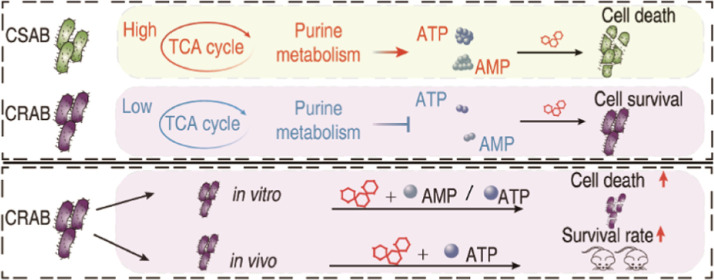
Abstract
Carbapenem-resistant Acinetobacter baumannii (CRAB) strains are prevalent worldwide and represent a major threat to public health. However, treatment options for infections caused by CRAB are very limited as they are resistant to most of the commonly used antibiotics. Consequently, understanding the mechanisms underlying carbapenem resistance and restoring bacterial susceptibility to carbapenems hold immense importance. The present study used gas chromatography–mass spectrometry (GC–MS)-based metabolomics to investigate the metabolic mechanisms of antibiotic resistance in clinically isolated CRAB. Inactivation of the pyruvate cycle and purine metabolism is the most typical characteristic of CRAB. The CRAB exhibited a reduction in the activity of enzymes involved in the pyruvate cycle, proton motive force, and ATP levels. This decline in central carbon metabolism resulted in a decrease in the metabolic flux of the α-ketoglutarate-glutamate-glutamine pathway toward purine metabolism, ultimately leading to a decline in adenine nucleotide interconversion. Exogenous adenosine monophosphate (AMP) and adenosine triphosphate (ATP) enhance the killing efficacy of Meropenem against CRAB. The combination of ATP and Meropenem also has a synergistic effect on eliminating CRAB persisters and the biofilm, as well as protecting mice against peritonitis-sepsis. This study presents a novel therapeutic modality to treat infections caused by CRAB based on the metabolism reprogramming strategy.
Keywords: carbapenem-resistant A. baumannii, reprogramming metabolomics, Meropenem, ATP, TCA cycle
Acinetobacter baumannii is an opportunistic Gram-negative pathogen that causes a wide range of severe nosocomial infections, including hospital-acquired pneumonia, ventilator-associated pneumonia, septicemia, urinary tract infections, meningitis, and gastrointestinal infections.1 It mostly infects patients undergoing invasive procedures and underlying debilitating conditions in the intensive care unit, thus prolonging hospital stays and increasing mortality.2,3 More importantly, multidrug-resistant A. baumannii causes a more severe situation because it is not sensitive to at least three classes of antibiotics. At this time, carbapenems, either as monotherapy or in combination with other pharmacological agents, were employed for the treatment of A. baumannii-induced infections.4
Carbapenems are β-lactam antibiotics that exhibit a wide range of antimicrobial activities and demonstrate exceptional efficacy. It is often used to treat patients with severe infections triggered by multidrug-resistant organisms.5 With increasing use, especially inappropriate and irrational use, however, an increase in carbapenem-resistant A. baumannii (CRAB) has become a major concern worldwide.6,7 In China, the occurrence rate of CRAB ranges from 60 to 70% in clinical strains obtained from diverse specimens.8 Moreover, a recent systematic review has reported that 90% of clinically isolated A. baumannii strains are CRAB in some areas surrounding the Mediterranean.9 CRAB tends to be resistant to almost all other antibiotics. Therefore, restoring carbapenem killing is especially important for eliminating A. baumannii.
Recently, reprogramming metabolomics has emerged as a potent approach to reveal the metabolic mechanisms of antibiotic resistance and develop new treatment strategies to restore the susceptibility of existing antibiotics that are already resistant to combating antibiotic-resistant bacteria.10 The susceptibility of bacteria to antibiotics is contingent upon their metabolic state, and the optimization of antibiotic effectiveness can be achieved through the manipulation of bacterial metabolic states.11 Reprogramming metabolomics identifies key biomarkers by understanding the metabolic mechanisms of antibiotic resistance, uses the key biomarkers to reverse the antibiotic-resistant state into an antibiotic-sensitive state for elevating antibiotics that already are resistant and explores the reprogramming mechanisms.12 Based on metabolic reprogramming, programming metabolites like glucose, alanine, or glutamate are used to reverse the antibiotic resistance of Edwardsiella tarda, Escherichia coli, Vibrio alginolyticus, or Pseudomonas aeruginosa by promoting the pyruvate cycle (the P cycle), increasing the production of NADH and proton motive force (PMF), and stimulating aminoglycoside uptake.13−15 Further evidence shows that glutamine potentiates ampicillin killing in multidrug-resistant E. coli(16) and glucose and manganese ions delay the transition of bacterial tolerance to resistance to ampicillin.17
The metabolomics of antibiotic-resistant A. baumannii have been reported in recent years,18 however, the use of a reprogramming metabolomics approach to restore the susceptibility of existing antibiotics against the bacteria has not been studied. Furthermore, information regarding whether the reprogramming metabolomics approach could restore carbapenem killing is not available. Here, GC–MS-based metabolomics was used to compare clinically isolated carbapenem-susceptible A. baumannii (CSAB) and CRAB. This leads to the findings that downregulation of the pyruvate cycle (the P cycle) and purine metabolism in CRAB and elevation of Meropenem-mediated killing of CRAB by exogenous AMP and ATP.
Results
Phenotypes of Clinically-Isolated CSAB and CRAB
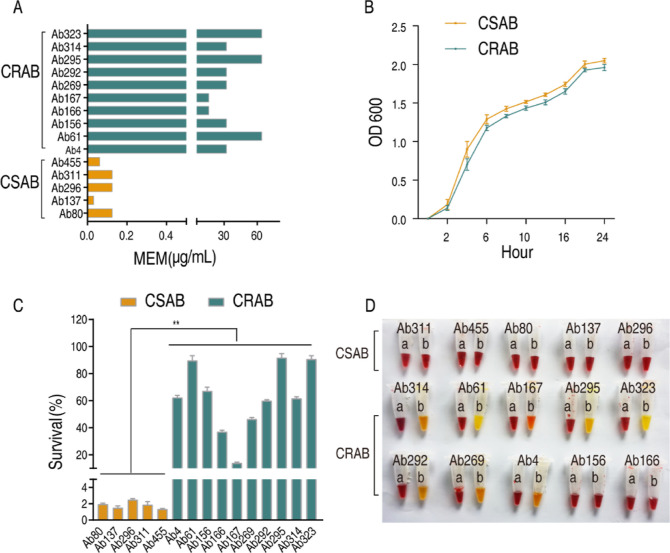
To validate CSAB and CRAB obtained from the hospital, the minimum inhibitory concentration (MIC) against Meropenem was measured in 5 strains of CSAB and 10 strains of CRAB. The MIC values ranged between 0.03125 and 0.125 μg/mL in CSAB and 16–64 μg/mL in CRAB (Figure 1A), indicating the reliability of these strains to Meropenem sensitivity/resistance. Additionally, the MICs of these bacteria against other various antibiotics were assessed, and then the bacteria were categorized according to drug resistance characteristics as MDR or XDR. The results are shown in Table S1. Then, growth curve, survival capability, and carbapenemase were detected in these strains. The growth rate of CRAB closely approximates that of CSAB (Figures 1B and S1). Survival capability was higher in CRAB than in CSAB when they were incubated in LB medium with 8 μg/mL Meropenem for 6 h (Figure 1C). The CarbAcineto NP method was used to detect carbapenemase, which is one of the most significant mechanisms of carbapenem resistance in A. baumannii. As expected, 80% of CRAB showed the activity of carbapenemase, whereas all CSAB was negative for this enzyme (Figure 1D). These results indicate that CRAB and CSAB have differential drug-resistant phenotypes.
Figure 1.
Phenotypes of clinically isolated CSAB and CRAB. (A) Minimal inhibitory concentration (MIC) of clinically isolated A. baumannii to Meropenem. (B) Growth curves of CSAB and CRAB. (C) Survival capability of CSAB and CRAB under the indicated concentration of Meropenem (8 μg/mL). (D) Carbapenemase was detected using the CarbAcineto NP method. Results are displayed as means ± SD. Significant differences are identified **P < 0.01.
Metabolic Profile of Clinically Isolated CSAB and CRAB
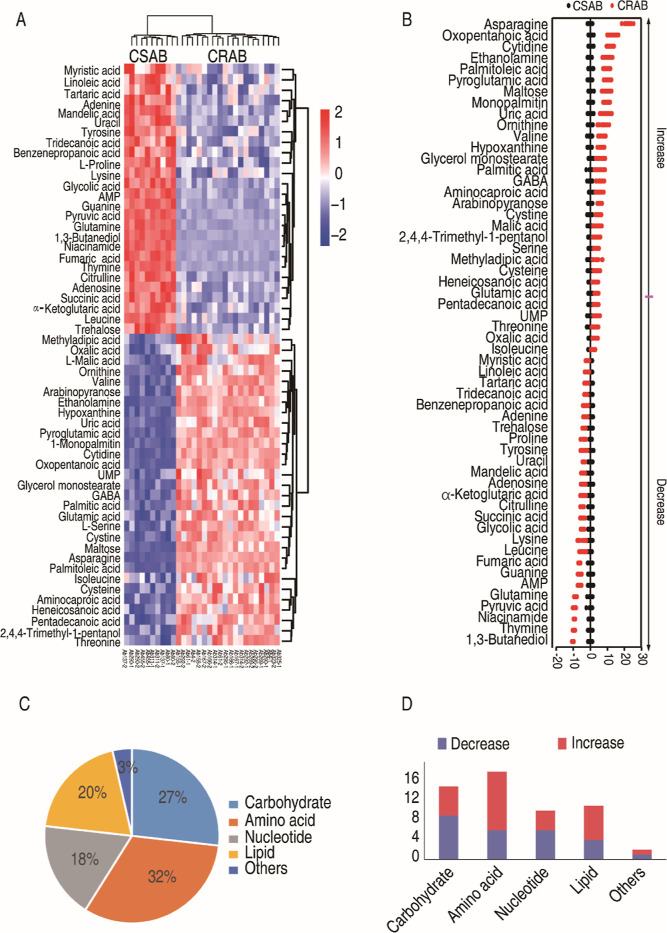
The metabolic profiles of 5 strains of CSAB and 10 strains of CRAB were investigated using GC–MS-based metabolomics. For each sample, we performed two technical replicates and determined the correlation coefficients between them. The correlation coefficients for the technical replicates ranged from 0.96 to 0.99 (Figure S2B), demonstrating the high reliability of the technical repetitions and the stability of the instrument platform. In each sample, a cumulative count of 203 aligned peaks was acquired (Figure S2A). After the internal standard ribitol was eliminated and any known artificial peaks were excluded, the compounds were integrated, resulting in the identification of 109 metabolites in each sample. The metabolites were categorized into five distinct groups, specifically carbohydrate (33%), amino acid (19%), nucleotide (10%), lipid (19%), and others (19%) (Figure S2C). As shown in the heat map of the total metabolites, CRAB and CSAB were clustered independently (Figure S2D). These results suggest that CRAB and CSAB exhibit distinct metabolic profiles. The Kruskal–Wallis test (P < 0.05) was employed to ascertain metabolites exhibiting differential abundance between CRAB and CSAB. Out of the 109 metabolites that were examined, a total of 56 metabolites exhibited notable variations in their levels of abundance (Figure 2A). Z scores were calculated to compare the relative abundance of metabolites based on CSAB. The Z scores ranged from −10.1 to 21.9. Among the 56 differential abundances of metabolites, 26 were down-regulated and 30 were up-regulated in CRAB (Figure 2B). They were classified as 27% carbohydrate, 32% amino acid, 18% nucleotide, 20% lipid, and 3% others (Figure 2C). The quantities of downregulated and upregulated metabolites within each category are presented in Figure 2D. These results indicate that CRAB displays specific metabolic characteristics that may contribute to carbapenem resistance.
Figure 2.
Differential metabolomes between clinically isolated CSAB and CRAB. (A) Heat maps of the differential metabolites (row). Blue and red colors represent low to high abundance relative to the median value (see color scale). (B) Plot of the distribution of the Z score. Each point corresponds to a single technical replicate for a specific metabolite (black, CSAB; red, CRAB). (C) Categories of the differential metabolites in clinically isolated CRAB. (D) The quantities of downregulated and upregulated metabolites in the CRAB group compared to the CSAB group.
Inactivation of the P Cycle and Purine Metabolism Contributes to Carbapenem Resistance in Clinically Isolated A. baumannii
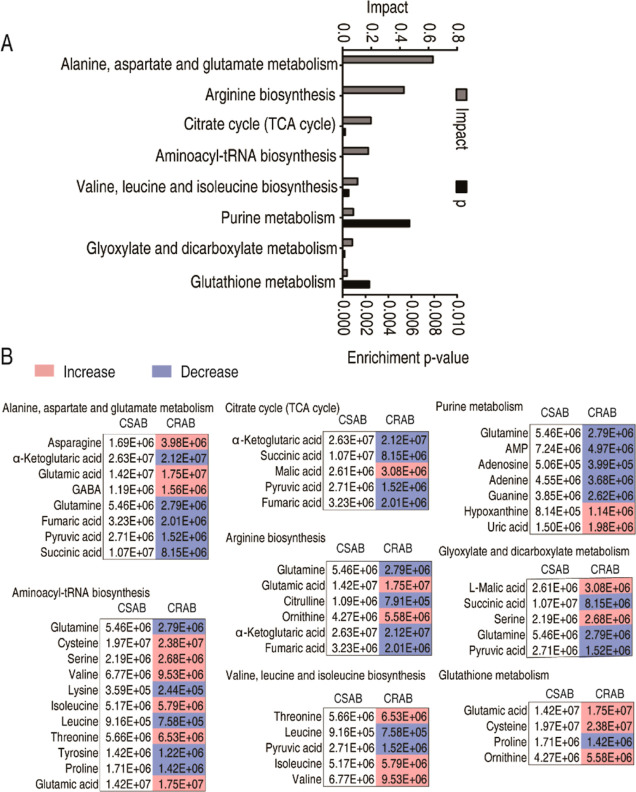
MetaboAnalyst 5.0 was used to enrich metabolic pathways. A total of 8 metabolic pathways were determined (Figure 3A). The most impactful pathway was alanine, aspartate, and glutamate metabolism, followed by arginine biosynthesis; citrate cycle (TCA cycle); aminoacyl-tRNA biosynthesis; valine, leucine, and isoleucine biosynthesis; purine metabolism; glyoxylate and dicarboxylate metabolism; and glutathione metabolism. The abundance of metabolites in enriched pathways is shown in Figure 3B. Among them, most of the metabolites in the TCA cycle, purine metabolism, and arginine biosynthesis were reduced. Notably, the TCA cycle provides sources for purine metabolism and arginine biosynthesis, suggesting that the reduced TCA cycle is responsible for the reduction in purine metabolism and arginine biosynthesis. A recent study has proven that the P cycle rather than the TCA cycle provides respiratory energy for bacteria, and the P cycle consists of the TCA cycle plus the phosphoenolpyruvate (PEP)–pyruvate–AcCoA pathway.19 Therefore, the TCA cycle is replaced with the P cycle in the following study.
Figure 3.
Analysis of the pathway enrichment in clinically isolated CRAB. (A) Pathway enrichment of differential metabolites. (B) Abundance of metabolites in the enriched pathway. The metabolites in the CRAB group were compared to those in the CSAB group. Blue and light red colors represent low and high abundance, respectively.
Identification of Crucial Biomarkers in CRAB
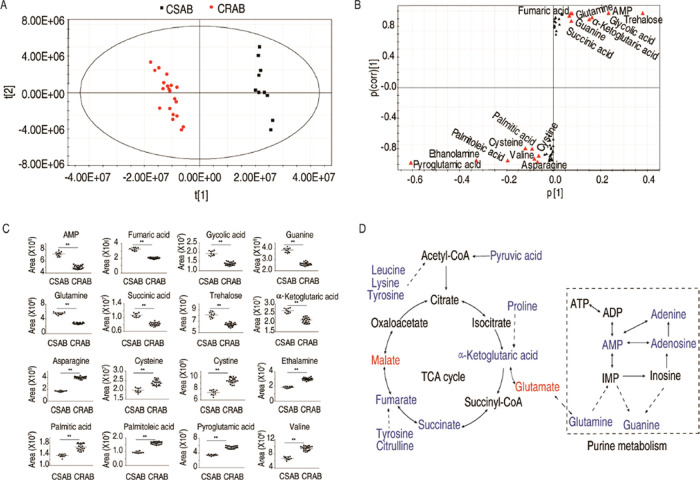
After obtaining the differential metabolites and related pathways through the metabolomics analysis, we identified crucial biomarkers representing the differential metabolomes of CRAB by utilizing orthogonal partial least-squares discriminant analysis (OPLS-DA). Principal component analysis (PCA) revealed that component t [1] was able to differentiate CRAB group from the CSAB group (Figure 4A). The S-plot analysis identified 16 discriminant variables (AMP, asparagine, cysteine, cystine, ethanolamine, fumarate, glycolic acid, guanine, palmitic acid, palmitoleic acid, pyroglutamic acid, glutamine, succinic acid, trehalose, valine, and α-ketoglutarate) (Figure 4B). The abundance of these potential biomarkers was compared by using a scatter plot (Figure 4C). A total of 8 substances were down-regulated, 6 of which belonged to the P cycle (fumaric acid, succinic acid, and α-ketoglutaric acid) and purine metabolism (AMP, guanine, and glutamine) (Figure 4D). The aforementioned findings suggest that the reduced P cycle and purine metabolism are the most characteristic features in CRAB.
Figure 4.
Identification of the crucial biomarkers. (A) Principal component analysis. Each dotted line represents one technique replicate. (B) S-plot generated from OPLS-DA. Potential biomarkers with predictive component p [1] and correlation p(corr) [1] greater than or equal to 0.05 and 0.5 are highlighted in red triangle. (C) Scatter maps are for crucial biomarkers. (D) Global view for metabolite abundance alteration in CRAB. Blue and red represent low to high abundance of metabolites.
Reduced P Cycle Is Responsible for the Reduced Purine Metabolism
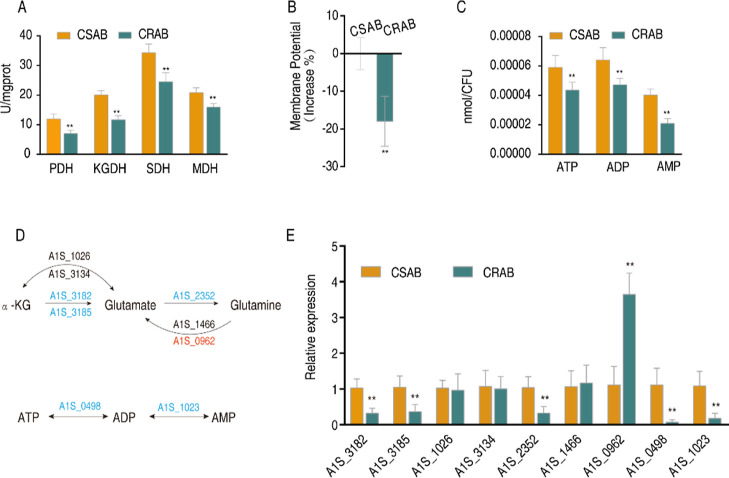
To confirm that the reduced purine metabolism is attributed to the reduced P cycle, first we validated the inactivation of the P cycle through measuring the activity of four key enzymes in the P cycle and its related products, proton motive force (PMF) and ATP. This is because NADH generated by the P cycle powers oxidative phosphorylation to generate PMF, which promotes the synthesis of ATP. The activities of pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), succinate dehydrogenase (SDH), and malate dehydrogenase (MDH) were lower in CRAB than CSAB (Figure 5A). Consistently, lower PMF and ATP levels were measured in CRAB than CSAB (Figure 5B,C). Then, we investigated whether the P cycle limits sources for the purine metabolism. To do this, qRT-PCR was employed to quantify the expression levels of genes responsible for encoding glutamate synthase (A1S_3182 and A1S_3185, transforming α-ketoglutaric acid into glutamate), glutamate dehydrogenase (A1S_1026 and A1S_3134, transforming glutamate into α-ketoglutaric acid reversely), glutamine synthase (A1S_2352, transforming glutamate acid into glutamine), and glutaminase (A1S_1466 and A1S_0962, transforming glutamine into glutamate). Compared to CSAB, CRAB exhibited decreased expression of A1S_3182, A1S_3185, and A1S_2352 and increased expression of A1S_0962 (Figure 5D,E), suggesting that the flux of the P cycle to purine metabolism via the glutamate–glutamine metabolic pathway is impaired. Finally, we bridged the gap between the depressed AMP in the metabolomics approach and the decreased ATP in the P cycle analysis. Similar to ATP, the concentrations of ADP and AMP were lower in CRAB than in CSAB (Figure 5C). qRT-PCR showed that a lower expression of genes encoding adenylate kinase (A1S_1023, catalyzing the mutual transformation of AMP and ADP) and nucleoside-diphosphate kinase (A1S_0498, catalyzing the mutual transformation of ADP and ATP) is detected in CRAB than in CSAB (Figure 5D,E). Taken together, the inactivated P cycle is responsible for the reduced purine metabolism and further affects the transformation of adenine nucleotides.
Figure 5.
Inactivated P cycle is responsible for the reduced purine metabolism. (A) Enzyme activity in the P cycle in CSAB and CRAB. (B) Proton motive force (PMF) values of CSAB and CRAB. (C) ATP, ADP, and AMP level. (D,E) qRT-PCR for expression of genes in the α-ketoglutaric acid–glutamate–glutamine pathway and the AMP–ADP–ATP pathway. Blue and red represent downregulated and upregulated expressions of genes. Results are displayed as means ± SD. Significant differences are identified **P < 0.01.
ATP and AMP Promote Bacterial Sensitivity to Meropenem
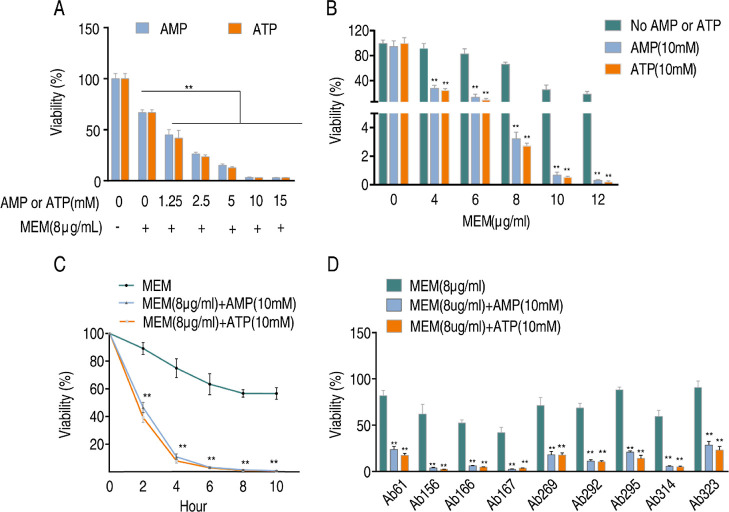
As clinically isolated CRAB showed decreased adenine nucleotide levels, this led us to speculate whether exogenous adenine nucleotides, such as AMP or ATP, could reverse the metabolic state and restore the bacterial susceptibility to carbapenems. To test this assumption, we examined the susceptibility of Ab4, a CRAB strain, to Meropenem in the presence or absence of AMP or ATP using an antibiotic bactericidal assay. As anticipated, the addition of exogenous AMP or ATP did not affect the growth of A. baumannii but promoted Meropenem-mediated killing of the clinically isolated CRAB strain in an ATP and AMP dose-dependent manner (Figure 6A). Furthermore, antibiotic concentration and time-effect experiments were performed for AMP and ATP. The results demonstrated that the killing efficacy increased with higher doses of Meropenem and longer incubation periods (Figure 6B,C). The synergistic effect was also observed in the other 9 CRAB strains (Figure 6D). To further verify the above results, we tested the killing efficacy on 10 additional CRAB strains (MIC for Meropenem ranging from 16 to 64 μg/mL), and the synergistic effect was still observed (Figure S3A,B). Above all, these findings suggest that the synergistic use of AMP or ATP with Meropenem is a feasible approach to combating CRAB, even in strains that have the ability to hydrolyze carbapenems (Figure 1D).
Figure 6.
AMP and ATP promote the bacterial sensitivity to Meropenem. (A) Viability of Ab4 in indicated concentrations of AMP or ATP plus Meropenem. (B) Viability of Ab4 in the indicated concentrations of Meropenem plus AMP or ATP. (C) Viability of Ab4 at specific time points after treatment with Meropenem plus AMP or ATP. (D) Viability of another 9 CRAB strains after treatment with Meropenem plus AMP or ATP. Results are displayed as means ± SD. Significant differences are identified **P < 0.01.
ATP Enhances the Killing Efficacy of Meropenem against CRAB Persisters, Biofilm, and Systemic Infections
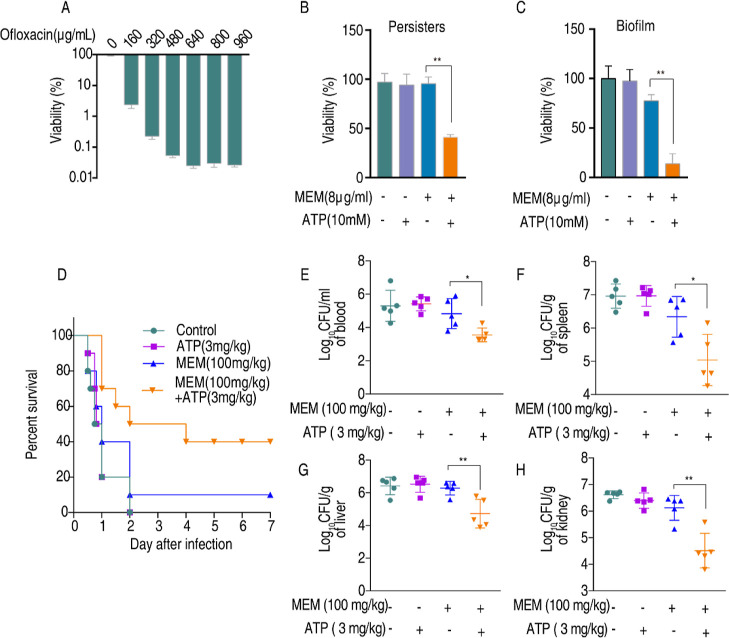
A. baumannii persisters and biofilms are the main cause of recurrent infections. We thus tested the synergistic effect of ATP and Meropenem on Ab4 persisters and biofilm. To do this, persisters were generated by exposing bacteria to increasing concentrations of ofloxacin ranging from 0 to 960 μg/mL. Bacterial survival was decreased with increasing concentrations of ofloxacin until 640 μg/mL ofloxacin was used (Figure 7A), suggesting the remaining cells were persisters in the presence of 640–960 μg/mL ofloxacin. Exogenous ATP enabled the killing of Meropenem-resistant persisters by Meropenem (Figure 7B). The combination of Meropenem and ATP also led to a decrease in the viability of the biofilm (Figure 7C). We further tested the synergistic action of Meropenem with ATP in a mouse model. Mice were infected with CRAB Ab4 via intraperitoneal injection and then received treatment with Meropenem (100 mg/kg) either alone or in combination with ATP (3 mg/kg) 1 h after injection. The survival rate and bacterial loads in various tissues were monitored. All mice treated with saline or ATP died within 48 h and only 10% of mice survived after the injection of Meropenem alone. However, when Meropenem was administered in combination with ATP, 40% of these mice survived (Figure 7D). The bacterial loads in the blood, spleen, liver, and kidneys decreased after the synergy of Meropenem with ATP (Figure 7E–H). These results demonstrate that the combination of Meropenem and ATP is an effective way to eliminate persisters and biofilm. The synergistic effect in mouse peritoneal infection models suggests that concurrent administration of ATP alongside Meropenem represents a viable strategy for combating infections caused by CRAB.
Figure 7.
ATP potentiates Meropenem combat against CRAB persisters, biofilm, and systemic infections. (A) Viability of Ab4 after incubation with an increasing concentration of ofloxacin (0–960 μg/mL). (B) Viability of Ab4 persists in the presence or absence of Meropenem plus ATP for 6 h in vitro. (C) Viability of the Ab4 biofilm in the presence or absence of Meropenem and ATP for 6 h in vitro. (D) Percent survival of mice intraperitoneally challenged with 7 × 107 cfu Ab4 and then treated with saline solution, ATP (3 mg/kg), Meropenem (100 mg/kg), or both 1 h after infection (n = 10 per group). (E) Bacterial load in mice blood, spleen (F), liver (G), and kidney (H) after treatment with saline solution, ATP (3 mg/kg), Meropenem (100 mg/kg), or both for 6 h (n = 5 per group). Results are displayed as means ± SD. Significant are identified *P < 0.05 and **P < 0.01.
Discussion
The widespread use of carbapenems has led to a significant increase in the global prevalence of CRAB.20,21 Conventional resistance mechanisms, decreased membrane permeability, overexpressed efflux, and altered penicillin-binding protein contribute to the resistance of CRAB, but the main resistance mechanism of CRAB is thought to be the hydrolysis of carbapenems by carbapenemases.22 To overcome the carbapenemase-mediated mechanism, a combination of a β-lactam/β-lactamase inhibitor approach is developed.23 However, this approach does not cover all carbapenemases. Moreover, resistance to β-lactamase inhibitors is reported in CRAB and extensively drug-resistant (XDR) A. baumannii.24,25 Hence, there is an urgent need to explore new methods for treating CRAB infections. The present study uses the recently developed reprogramming metabolomics approach to explore a method for potentiating Meropenem to kill CRAB. This leads to the identification of ATP and AMP as metabolome-reprogramming molecules. When Meropenem and ATP or AMP are synergistically used, killing efficacy for CRAB is promoted. Our results indicate that the recently developed reprogramming metabolomics approach is effective in eliminating CRAB and that ATP and AMP can potentiate Meropenem-mediated killing.
Modulating the metabolic state of bacteria is a potential strategy to combat antibiotic-resistant bacteria because of the close relationship between changes in the bacterial state and the development of antibiotic resistance.26 A previous study utilizing transcriptomics and proteomics analysis showed that A. baumannii exhibits an increased ability to catabolize phenylacetic acid under subinhibitory concentrations of trimethoprim and sulfamethoxazole and increase susceptibility to antibiotics when this process is interfered.27 It was also demonstrated that long-chain polyunsaturated fatty acids affect the structure and function of the efflux system, thereby slowing down the development of tetracycline resistance in A. baumannii.28 However, it is not yet clear whether these metabolites have therapeutic effects for multidrug-resistant A. baumannii. Fortunately, recently developed reprogramming metabolomics shows that crucial biomarkers could reverse biological phenotypes, including antibiotic resistance.29−31 While a depressed nitrite-dependent NO biosynthesis is determined in cefoperazone-sulbactam-resistant P. aeruginosa, the addition of nitrite or nitrate significantly enhances the bactericidal activity of cefoperazone-sulbactam.32 Glutamine, a depressed metabolite in multidrug-resistant E. coli, potentiates conventional antibiotics to kill S2-lac that harbors a plasmid expressing a β-lactamase via a glutamine-inosine-CpxA/CpxR-OmpF regulatory pathway. The aforementioned findings suggest that reprogramming metabolomics is a feasible approach to exploring the metabolic profile of clinically isolated CRAB and eventually identifying a potential biomarker that increases the susceptibility of CRAB to carbapenems.
To carry out the reprogramming metabolomics approach, we conducted a metabolic comparison between CRAB and CSAB. The metabolic profile of CRAB is distinctly separated from that of CSAB, supporting the conclusion that antibiotic-resistant and antibiotic-susceptible bacteria exhibit differential metabolomes. The carbapenem-resistant metabolome is characterized by the downregulation of most metabolites in the enriched P cycle and purine metabolism and the identification of these downregulated metabolites as biomarkers. The P cycle generates a significant amount of NADH/FADH2, which supplies electrons for oxidative phosphorylation, while purine metabolism provides substrates for ATP synthases. The inactivation of the P cycle and purine metabolism implies a weak ATP synthesis capacity, which is a prominent metabolic characteristic of CRAB. To substantiate this hypothesis, we conduct qRT-PCR, enzyme activity measurement, and purine nucleotide content detection. The CRAB samples exhibit reduced enzyme activity in the P cycle as well as a decreased PMF level. Additionally, we observe downregulated gene expression in the α-ketoglutaric acid–glutamate–glutamine and AMP–ADP–ATP pathways, which ultimately resulted in lower ATP contents within CRAB.
ATP is the main energy carrier that is essential for cellular metabolism in living cells. A recent study has indicated that a decrease in cellular ATP levels results in a reduction of antibiotic target activity and leads to the formation of persisters.33 The cellular ATP level regulates the bacterial dormancy depth.34 However, the role of extracellular ATP in bacteria is still unclear. Li et al.35 found that ATP treatment increases antibiotic production and morphological differentiation of Streptomyces coelicolor without affecting its growth. Tatano et al.36 demonstrated that ATP exhibits antimicrobial action in various bacteria, including Staphylococcus, Pseudomonas, and Mycobacteria, by inhibiting iron uptake. According to another study, ATP has been found to impede the proliferation of rapidly multiplying microorganisms, such as Pseudomonas syringae, while simultaneously fostering the growth of actinomycetes within compost.37 The growth of A. baumannii appears to be unaffected by treatment with exogenous ATP, which is consistent with previous research.38 It seems that the effect of exogenous ATP on bacterial growth is strain-specific.
ATP and AMP are nucleotides that play important roles in cellular metabolism and energy production.39 We show that both ATP and AMP potentiate Meropenem to kill CRAB without affecting the growth of A. baumannii. Moreover, the combination of Meropenem and ATP exhibits a synergistic therapeutic effect against persisters, biofilms, and systemic inflammation in a peritonitis-sepsis model. ATP is considered a potential therapeutic molecule against various diseases, including paroxysmal supraventricular tachycardia, Alzheimer’s disease, osteoarthritis pain, and depression.40 The combination of Meropenem with ATP potentially offers an additional therapeutic option for treating CRAB infections. Recent studies have reported that metabolites such as lysine and oleanolic acid enhance the killing efficacy of aminoglycosides against multidrug-resistant A. baumannii in a PMF-dependent manner.41,42 Unfortunately, the clinical use of aminoglycosides is limited by their renal toxicity.43 This is the first time that we have shown the metabolites that reverse Meropenem resistance in clinical CRAB isolates. Further investigations are necessary to understand the mechanism by which ATP and AMP enhance the killing potency of Meropenem.
Conclusions
In summary, GC–MS-based metabolomics was adopted to explore the metabolic profile of CRAB and CSAB, where the inactivation of the P cycle and purine metabolism are identified as the most characteristic features of CRAB, which leads to decreased adenine nucleotide levels. Exogenous ATP and AMP potentiate Meropenem to kill clinically isolated CRAB. This study establishes a novel strategy for eradicating bacterial infections and highlights the potential of ATP and AMP as promising adjuvant candidates for Meropenem in combating CRAB.
Materials and Methods
Bacterial Strains and Culture Conditions
The carbapenem-susceptible A. baumannii (CSAB) and carbapenem-resistant A. baumannii (CRAB) strains employed in the experiment were acquired from the respiratory tract specimens of hospitalized patients in our hospital. These strains were meticulously preserved at a temperature of −80 °C. For the experiment, strains obtained from glycerol stocks were streaked onto LB agar plates, and subsequently, single clones were inoculated into fresh LB medium containing 1% bacterial peptone, 0.5% yeast extract, and 1% sodium chloride. The incubation process was conducted at a temperature of 37 °C with continuous shaking at 200 rpm. All strains used in the experiment are listed in Table S2.
Measurement of MIC
The MIC was determined using microdilution in accordance with the guidelines provided by the Clinical and Laboratory Standards Institute.44 In brief, bacteria that had been saturated overnight were transferred to a fresh LB medium in a ratio of 1:100 and cultivated until the optical density at 600 nm (OD600) reached 0.5. Then, bacteria equivalent to 0.5 × 105 cfu/mL were exposed to a series of concentration of antimicrobial agents diluted by LB medium in 96-well cell culture plates and incubated at 37 °C for 16 h. The lowest antibiotic concentration that inhibited visible growth was defined as the MIC. Three biological replicates were performed.
Measurement of the Growth Curve
Overnight cultures of bacteria were inoculated into 250 mL of fresh LB medium at a dilution ratio of 1:100. Then, bacteria were transferred into sterilized test tubes at a volume of 5 mL per tube and incubated at 37 °C while being shaken at 200 rpm. OD600 values were recorded at specific time intervals. The growth curves were constructed by using the OD600 values obtained at various time intervals for the bacteria.
Survival Capability
Tests of survival capability were conducted as previously reported.45 The overnight-saturated bacteria were diluted 1:1000 into test tubes containing 5 mL of fresh LB medium. Each strain was transferred to 6 separate tubes, with 3 tubes devoid of antibiotics and 3 tubes containing antibiotics (Meropenem 8 μg/mL). After incubation for 6 h, the OD600 was measured. The survival rate was determined by calculating the ratio of the OD values between strains with and without antibiotics.
CarbAcineto NP Test
The method of the CarbAcineto NP test refers to the previous research.46 In brief, a fully calibrated loop of bacterial inoculum was resuspended into two Eppendorf tubes containing 100 μL of 5 M NaCl. In tube a, 100 μL of the revealing solution (composed of 16.6 mL of ddH2O, 2 mL of 0.5% phenol red solution, and 180 μL of 10 mM zinc sulfate heptahydrate, pH 7.8) was added. In tube b, an additional imipenem (6 mg/mL) was added. Then, the tubes were subjected to incubation at 37 °C for a maximum duration of 2 h. The interpretation of the results was as follows: the presence of a red color in both tubes signifies a negative carbapenemase outcome, while a red color in tube a and a yellow/orange color in tube b indicate a positive carbapenemase outcome. In the event that both tubes exhibit a yellow-orange color, the result is deemed inconclusive.
Metabolomic Profiling
Metabolomic samples were prepared according to a previous method. Briefly, stationary-phase bacteria were diluted 1:100 with fresh LB medium and recultivated until an OD600 of 1.0. An aliquot of 10 mL of the cultures was then collected and suspended in 1 mL of precooled methanol (−80 °C) for the extraction of cellular metabolites, with 10 μL of 0.1 mg/mL ribitol (Sigma-Aldrich) serving as an internal standard. Cells were then lysed by sonication for 5 min over an ice-water mixture and centrifuged at 12,000 rpm at 4 °C for 10 min. Following this, 500 μL of supernatants were removed and dried using a rotary vacuum centrifuge device (Labconco) at 37 °C. A total of 80 μL of 20 mg/mL oxime hydrochloride (Sigma-Aldrich) in pyridine was added to protect the carbonyl moieties of the samples through methoximation for 90 min at 37 °C. After that, 80 μL of N-methyl-N-trimethylsilytrifluoroacetamide (MSTFA, Sigma-Aldrich) was used for derivatization at 37 °C for 30 min. The obtained samples were detected by an Agilent 7890A GC and an Agilent 5975C inert XL mass selector (Agilent Technologies).
GC–MS Data Analysis
The mass fragmentation spectrum in XCalibur software (Thermo Fisher; version 2.1) was compared to the National Institute of Standards and Technology (NIST) Mass Spectral Library to identify the metabolites. A metabolite was deemed statistically significant if its p value was less than 0.05. Hierarchical clustering was performed in the R platform. Principal component analysis and orthogonal partial least-squares discriminant analysis were conducted using SIMCA-P + 12.0 software (Umetrics, Umea, Sweden). Additionally, the enrichment of pathways associated with differential metabolites was carried out using MetaboAnalyst version 5.0 (http://www.metaboanalyst.ca).
Measurement of Membrane Potential
Membrane potential was determined by referring to the previous report.47 The procedure involved serial dilution of bacterial samples to approximately 106 cfu/mL, followed by the addition of 10 μL of 3 mM 3,3′-diethyloxacarbocyanine iodide ([DIOC2(3), Invitrogen]) to an Eppendorf tube containing 1 mL of the diluted bacterial solution. After incubation at 37 °C for 30 min, a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) was employed for detection, with the excitation wavelengths for both green and red fluorescence set at 488 nm and the emission wavelengths set at 530 and 610 nm, respectively. The determination of the membrane potential was based on the fluorescence intensity ratio between red and green.
Measurement of Enzyme Activity
To assess the activities of enzymes in the P cycle, 30 mL of the bacterial cultures of OD 1.0 were obtained and crushed by sonication in 1 mL of PBS for 5 min (2 or 3 s intervals and 35% intensity). Following centrifugation at 12,000 rpm for 10 min, the supernatant was collected to quantify the protein concentration using a BCA protein concentration determination kit (Beyotime Inc., China). A 200 μg portion of protein was utilized for the assessment of enzyme activity. The reaction mixture for PDH and KGDH included 0.15 mM 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 2.5 mM MgCl2, 0.5 mM phenazine methosulfate (PMS), 0.2 mM thiamine pyrophosphate (TPP), 50 mM PBS, 2 mM sodium pyruvate/5 mM α-ketoglutaric acid potassium salt, and ddH2O to a final volume of 200 μL. The reaction mixture for SDH and MDH consisted of 0.15 mM MTT, 1 mM PMS, 50 mM PBS, 2 mM sodium succinate/50 mM sodium malate, and ddH2O to a final volume of 200 μL. Following incubation at 37 °C for the specified duration (5 min for PDH and SDH, 20 min for KGDH and MDH), the absorbance at OD566 was measured. Enzyme activity was determined by using a standard curve.
Measurement of Adenine Nucleotide Levels
In summary, bacteria were resuspended in 600 μL of 80 °C absolute ethanol and incubated in an 80 °C water bath for 10 min. Subsequently, 1.4 mL of buffer solution (composed of 2.5 mL of 1 M Tris, 5 mL of 100 mM MgSO4, 1 mL of 100 mM EDTA, 41.5 mL of ddH2O, and 300 μL of 5 M NaOH) was added, followed by centrifugation at 12,000 rpm for 2 min. The resulting supernatant was utilized for the determination of adenine nucleotide through a luciferin–luciferase bioluminescence assay. To measure ATP, a 10 μL reaction buffer was added, consisting of 3.75 mL of 1 M Tris, 1.25 mL of 200 mM MgCl2, 6.25 mL of 100 mM KCl, and 37.5 mL of ddH2O, and adjusted to a pH of 7.75. No ATP-generating enzyme was included in this buffer. To measure ADP, a reaction buffer containing pyruvate kinase (Sigma-Aldrich) and phosphoenolpyruvate (Sigma-Aldrich) was added. To measure AMP, a reaction buffer containing pyruvate kinase, phosphoenolpyruvate, and adenylate kinase (Sigma-Aldrich) was added. The quantity of ADP was determined by subtracting the ATP value from the combined ATP + ADP value, while the quantity of AMP was computed based on the disparity between the ATP + ADP + AMP content and the ATP + ADP content.
Quantitative Real-Time PCR
The RNA extract from the bacteria (1 mL, OD600 = 1.0) was prepared by lysis with TRIzol Reagent (Invitrogen Life Technologies), chloroform separation, and isopropyl alcohol precipitation. Total RNA (1 μg) was subjected to RT-PCR using an EvoM-MLV RT kit with gDNA Clean for qPCR II (Accurate Biotechnology). Primers used in qPCR are listed in Table S3. The qRT-PCR was carried out using the SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology) on a 384-well optical plate with a final reaction volume of 10 μL. The reaction was performed on a LightCycler 480 system (Roche, Germany). The cycling parameters were as follows: 95 °C for 30 s to activate the polymerase, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing at 60 °C for 30 s. Data was calculated by using the 2–ΔΔCT method with 16S rRNA as a reference.
Antibiotic Bactericidal Assay
The antibacterial assay was conducted following previously established protocols. A single colony was inoculated into 100 mL of LB medium in 250 mL flasks and grown for 16 h at 37 °C. The bacterial culture was harvested and centrifuged at 8000 rpm for 5 min. The resulting pellets were washed three times with saline, followed by resuspension in M9 buffer containing NaAc (10 mM), MgSO4 (2 mM), and CaCl2 (0.1 mM) and adjusted to an OD600 of 0.2. The samples were then supplemented with Meropenem (Solarbio) and AMP or ATP (Sigma-Aldrich), followed by incubation at 37 °C for 6 h. To determine bacterial counts, 100 μL of the samples underwent a 10-fold serial dilution, and subsequently, 5 μL of each dilution was applied onto 2% LB agar plates for cultivation at 37 °C for 18 h. The viability was assessed by calculating the ratio of clone numbers observed in the treated group compared with the control group.
Persister Assay
Persisters were generated and isolated, as described by Allison.48 Briefly, a single clone of Ab4 was grown overnight to saturation. Samples were collected, subsequently washed with saline 3 times, and then subjected to centrifugation at 8000 rpm for 5 min. The resulting pellets were resuspended in PBS at an OD600 of 0.8 and incubated with increasing concentrations (0–960 μg/mL) of ofloxacin (Sangon Biotech) for 4 h. The remaining cells that could not be eradicated by increasing the concentration of ofloxacin were persisters. After washing with saline three times, the persisters were resuspended in M9 medium and adjusted OD600 = 0.2. 5 mL of the persisters plus Meropenem with or without ATP was added, and the samples were incubated at 37 °C for 6 h. 100 μL aliquots of the samples were removed and serially diluted with saline solution. 5 μL aliquots of the diluted samples were spot-plated onto LB agar plates to count the colonies and determine viability.
Biofilm Assay
A biofilm assay was performed, as previously described. Stationary-phase bacteria were diluted 1:100 in an EP tube containing 1 mL of fresh LB medium, and then 6 mm PE50 catheters (0.58 mm × 0.96 mm) were added. Samples were incubated at 37 °C for 3 days, with culture medium being replaced every 24 h. The resulting catheters were washed with saline 5 times to remove the unattached and planktonic bacteria. Then M9 medium plus Meropenem with or without ATP was added and incubated at 37 °C for 6 h. Samples were then sonicated in 1 mL of saline by an ultrasonic cleaner (Elmasonic) at 40 kHz for 30 min to detach the biofilm cells. Serial dilutions and spot plating were performed, and the viability was calculated, as described above.
Mouse Infection Assay
Male BALB/c mice were obtained from the Animal Center of Sun Yat-sen University (Guangzhou, China). They were housed in rooms with suitable humidity and temperature and acclimatized for 1 week. To investigate the potential impact of exogenous ATP and Meropenem on a murine model of septic infection, a total of 40 mice were intraperitoneally administered with 0.2 mL of log-phase bacteria (Ab4, equivalent to 7 × 107 cfu) suspended in a saline solution containing 5% mucin. One hour later, they were treated with saline (n = 10), 3 mg/kg ATP (n = 10), 100 mg/kg Meropenem (n = 10), and 3 mg/kg ATP plus 100 mg/kg Meropenem (n = 10). Survival status was monitored over a period of seven consecutive days. To further assess the bacterial loads in mice after treatment with ATP and/or Meropenem, 20 mice were infected with A. baumannii, as described above, and divided into 4 groups, with 5 mice in each group. The blood, spleen, kidney, and liver were separately collected and homogenized after treatment for 6 h. The supernatant was serially diluted with a saline solution and spot-plated onto LB agar plates to quantify viable bacterial cells.
Statistical Analysis
Statistical analysis was conducted by using IBM SPSS Statistics 20.0 software. The data are presented as means ± SD. An unpaired t-test was employed for statistical analysis in the in vitro study, while the Mann–Whitney U test was used for the in vivo study. All experiments were performed with a minimum of three biological replicates. Adobe Illustrator CS6 and GraphPad Prism 8 were utilized for figure editing.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.3c00480.
Growth curves and metabolic profile of A. baumannii; antibacterial assay of AMP or ATP with Meropenem for 10 additional CRAB strains; MIC of A. baumannii to other antibiotics; all bacteria strains used in the experiment; and primers for qPCR (PDF)
Author Contributions
X.L., D.F., and J.Z. contributed equally. T.Z. conceived and designed the research. X.L., D.F., and J.Z. conducted experiments and wrote the manuscript. W.W., W.Z., and W.G. analyzed the data. M.J. contributed to analytical tools. X.P. and H.L. revised the manuscript.
This study received financial support from the National Natural Science Foundation of China (grant number: 82170014) and the Guangdong Basic and Applied Basic Research Foundation (grant number: 2022A1515220008).
All data sets generated and analyzed during this study are included in the article; further inquiries can be directed to the corresponding author.
The authors declare no competing financial interest.
Notes
Ethical approval: The animal studies carried out in this manuscript were granted approval by the Institutional Animal Care and Use Committee of Sun Yat-sen University (approval number: SYSU-IACUC-2020-B126716).
Supplementary Material
References
- Antunes L. C.; Visca P.; Towner K. J. Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- Blanco N.; Harris A. D.; Rock C.; Johnson J. K.; Pineles L.; Bonomo R. A.; Srinivasan A.; Pettigrew M. M.; Thom K. A. the CDC Epicenters Program. Risk Factors and Outcomes Associated with Multidrug-Resistant Acinetobacter baumannii upon Intensive Care Unit Admission. Antimicrob. Agents Chemother. 2018, 62, e01631 10.1128/aac.01631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D. Y.; Zhou Y. Q.; Zou X. L.; Zhou M.; Zhu J. X.; Wang Y. H.; Zhang T. T. Differences in microbial etiology between hospital-acquired pneumonia and ventilator-associated pneumonia: a single-center retrospective study in Guang Zhou. Infect. Drug Resist. 2019, 12, 993–1000. 10.2147/IDR.S204671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S.; Al-Saryi N.; Al-Kadmy I.; Aziz S. N. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 2021, 48, 6987–6998. 10.1007/s11033-021-06690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrier J.; Timsit J. F. Carbapenem use in critically ill patients. Curr. Opin. Infect. Dis. 2020, 33, 86–91. 10.1097/QCO.0000000000000622. [DOI] [PubMed] [Google Scholar]
- Murray C. J. L.; et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. S.; Bonomo R. A.; Tolmasky M. E. Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 2020, 10, 720. 10.3390/biom10050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Huang Y.; Zhang T. T.; Cao B.; Wang H.; Zhuo C.; Ye F.; Su X.; Fan H.; Xu J. F.; Zhang J.; Lai G. X.; She D. Y.; Zhang X. Y.; He B.; He L. X.; Liu Y. N.; Qu J. M. Chinese guidelines for the diagnosis and treatment of hospital-acquired pneumonia and ventilator-associated pneumonia in adults (2018 Edition). J. Thorac. Dis. 2019, 11, 2581–2616. 10.21037/jtd.2019.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; McClean S. Mapping Global Prevalence of Acinetobacter baumannii and Recent Vaccine Development to Tackle It. Vaccines 2021, 9, 570. 10.3390/vaccines9060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B.; Li H.; Peng X. X. Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell 2015, 6, 628–637. 10.1007/s13238-015-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J. M.; Lopatkin A. J.; Lobritz M. A.; Collins J. J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. 10.1016/j.cmet.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B.; Su Y. B.; Li H.; Han Y.; Guo C.; Tian Y. M.; Peng X. X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 2015, 21, 249–262. 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Yang M. J.; Peng B.; Peng X. X.; Li H. Reduced ROS-mediated antibiotic resistance and its reverting by glucose in Vibrio alginolyticus. Environ. Microbiol. 2020, 22, 4367–4380. 10.1111/1462-2920.15085. [DOI] [PubMed] [Google Scholar]
- Tang X. K.; Su Y. B.; Ye H. Q.; Dai Z. Y.; Yi H.; Yang K. X.; Zhang T. T.; Chen Z. G. Glucose-Potentiated Amikacin Killing of Cefoperazone/Sulbactam Resistant Pseudomonas aeruginosa. Front. Microbiol. 2022, 12, 800442. 10.3389/fmicb.2021.800442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. B.; Peng B.; Han Y.; Li H.; Peng X. X. Fructose restores susceptibility of multidrug-resistant Edwardsiella tarda to kanamycin. J. Proteome Res. 2015, 14, 1612–1620. 10.1021/pr501285f. [DOI] [PubMed] [Google Scholar]
- Zhao X. L.; Chen Z. G.; Yang T. C.; Jiang M.; Wang J.; Cheng Z. X.; Yang M. J.; Zhu J. X.; Zhang T. T.; Li H.; Peng B.; Peng X. X. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 2021, 13, eabj0716 10.1126/scitranslmed.abj0716. [DOI] [PubMed] [Google Scholar]
- Jiang M.; Su Y. B.; Ye J. Z.; Li H.; Kuang S. F.; Wu J. H.; Li S. H.; Peng X. X.; Peng B. Ampicillin-controlled glucose metabolism manipulates the transition from tolerance to resistance in bacteria. Sci. Adv. 2023, 9, eade8582 10.1126/sciadv.ade8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamad Maifiah M. H.; Cheah S. E.; Johnson M. D.; Han M. L.; Boyce J. D.; Thamlikitkul V.; Forrest A.; Kaye K. S.; Hertzog P.; Purcell A. W.; Song J.; Velkov T.; Creek D. J.; Li J. Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci. Rep. 2016, 6, 22287. 10.1038/srep22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y. B.; Peng B.; Li H.; Cheng Z. X.; Zhang T. T.; Zhu J. X.; Li D.; Li M. Y.; Ye J. Z.; Du C. C.; Zhang S.; Zhao X. L.; Yang M. J.; Peng X. X. Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, E1578 10.1073/pnas.1714645115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versporten A.; Zarb P.; Caniaux I.; Gros M. F.; Drapier N.; Miller M.; Jarlier V.; Nathwani D.; Goossens H.; Koraqi A.; et al. Antimicrobial consumption and resistance in adult hospital inpatients in 53 countries: results of an internet-based global point prevalence survey. Lancet Global Health 2018, 6, e619–e629. 10.1016/s2214-109x(18)30186-4. [DOI] [PubMed] [Google Scholar]
- Nguyen M.; Joshi S. G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: a scientific review. J. Appl. Microbiol. 2021, 131, 2715–2738. 10.1111/jam.15130. [DOI] [PubMed] [Google Scholar]
- Kyriakidis I.; Vasileiou E.; Pana Z. D.; Tragiannidis A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. 10.3390/pathogens10030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou Zerdan M.; Al Hassan S.; Shaker W.; El Hajjar R.; Allam S.; Bou Zerdan M.; Naji A.; Zeineddine N. Carbapenemase Inhibitors: Updates on Developments in 2021. J. Clin. Med. Res. 2022, 14, 251–259. 10.14740/jocmr4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-López F.; Martínez-Meléndez A.; Morfin-Otero R.; Rodriguez-Noriega E.; Maldonado-Garza H. J.; Garza-González E. Efficacy and In Vitro Activity of Novel Antibiotics for Infections with Carbapenem-Resistant Gram-Negative Pathogens. Front. Cell. Infect. Microbiol. 2022, 12, 884365. 10.3389/fcimb.2022.884365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naha A.; Vijayakumar S.; Lal B.; Shankar B. A.; Chandran S.; Ramaiah S.; Veeraraghavan B.; Anbarasu A. Genome sequencing and molecular characterisation of XDR Acinetobacter baumannii reveal complexities in resistance: Novel combination of sulbactam-durlobactam holds promise for therapeutic intervention. J. Cell. Biochem. 2021, 122, 1946–1957. 10.1002/jcb.30156. [DOI] [PubMed] [Google Scholar]
- Li L.; Su Y. B.; Peng B.; Peng X. X.; Li H. Metabolic mechanism of colistin resistance and its reverting in Vibrio alginolyticus. Environ. Microbiol. 2020, 22, 4295–4313. 10.1111/1462-2920.15021. [DOI] [PubMed] [Google Scholar]
- Hooppaw A. J.; McGuffey J. C.; Di Venanzio G.; Ortiz-Marquez J. C.; Weber B. S.; Lightly T. J.; van Opijnen T.; Scott N. E.; Cardona S. T.; Feldman M. F. The Phenylacetic Acid Catabolic Pathway Regulates Antibiotic and Oxidative Stress Responses in Acinetobacter. mBio 2022, 13, e0186321 10.1128/mbio.01863-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang M.; Adams F. G.; Hassan K. A.; Eijkelkamp B. A. The Impact of Omega-3 Fatty Acids on the Evolution of Acinetobacter baumannii Drug Resistance. Microbiol. Spectrum 2021, 9, e0145521 10.1128/spectrum.01455-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou T. S.; Wu J. H.; Chen X. W.; Chen Z. G.; Zheng J.; Peng B. Exogenous glycine promotes oxidation of glutathione and restores sensitivity of bacterial pathogens to serum-induced cell death. Redox Biol. 2022, 58, 102512. 10.1016/j.redox.2022.102512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M.; Kuang S. F.; Lai S. S.; Zhang S.; Yang J.; Peng B.; Peng X. X.; Chen Z. G.; Li H. Na+-NQR Confers Aminoglycoside Resistance via the Regulation of l-Alanine Metabolism. mBio 2020, 11, e02086 10.1128/mbio.02086-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. X.; Guo C.; Chen Z. G.; Yang T. C.; Zhang J. Y.; Wang J.; Zhu J. X.; Li D.; Zhang T. T.; Li H.; Peng B.; Peng X. X. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. Commun. 2019, 10, 3325. 10.1038/s41467-019-11129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S. F.; Li X.; Feng D. Y.; Wu W. B.; Li H.; Peng B.; Peng X. X.; Chen Z. G.; Zhang T. T. Nitrite Promotes ROS Production to Potentiate Cefoperazone-Sulbactam-Mediated Elimination to Lab-Evolved and Clinical-Evolved Pseudomonas aeruginosa. Microbiol. Spectrum 2022, 10, e232721 10.1128/spectrum.02327-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y.; Brown Gandt A.; Rowe S. E.; Deisinger J. P.; Conlon B. P.; Lewis K. ATP-Dependent Persister Formation in Escherichia coli. mBio 2017, 8, e02267 10.1128/mbio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y.; Li Y.; Jin X.; Tian T.; Ma Q.; Zhao Z.; Lin S. Y.; Chen Z.; Li B.; Yao G.; Leake M. C.; Lo C. J.; Bai F. ATP-Dependent Dynamic Protein Aggregation Regulates Bacterial Dormancy Depth Critical for Antibiotic Tolerance. Mol. Cell 2019, 73, 143–156.e4. 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- Li M.; Kim T. J.; Kwon H. J.; Suh J. W. Effects of extracellular ATP on the physiology of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2008, 286, 24–31. 10.1111/j.1574-6968.2008.01248.x. [DOI] [PubMed] [Google Scholar]
- Tatano Y.; Kanehiro Y.; Sano C.; Shimizu T.; Tomioka H. ATP exhibits antimicrobial action by inhibiting bacterial utilization of ferric ions. Sci. Rep. 2015, 5, 8610. 10.1038/srep08610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Lee S. K.; Yang S. H.; Ko J. H.; Han J. S.; Kim T. J.; Suh J. W. ATP modulates the growth of specific microbial strains. Curr. Microbiol. 2011, 62, 84–89. 10.1007/s00284-010-9677-3. [DOI] [PubMed] [Google Scholar]
- Xi C.; Wu J. dATP/ATP, a multifunctional nucleotide, stimulates bacterial cell lysis, extracellular DNA release and biofilm development. PLoS One 2010, 5, e13355 10.1371/journal.pone.0013355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man Z.; Guo J.; Zhang Y.; Cai Z. Regulation of intracellular ATP supply and its application in industrial biotechnology. Crit. Rev. Biotechnol. 2020, 40, 1151–1162. 10.1080/07388551.2020.1813071. [DOI] [PubMed] [Google Scholar]
- Suwara J.; Radzikowska-Cieciura E.; Chworos A.; Pawlowska R. The ATP-dependent Pathways and Human Diseases. Curr. Med. Chem. 2023, 30, 1232–1255. 10.2174/0929867329666220322104552. [DOI] [PubMed] [Google Scholar]
- Shin B.; Park W. Synergistic Effect of Oleanolic Acid on Aminoglycoside Antibiotics against Acinetobacter baumannii. PLoS One 2015, 10, e0137751 10.1371/journal.pone.0137751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W.; Fu T.; Zhang Z.; Jiang X.; Xie J.; Sun H.; Hu P.; Ren H.; Zhou P.; Liu Q.; Long Q. L-lysine potentiates aminoglycosides against Acinetobacter baumannii via regulation of proton motive force and antibiotics uptake. Emerging Microbes Infect. 2020, 9, 639–650. 10.1080/22221751.2020.1740611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika N. T.; Garneau-Tsodikova S. A review of patents (2011–2015) towards combating resistance to and toxicity of aminoglycosides. MedChemComm 2016, 7, 50–68. 10.1039/C5MD00453E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. CLSI Document M07-A9, 9th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, 2012.
- Li H.; Lin X. M.; Wang S. Y.; Peng X. X. Identification and antibody-therapeutic targeting of chloramphenicol-resistant outer membrane proteins in Escherichia coli. J. Proteome Res. 2007, 6, 3628–3636. 10.1021/pr070307y. [DOI] [PubMed] [Google Scholar]
- Dortet L.; Poirel L.; Errera C.; Nordmann P. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J. Clin. Microbiol. 2014, 52, 2359–2364. 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M.; Li X.; Xie C. L.; Chen P.; Luo W.; Lin C. X.; Wang Q.; Shu D. M.; Luo C. L.; Qu H.; Ji J. Fructose enabled killing of antibiotic-resistant Salmonella Enteritidis by gentamicin: Insight from reprogramming metabolomics. Int. J. Antimicrob. Agents 2023, 62, 106907. 10.1016/j.ijantimicag.2023.106907. [DOI] [PubMed] [Google Scholar]
- Allison K. R.; Brynildsen M. P.; Collins J. J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 2011, 473, 216–220. 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.