Abstract
Objective
Cholangiocarcinoma (CHOL) is a malignant disease that affects the digestive tract, and it is characterized by a poor prognosis. This research sought to explore the involvement of cuproptosis-related lncRNAs (CRLs) in the prognostic prediction and immune infiltration of cholangiocarcinoma.
Methods
The expression profiles and clinical data of CHOL patients were acquired from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases, and CRLs were defined via co-expression analysis. Two molecular clusters distinguished by cuproptosis-related genes (CRGs) were produced. Then a risk signature consisted by four CRLs was formed, and all samples were separated into low- and high-risk groups using a risk score. Kaplan–Meier survival analysis, principal component analysis, differentially expressed analysis, immune cell infiltration analysis, and sensitivities analysis of chemotherapy drugs were conducted between the two groups. Simultaneously, the expression values of four lncRNAs confirmed by real-time PCR in our own 20 CHOL samples were brought into the risk model.
Results
The CHOL samples could be differentiated into two molecular clusters, which displayed contrasting survival times. Additionally, patients with higher risk scores had significantly worse prognosis compared to those in the low-risk group. Furthermore, both immune infiltration and enrichment analysis revealed significant discrepancies in the tumor immune microenvironment (TIME) between different risk groups. Moreover, the predictive power and the correlation with CA19-9 and CEA of risk signature were validated in our own samples.
Conclusion
We developed a risk signature which could serve as an independent prognostic factor and offer a promising prediction for not only prognosis but also TIME in CHOL patients.
Keywords: Cholangiocarcinoma, Cuproptosis, Bioinformatics, Risk signature, lncRNAs
2. Introduction
Cholangiocarcinoma (CHOL) is an extremely malignant disease that occurring from bile duct, which includes intrahepatic distal, perihilar, and intrahepatic cholangiocarcinoma (dCCA, pCCA and iCCA) [1]. The reported incidence rate of CHOL worldwide has steadily increased recently [2]. Although surgery is an effective treatment, many patients metastasize when diagnosed [3]. Consequently, the prognosis for CHOL is dismal, with advanced patients having an approximately 5 % 5-year survival rate [4]. However, even in patients with the same clinical stage and grade, the prognosis is distinctly different. Therefore, to improve the treatment effect, there is an urgent need to distinguish effective tumor characteristics in clinical settings and facilitate the customization of personalized regimens.
In recent years, gene sequencing technology has attracted increasingly more attention in cancer research, and bringing great interest in constructing a gene signature to stratify cancer patients [[5], [6], [7]]. Cuproptosis is a copper-dependent cell death mechanism and represents a newly discovered form of regulated cell death (RCD) [8]. As reported, cuproptosis is initiated by the direct binding of copper with the lipoylated components of the tricarboxylic acid (TCA) cycle [9,10]. At present, whether cuproptosis participates in the progress of cancer is still unclear [11]. Thus, exploring the prospective clinical value of cuproptosis in CHOL is highly significant.
Long non-coding RNAs (lncRNAs), which consist of more than 200 base pairs (bp), are unable to encode proteins. With continuous investigation and development of tumor molecular biology, lncRNAs have been identified as biomarkers or oncogenes in multiple human cancers [12,13]. Numerous studies have consistently explored the involvement of lncRNAs in tumor occurrence, progression, metastasis, and drug resistance [[14], [15], [16]]. Zhu et al. proved that lncRNA TTN-AS1 promotes the tumor progression in CHOL [17]. Moreover, Zhang et al. comprehensively investigated that lncRNA-CCAT1 accelerates invasion and migration in iCCA [18]. However, the comprehensive analysis of CHOL related lncRNAs is still insufficient.
In this research, we downloaded data of CHOL patients from public datasets, which included expression and clinical information. Next, we conducted co-expression analysis with 19 CRGs to identify CRLs. Afterward, by utilizing univariate Cox analysis, LASSO regression analysis and multivariate Cox regression analysis, four lncRNAs (C10orf55, NOP14-AS1, PAXIP1-AS1 and WAC-AS1) were left to construct the risk model. Upon validation, the risk model demonstrated great accuracy as an independent predictor. Additionally, the immune cell infiltration analysis revealed substantial disparities in TIME between different risk groups. Furthermore, to test the steadiness and accuracy of this risk signature, we verified it in CHOL samples from our own cohort. In conclusion, this CRL signature provided an accurate prediction model for risk stratification in CHOL patients, which may be helpful to customize the personalized regimens to improve the treatment effect in clinical.
3. Material and METHODS
3.1. Source and preprocessing of expression data
Transcriptome data and ancillary clinical information were downloaded from two databases: the TCGA-CHOL data set (https://www.tcga.org) and the GSE107943 data set (https://www.ncbi.nlm.nih.gov/geo). The clinical data for these two datasets included expression profiles, gender, stage, and overall survival. The expression profiles from these two datasets were merged and batch effect was eliminated. The values of gene expression were transferred to Transcripts Per Kilobase of exon model per Million mapped reads (TPM), and the log2 function was used. When single gene had multiple probes, the average value was calculated. Finally, 65 samples of CHOL were acquired for further analysis.
3.2. Consensus unsupervised clustering
In the analysis, we employed the ‘ConsensusClusterPlus’ R package along with the K-Means algorithm for conducting unsupervised cluster analysis. The selection of k value, was determined using the proportion of ambiguous clustering. Subsequently, ‘limma’ package was utilized to determine the differentially expressed genes (DEGs). Criteria used to identify DEGs were based on a |log2FoldChange|>1 and p < 0.05. This allowed us to narrow down the genes that exhibited significant changes in expression levels between the different clusters.
3.3. Identification of the CRLs
To obtain the candidate CRLs, we obtained CRGs from previous literature (shown in Supplementary Table S1). From this list, we extracted the expression data for these genes. Next, we defined the CRLs via Pearson's correlation coefficient. We applied a criterion of |Pearson R| > 0.35 and a p-value <0.001, as recommended by Ref[19]. Using this criterion, we identified a total of 214 CRLs which exhibited co-expression with CRGs. These lncRNAs were further studied in this work.
3.4. Construction and validation of the risk model
The CHOL samples were equally separated into two sets: training and validation sets. To determine the lncRNAs correlated to prognosis, we employed the univariate Cox regression analysis in training set (p < 0.05). Then, we employed a Least Absolute Shrinkage and Selection Operator (LASSO) regression analysis, followed by multivariate Cox regression analysis to construct and optimize the risk model. Consequently, the formula followed:
| risk score = Ʃ [Exp (lncRNA) × coef (lncRNA)] |
The risk scores of all samples in training and validation sets were calculated according to the aforementioned formula. Then, the samples were classified into two groups, high- and low-risk groups, based on the median risk score. Subsequently, Kaplan-Meier (KM) survival analysis was performed between the two groups via 'survival' and 'KMsurv' packages, while receiver operating characteristic (ROC) curves were generated using the 'survivalROC' package. To validate the reliability and accuracy of the risk signature, an internal validation set and the entire cohort were utilized.
3.5. Differential expression analysis
The identification of DEG were confirmed using the 'limma' package. The filtering criteria were set to |log2FoldChange| > 1 and an adjusted p-value <0.05. Then we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis to investigate the pathways and biological functions enriched in DEGs. These analyses were conducted by 'clusterprofiler' package. In addition, the 'GSEAbase' package was employed to run single-sample gene set enrichment analysis (ssGSEA) [20]. A significance level of FDR <0.05 was thought statistically significant. The 'org.Hs.e.g.db' R packages was also utilized in this process [21].
3.6. Principal component analysis
Principal component analysis (PCA) was conducted to reduce dimensionality and extract features from the two groups. ‘scatterplot3d’ was utilized during this process [22].
3.7. Establishment of nomogram and calibration curves
A nomogram was established through integrating relevant clinical features with risk score. This nomogram aimed to establish a practical and quantitative approach for prognostic prediction in CHOL patients. Subsequently, calibration curves were used to confirm the reliability and stability of the established nomogram.
3.8. Analysis of immune cell infiltration in tumor
To compare the disparities in the TIME in two groups, proportions and enrichment scores of immune and stroma cells within TIME in CHOL samples were calculated. The "ESTIMATE" package (https://bioinformatics.mdanderson.org/estimate) was performed initially to calculate the tumor purity and infiltrating cells. This assessment revealed differences in the TIME between the two groups [23]. Next, we utilized CIBERSORT (https://cibersort.stanford.edu) to calculate the levels of infiltrating immune cells in every sample [24]. Moreover, expression values of key immune checkpoint molecules (ICMs) were evaluated between different risk groups.
3.9. Prediction of chemotherapeutic drugs sensitivity
To test the sensitivity of chemotherapeutics drugs applied in CHOL treatment, Wilcoxon signed-rank test was employed in the two groups. The ‘pRRophetic’ (http://www.cancerrxgene.org) was utilized in this analysis [25]. P < 0.001 was considered a statistical significance.
3.10. Quantitative real-time PCR
TRIzol (Takara Bio, Dalian, China) was used to isolate RNA from tumor samples. RNA was then transcribed to cDNA through the Prime Script RT Master Mix reagent (Takara Bio, Dalian, China). FastStart Universal SYBR Green Master (Roche, Basel, Switzerland) was utilized for Quantitative real-time PCR (qRT-PCR) and a 7500 Real-time PCR system was employed. The recommended thermal cycling settings were as follows: one initial cycle at 95 °C for 10 min, after that, 40 cycles of 15 s at 95 °C and 60 s at 60 °C followed. The expression level of each lncRNA was calculated by the 2−ΔΔct method which was described in previous study. The internal reference gene used was GAPDH [26]. The primer sequences we used in qRT-PCR were shown in the Supplementary Table S2.
3.11. Human cholangiocarcinoma samples
The primary cholangiocarcinoma tumor tissues were obtained from the cholangiocarcinoma patients treated at Department of Biliary-Pancreatic Surgery, Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine from January 2018 to June 2020.
The pathological information of the samples was confirmed by the pathologist, and the overall survival time was calculated from the date of surgery to cholangiocarcinoma-related death. This study adhered to the Declaration of Helsinki guidelines, obtained approval from the Medical Ethics Committee of Ren Ji Hospital, and secured informed consent from the patients.
3.12. Statistical analysis
R software (version 4.0.2) was utilized for statistical analysis in this study (R; https://www.r-project.org). A two-sample independent t-test was adopted to compare continuous variables. To determine differences between the two groups, Student's t-test was employed. Kaplan-Meier curve and Log-rank test were employed in comparing the differences in survival time between the high-risk and low-risk groups. A significance level of p < 0.05 was considered statistically significant.
4. Results
4.1. Data collection and identification of cuproptosis-related clusters in cholangiocarcinoma
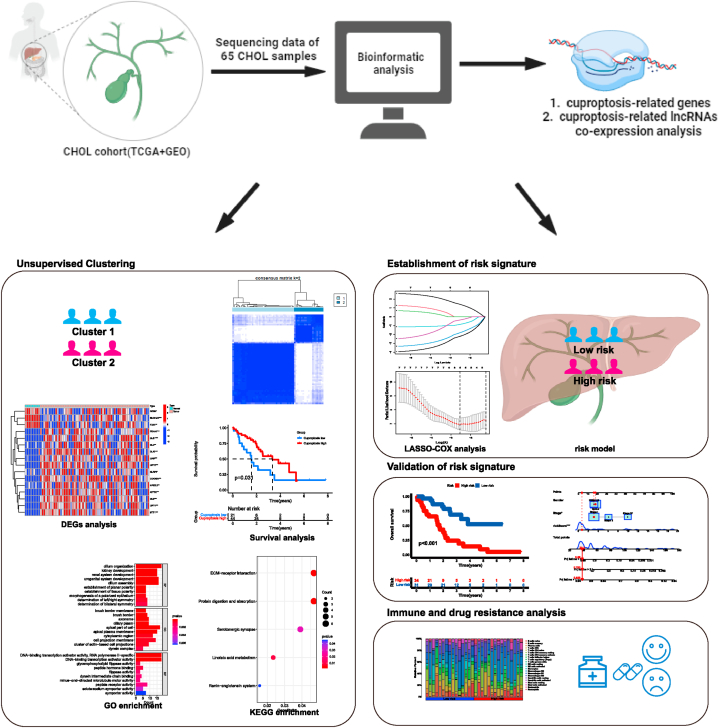
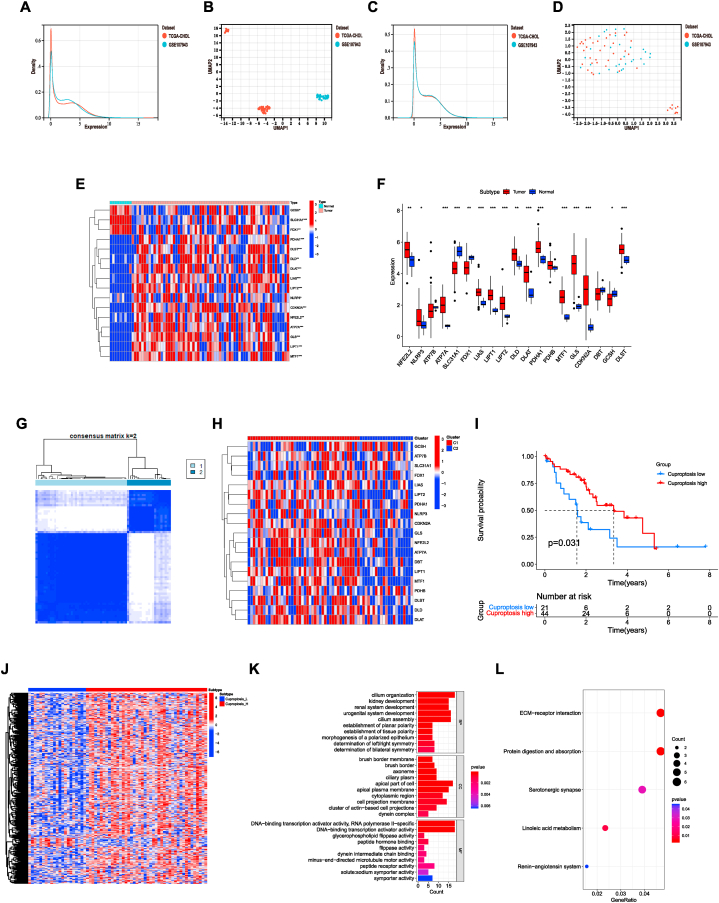
The flow chart detailing the methodology of this study was presented in Fig. 1. The expression data of CHOL samples were acquired from two datasets: the TCGA-CHOL dataset and the GSE107943 dataset. With the preprocessing mentioned above, the two expression profiles were merged and the batch effect was removed (Fig. 2A–D). Finally, a total of 74 samples comprising 65 tumor samples and 9 normal samples were collected. We initially explored the expression patterns of 19 CRGs in both CHOL samples and normal samples. As presented, the most genes, including PDHA1, DLST, DLD, DLAT, LIAS, LIPT2, NLRP3, CDKN2A, NFE2L2, ATP7A, GLS, LIPT1, and MTF1 were highly expressed in tumor samples (Fig. 2E–F). Furthermore, cuproptosis-related clusters of CHOL were identified by consensus clustering (Fig. 2G–H). As a result, the cluster 2 (C2) showed a high CRGs expression level, while the cluster 1 (C1) was identified as a cuproptosis-low cluster. Survival analysis revealed a remarkable difference between two clusters. In general, the cuproptosis-low cluster exhibited a dismal prognosis compared to the cuproptosis-high cluster (Fig. 2I). Furthermore, DEGs between the two clusters were analyzed. Identified 422 DEGs were obtained and enrichment analysis was conducted (Fig. 2J). The results suggested that these DEGs were significantly enriched in DNA-binding transcription activator activity, cell projection membrane, peptide hormone binding in GO enrichment, and ECM-receptor interaction in KEGG enrichment analysis (Fig. 2K-L).
Fig. 1.
Workflow of this study.
Fig. 2.
Identify the cuproptosis-related clusters in cholangiocarcinoma. (A–B), The expression level of samples from two databases before removing the batch effect. (C–D), The expression level of samples from two databases after removing the batch effect. (E–F), The expression patterns of CRGs in CHOL samples and normal samples. (G), The heatmap of consensus clustering solution (k = 2) in CHOL samples. (H), The expression of CRGs in different clusters. (I), Kaplan–Meier curves of OS in cuproptosis-high and cuproptosis-low clusters. (J), The heatmap of DEGs between the two clusters. (K–L), GO (K) and KEGG (L) enrichment analysis. ***p < 0.001; **p < 0.01; *p < 0.05.
4.2. Identification of CRLs and construction of a risk signature
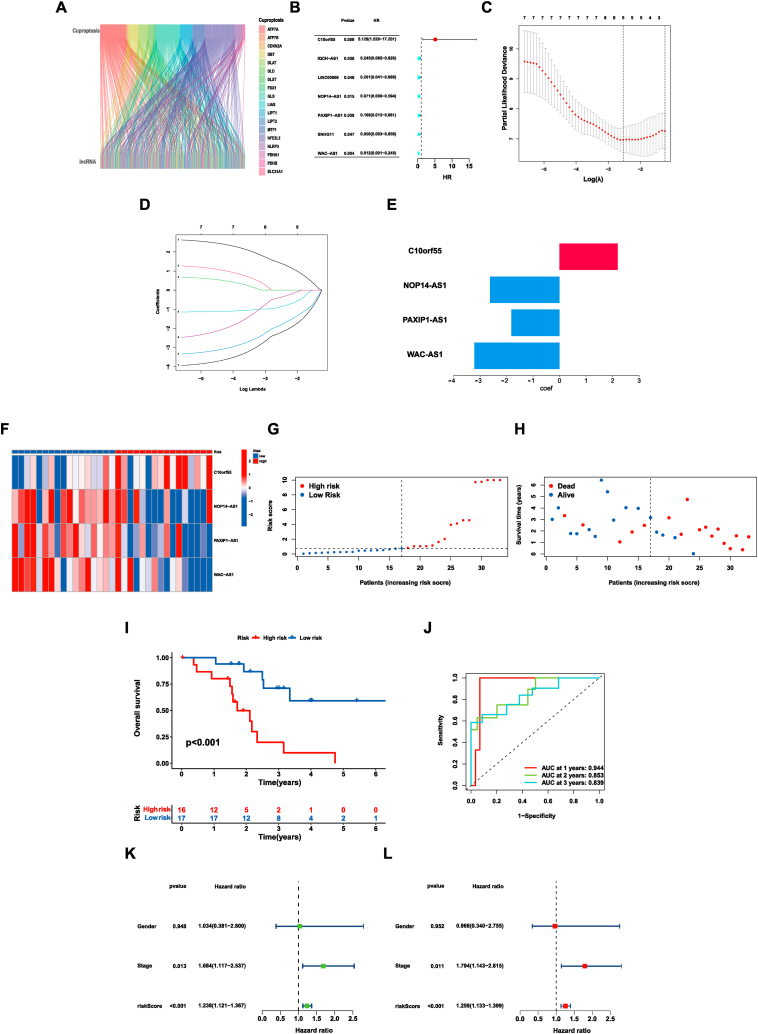
To identify lncRNAs associated with cuproptosis, we gathered 19 CRGs from published literature. Next, we extracted the expression levels of these genes from the previously obtained expression profile. Pearson's correlation coefficient was then used, with a criterion which was described in the method. Totally 214 CRLs were identified (Fig. 3A). After obtaining clinical data, a total of 65 CHOL samples were separated into two sets: a training set and a validation set. In the training group, utilizing univariate Cox analysis, a total of 7 CRLs were identified as prognosis-related lncRNAs (Fig. 3B). Afterward, the LASSO regression analysis was conducted to prevent overfitting. As a result, five lncRNAs related to cuproptosis were left for further analysis (Fig. 3C–D). Subsequently, the multivariate Cox regression analysis was utilized to identify independent indicators among these lncRNAs in the training group. Ultimately, four lncRNAs (C10orf55, NOP14-AS1, PAXIP1-AS1 and WAC-AS1) remained to establish a risk signature associated with the prognosis of CHOL samples (Fig. 3E, Table 1). Among them, NOP14-AS1, PAXIP1-AS1, and WAC-AS1 were found to be protective factors, and the samples with higher expression of them had a better prognosis. While C10orf55 was a poor prognostic factor, the samples with higher expression of C10orf55 had a worse prognosis. Consequently, a four-lncRNA risk signature was established for prognostic prediction of CHOL patients. The risk score was calculated by the expression levels of the four lncRNAs and the corresponding multivariate Cox regression coefficients.
| Risk score = 2.196 × (C10orf55)+ −2.621 × Exp(NOP14-AS1)+ −1.817 × Exp(PAXIP1-AS1)+ −3.216 × Exp(WAC-AS1). |
Fig. 3.
The prognostic value of four cuproptosis-related lncRNAs in the training group. (A), The Sankey diagram of CRGs and CRLs. (B), The results of the Univariate Cox regression analysis. (C), Cross-validation plot for the results of LASSO regression analysis. (D), Plots for LASSO expression coefficients of the CRLs. (E), The coefficient of four CRLs. (F), The expression levels of four lncRNAs in every sample. The change from blue to red means the rise of expression. (G), The distribution of the risk scores in the training group. (H), The correlation of risk score and survival status in the training group. (I), Kaplan–Meier overall survival curve of samples in the low- and high-risk groups. (J), ROC curves of the risk signature in the training group. (K), Univariate Cox regression analysis of risk score and other clinical features in CHOL patients. (L), Multivariate Cox regression analysis of risk score and other clinical features in CHOL patients. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
The results of Multivariate Cox regression analysis.
| Id | Coef | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| C10orf55 | 2.20 | 5.128 | 1.529 | 17.201 | 0.008 |
| NOP14-AS1 | −2.62 | 0.071 | 0.009 | 0.594 | 0.015 |
| PAXIP1-AS1 | −1.82 | 0.108 | 0.013 | 0.881 | 0.038 |
| WAC-AS1 | −3.22 | 0.012 | 0.001 | 0.248 | 0.004 |
By employing the median risk score as a threshold, we categorized all samples in both the training and validation sets into high- and low-risk groups.
4.3. The risk signature exhibited an association with the prognosis of CHOL patients in the training set
The risk scores for all sample were calculated using the aforementioned formula. The distribution of these four lncRNAs was indicated in the heat map (Fig. 3F). The increase in the risk score was found to have a closely correlation with a decrease in the number of surviving patients and an increase in the number of deceased samples (Fig. 3G–H). Furthermore, the overall survival (OS) time of patients with higher risk scores was shorter compared to that in the group of low-risk (p < 0.001) (Fig. 3I). Besides, the accuracy and reliability of the signature in prognosis prediction was proved by the ROC curves (AUC = 0.944, 0.853, 0.839 for 1-, 2-, and 3-year) (Fig. 3J). To validate whether the signature impacted the prognosis of CHOL patients independently, Cox regression analyses were performed. The results supported that the risk signature acted as a significant, independent prognostic factor, strongly influencing the outcomes of CHOL patients (p < 0.001) (Fig. 3K-L).
4.4. Verification of the risk signature in the validation set and all samples
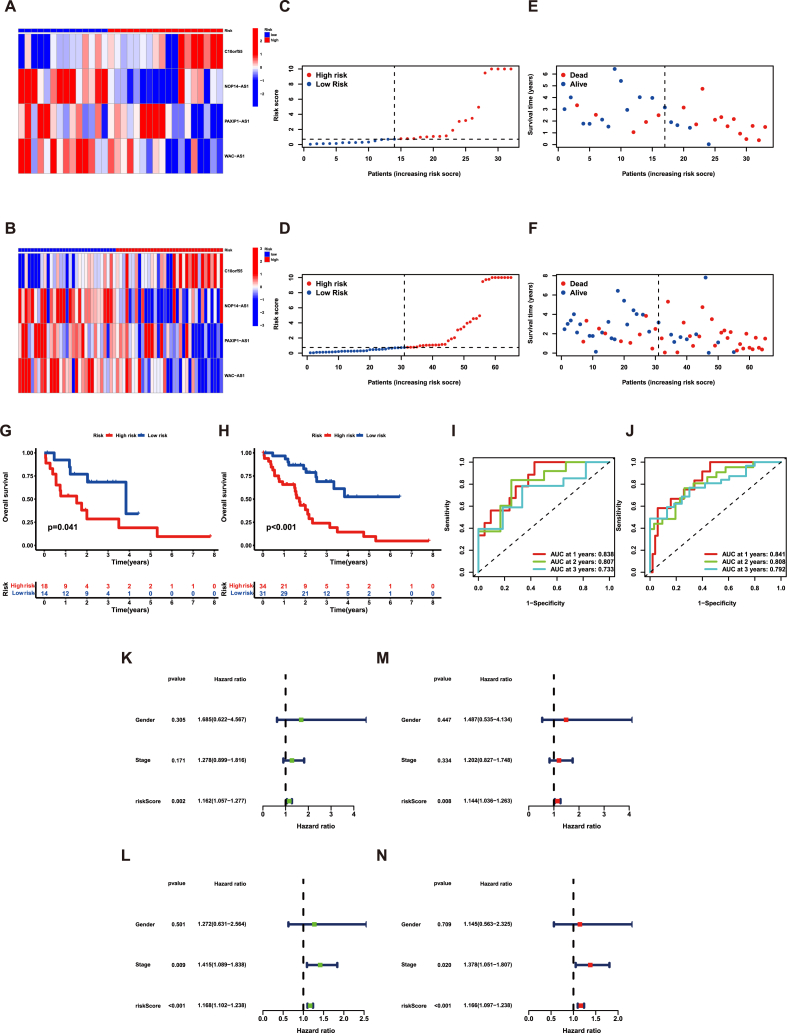
To enhance the reliability of the risk model, verification was made in both the validation set and entire cohort. Similar to the training set, the results presented the distributions of the 4 lncRNAs and the risk scores. We observed that when the risk score increased, survival samples decreased while death samples increased (Fig. 4A–F). In accordance with the training cohort, the group of high-risk had a shorter OS time compared to the other group (Fig. 4G–H). In addition, the ROC curves indicated that the risk signature had a high level of accuracy in predicting the prognosis, both in the validation group and across all samples (Fig. 4I–J). Moreover, in line with the training group, the results supported that the risk signature had ability to predict the prognosis for CHOL patients independently in both the validation set and entire cohort (Fig. 4K-N). These findings further validated the significance of the risk signature in predicting outcomes for CHOL patient.
Fig. 4.
The prognostic value of four cuproptosis-related lncRNAs in the validation set and all samples. The expression levels of four lncRNAs in the (A) validation set and (B) all samples. The change from blue to red means the rise of expression. The distribution of the risk scores in the (C) validation set and (D) all samples. The correlation of risk scores and survival status in the (E) validation set and (F) all samples. Kaplan–Meier overall survival curve of samples in the (G) validation set and (H) all samples. ROC curves of the risk signature in the (I) validation set and (J) all samples. The Univariate Cox regression analysis of risk score and other clinical features in the (K) validation set and (L) all samples. The Multivariate Cox regression analysis of risk score and other clinical features in the (M) validation set and (N) all samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4.5. Comparison of predictive capacity between risk signature and other clinical characteristics
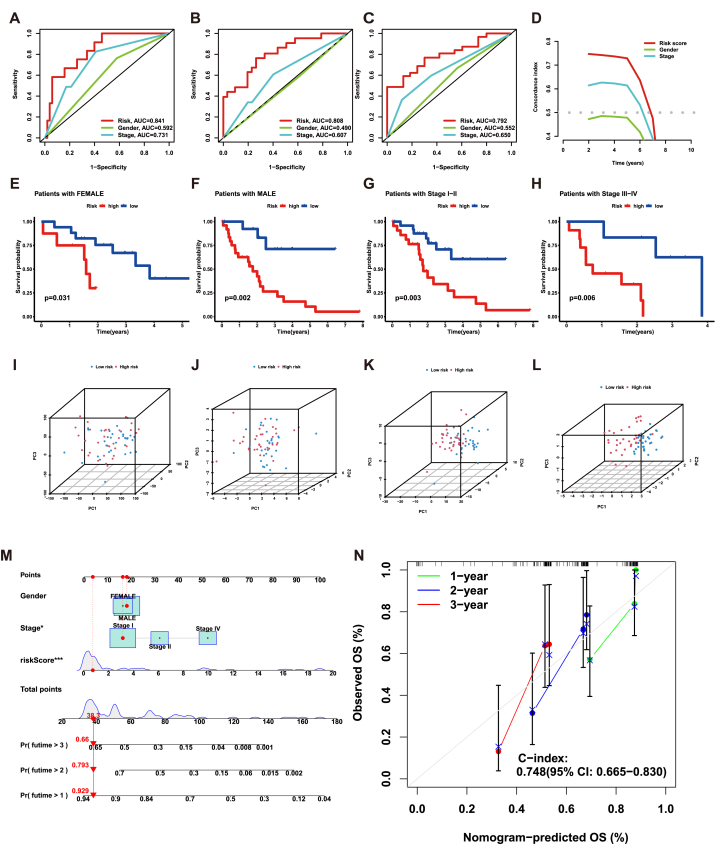
To compare the capabilities of prognosis prediction between risk signature and other clinicopathological characteristics, we conducted ROC curve analysis. The analysis suggested that the signature was confirmed to be the best predictor compared with others (Fig. 5A–C). Furthermore, these results were further verified by the c-index (Fig. 5D). Kaplan-Meier survival analysis within each subtype was conducted to assess the stability of risk signature in prognostic prediction across different patient subgroups. Regardless of the gender or stage of patients, the risk signature still had great performance in distinguishing the remarkable difference in OS time between groups with different risk scores (Fig. 5E–H). Subsequently, PCA was utilized to evaluate the grouping ability of the risk signature. Using whole-genome expression profiles, 19 CRGs, 214 CRLs, and the risk signature, the results revealed a significant distinction between different risk groups. It suggested that the risk signature effectively stratified the samples into two distinct groups (Fig. 5I-L). Finally, the establishment of a nomogram offered a practical and quantitative approach for prognostic prediction in CHOL patients (Fig. 5M). To assess the reliability and stability of the nomogram, we utilized calibrate curves, which revealed that the predicted survival rates from the nomogram closely aligned the actual survival rates (Fig. 5N). Collectively, these results suggested that the signature developed in this study had stable and reliable capabilities in prognostic prediction for CHOL patients.
Fig. 5.
Validate the stability of the risk signature. (A–C), ROC curves of 1-, 2-, 3-year overall survival for risk score and other clinical features. (D), The concordance index (c-index) for the risk score and other clinical features. (E–H), The Kaplan-Meier survival curves of risk signature in different subtypes of patients. (I), PCA of all genes. (J), PCA of CRGs. (K), PCA of CRLs. (L), PCA of risk score. (M), The nomogram constructed by risk score and other clinical features. (N), 1-, 2-, 3-year overall survival calibration curves of all samples. ***p < 0.001; **p < 0.01; *p < 0.05.
4.6. Expression difference analysis
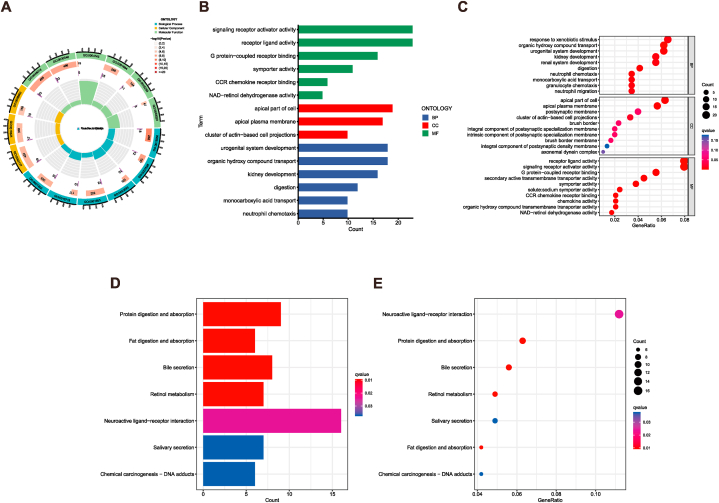
Initially, the DEGs were screened between the two risk groups using a significance criterion mentioned in the method. Then, to uncover the expression differences and biological processes associated with the risk model, GO and KEGG enrichment analysis were employed. The GO analysis revealed that the identified DEGs between the two groups enriched in multiple immune-associated processes, such as neutrophil migration, neutrophil chemotaxis, granulocyte chemotaxis, chemokine activity, and CCR chemokine receptor binding (Fig. 6A–C). In contrast, neuroactive ligand−receptor interaction, bile secretion, and other signaling pathways were enriched in the KEGG enrichment analysis (Fig. 6D–E). These findings provided evidence that the identified risk signature could act as a predictive tool of immune response for CHOL patients.
Fig. 6.
Functional enrichment analysis in low- and high-risk groups. (A–C), Gene Ontology (GO) enrichment analysis of DEGs in two groups. (D–E), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of DEGs in two groups.
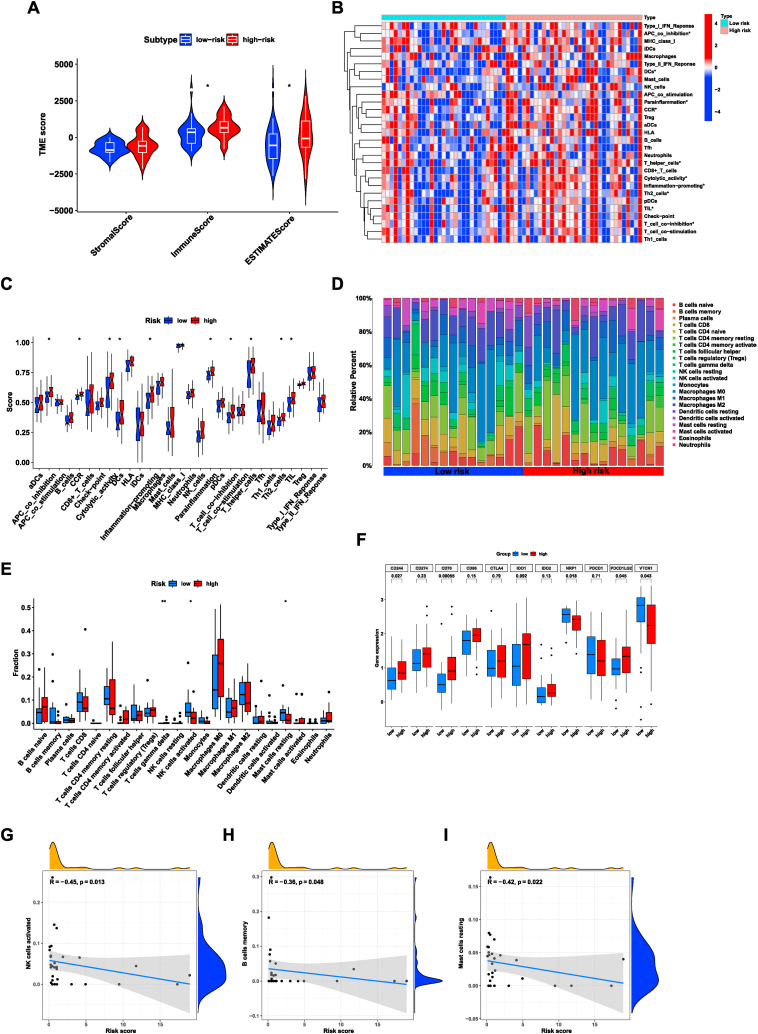
4.7. Correlation between risk signature and immune infiltration
TIME has been shown to exert a critical influence on the progression of cholangiocarcinoma [[27], [28], [29]]. To further investigate the characteristics of the TIME in CHOL samples, several analyses were conducted. Firstly, we employed the ‘ESTIMATE’ algorithm to evaluate the differences in immune-related scores among two groups. The findings revealed that lower estimate and immune scores existed in the group of low-risk (Fig. 7A). Moreover, we utilized the ssGSEA to compute the enrichment of several immune functions for all samples. The results suggested that several immune functions such as parainflammation, inflammation-promoting, cytolytic activity, and APC co-inhibition were more active in the patients with higher risk scores (Fig. 7B–C). Furthermore, the CIBERSORT package was conducted to investigate the differences in the composition of immune cell subtypes in two groups. The results revealed a higher proportion of T cells gamma delta in the group of high-risk. However, Mast cells resting and activated NK cells were the opposite (Fig. 7D–E). Besides, the proportions of Mast cells resting, activated NK cells, and B cells memory were negatively correlated with risk score (Fig. 7G–I). Overall, these findings indicated a substantial disparity in the TIME between different risk groups.
Fig. 7.
The relationship between tumor immune microenvironment and the risk signature. (A), The relationship between risk score and immune-related scores in CHOL samples. (B–C), Comparison of immune-related pathways between the low- and high-risk groups. (D–E), The proportion of infiltrating immune cells between the high- and low-risk groups calculated by CIBERSORT algorithm. (F), Comparison of the expression levels of immune checkpoint key molecules in the low- and high-risk groups. (G–I), Correlation between the risk score and immune cells. ***p < 0.001; **p < 0.01; *p < 0.05.
The advancement of ICM research has revolutionized the clinical decision-making process in cancer oncology by introducing immune checkpoint inhibitors [[30], [31], [32]]. Subsequently, we selected certain ICMs, and the relative expression of them were examined among different risk groups. The analysis uncovered a noteworthy association between the risk signature and ICMs. The results demonstrated that CD244, CD70, and PDCD1LG2 were elevated in the group with higher risk score. On the other hand, NRP1 and VTCN1 exhibited opposite trends, suggesting downregulation in the high-risk group (Fig. 7F). Therefore, the association between the risk model and the expression of ICMs provided valuable insights into the potential utilities of the risk model in predicting the response to immune checkpoint blockade therapies in CHOL patients.
4.8. Investigation of the association between the risk signature and sensitivities of chemotherapy drugs
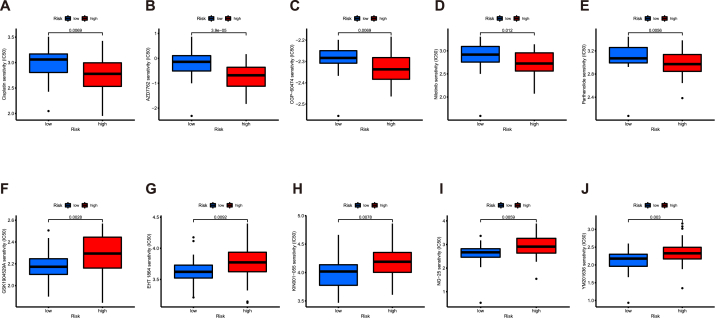
In recent years, the introduction of numerous anti-tumor drugs has made chemotherapy a preferred treatment option in many clinical scenarios. However, the availabilities of the drugs are limited. Besides, because of the existence of drug resistance, many anti-tumor drugs fail in the anti-tumor process [33]. Therefore, it is crucial to screen the potential chemotherapy drugs for the different patients, which could ensure great responses to the chemotherapy and improve long-term quality of life [34]. Thus, pRRophetic algorithm was utilized to screen the drugs with different sensitivities in two groups. As presented in the result, patients with higher risk scores exhibited greater sensitivity to these drugs (Cisplatin, AZD7762, CGP-60474, Nilotinib, Parthenolide), while patients with lower risk scores displayed lower IC50 values for the remaining drugs (GSK1904529A, EHT 1864, KIN001-055, NG-25, YM201636) (Fig. 8A–J). These findings may offer valuable insights for the future application of these drugs in a clinical setting.
Fig. 8.
Verification of the risk signature and chemotherapy drug sensitivity. (A–J), 10 common chemical drugs which exhibit different IC50 in two groups.
4.9. Validating the prognostic prediction capacity of the risk signature using our own samples
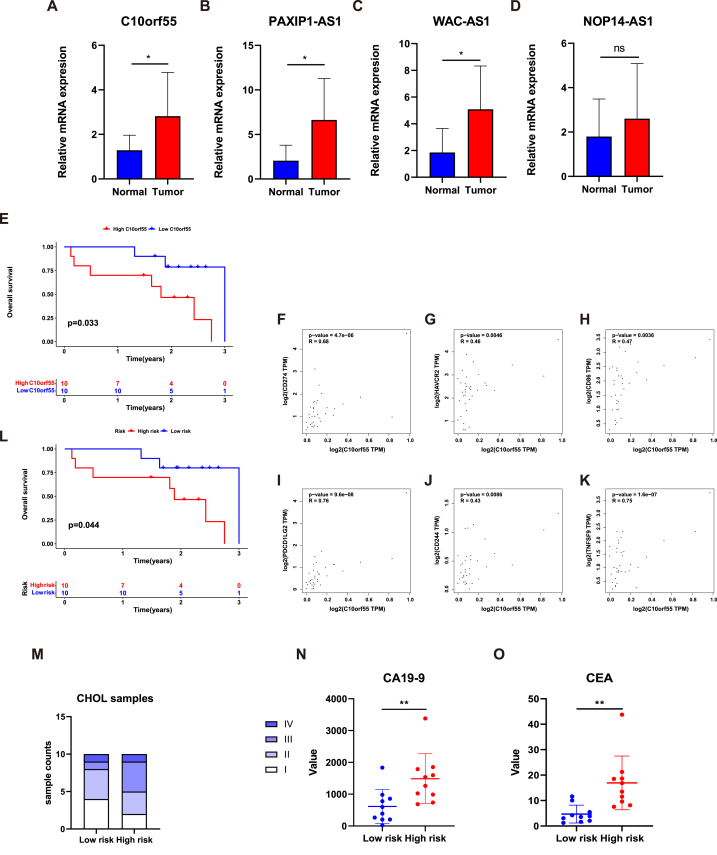
Initially, the expression level of each lncRNA was detected by qRT-PCR. The results revealed significantly higher expression value of C10orf55, PAXIP1-AS1, and WAC-AS1 in CHOL samples compared to normal tissues. (Fig. 9A–D). In this study, C10orf55 was identified as a risk factor, while PAXIP1-AS1 and WAC-AS1 were identified as protective factors. Hence, the reasons underlying the elevated expression of PAXIP1-AS1 and WAC-AS1 in tumor samples may be complex. The roles played by PAXIP1-AS1 and WAC-AS1 in CHOL are multifaceted, and additional experiments are necessary to illustrate the potential effects of C10orf55, PAXIP1-AS1 and WAC-AS1 in CHOL in the future. Additionally, we found that C10orf55 may serve as an indicator for prognosis prediction in our own samples. The higher the expression level of C10orf55, the worse the prognosis of CHOL patients (Fig. 9E) (p = 0.033). Moreover, we validated the correlation between the C10orf55 and key ICMs to elaborate the impact of C10orf55 on the immunotherapy of CHOL. As presented in the results, the expression of C10orf55 was significantly related with CD274 (p = 4.7e-06, R = 0.68), HAVCR2 (p = 0.0046, R = 0.46), CD86 (p = 0.0036, R = 0.47), PDCD1LG2 (p = 9.6e-08, R = 0.76), CD244 (p = 0.0086, R = 0.43), and TNFSF9 (p = 1.6e-07, R = 0.75) (Fig. 9F–K). These results indicated that C10orf55 could serve as a potential target in immunotherapy in CHOL. Furthermore, we also validated the predictive capacity of the signature in 20 CHOL samples from Ren Ji hospital.
Fig. 9.
Validation of the four lncRNAs and the risk signature in our tumor samples. (A–D), Comparison of the expression levels of four lncRNAs in normal and tumor samples. (E), Kaplan–Meier survival curve of the high expression of C10orf55 and the low expression of C10orf55 in our samples. (F–K), The expression pattern of C10orf55 and several immune checkpoint molecules in CHOL samples. (L), Kaplan–Meier survival curve of the high-risk group and the low-risk group in our samples. (M), The differences of tumor stage between the high- and low-risk groups. (N–O), The differences of tumor markers (CA19-9 and CEA) between the high- and low-risk groups. ***p < 0.001; **p < 0.01; *p < 0.05.
The expression level of each lncRNA, detected by qRT-PCR, was integrated into the previously described formula. Then the samples were categorized into high- and low-risk groups similarly.
Consistent to the previous findings, patients belonging to the group of high-risk demonstrated significantly shorter OS times compared to those belonging to the other group (Fig. 9L) (p = 0.044). Besides, we conducted a comparison of clinical data, including stage, CEA, and CA19-9, between different risk groups (Fig. 9M − O). The results revealed that the levels of CEA and CA19-9 were higher in the group of high-risk. In conclusion, the signature had great accuracy and robustness not only in the database, but also in our own patients.
5. Discussion
Cholangiocarcinoma (CHOL) is an extremely malignant disease with unfortunate prognosis. Although studies on cholangiocarcinoma have increased in recent years, the potential mechanism underlying the occurrence and development of this disease remains unclear [35,36]. Surgical resection, which involves the removal of the tumor and surrounding healthy tissue, remains the primary treatment strategy for CHOL. However, most of the patients with CHOL are unqualified for surgical operation because they are diagnosed at advanced stages. Therefore, the discovery and identification of new biomarkers related to disease prognosis and stage are of significant importance. Recently, Carbohydrate antigen 19-9 (CA19-9), as well as cancer antigen 125 (CA125) have been acknowledged as early biomarkers of CHOL in clinical work [37,38]. Nevertheless, it is disappointing that their specificity is not satisfactory [39,40]. CA19-9 and CA125 will also increase when other abdominal tumors occur. Consequently, identifying a new biomarker and integrating it into clinical practice for CHOL patients is urgent.
Cell death is a fundamental process in maintaining organ development and tissue homeostasis [41]. Cancerous cells always have the characteristic of uncontrolled proliferation because of confusion about cell death procedures. Cell death is classified into two categories based on the speed of occurrence: accidental cell death (ACD) and RCD [42]. Cuproptosis, a form of RCD triggered by excess intracellular copper, has garnered significant research attention. However, how cuproptosis participates in tumorigenesis and progression remains unclear.
Previous studies have shown promising results in developing prognostic signatures for CHOL patients. Zhang et al. established a four-gene prognostic signature using ferroptosis-related genes for CHOL and photodynamic therapy [5]. Xie et al. reported a signature based on lncRNAs for survival prediction and risk stratification in CHOL [7]. Another study identified a metabolism-related signature to deeply explore the target genes and potential mechanism in intrahepatic cholangiocarcinoma [43]. However, whether cuproptosis is also a great indicator for prognostic prediction in CHOL was not studied. The primary purpose of this research was to find and validate a risk signature for CHOL prognosis using CRLs, and assess the accuracy and reliability of this signature by ROC curve analysis. This signature was validated in the validation set and all samples, which confirmed the signature served as an independent prognostic factor. It also made the risk signature as a crucial piece of information in clinical decision-making.
Recently, numerous pieces of research have focused on the relationship between antitumor immunity and the progression of tumors. Plenty of studies highlighted the crucial role of the TIME in both tumor development and patient prognosis. Understanding the characteristics of the TIME is crucial for developing effective clinical therapies. TIME and the role of tumor-infiltrating immune cells have become major areas of research in the context of CHOL as well. A study states clearly that LAIR2 and PNOC are associated with higher immune infiltration status in CHOL [44]. Furthermore, extensive research has uncovered the influences of lncRNAs on infiltrating immune cells. Xu et al. revealed that SATB2-AS1 affected the TIME in colorectal cancer [45]. The study of Sun et al. suggested that LINC00301 was associated with the immunosuppressive microenvironment in lung cancer [46]. Therefore, we speculated that tumor-infiltrating immune cell subtypes were closely related to lncRNAs. In our research, a relationship was established between the infiltrating immune cells and risk signature.
To further investigate the association between risk model and the drug sensitivity, we employed the pRRophetic algorithm. As a result, we identified multiple drugs that exhibited differing sensitivities between the two groups. Cisplatin has been used for treating numerous human cancers including lung cancer, ovarian cancer and CHOL. The mechanism of it could be explained as interfering the process of DNA repair and finally inducing apoptosis in tumor cells [47]. CHK1 was reported to be a cell cycle checkpoint, which protected the tumor cells from apoptosis in p53-deficient situation [48]. AZD7762 is known for its strong ability to inhibit CHK1, which showed a lower IC50 in the patients with higher risk scores. Research has identified the oncogenic role of NF-κB signaling, so the NF-κB signaling becomes the new target to inhibit the progression of cancers in the recent years [49]. Parthenolide exhibited anti-tumor activity by inhibiting NF-κB activation, which showed a lower IC50 in the patients with higher risk scores. Among the other potential drugs, the GSK1904529A, an inhibitor of insulin-like growth factor-1 receptor (IGF-1R) and insulin receptor (IR), and EHT 1864, which is an inhibitor of Rac family small GTPases, were more sensitive to the patients with lower risk scores. Substantially, the incorporation of risk model into personalized medicine allows healthcare providers to make more informed treatment decisions tailored to individual patients.
Despite the increasing identification of various lncRNAs in tumors development, the complete function of lncRNAs has not been deeply studied. Even if lncRNAs cannot encode proteins, their significance in cancer progression is comparable to that of protein-coding genes. Similarly, the use of lncRNAs as potential biomarkers has been extensively explored in various studies to predict patient survival. These findings provided further insights into the critical role played by lncRNAs in tumor development and progression. Our research successfully identified a four-lncRNA signature consisting of C10orf55, NOP14-AS1, PAXIP1-AS1, and WAC-AS1. This signature has shown a closely correlation with the prognosis of CHOL patients. In previous studies, C10orf55 was incorporated into the signature, enabling the prediction of relapse in cytogenetically normal acute myeloid leukemia [50]. LncRNA NOP14-AS1 was found to promote tongue squamous cell carcinoma progression [51]. Moreover, PAXIP1-AS1 was found to facilitate the progression of both glioma and ovarian cancer [52,53]. Similarly, Xia et al. discovered that WAC-AS1 participated in the progression of hepatocellular carcinoma [54]. In this study, we confirmed the predictive power of these four lncRNAs in our own CHOL samples, which proved the accuracy and robustness of the signature. However, the precise mechanism of these lncRNAs in the development of CHOL remains unclear and requires further investigation.
Additionally, some limitations still exist. Firstly, as the data were obtained from public datasets, there may be inherent limitations in data quality and consistency. Secondly, our study was based on retrospective samples. Therefore, more studies are necessary to prove the reliability of this signature through the inclusion of additional prospective cohorts. Additionally, it is important to provide more evidence by which these four lncRNAs promote to the tumorigenesis and progression of CHOL.
Ethical statement
This study was approved under the number 2018-076 assigned by the Research Ethics Committee of Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine.
Data availability statement
The data used in this study are all available from the TCGA database (https://portal.gdc.cancer.gov/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/).
CRediT authorship contribution statement
Hong-Fei Yao: Data curation, Formal analysis, Software, Writing – original draft. Min He: Data curation, Formal analysis, Writing – original draft. Yu-Heng Zhu: Data curation, Formal analysis, Writing – original draft. Bo Zhang: Formal analysis, Writing – review & editing. Peng-Cheng Chen: Formal analysis, Writing – review & editing. Yan-Miao Huo: Data curation, Resources. Jun-Feng Zhang: Conceptualization, Methodology, Supervision, Writing – original draft. Chao Yang: Conceptualization, Methodology, Supervision, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Table S1.
| Cuproptosis-related genes |
|---|
| NFE2L2 NLRP3 ATP7B ATP7A SLC31A1 FDX1 LIAS LIPT1 LIPT2 DLD DLAT PDHA1 PDHB MTF1 GLS CDKN2A DBT GCSH DLST |
Table S2.
| Gene | Primers sequences | Amplicon size | Accession no. |
|---|---|---|---|
| GAPDH forward GAPDH reverse |
5′-CAGGAGGCATTGCTGATGAT-3′ 5′-GAAGGCTGGGGCTCATTT-3′ |
138 | NM_002046 |
| C10orf55 forward C10orf55 reverse |
5′-ATTCGGGAGGAGGCTTCATCA-3′ 5′-TGAGAACTAGATACGAACAGGGT-3′ |
127 | NR_160937 |
| NOP14-AS1 forward NOP14-AS1 reverse |
5′-GTCGTAGTCTGGTTAATCCTT-3′ 5′-ACTGAACTTGTGTCCTTAGC-3′ |
178 | NR_015453 |
| PAXIP1-AS1 forward PAXIP1-AS1 reverse |
5′-ACGCCTGTAATCCTATCCT-3′ 5′-CCGAGTTCAAGCCATTCT-3′ |
190 | NR_028090 |
| WAC-AS1 forward WAC-AS1 reverse |
5′-GACAGAGCGAGATTCCATC-3′ 5′-AGAGTATGACAACAGCAACA-3′ |
190 | NR_033805 |
Contributor Information
Yan-Miao Huo, Email: huoyanmiao@126.com.
Jun-Feng Zhang, Email: zhangjunfeng@renji.com.
Chao Yang, Email: zhaoyang75@hotmail.com.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Rizvi S., Khan S.A., Hallemeier C.L., et al. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018;15(2):95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brindley P.J., Bachini M., Ilyas S.I., et al. Cholangiocarcinoma. Nature reviews Disease primers. 2021;7(1):65. doi: 10.1038/s41572-021-00300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y.T., Wang J.J., Luu M., et al. Comparison of clinical features and outcomes between intrahepatic cholangiocarcinoma and hepatocellular carcinoma in the United States. Hepatology. 2021;74(5):2622–2632. doi: 10.1002/hep.32007. [DOI] [PubMed] [Google Scholar]
- 4.Kelley R.K., Bridgewater J., Gores G.J., et al. Systemic therapies for intrahepatic cholangiocarcinoma. J. Hepatol. 2020;72(2):353–363. doi: 10.1016/j.jhep.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z.J., Huang Y.P., Li X.X., et al. A novel ferroptosis-related 4-gene prognostic signature for cholangiocarcinoma and photodynamic therapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.747445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., Chen X., Jiang Z. Immune infiltration and a ferroptosis-related gene signature for predicting the prognosis of patients with cholangiocarcinoma. Am. J. Tourism Res. 2022;14(2):1204–1219. [PMC free article] [PubMed] [Google Scholar]
- 7.Xie X., Wang Y., Zhang S., et al. A novel five-lncRNA signature panel improves high-risk survival prediction in patients with cholangiocarcinoma. Aging. 2021;13(2):2959–2981. doi: 10.18632/aging.202446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsvetkov P., Coy S., Petrova B., et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Min J., Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Targeted Ther. 2022;7(1):378. doi: 10.1038/s41392-022-01229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D., Tian Z., Zhang P., et al. The molecular mechanisms of cuproptosis and its relevance to cardiovascular disease. Biomed. Pharmacother. 2023;163 doi: 10.1016/j.biopha.2023.114830. [DOI] [PubMed] [Google Scholar]
- 11.Xie J., Yang Y., Gao Y., et al. Cuproptosis: mechanisms and links with cancers. Mol. Cancer. 2023;22(1):46. doi: 10.1186/s12943-023-01732-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheykhhasan M., Ahmadyousefi Y., Seyedebrahimi R., et al. DLX6-AS1: a putative lncRNA candidate in multiple human cancers. Expet Rev. Mol. Med. 2021;23:e17. doi: 10.1017/erm.2021.17. [DOI] [PubMed] [Google Scholar]
- 13.Sheykhhasan M., Tanzadehpanah H., Ahmadieh Yazdi A., et al. FLVCR1-AS1 and FBXL19-AS1: two putative lncRNA candidates in multiple human cancers. Non-coding RNA. 2022;9(1) doi: 10.3390/ncrna9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCabe E.M., Rasmussen T.P. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin. Cancer Biol. 2021;75:38–48. doi: 10.1016/j.semcancer.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Malakoti F., Targhazeh N., Karimzadeh H., et al. Multiple function of lncRNA MALAT1 in cancer occurrence and progression. Chem. Biol. Drug Des. 2023;101(5):1113–1137. doi: 10.1111/cbdd.14006. [DOI] [PubMed] [Google Scholar]
- 16.Wei L., Sun J., Zhang N., et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol. Cancer. 2020;19(1):62. doi: 10.1186/s12943-020-01185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H., Zhai B., He C., et al. LncRNA TTN-AS1 promotes the progression of cholangiocarcinoma via the miR-320a/neuropilin-1 axis. Cell Death Dis. 2020;11(8):637. doi: 10.1038/s41419-020-02896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S., Xiao J., Chai Y., et al. LncRNA-CCAT1 promotes migration, invasion, and EMT in intrahepatic cholangiocarcinoma through suppressing miR-152. Dig. Dis. Sci. 2017;62(11):3050–3058. doi: 10.1007/s10620-017-4759-8. [DOI] [PubMed] [Google Scholar]
- 19.Schober P., Boer C., Schwarte L.A. Correlation coefficients: appropriate use and interpretation. Anesth. Analg. 2018;126(5):1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- 20.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S., Kang D., Huo Z., et al. Meta-analytic principal component analysis in integrative omics application. Bioinformatics. 2018;34(8):1321–1328. doi: 10.1093/bioinformatics/btx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K., Shahmoradgoli M., Martínez E., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahaki H., Jabarivasal N., Sardarian K., et al. The effects of extremely low-frequency electromagnetic fields on c-Maf, STAT6, and RORα expressions in spleen and thymus of rat. Electromagn. Biol. Med. 2019;38(2):177–183. doi: 10.1080/15368378.2019.1608832. [DOI] [PubMed] [Google Scholar]
- 27.Diggs L.P., Ruf B., Ma C., et al. CD40-mediated immune cell activation enhances response to anti-PD-1 in murine intrahepatic cholangiocarcinoma. J. Hepatol. 2021;74(5):1145–1154. doi: 10.1016/j.jhep.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y.H., Zhang C.Z., Huang Q.S., et al. Clinicopathologic features, tumor immune microenvironment and genomic landscape of Epstein-Barr virus-associated intrahepatic cholangiocarcinoma. J. Hepatol. 2021;74(4):838–849. doi: 10.1016/j.jhep.2020.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Fabris L., Sato K., Alpini G., et al. The tumor microenvironment in cholangiocarcinoma progression. Hepatology. 2021;73(1):75–85. doi: 10.1002/hep.31410. Suppl 1(Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagchi S., Yuan R., Engleman E.G. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annual review of pathology. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 31.Galluzzi L., Humeau J., Buqué A., et al. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2020;17(12):725–741. doi: 10.1038/s41571-020-0413-z. [DOI] [PubMed] [Google Scholar]
- 32.Gutiérrez-Larrañaga M., González-López E., Roa-Bautista A., et al. Immune checkpoint inhibitors: the emerging cornerstone in cholangiocarcinoma therapy? Liver Cancer. 2021;10(6):545–560. doi: 10.1159/000518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afshar S., Sedighi Pashaki A., Najafi R., et al. Cross-resistance of acquired radioresistant colorectal cancer cell line to gefitinib and regorafenib. Iran. J. Med. Sci. 2020;45(1):50–58. doi: 10.30476/ijms.2019.44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manoochehri H., Jalali A., Tanzadehpanah H., et al. Identification of key gene targets for sensitizing colorectal cancer to chemoradiation: an integrative network analysis on multiple transcriptomics data. J. Gastrointest. Cancer. 2022;53(3):649–668. doi: 10.1007/s12029-021-00690-2. [DOI] [PubMed] [Google Scholar]
- 35.Bergquist A., von Seth E. Epidemiology of cholangiocarcinoma. Best Pract. Res. Clin. Gastroenterol. 2015;29(2):221–232. doi: 10.1016/j.bpg.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Jusakul A., Cutcutache I., Yong C.H., et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7(10):1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto R., Sugiura T., Ashida R., et al. Prognostic value of carbohydrate antigen 19-9 and the surgical margin in extrahepatic cholangiocarcinoma. Annals of gastroenterological surgery. 2022;6(2):307–315. doi: 10.1002/ags3.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Z.L., Ou Y.J., Dai H.S., et al. Elevated preoperative CA125 levels predicts poor prognosis of hilar cholangiocarcinoma receiving radical surgery. Clinics and research in hepatology and gastroenterology. 2021;45(6) doi: 10.1016/j.clinre.2021.101695. [DOI] [PubMed] [Google Scholar]
- 39.Patel A.H., Harnois D.M., Klee G.G., et al. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am. J. Gastroenterol. 2000;95(1):204–207. doi: 10.1111/j.1572-0241.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 40.Loosen S.H., Roderburg C., Kauertz K.L., et al. CEA but not CA19-9 is an independent prognostic factor in patients undergoing resection of cholangiocarcinoma. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-17175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Günther C., Neumann H., Neurath M.F., et al. Apoptosis, necrosis and necroptosis: cell death regulation in the intestinal epithelium. Gut. 2013;62(7):1062–1071. doi: 10.1136/gutjnl-2011-301364. [DOI] [PubMed] [Google Scholar]
- 42.Galluzzi L., Vitale I., Aaronson S.A., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou W., Wang Z., Wang F., et al. A metabolism-related 4-lncRNA prognostic signature and corresponding mechanisms in intrahepatic cholangiocarcinoma. BMC Cancer. 2021;21(1):608. doi: 10.1186/s12885-021-08322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z., Yu M., Yan J., et al. PNOC expressed by B cells in cholangiocarcinoma was survival related and LAIR2 could Be a T cell exhaustion biomarker in tumor microenvironment: characterization of immune microenvironment combining single-cell and bulk sequencing technology. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.647209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu M., Xu X., Pan B., et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol. Cancer. 2019;18(1):135. doi: 10.1186/s12943-019-1063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C.C., Zhu W., Li S.J., et al. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome Med. 2020;12(1):77. doi: 10.1186/s13073-020-00773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dasari S., Tchounwou P.B. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma C.X., Janetka J.W., Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol. Med. 2011;17(2):88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diéguez-Martínez N., Espinosa-Gil S., Yoldi G., et al. The ERK5/NF-κB signaling pathway targets endometrial cancer proliferation and survival. Cell. Mol. Life Sci. : CMLS. 2022;79(10):524. doi: 10.1007/s00018-022-04541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker C.J., Mrózek K., Ozer H.G., et al. Gene expression signature predicts relapse in adult patients with cytogenetically normal acute myeloid leukemia. Blood advances. 2021;5(5):1474–1482. doi: 10.1182/bloodadvances.2020003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Fan S., Liu S., et al. LncRNA NOP14-AS1 promotes tongue squamous cell carcinoma progression by targeting MicroRNA-665/HMGB3 Axis. Cancer Manag. Res. 2021;13:2821–2834. doi: 10.2147/CMAR.S293322. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Xu H., Zhao G., Zhang Y., et al. Long non-coding RNA PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression. J. Exp. Clin. Cancer Res. 2019;38(1):486. doi: 10.1186/s13046-019-1474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma Y., Zheng W. H3K27ac-induced lncRNA PAXIP1-AS1 promotes cell proliferation, migration, EMT and apoptosis in ovarian cancer by targeting miR-6744-5p/PCBP2 axis. J. Ovarian Res. 2021;14(1):76. doi: 10.1186/s13048-021-00822-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Xia X., Zhang H., Xia P., et al. Identification of glycolysis-related lncRNAs and the novel lncRNA WAC-AS1 promotes glycolysis and tumor progression in hepatocellular carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.733595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are all available from the TCGA database (https://portal.gdc.cancer.gov/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/).