pmtA in vitro and in vivo (5).
In addition to PC and PE, the Brucella envelope also contains a significant fraction of OLs (94, 122). Prior to studies in Brucella, OL synthesis had been characterized in S. meliloti, where two genes, olsA and olsB, are required for OL synthesis (46, 131). Brucella contains homologs of these genes and synthesizes OL through a two-step pathway (94). Deletion of OL synthesis did not impact membrane permeability, susceptibility to antimicrobial peptides, or infection in an in vitro model (94). Moreover, a B. abortus strain lacking OLs did not induce an inflammatory response in mice that differed from wild type. These results suggest that Brucella OLs have little impact on B. abortus envelope stress resistance, infection, or host immunity. Nonetheless, the abundance of OLs in Brucella membranes suggests these molecules have an important role in some context.
THE PEPTIDOGLYCAN CELL WALL
As a gram-negative bacterium grows, it builds new peptidoglycan cell wall in the periplasmic space. At the point of division, the cell must cleave and reanneal the peptidoglycan wall that surrounds it. This highly complex process requires exquisite spatiotemporal coordination of a multitude of proteins on a submicron scale and has been the subject of intense study for many years (104). E. coli, C. crescentus, and many other model bacteria grow laterally and assemble the division site (i.e., the divisome) at the center of the cell after a certain period of growth. The Rhizobiales (including Brucella) grow by budding, a process in which new peptidoglycan material is added to one cell pole (17) (Figure 1a). The mechanism by which Brucella and other Rhizobiales localize the peptidoglycan biosynthesis machinery to one pole and maintain cell envelope integrity during the process of new cell addition and division remains largely undefined (124).
Penicillin-binding proteins (PBPs) are critical for peptidoglycan cell wall synthesis, and a transposon screen of B. abortus in complex medium uncovered Pbp1a (Bab1_0932) and FtsI as the only two essential PBPs of the seven encoded in the genome (116). As expected, this study also identified numerous divisome (fts) and cell division (tol-pal) genes as essential for growth. Screening this same transposon library in macrophages identified a set of genes that were conditionally essential in the intracellular niche. Among these was the histidine biosynthesis gene, hisB, which exhibited a surprising cell chaining/division defect inside mammalian cells resulting from uncleaved peptidoglycan at the cell division site (103). The hisB division phenotype could be suppressed by overexpression of either DipM or CdlP, each of which contains a peptidoglycan-binding LysM domain and is predicted to function as a metallopeptidase. This targeted suppressor approach thus identified two putative peptidoglycan metallopeptidases involved in Brucella cell division, though the exact connection between histidine metabolism, peptidoglycan cleavage, and division remains undefined. Cell wall metabolism and cell division are a relatively new area of investigation in Brucella, and future studies are certain to elucidate interesting connections between central metabolism and division.
CONSERVED CELL ENVELOPE REGULATION SYSTEMS
When infecting a host, the Brucella cell must withstand many host-derived stressors to survive. It is the cell envelope that meets this complex host assault. Accordingly, the composition of the envelope is highly regulated by multiple stress response systems. In this section, we discuss select Brucella envelope regulators that are conserved in the Alphaproteobacteria.
The General Stress Response System
The general stress response (GSR) system of Alphaproteobacteria regulates large-scale changes in gene expression that confer resistance to a range of environmental stressors (40, 42). Activation of the B. abortus GSR involves stress-dependent phosphorylation of the anti-anti-sigma protein PhyR (68, 69, 119). PhyR phosphorylation promotes its binding to the anti-σE1 protein, NepR (80), thereby releasing the alternative sigma factor σE1 (also known as ecfG) to regulate transcription of dozens of genes (Figure 5a). Studies in B. abortus and several other Alphaproteobacteria provide evidence that multiple HWE-family sensor kinases (60) coordinately regulate PhyR phosphorylation and GSR activation (Figure 5a), which may explain how this regulatory system is able to respond to such a diverse range of stress conditions (40, 42).
Figure 5.
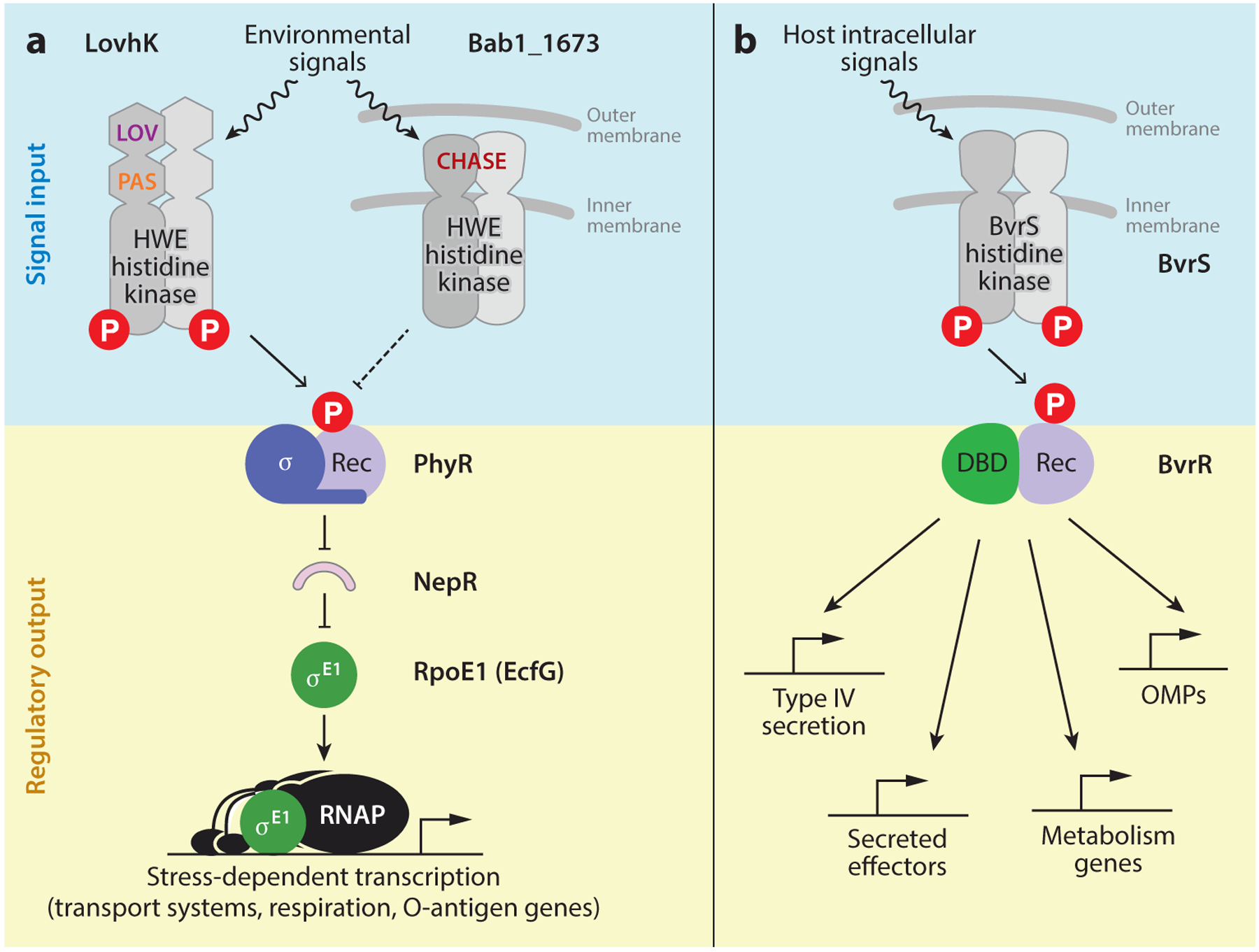
(a) The GSR signaling pathway in Alphaproteobacteria is activated by environmental signals and integrates features of two-component and sigma factor–dependent regulation of gene expression. Studies of this pathway in Brucella abortus support a model in which phosphorylation of PhyR is controlled by two HWE-family sensor kinases: LovhK and Bab1_1673. PhyR phosphorylation promotes its binding to NepR, which releases σE1 to control transcription of select cell envelope genes that influence in vitro stress survival and host infection. The LOV and PAS sensory domains of the cytoplasmic GSR-activating kinase, LovhK, are labeled. The periplasmic CHASE family sensor domain of the transmembrane GSR-repressive kinase, Bab1_1673, is labeled. Solid lines indicate a direct interaction; dashed lines indicate interactions that may be direct or indirect. (b) Brucella spp. BvrR/BvrS is a conserved, archetypal two-component system that is a critical regulator of infection. In response to detection of intracellular signals during infection, the histidine kinase BvrS phosphorylates BvrR, which then regulates transcription of a suite of cell envelope and metabolic genes important for intracellular survival and replication. Abbreviations: DBD, DNA-binding domain; GSR, general stress response; REC, REC, receiver domain; RNAP, RNA polymerase; OMP, outer membrane protein.
Deletion of GSR regulatory genes in B. abortus and B. melitensis results in a variable replication/survival defect in mice depending on the particular gene deletion and the genetic background of the animal host (53, 68, 69). The transcription of numerous B. abortus membrane transport systems is strongly activated by σE1, including an RND-family efflux system, a CrcB-family transporter, ABC-type transporters, and a major facilitator family transporter (68). Thus B. abortus remodels the transport capabilities of its envelope when the GSR system is activated. In addition, transcription of the cydABX high-affinity terminal oxidase genes and the O-polysaccharide biosynthesis gene, pgm, is activated by σE1, as are multiple hypothetical transmembrane DUF proteins. In B. melitensis, deletion of the GSR-activating kinase lovhK is reported to activate expression of the type IV secretion system (virB) and select flagellar genes (53). Together, these studies provide evidence that the GSR system is a major regulator of the Brucella cell envelope.
The BvrR-BvrS Envelope Homeostasis System
The BvrR-BvrS two-component regulatory system is conserved in the Alphaproteobacteria, where it has important functions in host-bacteria interactions (22, 83). In Brucella, this system is a key determinant of infection in in vitro and in vivo models (78, 114). B. abortus bvrR-bvrS directly and indirectly regulates the type IV secretion system (86), contributes to homeostasis of membranes, and regulates lipid A acylation and the expression of periplasmic and outer membrane proteins (55, 74, 82) (Figure 5b). The general cell envelope defect of strains harboring mutations in this two-component system is evident in the fact that deletion of the envelope integrity protein eipA is synthetically lethal with bvrR deletion in B. abortus (62). Recent ChIP-seq analysis of B. abortus BvrR demonstrates that this regulator not only directly binds the promoter regions of type IV secretion genes (virB), genes for secreted effectors, and genes that function in multiple aspects of cell envelope homeostasis but also binds promoters of multiple genes with roles in central metabolism (102). This study provides evidence for a broader regulatory role for the BvrR-BvrS system in Brucella cell envelope biology and metabolism than has been previously appreciated.
Ros/MucR
Like the BvrR-BvrS system, the zinc finger transcription factor Ros/MucR is conserved in the Alphaproteobacteria (1, 7, 23, 65), where it has an established role in virulence gene regulation and plant-rhizobia interactions (6, 14, 23). Deletion of this regulator in multiple Brucella species results in alterations in cell envelope properties (21, 88, 121) that, in B. melitensis, lead to sensitivity to a range of cell envelope stressors in vitro. Studies in B. melitensis further provide evidence that MucR regulates modification of lipid A core and regulates transcription of flagellar genes via the flagellar regulator protein FtcR (88). Brucella mucR can functionally complement an S. meliloti mucR deletion mutant (88), providing an additional example of how a conserved set of regulatory proteins underpins host-microbe interactions in Alphaproteobacteria.
CONCLUSIONS AND OUTLOOK
Brucella spp. are important animal pathogens that have been studied for over a century. Since their discovery, we have gained significant understanding of animal host responses to infection and the Brucella genetic factors that determine infection and pathogenesis. Molecular components of the cell envelope play a crucial role in host interactions, and subtle differences in cell envelope composition or regulation likely contribute to the fascinating animal host preferences exhibited by the highly related Brucella species. Deciphering the genetic basis of host preference is an interesting and exciting area of investigation that may be advanced by combining pangenome analyses with omics-based approaches to analyze cell envelope lipids, polysaccharides, and proteins.
Brucella has emerged as a powerful comparative model for the investigation of cell cycle, polar cell development, and cell division in gram-negative bacteria. The development of fluorescent d-amino acids for study of peptidoglycan synthesis was instrumental in demonstrating that cell growth in Brucella is unipolar (73), and this tool will be useful as the community works to decipher the molecular mechanism of polar cell growth in Brucella and other Rhizobiales. Select genes for other well-studied polar surface structures in Alphaproteobacteria, such as unipolar polysaccharide (UPP) (63, 93), are conserved in Brucella. It is not known whether Brucella spp. elaborate a UPP, but it seems likely that cell cycle–regulated polar envelope structures—perhaps including UPP— contribute to the interesting link between the developmental state of the Brucella cell and the process of host infection (33).
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of General Medical Science of the National Institutes of Health under award number R35GM131762 to S.C.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Glossary
- LPS
lipopolysaccharide
- S-LPS
smooth lipopolysaccharide
- R-LPS
rough lipopolysaccharide
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
LITERATURE CITED
- 1.Adhikari S, Erill I, Curtis PD. 2021. Transcriptional rewiring of the GcrA/CcrM bacterial epigenetic regulatory system in closely related bacteria. PLOS Genet. 17:e1009433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aktas M, Wessel M, Hacker S, Klusener S, Gleichenhagen J, Narberhaus F. 2010. Phosphatidylcholine biosynthesis and its significance in bacteria interacting with eukaryotic cells. Eur. J. Cell Biol 89:888–94 [DOI] [PubMed] [Google Scholar]
- 3.Al Dahouk S, Kohler S, Occhialini A, Jimenez de Bagues MP, Hammerl JA, et al. 2017. Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci. Rep 7:44420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anwari K, Webb CT, Poggio S, Perry AJ, Belousoff M, et al. 2012. The evolution of new lipoprotein subunits of the bacterial outer membrane BAM complex. Mol. Microbiol 84:832–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragon-Aranda B, Palacios-Chaves L, Salvador-Bescos M, de Miguel MJ, Munoz PM, et al. 2021. The phospholipid N-methyltransferase and phosphatidylcholine synthase pathways and the ChoXWV choline uptake system involved in phosphatidylcholine synthesis are widely conserved in most, but not all Brucella species. Front. Microbiol 12:614243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araujo RS, Robleto EA, Handelsman J. 1994. A hydrophobic mutant of Rhizobium etli altered in nodulation competitiveness and growth in the rhizosphere. Appl. Environ. Microbiol 60:1430–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ardissone S, Fumeaux C, Berge M, Beaussart A, Theraulaz L, et al. 2014. Cell cycle constraints on capsulation and bacteriophage susceptibility. eLife 3:e03587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asmar AT, Collet JF. 2018. Lpp, the Braun lipoprotein, turns 50—major achievements and remaining issues. FEMS Microbiol. Lett 365(18):fny199. [DOI] [PubMed] [Google Scholar]
- 9.Bang B 1897. Die Aetiologie des seuchenhaften (“infectiösen”) Verwerfens. Z. Tiermedizin 1:241–78 [Google Scholar]
- 10.Barquero-Calvo E, Chaves-Olarte E, Weiss DS, Guzman-Verri C, Chacon-Diaz C, et al. 2007. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLOS ONE 2:e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batut J, Andersson SG, O’Callaghan D. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol 2:933–45 [DOI] [PubMed] [Google Scholar]
- 12.Bhat UR, Carlson RW, Busch M, Mayer H. 1991. Distribution and phylogenetic significance of 27-hydroxy-octacosanoic acid in lipopolysaccharides from bacteria belonging to the alpha-2 subgroup of Proteobacteria. Int. J. Syst. Bacteriol 41:213–17 [DOI] [PubMed] [Google Scholar]
- 13.Bialer MG, Ruiz-Ranwez V, Sycz G, Estein SM, Russo DM, et al. 2019. MapB, the Brucella suis TamB homologue, is involved in cell envelope biogenesis, cell division and virulence. Sci. Rep 9(1):2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bittinger MA, Milner JL, Saville BJ, Handelsman J. 1997. rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol. Plant Microbe Interact 10:180–86 [DOI] [PubMed] [Google Scholar]
- 15.Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, et al. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49:D344–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun V, Bosch V. 1972. Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur. J. Biochem 28:51–69 [DOI] [PubMed] [Google Scholar]
- 17.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, et al. 2012. Polar growth in the Alphaproteobacterial order Rhizobiales. PNAS 109:1697–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruce D 1887. Note on the discovery of a microorganism in Malta fever. Practitioner 39:161–70 [Google Scholar]
- 19.Bukata L, Altabe S, de Mendoza D, Ugalde RA, Comerci DJ. 2008. Phosphatidylethanolamine synthesis is required for optimal virulence of Brucella abortus. J. Bacteriol 190:8197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casabuono AC, Czibener C, Del Giudice MG, Valguarnera E, Ugalde JE, Couto AS. 2017. New features in the lipid A structure of Brucella suis and Brucella abortus lipopolysaccharide. J. Am. Soc. Mass Spectrom 28:2716–23 [DOI] [PubMed] [Google Scholar]
- 21.Caswell CC, Elhassanny AE, Planchin EE, Roux CM, Weeks-Gorospe JN, et al. 2013. Diverse genetic regulon of the virulence-associated transcriptional regulator MucR in Brucella abortus 2308. Infect. Immun 81:1040–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charles TC, Nester EW. 1993. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol 175:6614–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou AY, Archdeacon J, Kado CI. 1998. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. PNAS 95:5293–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cloeckaert A, Vizcaino N, Paquet JY, Bowden RA, Elzer PH. 2002. Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol 90:229–47 [DOI] [PubMed] [Google Scholar]
- 25.Cloeckaert A, Zygmunt MS, de Wergifosse P, Dubray G, Limet JN. 1992. Demonstration of peptidoglycan-associated Brucella outer-membrane proteins by use of monoclonal antibodies. J. Gen. Microbiol 138:1543–50 [DOI] [PubMed] [Google Scholar]
- 26.Coloma-Rivero RF, Flores-Concha M, Molina RE, Soto-Shara R, Cartes A, Onate AA. 2021. Brucella and its hidden flagellar system. Microorganisms 10:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comerci DJ, Altabe S, de Mendoza D, Ugalde RA. 2006. Brucella abortus synthesizes phosphatidylcholine from choline provided by the host. J. Bacteriol 188:1929–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conde-Alvarez R, Arce-Gorvel V, Iriarte M, Mancek-Keber M, Barquero-Calvo E, et al. 2012. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLOS Pathog. 8:e1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conde-Alvarez R, Palacios-Chaves L, Gil-Ramirez Y, Salvador-Bescos M, Barcena-Varela M, et al. 2017. Identification of lptA, lpxE, and lpxO, three genes involved in the remodeling of Brucella cell envelope. Front. Microbiol 8:2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conde-Alvarez R, Grilló MJ, Salcedo SP, De Miguel MJ, Fugier E, et al. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell. Microbiol 8:1322–35 [DOI] [PubMed] [Google Scholar]
- 31.Conner JG, Zamorano-Sanchez D, Park JH, Sondermann H, Yildiz FH. 2017. The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol 36:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corbel MJ. 1997. Brucellosis: an overview. Emerg. Infect. Dis 3:213–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Bolle X, Crosson S, Matroule JY, Letesson JJ. 2015. Brucella abortus cell cycle and infection are coordinated. Trends Microbiol. 23:812–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. 2015. TRANSIT–a software tool for Himar1 TnSeq analysis. PLOS Comput. Biol 11:e1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeJesus MA, Ioerger TR. 2015. Capturing uncertainty by modeling local transposon insertion frequencies improves discrimination of essential genes. IEEE/ACM Trans. Comput. Biol. Bioinform 12:92–102 [DOI] [PubMed] [Google Scholar]
- 36.DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, et al. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. PNAS 99:443–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duenas AI, Orduna A, Crespo MS, Garcia-Rodriguez C. 2004. Interaction of endotoxins with Toll-like receptor 4 correlates with their endotoxic potential and may explain the proinflammatory effect of Brucella spp. LPS. Int. Immunol 16:1467–75 [DOI] [PubMed] [Google Scholar]
- 38.Eswara PJ, Ramamurthi KS. 2017. Bacterial cell division: nonmodels poised to take the spotlight. Annu. Rev. Microbiol 71:393–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferguson GP, Datta A, Baumgartner J, Roop RM, Carlson RW, Walker GC. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. PNAS 101:5012–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiebig A, Herrou J, Willett J, Crosson S. 2015. General stress signaling in the Alphaproteobacteria. Annu. Rev. Genet 49:603–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontana C, Conde-Alvarez R, Stahle J, Holst O, Iriarte M, et al. 2016. Structural studies of lipopolysaccharide-defective mutants from Brucella melitensis identify a core oligosaccharide critical in virulence. J. Biol. Chem 291:7727–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francez-Charlot A, Kaczmarczyk A, Fischer HM, Vorholt JA. 2015. The general stress response in Alphaproteobacteria. Trends Microbiol. 23:164–71 [DOI] [PubMed] [Google Scholar]
- 43.Francis N, Poncin K, Fioravanti A, Vassen V, Willemart K, et al. 2017. CtrA controls cell division and outer membrane composition of the pathogen Brucella abortus. Mol. Microbiol 103:780–97 [DOI] [PubMed] [Google Scholar]
- 44.Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, et al. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 7:687–98 [DOI] [PubMed] [Google Scholar]
- 45.Gamazo C, Moriyon I. 1987. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect. Immun 55:609–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao JL, Weissenmayer B, Taylor AM, Thomas-Oates J, Lopez-Lara IM, Geiger O. 2004. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol. Microbiol 53:1757–70 [DOI] [PubMed] [Google Scholar]
- 47.Giambartolomei GH, Zwerdling A, Cassataro J, Bruno L, Fossati CA, Philipp MT. 2004. Lipoproteins, not lipopolysaccharide, are the key mediators of the proinflammatory response elicited by heat-killed Brucella abortus. J. Immunol 173:4635–42 [DOI] [PubMed] [Google Scholar]
- 48.Gil-Ramirez Y, Conde-Alvarez R, Palacios-Chaves L, Zuniga-Ripa A, Grillo MJ, et al. 2014. The identification of wadB, a new glycosyltransferase gene, confirms the branched structure and the role in virulence of the lipopolysaccharide core of Brucella abortus. Microb. Pathog 73:53–59 [DOI] [PubMed] [Google Scholar]
- 49.Godessart P, Lannoy A, Dieu M, Van der Verren SE, Soumillion P, et al. 2021. b-Barrels covalently link peptidoglycan and the outer membrane in the a-proteobacterium Brucella abortus. Nat. Microbiol 6:27–33 [DOI] [PubMed] [Google Scholar]
- 50.Godfroid F, Taminiau B, Danese I, Denoel P, Tibor A, et al. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun 66:5485–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez-Miguel MJ, Moriyon I. 1986. Demonstration of a peptidoglycan-linked lipoprotein and characterization of its trypsin fragment in the outer membrane of Brucella spp. Infect. Immun 53:678–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goolab S, Roth RL, van Heerden H, Crampton MC. 2015. Analyzing the molecular mechanism of lipoprotein localization in Brucella. Front. Microbiol 6:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gourley CR, Petersen E, Harms J, Splitter G. 2015. Decreased in vivo virulence and altered gene expression by a Brucella melitensis light-sensing histidine kinase mutant. Pathog. Dis 73:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grabowicz M 2019. Lipoproteins and their trafficking to the outer membrane. EcoSal Plus 8. 10.1128/ecosalplus.ESP-0038-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzman-Verri C, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, et al. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. PNAS 99:12375–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallez R, Mignolet J, Van Mullem V, Wery M, Vandenhaute J, et al. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 26:1444–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hantke K, Braun V. 1973. Covalent binding of lipid to protein: diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur. J. Biochem 34:284–96 [DOI] [PubMed] [Google Scholar]
- 58.Heinz E, Selkrig J, Belousoff MJ, Lithgow T. 2015. Evolution of the translocation and assembly module (TAM). Genome Biol. Evol 7:1628–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrmann CK, Bukata L, Melli L, Marchesini MI, Caramelo JJ, Comerci DJ. 2013. Identification and characterization of a high-affinity choline uptake system of Brucella abortus. J. Bacteriol 195:493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrou J, Crosson S, Fiebig A. 2017. Structure and function of HWE/HisKA2-family sensor histidine kinases. Curr. Opin. Microbiol 36:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herrou J, Willett JW, Fiebig A, Czyz DM, Cheng JX, et al. 2019. Brucella periplasmic protein EipB is a molecular determinant of cell envelope integrity and virulence. J. Bacteriol 201:e00134–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herrou J, Willett JW, Fiebig A, Varesio LM, Czyz DM, et al. 2019. Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus. Mol. Microbiol 111:637–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hershey DM, Porfirio S, Black I, Jaehrig B, Heiss C, et al. 2019. Composition of the holdfast polysaccharide from Caulobacter crescentus. J. Bacteriol 201:e00276–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howell M, Brown PJ. 2016. Building the bacterial cell wall at the pole. Curr. Opin. Microbiol 34:53–59 [DOI] [PubMed] [Google Scholar]
- 65.Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, et al. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact 8:267–77 [DOI] [PubMed] [Google Scholar]
- 66.Kerrinnes T, Young BM, Leon C, Roux CM, Tran L, et al. 2015. Phospholipase A1 modulates the cell envelope phospholipid content of Brucella melitensis, contributing to polymyxin resistance and pathogenicity. Antimicrob. Agents Chemother 59:6717–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khan M, Harms JS, Marim FM, Armon L, Hall CL, et al. 2016. The bacterial second messenger cyclic di-GMP regulates Brucella pathogenesis and leads to altered host immune response. Infect. Immun 84:3458–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim HS, Caswell CC, Foreman R, Roop RM, Crosson S. 2013. The Brucella abortus general stress response system regulates chronic mammalian infection and is controlled by phosphorylation and proteolysis. J. Biol. Chem 288:13906–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim HS, Willett JW, Jain-Gupta N, Fiebig A, Crosson S. 2014. The Brucella abortus virulence regulator, LovhK, is a sensor kinase in the general stress response signalling pathway. Mol. Microbiol 94:913–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleanthous C, Armitage JP. 2015. The bacterial cell envelope. Philos. Trans. R. Soc. Lond 370(1679):20150019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krol E, Schaper S, Becker A. 2020. Cyclic di-GMP signaling controlling the free-living lifestyle of alpha-proteobacterial rhizobia. Biol. Chem 401:1335–48 [DOI] [PubMed] [Google Scholar]
- 72.Kubler-Kielb J, Vinogradov E. 2013. The study of the core part and non-repeating elements of the O-antigen of Brucella lipopolysaccharide. Carbohydr. Res 366:33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, et al. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem. Int. Ed. Engl 51:12519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lamontagne J, Butler H, Chaves-Olarte E, Hunter J, Schirm M, et al. 2007. Extensive cell envelope modulation is associated with virulence in Brucella abortus. J. Proteome Res 6:1519–29 [DOI] [PubMed] [Google Scholar]
- 75.Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, et al. 2009. Intracellular adaptation of Brucella abortus. J. Proteome Res 8:1594–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lapaque N, Moriyon I, Moreno E, Gorvel JP. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol 8:60–66 [DOI] [PubMed] [Google Scholar]
- 77.Lasker K, Mann TH, Shapiro L. 2016. An intracellular compass spatially coordinates cell cycle modules in Caulobacter crescentus. Curr. Opin. Microbiol 33:131–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lopez-Goni I, Guzman-Verri C, Manterola L, Sola-Landa A, Moriyon I, Moreno E. 2002. Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet. Microbiol 90:329–39 [DOI] [PubMed] [Google Scholar]
- 79.Lori C, Ozaki S, Steiner S, Bohm R, Abel S, et al. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–39 [DOI] [PubMed] [Google Scholar]
- 80.Luebke JL, Eaton DS, Sachleben JR, Crosson S. 2018. Allosteric control of a bacterial stress response system by an anti-sigma factor. Mol. Microbiol 107:164–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malinverni JC, Silhavy TJ. 2011. Assembly of outer membrane b-barrel proteins: the Bam complex. EcoSal Plus 4. 10.1128/ecosalplus.4.3.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manterola L, Moriyon I, Moreno E, Sola-Landa A, Weiss DS, et al. 2005. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol 187:5631–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mantis NJ, Winans SC. 1993. The chromosomal response regulatory gene chvI of Agrobacterium tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. J. Bacteriol 175:6626–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez de Tejada G, Pizarro-Cerda J, Moreno E, Moriyon I. 1995. The outer membranes of Brucella spp. are resistant to bactericidal cationic peptides. Infect. Immun 63:3054–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinez-Morales F, Schobert M, Lopez-Lara IM, Geiger O. 2003. Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology 149:3461–71 [DOI] [PubMed] [Google Scholar]
- 86.Martinez-Nunez C, Altamirano-Silva P, Alvarado-Guillen F, Moreno E, Guzman-Verri C, Chaves-Olarte E. 2010. The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J. Bacteriol 192:5603–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meikle PJ, Perry MB, Cherwonogrodzky JW, Bundle DR. 1989. Fine structure of A and M antigens from Brucella biovars. Infect. Immun 57:2820–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mirabella A, Terwagne M, Zygmunt MS, Cloeckaert A, De Bolle X, Letesson JJ. 2013. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J. Bacteriol 195:453–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Monreal D, Grillo MJ, Gonzalez D, Marin CM, De Miguel MJ, et al. 2003. Characterization of Brucella abortus O-polysaccharide and core lipopolysaccharide mutants and demonstration that a complete core is required for rough vaccines to be efficient against Brucella abortus and Brucella ovis in the mouse model. Infect. Immun 71:3261–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moreno E, Cloeckaert A, Moriyon I. 2002. Brucella evolution and taxonomy. Vet. Microbiol 90:209–27 [DOI] [PubMed] [Google Scholar]
- 91.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol 172:3569–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu. Rev. Microbiol 65:239–59 [DOI] [PubMed] [Google Scholar]
- 93.Onyeziri MC, Hardy GG, Natarajan R, Xu J, Reynolds IP, et al. 2022. Dual adhesive unipolar polysaccharides synthesized by overlapping biosynthetic pathways in Agrobacterium tumefaciens. Mol. Microbiol 117:1023–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palacios-Chaves L, Conde-Alvarez R, Gil-Ramirez Y, Zuniga-Ripa A, Barquero-Calvo E, et al. 2011. Brucella abortus ornithine lipids are dispensable outer membrane components devoid of a marked pathogen-associated molecular pattern. PLOS ONE 6:e16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Panis G, Murray SR, Viollier PH. 2015. Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol. Rev 39:120–33 [DOI] [PubMed] [Google Scholar]
- 96.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect. Dis 6:91–99 [DOI] [PubMed] [Google Scholar]
- 97.Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G. 2011. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J. Bacteriol 193:5683–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poncin K, Gillet S, De Bolle X. 2018. Learning from the master: targets and functions of the CtrA response regulator in Brucella abortus and other alpha-proteobacteria. FEMS Microbiol. Rev 42:500–13 [DOI] [PubMed] [Google Scholar]
- 99.Quilici G, Berardi A, Fabris C, Ghitti M, Punta M, et al. 2022. Solution structure of the BPSL1445 protein of Burkholderia pseudomallei reveals the SYLF domain three-dimensional fold. ACS Chem. Biol 17:230–39 [DOI] [PubMed] [Google Scholar]
- 100.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reboul A, Carlier E, Stubbe FX, Barbieux E, Demars A, et al. 2021. PdeA is required for the rod shape morphology of Brucella abortus. Mol. Microbiol 116:1449–63 [DOI] [PubMed] [Google Scholar]
- 102.Rivas-Solano O, Van der Henst M, Castillo-Zeledon A, Suarez-Esquivel M, Munoz-Vargas L, et al. 2022. The regulon of Brucella abortus two-component system BvrR/BvrS reveals the coordination of metabolic pathways required for intracellular life. PLOS ONE 17:e0274397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roba A, Carlier E, Godessart P, Naili C, De Bolle X. 2022. Histidine auxotroph mutant is defective for cell separation and allows the identification of crucial factors for cell division in Brucella abortus. Mol. Microbiol 118:145–54 [DOI] [PubMed] [Google Scholar]
- 104.Rohs PDA, Bernhardt TG. 2021. Growth and division of the peptidoglycan matrix. Annu. Rev. Microbiol 75:315–36 [DOI] [PubMed] [Google Scholar]
- 105.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev 77:1–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roop RM 2nd, Barton IS, Hopersberger D, Martin DW. 2021. Uncovering the hidden credentials of Brucella virulence. Microbiol. Mol. Biol. Rev 85:e00021–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sandoz KM, Moore RA, Beare PA, Patel AV, Smith RE, et al. 2021. b-Barrel proteins tether the outer membrane in many Gram-negative bacteria. Nat. Microbiol 6:19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schaper S, Yau HCL, Krol E, Skotnicka D, Heimerl T, et al. 2018. Seven-transmembrane receptor protein RgsP and cell wall-binding protein RgsM promote unipolar growth in Rhizobiales. PLOS Genet. 14:e1007594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scholz H, Banai M, Cloeckaert A, Kampfer P, Whatmore A. 2018. Brucella. In Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Trujillo ME, Dedysh S, DeVos P, Hedlund B, Kämpfer P, et al. Hoboken, NJ: John Wiley. 10.1002/9781118960608.gbm00807.pub2 [DOI] [Google Scholar]
- 110.Servais C, Vassen V, Verhaeghe A, Kuster NS, Carlier E, et al. 2022. Lipopolysaccharide synthesis and traffic in the envelope of the pathogen Brucella abortus. bioRxiv 2022.05.19.492625, May 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, et al. 2014. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. PNAS 111:14229–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sohlenkamp C, Lopez-Lara IM, Geiger O. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res 42:115–62 [DOI] [PubMed] [Google Scholar]
- 114.Sola-Landa A, Pizarro-Cerda J, Grillo MJ, Moreno E, Moriyon I, et al. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol 29:125–38 [DOI] [PubMed] [Google Scholar]
- 115.Sperandeo P, Deho G, Polissi A. 2009. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791:594–602 [DOI] [PubMed] [Google Scholar]
- 116.Sternon JF, Godessart P, Goncalves de Freitas R, Van der Henst M, Poncin K, et al. 2018. Transposon sequencing of Brucella abortus uncovers essential genes for growth in vitro and inside macrophages. Infect. Immun 86:e00312–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stranahan LW, Arenas-Gamboa AM. 2021. When the going gets rough: the significance of Brucella lipopolysaccharide phenotype in host-pathogen interactions. Front. Microbiol 12:713157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sutcliffe IC, Harrington DJ, Hutchings MI. 2012. A phylum level analysis reveals lipoprotein biosynthesis to be a fundamental property of bacteria. Protein Cell 3:163–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sycz G, Carrica MC, Tseng TS, Bogomolni RA, Briggs WR, et al. 2015. LOV histidine kinase modulates the general stress response system and affects the virB operon expression in Brucella abortus. PLOS ONE 10:e0124058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tan Y, Kagan JC. 2014. A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol. Cell 54:212–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tartilan-Choya B, Sidhu-Munoz RS, Vizcaino N. 2021. The transcriptional regulator MucR, but not its controlled acid-activated chaperone HdeA, is essential for virulence and modulates surface architecture and properties in Brucella ovis PA. Front. Vet. Sci 8:814752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thiele OW, Schwinn G. 1973. The free lipids of Brucella melitensis and Bordetella pertussis. Eur. J. Biochem 34:333–44 [DOI] [PubMed] [Google Scholar]
- 123.Valguarnera E, Spera JM, Czibener C, Fulgenzi FR, Casabuono AC, et al. 2018. RomA, a periplasmic protein involved in the synthesis of the lipopolysaccharide, tunes down the inflammatory response triggered by Brucella. J. Infect. Dis 217:1257–66 [DOI] [PubMed] [Google Scholar]
- 124.Van der Henst M, De Bolle X. 2022. Brucella abortus, a pathogenic rhizobiale with a complex cell cycle. In Cell Cycle Regulation and Development in Alphaproteobacteria, ed. Biondi EG, pp. 287–301. Cham, Switz.: Springer [Google Scholar]
- 125.van Teeseling MCF, Thanbichler M. 2020. Generating asymmetry in a changing environment: cell cycle regulation in dimorphic alphaproteobacteria. Biol. Chem 401:1349–63 [DOI] [PubMed] [Google Scholar]
- 126.Varesio LM, Willett JW, Fiebig A, Crosson S. 2019. A carbonic anhydrase pseudogene sensitizes select Brucella lineages to low CO2 tension. J. Bacteriol 201:e00509–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vassen V, Valotteau C, Feuillie C, Formosa-Dague C, Dufrene YF, De Bolle X. 2019. Localized incorporation of outer membrane components in the pathogen Brucella abortus. EMBO J. 38:e100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Verger JM, Grimont F, Grimont PAD, Grayon M. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic-acid hybridization. Int. J. Syst. Bacteriol 35:292–95 [Google Scholar]
- 129.Verstreate DR, Creasy MT, Caveney NT, Baldwin CL, Blab MW, Winter AJ. 1982. Outer membrane proteins of Brucella abortus: isolation and characterization. Infect. Immun 35:979–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wattam AR, Inzana TJ, Williams KP, Mane SP, Shukla M, et al. 2012. Comparative genomics of early-diverging Brucella strains reveals a novel lipopolysaccharide biosynthesis pathway. mBio 3:e00246–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weissenmayer B, Gao JL, Lopez-Lara IM, Geiger O. 2002. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol 45:721–33 [DOI] [PubMed] [Google Scholar]
- 132.Whatmore AM. 2009. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol 9:1168–84 [DOI] [PubMed] [Google Scholar]
- 133.Whatmore AM, Foster JT. 2021. Emerging diversity and ongoing expansion of the genus Brucella. Infect. Genet. Evol 92:104865. [DOI] [PubMed] [Google Scholar]
- 134.Willett JW, Herrou J, Briegel A, Rotskoff G, Crosson S. 2015. Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen. PNAS 112:E3709–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu Q, Pei J, Turse C, Ficht TA. 2006. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 6:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xue S, Biondi EG. 2019. Coordination of symbiosis and cell cycle functions in Sinorhizobium meliloti. Biochim. Biophys. Acta Gene Regul. Mech 1862:691–96 [DOI] [PubMed] [Google Scholar]
- 137.Zhao Y, Arce-Gorvel V, Conde-Alvarez R, Moriyon I, Gorvel JP. 2018. Vaccine development targeting lipopolysaccharide structure modification. Microbes Infect. 20:455–60 [DOI] [PubMed] [Google Scholar]
- 138.Zygmunt MS, Jacques I, Bernardet N, Cloeckaert A. 2012. Lipopolysaccharide heterogeneity in the atypical group of novel emerging Brucella species. Clin. Vaccine Immunol 19:1370–73 [DOI] [PMC free article] [PubMed] [Google Scholar]


