Abstract
Purpose of Review
Type 1 interferons (IFN-I) are of increasing interest across a wide range of autoimmune rheumatic diseases. Historically, research into their role in rheumatoid arthritis (RA) has been relatively neglected, but recent work continues to highlight a potential contribution to RA pathophysiology.
Recent Findings
We emphasise the importance of disease stage when examining IFN-I in RA and provide an overview on how IFN-I may have a direct role on a variety of relevant cellular functions. We explore how clinical trajectory may be influenced by increased IFN-I signalling, and also, the limitations of scores composed of interferon response genes. Relevant environmental triggers and inheritable RA genetic risk relating to IFN-I signalling are explored with emphasis on intriguing data potentially linking IFN-I exposure, epigenetic changes, and disease relevant processes.
Summary
Whilst these data cumulatively illustrate a likely role for IFN-I in RA, they also highlight the knowledge gaps, particularly in populations at risk for RA, and suggest directions for future research to both better understand IFN-I biology and inform targeted therapeutic strategies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11926-023-01125-6.
Keywords: Rheumatoid arthritis, Early rheumatoid arthritis, Type 1 interferons, Interferon gene signature, Biomarkers
Introduction
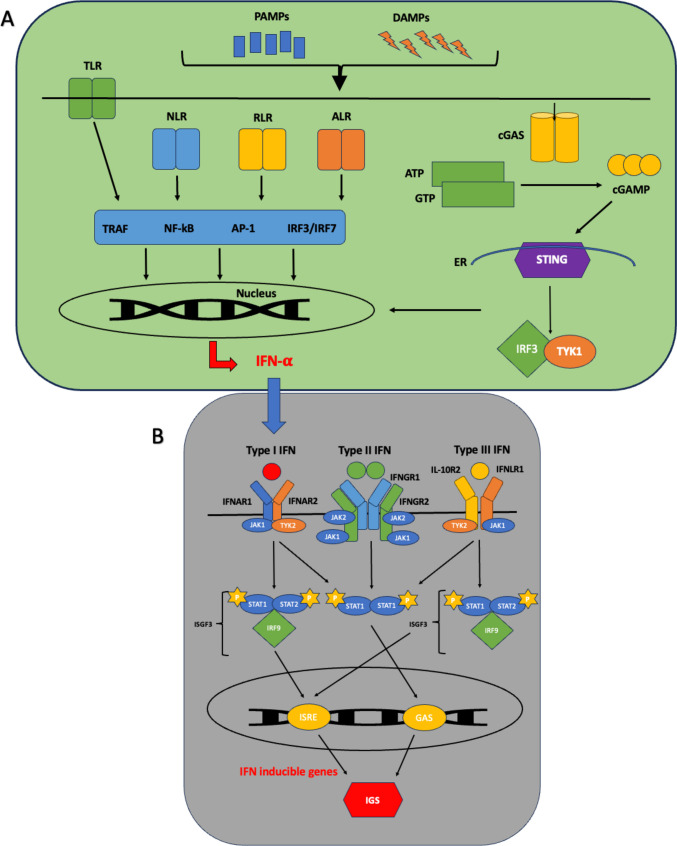
Interferons (IFN) are a widely expressed family of cytokines. They are categorised, based on their receptor signalling, into types I, II, and III [1]. IFN-I signal via a heterodimeric receptor composed of two distinct multi-chain structures, IFN-α receptor 1 and 2 (IFNAR-1 and IFNAR-2). The former is constitutively associated with tyrosine kinase 2 (TYK2) and the latter associated with Janus Kinase 1 (JAK1) [2]. IFN-I are produced as part of the innate immune response to infection and possess potent antiviral effects [2]. Triggers of IFN-I production and subsequent downstream signalling have been recently reviewed in [3] and is summarised in Fig. 1. Similarly, IFN-II and IFN-III signal via their own unique heterodimeric receptors composed of IFN-γ receptors 1 and 2 (IFNGR-1 and IFNGR-2), IFNLR1 (IFN lambda receptor-1), and IL-10R2 (interleukin-10 receptor 2) subunits, respectively [4••]. Both of which subsequently lead to downstream signalling and potential induction of interferon inducible genes. In this review, we explore what role IFN-I, particularly IFN-α, may play in rheumatoid arthritis (RA) pathophysiology.
Fig. 1.
Schematic of interferon (IFN) triggers and downstream signalling pathways. A The production of IFN-I can occur following recognition of pathogen-associated molecular patterns (PAMPs), often associated with foreign bacteria or viruses, such as cytosolic DNA and double stranded RNA. These are detected by pattern recognition receptors (PRRs) which comprise of a large repertoire of germline-encoded receptors. These PRRs can be divided into subclasses including cell surface toll-like receptors (TLRs), cytosolic nod-like receptors (NLRs), retinoic acid inducible gene I receptors (RLRs), AIM2 like receptors (ALRs), and cGAS-STING pathway. Recognition of damage-associated molecular patterns (DAMPs) or PAMPS by PRRs results in transcription factor activation, such as TRAF (tumour necrosis factor receptor-associated factor), NF-kB nuclear factor kappa B, activating protein-1 (AP-1), and interferon regulatory factors (IRFs), STING (stimulator of interferon genes), and TBK1 (tank binding kinase 1), all involved in the transcription of IFN-I genes. B IFNs are categorised based on their receptor signalling, into IFN-I, IFN-II, and IFN-III. IFN-I signal via a heterodimeric receptor composed of two distinct multi-chain structures, IFN-α receptor 1 and 2 (IFNAR-1 and IFNAR-2) subunits. IFNAR associates with Janus Kinases (JAKs), with the former constitutively associated with JAK1 and the latter associated with tyrosine kinase 2 (TYK2). In response to ligand binding, these JAKs undergo activation and phosphorylate two latent transcription factors, signal transducers, and activators of transcription 1 and 2 (STAT1 and STAT2), resulting in their activation and subsequent heterodimer formation. This binds with IRF9 (IFN regulatory factor 9) or p48 to form a multi-component transcription complex called interferon-stimulated gene factor 3 (ISGF3). This complex translocates to the nucleus and binds to specific sites called IFN-stimulated response elements (ISREs), leading to the transcriptional induction of several IRGs ultimately responsible for IFN-I’s antiviral and immunomodulatory properties. The phosphorylated STAT proteins can alternatively form STAT1-STAT1 homodimers which bind gamma-activated sequences (GASs) to induce pro-inflammatory genes. As IFN-II can also signal via this alternative route (via their own heterodimeric receptor, composed of IFNGR1 and IFNGR2 subunits and associated with JAK1 and JAK 2 signalling), there can be a crossover between IFN-I and IFN-II signalling. Finally, IFN-III signals via its own heterodimeric receptor composed of IL-10R2 and IFNLR1 subunits, associated with the activation of TYK2 and JAK1, respectively. This can result in the formation and activation of STAT1-STAT2 heterodimers which associate with IRF9 to form ISGF3 complexes, with subsequent signalling as per IFN-I. AP-1, activating protein-1; DNA, deoxyribonucleic acid; ER, endoplasmic reticulum; NF-kB, nuclear factor kappa-light-chain-enhancer of activated B cells; NLR, nod-like receptor; P, phosphate; RLR, rig-I-like receptor; RNA, ribonucleic acid; TRAF, tumour necrosis factor receptor-associated factor.
The Interferon Gene Signature (IGS)
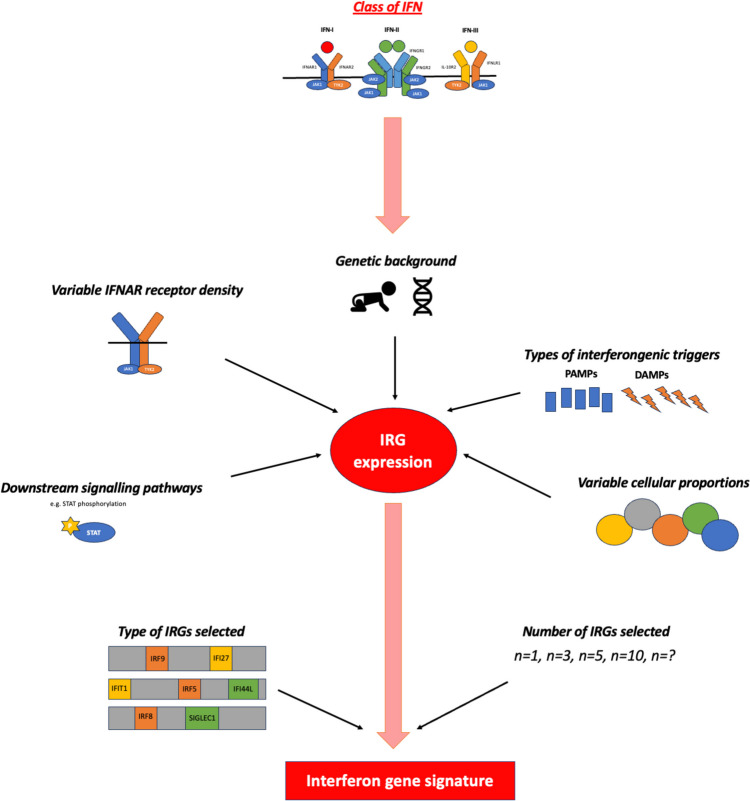
Measuring IFN-α protein in vivo has been historically challenging due to low circulating levels being frequently below the detection thresholds of standard assays. A solution was to infer IFN-I exposure, and hence levels, by measuring transcripts that reflect interferon stimulated or response genes (IRGs), and their cumulative expression was termed the interferon gene signature (IGS) (see Fig. 2). However, there are over 2000 IRGs and which IRGs are chosen to generate an IGS is lacking consensus across studies [5••]. Despite this, an IGS is widely reported in autoimmune rheumatic diseases, and there are mutual IRGs increased in RA and other rheumatic diseases [6]. Nevertheless, some propose an exclusive and highly diverse IRG transcriptional profile in RA peripheral whole blood [7] as well as in synovial biopsy samples [8], distinct from that found in SLE. However, IRG expression, and subsequently the calculated IGS, may vary between different cell types, suggesting that differences seen amongst related autoimmune diseases could be secondary to different immune cell proportions and signalling pathway activation [9]. Indeed, variation is seen in flow cytometry detected STAT class phosphorylation in CD4+ T cells, CD8+ T cells, B cells, and monocytes following IFN-I stimulation [10].
Fig. 2.
Figure highlighting factors that may influence interferon response gene (IRG) expression, as well as additional aspects that can influence the subsequent calculation of the interferon gene signature (IGS). Primarily, class of IFN will dictate IRG expression and thus the resulting IGS calculated, however, additional contributory factors, for example genetic background or IFNAR expression, are highlighted. DAMP, damage associated molecular patterns; IFNAR, IFN alpha receptor; IFNGR, IFN gamma receptor; IFNLR, IFN lambda receptor; IGS, interferon gene signature; IRG, interferon response gene; PAMP, pattern associated molecular pattern; STAT, signal transducer and activators of transcription
As IFN-I, IFN-II, or even IFN-III can induce IRGs (see Fig. 1), there has been a historical lack of clarity as to which IFN class was responsible for the IGS in RA. Indeed, it remains a controversial topic as IRG expression may be modulated by additional stimuli, such as TNF-α, with variable effects reported in monocytes vs T cells [11]. Nevertheless, in established RA, there was reportedly equal contribution of IFN-α and IFN-β to the whole blood IGS vs IFN-α exposure being dominant in SLE [9]. However, in a cohort of nearly 200 early drug naïve RA patients, circulating IFN-α protein and not IFN-β, IFN-II, or IFN-III nor any other circulating inflammatory cytokine uniquely correlated with the whole blood IGS [12••]. This work remains to be validated, and reported differences may reflect disease stages, but does implicate predominantly IFN-α with the IGS in early RA.
Despite these caveats regarding its calculation, the IGS remains a useful tool in dissecting the role of IFN-I in RA, as explored below.
The IGS by Disease Phase
It is increasingly appreciated that disease processes in early RA are likely to be distinct from established RA. In early RA, a raised IGS (MxA, OAS1, ISG15, IFI44L, IFI6) was more prevalent compared with established RA, approximately 50% vs 20% of patients, respectively [13], and fell with the initiation of therapy [12••, 13]. Therapeutics may contribute to a reduced incidence in established RA as glucocorticoids, as well as disease modifying anti-rheumatoid drugs (DMARDs), can modify the IGS [14]. Notably, this increase in early RA persists even after accounting for potential confounders such as disease stage dependant variation in cell subset proportions [15].
Corroborating the raised IGS noted at disease onset, there is emerging data that IFNs may contribute to the transition from preclinical to sustained clinical disease. In ontology studies and network pathway analyses, the IGS distinguished DMARD-naïve early arthritis patients that developed a persistent inflammatory arthritis from those that had a self-limiting course [16]. In ACPA+ arthralgia populations, i.e. those who are at risk for developing RA, an IGS increases the chance of progression to synovitis, and its inclusion in outcome models improved its predictive capacity [17, 18]. Even in healthy asymptomatic CCP+ individuals, there was evidence of increased IFN-α signalling which mirrored what was seen in early RA cohorts, and this, with other parameters, was able to differentiate progressors with a median of 4.1 years before symptom onset, from controls [19, 20•]. In seropositive and seronegative RA, as well as in high-risk seropositive arthralgia patients, there was an overlap in circulating cytokine profiles with IFN-α, as well as IL-5, and TNF-α upregulated up to 50% in seropositive arthralgia and seropositive RA patients but not in seronegative RA [21] with an odds ratio (OR) of 21 for RA development in seropositive arthralgia patients [18, 21].
Clinical Characteristics and the IGS
There has been conflicting evidence around the impact of an IGS/IFN-I signalling on autoantibody production in RA. In established RA, there is a significant correlation between the IGS and ACPA titres and anti-carbamylated protein (anti-CarP) antibodies as well as with genes linked to B cell differentiation and antibody production [22]. Conversely, others found no relation between the IGS and the presence and/or titres of ACPA and RF in established disease [23]. Similarly, a 2016 systematic analysis, involving patients with established RA, found that there was no difference seen in the IGS between ACPA negative and ACPA positive patients [24]. Conversely, rheumatoid factor (RF) demonstrated a positive association between either the IGS or circulating IFN-α levels in both established and early RA as well as across several autoimmune rheumatic diseases [12••, 13, 25]. These differences may reflect disease stage but may also reflect variability in the IRGs chosen to represent the IGS, with some using a combination of 19 IRGs [24] and others using only 6 (IFI27, IFI44L, IFIT1, ISG15, RSAD2 and SIGLEC1) [25] for example.
Multiple observational studies in established RA have found no association between the IGS and disease activity [13, 24]. This contrasts with early drug naïve RA where in a number of prospective observational studies, a higher IGS in early drug naïve RA, were associated with increased baseline disease activity as well as a poorer response to initial therapies [12, 13, 15] which was validated in additional cohorts for specific IGSs [26].
RA, including early disease, is a risk factor for cardiovascular disease (CVD). [27, 28]. In mouse and human in vitro models, IFN-α stimulation of macrophages resulted in significant metabolic rewiring with over 500 metabolic genes, including those related to key processes in the pathophysiology of CVD such as glycolysis, oxidative phosphorylation, fatty acid synthesis, and lipid metabolism [29]. In addition, endothelial progenitor cells (EPCs), involved in vasculogenesis and repair, have impaired function in RA, with IFN-α implicated in both in vitro and in vivo studies [30–32]. In murine lupus models, prolonged and enhanced IFN-I exposure significantly reduced EPC numbers, with acute exposure affecting only EPC differentiation but not the cellular number [30]. Finally, IFN-α may also influence CVD by promoting insulin resistance given, as early as the 1980s, IFN-α was shown to impair glucose tolerance and insulin sensitivity [33] with reversal of this effect in IFNAR-/- mouse models [34].
IFN-I and Its Effects on Cellular Function
B and T Cells
IFN-I can widely influence B cell activity which may contribute to RA pathophysiology, for example, by supporting B cell survival via increased monocyte B-lymphocyte stimulator (BLyS) production, by direct stimulation of B cells, and indirectly through T cell and Dendritic Cells (DCs) stimulation [35]. Prolonged B cell survival can lead to increased differentiation into memory and plasma cells, immunoglobulin isotype switching, and autoantibody formation [36, 37]. Furthermore, IFN-α modifies the plasma cell transcriptome towards a proinflammatory phenotype [38]. IFN-I regulates BCR signalling, specifically via IFN-αR, which in turn may promote pathways involved in antibody formation and germinal centre development in murine models [39]. IFN-I can also influence the differentiation of CD4+ T cells towards a Th1 response [40], fostering B cell activation and subsequent activity [41]. IFN-I also promotes CD8+ T cell survival and CD8+ cytotoxic T cell activity as well as prolonging the proliferation and expansion of CD8+ antigen specific T cells via inhibition of apoptosis [42].
Dendritic Cells
Dendritic cells (DCs) upregulate HLA-DR, CD40, CD80, and CD86 expression upon IFN-I exposure [43]. DC maturation and enhanced antigen presentation, in the context of increased co-stimulatory molecules, can result in the induction of autoimmunity in predisposed individuals via self-antigen presentation to low affinity autoreactive T cells [44]. In SLE susceptible mice, IFN-I-treated DCs showed relative apoptosis resistance, this activated DC longevity potentially contributing to the development of autoimmunity [45]. Conversely, in early drug naive RA, there was no association between the IGS and circulating CD1c or pDC frequency, but there was an inverse association with CD141+ DC frequency [46]. This highlights the DC subset dependant complexity of IFN-I signalling in vivo.
Monocytes
Classical and non-classical monocytes have been implicated in RA pathogenesis [47]. How the IGS affects monocyte function in vivo in RA remains to be fully examined but, when exposed to IFN-Is in vitro, monocytes upregulate TLR7 and IRF expression, resulting in increased responsiveness to subsequent immunostimulatory ligands [48]. IFN-I exposure also increases expression of CD40, CD80, and CD86 and HLA-DR, ultimately promoting differentiation into a monocyte-derived dendritic cell, or mo-DC, with high capacity for antigen presentation [43, 49]. Mo-DCs are also known to be increased in the RA synovial compartment and promote Th17 differentiation [50]. However, as with DCs, what happens in vivo may be subset dependant as highlighted by enhanced responsiveness to IFN-α in murine proinflammatory monocytes secondary to increased IFNAR expression when compared with anti-inflammatory monocytes [51].
Neutrophils
Neutrophils are one of the first cell types to enter the RA joint and may play an important role in the development and progression of RA [52]. They are a major contributor to the whole blood IGS in RA, attributed to their uniquely upregulated IFNAR expression, a phenomenon not seen in either healthy controls or RA PBMCs [53•]. Indeed, next generation sequencing of isolated blood neutrophils has found significantly upregulated IRGs in RA neutrophils compared to healthy controls [54]. How this increased sensitivity to IFN-I influences neutrophil function is being explored, but, intriguingly, the pathogenic phenotype proposed for RA consists of delayed neutrophil apoptosis, increased ROS production and chemokine expression which, in part, can be recapitulated by IFN-I exposure in vitro [55•].
Fibroblasts
Synovial fibroblasts are resident cells in the stroma of joints [56], and we recently demonstrated comparable IFN-α levels in serum and early RA synovial fluid [12••]. In RA, these fibroblasts have an activated phenotype, characterised by resistance to apoptosis, and increased proliferation and production of inflammatory mediators that promote immune cell differentiation and survival [57]. Histology and RNA sequencing of early RA synovial tissue demonstrated three distinct pathotypes: fibroblastic pauci-immune, macrophage-rich diffuse myeloid, and a lympho-myeloid pathotype [58••]. In the lympho-myeloid pathotype, seven out of the eight differentially expressed blood transcripts in synovial versus whole blood were IFN-I responses genes (IFI27, ISG15, IFI44L, OASL, USP18, RSAD2, LY6E) [58•]. In addition, a pathogenic subset of sub-lining fibroblasts (THY1+HLA−DRhigh) have increased IRG expression [59]. However, this may not directly be secondary to IFN-I as TNF-α induced signalling co-opts the mTOR pathway to shift fibroblast like synoviocytes towards an IFN response [60] which has been shown to be via secondary autocrine production of IFNβ and subsequent activation of the IRF1-IFNβ-IFNAR-JAK-STAT1 axis [61]. Nevertheless, the role of IFN-α on fibroblast function in RA remains an important research question.
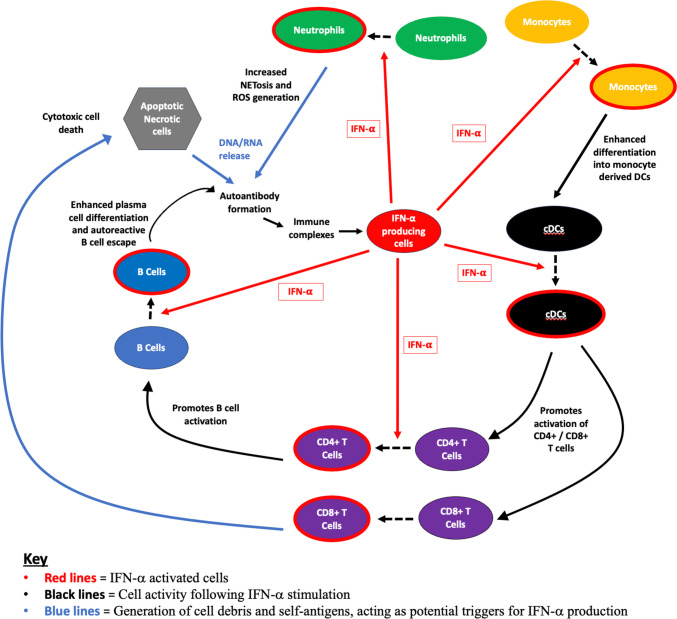
Cumulatively, these effects are likely to contribute to a highly activated and potentially autoimmune prone phenotype as summarised in Fig. 3.
Fig. 3.
Schematic depicting interaction of cellular subsets in the presence of IFN-I. IFN-α influences the activity of surrounding innate and adaptive immune cells. It remains unknown what initially triggers the cascade of IFN production; however, it has been suggested that the generation of DNA/RNA via cell death pathways including apoptosis, necrosis, and NETosis (with subsequent ROS generation) plays a role. Exposure to these self-antigens increases the risk of developing autoantibodies, which form immune complexes that have potential to interact with IFN-producing cells to enhance further IFN-I production. Monocytes develop an inflammatory phenotype and activated cDCs promote activation of CD4+ and CD8+ T cell subsets. These T cells themselves upon exposure to IFN-I can further enhance B cell activation and mediation of cell death, respectively. cDCs, conventional dendritic cells; IFN-𝛂, interferon-𝛂; NET, neutrophil extracellular traps; ROS, reactive oxygen species
Source of IFN-I in RA
pDCs, particularly in their immature state, are the main IFN-I producing cell, however, whether they are the primary source of IFN-α in RA remains unclear. In SLE, there is an element of so-called pDC fatigue, where the ability of the pDC to produce IFN-I reduces and other cells take over production [62]. In early RA, circulating pDCs were not the primary source of IFNA transcript, with comparable expression in circulating lymphocytes. However, circulating pDC numbers were reduced with increased CCR7 expression inferring increased migration to the synovial compartment and target tissue [46]. Indeed, in established RA patients, the synovial compartment has increased numbers of pDCs with reduced numbers seen in peripheral blood. However, those that remained in the circulation were immature with inferred increased IFN-I producing capacity [63]. Nevertheless, RA synovial pDCs are potent producers of IFN-α [64] and, in mice, intraarticular transfer of IFN-I producing dendritic cells was sufficient to propagate a persistent inflammatory arthritis [65].
Conversely, after arthritogenic serum transfer in K/BxN serum-induced arthritis, collagen-induced arthritis, and human TNF transgene insertion, only pDC deficient mice showed exacerbations of symptoms and signs of inflammatory arthritis [66] and topical imiquimod, a TLR7 agonist, increased pDC recruitment and activity which subsequently improved arthritis [66]. Furthermore, transcriptomic analysis of circulating pDCs in early RA suggested enhanced tolerogenic function [46]. These discrepancies may arise due to the complexities of DC development [67] and cellular differences across species. Given their relative paucity in vivo, pDCs have been relatively neglected in RA research; however, better understanding of their complexity, particularly in relation to any location specific function, will help inform their role in RA and role in IFN-I production.
Potential Triggers of IFN Production
What drives the observed increased IGS/IFN-α in RA remains unclear; however, triggers may include viral infections or microbial DNA or antigen fragments, with these elements repeatedly reported in the joints of RA patients [68–70]. Retroelements are non-protein encoding portions of DNA derived from ancient transposable elements, such as retroviruses, that have been historically incorporated into the genome. Their activity can trigger intracellular viral sensors and thus promote local IFN-I production [71]. In SLE and primary Sjogren’s syndrome, increased retrotransposon activity in disease relevant tissue associated with increased local IFN-α production [72•], and, in established RA synovium, there is also increased retroelement expression [73, 74]. Furthermore, in a subgroup of RA patients, a transcriptional profile was documented, reminiscent of a viral infection, which associated with both IFN-I signalling as well as increased ACPA titres [75, 76]. How these retroelements may influence IFN-I production in RA remains to be seen.
Cell-free nucleic acids have been extensively implicated in IFN-I generation in SLE [77] and monogenic interferonopathies [78]. Mouse models with DNA clearance defects develop autoantibody-mediated chronic polyarthritis, resembling human RA [79]. This corroborates RA human observational data, where evidence of raised levels of circulating cell -free DNA have been found in both peripheral blood [80, 81] and synovial fluid [82]. Although direct links with IFN-I were not made in these human RA studies, a similar mechanism to that described in SLE may be present.
Neutrophils, found in high numbers in RA synovium, can undergo NETosis, a unique form of cell death which has been proposed as a potential trigger for IFN-I production [83]. DNA from these NETs form complexes with antimicrobial peptides including LL37, secretory leukocyte protease inhibitor (SLPI), or with immunoglobulins to form immune complexes which facilitate pDC TLR7/9 signalling ultimately culminating in IFN-α production [84–86]. In RA, links between NETs and ACPA have been reported [87, 88] and known pathogenic cytokines in RA, such as TNF-α and IL-17A, as well as IFN-a itself [89], can also induce NETosis, potentially creating a vicious cycle of inflammation and disease activity [88].
Other potential triggers include lifestyle and environmental factors. An inverse correlation between physical activity and IFN-I signalling has been reported [90]. In addition, physical activity was associated with downregulation of TLR and IL-17R signalling and reduced inflammatory cytokines production, including IFN-I [90].
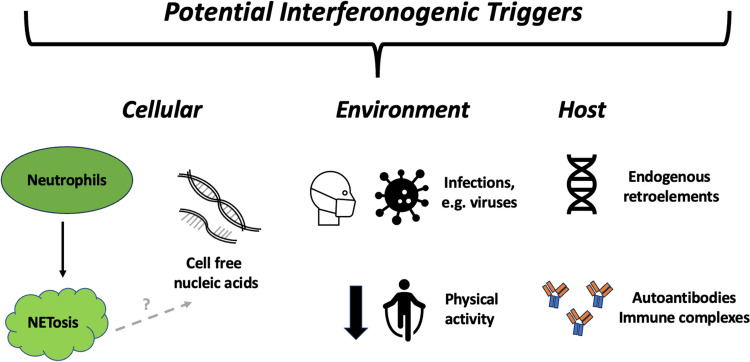
Potential triggers are summarised in Fig. 4; however, much of the above involves extrapolation from other diseases, such as SLE, and caveats exist including differences in IRG expression and genetic risk between these diseases [91]. Further work is needed to explore these pathways in RA specifically.
Fig. 4.
Figure depicting some potential triggers of IFN-α production. Here, potential triggers are split into three subtypes: (1) cellular comprising of neutrophils, (2) environmental including infections increasing IFN-α production via cellular death and debris and a reduction in physical activity reportedly linked to increased IFN-α levels, and (3) proposed non-cellular host triggers including endogenous retroelement activity and the development of autoantibodies or immune complexes resulting in increased IFN-α production
Heritable Genetic Risk and IFN-I Signalling
As genome-wide association studies (GWAS), and relevant data sets, become more available, numerous single nucleotide polymorphisms (SNPs) have been identified as contributing to the genetic risk of RA. Interestingly, a number of these SNPs are in genes related to the IFN-I response pathway including DNA-sensing proteins, toll-like receptors, and JAK-STAT protein mediators. These are summarised in Table 1. However, the functional consequences of these polymorphisms in RA with regards to IFN-I production or signalling are yet to be elucidated. Nevertheless, an overlap of certain at-risk genes associated with increased IFN-I signalling in SLE has also been linked to RA, for example SNPs in IRF5, STAT4, and PTPN22 [92]. Some of these RA risk SNPs, such as IRF5 polymorphisms, associate with more severe or erosive disease [93, 94], which may corroborate the clinical refractory disease phenotype observed in IGS high early RA patients [12••]. Further work is needed to elucidate both the role of IFN-I on susceptible genetic backgrounds as well as the contribution of these SNPs to IFN-I production.
Table 1.
Known single nucleotide polymorphisms (SNPs) associated with RA genetic risk and how their function may affect IFN-I biology
| Gene with known RA risk variant | Role in IFN biology | Reference* |
|---|---|---|
| TNFAIP3 (A20) | • NF-κB and other A20-regulated signalling molecules can induce IFN-I | 1 |
| PADI4 | • PADI4 knockouts resulted in reduced IFN-I responses | 2 |
| STAT4 | • STAT4 promotes RIG-I signalling independent of its classical activation pathway and promotes IFN-β production in myeloid innate cells | 3 |
| CD40 | • CD40 can enhance STING-mediated IFN-I responses | 4 |
| UBE2L3 | • UBE2L3 shown to negatively regulate IFN-I expression | 5 |
| IFNAR1/IFNGR2 | • Encodes signalling receptors for IFN-I and IFN-II | 6 |
| ETS1 | • ETS-1 suggested in SLE patient studies to be associated with IFN-I and to negatively regulate ISG3 and ISRE binding sites | 7,8 |
| PVT1 |
• PVT1 negative feedback mediator for IFN-I signalling via STAT1 interaction and subsequent reduction of its phosphorylation. • IFN-α stimulation shown to upregulate PVT1 RNA expression |
9,10 |
| CDK6 | • CKD6 regulates IFN-I signalling negative feedback loops | 11 |
| ETV7 | • ETV7 negatively regulates IFN-I signalling | 12 |
| EOMES | • EOMES expression is driven by IFN-I signalling in CD8+ T cells which leads to regulation of memory-like CD8+ T cell homeostasis and function | 13 |
| TYK2 | • TYK2 required for IFN-I induced activation of transcription factors STAT1-4 and downstream signalling | 14 |
| IRF8 | • Regulates IFN-I production | 15 |
| Runx1 | • RUNX1 upregulates IFNs and IRGs via IFN-I signalling | 16 |
| RCAN1 | • RCAN1 protein stability negatively affected by IFN-α treatment via STAT2 activation | 17 |
| GATA3 |
• GATA3 overexpression promotes IFN-I expression • IFN- α/β treatment suppresses GATA3 expression |
18,19 |
| DDX6 |
• DDX6 regulates RIG-I mediated IFN-I signalling • DDX6 depletion leads to increased IRG expression |
20,21 |
| PRDM1 |
• Deletion results in impaired IFN-I production • Control IKKα and IRF7 activation via direct suppression of Irak3, a negative regulator of TLR signalling |
23 |
| IRF5 |
• IRF5 shown to a positive regulator of IFN-I signalling • Risk haplotype of IRF5 associated with SLE and with increased IFN-I production |
24 |
*Separate reference list in supplementary file 1.
IFN-I and Epigenetics
Epigenetic changes are modifications that regulate genome activity, independent of DNA sequence. This occurs via molecular factors and processes, such as DNA methylation of CPG sites or chromatin conformational changes, which subsequently modulate transcription. They are frequently triggered by environmental factors or exposure to inflammatory stimuli, such as cytokines. Methylation changes are noted early in RA progression and vary by cell subset [95••]. Furthermore, differential methylation has been implicated in initial response to methotrexate in early drug naïve RA patients as well as to certain biologics in established disease [95••, 96–98], and these processes are emerging as important modifiers of RA clinical progression and phenotype [99].
Analysis of B and CD4 T cells from early drug naïve RA patients demonstrated differentially methylated CPG sites at disease relevant genes, such as PARP9, STAT1, and EPSTI between IGS high and low patients. It also implicated altered transcription factor binding, cumulatively promoting increased lymphocyte activation, and a proliferative phenotype in the IGS high cohort [12••]. These data suggest that these changes may be IFN-α induced, and negatively influence clinical trajectory. In undifferentiated arthritis (UA) monocytes, methylation changes, which associated with disease progression and a poor prognosis, were partially recapitulated by monocyte exposure to IFN-α [100•]. Furthermore, IFN-α treatment causes methylation changes in monocytes similar to those seen in established RA, which in vivo were themselves associated with increased disease activity [101]. Intriguingly, in models of type 1 diabetes, where IFN-I plays a key part in disease initiation, exposure to IFN-α triggered increased TET2 expression. This prompted hypomethylation changes in genes controlling inflammatory and immune pathways, ultimately resulting in their increased expression and disease acceleration [102]. TET proteins are key players in demethylation and are also increased in early drug naïve RA circulating lymphocytes [103], however whether this is secondary to IFN-α is unknown.
It is important to acknowledge that CPG sites in IRGs themselves are frequently hypomethylated in autoimmune conditions, including RA [104, 105]. In twin studies of CD4 T cells, hypomethylation of IRGs IFIT1, IRF7, MX1, OAS1, USP18, RSAD2, and IFI44L has even been proposed as biomarkers of progression to RA [104]. This questions whether the IGS could be an artefact of altered gene expression secondary to hypomethylation caused by other circulating inflammatory cytokines, such as IL6, or due to increased IFN-α signalling itself. IFN-α protein is increased in early RA and uniquely correlates with the IGS [12••], so the reality is likely to involve both mechanisms.
Although less extensively investigated, IFN-I-associated chromatin conformational changes may also be relevant to RA pathophysiology. There is variation in chromatin accessibility in RA synovial fibroblasts which is likely influenced by the synovial environment [106], where IFN-α is known to be present [12••]. In early RA, chromatin conformation changes in IFNAR2 were associated with poorer outcomes [107]. Furthermore, monocytes stimulated with IFN-α have increased trimethylated histone H3 Lys 4 (H3K4me3) which enhances transcription at promotors of genes that encode inflammatory mediators. In a more representative in vivo environment, incubation of monocytes with both IFN-α and TNF-α was associated with increased H3K4me3 that reduced monocyte tolerization to LPS and promoted an enhanced response to subsequent environmental challenges [108]. This intriguingly implicates IFN-I, and chromatin-mediated modifications, with the induction of inflammatory genes beyond canonical IRGs. Indeed, instances where prior exposure to IFN-α can influence cellular response to additional stimuli are increasingly being reported [102, 109–111] and remain a potential mechanism whereby IFN-I can influence disease development in RA.
The IGS/IFN as a Therapeutic Target
Anifrolumab targets IFNAR1 and therefore blocks IFN-α and IFN-β signalling [112]. In a pilot trial, seven established RA patients, all with a high IGS (IFI27, IFI44, IFI44L and RSAD2), and active diseases were randomised to anifrolumab or placebo [113•]. The primary endpoint of an American College of Rheumatology (ACR) response of ≥ 20% after 24 weeks was achieved in patients receiving anifrolumab although only one patient in each arm completed the study despite a safety profile similar to that reported in SLE [114]. Reasons for early discontinuation in the treatment group included lack of efficacy, hypersensitivity reaction, and infection whilst the control group participants stopped due to insufficient therapeutic response [113•]. Larger trials are needed to assess the efficacy of this drug in RA.
Alternatively, JAK inhibitors (JAKi) suppress phosphorylation of STAT and thus affect downstream IFN signalling and reduce IRG expression [92]. Indeed, in vitro JAKi reduce IFN-I driven plasmablast differentiation [115], synovial BAFF expression, monocyte-derived DCs costimulatory molecule CD80/CD86 expression, and T cell differentiation into Th1 and Th17 cells [115, 116] [61]. The efficacy of JAKi in the treatment of established RA has been widely reported [117], although how the IGS impacts on its effect has not been comprehensively examined. However, analysis of baricitinib SLE trial data demonstrated that clinical effect was independent of IGS reduction [118].
Other inhibitors of downstream IFN-I signalling include a novel small molecule selective for JAK3/JAK1/TBK1 (tank-1 binding kinase), which, in mouse models, suppressed IFN-I production and osteoclast formation via TBK1 inhibition [119]. Autoantibody dependent collagen-induced arthritis mice models confirmed the clinical benefit of TBK1 inhibition [120, 121] and TBK1 deficient mice have reduced IRG and protein expression [122]. These findings are yet to be reproduced in human studies, but given the interest in cancer regarding TBK1 inhibition [123], this may provide a novel therapeutic approach.
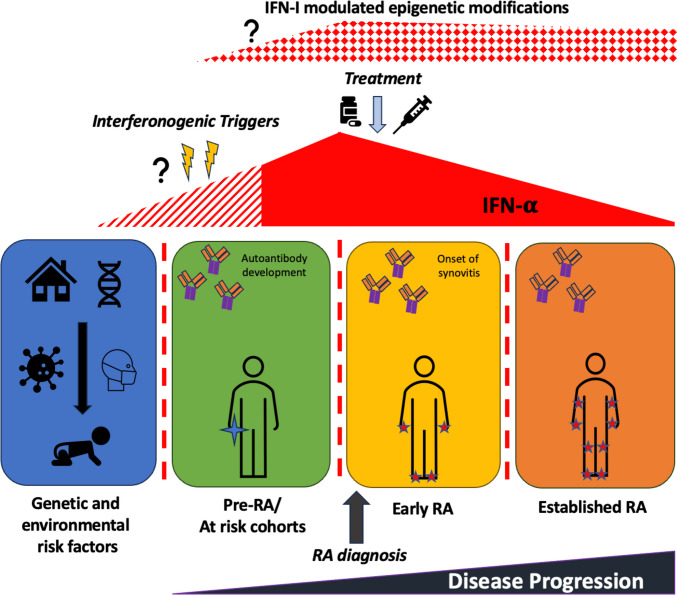
Conclusions
There is growing evidence that IFN-α plays an important role in early RA pathophysiology and Fig. 5 summarises a working paradigm on IFN-α influencing RA progression. However, the triggers of IFN-α production and its timing in relation to early immune dysregulation or symptom onset remain unclear. Further work focusing on early disease or at-risk populations with a focus on genetic and epigenetic factors is likely to be informative. Despite mechanistic uncertainties, there is clear rationale to further test IFN-α targeting therapies in early RA, potentially using the IGS as a theragnostic biomarker, or to use the IGS as a biomarker for more intensive initial therapy. The heterogeneity and variety of IGSs remain challenging with regard to clinical utility, but recent progress in the international community on IGS stratification and uniform application of standardised measures of IFN-I signalling is encouraging [4••, 5••, 124, 125], and its use in this capacity may be on the horizon.
Fig. 5.
Schematic depicting associations between RA disease progression and IFN-𝛂 levels over time. There is increasing evidence that IFN-I is increased at RA disease onset and in at-risk cohorts. Proposed triggers include environmental influences including infection on the background of genetic risk; however, when these events may occur in relation to disease onset or initial immune dysfunction, with regards to autoantibody generation, is unclear. There is emerging evidence that this IFN-α exposure in early RA populations may cause potentially pathogenic epigenetic changes in key cellular subsets which could persist into established disease. IFN-𝛂, interferon-𝛂; RA, rheumatoid arthritis
Supplementary Information
Funding
This work was supported by the Research into Inflammatory Arthritis Centre Versus Arthritis (RACE) (grant number 22072) and the National Institute for Health and Care Research (NIHR) Newcastle Biomedical Research Centre for Ageing and Long-Term Conditions; views expressed are the authors’ and not necessarily those of the National Health Service, the National Institute of Health and Care Research, or the Department of Health.
Compliance with Ethical Standards
Conflict of Interest
JDI discloses research grants from Pfizer, Janssen, and GSK; conference support from Eli Lilly and Gilead; speaker/consulting fees from AbbVie, BMS, Gilead, Roche, and UCB. FAHC discloses speaker fees from AstraZeneca. The remaining authors have no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Armstrong MAL. A Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Front Immunol. 2016;7:609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 3.Mesev EV, LeDesma RA, Ploss A. Decoding type I and III interferon signalling during viral infection. Nat Microbiol. 2019;4(6):914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.•• Cooles FA, Isaacs JD. The interferon gene signature as a clinically relevant biomarker in autoimmune rheumatic disease. Lancet Rheumatol. 2021;4(1) 10.1016/S2665-9913(21)00254-X. A recent review exploring how a raised IGS can function as a clinically relevant biomarker in rheumatic diseases. [DOI] [PubMed]
- 5.•• Rodriguez-Carrio J, Burska A, Conaghan PG, Dik WA, Biesen R, Eloranta ML, et al. EULAR points to consider for the measurement, reporting and application of IFN-I pathway activation assays in clinical research and practice. Ann Rheum Dis. 2023;82(6):754–62. 10.1136/ard-2022-223628. A thorough review of using IFN-I assays in clinical research and their clinical utility. [DOI] [PubMed]
- 6.Reynier F, Petit F, Paye M, Turrel-Davin F, Imbert PE, Hot A, et al. Importance of correlation between gene expression levels: application to the type I interferon signature in rheumatoid arthritis. PLoS One. 2011;6(10):e24828. doi: 10.1371/journal.pone.0024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smiljanovic B, Grun JR, Biesen R, Schulte-Wrede U, Baumgrass R, Stuhlmuller B, et al. The multifaceted balance of TNF-alpha and type I/II interferon responses in SLE and RA: how monocytes manage the impact of cytokines. J Mol Med (Berl). 2012;90(11):1295–1309. doi: 10.1007/s00109-012-0907-y. [DOI] [PubMed] [Google Scholar]
- 8.Nzeusseu Toukap A, Galant C, Theate I, Maudoux AL, Lories RJ, Houssiau FA, et al. Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum. 2007;56(5):1579–1588. doi: 10.1002/art.22578. [DOI] [PubMed] [Google Scholar]
- 9.de Jong TD, Vosslamber S, Mantel E, de Ridder S, Wesseling JG, van der Pouw Kraan TC, et al. Physiological evidence for diversification of IFNalpha- and IFNbeta-mediated response programs in different autoimmune diseases. Arthritis Res Ther. 2016;18:49. doi: 10.1186/s13075-016-0946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Boxel-Dezaire AH, Zula JA, Xu Y, Ransohoff RM, Jacobberger JW, Stark GR. Major differences in the responses of primary human leukocyte subsets to IFN-beta. J Immunol. 2010;185(10):5888–5899. doi: 10.4049/jimmunol.0902314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henig N, Avidan N, Mandel I, Staun-Ram E, Ginzburg E, Paperna T, et al. Interferon-beta induces distinct gene expression response patterns in human monocytes versus T cells. PLoS One. 2013;8(4):e62366. doi: 10.1371/journal.pone.0062366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.•• FAH C, Tarn J, Lendrem DW, Naamane N, Lin CM, Millar B, et al. Interferon-alpha-mediated therapeutic resistance in early rheumatoid arthritis implicates epigenetic reprogramming. Ann Rheum Dis. 2022;81(9):1214–23. 10.1136/annrheumdis-2022-222370. This study used a multicentre inception cohort of early RA patients to demonstrate the IGS as a robust prognostic biomarker for early RA, specifically with regards to the IGS as a reflection of circulating IFN-α protein and influencer of epigenome. [DOI] [PMC free article] [PubMed]
- 13.Cooles FAH, Anderson AE, Lendrem DW, Norris J, Pratt AG, Hilkens CMU, et al. The interferon gene signature is increased in patients with early treatment-naive rheumatoid arthritis and predicts a poorer response to initial therapy. J Allergy Clin Immunol. 2018;141(1):445–8 e4. doi: 10.1016/j.jaci.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong TD, Sellam J, Agca R, Vosslamber S, Witte BI, Tsang ASM, et al. A multi-parameter response prediction model for rituximab in rheumatoid arthritis. Joint Bone Spine. 2018;85(2):219–226. doi: 10.1016/j.jbspin.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Carrio J, Alperi-Lopez M, Lopez P, Ballina-Garcia FJ, Suarez A. Heterogeneity of the type I interferon signature in rheumatoid arthritis: a potential limitation for its use as a clinical biomarker. Front Immunol. 2017;8:2007. doi: 10.3389/fimmu.2017.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seyhan AA, Gregory B, Cribbs AP, Bhalara S, Li Y, Loreth C, et al. Novel biomarkers of a peripheral blood interferon signature associated with drug-naive early arthritis patients distinguish persistent from self-limiting disease course. Sci Rep. 2020;10(1):8830. doi: 10.1038/s41598-020-63757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubbers J, Brink M, van de Stadt LA, Vosslamber S, Wesseling JG, van Schaardenburg D, et al. The type I IFN signature as a biomarker of preclinical rheumatoid arthritis. Ann Rheum Dis. 2013;72(5):776–780. doi: 10.1136/annrheumdis-2012-202753. [DOI] [PubMed] [Google Scholar]
- 18.van Baarsen LG, Bos WH, Rustenburg F, van der Pouw Kraan TC, Wolbink GJ, Dijkmans BA, et al. Gene expression profiling in autoantibody-positive patients with arthralgia predicts development of arthritis. Arthritis Rheum. 2010;62(3):694–704. doi: 10.1002/art.27294. [DOI] [PubMed] [Google Scholar]
- 19.Macias-Segura N, Castaneda-Delgado JE, Bastian Y, Santiago-Algarra D, Castillo-Ortiz JD, Aleman-Navarro AL, et al. Transcriptional signature associated with early rheumatoid arthritis and healthy individuals at high risk to develop the disease. PLoS One. 2018;13(3):e0194205. doi: 10.1371/journal.pone.0194205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.• Brink M, Lundquist A, Alexeyenko A, Lejon K, Rantapaa-Dahlqvist S. Protein profiling and network enrichment analysis in individuals before and after the onset of rheumatoid arthritis. Arthritis Res Ther. 2019;21(1):288. 10.1186/s13075-019-2066-9. This study confirmed the importance of IFN-α signalling in early RA using network enrichment analysis. [DOI] [PMC free article] [PubMed]
- 21.Chalan P, Bijzet J, van den Berg A, Kluiver J, Kroesen BJ, Boots AM, et al. Analysis of serum immune markers in seropositive and seronegative rheumatoid arthritis and in high-risk seropositive arthralgia patients. Sci Rep. 2016;6:26021. doi: 10.1038/srep26021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castaneda-Delgado JE, Bastian-Hernandez Y, Macias-Segura N, Santiago-Algarra D, Castillo-Ortiz JD, Aleman-Navarro AL, et al. Type I interferon gene response is increased in early and established rheumatoid arthritis and correlates with autoantibody production. Front Immunol. 2017;8:285. doi: 10.3389/fimmu.2017.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantaert T, van Baarsen LG, Wijbrandts CA, Thurlings RM, van de Sande MG, Bos C, et al. Type I interferons have no major influence on humoral autoimmunity in rheumatoid arthritis. Rheumatology (Oxford). 2010;49(1):156–166. doi: 10.1093/rheumatology/kep345. [DOI] [PubMed] [Google Scholar]
- 24.de Jong TD, Blits M, de Ridder S, Vosslamber S, Wolbink G, Nurmohamed MT, et al. Type I interferon response gene expression in established rheumatoid arthritis is not associated with clinical parameters. Arthritis Res Ther. 2016;18(1):290. doi: 10.1186/s13075-016-1191-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds JA, Briggs TA, Rice GI, Darmalinggam S, Bondet V, Bruce E, et al. Type I interferon in patients with systemic autoimmune rheumatic disease is associated with haematological abnormalities and specific autoantibody profiles. Arthritis Res Ther. 2019;21(1):147. doi: 10.1186/s13075-019-1929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plant D, Maciejewski M, Smith S, Nair N, Maximising therapeutic utility in rheumatoid arthritis consortium tRSG. Hyrich K, et al. Profiling of gene expression biomarkers as a classifier of methotrexate nonresponse in patients with rheumatoid arthritis. Arthritis Rheumatol. 2019;71(5):678–684. doi: 10.1002/art.40810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visseren FLJ, Mach F, Smulders YM, Carballo D, Koskinas KC, Back M, et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice. Eur Heart J. 2021;42(34):3227–3337. doi: 10.1093/eurheartj/ehab484. [DOI] [PubMed] [Google Scholar]
- 28.Hansildaar R, Vedder D, Baniaamam M, Tausche AK, Gerritsen M, Nurmohamed MT. Cardiovascular risk in inflammatory arthritis: rheumatoid arthritis and gout. Lancet Rheumatol. 2021;3(1):e58–e70. doi: 10.1016/S2665-9913(20)30221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed D, Jaworski A, Roy D, Willmore W, Golshani A, Cassol E. Transcriptional profiling suggests extensive metabolic rewiring of human and mouse macrophages during early interferon alpha responses. Mediators Inflamm. 2018;2018:5906819. doi: 10.1155/2018/5906819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thacker SG, Zhao W, Smith CK, Luo W, Wang H, Vivekanandan-Giri A, et al. Type I interferons modulate vascular function, repair, thrombosis, and plaque progression in murine models of lupus and atherosclerosis. Arthritis Rheum. 2012;64(9):2975–2985. doi: 10.1002/art.34504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Carrio J, de Paz B, Lopez P, Prado C, Alperi-Lopez M, Ballina-Garcia FJ, et al. IFNalpha serum levels are associated with endothelial progenitor cells imbalance and disease features in rheumatoid arthritis patients. PLoS One. 2014;9(1):e86069. doi: 10.1371/journal.pone.0086069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Carrio J, Prado C, de Paz B, Lopez P, Gomez J, Alperi-Lopez M, et al. Circulating endothelial cells and their progenitors in systemic lupus erythematosus and early rheumatoid arthritis patients. Rheumatology (Oxford). 2012;51(10):1775–1784. doi: 10.1093/rheumatology/kes152. [DOI] [PubMed] [Google Scholar]
- 33.Koivisto VA, Pelkonen R, Cantell K. Effect of interferon on glucose tolerance and insulin sensitivity. Diabetes. 1989;38(5):641–647. doi: 10.2337/diab.38.5.641. [DOI] [PubMed] [Google Scholar]
- 34.McCabe KM, Hsieh J, Thomas DG, Molusky MM, Tascau L, Feranil JB, et al. Antisense oligonucleotide treatment produces a type I interferon response that protects against diet-induced obesity. Mol Metab. 2020;34:146–156. doi: 10.1016/j.molmet.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seyler TM, Park YW, Takemura S, Bram RJ, Kurtin PJ, Goronzy JJ, et al. BLyS and APRIL in rheumatoid arthritis. J Clin Invest. 2005;115(11):3083–3092. doi: 10.1172/JCI25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson SW, Kolhatkar NS, Rawlings DJ. B cells take the front seat: dysregulated B cell signals orchestrate loss of tolerance and autoantibody production. Curr Opin Immunol. 2015;33:70–77. doi: 10.1016/j.coi.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3(9):822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Care MA, Stephenson SJ, Barnes NA, Fan I, Zougman A, El-Sherbiny YM, et al. Network analysis identifies proinflammatory plasma cell polarization for secretion of ISG15 in human autoimmunity. J Immunol. 2016;197(4):1447–1459. doi: 10.4049/jimmunol.1600624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domeier PP, Chodisetti SB, Schell SL, Kawasawa YI, Fasnacht MJ, Soni C, et al. B-cell-intrinsic type 1 interferon signaling is crucial for loss of tolerance and the development of autoreactive B cells. Cell Rep. 2018;24(2):406–418. doi: 10.1016/j.celrep.2018.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jarry A, Malard F, Bou-Hanna C, Meurette G, Mohty M, Mosnier JF, et al. Interferon-alpha promotes Th1 response and epithelial apoptosis via inflammasome activation in human intestinal mucosa. Cell Mol Gastroenterol Hepatol. 2017;3(1):72–81. doi: 10.1016/j.jcmgh.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenbarth SC, Baumjohann D, Craft J, Fazilleau N, Ma CS, Tangye SG, et al. CD4(+) T cells that help B cells - a proposal for uniform nomenclature. Trends Immunol. 2021;42(8):658–669. doi: 10.1016/j.it.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202(5):637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13(5):543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12(2):125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen M, Wang YH, Wang Y, Huang L, Sandoval H, Liu YJ, et al. Dendritic cell apoptosis in the maintenance of immune tolerance. Science. 2006;311(5764):1160–1164. doi: 10.1126/science.1122545. [DOI] [PubMed] [Google Scholar]
- 46.Cooles FAH, Anderson AE, Skelton A, Pratt AG, Kurowska-Stolarska MS, McInnes I, et al. Phenotypic and transcriptomic analysis of peripheral blood plasmacytoid and conventional dendritic cells in early drug naive rheumatoid arthritis. Front Immunol. 2018;9:755. doi: 10.3389/fimmu.2018.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wampler Muskardin TL, Fan W, Jin Z, Jensen MA, Dorschner JM, Ghodke-Puranik Y, et al. Distinct single cell gene expression in peripheral blood monocytes correlates with tumor necrosis factor inhibitor treatment response groups defined by type I interferon in rheumatoid arthritis. Front Immunol. 2020;11:1384. doi: 10.3389/fimmu.2020.01384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obermoser G, Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19(9):1012–1019. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korthals M, Safaian N, Kronenwett R, Maihofer D, Schott M, Papewalis C, et al. Monocyte derived dendritic cells generated by IFN-alpha acquire mature dendritic and natural killer cell properties as shown by gene expression analysis. J Transl Med. 2007;5:46. doi: 10.1186/1479-5876-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coutant F. Shaping of monocyte-derived dendritic cell development and function by environmental factors in rheumatoid arthritis. Int J Mol Sci. 2021;22(24). 10.3390/ijms222413670. [DOI] [PMC free article] [PubMed]
- 51.Han S, Zhuang H, Lee PY, Li M, Yang L, Nigrovic PA, et al. Differential responsiveness of monocyte and macrophage subsets to interferon. Arthritis Rheumatol. 2020;72(1):100–113. doi: 10.1002/art.41072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edilova MI, Akram A, Abdul-Sater AA. Innate immunity drives pathogenesis of rheumatoid arthritis. Biomed J. 2021;44(2):172–182. doi: 10.1016/j.bj.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.• de Jong TD, Lubbers J, Turk S, Vosslamber S, Mantel E, Bontkes HJ, et al. The type I interferon signature in leukocyte subsets from peripheral blood of patients with early arthritis: a major contribution by granulocytes. Arthritis Res Ther. 2016;18:165. 10.1186/s13075-016-1065-3. This study highlighted that polymorphonuclear neutrophils are the main contributors to the whole blood type I IFN signature in patients with early RA. [DOI] [PMC free article] [PubMed]
- 54.Wright HL, Thomas HB, Moots RJ, Edwards SW. Interferon gene expression signature in rheumatoid arthritis neutrophils correlates with a good response to TNFi therapy. Rheumatology (Oxford). 2015;54(1):188–193. doi: 10.1093/rheumatology/keu299. [DOI] [PubMed] [Google Scholar]
- 55.• Glennon-Alty L, Moots RJ, Edwards SW, Wright HL. Type I interferon regulates cytokine-delayed neutrophil apoptosis, reactive oxygen species production and chemokine expression. Clin Exp Immunol. 2021;203(2):151–9. 10.1111/cei.13525. This study demonstrated how type I IFNs can alter neutrophil responses in healthy controls. [DOI] [PMC free article] [PubMed]
- 56.Nemeth T, Nagy G, Pap T. Synovial fibroblasts as potential drug targets in rheumatoid arthritis, where do we stand and where shall we go? Ann Rheum Dis. 2022;81(8):1055–1064. doi: 10.1136/annrheumdis-2021-222021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mousavi MJ, Karami J, Aslani S, Tahmasebi MN, Vaziri AS, Jamshidi A, et al. Transformation of fibroblast-like synoviocytes in rheumatoid arthritis; from a friend to foe. Auto Immun Highlights. 2021;12(1):3. doi: 10.1186/s13317-020-00145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.•• Lewis MJ, Barnes MR, Blighe K, Goldmann K, Rana S, Hackney JA, et al. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 2019;28(9):2455–70 e5. 10.1016/j.celrep.2019.07.091. This study dissected gene signatures in RA, which identified transcriptional subgroups in the synovium, suggestive of divergent pathogenic pathways. [DOI] [PMC free article] [PubMed]
- 59.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20(7):928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karonitsch T, Kandasamy RK, Kartnig F, Herdy B, Dalwigk K, Niederreiter B, et al. mTOR senses environmental cues to shape the fibroblast-like synoviocyte response to inflammation. Cell Rep. 2018;23(7):2157–2167. doi: 10.1016/j.celrep.2018.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonelli M, Dalwigk K, Platzer A, Olmos Calvo I, Hayer S, Niederreiter B, et al. IRF1 is critical for the TNF-driven interferon response in rheumatoid fibroblast-like synoviocytes : JAKinibs suppress the interferon response in RA-FLSs. Exp Mol Med. 2019;51(7):1–11. doi: 10.1038/s12276-019-0267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwok SK, Lee JY, Park SH, Cho ML, Min SY, Park SH, et al. Dysfunctional interferon-alpha production by peripheral plasmacytoid dendritic cells upon Toll-like receptor-9 stimulation in patients with systemic lupus erythematosus. Arthritis Res Ther. 2008;10(2):R29. doi: 10.1186/ar2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jongbloed SL, Lebre MC, Fraser AR, Gracie JA, Sturrock RD, Tak PP, et al. Enumeration and phenotypical analysis of distinct dendritic cell subsets in psoriatic arthritis and rheumatoid arthritis. Arthritis Res Ther. 2006;8(1):R15. doi: 10.1186/ar1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebre MC, Jongbloed SL, Tas SW, Smeets TJ, McInnes IB, Tak PP. Rheumatoid arthritis synovium contains two subsets of CD83-DC-LAMP- dendritic cells with distinct cytokine profiles. Am J Pathol. 2008;172(4):940–950. doi: 10.2353/ajpath.2008.070703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narendra SC, Chalise JP, Hook N, Magnusson M. Dendritic cells activated by double-stranded RNA induce arthritis via autocrine type I IFN signaling. J Leukoc Biol. 2014;95(4):661–666. doi: 10.1189/jlb.0613320. [DOI] [PubMed] [Google Scholar]
- 66.Nehmar R, Alsaleh G, Voisin B, Flacher V, Mariotte A, Saferding V, et al. Therapeutic modulation of plasmacytoid dendritic cells in experimental arthritis. Arthritis Rheumatol. 2017;69(11):2124–2135. doi: 10.1002/art.40225. [DOI] [PubMed] [Google Scholar]
- 67.Yamada S, Nagafuchi Y, Wang M, Ota M, Hatano H, Takeshima Y, et al. Immunomics analysis of rheumatoid arthritis identified precursor dendritic cells as a key cell subset of treatment resistance. Ann Rheum Dis. 2023;82(6):809–819. doi: 10.1136/ard-2022-223645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Y, Chen B, Li S, Yang L, Zhu D, Wang Y, et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci Rep. 2018;8(1):14305. doi: 10.1038/s41598-018-32675-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Heijden IM, Wilbrink B, Tchetverikov I, Schrijver IA, Schouls LM, Hazenberg MP, et al. Presence of bacterial DNA and bacterial peptidoglycans in joints of patients with rheumatoid arthritis and other arthritides. Arthritis Rheum. 2000;43(3):593–598. doi: 10.1002/1529-0131(200003)43:3<593::AID-ANR16>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 70.Chen T, Rimpilainen M, Luukkainen R, Mottonen T, Yli-Jama T, Jalava J, et al. Bacterial components in the synovial tissue of patients with advanced rheumatoid arthritis or osteoarthritis: analysis with gas chromatography-mass spectrometry and pan-bacterial polymerase chain reaction. Arthritis Rheum. 2003;49(3):328–334. doi: 10.1002/art.11119. [DOI] [PubMed] [Google Scholar]
- 71.Kassiotis G. The immunological conundrum of endogenous retroelements. Annu Rev Immunol. 2023;41:99–125. doi: 10.1146/annurev-immunol-101721-033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.• Mavragani CP, Nezos A, Sagalovskiy I, Seshan S, Kirou KA, Crow MK. Defective regulation of L1 endogenous retroelements in primary Sjogren's syndrome and systemic lupus erythematosus: role of methylating enzymes. J Autoimmun. 2018;88:75–82. 10.1016/j.jaut.2017.10.004. This study demonstrated the role of altered methylation mechanisms, including retroelement expression, in the pathogenesis of systemic autoimmune disorders. [DOI] [PMC free article] [PubMed]
- 73.Neidhart M, Rethage J, Kuchen S, Kunzler P, Crowl RM, Billingham ME, et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 2000;43(12):2634–2647. doi: 10.1002/1529-0131(200012)43:12<2634::AID-ANR3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 74.Ali M, Veale DJ, Reece RJ, Quinn M, Henshaw K, Zanders ED, et al. Overexpression of transcripts containing LINE-1 in the synovia of patients with rheumatoid arthritis. Ann Rheum Dis. 2003;62(7):663–666. doi: 10.1136/ard.62.7.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Pouw Kraan TC, van Baarsen LG, Wijbrandts CA, Voskuyl AE, Rustenburg F, Baggen JM, et al. Expression of a pathogen-response program in peripheral blood cells defines a subgroup of rheumatoid arthritis patients. Genes Immun. 2008;9(1):16–22. doi: 10.1038/sj.gene.6364438. [DOI] [PubMed] [Google Scholar]
- 76.Mavragani CP, Sagalovskiy I, Guo Q, Nezos A, Kapsogeorgou EK, Lu P, et al. Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease. Arthritis Rheumatol. 2016;68(11):2686–2696. doi: 10.1002/art.39795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crow MK. Pathogenesis of systemic lupus erythematosus: risks, mechanisms and therapeutic targets. Ann Rheum Dis. 2023;82(8):999–1014. doi: 10.1136/ard-2022-223741. [DOI] [PubMed] [Google Scholar]
- 78.Muzes G, Bohusne Barta B, Szabo O, Horgas V, Sipos F. Cell-free dna in the pathogenesis and therapy of non-infectious inflammations and tumors. Biomedicines. 2022;10(11). 10.3390/biomedicines10112853. [DOI] [PMC free article] [PubMed]
- 79.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443(7114):998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 80.Rykova E, Sizikov A, Roggenbuck D, Antonenko O, Bryzgalov L, Morozkin E, et al. Circulating DNA in rheumatoid arthritis: pathological changes and association with clinically used serological markers. Arthritis Res Ther. 2017;19(1):85. doi: 10.1186/s13075-017-1295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hashimoto T, Yoshida K, Hashimoto N, Nakai A, Kaneshiro K, Suzuki K, et al. Circulating cell free DNA: a marker to predict the therapeutic response for biological DMARDs in rheumatoid arthritis. Int J Rheum Dis. 2017;20(6):722–730. doi: 10.1111/1756-185X.12959. [DOI] [PubMed] [Google Scholar]
- 82.Dong C, Liu Y, Sun C, Liang H, Dai L, Shen J, et al. Identification of specific joint-inflammatogenic cell-free dna molecules from synovial fluids of patients with rheumatoid arthritis. Front Immunol. 2020;11:662. doi: 10.3389/fimmu.2020.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song W, Ye J, Pan N, Tan C, Herrmann M. Neutrophil extracellular traps tied to rheumatoid arthritis: points to ponder. Front Immunol. 2020;11:578129. doi: 10.3389/fimmu.2020.578129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol. 2013;190(3):1217–1226. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guiducci C, Tripodo C, Gong M, Sangaletti S, Colombo MP, Coffman RL, et al. Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. J Exp Med. 2010;207(13):2931–2942. doi: 10.1084/jem.20101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu S, Peng W, Liang X, Wang W. Anti-citrullinated protein antibodies are associated with neutrophil extracellular trap formation in rheumatoid arthritis. J Clin Lab Anal. 2021;35(3):e23662. doi: 10.1002/jcla.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5(178):178ra40. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng Y, Wu X, Zhang S, Deng C, Zhao L, Wang M, et al. The potential roles of type I interferon activated neutrophils and neutrophil extracellular traps (NETs) in the pathogenesis of primary Sjogren's syndrome. Arthritis Res Ther. 2022;24(1):170. doi: 10.1186/s13075-022-02860-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patterson SL, Sun S, Rychkov D, Katz P, Tsitsiklis A, Nakamura MC, et al. Physical activity associates with lower systemic inflammatory gene expression in rheumatoid arthritis. J Rheumatol. 2022;49(12):1320–1327. doi: 10.3899/jrheum.220050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3(73):73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muskardin TLW, Niewold TB. Type I interferon in rheumatic diseases. Nat Rev Rheumatol. 2018;14(4):214–228. doi: 10.1038/nrrheum.2018.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dawidowicz K, Allanore Y, Guedj M, Pierlot C, Bombardieri S, Balsa A, et al. The interferon regulatory factor 5 gene confers susceptibility to rheumatoid arthritis and influences its erosive phenotype. Ann Rheum Dis. 2011;70(1):117–121. doi: 10.1136/ard.2010.129171. [DOI] [PubMed] [Google Scholar]
- 94.van der Helm-van Mil AH, Huizinga TW. Advances in the genetics of rheumatoid arthritis point to subclassification into distinct disease subsets. Arthritis Res Ther. 2008;10(2):205. doi: 10.1186/ar2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.•• Adams C, Nair N, Plant D, Verstappen SMM, Quach HL, Quach DL, et al. Identification of cell-specific differential DNA methylation associated with methotrexate treatment response in rheumatoid arthritis. Arthritis Rheumatol. 2023. 10.1002/art.42464. This study highlighted cell-specific changes in DNA methylation that were associated with MTX treatment response in RA patients. [DOI] [PMC free article] [PubMed]
- 96.Plant D, Webster A, Nair N, Oliver J, Smith SL, Eyre S, et al. Differential methylation as a biomarker of response to etanercept in patients with rheumatoid arthritis. Arthritis Rheumatol. 2016;68(6):1353–1360. doi: 10.1002/art.39590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nair N, Plant D, Verstappen SM, Isaacs JD, Morgan AW, Hyrich KL, et al. Differential DNA methylation correlates with response to methotrexate in rheumatoid arthritis. Rheumatology (Oxford). 2020;59(6):1364–1371. doi: 10.1093/rheumatology/kez411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gosselt HR, van Zelst BD, de Rotte M, Hazes JMW, de Jonge R, Heil SG. Higher baseline global leukocyte DNA methylation is associated with MTX non-response in early RA patients. Arthritis Res Ther. 2019;21(1):157. doi: 10.1186/s13075-019-1936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang C, Li D, Teng D, Zhou Y, Zhang L, Zhong Z, et al. Epigenetic regulation in the pathogenesis of rheumatoid arthritis. Front Immunol. 2022;13:859400. doi: 10.3389/fimmu.2022.859400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.• de la Calle-Fabregat C, Rodriguez-Ubreva J, Ciudad L, Ramirez J, Celis R, Azuaga AB, et al. The synovial and blood monocyte DNA methylomes mirror prognosis, evolution, and treatment in early arthritis. JCI Insight. 2022;7(9) 10.1172/jci.insight.158783. This study identified differences in DNA methylation profiles between undifferentiated arthritis and healthy controls, highlighting its potential use as a prognostic biomarker as well as marker of disease activity and treatment efficacy in early inflammatory arthritis. [DOI] [PMC free article] [PubMed]
- 101.Rodriguez-Ubreva J, de la Calle-Fabregat C, Li T, Ciudad L, Ballestar ML, Catala-Moll F, et al. Inflammatory cytokines shape a changing DNA methylome in monocytes mirroring disease activity in rheumatoid arthritis. Ann Rheum Dis. 2019;78(11):1505–1516. doi: 10.1136/annrheumdis-2019-215355. [DOI] [PubMed] [Google Scholar]
- 102.Stefan-Lifshitz M, Karakose E, Cui L, Ettela A, Yi Z, Zhang W, et al. Epigenetic modulation of beta cells by interferon-alpha via PNPT1/mir-26a/TET2 triggers autoimmune diabetes. JCI Insight. 2019;4(5). 10.1172/jci.insight.126663. [DOI] [PMC free article] [PubMed]
- 103.de Andres MC, Perez-Pampin E, Calaza M, Santaclara FJ, Ortea I, Gomez-Reino JJ, et al. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis Res Ther. 2015;17(1):233. doi: 10.1186/s13075-015-0748-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Svendsen AJ, Gervin K, Lyle R, Christiansen L, Kyvik K, Junker P, et al. Differentially methylated dna regions in monozygotic twin pairs discordant for rheumatoid arthritis: an epigenome-wide study. Front Immunol. 2016;7:510. doi: 10.3389/fimmu.2016.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen S, Pu W, Guo S, Jin L, He D, Wang J. Genome-wide DNA methylation profiles reveal common epigenetic patterns of interferon-related genes in multiple autoimmune diseases. Front Genet. 2019;10:223. doi: 10.3389/fgene.2019.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weinand K, Sakaue S, Nathan A, Jonsson AH, Zhang F, Watts GFM, et al. The chromatin landscape of pathogenic transcriptional cell states in rheumatoid arthritis. bioRxiv. 2023. 10.1101/2023.04.07.536026. [DOI] [PMC free article] [PubMed]
- 107.Carini C, Hunter E, Scottish Early Rheumatoid Arthritis Inception cohort I. Ramadass AS, Green J, Akoulitchev A, et al. Chromosome conformation signatures define predictive markers of inadequate response to methotrexate in early rheumatoid arthritis. J Transl Med. 2018;16(1):18. doi: 10.1186/s12967-018-1387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park SH, Kang K, Giannopoulou E, Qiao Y, Kang K, Kim G, et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat Immunol. 2017;18(10):1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Akita K, Yasaka K, Shirai T, Ishii T, Harigae H, Fujii H. Interferon alpha enhances B cell activation associated with FOXM1 induction: potential novel therapeutic strategy for targeting the plasmablasts of systemic lupus erythematosus. Front Immunol. 2020;11:498703. doi: 10.3389/fimmu.2020.498703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zahalka S, Starkl P, Watzenboeck ML, Farhat A, Radhouani M, Deckert F, et al. Trained immunity of alveolar macrophages requires metabolic rewiring and type 1 interferon signaling. Mucosal Immunol. 2022;15(5):896–907. doi: 10.1038/s41385-022-00528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiu J, Xu B, Ye D, Ren D, Wang S, Benci JL, et al. Cancer cells resistant to immune checkpoint blockade acquire interferon-associated epigenetic memory to sustain T cell dysfunction. Nat Cancer. 2023;4(1):43–61. doi: 10.1038/s43018-022-00490-y. [DOI] [PubMed] [Google Scholar]
- 112.Casey KA, Guo X, Smith MA, Wang S, Sinibaldi D, Sanjuan MA, et al. Type I interferon receptor blockade with anifrolumab corrects innate and adaptive immune perturbations of SLE. Lupus Sci Med. 2018;5(1):e000286. doi: 10.1136/lupus-2018-000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.• Karonitsch T, Yeghiazaryan L, Lackner A, Brezinsek HP, Stamm TA, Konig F, et al. Targeting type I interferon (IFN) signalling in patients with RA with a high type I IFN gene signature. RMD Open. 2022;8(2). 10.1136/rmdopen-2022-002525. This was a randomised, double-blind, placebo-controlled, multicentre pilot trial that recruited patients with active RA and a high type I IGS to receive either the type I IFN receptor blocking antibody anifrolumab or placebo. [DOI] [PMC free article] [PubMed]
- 114.Morand EF, Furie R, Tanaka Y, Bruce IN, Askanase AD, Richez C, et al. Trial of anifrolumab in active systemic lupus erythematosus. N Engl J Med. 2020;382(3):211–221. doi: 10.1056/NEJMoa1912196. [DOI] [PubMed] [Google Scholar]
- 115.Kubo S, Nakayamada S, Sakata K, Kitanaga Y, Ma X, Lee S, et al. Janus kinase inhibitor baricitinib modulates human innate and adaptive immune system. Front Immunol. 2018;9:1510. doi: 10.3389/fimmu.2018.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kubo S, Yamaoka K, Kondo M, Yamagata K, Zhao J, Iwata S, et al. The JAK inhibitor, tofacitinib, reduces the T cell stimulatory capacity of human monocyte-derived dendritic cells. Ann Rheum Dis. 2014;73(12):2192–2198. doi: 10.1136/annrheumdis-2013-203756. [DOI] [PubMed] [Google Scholar]
- 117.Angelini J, Talotta R, Roncato R, Fornasier G, Barbiero G, Dal Cin L, et al. JAK-inhibitors for the treatment of rheumatoid arthritis: a focus on the present and an outlook on the future. Biomolecules. 2020;10(7). 10.3390/biom10071002. [DOI] [PMC free article] [PubMed]
- 118.Dörner T, Tanaka Y, Petri M, Smolen JS, Dow ER, Higgs RE, Benschop RJ, Abel A, Silk ME, de Bono S, Hoffman RW. 185 Baricitinib-associated changes in type I interferon gene signature during a 24-week phase 2 clinical SLE trial. Lupus. Science & Medicine. 2019:6. 10.1136/lupus-2019-lsm.185.
- 119.Shan S, Zhou Y, Yu J, Yang Q, Pan D, Wang Y, et al. Therapeutic treatment of a novel selective JAK3/JAK1/TBK1 inhibitor, CS12192, in rat and mouse models of rheumatoid arthritis. Int Immunopharmacol. 2019;77:105914. doi: 10.1016/j.intimp.2019.105914. [DOI] [PubMed] [Google Scholar]
- 120.Louis C, Ngo D, D'Silva DB, Hansen J, Phillipson L, Jousset H, et al. Therapeutic effects of a TANK-binding kinase 1 inhibitor in germinal center-driven collagen-induced arthritis. Arthritis Rheumatol. 2019;71(1):50–62. doi: 10.1002/art.40670. [DOI] [PubMed] [Google Scholar]
- 121.Fang Z, Hu Y, Dai J, He L, He J, Xu B, et al. CS12192, a novel JAK3/JAK1/TBK1 inhibitor, synergistically enhances the anti-inflammation effect of methotrexate in a rat model of rheumatoid arthritis. Int J Mol Sci. 2022;23(21). 10.3390/ijms232113394. [DOI] [PMC free article] [PubMed]
- 122.Hammaker D, Boyle DL, Firestein GS. Synoviocyte innate immune responses: TANK-binding kinase-1 as a potential therapeutic target in rheumatoid arthritis. Rheumatology (Oxford). 2012;51(4):610–618. doi: 10.1093/rheumatology/ker154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun Y, Revach OY, Anderson S, Kessler EA, Wolfe CH, Jenney A, et al. Targeting TBK1 to overcome resistance to cancer immunotherapy. Nature. 2023;615(7950):158–167. doi: 10.1038/s41586-023-05704-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carter LM, Alase A, Wigston Z, Psarras A, Burska A, Sutton E, et al. Gene expression and autoantibody analysis revealing distinct ancestry-specific profiles associated with response to rituximab in refractory systemic lupus erythematosus. Arthritis Rheumatol. 2023;75(5):697–710. doi: 10.1002/art.42404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Iwasaki T, Watanabe R, Ito H, Fujii T, Okuma K, Oku T, et al. Dynamics of type I and type II interferon signature determines responsiveness to anti-TNF therapy in rheumatoid arthritis. Front Immunol. 2022;13:901437. doi: 10.3389/fimmu.2022.901437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.