Abstract
The eukaryotic protein synthesis inhibitor cycloheximid has been used by many investigators to selectively radiolabel intracellular bacteria. Although cycloheximide has no direct effect on bacterial gene expression, there are concerns that long-term inhibition of the host cell protein synthesis may have secondary effects on bacterial gene expression. Therefore, prior to further identification and cloning of the macrophage-induced (MI) genes of Legionella pneumophila, the effects of cycloheximide on L. pneumophila-infected U937 cells were evaluated by transmission electron microscopy. Inhibition of protein synthesis of the host cell for 6 h had no major effect on the ultrastructure of the host cell, on the formation of rough endoplasmic reticulum-surrounded replicative phagosome, or on initiation of intracellular bacterial replication. In contrast, by 15 h of cycloheximide treatment, there was profound deterioration in the host cell as well as in the phagosome. To examine protein synthesis by L. pneumophila during the intracellular infection, U937 macrophage-like cells were infected with L. pneumophila, and intracellular bacteria were radiolabeled during a 2-h cycloheximide treatment or following 12 h of cycloheximide treatment. Comparison by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the protein profile of radiolabeled in vitro-grown L. pneumophila to that of intracellularly radiolabeled bacteria showed that 23 proteins were induced in response to the intracellular environment during 2 h of inhibition of host cell protein biosynthesis. Twelve MI proteins of L. pneumophila were artifactually induced due to prolonged inhibition of the host cell protein synthesis. The gene encoding a 20-kDa MI protein was cloned by a reverse genetics technique. Sequence analysis showed that the cloned gene encoded a protein that was 80% similar to the enzyme inorganic pyrophosphatase. Studies of promoter fusion to a promoterless lacZ gene showed that compared to in vitro-grown bacteria, expression of the pyrophosphatase gene (ppa) was induced fourfold throughout the intracellular infection. There was no detectable induction in transcription of the ppa promoter during exposure to stress stimuli in vitro. The ppa gene of L. pneumophila is the first example of a regulated ppa gene which is selectively induced during intracellular infection and which may reflect enhanced capabilities of macromolecular biosynthesis by intracellular L. pneumophila. The data indicate caution in the long-term use of inhibition of host cell protein synthesis to selectively examine gene expression by intracellular bacteria.
Legionella pneumophila is a ubiquitous organism in the natural aquatic environment where it is amplified by intracellular multiplication within protozoa (reviewed in reference 17). The ability of L. pneumophila to cause Legionnaires’ disease is dependent on its capacity to survive and to multiply within host alveolar macrophages and possibly alveolar epithelial cells (14, 21, 35).
Part of the uptake of L. pneumophila by macrophages is mediated by the complement receptor (36). We have recently shown that uptake of L. pneumophila by the protozoan Hartmanella vermiformis is mediated by a β2 integrin-like galactose lectin, which undergoes a time-dependent tyrosine dephosphorylation upon bacterial attachment (46). Although the mechanisms of L. pneumophila uptake by macrophages and by protozoa are different (5, 25), the intracellular life cycles of the bacterium within mammalian macrophages and within protozoa are highly similar (2, 18a, 19). In both host cells, the bacterium replicates in a phagosome that is inhibited from maturation through the endosomal-lysosomal pathway and is surrounded by the rough endoplasmic reticulum (RER) (2, 10, 19, 20, 43).
Pathogenic bacteria such as L. pneumophila and Salmonella typhimurium respond and adapt to the various local environmental conditions that they encounter by coordinate regulation of gene expression (1, 3, 16, 42). Upon phagocytosis and during intracellular survival, facultative intracellular pathogens are exposed to a complex mixture of stimuli, and they respond to these stimuli by a profound alteration in gene expression (1, 3, 16). These phenotypic modulations allow intracellular bacteria to survive and adapt to environmental conditions that may be encountered intracellularly, including nutrient limitation, temperature, oxidative injury, osmolarity, pH, and other stimuli (6, 33, 41). The phenotypic alteration by intracellular pathogens is controlled by multiple regulons and is most probably a simultaneous response to a complex combination of unknown stimuli. Characterization of the bacterial macrophage-induced (MI) genes and examination of the kinetics of their expression during the intracellular infection will facilitate characterization of the phagosomal microenvironment that the organisms are exposed to within the host cell. We and others have shown that intracellular L. pneumophila manifests a stress response to the intracellular environment (3, 6, 16). We have recently shown that the global stress gene (gspA) of L. pneumophila is induced in response to in vitro stress stimuli and is also induced throughout the intracellular infection period (4, 6). Differential expression of gspA by a ς32-regulated promoter throughout the intracellular infection is indicative of a continuous stress response manifested by intracellular bacteria (6). Besides the evidence of exposure of intracellular L. pneumophila to unidentified stress stimuli (3, 6, 16), the intracellular niche of L. pneumophila is not well identified.
To start understanding the nature of the microenvironment within the replicative vacuole and the biochemical signals that intracellular L. pneumophila is exposed to, the technique of reverse genetics was utilized to isolate and characterize one of the 23 L. pneumophila MI genes that are specifically induced during intracellular infection.
MATERIALS AND METHODS
Bacterial strains and vectors.
The virulent AA100 strain of L. pneumophila has been described previously (3, 15). Escherichia coli XL1-Blue was used as the host for the genomic library. E. coli K-12 derivative strain χ2981 is an asd insertion mutant that renders it auxotrophic for diaminopimelic acid (7).
The plasmids pBluescript and pBC were purchased from Stratagene (La Jolla, Calif.). The plasmid pUC-4K was purchased from Pharmacia Biotechnologies (Piscataway, N.J.), and was the source of the kanamycin resistance gene in generating the insertional mutations. The single-copy plasmid pEU730 is a spectinomycin-resistant plasmid containing a promoterless lacZ gene, kindly provided by R. Perry (University of Kentucky, Lexington) (18). Bacteria harboring pEU730 or its derivative (pPPZ) were maintained by growth on agar plates supplemented with 0.5 mg of spectinomycin per ml. The high concentration of spectinomycin is required to select for extrachromosomal locus on the plasmid, since L. pneumophila contains a chromososmal spectinomycin resistance gene (12a). The plasmid pBOC20 (kindly provided by N. Cianciotto, Northwestern University, Chicago, Ill.) is a chloramphenicol-resistant plasmid that contains oriT and the sacB gene, which is lethal to L. pneumophila grown in the presence of sucrose (13). The conjugative plasmid pRK212.1 was used as a helper plasmid to mobilize the pBOC20 vector and its recombinant derivatives by triparental conjugation into L. pneumophila (8).
DNA manipulations.
Chromosomal DNA preparations, transfection, in situ colony hybridization, end radiolabeling of oligonucleotides, restriction enzyme digestion, and DNA ligation were performed as described elsewhere (39) unless specified. Restriction enzymes, T4 polynucleotide kinase, and T4 DNA ligase were obtained from Bethesda Research Laboratories (BRL) (Gaithersburg, Md.).
Plasmid DNA preparations were done by using the QIAGEN (Chatsworth, Calif.) plasmid kit according to manufacturer’s recommendations. Transformations were carried out by electroporation with a Bio-Rad (Hercules, Calif.) gene pulser as recommended by the manufacturer. Purification of DNA fragments from agarose gels for subcloning or labeling for Southern hybridization was done by using a QIAEX kit (QIAGEN) according to manufacturer’s recommendations. Transfer of DNA from agarose gels onto membranes, fluorescein labeling of DNA probes, hybridizations, and detection were done as described previously (4). For DNA sequencing, the dideoxy chain termination method of Sanger et al. (40) was employed. Oligonucleotide primers were synthesized commercially (BRL).
Tissue culture.
Macrophage-like U937 cells were maintained at 37°C and 5% CO2 in RPMI 1640 tissue culture medium supplemented with 10% heat-inactivated fetal calf serum (Sigma Chemical Co., St. Louis, Mo.). Prior to infection, the cells were differentiated with phorbol 12-myristate 13-acetate for 48 h, as described previously (3). Differentiated cells are nonreplicative adherent macrophage-like cells. Monolayers were washed three times with the tissue culture medium prior to infection. For infection of monolayers, L. pneumophila grown for 48 h at 37°C on BCYE agar plates were resuspended in RPMI 1640. The infection was carried out as described for each experiment.
Transmission electron microscopy.
U937 cell monolayers were treated with cycloheximide for 30 min prior to the beginning of the infection by L. pneumophila at a multiplicity of infection (MOI) of 10, followed by extensive washing of extracellular bacteria with tissue culture medium. To examine phagocytosis, infection was carried out for 3 min at an MOI of 500. At several time intervals, infected U937 cell monolayers were fixed in 3.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 1 h on ice followed by 1% OsO4 in 0.1 M phosphate buffer for 1 h on ice. Dehydration was accomplished by serial exposure to ethanol (50 to 100%) for 10 min at room temperature. The cells were embedded in Eponate 12 resin (Ted Pella, Redding, Calif.) according to the manufacturer’s recommendations. Ultrathin sections were stained with uranyl acetate followed by lead citrate and examined with a Hitachi model H-7000/STEM electron microscope at 75 kV.
Radiolabeling of bacteria and two-dimensional polyacrylamide gel electrophoresis (PAGE).
A modification of a previously reported protocol (3) was used to radiolabel intracellular L. pneumophila. Monolayers of differentiated U937 cells were infected for 1 h with L. pneumophila AA100 resuspended in RPMI 1640 (Gibco, Gaithersburg, Md.) at a bacterium-to-U937 ratio of 10:1. The monolayers were subsequently washed and treated with gentamicin (50 μg/ml) for 1 h to kill extracellular bacteria. Two sets of infected monolayers were used; in one set, cycloheximide (200 μg/ml) was added immediately to the monolayers after the gentamicin treatment, while for the second infected monolayers, cycloheximide was added after 12 h of the infection, prior to radiolabeling of intracellular bacteria. [35S]methionine (150 μCi/ml) was added to both sets of infected monolayers for an additional 2 h to permit incorporation of the label. Uninoculated U937 cells monolayers and monolayers infected with heat-killed L. pneumophila were also radiolabeled under the same experimental conditions to confirm the complete inhibition of protein synthesis of the macrophage. Intracellular radiolabeled organisms were harvested as described previously (3). For comparison, bacteria were radiolabeled in vitro in defined medium during mid-log growth phase in a manner similar to that of intracellular bacteria, exactly as described previously (3).
Cell extracts of the radiolabeled bacterial pellet were prepared by the methods of O’Farrel, as described previously (3). Equal amounts of tricarboxylic acid-precipitated radiolabeled proteins were subjected to equilibrium isoelectric focusing prior to separation in the second dimension on sodium dodecyl sulfate (SDS)–11.5% PAGE slab gels. After electrophoresis, the gels were dried and autoradiographed by using Kodak BIO-MAX film.
Protein isolation and partial N-terminal sequence analysis.
The protein spot on two-dimensional SDS-PAGE corresponding to the MI protein was isolated for N-terminal sequencing, as described previously (3). Briefly, after electrophoresis gels were soaked in transfer buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid–10% methanol (pH 11.0)] for 10 min and the proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon; Millipore) for 30 min at 50 V (150 to 170 mA) in a Trans-Blot cell (Bio-Rad, Richmond, Calif.). The membranes were stained with Coomassie brilliant blue R-250 (Sigma), and the stained protein spots were subjected to direct, automated Edman degradation.
Cloning.
A degenerate oligonucleotide corresponding to the amino acid sequence of the N terminus of the MI protein was synthesized (BRL). The sequence of the degenerate oligonucleotide was 5′-GGI(C/A)GIGA(T/C)GTICCIAA, where I is inosine. The oligonucleotide was radiolabeled and used as a probe for in situ colony hybridizations to screen the L. pneumophila partial genomic library (39). The partial genomic library was constructed by ligating gel-purified 2- to 2.5-kb EcoRI fragments into plasmid pBC and transformed into E. coli. The in situ colony hybridizations were performed exactly as described previously (4).
Transposon mutagenesis.
To facilitate subsequent allelic exchanges of insertion mutations in the cloned 2.2-kb insert, it was subcloned into plasmid pBOC20 to generate pJA2a. Transposon mutagenesis of pJA2 was performed by using lambda 1105 containing mini-Tn10-kan, exactly as described previously (7). Selected insertions were introduced into L. pneumophila AA100 by triparental conjugation (8).
Primer extension.
Total RNA was isolated from BCYE-grown L. pneumophila and from intracellular bacteria in U937 monolayers, exactly as described previously (7). The isolated RNA was used immediately or stored at −20°C. For 5′ primer extensions, an oligonucleotide with the sequence 5′-CTTCATACTTTACAGGTTCTCCGTGCATAG was used, and the reactions were carried out exactly as described previously (4).
In vitro transcription-translation.
A coupled in vitro transcription-translation system was used to examine the polypeptides expressed by pJA2 (Promega Biotech, Madison, Wis.). [35S]methionine was used to radiolabel the polypeptides. The reaction products were resolved by SDS-PAGE and autoradiography.
Fusion of the ppa promoter to a promoterless lacZ gene and β-galactosidase (β-Gal) assays.
The sequence encompassing the promoter region upstream of the initiation codon of ppa was amplified by PCR with the primers 5′-CGCTTAATTAAGTGGTAAACTTCCTGCCC and 5′-GCGGGTACCTTACGCACCCTTAATAAT. The PCR reaction was carried out in the presence of 2.5 mM Mg2+–0.25 mM deoxynucleoside triphosphates–0.2 μg of each primer–50 ng of the template PJA2a–2.5 U of Taq polymerase (Stratagene) in a 100-μl reaction mixture. The PCR protocol included one step of denaturation at 94°C for 5 min, 5 cycles of annealing at 48°C for 1 min, and extension at 72°C for 30 s followed by 25 cycles of annealing at 58°C and extension at 72°C for 1 min each. The PCR product was purified from an agarose gel and directionally cloned into the PacI-KpnI sites of the single-copy plasmid pEU730 (18), which places the promoterless lacZ gene under the control of the ppa promoter. The in-frame construct was confirmed by DNA sequencing of the junction. The recombinant was designated pPPZ and electroporated into L. pneumophila.
β-Gal expression was monitored during several stages of growth in vitro. Culture aliquots for each β-Gal determination were plated to determine the number of CFU per milliliter, and thus, β-Gal activity could be expressed as units per CFU (34). To assess β-Gal activity in response to stress stimuli, mid-log-phase L. pneumophila grown in BYE (optical density at 550 nm of 0.6 to 0.7) was exposed to stress stimuli, as described previously (3). Bacteria were pelleted immediately after exposure to the stress stimuli and resuspended in BYE for determination of β-Gal activity.
To determine β-Gal activity from intracellularly grown L. pneumophila at several stages of the intracellular infection, monolayers of differentiated U937 cells were infected with L. pneumophila harboring pPPZ or pEU730. The infection was accomplished with an MOI of 10 for 1 h followed by three washes with tissue culture medium to remove most of the extracellular bacteria. Gentamicin treatment (50 μg/ml) was done for 1 h to kill any remaining extracellular bacteria, followed by three washes with tissue culture medium. At several time intervals, monolayers were washed three times with tissue culture medium and lysed hypotonically with distilled water. Aliquots of the lysate were diluted and plated to determine the number of intracellular CFU per milliliter. β-Gal activity was determined for each lysate and calculated in terms of Miller units per CFU (34). To determine whether the β-Gal assay was affected by debris of the lysed U937 cells, mid-log-phase BYE-grown L. pneumophila was pelleted and resuspended in a lysate of uninfected monolayers for 30 to 60 min prior to measurement of β-Gal activity.
Nucleotide sequence accession number.
The nucleotide sequence of the predicted open reading frame encoding the 178-amino-acid protein identified in this study has been assigned GenBank accession no. AFO31464.
RESULTS
Effects of long-term inhibition of host cell protein biosynthesis on phagosomal ultrastructure.
Cycloheximide is a protein synthesis inhibitor of eukaryotic cells that has been used by many investigators to examine protein synthesis by intracellular bacterial pathogens (1, 3). However, the long-term use of this inhibitor may have secondary effects on bacterial gene expression due to possible physiological changes in the host cell. To examine this possibility, the effect of inhibition of protein synthesis of the host cell was examined at several times during the intracellular infection of the U937 macrophage-like cells. In these experiments, cycloheximide was added to the monolayers 30 min prior to the infection. At several intervals (3 min and 1 h, 2, 4, 6, 8, and 15 h), the monolayers were fixed and processed for transmission electron microscopy. Control untreated, infected monolayers were also examined. Complete inhibition of host cell protein synthesis was confirmed by the failure of uninfected U937 cells to incorporate [35S]methionine compared to that of untreated cells (data not shown). Compared to that in untreated infected cells, there was no detectable effect of cycloheximide on recruitment of the host cell organelles around the L. pneumophila phagosome or on formation of the RER-surrounded replicative vacuole during the first 6 h of treatment (Fig. 1) (19, 43). Inhibition of host cell protein synthesis for 8 h did not have any detectable effect on the ability of L. pneumophila to initiate bacterium replication as detected by transmission electron microscopy at 8 h postinfection (Fig. 1 and 2A) and by the number of intracellular bacteria, which had increased for at least two generations (data not shown). There were no major ultrastructural alterations in the host cell during the first 8 h of inhibition of protein synthesis, with the exception of increased frequency of fusion of cellular vesicles to the phagosome, and there may have been some minor changes in the host cell at 6 h as well (Fig. 1 and 2). These data showed that inhibition of protein synthesis of the host cell for 6 h during infection did not have a major effect on the ultrastructure of the replicative vacuole or the rest of the infected cell.
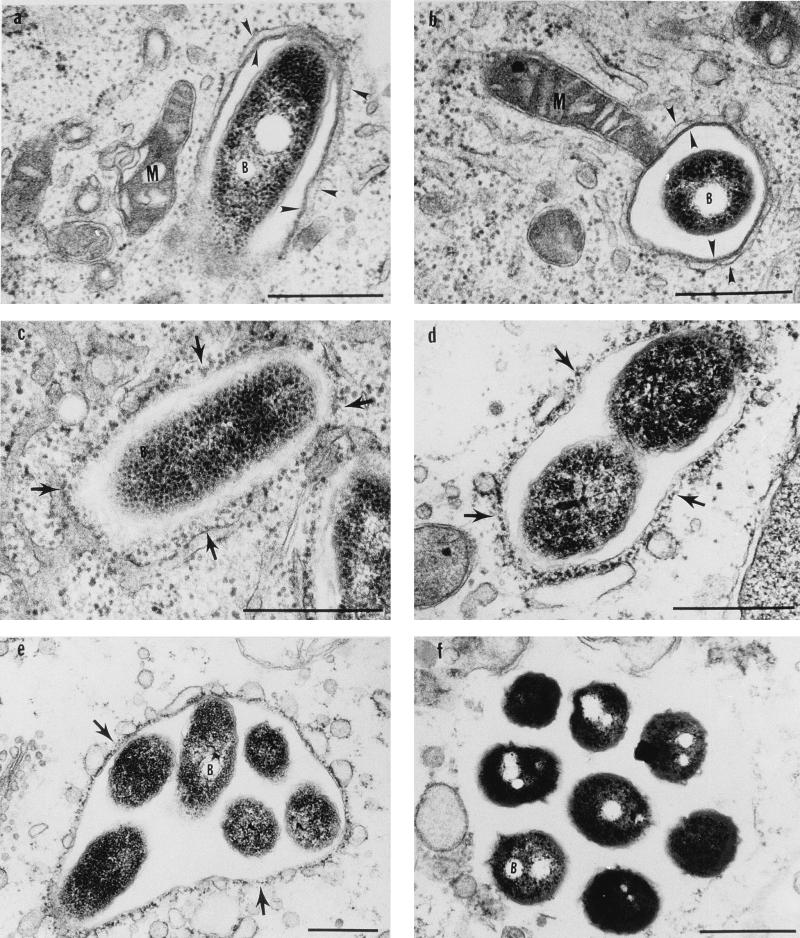
FIG. 1.
Electron micrographs of infected U937 cells during inhibition of host cell protein synthesis at 1 h (a), 2 h (b), 4 h (c), 6 h (d), 8 h (e), and 15-h (f) postinfection. Arrowheads indicate the smooth multilayer phagosomal membrane, arrows indicate the RER surrounded phagosome, M indicates mitochondria, and B indicates bacteria. Note the vesicles making contact and possibly fusing with the phagosomal membrane at 8 h in panel e. Note the absence of a defined membrane around the phagosome at 15 h in panel f. Bars, 0.5 μm.
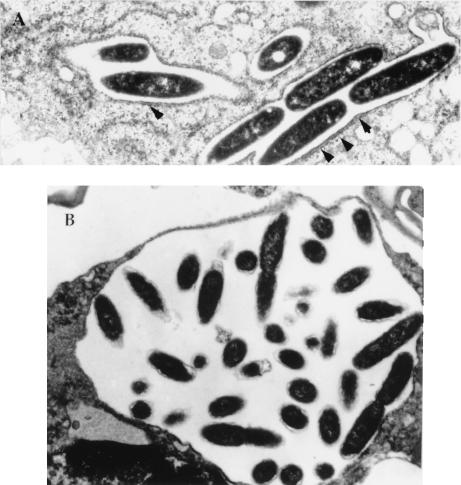
FIG. 2.
Electron micrographs of untreated infected U937 cells at 8 h (A) and 15 h (B) postinfection. Arrowheads indicate the RER-surrounded phagosome.
In contrast, after 15 h of inhibition of protein synthesis of the host cell, there was a clear evidence of deterioration of the ultrastructure of the replicative vacuole and the rest of the host cell as well (Fig. 1f and 2B). Although L. pneumophila appeared to be in clusters, there was no definitive visible phagosomal membrane and no visible RER around the vacuole. These observations indicated that long-term inhibition of host cell protein synthesis had dramatic effects on cell physiology and ultrastructure. These changes may have a secondary effect on bacterial gene expression. The data indicated that caution should be taken with interpretation of data obtained with the long-term use of this inhibitor to examine gene expression of other slow-growing intracellular bacterial pathogens, such as Mycobacterium tuberculosis, for which bacterial protein synthesis has been examined after 23 h of cycloheximid treatment of infected cells (31).
Protein synthesis by intracellular L. pneumophila.
The protein profile of radiolabeled intracellular L. pneumophila has been previously reported (3). In that report, the U937 macrophage-like cells were treated with cycloheximide for more than 20 h during radiolabeling of intracellular bacteria. Since inhibition of protein synthesis of the host cell for 15 h had profound effects on the host cell biology, we reevaluated protein synthesis by intracellular L. pneumophila radiolabeled during cycloheximide treatment of infected cells. In the previous study cycloheximide was added to the monolayers at the beginning of the infection, and intracellular bacteria were radiolabeled at mid-log phase (3). In this study, mid-log-phase intracellular bacteria were radiolabeled for 2 h in U937 cells treated with cycloheximide at the beginning of the infection or just prior to the 2 h radiolabeling of intracellular bacteria. Examination of infected cells at the end of the cycloheximide treatment showed that in contrast to cellular deteriorations after prolonged inhibition of protein synthesis of the host cell, this short-term treatment did not have any detectable effect on the ultrastructural characteristics of the replicative phagosome or on multiplication of intracellular bacteria (data not shown).
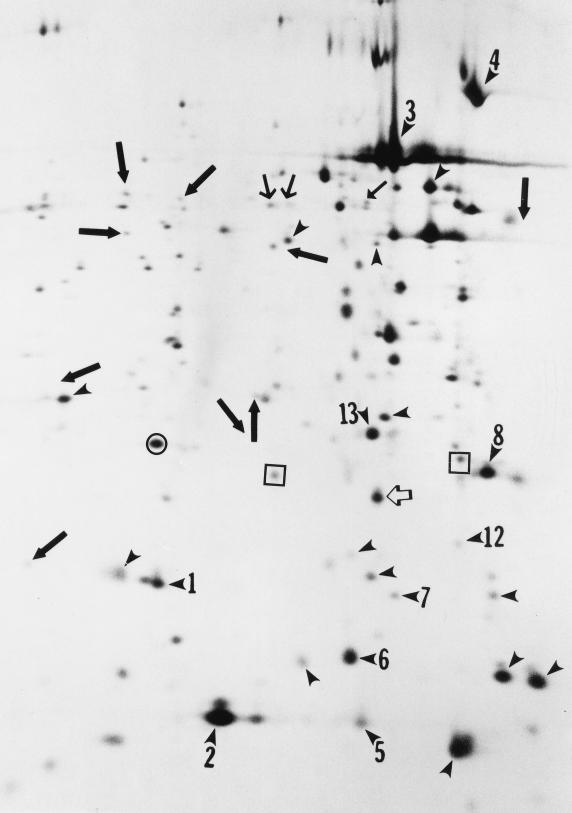
Extracts of intracellular radiolabeled L. pneumophila were subjected to two-dimensional SDS-PAGE, and the protein profile was compared to that of in vitro-grown bacteria radiolabeled during the same growth phase. Equal amounts of radioactivity (in counts per minute) were loaded for intracellular and in vitro-grown bacteria. Figure 3 shows the protein profile of radiolabeled intracellular L. pneumophila in which cycloheximide was added just prior to radiolabeling of intracellular bacteria. Compared to the protein profile of in vitro-grown organisms (3), the spots marked by arrowheads (Fig. 3) indicate the 23 proteins that were induced in response to the intracellular environment (3). In contrast to the detection of 35 MI proteins in intracellular L. pneumophila following 12 h of cycloheximide treatment of infected cells (3), there was an induction of only 23 of the 35 MI proteins induced during prolonged inhibition of the host cell protein biosynthesis (3). There was no detectable difference in the repressed proteins. It has been previously shown that there is no detectable difference in the protein profiles of intracellular bacteria labeled at 10 to 12 h postinfection compared to that radiolabeled at 20 to 22 h postinfection (3). Thus, long-term inhibition of protein synthesis of the host cell had a secondary effect of artifactual induction of expression of at least 12 L. pneumophila proteins (marked by small and large bold arrows in Fig. 3).
FIG. 3.
Autoradiograph of two-dimensional SDS-PAGE of L. pneumophila proteins radiolabeled intracellularly in U937 cells during 2 h of cycloheximide treatment and compared to the protein profile of intracellular bacteria radiolabeled during the same growth phase after 12 h of inhibition of host cell protein synthesis and to in vitro-grown bacteria during the same growth phase (3). The alkaline-to-acid gradient is from left to right. Arrowheads indicate the proteins that were induced in response to the intracellular environment. Closed arrows indicate the proteins that were artifactually induced as a result of prolonged inhibition of host cell protein synthesis (3). The open arrow indicates the 20-kDa PPase that is uniquely induced in response to the intracellular environment. The spots in squares represent the two major spots shown in Fig. 4 on both sides of the 20 kDa PPase. The spot in the circle represents the protein to which the levels of 20-kDa PPase was normalized.
Cloning of one of the MI genes of L. pneumophila.
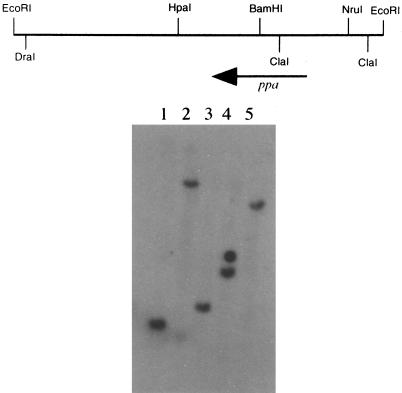
The microenvironment of the RER-surrounded phagosome containing L. pneumophila and the biochemical signals to which the bacteria are exposed in their specialized and unique niche are not known. To start understanding this microenvironment, one of the MI genes that was specifically induced during the intracellular infection and not induced by in vitro stress stimuli was cloned and characterized (3). The protein spot marked by an open arrow (Fig. 3 and 4), which was shown by phosphorimaging semiquantitation to be induced by approximately 3.4-fold, was subjected to N-terminal amino acid sequencing by Edman degradation. The sequence obtained was SLMEIPSGRDVPNEVNVIIE, with serine as the N-terminal amino acid. A degenerate oligonucleotide corresponding to the underlined sequence was synthesized. In Southern hybridizations, the probe hybridized to single 1.8-kb BamHI, 11-kb ClaI, 2.2-kb EcoRI, 3.2-kb EcoRV, and 8-kb HindIII fragments of L. pneumophila chromosomal DNA (Fig. 5). A partial genomic library of the ∼2.2-kb EcoRI fragments was constructed in plasmid pBC. Screening was performed by in situ colony hybridizations with the degenerate oligonucleotide as a probe. Two positive clones were isolated, and one clone, designated pJA2, was chosen for further analysis (Fig. 5).
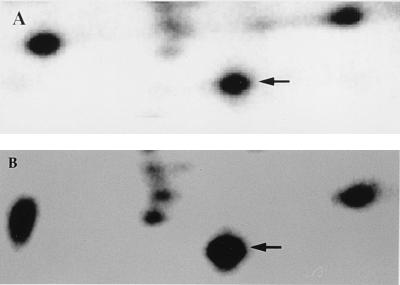
FIG. 4.
Magnification of portions of two-dimensional gels to illustrate the protein level of the PPase protein expressed in vitro-grown bacteria (A) compared to that expressed by intracellular bacteria (B). The arrows indicate the 20-kDa PPase that is uniquely induced in response to the intracellular environment. Although three spots on the left of PPase seem to be slightly induced in this gel, they were not reproducible in multiple experiments.
FIG. 5.
(Top) Restriction map of pJA2 insert; (bottom) Southern hybridization of chromosomal DNA of L. pneumophila digested and probed with the degenerate oligonucleotide corresponding to the N-terminus sequence of the 20-kDa MI protein. Lanes: 1, 1.8-kb BamHI; 2, 11-kb ClaI; 3, 2.2-kb EcoRI; 4, 3.2-kb EcoRV; 5, 8-kb HindIII. The second signal in lane 4 is an artifact. The sizes were based on a 1-kb ladder (BRL).
Sequence analysis.
The degenerate oligonucleotide corresponding to the N-terminal sequence was used for an initial sequencing reaction, and subsequent oligonucleotides were synthesized. The DNA sequence revealed the presence of a predicted open reading frame consisting of 534 nucleotides encoding a 178-amino-acid protein (Gen Bank accession no. AF031464. There was a Shine-Dalgarno sequence 8 nucleotides upstream of the initiation codon, strongly suggesting that this initiation codon represented a translational initiation site.
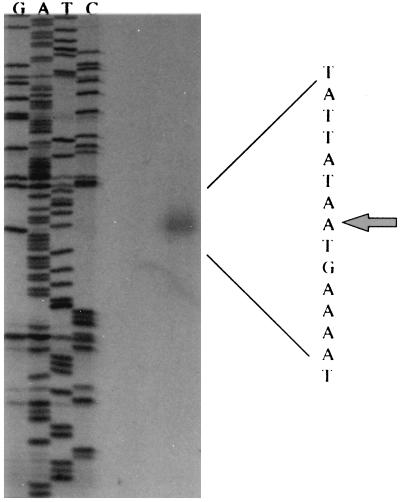
To identify the promoter regulating transcription of the MI gene, we identified the transcriptional initiation site by 5′ primer extension analysis. By comparing the synthesized cDNA to the DNA from a sequencing reaction primed with the same oligonucleotide, the 5′ primer end of the cDNA was mapped to 30 nucleotides upstream of the initiation codon (Fig. 6). Analysis of the DNA sequence upstream from the transcriptional start site suggested the presence of potential -10 and -35 regions that appeared to be similar to ς70-regulated promoters.
FIG. 6.
Primer extension analysis of ppa transcript. A radiolabeled oligonucleotide was annealed to RNA isolated from BYE-grown bacteria, and reverse transcriptase was added to produce cDNA. The same oligonucleotide was used to prime the dideoxy sequencing reaction of pJA2 DNA. The letters above each lane indicate the dideoxy nucleotide used to terminate each reaction. The arrow indicates the 5′ end of the mRNA specie corresponding to nucleotide 140.
Translation of the predicted open reading frame revealed that it encoded a protein with a predicated molecular mass of 20 kDa and an estimated isoelectric point of 5.22. These results are in agreement with the size of the protein and its migration in the acidic portion of the two-dimensional SDS-PAGE (Fig. 3). Expression of the 20-kDa MI protein by the cloned fragment was confirmed by in vitro transcription-translation, which showed synthesis of the 20-kDa polypeptide encoded by pJA2 compared to the pBC vector (Fig. 7). Furthermore, the N-terminal sequence derived by Edman degradation completely matched the predicted N-terminal sequence of the open reading frame. It is possible that the protein is processed posttranslationally to remove the N-terminal methionine from the mature protein. These data confirmed that the predicted open reading frame encoded the 20-kDa MI protein.
FIG. 7.
In vitro transcription-translation of pJA2 (lane 2) and the vector pBC (lane 1). The arrow indicates the 20-kDa 35S-labeled protein. Sizes were estimated based on Bio-Rad low-molecular-mass prestained protein standards.
Alignment with sequences in GenBank showed that the MI protein was 62% identical and 80% similar to the inorganic pyrophosphatase (PPase) of E. coli (Fig. 8) (27). Similar identities to inorganic pyrophosphatase of other organisms were also observed (data not shown). Similarly, the N-terminal sequence of the mature PPase of E. coli is also posttranslationally processed by an aminopeptidase to remove the N-terminal methionine (27).
FIG. 8.
Alignment of L. pneumophila (Lpn) PPase to the E. coli (Ec) homolog. Conservative substitutions are indicated by colons. The amino acid residues that have been shown to be crucial for structural and catalytic activity of the enzyme in E. coli (see Discussion) are indicated in bold.
Construction of null mutation in ppa.
A mini-Tn10::kan was used to construct an insertion mutation in the ppa gene in pJA2a. Three different insertions within the gene and many insertions in the sequences flanking the gene were obtained. The mutagenized plasmids were introduced into L. pneumophila by triparental conjugation (see Materials and Methods). Allelic exchange was achieved by selection on kanamycin-containing plates in the presence of sucrose. Allelic exchange was achieved for all four insertions (attempted) in the flanking regions of the ppa gene at a frequency of approximately 10−4 to 10−5. In contrast, many trials for allelic exchange of the three different insertion mutations within the ppa gene were unsuccessful, with the exception of occasional integration of the whole plasmid into the chromosome, creating a merodiploid (data not shown). These allelic exchanges are performed routinely in our laboratory (6, 7), and this is the first example of our inability to achieve the proper allelic exchange. Although these data suggested but did not confirm that PPase was essential to L. pneumophila, the observations that PPase is essential for the viability of both E. coli and yeast and our inability to construct a mutation in ppa suggested that this gene is probably essential for the viability of L. pneumophila (12, 32). Considering the essential and ubiquitous role of this protein in macromolecular biosynthesis (see Discussion), it is not surprising that it would be essential for all living organisms.
Induction in transcription of the ppa gene of L. pneumophila within macrophages.
The PPase gene (ppa) of E. coli is constitutively expressed and is not regulated in vitro. Interestingly, synthesis of the 20-kDa MI protein corresponding to PPase of L. pneumophila was induced in response to the intracellular environment but was not induced upon exposure to in vitro stress stimuli (3). These observations indicated that certain unique stimuli within the phagosomal microenvironment triggered expression of the 20-kDa protein. To examine the kinetics of transcription of ppa throughout the intracellular infection, we constructed a fusion of the ppa promoter to a promoterless lacZ gene. The promoter fusion construct (pPPZ) or the vector (pEU730) was introduced into L. pneumophila.
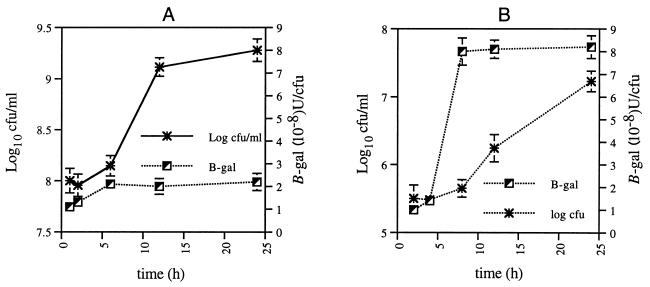
Monolayers of differentiated U937 cells were infected with L. pneumophila harboring pPPZ or pEU730, followed by gentamicin treatment to kill extracellular bacteria, and further incubated without gentamicin (see Materials and Methods). At several times, monolayers were lysed hypotonically and β-Gal activity was determined (34). To determine whether the β-Gal assay was affected by debris of the lysed U937 cells, BYE-grown L. pneumophila in growth phases similar to those of intracellular bacteria was pelleted and resuspended in a lysate of uninfected monolayers, prior to measurement of β-Gal activity. Compared to that of in vitro-grown bacteria, transcription of the ppa gene was induced during intracellular logarithmic replication by approximately fourfold (Fig. 9). There was an induction in ppa transcription upon entry to the logarithmic phase by approximately two- and eightfold by in vitro-grown and intracellular bacteria, respectively. These data indicated that expression of ppa was induced in response to the intracellular microenvironment. Since the density of the bacteria may affect the level of expression through the lacZ fusion, the β-Gal levels were evaluated for in vitro cultures of different densities (105 to 109 CFU/ml), and the density of the culture had no detectable effect on the expression of β-Gal per CFU (data not shown).
FIG. 9.
Kinetics of β-Gal expression by in vitro-grown (A) and macrophage-grown (B) L. pneumophila harboring a ppa promoter (pPPZ) fused to the promoterless lacZ gene of pEU730. Data are expressed as the mean β-Gal activity for triplicate samples (34). L. pneumophila harboring the vector pEU730 did not show any detectable β-Gal activity (data not shown).
In contrast to the intracellular induction of the ppa promoter, there was no induction in expression following exposure of L. pneumophila to heat shock, osmotic shock, acid shock, and oxidative stress (data not shown). These data further confirmed the stable expression of ppa upon exposure to stress stimuli and further showed that ppa was specifically induced in response to the intracellular environment of the host cell.
DISCUSSION
Radiolabeling of intracellular bacterial pathogens during a period of inhibition of host cell protein synthesis has been used by many investigators to examine bacterial gene expression during intracellular infection (1, 3). Cycloheximide has no direct effect on bacterial viability, growth rate, or pattern of protein synthesis in vitro (1, 3). However, prior to further cloning of L. pneumophila MI genes, the effects of this inhibitor on the host cell physiology were evaluated to ensure that some L. pneumophila MI proteins were not artifactually induced as a secondary effect of prolonged inhibition of protein synthesis of the host cell. Transmission electron microscopy was used to evaluate the effects of this inhibitor on the intracellular infection by L. pneumophila and on the ability of the bacterium to form the RER-surrounded replicative phagosome. Inhibition of protein synthesis of L. pneumophila-infected U937 cells for up to 6 h did not cause any detectable structural changes in the host cell. By 15 h posttreatment, there was clear evidence of dramatic structural deteriorations in the host cell and in the vacuole containing intracellular bacteria. These detectable structural deteriorations in the host cell caused a secondary effect of an artifactual induction of 12 bacterial proteins. Our data caution that extended use of this inhibitor to selectively radiolabel intracellular bacterial pathogens (3, 31) has profound effects on the host cell physiology, which may lead to changes in bacterial gene expression. It is recommended that to exclude any possible secondary effects on expression by intracellular bacteria, this inhibitor not to be used for more than 6 h.
Formation of the RER-surrounded replicative phagosome of L. pneumophila was not affected by inhibition of protein synthesis of the host cell. These data indicated that formation of the replicative vacuole is dictated at the biochemical level of signal transduction and that new host cell proteins are not required for this process. Bacterial replication within the RER-surrounded phagosome was also independent of the host cell protein synthesis. The data also excluded the possibility that degradation of newly synthesized proteins is required for bacterial replication.
Expansion of the phagosome was also independent of host cell protein synthesis. The data suggested that expansion of the membrane of the replicative phagosome was due to incorporation of host cell vesicles of presynthesized proteins or of bacterial components or both. The incorporation of bacterial components into the phagosomal membrane has been described previously. For example, protein antigens of the intracellular pathogens Chlamydia trachomatis and Chlamydia psittaci are incorporated into the phagosomal membrane (37, 44). Incorporation of bacterial proteins into the phagosomal membrane by L. pneumophila has yet to be determined. Such incorporation may have significant effects on the biochemical interaction between L. pneumophila and the host cell.
The PPase of L. pneumophila was 80% similar to the E. coli PPase (EC 3.6.1.1). The “housekeeping” function of PPase may explain its highly conserved domains that are involved in catalytic and structural integrity. The PPase of E. coli has been extensively characterized at the biochemical, structural, and ultrastructural levels. Many residues of the PPase have been shown to be critical for activity and structural integrity of the enzyme. The E. coli PPase is composed of six identical subunits (47) arranged in an octahedral configuration. Residues that are involved in structural stability of the homohexamer (H136 and H140) (9, 45), in binding to the Mg2+ cofactor (D65, D70, N24, D26, E98, Y141, and D102) (23, 28, 29, 38), and in catalytic activity of the enzyme (K29, R43, K142, D97, D102, Y55, and E20) (24, 28, 38) are all conserved in L. pneumophila. X-ray crystallography of the three-dimensional structure of the E. coli PPase clearly showed the presence of a group of functionally important residues that are conserved among all identified PPases (22). The consensus sequence is D-hyd-D-P-ali-D-ali-ali (hyd is a small hydrophilic residue like S, G, or N; ali, an aliphatic hydrophobic residue like C, I, L, M, or V) (22). All of these residues are also conserved in the L. pneumophila PPase (D-G-D-P-V-D-V-L). These data clearly demonstrated that the 20-kDa MI protein of L. pneumophila is a PPase. These data further confirmed the highly conserved nature of this enzyme, which is underlined by its vital housekeeping functions in macromolecular biosynthesis.
The data suggested that the PPase may be essential for viability of L. pneumophila. PPase is essential for survival in E. coli as well as in yeast (12, 32). It catalyzes reversible transfer of the phosphoryl group from pyrophosphate to water, providing a thermodynamic pull for essential biosynthetic processes such as tRNA charging and DNA, RNA, protein, and polysaccharide biosynthesis (reviewed in reference 26). It may also have an important role in evolutionary events by affecting the accuracy by which DNA molecules are copied during chromosomal replication (30). Therefore, it may not be surprising that this enzyme is essential for viability (12, 32).
The predicted sequence of the ppa promoter had some similarity to ς70-regulated promoters. Induction in transcription of ppa by intracellular L. pneumophila indicated the presence of other factors and/or other unidentified upstream sequences that are involved in transcriptional regulation of this gene. Mutational analysis of the upstream region may provide an explanation of the inducible nature of this gene and whether this induction is essential for survival of L. pneumophila within the intracellular environment.
Although the levels of PPase have been shown to be constitutive for E. coli (11), regulation in expression of none of the characterized PPase genes has been examined. L. pneumophila ppa is the first example of a regulated ppa gene during growth in a unique niche. Transcription of L. pneumophila ppa was specifically induced during intracellular multiplication but was not induced in response to in vitro stress stimuli. It is expected that actively growing organisms would require a higher rate of macromolecular biosynthesis including that of DNA, RNA, protein, and polysaccharides. Since pyrophosphate is a by-product of these reactions, higher levels of PPase activity would be required to provide the thermodynamic pull for these biosynthetic reactions. The short generation time of intracellular L. pneumophila compared to that of in vitro-grown bacteria in rich medium (21) indicates that intracellular L. pneumophila undergoes macromolecular biosynthesis at a higher rate than do in vitro-grown bacteria. These observations suggest that for replication of L. pneumophila, the microenvironment within macrophages is a more favorable niche than is rich medium. Whether enhanced macromolecular biosynthesis is the only factor required for induction of ppa transcription by intracellular bacteria or whether ppa induction is triggered by other signals to which intracellular L. pneumophila is exposed is still to be determined. Future studies to examine regulation of expression of ppa by other intracellular pathogens would shed light on whether induction of ppa in the intracellular environment is a general phenomenon among intracellular pathogens.
In summary, short-term radiolabeling of intracellular bacteria during inhibition of protein synthesis of the host cell does not have any detectable effect on the host cell biology, on the formation of the RER-surrounded replicative phagosome, or on replication of L. pneumophila. One of the MI proteins was identified as PPase, a protein highly conserved through evolution. The ppa gene of L. pneumophila is regulated intracellularly and is the first example of a regulated ppa gene. Regulation of ppa during intracellular infection may reflect enhanced capabilities of macromolecular biosynthesis by intracellular L. pneumophila compared to those of in vitro-grown bacteria.
ACKNOWLEDGMENTS
Y.A.K. is supported by Public Health Service grant no. 1R29AI38410.
I thank R. Perry for the pEU730 plasmid gift, J. Abbott for technical assistance, members of my laboratory for their valuable comments on the manuscript, and Mary G. Engle for her valuable electron microscopy expertise.
REFERENCES
- 1.Abshire K Z, Neidhardt F C. Analysis of proteins synthesized by Salmonella typhimurium during growth within host macrophage. J Bacteriol. 1993;175:3734–3743. doi: 10.1128/jb.175.12.3734-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress stimuli. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Fields B S, Engleberg N C. Protein expression by the protozoan Hartmannella vermiformis upon contact with its bacterial parasite Legionella pneumophila. Infect Immun. 1994;62:1860–1866. doi: 10.1128/iai.62.5.1860-1866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu Kwaik Y, Gao L-Y, Harb O S, Stone B J. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol Microbiol. 1997;24:629–642. doi: 10.1046/j.1365-2958.1997.3661739.x. [DOI] [PubMed] [Google Scholar]
- 7.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 8.Arroyo J, Hurley M C, Wolf M, McClain M S, Eisenstein B I, Engleberg N C. Shuttle mutagenesis of Legionella pneumophila: identification of a gene associated with host cell cytopathicity. Infect Immun. 1994;62:4075–4080. doi: 10.1128/iai.62.9.4075-4080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baykov A A, Dudarenkov V Y, Kapyla J, Salminen T, Hyytia T, Kasho V N, Husgafvel S, Cooperman B S, Goldman A, Lahti R. Dissociation of the hexameric Escherichia coli inorganic pyrophosphatase into trimers on His136Gln or His140Gln substitution and its effect on enzyme catalytic properties. J Biol Chem. 1995;270:30804–30812. doi: 10.1074/jbc.270.51.30804. [DOI] [PubMed] [Google Scholar]
- 10.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba catellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton P M, Hall D C, Josse J. Constitutive inorganic pyrophosphatase of Escherichia coli: chemical studies of protein structure. J Biol Chem. 1970;245:4346–4352. [PubMed] [Google Scholar]
- 12.Chen J, Brevet A, Fromant M, Leveque F, Schmitter J-M, Blanquet S, Plateau P. Pyrophosphatase is essential for growth of Escherichia coli. J Bacteriol. 1990;172:5686–5689. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Cianciotto, N. Personal communication.
- 13.Cianciotto N P, Long R, Eisenstein B I, Engleberg N C. Site-specific mutagenesis in Legionella pneumophila by allelic exchange using counterselectable ColE1 vectors. FEMS Microbiol Lett. 1988;56:203–208. [Google Scholar]
- 14.Cianciotto N P, Stamos J K, Kamp D W. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr Microbiol. 1995;30:247–250. doi: 10.1007/BF00293641. [DOI] [PubMed] [Google Scholar]
- 15.Edelstein P H, Brenner D J, Moss C W, Steigerwalt A G, Francis E M, George W L. Legionella wadsworthii species nova: a cause of human pneumonia. Ann Intern Med. 1982;97:809–813. doi: 10.7326/0003-4819-97-6-809. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez R C, Logan S, Lee S H S, Hoffman P S. Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlated with virulence. Infect Immun. 1996;64:1968–1976. doi: 10.1128/iai.64.6.1968-1976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 18.Froehlich B, Husmann L, Caron J, Scott J R. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Gao L-Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwitz M A. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz M A. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz M A, Silverstein S C. Legionnaires’ disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J Clin Invest. 1980;66:441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kankare J, Neal G S, Salminen T, Glumhoff T, Cooperman B S, Lahti R, Goldman A. The structure of E. coli soluble inorganic pyrophosphatase at 2.7 A resolution. Protein Eng. 1994;7:823–830. doi: 10.1093/protein/7.7.823. [DOI] [PubMed] [Google Scholar]
- 23.Kankare J, Salminen T, Lahti R, Cooperman B S, Baykov A A, Goldman A. Crystallographic identification of metal-binding sites in Escherichia coli inorganic pyrophosphatase. Biochemistry. 1996;35:4670–4677. doi: 10.1021/bi952637e. [DOI] [PubMed] [Google Scholar]
- 24.Kapyla J, Hyytia T, Lahti R, Goldman A, Baykov A A, Cooperman B S. Effect of D97E substitution on the kinetic and thermodynamic properties of Escherichia coli inorganic pyrophosphatase. Biochemistry. 1995;34:792–800. doi: 10.1021/bi00003a012. [DOI] [PubMed] [Google Scholar]
- 25.King C H, Fields B S, Shotts E B, Jr, White E H. Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect Immun. 1991;59:758–763. doi: 10.1128/iai.59.3.758-763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahti R. Microbial inorganic pyrophosphatases. Microbiol Rev. 1983;47:169–179. doi: 10.1128/mr.47.2.169-178.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahti R, Pitkaranta T, Valve E, Ilta I, Kukko-Kalske E, Heinonen J. Cloning and characterization of the gene encoding inorganic pyrophosphatase of Escherichia coli K-12. J Bacteriol. 1988;170:5901–5907. doi: 10.1128/jb.170.12.5901-5907.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lahti R, Pohjanoksa K, Pitkaranta T, Heikinheimo P, Salminen T, Meyer P, Heinonen J. A site-directed mutagenesis study on Escherichia coli inorganic pyrophosphatase. Glutamic acid-98 and lysine-104 are important for structural integrity, whereas aspartic acids-97 and -102 are essential for catalytic activity. Biochemistry. 1990;29:5761–5766. doi: 10.1021/bi00476a017. [DOI] [PubMed] [Google Scholar]
- 29.Lahti R, Salminen T, Latonen S, Heikinheimo P, Pohjanoska K, Heinonen J. Genetic engineering of Escherichia coli inorganic pyrophosphatase: Tyr55 and Tyr141 are important for the structural integrity. Eur J Biochem. 1991;198:293–297. doi: 10.1111/j.1432-1033.1991.tb16015.x. [DOI] [PubMed] [Google Scholar]
- 30.Lecomte P, Doupleday O P, Radman M. Evidence for an intermediate in DNA synthesis involving pyrophosphate exchange. A possible role in fidelity. J Mol Biol. 1986;189:643–652. doi: 10.1016/0022-2836(86)90494-8. [DOI] [PubMed] [Google Scholar]
- 31.Lee B Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Invest. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundin M, Baltscheffsky H, Ronne H. Yeast PPA2 gene encodes a mitochondrial inorganic pyrophosphatase that is essential for mitochondrial function. J Biol Chem. 1991;266:12168–12172. [PubMed] [Google Scholar]
- 33.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 35.Mody C H, Paine III R, Shahrabadi M S, Simon R H, Pearlman E, Eisenstein B I, Toews G B. Legionella pneumophila replicates within rat alveolar epithelial cells. J Infect Dis. 1993;167:1138–1145. doi: 10.1093/infdis/167.5.1138. [DOI] [PubMed] [Google Scholar]
- 36.Payne N R, Horwitz M A. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J Exp Med. 1987;166:1377–1389. doi: 10.1084/jem.166.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rockey D D, Rosquist J L. Protein antigens of Chlamydia psittaci present in infected cells but not detected in the infectious elementary body. Infect Immun. 1994;62:106–112. doi: 10.1128/iai.62.1.106-112.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salminen T, Kapyla J, Heikinheimo P, Kankare J, Goldman A, Heinonen J, Baykov A A, Cooperman B S, Lahti R. Structure and function analysis of Escherichia coli inorganic pyrophosphatase: is a hydroxide ion the key to catalysis? Biochemistry. 1995;34:782–791. doi: 10.1021/bi00003a011. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorger K P. Heat shock factor and heat shock response. Cell. 1991;65:363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- 42.Susa M, Hacker J, Marre R. De novo synthesis of Legionella pneumophila antigens during intracellular growth in phagocytic cells. Infect Immun. 1996;64:1679–1684. doi: 10.1128/iai.64.5.1679-1684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson M S, Isberg R R. Formation of the Legionella pneumophila replicative phagosome. Infect Agents Dis. 1995;2:269–271. [PubMed] [Google Scholar]
- 44.Taraska T, Ward D M, Ajioka R S, Wyrick P B, Davis-Kaplan S R, Davis C H, Kaplan J. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect Immun. 1996;64:3713–3727. doi: 10.1128/iai.64.9.3713-3727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velichko I, Volk S E, Dudarenkov V Y, Magretova N N, Chernyak V Y, Goldman A, Cooperman B S, Lahti R, Baykov A A. Cold lability of the mutant forms of Escherichia coli inorganic pyrophosphatase. FEBS Lett. 1995;359:20–22. doi: 10.1016/0014-5793(95)00003-r. [DOI] [PubMed] [Google Scholar]
- 46.Venkataraman C, Haack B J, Bondada S, Abu Kwaik Y. Identification of a Gal/GalNAc lectin in the protozoan Hartmanella vermiformis as a potential receptor for attachment and invasion by the Legionnaires’ disease bacterium, Legionella pneumophila. J Exp Med. 1997;186:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong S C, Hall D C, Josse J. Constitutive inorganic pyrophosphatase of Escherichia coli. III. Molecular weight and physical properties of the enzyme and its subunits. J Biol Chem. 1970;245:4335–4345. [PubMed] [Google Scholar]